Abstract
CD8 T cells and natural killer (NK) cells, central cellular components of immune responses against pathogens and cancer, rely on IL-15 for homeostasis. Here we show that IL-15 also mediates homeostatic priming of CD8 T cells for antigen-stimulated activation, which is controlled by a deubiquitinase, Otub1. IL-15 mediates membrane recruitment of Otub1, which inhibits ubiquitin-dependent activation of AKT, a pivotal kinase for T cell activation and metabolism. Otub1 deficiency in mice causes aberrant responses of CD8 T cells to IL-15, rendering naive CD8 T cells hyper-sensitive to antigen stimulation characterized by enhanced metabolic reprograming and effector functions. Otub1 also controls the maturation and activation of NK cells. Consistently, Otub1 deletion profoundly enhances anticancer immunity through unleashing the activity of CD8 T cells and NK cells. These findings suggest that Otub1 controls the activation of CD8 T cells and NK cells by functioning as a checkpoint of IL-15-mediated priming.
Introduction
CD8 T cells and natural killer (NK) cells are major cytotoxic effector cells of the immune system responsible for destruction of pathogen-infected cells and cancer cells1, 2. CD8 T cells detect specific antigens via the T cell receptor (TCR), while NK cells are innate lymphocytes that use different receptors for sensing target cells. These effector cells also function in different phases of an immune response, with NK cells acting in the early phase of innate immunity and CD8 T cells acting in the late phase of adaptive immunity. NK cells also play an important role in regulating T cell responses3. Thus, CD8 T cells and NK cells are considered complementary cytotoxic effectors and have been actively explored for cancer immunotherapy4.
A common feature of CD8 T cells and NK cells is their dependence on the cytokine IL-15 for homeostasis5, 6. IL-15 is a member of common gamma-chain (γc) family cytokines that functions through the IL-15 receptor (IL-15R) complex, composed of IL-15Rα, IL-15Rβ (also called IL-2Rβ or CD122), and γc (also called CD132). IL-15 induces signaling via a transpresentation mechanism, in which IL-15Rα binds to IL-15 and transpresents IL-15 to the IL-15R β/γ complex on responding cells6. Under physiological conditions, IL-15 is specifically required for the homeostasis of CD8 T cells and NK cells that express high levels of IL-15R βγ heterodimer7, 8. Exogenously administered IL-15 can also promote activation of CD8 T cells and NK cells and, therefore, has been exploited as an adjuvant for cancer immunotherapies9, 10, 11. However, the physiological function of IL-15 in regulating the activation of CD8 T cells and NK cells is poorly defined, and how the signal transduction from IL-15R is regulated is also elusive.
Ubiquitination has become a crucial mechanism that regulates diverse biological processes, including immune responses12. Ubiquitination is a reversible reaction counter-regulated by ubiquitinating enzymes and deubiquitinases (DUBs)13. In vitro studies identified an atypical DUB, Otub1, which can both directly cleave ubiquitin chains from target proteins and indirectly inhibit ubiquitination via blocking the function of specific ubiquitin-conjugating enzymes (E2s), including the K63-specific E2 Ubc1314, 15, 16, 17. However, the in vivo physiological function of Otub1 has been poorly defined. In the present study, we identified Otub1 as a pivotal regulator of IL-15R signaling and homeostasis of CD8 T cells and NK cells. Otub1 controls IL-15-stimulated activation of AKT, a pivotal kinase for T cell activation, metabolism, and effector functions18, 19, 20. Our results suggest that Otub1 also controls the activation and function of CD8 T cells and NK cells in immune responses against infections and cancer.
Results
T cell-specific Otub1 deficiency causes aberrant activation of CD8 T cells
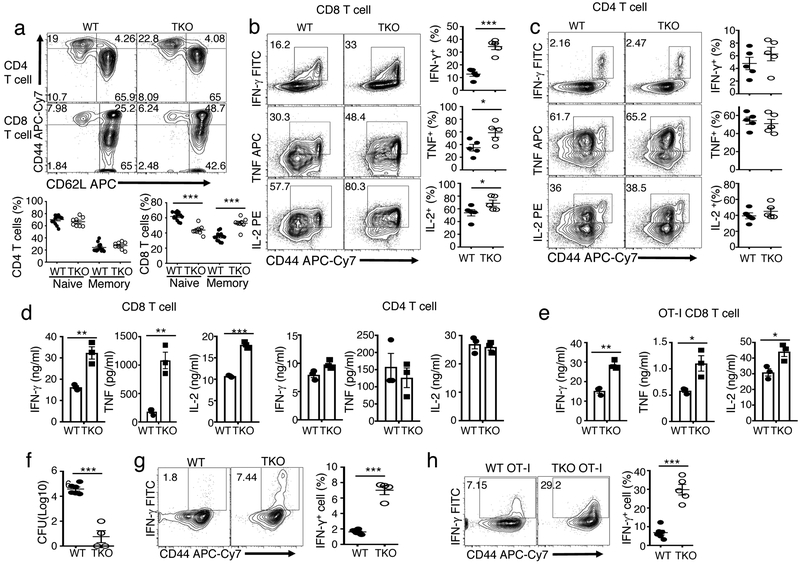
To study the function of Otub1 in T cells, we generated Otub1 T cell conditional knockout (TKO) mice (Supplementary Fig. 1a-c). The Otub1-TKO mice had normal frequencies of thymocyte and peripheral T cell populations (Supplementary Fig. 1d,e). However, they had increased frequencies of effector/memory-like (CD44hi) CD8 T cells producing effector cytokines, IFN-γ, TNF, and IL-2 (Fig. 1a,b). Although Otub1 was similarly expressed in CD4 and CD8 T cells (data not shown), Otub1 deficiency did not increase the frequency of CD4 effector/memory T cells (Fig. 1a,c). The Otub1-TKO and wildtype (WT) mice had comparable frequencies of regulatory T cells (Treg cells), and the Otub1-deficient Treg cells were fully functional in suppressing naive CD4 T cells (Supplementary Fig. 2a-c). Mixed bone marrow adoptive transfer studies revealed that the Otub1-TKO CD8 T cells had increased frequencies of effector/memory-like population than WT CD8 T cells even in the same recipient mice (Supplementary Fig. 2d,e), suggesting a cell-intrinsic role for Otub1 in maintaining CD8 T cell homeostasis. Furthermore, the Otub1-deficient naive CD8 T cells were hyper-responsive to in vitro activation (Fig. 1d). Similar results were obtained with naive CD8 T cells from OT-I mice, producing CD8 T cells with a recombinant TCR specific for the chicken ovalbumin (OVA) peptide SIINFEKL21 (Fig. 1e). In contrast, Otub1 deficiency had no effect on naive CD4 T cell activation (Fig. 1d).
Figure 1.
Otub1 regulates the homeostasis and activation of CD8 T cells. a, Flow cytometric analysis of naive (CD44loCD62Lhi) and memory (CD44hiCD62Llo) CD4 T cells and naive (CD44lo) and memory (CD44hi) CD8 T cells in the spleen of WT and Otub1-TKO (TKO) mice. Data are presented as a representative plot (upper) and summary graphs (lower) based on multiple mice (WT, n=13; TKO, n=8). b,c, Flow cytometric analysis of intracellular IFN-γ, TNF and IL-2 in WT and Otub1-TKO splenic CD8 T cells (b) or CD4 T cells (c), stimulated for 4 h with PMA and ionomycin in the presence of monensin. Data are presented as representative plots (left) and summary graphs (right) based multiple mice (WT, n=5; TKO, n=5). d,e, ELISA of the indicated cytokines in the culture supernatant of naive CD8 and CD4 T cells (d) or OT-I CD8 T cells (e) purified from the spleen of young (6 wk, n=4) WT and Otub1-TKO mice and stimulated for 66 h with plate-bound anti-CD3 (1 μg/ml) and anti-CD28 (1 μg/ml). f-h, Liver bacteria titer (f) and flow cytometric analysis of IFN-γ-producing CD8 effector T cell frequency in OVA257-264-stimulated splenic T cells (g,h) derived from WT and Otub1-TKO mice (g, WT: n=6; TKO: n=4) or WT OT-I and TKO OT-I mice (h, WT OT-I: n=6; TKO OT-I: n=5) infected with LM-OVA for 7 days. Data summarize three (b-h), or five (a), independent experiments. Summary graphs are presented as mean±s.e.m. with P values being determined by two-tailed Student’s t-test. *< 0.05, **<0.01, ***<0.0001. Numbers in quadrants indicate percentage of cells.
To examine the in vivo function of Otub1, we employed a bacterial infection model using a recombinant Listeria monocytogenes strain expressing chicken ovalbumin, LM-OVA. The Otub1-TKO mice displayed markedly enhanced immune responses against LM-OVA infection, as demonstrated by reduced liver bacterial load and increased frequencies of antigen-specific CD8 effector T cells producing IFN-γ (Fig. 1f,g). Similar results were obtained using WT and Otub1-TKO OT-I mice producing OVA-specific CD8 T cells (Fig. 1h). These results suggest that Otub1 maintains CD8 T cell homeostasis and negatively regulates CD8 T cell activation.
Otub1 regulates CD8 T cell responses to IL-15
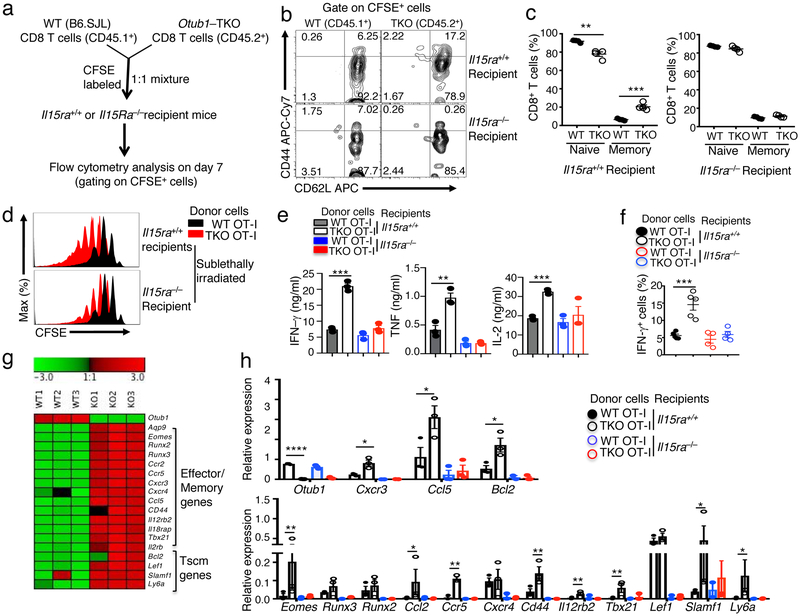
The γc family cytokines IL-7 and IL-15 are important for T cell homeostasis5, 22. While IL-7 regulates both CD4 and CD8 T cells, IL-15 is particularly important for regulating CD8 T cells that express high levels of IL-15Rβ and γc7, 8. Since Otub1 deficiency had selective effect on CD8 T cells (Fig. 1a), we tested whether Otub1 played a role in regulating CD8 T cell responses to IL-15 by performing mixed CD8 T cell transfer using Il15ra+/+ or Il15ra−/− recipient mice (Fig. 2a). Since IL-15Rα is required for IL-15 transpresentation, T cells transferred to the Il15ra−/− mice are defective in IL-15 stimulation23, 24. In the Il15ra+/+ recipients, Otub1-TKO CD8 T cells had much higher frequencies of memory-like T cells than WT CD8 T cells (Fig. 2b,c). However, this phenotype was no longer significant in Il15ra−/− recipients, suggesting a role for Otub1 in controlling CD8 T cell responses to IL-15 (Fig. 2b,c).
