Abstract
BACKGROUND/OBJECTIVES:
Acute pancreatitis (AP) is emerging in pediatrics. A subset of children with AP progresses to acute recurrent pancreatitis (ARP) and chronic pancreatitis (CP). The role of extensive gene testing in the progression has not been investigated previously. We have followed children enrolled in the registry and at our center for progression to ARP and CP after the first attack.
METHODS:
This study utilizes an extensive gene sequencing panel as a platform to evaluate the role of genetics in first attack AP, and the progression over time, from first attack to ARP and CP in children.
RESULTS:
Genes, with corresponding variants were involved in the 3 groups studied: AP, ARP and CP. We have shown that the presence of gene variants from the eight tested genes is enriched in the CP group compared to the AP and ARP groups. The presence of more than one gene was associated with CP (p=0.01). SPINK1 mutation(s) was significantly associated with faster progression to ARP, (p=0.04). Having a variant from CFTR, SPINK1 or PRSS1, was associated with the faster progression from AP to CP over time (p<0.05).
CONCLUSIONS:
This study shows that genetics have a significant role in progression to ARP and CP from the first attack of pancreatitis.
Keywords: pediatric pancreatitis, extensive genetic testing
INTRODUCTION
Pediatric acute pancreatitis (AP) is on the rise, with an incidence approaching adult AP. 1 A subset of children develop acute recurrent pancreatitis (ARP) and chronic pancreatitis (CP) after the first attack. 2 Studies in pediatric AP have been limited in sample size and the retrospective designs. 3 Studies that follow patients from first attack onwards, are lacking in children. Earlier studies in disease have classified genetic alterations into pathologic pathways that eventually lead to pancreatic inflammation and the clinical picture of pancreatitis. Those pathways are divided into trypsin- dependent, ductal and misfolding dependent pathways. The pathology involved in the mechanisms has been elucidated to the gene level which makes genetic studies very relevant for our understanding mechanistically and eventually would facilitate venues for therapies. 4 Genetics as a risk factor were shown to be involved in a significant portion of cases of ARP and CP in childhood.5, 6 Family history of pancreatitis have been linked to earlier disease.7-9 Genetics as a risk factor of all AP cases, including the first attack, have not been well studied. It is important to study pediatric AP as a distinct entity from adult AP as etiologies and outcomes of the two entities vary greatly. In children AP results from a host of etiologies: systemic, viral, anatomic causes, gallstone, genetic risk factors, and trauma. 3, 10, 11 However, in adults AP is mostly associated with alcohol and gallstones. 12, 13
Given that ARP and CP have a significant disease burden for children and families14, studying factors that lead to ARP and CP is important to help advance knowledge, and design therapies that can halt disease progression. Investigating the role of genetics in AP, from the first attack to the progression to ARP and CP has not been elucidated before. We have developed and validated a high-throughput sequencing panel consisting of eight genes implicated in pancreatitis risk. We have established a prospective AP registry and biorepository at Cincinnati Children’s Hospital Medical Center (CCHMC) to enable epidemiologic studies of AP in children. This proposed study leverages our previous work in a novel design that allows for systematic longitudinal follow-up of AP and for the first time the generation of clinical and genetic models to predict AP complications.15 In this paper we cover clinical and genetic factors involved in first attack AP in children who were followed for the study duration without a recurrence, and children who developed ARP/CP during the study period.
METHODS
Patient selection: included patients from our prospective registry and a historic cohort from patients presenting at CCHMC with first attack of AP, we have collected clinical data for the entire duration of the study, and in case of a second attack separated more than one month from previous, a diagnosis of ARP was given. Data collected included patient demographics, presentation, hospital course, risk factors, family history, and repeated attacks. Children presenting to our hospital prior to December 2016 with at least one attack of pancreatitis and available DNA samples either with the biobank or through our registry were included. One group comprised AP and no recurrence, children who progressed from AP to ARP or CP during the follow up period were included to be the comparison group. Patients with known diagnosis of ARP or CP were also included in the comparison group. Patients with AP, ARP or CP were defined as per the INSPPIRE criteria.2 Research was approved by the CCHMC Institutional Review Board (IRB# 2012-4050 and IRB# 2010-2039).
