Abstract
RNA processing mutants have been broadly implicated in genome stability, but mechanistic links are often unclear. Two predominant models have emerged: one involving changes in gene expression that perturb other genome maintenance factors and another in which genotoxic DNA:RNA hybrids, called R-loops, impair DNA replication. Here we characterize genome instability phenotypes in yeast splicing factor mutants and find that mitotic defects, and in some cases R-loop accumulation, are causes of genome instability. In both cases, alterations in gene expression, rather than direct cis effects, are likely to contribute to instability. Genome instability in splicing mutants is exacerbated by loss of the spindle-assembly checkpoint protein Mad1. Moreover, removal of the intron from the α-tubulin gene TUB1 restores genome integrity. Thus, differing penetrance and selective effects on the transcriptome can lead to a range of phenotypes in conditional mutants of the spliceosome, including multiple routes to genome instability.
INTRODUCTION
Genome stability maintenance is a complex process involving the coordination of essentially all DNA transactions, including transcription, chromatin state, DNA replication, DNA repair, and mitosis. Regulation of genome stability is critical to prevent cancer (Hanahan and Weinberg, 2011). While screens across model organisms and human cells have implicated numerous genes as regulators of genome maintenance, in many cases we do not understand the mechanism of action (Paulsen et al., 2009; Stirling et al., 2011).
Defects in RNA processing have been implicated in genome instability across species and in both human cancer and repeat-expansion diseases (Richard and Manley, 2017). Indeed, RNA splicing factors are frequently mutated in cancers where they shift gene expression landscapes, favoring oncogenesis (Darman et al., 2015; Dolatshad et al., 2015; Joshi et al., 2017). Previous work has suggested that loss of splicing factors like pre-mRNA-splicing factor 2/alternative splicing factor (SF2/ASF) (Li and Manley, 2005), or treatment with splicing inhibitors (Wan et al., 2015), causes the accumulation of DNA:RNA hybrids in genomic DNA. These three-stranded R-loop structures contribute to genome instability by exposing single-stranded DNA (ssDNA) and by blocking replication forks, causing replication stress induced genome instability (Aguilera and Garcia-Muse, 2012; Chan et al., 2014b). Indeed, splicing has been ascribed a protective role in genome maintenance in yeast (Bonnet et al., 2017). More recently, cancer-associated mutations in splicing factors such as SRSF2 and U2AF1 have been attributed roles in R-loop prevention related to genome instability in myelodysplastic syndromes (Chen et al., 2018). These data compound other observations indicating that transcription termination (Skourti-Stathaki et al., 2011), 3′-end processing (Stirling et al., 2012), and mRNA packaging and export mutants (Gomez-Gonzalez et al., 2011) together create a robust R-loop prevention system.
Other data have suggested that splicing factor disruption, such as loss of CDK12, causes changes in gene expression, which reduce the activity of canonical genome maintenance factors (Blazek et al., 2011). Indeed, cancer-associated mutations in splicing factor SF3B1 have been shown to disrupt splicing of DNA damage response–related transcripts in myelodysplastic syndromes (Dolatshad et al., 2015). There is little evidence for gene expression changes driving genome instability in yeast splicing mutants, but cell-cycle delays, possibly linked to genome instability, have been previously connected to defective tubulin mRNA splicing in some mutants (Burns et al., 2002). Whether altered gene expression, R-loops, or both contribute to genome instability in splicing mutants is unclear.
We previously identified many spliceosome components whose disruption in yeast leads to genome instability (Stirling et al., 2011). Subsequent work found that only a handful of these genome destabilizing splicing mutants caused detectable increases in bulk R-loop levels (Chan et al., 2014a). Importantly, only 5% of yeast genes encode introns reducing the complexity of interpreting specific splicing changes as drivers of genome instability (Parenteau et al., 2008). Here we set out to test the contribution of R-loops versus gene expression changes in splicing-loss induced genome instability in yeast. While we observe evidence of R-loop induced DNA damage in a mutant allele of SNU114, all splicing mutants appear to cause aberrant splicing of the α-tubulin transcript from the TUB1 gene. A mitotic defect arising from Tub1 depletion is therefore a common driver of chromosome loss in yeast splicing mutants.