Figure 2.
Otub1 controls IL-15-mediated homeostatic responses and priming of CD8 T cells. a-c, Schematic of experimental design (a), a representative plot (b), and summary graph (c) of flow cytometric analyses of memory (CD44hi) and naive (CD44lo) CD8 T cells from Il15ra+/+ or Il15ra−/− recipient mice 7 days after adoptive transfer with carboxyfluorescein succinimidyl ester (CFSE)-labeled WT and Otub1-TKO naive CD8 T cells. The WT and Otub1-TKO CD8 T cells were detected as CFSE+ cells and distinguished based on CD45 congenic marker (WT: CD45.1+; TKO: CD45.2+). d, Cell proliferation assays (based on CFSE dilution) of WT and Otub1-TKO OT-I cells isolated from sublethally irradiated Il15ra+/+ or Il15ra−/− recipient mice 8 days after adoptive transfer with a mixture (1:1 ratio, 12 × 106 cells) of CFSE-labeled WT OT-I (CD45.1+CD45.2+) and Otub1-TKO OT-I (TKO OT-I; CD45.2+) cells. e,f, ELISA (e) and intracellular IFN-γ flow cytometric analysis (f) of WT and Otub1-TKO OT-I cells isolated from Il15ra+/+ or Il15ra−/− recipient mice 7 days after being adoptively transferred with a mixture (1:1 ratio, 6 × 106 cells) of CFSE-labeled WT OT-I (CD45.1+CD45.2+) and Otub1-TKO OT-I (TKO OT-I; CD45.2+) cells (n=4 for Il15ra+/+ and Il15ra−/− recipients in e). g, Heatmap showing a list of effector/memory-related genes and stem memory T cell (Tscm) genes from RNA sequencing analysis of untreated WT and Otub1-TKO naive OT-I CD8 T cells freshly isolated from young mice (6 wk). h, qRT-PCR analysis of the indicated genes in WT and Otub1-TKO OT-I cells freshly isolated from Il15ra+/+ and Il15ra−/− recipient mice 7 days after being adoptively transferred with a mixture (1:1 ratio, 6×106 cells) of CFSE-labeled WT OT-I (CD45.1+CD45.2+) and TKO OT-I (CD45.2+) cells (WT recipients: n=4; Il15ra−/− recipients: n=5). Data are representative of one experiment (g) or summarize three (b-f, h) independent experiments. Summary data are mean±s.e.m. with P values being determined by two-tailed Student’s t-test. *< 0.05, **<0.01, ***<0.001, ****<0.0001.
We also examined the effect of Otub1 deficiency on IL-15-mediated CD8 T cell proliferation under lymphopenic conditions. We used OT-I CD8 T cells, since the OT-I TCR does not respond to commensal antigens, the OT-I T cell expansion is mediated by homeostatic cytokines, predominantly IL-7 and IL-155, 25. WT OT-I T cells proliferated to a similar level in Il15ra+/+ and Il15ra−/− recipient mice (Fig. 2d), consistent with the involvement of both IL-7 and IL-15 in mediating lymphopenic T cell proliferation5, 7, 25. However, the hyper-proliferation of the Otub1-TKO OT-I T cells was critically dependent on IL-15, since it was largely eliminated in the Il15ra−/− recipient mice (Fig. 2d). These results further emphasized a crucial role for Otub1 in controlling CD8 T cell responses to the homeostatic cytokine IL-15.
IL-15 primes CD8 T cells for activation under the control of Otub1
Our finding that Otub1 deficiency promoted the activation of CD8 T cells by TCR-CD28 signals indicated that homeostatic exposure of CD8 T cells to IL-15 might prime them for activation by antigens. In further support of this idea, the hyper-responsive phenotype of Otub1-TKO CD8 T cells was detected in Il15ra+/+, but not Il15ra−/−, background (Supplementary Fig. 3a). Furthermore, in a T cell adoptive transfer experiment, Otub1-TKO OT-I CD8 T cells isolated from Il15ra+/+ recipients, but not Il15ra−/− recipients, displayed the hyper-activation phenotype (Fig. 2e). As an in vivo model, we performed LM-OVA infection using Il15ra+/+ or Il15ra−/− mice adaptively transferred with a mixture of WT and Otub1-TKO naive OT-I CD8 T cells (Supplementary Fig. 3b,c). In Il15ra+/+ recipients, the Otub1-TKO OT-I T cells displayed a much stronger response to LM-OVA infection than the WT OT-I T cells, but this phenotype was not detected in the Il15ra−/− recipients (Fig. 2f and Supplementary Fig. 3d). Thus, Otub1 controls IL-15-mediated priming of CD8 T cells for antigen-specific responses both in vitro and in vivo.
RNA sequencing revealed that the Otub1-TKO naive OT-I T cells had upregulated expression of a large number of genes under homeostatic conditions (Supplementary Fig. 3e), including signatures associated with effector/memory functions and stem memory T cells (Tscm) (Fig. 2g). To examine whether this gene expression signature was dependent on IL-15 signaling, we performed qRT-PCR analysis using WT and Otub1-TKO CD8 T cells isolated from adoptively transferred Il15ra+/+ or Il15ra−/− recipient mice (Fig. 2h). Within the Il15ra+/+ recipient mice, the Otub1-TKO CD8 T cells displayed upregulated expression of almost all of the genes analyzed compared to the WT CD8 T cells (Fig. 2h). However, within the Il15ra−/− recipient mice, the WT and Otub1-TKO CD8 T cells no long displayed differences in gene expression, and both displayed reduced level of gene expression compared to CD8 T cells derived from the Il15ra+/+ recipient mice (Fig. 2h). Together, these results suggest that under homeostatic conditions, IL-15 primes CD8 T cells for responding to TCR-CD28 signals, which is negatively regulated by Otub1.
Otub1 also regulates NK cell maturation and activation
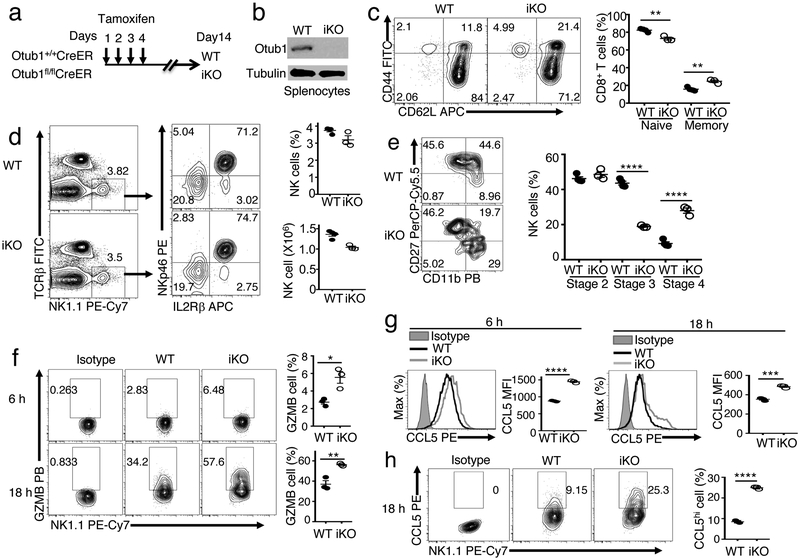
NK cells also express high levels of IL-15R β/γ heterodimer and rely on IL-15 for maturation and activation26. Based on surface expression of CD11b and CD27, NK cells can be divided into four maturation stages: stage 1 (CD11bloCD27lo), stage 2 (CD11bloCD27hi), stage 3 (CD11bhiCD27hi), and stage 4 (CD11bhiCD27lo), with progressive acquisition of effector functions27. IL-15 deficiency impairs generation of stage 3 and stage 4 NK cells, whereas IL-15 overexpression causes predominant accumulation of stage 4 NK cells28. To study the function of Otub1 in NK cell regulation, we inducibly deleted Otub1 in adult mice using a tamoxifen-inducible Cre (CreER) system (Fig. 3a,b). As expected from the Otub1-TKO result (Fig. 1a), Otub1 induced KO (Otub1-iKO) mice had increased frequencies of memory-like CD8 T cells (Fig. 3c). Importantly, although the Otub1 deletion had no effect on total NK cell number in the spleen, it markedly increased the frquency of stage 4 mature NK cells (CD11bhiCD27lo) and concomitantly reduced stage 3 NK cells (CD11bhiCD27hi) (Fig. 3d,e). Consistently, Otub1-iKO NK cells were hyper-responsive to cytokine-stimulated activation, detected based on production of Granzyme B and the chemokine CCL5 (Fig. 3f-h), mediating NK cell effector function and recruitment of type 1 conventional dendritic cells (cDC1), respectively29. These results suggest that Otub1 controls the maturation and activation of NK cells, further emphasizing the role of this DUB in regulating IL-15 responses.
Figure 3.
Otub1 controls the maturation and activation of NK cells. a,b, Schematic of experimental design for producing Otub1 tamoxifen-induced KO (iKO) and WT control mice (a) and immunoblot analysis of Otub1 in splenocytes of Otub1-iKO and WT mice (b). c-e, Flow cytometric analysis of the frequency of naive (CD44lo) and memory-like (CD44hi) CD8 T cells (c), NK cells (d), and maturation stage subpopulations of NK cells (stage 2: CD11bloCD27hi; stage 3: CD11bhiCD27hi; and stage 4: CD11bhiCD27lo). f-h, Flow cytometric analysis of intracellular granzyme B (f) and CCL5 (g,h) in WT or Otub1-iKO NK cells stimulated in vitro with IL-2 (5 ng/ml), IL-12 (10 ng/ml), and IL-18 (10 ng/ml) for the indicated time periods. The CCL5 results were presented as histogram (g) and dot plot (h). Data summarize two (b-e), three (f-h) independent experiments. Summary data are mean±s.e.m. with P values being determined by two-tailed Student’s t-test. *< 0.05, **<0.01, ***<0.001, ****<0.0001.