Genomic DNA:
Peripheral blood samples were collected on patients (consent was obtained from parents for children less than 18 years of age, and assent was obtained from children age 11 to 17). DNA was extracted from blood using FlexiGene DNA kits.
The Pancreas Gene Panel included 8 genes known to increase risk for pancreatitis; 16-18causing hereditary pancreatitis (CPA1,19 PRSS120), sporadic pancreatitis (SPINK1,21 CFTR,23 CLDN2,24 CTRC25) and other types of pancreatic disease or may be candidates for pancreatitis genes(SBDS,26 CASR 21, 22). DNA enrichment and sequencing. Gene sequencing was performed using the TruSeq Amplicon–Pancreas gene Panel (CCHMC and Illumina, San Diego, CA, USA). Data analysis/variant classification. The sequenced reads obtained from MiSeq were subjected to the Molecular Genetics Laboratory Next Generation Sequencing (NGS) data analysis. Fastq files were converted to fasta and the high quality sequence reads were filtered, simultaneously mapped to human genome reference sequence (NCBI build 37) and variant calls were made using NextGENe software V2.15 (Softgenetics, LLC, State College, PA). In silico tools were used for variants interpretation. Variants were classified based on the ACMG guidelines:27, 28 Pathogenic/likely pathogenic (prevalence in affected statistically increased over controls, amino acid change, functional studies supportive), variants of unknown clinical significance VUCS (with functional studies available, case series, increased incidence over the general population without a clear establishment with the disease group, i.e. does not meet pathogenic neither benign), and likely benign/benign (functional studies show no deleterious effects, nonsegregation with disease). Sanger confirmation was performed for those variants that were reportable: VUCS, likely pathogenic and pathogenic. From our preliminary studies, analysis of the MiSeq demonstrated 98% or more of nucleotides were sequenced at 20x coverage which is considered adequate coverage. Details of the gene coverage on the panel are in Supplementary Table 1.
Statistical analysis:
Data were analyzed using SAS®, version 9.4 (SAS Institute, Cary, NC). Due to sample sizes and the distribution of variables, continuous data were summarized as medians with 25th and 75th percentiles (interquartile range) while categorical data were summarized as frequency counts with percentages. Chi-square and Fisher’s exact tests were used as appropriate for group comparisons of categorical variables. For continuous data, nonparametric Wilcoxon Rank Sum tests and Kruskal-Wallis tests were used as appropriate to compare characteristics between groups. Kaplan-Meier survival curves were used to estimate the time to ARP and CP for genetic and etiology factors. Statistical significance was set a priori at α=0.05.
RESULTS
AP is defined by meeting two out of three criteria, abdominal pain, lipase and/or amylase at or above 3 times the upper limit of normal, or imaging finding of AP. Having more than one attack separated by one month pain free interval, as defined by INSPPIRE is ARP. CP is defined by imaging findings, and either pain that is suggestive of pancreatic origin, endocrine or exocrine insufficiency.2
The three groups comparisons:
One hundred eleven patients provided consent to have their gene testing performed, and have AP but no ARP during follow-up (n=54), ARP (n=31), CP (n=26) at time of study. Study follow-up time ranged from a median of 3.3 years for AP (no ARP group), to 4.7 years in ARP and 5.2 years in CP groups. Age and gender were similarly distributed in the AP, ARP and CP groups (Table 1). Genetic etiologies were more common in the CP group (31%) compared to the other two groups, 6% in ARP and 19% in AP without ARP, p=0.06. Etiologies of first attack AP in all patients belonging to the three groups are shown in Table 1.
Table 1.
Patient characteristics and etiologies of pancreatitis in AP, ARP and CP groups.