RESULTS AND DISCUSSION
Splicing factor mutations lead to chromosome instability
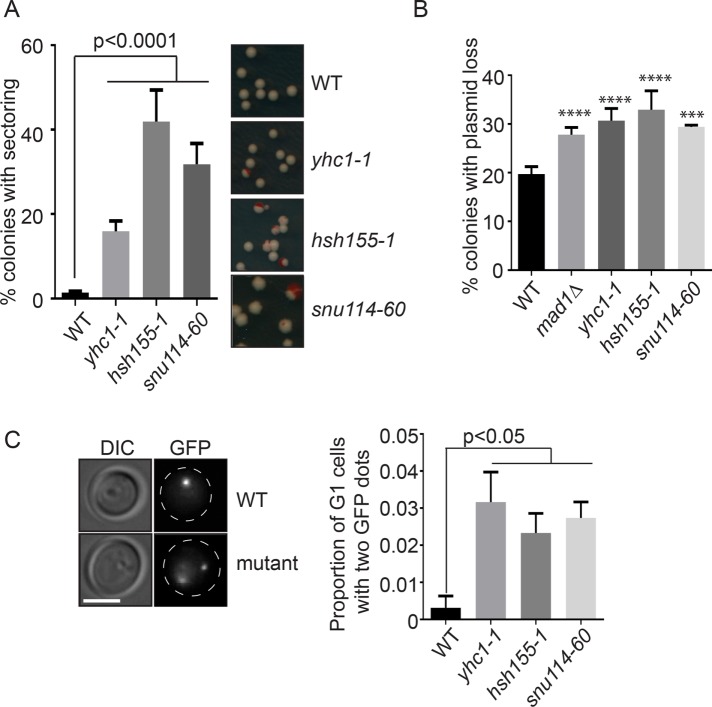
Previous screens have identified at least 25 splicing proteins that, when disrupted in yeast, lead to chromosomal instability (CIN) (Stirling et al., 2011). To begin to understand whether R-loops or other mechanisms drove genome instability, we conducted CIN assays in strains with mutations in each of the core snRNP complexes involved in establishing the splicing reaction (Measday and Stirling, 2016). We originally tested point mutations in HSH155 that were orthologous to those found in SF3B1 in human cancers; however, these alleles exhibited weak or no splicing defects and stable genomes, consistent with only minor effects seen in other studies (unpublished data) (Tang et al., 2016; Carrocci et al., 2017). Since each spliceosomal snRNP is essential, we instead used temperature-sensitive (ts) alleles of YHC1 (U1), HSH155 (U2), and SNU114 (U4/U6.U5), each of which had strong splicing defects as measured with a LacZ splicing reporter or at endogenously spliced gene RPL33B (Supplemental Figure S1, A and B). In all three mutants, we observed increases in artificial chromosome loss by the chromosome transmission fidelity (ctf) assay, which measures loss of an artificial chromosome fragment, confirming our previous findings (Stirling et al., 2011) (Figure 1A). We next measured the stability of a centromere (CEN) plasmid and found that splicing-defective cells have increased plasmid loss relative to wild type (WT) (Figure 1B). Since these two assays rely on the loss of episomes, we tested the stability of endogenous chromosomes by monitoring a strain with an integrated LacO array on ChrIII in cells expressing LacI-GFP to create a green fluorescent spot marking the chromosome. In haploid unbudded G1 cells, only a single GFP spot should be present; however, we observed that all splicing mutants tested showed an increased rate of gain of a LacI-GFP marked chromosome III, suggesting that a chromosome gain event has taken place (Figure 1C). Thus, multiple assays indicate splicing mutants led to a significant increase in chromosomal instability.
FIGURE 1:
Genome instability phenotypes of splicing mutants. (A) CTF phenotypes indicated by the percentage of sectored colonies. Right, representative images. (B) Plasmid loss frequency. ***p = 0.0005; ****p < 0.0001 (C) Frequency of unbudded cells with >1 GFP-marked LacO array. Left, representative images. Dashed lines indicate cell outlines; scale bar = 2 μm. Fisher’s exact test was used to calculate statistical significance. For all figures where applicable mean values with SEM bars are shown, n = 3.