Otub1 regulates AKT axis of IL-15 receptor signaling
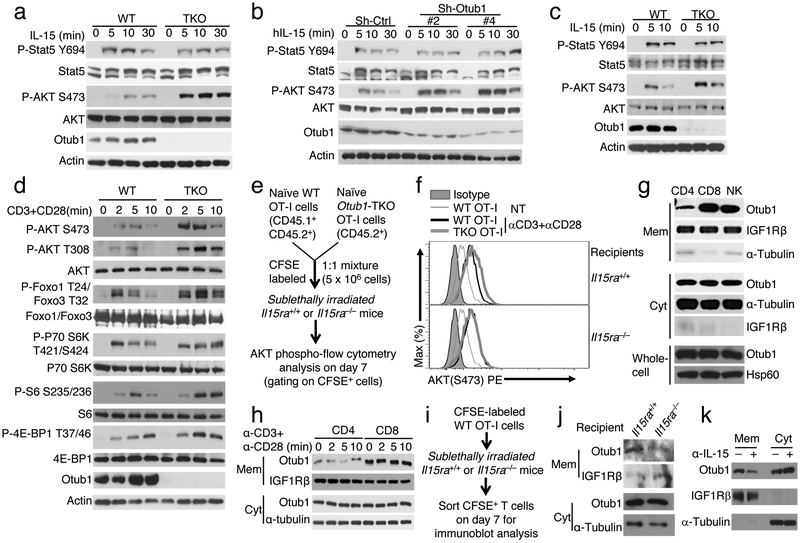
Stimulation of naive CD8 T cells with IL-15 triggered activation of the transcription factor STAT5 and the kinase AKT, as shown by their site-specific phosphorylations (Fig. 4a). Otub1 deficiency did not affect STAT5 activation but strikingly enhanced activation of AKT (Fig. 4a). AKT activation is mediated via its phosphorylation at threonine 308 (T308) and serine 473 (S473). AKT T308 phosphorylation is crucial for activation of the metabolic kinase mTORC1, whereas AKT S473 phosphorylation is required for phosphorylating and inactivating FOXO family of transcription factors, a mechanism that promotes CD8 T cell effector functions30, 31. The Otub1 deficiency enhanced IL-15-stimulated phosphorylation of AKT S473 as well as FOXO1 and FOXO3 (Fig. 4a, supplementary Fig. 4a). IL-15-stimulated AKT T308 phosphorylation was relatively weak, which required loading more cell lysates for clear detection (Supplementary Fig. 4a). Nevertheless, the AKT T308 phosphorylation was also enhanced in Otub1-deficient CD8 T cells (Supplementary Fig. 4a). Otub1 deficiency only had a weak effect on IL-2- and IL-7-stimulated AKT phosphorylation (Supplementary Fig. 4b). Notably, the receptors of IL-2 and IL-15 share two common subunits, IL-2/IL-15Rβ and γc, although these two cytokines display different biological functions32. Our finding also suggested signaling differences between these two closely related cytokines. The role of Otub1 in regulating IL-15-stimulated AKT activation was further demonstrated using an IL-15-responsive T cell line, 15R-KIT (human KIT-225 cell line stably transfected with IL-15Rα). Otub1 knockdown in 15R-KIT T cells strongly promoted IL-15-stimulated AKT phosphorylation (Fig. 4b). Furthermore, Otub1 deficiency in NK cells also profoundly enhanced IL-15-stimulated activation of AKT, but not activation of STAT5 (Fig. 4c). Thus, Otub1 controls the AKT axis of IL-15R signaling in both CD8 T cells and NK cells.
Figure 4.
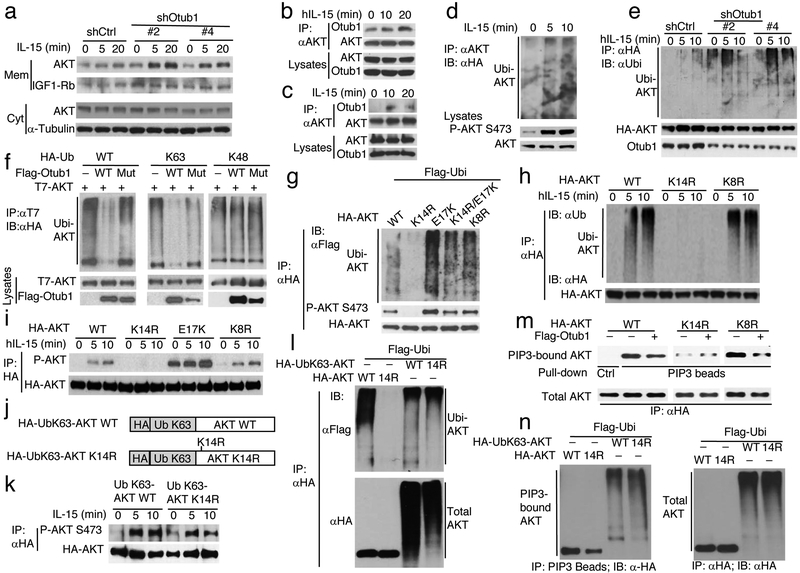
Otub1 controls AKT axis of IL-15R signaling and is located to membrane compartment in an IL-15-dependent manner. a-c, Immunoblot analyses of the indicated phosphorylated (P-) and total proteins in IL-15-stimulated CD8 T cells from 6-week old WT and Otub1-TKO OT-I mice (a), 15R-KIT cells transduced with either a control shRNA (sh-Ctrl) or two different Otub1-silencing shRNAs (Sh-Otub1) (b), or NK cells from tamoxifen-induced Otub1 KO (iKO) and WT control mice (NK cells were collected from 16 WT and 15 iKO mice). d, Immunoblot analyses of the indicated phosphorylated (P-) and total proteins in CD8 T cells from WT and Otub1-TKO OT-I mice (6 weeks old) stimulated with anti-CD3 plus anti-CD28. e,f, Schematic of experimental design (e) and representative plots (f) of flow cytometric analyses of S473-phosphorylated AKT in WT and Otub1-TKO OT-I cells sorted from Il15ra+/+ and Il15ra−/− recipients 7 days after being adoptively transferred with a mixture of CFSE-labeled WT OT-I and TKO OT-I CD8 T cells and stimulated in vitro with anti-CD3 plus anti-CD28 for 5 min. g,h, Immunoblot analysis of the indicated proteins in membrane (Mem) and cytosol (Cyt) fractions or whole-cell lysates (whole-cell) of untreated CD4 T, CD8 T, and NK cells (g) or anti-CD3/anti-CD28-stimulated CD4 and CD8 T cells (h). i,j Schematic of experimental design (i) and Immunoblot analysis of Otub1 and the indicated loading controls in membrane (Mem) and cytosol (Cyt) fractions of WT OT-I CD8 T cells sorted from Il15ra+/+ or Il15ra−/− recipients 7 days after adoptive transfer. k, Immunoblot analysis of Otub1, membrane protein IGF1Rβ, and cytoplasmic protein α-Tubulin in membrane (Mem) and cytoplasmic (Cyt) fractions of OT-I CD8 T cells sorted from WT OT-I mice injected (i.p.) with an IL-15 neutralizing antibody (200 mg/mouse) daily for three consecutive days. Data summarize two (c,f,h,j,k), three (b,d,g), or six(a) independent experiments.
Since the Otub1-deficient CD8 T cells were hyper-responsive to TCR-CD28 stimulation in vitro and antigen-specific responses in vivo (Fig. 1d-h), we examined the effect of Otub1 deletion on TCR signaling. Otub1 deficiency did not influence the phosphorylation of the protein tyrosine kinase Zap70, the adaptor protein SLP76, and the MAP kinase ERK (Supplementary Fig. 4c). However, Otub1 deficiency markedly enhanced TCR-CD28-stimulated activation of AKT and phosphorylation of several AKT downstream proteins, including the transcription factors Foxo1 and Foxo3 and the mTORC1 targets S6 kinase (S6K), ribosomal S6 protein, and 4E-BP1 (Fig. 4d). On the other hand, the Otub1 deficiency did not affect TCR-CD28-stimulated AKT signaling in CD4 T cells (Supplementary Fig. 4d), consistent with the finding that Otub1 controlled the activation of CD8, but not CD4, T cells (Fig. 1d).
To examine whether the TCR-CD28-stimulated AKT hyper-activation in Otub1-deficient CD8 T cells was due to IL-15 priming, we adoptively transferred WT or Otub1-TKO naive OT-I T cells to Il15ra+/+ or Il15ra−/− recipient mice and sorted the transferred T cells for AKT activation assay (Fig. 4e). The Otub1-TKO OT-I CD8 T cells isolated from Il15ra+/+, but not Il15ra−/−, recipient mice displayed hyper-activation of AKT (Fig. 4f), suggesting that IL-15 primes CD8 T cells for AKT axis of TCR-CD28 signaling under the control of Otub1. In an effort to further explore the mechanism by which Otub1 selectively regulates AKT signaling in CD8 T cells, we found that CD8, but not CD4, T cells contained abundant membrane-associated Otub1 (Fig. 4g). Like CD8 T cells, NK cells also contained a high level of membrane-associated Otub1 (Fig. 4g). The membrane association of Otub1 was not affected by TCR-CD28 signaling (Fig. 4h), but was critically dependent on IL-15 since it was diminished in CD8 T cells derived from Il-15Rα-deficient host in a T cell transfer study (Fig. 4i,j). Antibody-mediated IL-15 neutralization in WT OT-I mice also inhibited Otub1 membrane localization in CD8 T cells (Fig. 4k). Since AKT activation occurs in various membrane compartments33, these findings provide insight into the mechanism underlying the AKT-regulatory function of Otub1.
Otub1 inhibits K63 ubiquitination and PIP3-binding function of AKT
A key step in AKT activation is its recruitment to membrane compartments via interaction of its pleckstrin homology (PH) domain with the membrane lipid PIP334. Once in the membrane, AKT is phosphorylated T308 and S473 by PDK1 and mTORC2, respectively. We found that IL-15 stimulated membrane translocation of AKT, which was greatly enhanced by Otub1 knockdown (Fig. 5a). Otub1 knockdown had no obvious effect on the activity of AKT upstream regulators, PI3 kinase (PI3K) and PTEN (data not shown), which catalyze the forward and reverse PIP3 generation reactions, respectively35. Interestingly, AKT was physically associated with Otub1 in 15R-KIT cells, which was strongly enhanced upon IL-15 stimulation (Fig. 5b). In primary OT-I CD8 T cells, the AKT-Otub1 interaction was barely detectable at steady state but was strongly induced by IL-15 (Fig. 5c). The Otub1-AKT binding was also readily detected under transfection conditions (Supplementary Fig. 4e).
Figure 5.