| AP N=54 |
ARP N=31 |
CP N=26 |
P-value | |
|---|---|---|---|---|
| Gender (male) | 30 (56%) | 16 (52%) | 15 (58%) | 0.89 |
| Age 1st AP attack (years) | 13.0 (7.7-16.2) n=54 | 13.1 (9.7-16.1) n=31 | 9.4 (7.0-13.9) n=22 | 0.25 |
| Onset AP age | ||||
| Early (<6 yrs) | 10/54 (19%) | 5/31 (16%) | 8/26 (31%) | 0.34 |
| Age 2nd AP attack (years) | - | 14.5 (10.7-16.9) n=30 | 10.2 (7.9-14.4) n=21 | 0.07 |
| Time from AP to ARP (years) | - | 0.4 (0.2-1.3) n=30 | 0.5 (0.2-1.7) n=21 | 0.52 |
| Developed ARP within 12m of AP | - | 19/30 (63%) | 14/21 (67%) | - |
| Age CP (years) | - | - | 11.6 (7.9-14.4) n=25 | - |
| Time from AP to CP (years) | - | - | 1.6 (0.7-2.3) n=22 | - |
| Follow-up time (years) | 3.3 (2.5-3.9) n=54 | 4.7 (3.3-5.8) n=31 | 5.2 (4.3-6.2) n=25 | <0.0001 |
| Etiology (1st attack) | ||||
| Divisum | 1/52 (2%) | 4 (13%) | 2/25 (8%) | 0.08 |
| Biliary | 11/52 (21%) | 5 (16%) | 3/25 (12%) | 0.65 |
| Trauma | 5/52 (10%) | 1 (3%) | 1/25 (4%) | 0.60 |
| Genetic (Sanger confirmed) | 10 (19%) | 2 (6%) | 8 (31%) | 0.06 |
| Viral/systemic/toxic/drug | 22/52 (42%) | 16 (52%) | 6/25 (24%) | 0.11 |
| Metabolic | 1/52 (2%) | 1 (3%) | 1/25 (4%) | 0.80 |
| Idiopathic or other | 6/52 (12%) | 4 (13%) | 7/25 (28%) | 0.19 |
| Family history | 0.14 | |||
| None | 49 (91%) | 28 (90%) | 20/25 (80%) | |
| Pancreatitis | 2 (4%) | 2 (6%) | 5/25 (20%) | |
| Pancreatic cancer | 3 (6%) | 1 (3%) | 0 |
Data presented as n (%) or median (25th –75th percentile)
Genetic variants detected by TruSeq enrichment and Illumina sequencing, were tested in the eight genes involved in hereditary pancreatitis: CASR, CFTR, CLDN2, CPA1, CTRC, PRSS1, SBDS, and SPINK1. Twenty patients carried at least one genetic variant when tested for these genes; details of variant carriage are in Supplementary Table 2. The distribution of the following affected genes: CASR, CFTR, CPA1, CTRC, PRSS1 between AP, ARP and CP groups was not significantly different. However, the presence of SPINK1 gene involvement was associated with CP compared to the two other groups, p=0.003 (Table 2).
Table 2.
Type and number of genes involved in each group.
| AP Total N=54 |
ARP Total N=31 |
CP Total N=26 |
P-value | |
|---|---|---|---|---|
| Genetic Testing done, yes | 54 (100%) | 31 (100%) | 26 (100%) | |
| Genetic variants, yes | 10 (19%) | 2 (6%) | 8 (31%) | 0.06 |
| Genes | 11 genes | 2 genes | 12 genes | |
| CASR | 1 (2%) | 0 | 0 | 1.00 |
| CFTR | 6 (11%) | 1 (3%) | 5 (19%) | 0.15 |
| CPA1 | 2 (4%) | 1 (3%) | 0 | 1.00 |
| CTRC | 1 (2%) | 0 | 0 | 1.00 |
| PRSS1 | 1 (2%) | 0 | 3 (12%) | 0.06 |
| SPINK1 | 0 | 0 | 4 (15%) | 0.003 |
| More than one gene affected | 1 (2%) | 0 (0%) | 4 (15%) | 0.01 |
| Number of genes affected | - | |||
| 0 | 44 (81%) | 29 (94%) | 18 (69%) | |
| 1 | 9 (17%) | 2 (6%) | 4 (15%) | |
| 2 | 1 (2%) | 0 (0%) | 4 (15%) |
Having more than one gene affected from the panel significantly differed between the three groups with CP having the highest percentage of patients (15%) with more than one gene affected, p=0.01.
Progression from first attack to ARP:
When we studied the time from first attack to second attack of ARP, we observed that the number of genes involved (2 genes versus one gene or none) was not associated with the progression to ARP, p=0.52.