R-loop accumulation and associated DNA damage in snu114-60
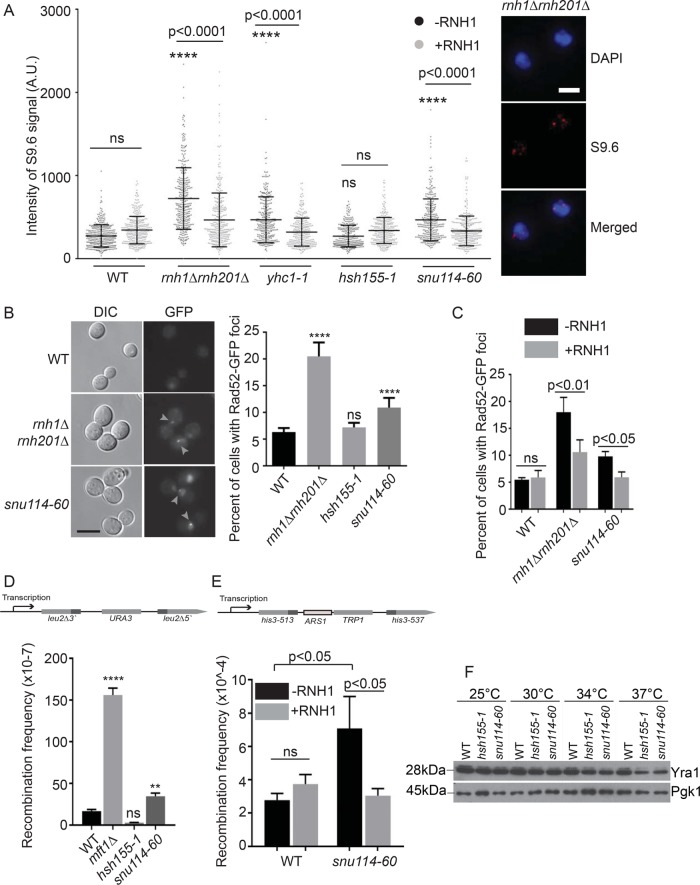
The CIN phenotypes observed in splicing mutants could arise by several mechanisms, including the formation of transcription coupled R-loops (Li and Manley, 2005; Bonnet et al., 2017). To test this model, we first performed chromosome spreads and used the S9.6 antibody to detect DNA:RNA hybrid levels in these spreads with immunofluorescence (Wahba et al., 2011). As reported previously, yhc1-1 and snu114-60 alleles showed higher levels of R-loop accumulation compared with WT, while the hsh155-1 allele had no increase in R-loops (Figure 2A) (Chan et al., 2014a). We elected to pursue comparison of snu114-60 and hsh155-1 since both alleles disrupt splicing and exhibit strong CIN phenotypes but have opposing effects on R-loop levels. In addition yhc1-1 cells are very sick across temperatures, making some assays technically unreliable. Consistent with the lack of R-loop accumulation in hsh155-1, when we analyzed Rad52-GFP foci, a marker of DNA damage repair that should increase if R-loops are driving genome instability, we found that only snu114-60 increased Rad52 foci frequency (Figure 2B). Importantly, this DNA damage phenotype could be partially suppressed by ectopic expression of RNaseH1 (RNH1), similarly to a rnh1Δrnh201Δ control strain (Figure 2C). To further test phenotypes known to correlate with aberrant R-loop levels, we used a plasmid-based direct repeat recombination system to test for hyperrecombination and again only snu114-60 showed a significant increase in recombination compared with WT (Figure 2D, mft1Δ is a hyperrecombination positive control [Chang et al., 2017]). We also tested the impact of snu114-60 on recombination at an integrated recombination substrate flanking a replication origin to assess the potential for replication-transcription conflicts that give rise to DNA damage and promote recombination. snu114-60 also causes hyperrecombination in this chromosomal context in a manner that was suppressed by RNH1 (Figure 2E). These data show that, while splicing alleles like snu114-60 contribute to R-loop accumulation and associated DNA damage, R-loops are not a unifying mechanism of genome instability across the various spliceosomal snRNP mutants (Chan et al., 2014a).