Otub1 inhibits K63 ubiquitination, PIP3-binding, and membrane translocation of AKT. a, Immunoblot analysis of AKT in membrane (Mem) and cytosol (Cyt) fractions of IL-15-stimulated 15R-KIT T cells transduced with either a control shRNA or two different Otub1 shRNAs. b,c, Co-immunoprecipitation analysis of endogenous Otub1-AKT interaction in IL-15-stimulated 15R-KIT T cells (b) and primary OT-I CD8 T cells (c). d, AKT ubiquitination analyses in IL-15-stimulated 15R-KIT T cells stably expressing HA-ubiquitin. e, AKT ubiquitination analysis in IL-15-stimulated Otub1-knockdown and control 15R-KIT T cells stably expressing HA-AKT. f, AKT ubiquitination analyses in HEK293T cells transiently transfected with HA-tagged WT, K63, or K48 ubiquitin in the presence (+) or absence (−) of the indicated expression vectors. Otub1 Mut harbors D88A/C91S mutations. g,h, Ubiquitination analysis of WT and mutant forms of AKT in transiently transfected HEK293 cells (g) or IL-15-stimulated 15R-KIT T cells stably expressing the indicated HA-AKT WT and mutants (h). i, Immunoblot analysis of phosphorylated (P) and total AKT immunoprecipitated from IL-15-stimulated 15R-KIT T cells stably expressing AKT WT and mutants. j,k, Schematic of ubiquitin K63 (UbK63)-AKT and UbK63-AKT K14R (j) and immunoblot analysis of their phosphorylation and total protein level immunoprecipitated from stably infected 15R-KIT T cells stimulated with IL-15 (k). l, Immunoblot analysis of ubiquitinated (upper) and total (lower) AKT or UbK63-AKT proteins immunoprecipitated from transiently transfected HEK293 cells. m, Immunoblot analysis of PIP3-bound (upper) and total (lower) HA-AKT proteins isolated by PIP3 bead-pull down (upper) and anti-HA IP (lower), respectively, from transiently transfected HEK293 cells. n, Immunoblot analysis of PIP3 bead-pull down (left) and anti-HA immunoprecipitated (right) AKT or UbK63-AKT proteins from transiently transfected HEK293 cells. Data summarize two (a,k), or three (b-I, l-n) independent experiments.
Since Otub1 is a DUB, we next examined whether Otub1 regulated the ubiquitination of AKT. IL-15 stimulated ubiquitination of AKT, which was enhanced upon Otub1 knockdown (Fig. 5d,e). Conversely, Otub1 overexpression inhibited AKT ubiquitination, which was efficient for K63-linked, but not K48-linked, polyubiquitin chains (Fig. 5f). A previous study identified three catalytic residues of Otub1: cysteine 91 (C91), aspartate 88 (D88), and histidine 265 (H265)36. We found that mutation of C91 only moderately inhibited the function of Otub1 (data not shown), but simultaneous mutations of D88 and C91 generated an Otub1 mutant, D88A/C91S, that was unable to inhibit AKT ubiquitination (Fig. 5f). WT Otub1, but not D88A/C91S, was also able to suppress AKT activation in reconstituted Otub1-deficient CD8 T cells and Otub1-knockdown 15R-KIT cells (Supplementary Fig. 4f,g), thus suggesting that Otub1-mediated inhibition of AKT K63 ubiquitination contributes to the negative regulation of AKT activation.
TRAF6 is known to mediate growth factor-induced AKT ubiquitination at K8 and K14 in cancer cells37. We found that mutation of K14 also abolished AKT ubiquitination under basal and IL-15-stimulated conditions (Fig. 5g,h). However, mutation of K8 had no effect on AKT ubiquitination (Fig. 5g,h). Consistently, mutation of K14, but not K8, abolished AKT phosphorylation (Fig. 5i), suggesting AKT K14 ubiquitination to mediate its activation by IL-15. To further assess the function of AKT K63 ubiquitination, we fused a K63 ubiquitin mutant (UbK63) with AKT or AKT K14R at the N-terminus close to residue K14 (Fig. 5j). The UbK63-AKT fusion protein behaved like AKT in responding to IL-15 for phosphorylation (Fig. 5k). Fusion of UbK63 to AKT K14R largely rescued its defect in IL-15-stimulated phosphorylation as well as in ubiquitination (Fig. 5k,l), suggesting that the fused UbK63 could serve as an acceptor ubiquitin for polyubiquitin chain formation and, thus, AKT activation.
AKT normally exists in a closed conformation due to the intramolecular interaction between its N-terminal PH domain and C-terminal kinase domain38. Since ubiquitination often causes conformation changes, we surmised that AKT ubiquitination might promote its PIP3-binding activity. While WT AKT and AKT K8R displayed strong PIP3-binding activity, the AKT K14R mutant was defective in PIP3 binding (Fig. 5m). Moreover, Otub1 strongly inhibited the PIP3-binding activity of AKT WT and AKT K8R, but it did not affect the residual PIP3-binding activity of K14R (Fig. 5m). Fusion of UbK63 to AKT K14R, which restored its ubiquitination (Fig. 5l), completely restored its PIP3-binding function (Fig. 5n). These results suggest that Otub1 deubiquitinates AKT to interfere with the PIP3-binding and membrane translocation of AKT, thereby inhibiting its phosphorylation and activation.
Otub1 regulates important gene signatures and metabolic programing in activated CD8 T cells
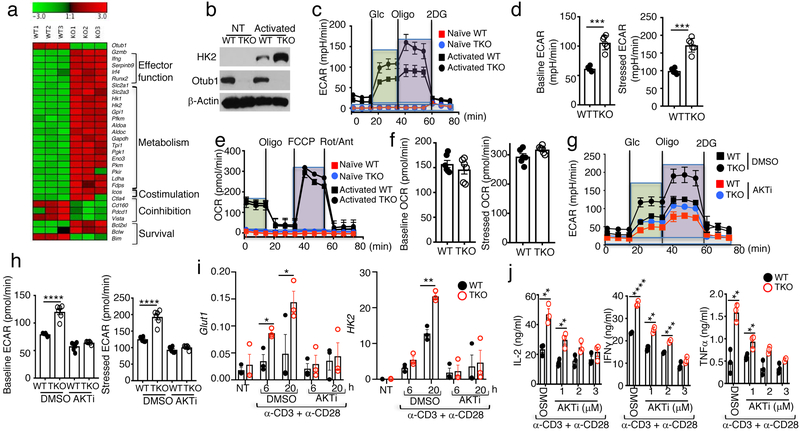
RNA sequencing analysis of in vitro activated CD8 T cells revealed that Otub1-deficient CD8 T cells had 1254 significantly upregulated and 297 significantly downregulated genes compared to WT CD8 T cells (Supplementary Fig. 5). The upregulated genes included those involved in activation and effector function or survival of CD8 T cells (Fig. 6a). The major down-regulated genes included those encoding a pro-apoptotic factor, Bim, and immune checkpoint molecules (Pd1, Vista, and CD160) (Fig. 6a). The most striking result was the upregulated expression of a metabolic gene signature in the Otub1-deficient CD8 T cells, particularly those involved in the glycolytic pathway, such as glucose transporter 1 (Glut1, also called Slc2a1) and hexokinase 2 (Hk2) (Fig. 6a and Supplementary Fig. 5). Immunoblot analyses confirmed the drastic upregulation of HK2, an enzyme catalyzing the first step of glycolytic pathway39, in Otub1-TKO CD8 T cells (Fig. 6b). These findings are intriguing, since metabolic reprograming is a hallmark of T cell activation and required for the function of effector T cells40, 41, 42.
Figure 6.
Otub1 regulates gene expression and glycolytic metabolism in activated CD8 T cells. a, Heatmap showing a list of differentially expressed genes from RNA sequencing analyses of WT and Otub1-TKO OT-I CD8 T cells activated for 24 h with plate-coated anti-CD3 (1 μg/ml) plus soluble anti-CD28 (1 μg/ml). b, Immunoblot analysis of HK2 in WT or Otub1-TKO naive OT-I CD8 T cells that were either not treated (NT) or stimulated with anti-CD3 plus anti-CD28 for 24 h (activated). c-f, Seahorse analysis of extracellular acidification rate (ECAR) under baseline (glucose injection) and stressed (oligomycin injection) conditions (c,d) and Seahorse analysis of oxygen consumption rate (OCR) under baseline (no treatment) and stressed (FCCP injection) conditions (e,f) in naive or anti-CD3/anti-CD28-activated (24 h) WT or Otub1-TKO naive OT-I CD8 T cells. Data are presented as a representative plot (c,e) and summary graphs (d,f). g,h, Searhorse analysis of extracellular acidification rate (ECAR) in WT or Otub1-TKO naive OT-I CD8 T cells that were activated with anti-CD3 plus anti-CD28 for 24 h in the presence of an AKT inhibitor (AKTi, 3 μM) or solvent control DMSO. Data are presented as a representative plot (g) and summary graphs (h). i,j, qRT-PCR analysis of Glut1 and Hk2 expression (i) and ELISA of the indicated cytokines in the culture supernatant (j) of WT or Otub1-TKO naive OT-I CD8 T cells that were either not treated (NT) or stimulated with anti-CD3 plus anti-CD28 in the presence of AKTi (3 μM)) or solvent control DMSO for the indicated time periods (i) or for 66 h (j). Data are representative of one (a), or summarize three (b-j) independent experiments. Summary data are mean±s.e.m. with P values being determined by two-tailed Student’s t-test. *< 0.05, **<0.01, ***<0.001, ****<0.0001.
We next performed Seahorse Extracellular Flux analyses to measure extracellular acidification rate (ECAR) and oxygen consumption rate (OCR), indicators of aerobic glycolysis and oxidative phosphorylation, respectively40. Compared to WT CD8 T cells, Otub1-deficient CD8 T cells had enhanced ECAR and maximum glycolytic capacity (stressed ECAR) under activated conditions (Fig. 6c,d). Unlike glycolysis, OCR was not significantly altered by the Otub1 deficiency (Fig. 6e,f). Otub1 appeared to regulate glycolysis through controlling AKT, since a selective AKT inhibitor (AKTi) erased the ECAR differences between WT and Otub1-TKO CD8 T cells (Fig. 6g,h). The AKT inhibitor also blocked TCR-CD28-stimulated hyper-expression of the glycolysis-regulatory genes, Glut1 and Hk2, and cytokine production in Otub1-TKO CD8 T cells (Fig. 6i,j). These results suggest that Otub1 controls glycolysis induction in activated CD8 T cells via a mechanism that involves regulation of AKT signaling.
Otub1 deficiency impairs CD8 T cell self-tolerance
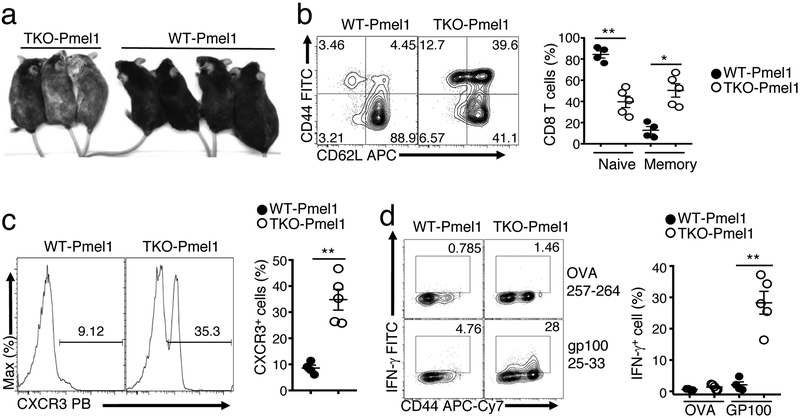
IL-15 is known to reduce the threshold of T cell activation and sensitizes CD8 T cells for responses to self-antigens10, 43. We examined the role of Otub1 in regulating CD8 T cell self-tolerance using a well-defined mouse model, Pmel1, producing CD8 T cells with a transgenic TCR specific for the melanocyte self-antigen, gp10044. The Pmel1 CD8 T cells are normally tolerant to the self-antigen gp100, and impaired self-tolerance causes a skin autoimmunity, vitiligo, characterized by hair depigmentation44, 45. Although WT Pmel1 mice only developed minor vitiligo up to 9 months of age, 100% of the Otub1-TKO Pmel1 mice developed severe vitiligo, starting from around 3 months of age and becoming more severe over time (Fig. 7a and data not shown). While the WT Pmel1 CD8 T cells were predominantly in a naive state, a large proportion of the Otub1-TKO Pmel1 CD8 T cells were activated, displaying CD44 and CXCR3 activation markers (Fig. 7b,c). Furthermore, the Otub1-TKO, but not WT, Pmel1 T cells responded to in vitro restimulation with the antigen gp100, for IFN-γ production (Fig. 7d). These results suggest that Otub1 controls CD8 T cell responses to microbial antigens and self-antigens in vivo.