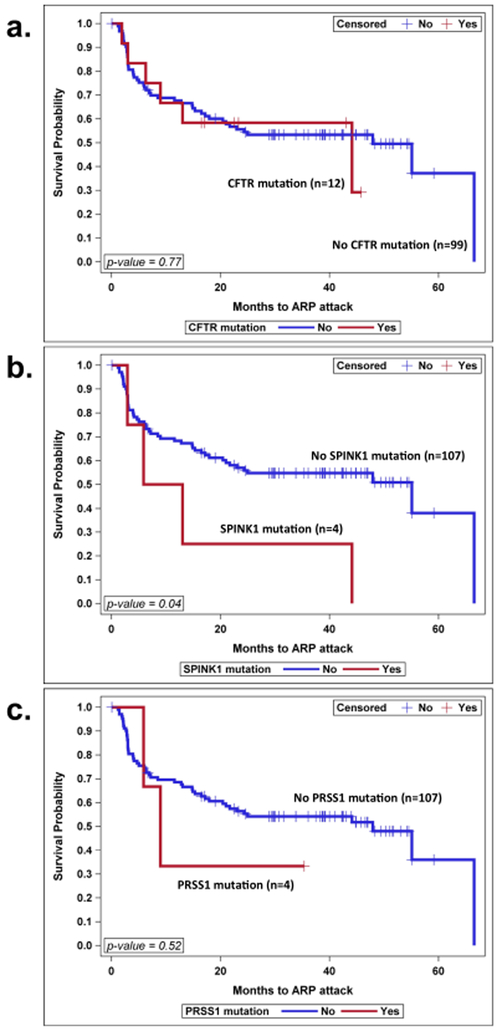
In terms of the type of genes involved in the progression to ARP, carrying a CFTR gene mutation(s), was not statistically significant in time to progression to ARP, p=0.77. PRSS1 mutation(s) was not statistically significant in progression to ARP, p=0.52, but having a SPINK1 mutation(s) was significantly associated with faster progression to ARP, p=0.04. Figure 1 (a, b and c).
Figure 1: Progression from AP to Second attack ARP over Time Differs per Genes Involved.
a. Progression in groups with or without CFTR gene involved, b. progression in groups with or without SPINK1 gene involved, c. progression with or without PRSS1 gene involved. Kaplan-Meier survival curves were used to analyze time to progression.
Progression from first attack to CP:
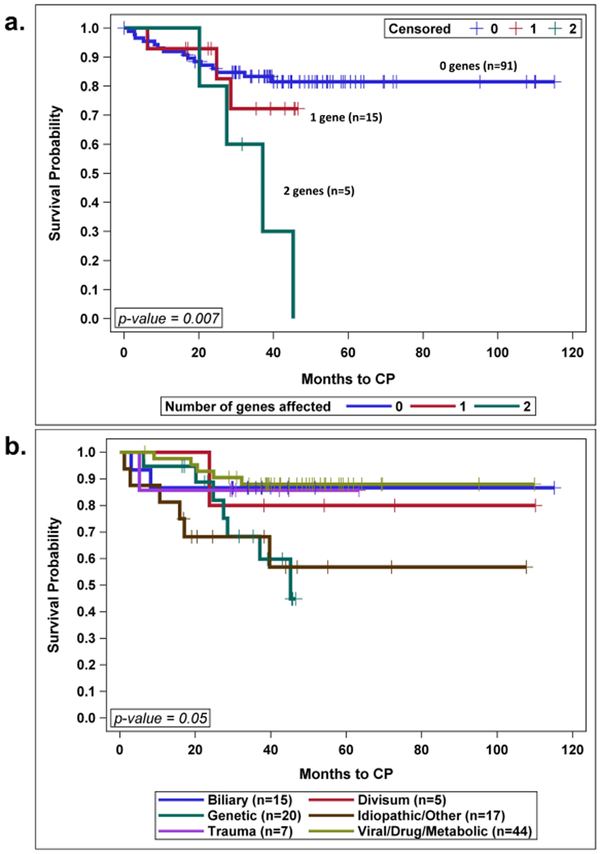
Having two genes involved was associated with faster progression to CP compared to having one or no genes involved, p=0.007, Figure 2a.
Figure 2: Progression to CP Over Time.
a. Progression to CP as a function of the number of genes involved. b. Progression rate to CP over time as a function of the etiology of the first attack. Kaplan-Meier survival curves were used to analyze time to progression.