FIGURE 2:
R-loop-associated genome instability in snu114-60. (A) s9.6 antibody staining intensities in chromosome spreads. ****p < 0.0001 relative to WT -RNH1. Right, representative images; scale bar = 2 μm. (B) Frequency of Rad52-GFP foci (highlighted with gray arrows), scale bar = 5 μm. (C) Suppression of Rad52-GFP foci by ectopic RNH1 expression. (D, E) Frequency of direct repeat recombination on the indicated plasmid (D) or integrated genomic reporter (E). Reporter construct schematics are presented above each panel. (F) Representative Western blot of Yra1 protein level in the indicated strains and temperatures. (A, C, E) one-way ANOVA, (B) Fisher’s exact test, and (D) Student’s t test. **p < 0.01; ****p < 0.0001.
It was puzzling that snu114-60 led to increased recombination in reporters that do not contain introns and thus presumably do not recruit the spliceosome in either WT or snu114-60 cells. This raised the possibility that the expression of an R-loop regulator was sensitive to disruption of Snu114 activity and that the snu114-60 mutation selectively depleted this factor. One candidate gene for this phenotype is YRA1 which encodes an RNA export protein whose function has been linked to transcription-associated recombination and R-loop formation (Jimeno et al., 2002; Gavalda et al., 2016; Garcia-Rubio et al., 2018). Measuring mRNA expression levels and splicing of YRA1 by quantitative reverse transcription PCR (RT-qPCR) indicated that YRA1 is overexpressed in both hsh155-1 and snu114-60 relative to WT control and that both alleles cause intron retention of YRA1 transcript (Supplemental Figure S1C). Both splicing mutants lead to lower levels of Yra1 protein by Western blot relative to WT control (Figure 2F), consistent with previous work showing YRA1 intron downregulating Yra1 expression in a splicing-dependent manner (Rodriguez-Navarro et al., 2002). While Yra1 loss could have caused R-loop accumulation, the similar amount of Yra1 depletion in hsh155-1 and snu114-60 suggests this is not the case. mRNA export is known to regulate R-loop-associated genome instability (Luna et al., 2005) and it is notable that several RNA export proteins are encoded by transcripts with complex splicing behavior. For example, DBP2, which encodes an RNA helicase and binding partner of Yra1, has the largest intron and its levels are also autoregulated by its intron (Barta and Iggo, 1995). Mtr2, another mRNA transport regulatory protein, is encoded by one of only a few Saccharomyces cerevisiae genes to exhibit alternative splice isoforms (Davis et al., 2000). Thus, while Yra1 protein levels alone are insufficient to explain R-loop–driven instability in snu114-60, it is possible that these other factors play a role. This awaits a more systematic study of proteome changes among R-loop regulators in splicing mutants.