Figure 7.
Otub1 deficiency promotes CD8 T cell responses to a self-antigen. a, A representative image of 9-month old WT and Otub1-TKO Pmel1 mice. 100% TKO Pmel1 (n=21) and 0% WT Pmel1 (n=18) mice develop severe vitiligo (hair depigmentation). b,c, Flow cytometric analysis of naive (CD44lo) and memory (CD44hi) T cell frequency (b) and CXCR3+ effector T cell frequency (c) of splenic CD8 T cells derived from WT Pmel1 and Otub1-TKO Pmel1 mice (WT, n=4; TKO, n=5). d, Flow cytometric analysis of IFN-γ-producing cells in WT and Otub1-TKO Pmel1 CD8 T cells stimulated for 6 h with GP10025–33 or control OVA257–264 peptide in the presence of monnesin (WT Pmel1, n=4, TKO Pmel1, n=5). Data are representative of one (a), or summarize three (b-j) independent experiments. Summary data are mean±s.e.m. with P values being determined by two-tailed Student’s t-test. *< 0.01, **<0.001.
Otub1 regulates anticancer immunity via both T cells and NK cells
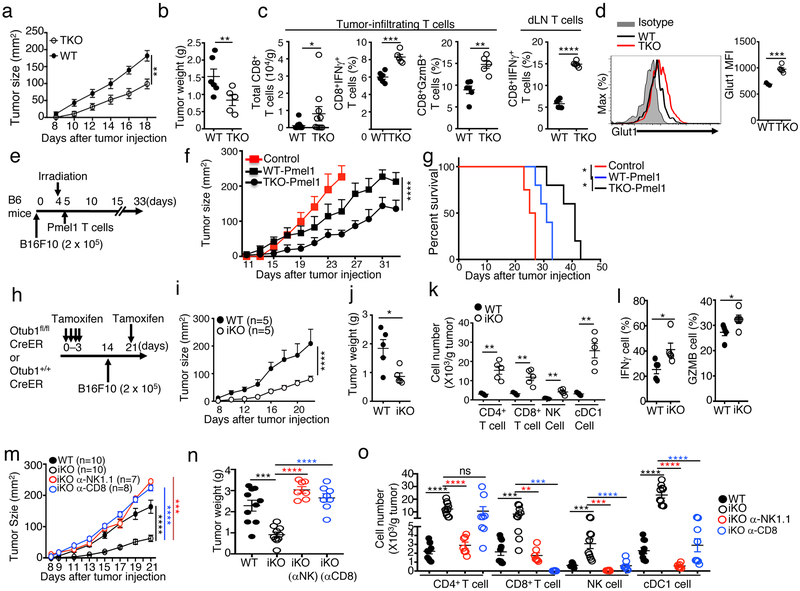
Although tolerance prevents autoimmunity, it poses a major obstacle to immune responses against cancer, and a general principle of cancer immunotherapy is to overcome immune tolerance46. Our finding that Otub1 controls the activation of CD8 T cells and NK cells, central components for cancer immunity1, 2, suggested a role for Otub1 in regulating antitumor immunity. We first tested the T cell-specific function of Otub1 by employing the Otub1-TKO mice and a murine melanoma model, B16-OVA (B16 cells expressing the surrogate antigen ovalbumin). Compared with WT mice, Otub1-TKO mice had significantly reduced tumor burden (Fig. 8a,b), coupled with increased frequencies of CD8 effector T cells producing IFN-γ and Granzyme B in both tumors and draining lymph nodes (Fig. 8c). Furthermore, the Otub1-TKO CD8 T cells expressed higher levels of Glut1 than WT CD8 T cells in tumor microenvironment (Fig. 8d), consistent with the role of Otub1 in regulating glycolysis (Fig. 6c,d).
Figure 8.
Otub1 regulates anticancer immunity. a-c, Tumor growth curve (a), day 18 tumor weight (b), and frequency of CD8 T cells and effector (IFN-γ+ and Granzyme B+) CD8 T cells (% of CD8 T cells) in the tumor and draining LN (dLN) (c) of WT or Otub1-TKO mice injected s.c. with 2 × 105 B16-OVA cells (WT, n=6; TKO, n=5). d, Flow cytometric analysis of Glut1 expression in tumor-infiltrating CD8 T cells. e-g, Schematic of experimental design (e), tumor growth curve (f), and Kaplan-Meier plot survival curve (g) of B6 mice that were inoculated with B16F10 melanoma cells and subsequently irradiated and injected with in vitro activated (with anti-CD3 plus anti-CD28 for 5 days) WT and Otub1-TKO Pmel1 T cells (6 × 105). Control mice were inoculated with B16F10 cells without irradiation and Pmel1 T cell injection. Control, n=4; WT Pmel1, n=5; TKO Pmel1, n=5. h-l, Schematic of experimental design (h), tumor growth curve (i), day 22 tumor weight (j), frequency of tumor-infiltrating immune cells (k), and frequency of tumor-infiltrating effector (IFN-γ+ and Granzyme B+) CD8 T cells (% of CD8 T cells) (l). m-o, Tumor growth curve (m), day 21 tumor weight (n), and frequency of tumor-infiltrating immune cells (o) in WT and Otub1-iKO (iKO) mice inoculated with B16F10 melanoma cells and, where indicated, injected with NK cell- and CD8 T cell-depletion antibodies (anti-NK1.1 and anti-CD8a) as depicted in Supplementary Fig. 6d). Data are representative of two (a-g), or three (h-o) independent experiments each with multiple biological replicates. Summary data are mean±s.e.m. with P values being determined by two-way ANOVA with Bonferroni’s post-test (a, f, i, m), two-tailed Student’s t-test (b-d, j-l, n-o), or Log-Rank (g). *< 0.05, **<0.01, ***<0.001, ****<0.0001.
To examine the therapeutic potential of targeting Otub1, we employed a mouse model of adoptive T cell therapy (ACT)47. We inoculated B6 mice with B16F10 melanoma cells and then treated the tumor-bearing mice by adoptive transfer of in vitro expanded CD8 T cells derived from WT or Otub1-TKO Pmel1 mice (Fig. 8e). The Pmel1 CD8 T cells recognize the tumor antigen gp100 expressed by B16F10 tumors. Compared to the WT Pmel1 CD8 T cells, the Otub1-TKO Pmel1 CD8 T cells were profoundly more effective in suppressing tumor growth and improving survival of the B16 tumor-bearing mice (Fig. 8f,g).
We next employed the Otub1-iKO model, in which Otub1 was inducibly deleted in adult mice in different cell types and challenged with B16F10 tumor cells (Fig. 8h). The Otub1-iKO mice had greatly reduced tumor burden compared to WT mice (Fig. 8i,j), associated with increased tumor-infiltrating CD8 T cells and NK cells as well as CD4 T cells and cDC1 cells (Fig. 8k). Moreover, tumor-infiltrating CD8 T cells in the Otub1-iKO mice contained a significantly higher frequency of effector cells expressing IFN-γ and Granzyme B (Fig. 8l). Similar results were obtained with the MC38 colon cancer model (Supplementary Fig. 6a-c). Antibody-mediated depletion of either CD8 T cells or NK cells impaired the potent anticancer immunity of Otub1-iKO mice, causing the increase of tumor burden to a level similar to or higher than that in WT mice (Fig. 8m,n, Supplementary Fig. 6d,e). NK cell depletion in Otub1-iKO mice drastically reduced the tumor-infiltrating cDC1 and CD4 T cells, whereas CD8 T cell depletion partially reduced the tumor-infiltrating cDC1, but not CD4 T, cells (Fig. 8o). These results suggest that hyper-activation of CD8 T cells and NK cells contributes to the strong anticancer immunity in the Otub1-iKO mice.
To assess the role of Otub1 in regulating antitumor immunity in human cancers, we analyzed cancer databases for potential correlation of Otub1 expression with T cell gene signature in tumors. Interestingly, our analysis of human skin cutaneous melanoma databases revealed a remarkable inverse correlation between Otub1 expression levels and the abundance of CD8 effector T cell gene signature as well as patient survival (Supplementary Fig. 7). Collectively, these results establish Otub1 as an important regulator of antitumor immunity and implicate Otub1 as a potential target for cancer immunotherapy.
Discussion
The results presented here suggest a ubiquitin-dependent mechanism that regulates IL-15R signaling and the IL-15-dependent homeostasis of CD8 T cells and NK cells and establish the DUB Otub1 as a crucial regulator. Otub1 controls IL-15-stimulated ubiquitination and activation of AKT, a kinase mediating the activation and metabolic reprograming of CD8 T cells. Despite the abundant expression of Otub1 in CD4 T cells, the Otub1 deficiency had no effect on the homeostasis of CD4 T cells. This cell type-specific function of Otub1 is explained by its role in regulating IL-15R signaling, which is specifically required for the homeostasis of CD8 T cells and NK cells7, 8, 26.
Our data suggest that homeostatic exposure of CD8 T cells to IL-15 serves as a crucial priming step for antigen-specific CD8 T cell activation, which is controlled by Otub1. T cell-specific deletion of Otub1 rendered CD8 T cells hyper-responsive to bacterial infections in vivo and to activation by TCR-CD28 signals in vitro. This phenotype was due to aberrant priming of the naive CD8 T cells by IL-15, since it was not detected in IL-15Rα-deficient mice. In CD8 T cells and NK cells, Otub1 is located to the membrane compartment. The membrane localization of Otub1 was dependent on IL-15 signaling, thus implicating Otub1 as a checkpoint of IL-15-mediated CD8 T cell priming. Since AKT activation occurs in various membrane compartments33, these findings suggest that the membrane localization of Otub1 may facilitate its role in regulating AKT activation.
Otub1 regulates different aspects of CD8 T cell activation and function. Otub1 deficiency sensitized CD8 T cells for activation by both TCR-CD28 stimuli and listeria infections and promoted generation of antigen-specific effector cells. The crucial role of Otub1 in regulating CD8 T cell responses was also revealed by the development of vitiligo in Otub1-TKO Pmel1 mice, which was due to aberrant CD8 T cell activation by the melanocyte self-antigen gp100. Another important function of Otub1 was to regulate the metabolic reprograming of activated CD8 T cells, an essential mechanism for supporting proliferation, effector cell generation and function40. This function of Otub1 is in line with its role in AKT regulation, since AKT is a master kinase mediating the activation, metabolism, and effector functions of CD8 T cells18, 19, 20.