First AP attack etiology affected progression to CP over time with genetics involvement, carrying the fastest rate of progression, p=0.05, Figure 2.b.
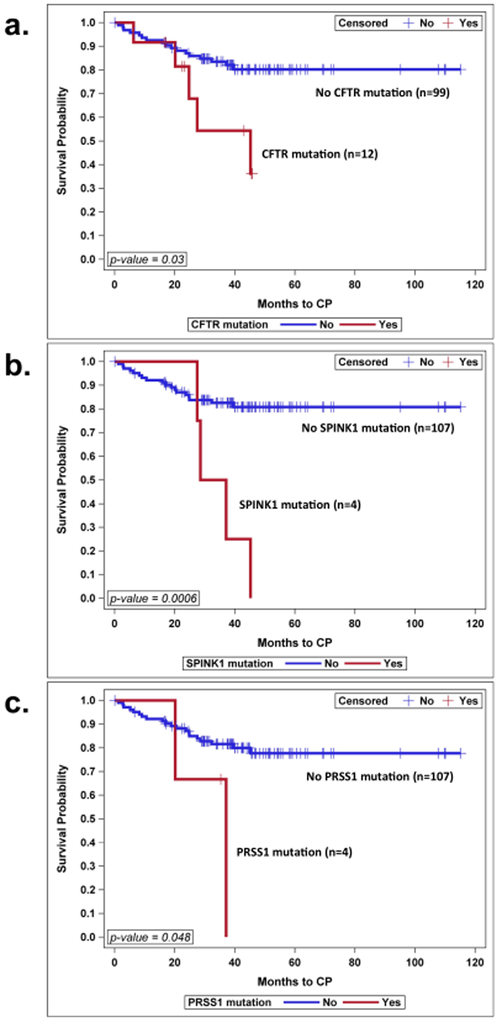
The progression to CP over time was faster in cases where genes were found positive for variants, compared to cases where gene variants were not present, in CFTR positive gene cases, p=0.03, SPINK1 positive gene cases, p=0.0006, and PRSS1 positive gene cases, p=0.048. Figure 3 (a, b and c).
Figure 3: Progression from AP to CP Over Time Differs per Genes Involved.
a. Progression in groups with or without CFTR gene involved, b. progression in groups with or without SPINK1 gene involved, c. progression with or without PRSS1 gene involved. Kaplan-Meier survival curves were used to analyze time to progression.
Sequencing results for other genes:
In this study we didn’t observe mutations in the following genes CLDN2 and SBDS for any of the patients tested.
DISCUSSION
This study evaluated the role of extensive NGS testing, using TruSeq enrichment followed by Illumina sequencing technology, to examine variants in genes known to be associated with hereditary pancreatitis. We applied the gene testing to 111 patients with pancreatitis, and found that 20 patients carried at least one genetic variant. The study is novel as it evaluated the role of genetics in patients who developed one attack of AP, and didn’t progress to ARP/CP over time (within the follow-up scope of this study), and compared those patients with the groups of ARP and CP. Another novel aspect of this study is that it used extensive genetic testing (more than one gene) and NGS as the most accepted technique in sequencing the genes known to be associated with hereditary pancreatitis in children with AP, ARP and CP.