Genetic interaction profiling reveals mitotic defects in hsh155-1
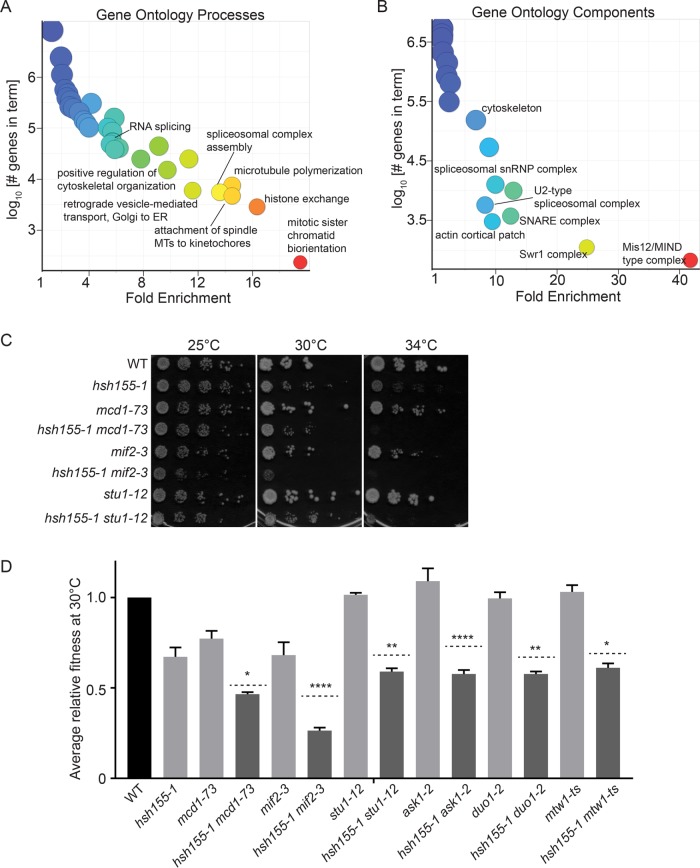
Knowing that R-loops likely do not account for genome instability in some splicing mutants (Chan et al., 2014a), we sought to understand common mechanisms. Mutations in HSH155 exhibited strong CIN phenotypes but showed no evidence of increased DNA damage or R-loops (Figure 2). We hypothesized that if a common mechanism of CIN existed for splicing mutants, it would be at play in hsh155-1 alleles. To determine this function, we performed a synthetic genetic array (SGA) screen using hsh155-1 as a query strain. This screen identified 102 negative and 103 positive genetic interaction candidates (Supplemental Table S1). A greater-than-expected number of essential intron-containing genes were negative interactors, consistent with a splicing defect enhancing phenotypes of these mutants (Supplemental Table S1). Positive interactions with proteasome subunits or translational apparatus could reflect stabilization of mutant Hsh155 protein leading to healthier cells (Supplemental Table S1) (van Leeuwen et al., 2016).
Analysis of gene ontology (GO) terms among negative genetic interactions highlighted expected groups such as spliceosomal complex assembly (13.6-fold enriched) and mRNA processing (5.5 fold) (Supplemental Table S2). Other potentially surprising GO terms such as retrograde vesicle-mediated transport, Golgi to ER (11.6 fold), and Golgi-associated vesicle (7.4 fold) (Supplemental Table S2) can be explained by the preponderance of intron-containing genes in this pathway (e.g., SNC1, BET1, SEC27, SFT1, SAR1, and YIP3 all encode introns that could cause a vesicle trafficking defect in splicing mutants). Dramatic enrichment for cytoskeletal processes among negative genetic interaction partners of hsh155-1 was also seen (Supplemental Tables S1 and S2) and was expected based on the enrichment of intron-containing genes in this pathway (e.g., ACT1, TUB1, MCM21, COF1, GIM5, CIN2, TUB3, and DYN2 encode introns) (Parenteau et al., 2008). The clearest direct connection to chromosome segregation also came from this analysis of GO terms in this group. Terms like attachment of spindle microtubules to kinetochore were highly enriched (19.5 fold) (Supplemental Table S2). GO term enrichments visualized with REVIGO (Supek et al., 2011) highlight this with processes like microtubule polymerization and mitotic sister chromatid biorientation and components like the Mis12/MIND type complex of the kinetochore (Figure 3, A and B).
FIGURE 3:
Genetic interaction network of hsh155-1. (A) GO biological process and (B) cellular component enrichments for hsh155-1 negative interactions (Supplemental Tables S1 and S2). Warmer colors associate with higher enrichment. (C, D) Validation of negative genetic interactions by (C) spot dilution assays and (D) growth curves. For D, light gray bars: fitness of single mutants; dashed lines: calculated expected fitness of double mutants using multiplicative model; dark gray bars: observed fitness of double mutants. Two-way ANOVA was used to calculate statistical significance between observed and expected fitness of double mutants. *p < 0.05; **p < 0.01; ****p < 0.0001.