Inducible deletion of Otub1 in adult mice greatly promoted tumor rejection, associated with increased tumor-infiltration with various immune cells, including CD8 T cells, NK cells, as well as CD4 T cells and cDC1 cells. Depletion of either NK cells or CD8 T cells impaired the anticancer immunity, erasing the differences between the WT and Otub1-iKO mice in tumor rejection. Antibody-mediated cell depletion studies revealed a crucial role for NK cells in mediating the recruitment of CD4 T cells and cDC1 cells in the Otub1-iKO mice. In an adoptive T cell therapy model, Otub1 deletion also profoundly enhanced the tumor-rejection activity of CD8 effector T cells, which was consistent with the role of Otub1 in regulating the metabolism and effector molecule expression of activated CD8 T cells. These findings implicate Otub1 as a potential drug target for cancer immunotherapy.
The role of ubiquitination in regulating IL-15R signaling has been poorly defined. Our present study demonstrated Otub1 as a DUB specifically regulating AKT axis of IL-15R signaling. A central step in AKT activation is its recruitment to the plasma membrane, where it is activated via S473 phosphorylation by mTORC2 and T308 phosphorylation by PDK148. The membrane recruitment of AKT involves its binding, via N-terminal PH domain, to the membrane phospho-lipid PIP3. Our data suggest that Otub1-mediated AKT deubiquitination attenuates its binding to PIP3. Notably, the ubiquitination site, K14, of AKT is located in its PH domain. It is thought that inactive AKT exists in a closed conformation due to intramolecular interaction between its N-terminal PH domain and C-terminal kinase domain38. We propose that ubiquitination of AKT in its PH domain may interfere with the intramolecular interaction, thereby facilitating the exposure of PH domain for PIP3 binding.
ONLINE METHODS
Mice
The Otub1-flox mice (in B6 genetic background) were generated using embryos obtained from The European Conditional Mouse Mutagenesis Program (EUCOMM, strain Otub1tm1a(EUCOMM)Hmgu). Otub1-flox mice were crossed with CD4Cre transgenic mice (both in B6 genetic background and from Jackson Laboratories) to produce age-matched Otub1+/+CD4Cre (named WT) and Otub1fl/flCD4Cre (named T cell-conditional Otub1 knockout or TKO) mice. The Otub1-flox mice were also crossed with ROSA26-CreER (Jackson Laboratories) to generate Otub1+/+CreER and Otub1fl/flCre-ER mice, which were then injected i.p. with tamoxifen (2 mg per mouse) in corn oil daily for four consecutive days to induce Cre function for generation of WT and induced Otub1 KO (iKO) mice. OT-I and Pmel1 TCR–transgenic mice, B6.SJL (CD45.1+), C57BL/6, Rag1-KO, and Il15ra-KO mice were from Jackson Laboratory. Experiments were performed with young adult (6–8 weeks) female and male mice except where indicated otherwise. All mice were in B6 genetic background and maintained in a specific pathogen–free facility of The University of Texas MD Anderson Cancer Center, and all animal experiments were done in accordance with protocols approved by the Institutional Animal Care and Use Committee of the University of Texas MD Anderson Cancer Center.
Cell lines
The HEK293T, B16F10, MC38 were from ATCC, and B16-OVA was provided by Qing Yi (Cleveland Clinic). The KIT225 T cell line stably transfected with IL-15Rα (15R-KIT)49 was provided by Dr. Sigrid Dubois (NCI/NIH) and cultured in RPMI 1640 medium supplemented with 10% FBS, antibiotics and human IL-2 (0.5 nM).
Plasmids, antibodies and reagents
pMIGR1-HA-AKT was generated by inserting human AKT1 cDNA into the EcoRI and BglII sites of the retrovirus vector pMIGR1 downstream of an HA tag, and the AKT mutants (K8R, K14R, E17K) were created by site-directed mutagenesis. The pcDNA3 expression vectors for Flag-tagged Otub1 and Otub1 C91S mutant were provided by Dr. Danuek Durocher (Lunenfeld-Tanenbaum Research Institute), and Flag-Otub1 C91S/D88A mutant was generated by site-directed mutagenesis. pPRIChp-Otub1-HA and pPRIChp-Otub1C91S/D88A-HA were generated by inserting human Otub1 and Otub1 C91S/D88A into the pPRIChp-HA retroviral vector (provided by Dr. Patrick Martin, University of Nice Sophia Antipolis). PRK5-HA-ubiquitin WT, K63, and K48 were obtained from Addgene (Plasmid #17608, #17605, #17606). Ubiquitin K63 and K48 harbor lysine-to-arginine substitutions at all lysines, except lysine 63 and lysine 48, respectively. pLenti puro HA-ubiquitin was obtained from Addgene (plasmid # 74218), and pLenti puro HA-Ub-AKT was generated by inserting human AKT1 cDNA into pLenti puro HA-ubiquitin immediately downstream of the ubiquitin cDNA. pLenti puro HA-Ub-AKT K14R was created by site-directed mutagenesis. pLenti puro HA-UbK63-AKT, and HA-UbK63-AKT K14R were generated by replacing WT ubiquitin with UbK63 in the pLenti HA-Ub-AKT and HA-Ub-AKT K14R vectors. T7-AKT was generated by inserting human AKT1 cDNA into the BamH1 and XbaI sites of T7-RelA vector (Addgene, #21984) to replace the RelA cDNA.
Functional grade anti–mouse (m) CD3ε (145–2C11) and anti-mCD28 (37.51) antibodies were from eBioscience. Goat anti-hamster IgG (H+L) was from Southern biotech. Mouse IL-15 monoclonal antibody (AIO.3) used for in vivo IL-15 neutralization was from eBioscience. Antibodies for AKT1 (B-1; used for immunoblotting assays), ERK½ (K-23), Ubiquitin (P4D1), SLP76 (H-300), Zap70 (1E7.2), P85α (B-9) and PTEN (A2B1) were from Santa Cruz Biotechnology. Anti-AKT (40D4; used for IP) was from Cell Signaling, and anti-Otub1 (EPR13028(B)) was from Abcam. Anti-Actin (C-4), and horseradish peroxidase–conjugated anti-Flag (M2) were from Sigma-Aldrich. Antibodies for phospho-AKT1 S473 (D9E), phospho-AKT1 T308 (C31E5E), phospho-FoxO1 Thr24/FoxO3a Thr32, phospho-S6K1 Thr421/Ser424, phospho-S6 Ser235/236 (D57.2.2E), phospho-Stat5 Tyr694 (C11C5), phosphor-SLP76 Ser376, phosphor-Zap70 Tyr329/Syk Tyr352, S6K1 (49D7), S6 (54D2), FoxO1 (C29H4), Foxo3a (75D8), HK2 (C64G5), α-Tubulin, IGF1Rβ (111a9), and Stat5 were from Cell Signaling Technology. Horseradish perxidase–conjugated anti-hemagglutinin (HA-7) was from Roche. The anti-CD8 (YTS169.4) and anti-NK1.1 (PK136) neutralizing antibodies were purchased from BioXCell.
Fluorescence-labeled antibodies for mCD4 (L3T4), mCD8 (53–6.7), mCD3 (145–2C11), CD44 (IM7), mCD62L (MEL-14), mTCRβ (H57–597), mCD45.1 (A20), mCD45.2 (104), mCXCR3 (CXCR3–173), mFoxp3 (FJK-16S), mCD45(30-F11), mNK1.1(PK136), mCD11c(N418), mMHCII(M5/114.15.2), mCD64(X54–5/7.1), mCD11b(M1/70), mIL-2 (JES6-SH4), mTNF (MP6-XT22) mGranzyme B (NGZB) and mIFN-γ (XMG1.2) were purchased from eBioscience. mCD24(M1/69) and mCD103(M290) were from BD and mCCL5(2E9/CCL5) was ordered from Biolegend. Glut1 (EPR3915) was from abcam.
Recombinant mouse IL-15, IL-2, IL-12, IL-18 and human IL-15 cytokines were from R&D. human IL-2 were requested from NCI. The ELISA reagents for mouse IL-2, TNF, IFN-γ were from eBioscience. PIP3 beads and ELISA kits for detecting the activity of PI3K and PTEN were from Echelon. The GP10025–33 and OVA257–264 were ordered from ANAspec. The AKT inhibitor ½ (AKTi) was from Calbiochem.
Flow cytometry analysis and cell sorting
Single-cell suspensions of splenocytes and lymph node cells were subjected to flow cytometry analysis and cell sorting as previously described50 using FACS fortessa and FACSAria (BD Biosciences). For intracellular cytokine staining (ICS) assays, T cells isolated from spleen, draining lymph nodes, or tumors of mice or from in vitro cultures were stimulated for 4 hours with PMA (50 ng/mL) and ionomycin (500 ng/mL) in the presence of monensin (10 μg/mL) during the last hour. The stimulated cells were fixed in 2% paraformaldehyde and permeablized in 0.5% saponin and then subjected to cytokine staining flow cytometry analyses. FACS data were analyzed in FlowJo 9.7.7 and proliferation index of CFSE labeled cells were calculated in FlowJo 10 proliferation modeling module. Gating strategies are summarized in Supplementary Fig. 8.
L. monocytogenes infection
Age- and sex-matched WT and KO mice (6–8 wk old) were infected i.v. with 1 × 105 colony-forming units of OVA-expressing recombinant L. monocytogenes (LM-OVA)51 (provided by Dr. Hao Shen, University of Pennsylvania). One day 7 post-infection, the mice were sacrificed for analysis of OVA-specific CD8 effector T cells in the spleen. Briefly, splenocytes were stimulated for 6 h with 10 μg/ml of OVA257–264 peptide (SIINFEKL, Genemed Synthesis), in the presence of a protein transport inhibitor, monensin, during the last hour, and then subjected to intracellular IFN-γ staining and flow cytometry analysis. 2 × 104 colony-forming units of LM-OVA were used to infect WT OT-I and Otub1-TKO OT-I mice. On day 7 post-infection, splenocytes were collected and stimulated for 6 h with OVA257–264 peptide (10 μg/ml), with monensin being added during the last hour, and then subjected to intracellular IFN-γ staining and flow cytometry analysis.
Tumor models
Age- and sex-matched WT and Otub1-TKO or WT and Otub1-iKO mice were injected s.c. with 2 × 105 murine melanoma cells B16F10 or B16-OVA or with 2 ×106 MC38 colon cancer cells and monitored for tumor growth. Mice were sacrificed and considered lethal when their tumor size reached 225 mm2 based on protocols approved by the Institutional Animal Care and Use Committee of the University of Texas MD Anderson. At the indicated time point, all mice were sacrificed for flow cytometric analysis of immune cells from both the draining lymph nodes and tumors. For CD8 T cell and NK cell depletion experiments, Age- and sex-matched WT and Otub1-iKO mice were inoculated s.c. with 2 × 105 B16F10 melanoma cells and also injected i.p. with an anti-CD8 (clone YTS169.4) and anti-NK1.1(clone PK136) neutralizing antibodies (100 μg) as depicted in Supplementary Fig. 6d.