The variants detected in our study are similar to those previously published in pediatrics, but add to our knowledge that cases of AP without progression to recurrent or chronic pancreatitis can have genes involved as well. Our patient population was similar to other published pediatric studies with pancreatitis presentation,29-32 however genes involved in AP without recurrence were not previously investigated. The positive genetic variant rate of 18% is lower than previous studies that ranged from 60-70% 6, 33 most likely given that our study included patients with the first attack of pancreatitis, without recurrence. We were interested in linking the different genotypes with the phenotypes of pancreatitis, to help us in following the clinical course of our patients. It is known from previous studies that genetics would affect the clinical manifestations of disease, for endocrine, exocrine, or pain patterns. 31, 33
The complex pathophysiology of CP likely involves the interplay of genetic and environmental factors leading to the fibrotic pancreatic destruction that differentiates patients with one attack compared to the groups that undergo further attacks and complete organ destruction eventually. This is manifested by fibrosis, destruction, ductal branching and calcifications; the late phenomena of CP.2 Diagnosing early CP remains challenging in pediatrics as in adults. Previous studies reported varying rates of genetic mutations associated with pancreatitis ranging from 30% to 73%.5, 6, 14, 33-35 Testing through gene panels holds promise to unravel the mystery of ARP and CP in children. Panels will increase efficiency and hopefully cut cost in gene testing. More over extensive testing, will allow studying gene to gene interactions in disease progression. In a small cohort of 50 patients, we found 11% of patients with more than one gene affected when tested for CFTR, SPINK1, PRSS1 and CTRC.33 A recent study of recurrent and CP patients showed genetic testing with NGS for a panel of 10 genes of interest: PRSS1, SPINK1, CFTR, CTRC, CASR, CTSB, KRT8, CLDN2, CPA1 and ATP8B1 to have variants in one or more of the genes of interest, with 18.8% having more than 1 gene affected.17 While this study had a small sample size, it highlights the importance of extensive gene testing with the expanded panels in cases of hereditary pancreatitis. The role of gene testing in first attack AP and how it affects progression wasn’t addressed in the previous designs.
While our study is novel, it has limitations. While it includes the most commonly known genes associated with pancreatitis, it does not involve all genes. Another limitation is in the fact that some of the findings such as in the SPINK1 gene may be related to the variant, rather than the gene itself, since all the positive findings were in the same risk allele in that case. Our data is generated from a single center, rather than covering a larger sample area, however as a quaternary referral center we capture patients from a wide geographic variation. Also, we are limited with the sample size.
Management of pancreatitis in children is mainly supportive, and focused on dealing with complications or sequalae of the disease. One day through discovery we can focus our efforts to reverse the trends of management to prevention. Our studies will allow identification of at-risk children and eventual development of interventions that can prevent these outcomes. As we advance our knowledge with known factors in ARP and CP, we will need more studies that are geared towards our clinical phenotypes, so that future studies can be tailored to halt progression from AP to ARP and CP in children.
Supplementary Material
Acknowledgments:
Funding: Maisam Abu-El-Haija was funded through an NIH grant R43 DK 105640-01. The funder provided support in the form of salaries for authors [MAH, CAV], but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. Disclaimer “The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health."
Footnotes
Conflicts of Interests: Dr. C. Alexander Valencia worked at Cincinnati Children’s when the study was conducted, he currently works at PerkinElmer genomics, Pittsburgh, PA, and this company has not provided any financial support for the study. Dr. Abu-El-Haija received in kind support from ChiroClin for research not related to this topic. All other authors declared no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Maisam Abu-El-Haija, Cincinnati Children’s Hospital Medical Center, Gastroenterology, Hepatology & Nutrition, 3333 Burnet Ave, Cincinnati, OH 45229, Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio.
C. Alexander Valencia, Cincinnati Children’s Hospital Medical Center during study period., Currently works at PerkinELmer Genomics, 250 Industry Drive, Pittsburgh, PA 15275.
Lindsey Hornung, Cincinnati Children’s Hospital Medical Center, Biostatistics and Epidemiology, 3333 Burnet Ave, Cincinnati, OH 45229.
Nour Youssef, Cincinnati Children’s Hospital, clinical rotation, LAU, School of Medicine, Lebanon.
Tyler Thompson, Cincinnati Children’s Hospital Medical Center, Gastrotenterology, Hepatology and Nutrition, 3333 Burnet Ave, Cincinnati, OH 45229.
Nathaniel W. Barasa, Research Assistant IV, Division of Human Genetics, Cincinnati Children’s Hospital Medical Center, Laboratory Genetics and Genomics, 3333 Burnet Ave, Cincinnati, OH 45229
Xinjian Wang, Division of Human Genetics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, Laboratory Genetics and Genomics, 3333 Burnet Ave, Cincinnati, OH 45229.
Lee A. Denson, Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, Ohio, Cincinnati Children’s Hospital Medical Center, Gastroenterology, Hepatology and Nutrition, 3333 Burnet Ave, Cincinnati, OH 45229
References:
- [1].Morinville VD, Barmada MM, Lowe ME. Increasing incidence of acute pancreatitis at an American pediatric tertiary care center: is greater awareness among physicians responsible? Pancreas. 2010;39:5–8. [DOI] [PubMed] [Google Scholar]
- [2].Morinville VD, Husain SZ, Bai H, Barth B, Alhosh R, Durie PR, et al. Definitions of pediatric pancreatitis and survey of present clinical practices. J Pediatr Gastroenterol Nutr. 2012;55:261–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bai HX, Lowe ME, Husain SZ. What have we learned about acute pancreatitis in children? J Pediatr Gastroenterol Nutr. 2011;52:262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mayerle J, Sendler M, Hegyi E, Beyer G, Lerch MM, Sahin-Toth M. Genetics and Pathophysiology of Pancreatitis. Gastroenterology. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Vue PM, McFann K, Narkewicz MR. Genetic Mutations in Pediatric Pancreatitis. Pancreas. 2016;45:992–6. [DOI] [PubMed] [Google Scholar]
- [6].Kumar S, Ooi CY, Werlin S, Abu-El-Haija M, Barth B, Bellin MD, et al. Risk Factors Associated With Pediatric Acute Recurrent and Chronic Pancreatitis: Lessons From INSPPIRE. JAMA Pediatr. 2016;170:562–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Giefer MJ, Lowe ME, Werlin SL, Zimmerman B, Wilschanski M, Troendle D, et al. Early-Onset Acute Recurrent and Chronic Pancreatitis Is Associated with PRSS1 or CTRC Gene Mutations. J Pediatr. 2017;186:95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jalaly NY, Moran RA, Fargahi F, Khashab MA, Kamal A, Lennon AM, et al. An Evaluation of Factors Associated With Pathogenic PRSS1, SPINK1, CTFR, and/or CTRC Genetic Variants in Patients With Idiopathic Pancreatitis. Am J Gastroenterol. 2017;112:1320–9. [DOI] [PubMed] [Google Scholar]
- [9].Pelaez-Luna M, Robles-Diaz G, Canizales-Quinteros S, Tusie-Luna MT. PRSS1 and SPINK1 mutations in idiopathic chronic and recurrent acute pancreatitis. World J Gastroenterol. 2014;20:11788–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Park AJ, Latif SU, Ahmad MU, Bultron G, Orabi AI, Bhandari V, et al. A comparison of presentation and management trends in acute pancreatitis between infants/toddlers and older children. J Pediatr Gastroenterol Nutr. 2010;51:167–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lopez MJ. The changing incidence of acute pancreatitis in children: a single-institution perspective. J Pediatr. 2002;140:622–4. [DOI] [PubMed] [Google Scholar]
- [12].Yadav D, Whitcomb DC. The role of alcohol and smoking in pancreatitis. Nat Rev Gastroenterol Hepatol. 2010;7:131–45. [DOI] [PubMed] [Google Scholar]
- [13].Wang GJ, Gao CF, Wei D, Wang C, Ding SQ. Acute pancreatitis: etiology and common pathogenesis. World J Gastroenterol. 2009;15:1427–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Schwarzenberg SJ, Bellin M, Husain SZ, Ahuja M, Barth B, Davis H, et al. Pediatric chronic pancreatitis is associated with genetic risk factors and substantial disease burden. J Pediatr. 2015;166:890–6.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Vujasinovic M, Tepes B, Makuc J, Rudolf S, Zaletel J, Vidmar T, et al. Pancreatic exocrine insufficiency, diabetes mellitus and serum nutritional markers after acute pancreatitis. World J Gastroenterol. 2014;20:18432–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Whitcomb DC. Genetic risk factors for pancreatic disorders. Gastroenterology. 2013;144:1292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xiao Y, Yuan W, Yu B, Guo Y, Xu X, Wang X, et al. Targeted Gene Next-Generation Sequencing in Chinese Children with Chronic Pancreatitis and Acute Recurrent Pancreatitis. J Pediatr. 2017;191:158–63 e3. [DOI] [PubMed] [Google Scholar]
- [18].Vitale DS, Abu-El-Haija M. Genetic Testing in Children with Recurrent and Chronic Pancreatitis. J Pediatr. 2017;191:10–1. [DOI] [PubMed] [Google Scholar]