Adoptive cell therapy (ACT) was performed using Pmel1 CD8 T cells recognizing the B16 melanoma antigen gp100. Briefly, splenocytes were isolated from WT Pmel1 or Otub1-TKO Pmel1 mice and stimulated in vitro using plate-coated anti-CD3 (1 μg/ml) and soluble anti-CD28 (1 μg/ml) antibodies. The culture was provided with mIL-2 (10 ng/ml) on day 2, and CD8 T cells were purified from the culture on day 5 and used for adoptive transfer experiment. To generate tumor-bearing mice, WT B6 mice were injected s.c. with B16F10 melanoma cells. After four days, the tumor-bearing mice were subjected to whole-body irradiation (500 rads, 137Cs irradiator) to induce lymphodepletion. One day after the irradiation, the mice were injected with the in vitro activated WT Pmel1 or Otub1-TKO Pmel1 CD8 T cells (6 × 105). Control mice were not irradiated or injected with Pmel1 T cells. Tumor size was measured every other day for the indicated time period.
Mixed bone marrow and mixed T cell adoptive transfer
Bone marrow cells (2 × 106) isolated from Otub1-TKO (CD45.2+) mice were mixed with bone marrow cells from WT B6.SJL (CD45.1+) mice in 1:1 ratio and adoptively transferred into irradiated (1000 rad) Rag1-KO mice. After 6 weeks, the bone marrow chimeric mice were sacrificed for analyzing the homeostasis of T cells derived from WT (B6.SJL) and Otub1-TKO bone marrows by flow cytometry based on the CD45.1 and CD45.2 congenital markers.
For mixed T cell transfer, WT (CD45.1+) and Otub1-TKO (CD45.2+) naive CD8 T cells (WT: CD45.1+; TKO: CD45.2+) or WT and Otub1-TKO naive OT-I CD8 T cells (WT OT-I: CD45.1+CD45.2+; TKO OT-I: CD45.2+) were labeled with CFSE dye, mixed in 1:1 ratio, and adoptively transferred into WT and Il15ra-KO mice. The transferred WT and Otub1-TKO CD8 T cells were analyzed by flow cytometry at the indicated time point. In some experiments, the Il15ra+/+ and Il15ra−/− recipient mice were sublethally irradiated (600 rads, 137Cs irradiator) to examine the role of IL-15 in mediating lymphopenic proliferation of CD8 T cells.
Metabolic Assays
OCR and ECAR were measured with an XF96 extracellular flux analyzer (Seahorse Bioscience) following the manufacturer’s instruction. Briefly, WT or Otub1-TKO CD8 naive T cells, either freshly isolated or in vitro activated with anti-CD3 plus anti-CD28 (for 24 h), were seeded in XF96 microplates (150,000 cells/well). The plates were quickly centrifuged to immobilize the cells. After incubation in a non-buffered assay medium (Seahorse Biosciences) in a non-CO2 incubator for 30 min, the cells were subjected to glycolysis assays with a XF glycolysis stress test kit (Seahorse Biosciences). Initial measurement of ECAR was done when cells were incubated in a glycolysis stress test medium without glucose to record the baseline. Glucose (10 mM) was then injected to induce ECAR, reflecting glycolysis rate under basal conditions. Subsequently, oligomycin (1μM) was injected to inhibit mitochondrial ATP production and shift the energy production to glycolysis, thereby measuring the maximum glycolytic capacity (also called stressed ECAR). Finally, a glucose analog, 2-deoxy-glucose (2-DG, 100 mM) was injected to inhibit glycolysis through targeting glucose hexokinase, resulting in decreased ECAR that served as a measure to confirm the glycolysis-dependence of the detected ECAR. Inhibitor studies were carried out by culturing the cells in 24-well plates (4 × 106 cells per well) in the presence of indicated concentrations of AKT½ inhibitor or DMSO.
The Mito stress test kit (Seahorse Biosciences) was used to measure OCR under different conditions. After initial measurement of baseline OCR, 1 μM oligomycin was injected to calculate ATP-linked respiration, followed by injection of the protonophore FCCP (0.25 μM) that uncoupled oxygen consumption from ATP production to obtain maximal OCR (also called stressed OCR). Lastly, 0.5 μM rotenone/antimycin A was injected to inhibit complex I and III and shut down ETC respiration for measuring non-mitochondrial respiration.
T-cell and NK cell purification and in vitro treatments
CD8 and CD4 T cells were isolated from splenocytes with anti-CD8- or anti-CD4-conjugated magnetic beads (Miltenyi), and naive CD8 or CD4 T cells were further purified by FACS sorting to get CD44loCD62Lhi population. The naive T cells were stimulated in replicate wells of 96-well plates (1 × 105 cells per well) for 66 h, and the culture supernatants were analyzed by ELISA (eBioScience). NK cells were isolated from splenocytes with NK cell isolation kit (Mietenyi). Purified NK cells were stimulated with IL2 (5 ng/ml), IL12 (10 ng/ml) and IL18 (10 ng/ml) for the indicated time periods and then subjected to flow cytometric analysis of intracellular granzyme B and CCL5.
RNA-sequencing analysis
Naïve CD8 T cells were isolated from the spleen of young (6–8 wk old) WT OT-I and Otub1-TKO OT-I mice and were either immediately lysed for RNA preparation or activated for 24 h with anti-CD3 (1μg/ml) plus anti-CD28 (1μg/ml). Total RNA was isolated with TRIzol (Invitrogen) and subjected to RNA-sequencing analysis using an Illumina sequencer in the Sequencing and Microarray Facility of the University of Texas MD Anderson Cancer Center. The raw reads were aligned to the mm10 reference genome (build mm10), using Tophat2 RNASeq alignment software. The mapping rate was 70% overall across all the samples in the dataset. HTseq-Count was used to quantify the gene expression counts from To-phat2 alignment files. Differential expression analysis was performed on the count data using R package DESeq2. P-values obtained from multiple binomial tests were adjusted using false discovery rate (Benjamini-Hochberg). Significant genes are defined by a Benjamini-Hochberg corrected p-value of cut-off of 0.05 and fold-change of at least two. RNA-sequencing data were analyzed by Genesis (http://genome.tugraz.at/) and multiplot (https://genepattern.broadinstitute.org/gp/pages/login.jsf ). RNA sequencing data were deposited to Gene Expression Omnibus with the accession code GSE126777.
Real-time quantitative PCR
RNA was extracted with TRIzol reagent from isolated WT OT-I or Otub1-TKO OT-I CD8 T cells. The RNA samples were subjected to quantitative PCR analyses using the SYBR regent (Bio-Rad). The expression of individual genes was calculated by a standard curve method and was normalized to the expression of Actb. Gene-specific primer sets used in this study (all for mouse genes) were listed in the following table.
| Actin | CGTGAAAAGATGACCCAGATCA | CACAGCCTGGATGGCTACGT | 71bp |
| Otub1 | GTAGCGACTCCGAAGGTGTT | ACCAGAGGATTCTGCACAGC | 100bp |
| Cxcr3 | AGCACCAGCCAAGCCATGTA | CGTAGGGAGAGGTGCTGTTTT | 97bp |
| Ccl5 | GCAGTCGTGTTTGTCACTCG | AGAGCAAGCAATGACAGGGA | 151bp |
| Bcl2 | TCTGTGCACTGTGCATCTCTC | GACTTGGTGCATGGAACACTG | 121bp |
| Eomes | TGAATGAACCTTCCAAGACTCAGA | GGTTATGGTCGATCTTTAGCTG | 108bp |
| Runx3 | TGTCAGCGTGCGACATGGCT | GAGTGAAGCGGCGGCTGGTG | 99bp |
| Runx2 | ATACCCCCTCGCTCTCTGTT | ACATAGGTCCCCATCTGCCT | 81bp |
| Ccl2 | GGGATCATCTTGCTGGTGAA | AGGTCCCTGTCATGCTTCTG | 127bp |
| Ccr5 | AGACATCCGTTCCCCCTACA | GCAGGGTGCTGACATACCAT | 107bp |
| Cxcr4 | CCATGGAACCGATCAGTGTGA | TTTTCATCCCGGAAGCAGGG | 106bp |
| Cd44 | CTCAGGAGCCCACAACGAGTGC | TCTGGGCTTCTTGCCTCTTGGGT | 78bp |
| Il12rb2 | CGGGAAGAGCTCTGGAGAACC | GCATTCTCTAACAGTCTGTGCC | 72bp |
| Tbx21 | GCCAGGGAACCGCTTATATG | GACGATCATCTGGGTCACATTGT | 136bp |
| Lef1 | TCATCACCTACAGCGACGAG | GGGTAGAAGGTGGGGATTTC | 104bp |
| Slamf1 | GCCTCTTATGCTTCAAACAACA | CAGCAGCATTGCCAAACAGT | 99bp |
| Ly6a | GAAACCCCTCCCTCTTCAGGA | AGGGCTGCACAGATAAAACTTC | 131bp |
| Hk2 | GATCGCCGGATTGGAACAGA | GGTCTAGCTGCTTAGCGTCC | 97bp |
| Glut1 | GCTGTGCTTATGGGCTTCTC | CACATACATGGGCACAAAGC | 114bp |
Retroviral and lentiviral infections
Retroviral particles were prepared using the indicated pMIGR1-GFP-based or pPRIChp-aHA-mCherry based expression vectors, as previously described50. For production of lentiviral particles, HEK293T cells were transfected (by calcium method) with pGIPZ lentiviral vectors encoding Otub1-specific shRNAs or a non-silencing control shRNA along with the packaging vectors psPAX2 and pMD2. 15R-KIT T cells were infected with the recombinant retroviruses or lentiviruses. After 48 h, the transduced cells were enriched by flow cytometric cell sorting based on GFP expression. For primary T cell infection, naive OT-I CD8 T cells were stimulated in 12-well plates for 24 h with plate-bound anti-CD3 (1ug/ml) plus anti-CD28 (1ug/ml) in the presence of 10 ng/ml IL-15 and 5 ng/ml IL-2 and then infected twice (at 48 h and 72 h) with retroviruses. 24 h after the second retroviral transduction, the infected T cells were starved in a low serum (0.5% FBS) medium overnight and then stimulated IL-15 (60 ng/ml) for signaling assays.
Immunoblot, co-immunoprecipitation, and ubiquitination assays
For immunoblot analysis of protein phosphorylation, naive CD4 and CD8 T cells or 15R-KIT T cell line cells were stimulated with IL-15 (60 ng/ml), IL-2 (60 ng/ml), or IL-7 (60 ng/ml) for the indicated time periods and lysed in a kinase cell lysis buffer supplemented with phosphatase inhibitors52. T cell stimulation with TCR and CD28 agonistic antibodies was performed using a crosslinking method52. Briefly, the cells were incubated on ice with anti-CD3 (2 μg/ml) and anti-CD28 (2 μg/ml), followed by crosslinking with goat anti-hamster Ig (25 μg/ml) for different time periods at 37ºC and then immediately lysed as described above for immunoblot assays.