- [19].Witt H, Beer S, Rosendahl J, Chen JM, Chandak GR, Masamune A, et al. Variants in CPA1 are strongly associated with early onset chronic pancreatitis. Nat Genet. 2013;45:1216–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Whitcomb DC, Gorry MC, Preston RA, Furey W, Sossenheimer MJ, Ulrich CD, et al. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat Genet. 1996;14:141–5. [DOI] [PubMed] [Google Scholar]
- [21].Felderbauer P, Klein W, Bulut K, Ansorge N, Dekomien G, Werner I, et al. Mutations in the calciumsensing receptor: a new genetic risk factor for chronic pancreatitis? Scand J Gastroenterol. 2006;41:343–8. [DOI] [PubMed] [Google Scholar]
- [22].Masson E, Chen JM, Ferec C. Overrepresentation of Rare CASR Coding Variants in a Sample of Young French Patients With Idiopathic Chronic Pancreatitis. Pancreas. 2015;44:996–8. [DOI] [PubMed] [Google Scholar]
- [23].LaRusch J, Jung J, General IJ, Lewis MD, Park HW, Brand RE, et al. Mechanisms of CFTR functional variants that impair regulated bicarbonate permeation and increase risk for pancreatitis but not for cystic fibrosis. PLoS Genet. 2014;10:e1004376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Whitcomb DC, LaRusch J, Krasinskas AM, Klei L, Smith JP, Brand RE, et al. Common genetic variants in the CLDN2 and PRSS1-PRSS2 loci alter risk for alcohol-related and sporadic pancreatitis. Nat Genet. 2012;44:1349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Beer S, Zhou J, Szabo A, Keiles S, Chandak GR, Witt H, et al. Comprehensive functional analysis of chymotrypsin C (CTRC) variants reveals distinct loss-of-function mechanisms associated with pancreatitis risk. Gut. 2013;62:1616–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, et al. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33:97–101. [DOI] [PubMed] [Google Scholar]
- [27].Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genetics in medicine : official journal of the American College of Medical Genetics. 2008;10:294–300. [DOI] [PubMed] [Google Scholar]
- [28].Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine : official journal of the American College of Medical Genetics. 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lucidi V, Alghisi F, Dall'Oglio L, D'Apice MR, Monti L, De Angelis P, et al. The etiology of acute recurrent pancreatitis in children: a challenge for pediatricians. Pancreas. 2011;40:517–21. [DOI] [PubMed] [Google Scholar]
- [30].Poddar U, Yachha SK, Mathias A, Choudhuri G. Genetic predisposition and its impact on natural history of idiopathic acute and acute recurrent pancreatitis in children. Digestive and liver disease : official journal of the Italian Society of Gastroenterology and the Italian Association for the Study of the Liver. 2015. [DOI] [PubMed] [Google Scholar]
- [31].Schwarzenberg SJ, Bellin M, Husain SZ, Ahuja M, Barth B, Davis H, et al. Pediatric chronic pancreatitis is associated with genetic risk factors and substantial disease burden. J Pediatr. 2015;166:890–6 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sobczynska-Tomaszewska A, Bak D, Oralewska B, Oracz G, Norek A, Czerska K, et al. Analysis of CFTR, SPINK1, PRSS1 and AAT mutations in children with acute or chronic pancreatitis. Journal of pediatric gastroenterology and nutrition. 2006;43:299–306. [DOI] [PubMed] [Google Scholar]
- [33].Palermo JJ, Lin TK, Hornung L, Valencia CA, Mathur A, Jackson K, et al. Genophenotypic Analysis of Pediatric Patients With Acute Recurrent and Chronic Pancreatitis. Pancreas. 2016;45:1347–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Masson E, Chen JM, Audrezet MP, Cooper DN, Ferec C. A conservative assessment of the major genetic causes of idiopathic chronic pancreatitis: data from a comprehensive analysis of PRSS1, SPINK1, CTRC and CFTR genes in 253 young French patients. PLoS One. 2013;8:e73522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wang W, Sun XT, Weng XL, Zhou DZ, Sun C, Xia T, et al. Comprehensive screening for PRSS1, SPINK1, CFTR, CTRC and CLDN2 gene mutations in Chinese paediatric patients with idiopathic chronic pancreatitis: a cohort study. BMJ Open. 2013;3:e003150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.