Co-immunoprecipitations was performed essentially as described53. Primary OT-I CD8 T cells or 15R-KIT T cell line cells were stimulated with IL-15 (80 ng/ml) for the indicated time periods and lysed in a kinase cell lysis buffer52. Cell lysates were immediately subjected to immunoprecipitation using the indicated antibodies followed by immunoblot analysis of the precipitated proteins. For ubiquitination assays, stimulated T cells or transiently transfected HEK293 cells were lysed in RIPA buffer [50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% (vol/vol) Nonidet P-40, 0.5% (vol/vol) sodium deoxycholate, and 1 mM EDTA] supplemented with 6M urea and 4 mM N-ethylmaleimide. Lysates were diluted 1 time with RIPA buffer and then subjected to AKT immunoprecipitation, followed by detection of ubiquitinated AKT by immunoblot. Full scans of blots for the gel figures are provided in Supplementary Fig. 9-12.
Membrane protein detection
Membrane and cytosol protein fractions were isolated from CD4 and CD8 T cells or NK cells with Mem-Per Plus Kit (Thermo Fisher) and subjected to immunoblot assays. To test the role of IL-15 in mediating Otub1 membrane localization, OT-I mice injected (i.p.) with a mouse IL-15 neutralizing antibody (AIO.3; 200 μg/mouse) daily for three times, and CD8 T cells were isolated on day 4 for preparing membrane and cytosol protein fractions. In some experiments, a T cell adoptive transfer approach was used. Briefly, OT-I CD8 T cells were labeled with CFSE and adoptively transferred into Il15ra+/+ or Il15ra−/− recipient mice. After 7 days, the OT-I CD8 T cells were isolated from recipient mice for membrane and cytosol protein preparations.
Immune signature and survival analysis of human cancer
To correlate the expression level of Otub1 with the level of CD8 effector T cells in human cancer, we collected 10 well-defined CD8 T cell-associated genes to form the immune signature. We downloaded SKCM tumor samples (n=458), including clinical and mRNA expression information, from http://www.oncolnc.org/ and submitted the compiled dataset to GenePattern (https://genepattern.broadinstitute.org/gp/pages/login.jsf ) to do unsupervised hierarchical clustering analysis. Survival data from different clusters were used to do Kaplan-Meier estimation in GraphPad Prism software.
Statistical analysis
For the tumor clinical scores, differences between groups were evaluated by two-way ANOVA with Bonferroni’s post-test. For survival, differences between groups were evaluated by Log-Rank test. Other statistical analyses were performed by two-tailed unpaired T test using the Prism software. P values less than 0.05 were considered significant, and the level of significance was indicated as *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
In our animal studies, 3–4 mice were required for each group based on our calculation to achieve a 2.3 fold change (effect size) in two-tailed T -test with 90% power and a significance level of 5%. All statistical tests justified as appropriate, and the data met the assumptions of the tests. The variance was similar between the groups that are being statistically compared.
Data availability
RNA sequencing datasets were deposited to Gene Expression Omnibus with the accession code GSE126777. Other datasets generated during the current study are available from the corresponding author on reasonable request. The human skin cutaneous melanoma datasets reported by other studies were downloaded from http://www.oncolnc.org/.
Supplementary Material
Acknowledgements
We thank Dr. S. Dubois and Q. Yi for cell lines; Drs. D. Durocher and P. Martin for plasmids. This study was supported by National Institutes of Health grant AI64639 (to S.-C.S.) and partially supported by the following grants: National Institutes of Health Nos. AI057555 and GM84459 (to S.-C.S.), No. AI121458–01A1 (to K.S.), and No AI145287 (to P.L.); Cancer Prevention & Research Institute of Texas No. RP160188 (to K.S.); Bridge Biotherapeutics, Inc (to S.-C.S.); Mission Therapeutics (to S.-C.S.); and Nektar Therapeutics (to K.S.). This study was also supported by the NIH/NCI grant P30 CA016672 and used the flow cytometry, sequencing and microarray, and animal facility of the CCSG-shared resources at The MD Anderson Cancer Center.
Footnotes
Competing financial interests statement
J.Y. is an employee of Therapeutic Tumor Microenvironment Strategies, Inc, S.-C.S. received research funding from Bridge Biotherapeutics, Inc and Mission Therapeutics, Inc, and the other authors declare no competing financial interests.
References
- 1.Durgeau A, Virk Y, Corgnac S & Mami-Chouaib F Recent Advances in Targeting CD8 T-Cell Immunity for More Effective Cancer Immunotherapy. Front Immunol 9, 14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiossone L, Dumas PY, Vienne M & Vivier E Natural killer cells and other innate lymphoid cells in cancer. Nat Rev Immunol 18, 671–688 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Crouse J, Xu HC, Lang PA & Oxenius A NK cells regulating T cell responses: mechanisms and outcome. Trends Immunol 36, 49–58 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg J & Huang J CD8(+) T Cells and NK Cells: Parallel and Complementary Soldiers of Immunotherapy. Curr Opin Chem Eng 19, 9–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Surh CD & Sprent J Homeostasis of naive and memory T cells. Immunity 29, 848–862 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Castillo EF & Schluns KS Regulating the immune system via IL-15 transpresentation. Cytokine 59, 479–490 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schluns KS, Kieper WC, Jameson SC & Lefrancois L Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat Immunol 1, 426–432 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Schluns KS & Lefrancois L Cytokine control of memory T-cell development and survival. Nat Rev Immunol 3, 269–279 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Liu K, Catalfamo M, Li Y, Henkart PA & Weng NP IL-15 mimics T cell receptor crosslinking in the induction of cellular proliferation, gene expression, and cytotoxicity in CD8+ memory T cells. Proc Natl Acad Sci U S A 99, 6192–6197 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshpande P et al. IL-7- and IL-15-mediated TCR sensitization enables T cell responses to self-antigens. J Immunol 190, 1416–1423 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teague RM et al. Interleukin-15 rescues tolerant CD8+ T cells for use in adoptive immunotherapy of established tumors. Nat Med 12, 335–341 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Hu H & Sun SC Ubiquitin signaling in immune responses. Cell Res. 26, 457–483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun SC Deubiquitylation and regulation of the immune response. Nat. Rev. Immunol. 8, 501–511 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juang YC et al. OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol Cell 45, 384–397 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakada S et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466, 941–946 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Wang T et al. Evidence for bidentate substrate binding as the basis for the K48 linkage specificity of otubain 1. J Mol Biol 386, 1011–1023 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiener R, Zhang X, Wang T & Wolberger C The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature 483, 618–622 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gubser PM et al. Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nat Immunol 14, 1064–1072 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Kim EH & Suresh M Role of PI3K/Akt signaling in memory CD8 T cell differentiation. Front Immunol 4, 20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cammann C et al. Early changes in the metabolic profile of activated CD8(+) T cells. BMC Cell Biol 17, 28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogquist KA et al. T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994). [DOI] [PubMed] [Google Scholar]
- 22.Lodolce JP, Burkett PR, Koka RM, Boone DL & Ma A Regulation of lymphoid homeostasis by interleukin-15. Cytokine Growth Factor Rev 13, 429–439 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Burkett PR et al. IL-15R alpha expression on CD8+ T cells is dispensable for T cell memory. Proc Natl Acad Sci U S A 100, 4724–4729 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schluns KS et al. Distinct cell types control lymphoid subset development by means of IL-15 and IL-15 receptor alpha expression. Proc Natl Acad Sci U S A 101, 5616–5621 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldrath AW et al. Cytokine requirements for acute and Basal homeostatic proliferation of naive and memory CD8+ T cells. J Exp Med 195, 1515–1522 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guillerey C, Huntington ND & Smyth MJ Targeting natural killer cells in cancer immunotherapy. Nat Immunol 17, 1025–1036 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Chiossone L et al. Maturation of mouse NK cells is a 4-stage developmental program. Blood 113, 5488–5496 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Polansky JK et al. High dose CD11c-driven IL15 is sufficient to drive NK cell maturation and anti-tumor activity in a trans-presentation independent manner. Sci Rep 6, 19699 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bottcher JP et al. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell 172, 1022–1037 e1014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vadlakonda L, Dash A, Pasupuleti M, Anil Kumar K & Reddanna P The Paradox of Akt-mTOR Interactions. Front Oncol 3, 165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim EH et al. Signal integration by Akt regulates CD8 T cell effector and memory differentiation. J Immunol 188, 4305–4314 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waldmann TA The shared and contrasting roles of IL2 and IL15 in the life and death of normal and neoplastic lymphocytes: implications for cancer therapy. Cancer Immunol Res 3, 219–227 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jethwa N et al. Endomembrane PtdIns(3,4,5)P3 activates the PI3K-Akt pathway. J Cell Sci 128, 3456–3465 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Cantley LC The phosphoinositide 3-kinase pathway. Science 296, 1655–1657 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Carnero A, Blanco-Aparicio C, Renner O, Link W & Leal JF The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets 8, 187–198 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Balakirev MY, Tcherniuk SO, Jaquinod M & Chroboczek J Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 4, 517–522 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang WL et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 325, 1134–1138 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calleja V, Laguerre M, Parker PJ & Larijani B Role of a novel PH-kinase domain interface in PKB/Akt regulation: structural mechanism for allosteric inhibition. PLoS Biol 7, e17 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts DJ & Miyamoto S Hexokinase II integrates energy metabolism and cellular protection: Akting on mitochondria and TORCing to autophagy. Cell Death Differ 22, 248–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearce EL, Poffenberger MC, Chang CH & Jones RG Fueling immunity: insights into metabolism and lymphocyte function. Science 342, 1242454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng Y, Delgoffe GM, Meyer CF, Chan W & Powell JD Anergic T cells are metabolically anergic. J Immunol 183, 6095–6101 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKinney EF & Smith KGC Metabolic exhaustion in infection, cancer and autoimmunity. Nat Immunol 19, 213–221 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Huang PL et al. Skeletal muscle interleukin 15 promotes CD8(+) T-cell function and autoimmune myositis. Skelet Muscle 5, 33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Overwijk WW et al. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J Exp Med 198, 569–580 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang P, Cote AL, de Vries VC, Usherwood EJ & Turk MJ Induction of postsurgical tumor immunity and T-cell memory by a poorly immunogenic tumor. Cancer Res 67, 6468–6476 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maueroder C et al. Tumor Immunotherapy: Lessons from Autoimmunity. Frontiers in immunology 5, 212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Restifo NP, Dudley ME & Rosenberg SA Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol 12, 269–281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mishra A, Sullivan L & Caligiuri MA Molecular pathways: interleukin-15 signaling in health and in cancer. Clin Cancer Res 20, 2044–2050 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dubois S, Mariner J, Waldmann TA & Tagaya Y IL-15Ralpha recycles and presents IL-15 In trans to neighboring cells. Immunity 17, 537–547 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Yu J et al. Regulation of T-cell activation and migration by the kinase TBK1 during neuroinflammation. Nat. Commun. 6, 6074 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pearce EL & Shen H Generation of CD8 T cell memory is regulated by IL-12. J Immunol 179, 2074–2081 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Reiley WW et al. Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J. Exp. Med. 204, 1475–1485 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao G, Harhaj EW & Sun SC NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol. Cell. 7, 401–409 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing datasets were deposited to Gene Expression Omnibus with the accession code GSE126777. Other datasets generated during the current study are available from the corresponding author on reasonable request. The human skin cutaneous melanoma datasets reported by other studies were downloaded from http://www.oncolnc.org/.