We validated by spot dilutions that hsh155-1 has negative interactions with mitotic genes like cohesin (MCD1), core kinetochore subunits (MIF2), and spindle regulators (STU1) (Figure 3C). We further used growth curves to identify subtle changes in growth in the hsh155-1 double mutants (Figure 3D). Analysis of published SGA profiles for snu114-60 and other splicing factors (www.thecellmap.org) also revealed genetic dependence on a functional mitotic apparatus (Costanzo et al., 2016), which encouraged us to explore mitotic defects further.
SNU114 and HSH155 mutants have mitotic defects
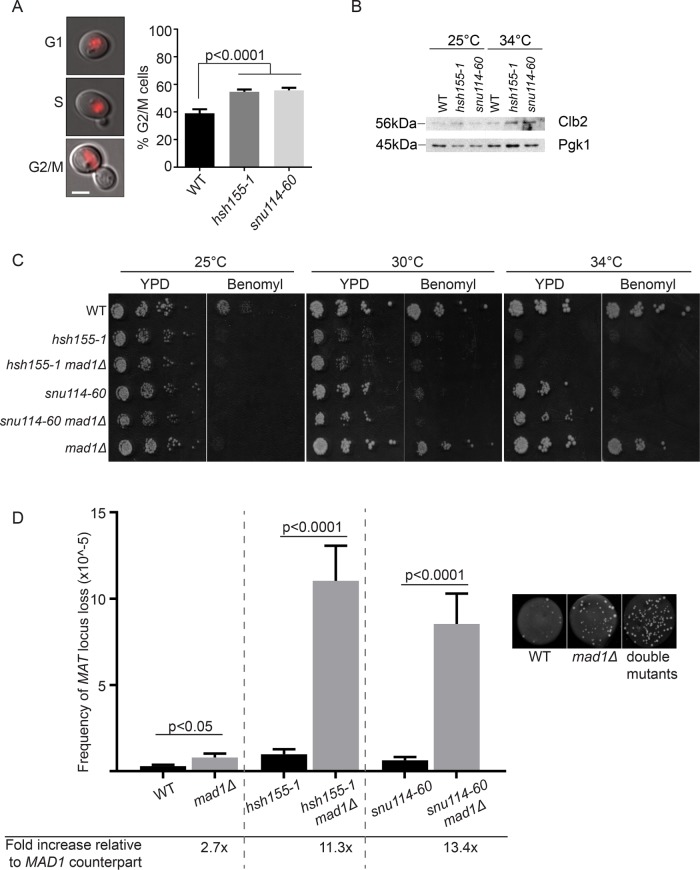
To test potential mitotic defects, we first measured the cell-cycle distribution of cells by budding index. After a shift to a nonpermissive temperature of 37°C, a small but significant proportion of the splicing mutant cells accumulated as large-budded G2/M cells, indicating a potential mitotic delay and defect (Figure 4A). To confirm these observations, we used α-factor to arrest cells in G1 and collected samples at 30, 90, and 150 min after release for measurement of DNA content. The results confirm a clear but subtle increase in 2N cells at 150 min postrelease (Supplemental Figure S1D), compared with the positive control ask1-2 that completely arrests in mitosis. This is consistent with the budding index data where the proportion of G2/M cells in mutants compared with wild type is only slightly elevated. To further confirm that cell-cycle progress is abnormal, we used Western blot analysis to measure levels of Clb2, a B-type cyclin that accumulates during G2 and M phases of the cell cycle (Amon et al., 1993). At the restrictive temperature, there is an increase of Clb2 protein levels in the splicing mutants, indicating that more cells are indeed in G2/M phases of the cell cycle (Figure 4B). These results complement the observed negative genetic interactions between hsh155-1 and genes with functions in spindle and kinetochore subunits (Figure 3). On the basis of the literature for other splicing alleles (Burns et al., 2002), we hypothesized that cell-cycle defects in hsh155-1 and snu114-60 were due to spindle defects activating the spindle assembly checkpoint (SAC). To test this, we deleted the SAC regulator MAD1 in each splicing mutant. Loss of MAD1 further sensitized hsh155-1 and snu114-60 alleles to the microtubule depolymerizing drug benomyl at semipermissive temperatures (Figure 4C). By quantifying the rate of endogenous chromosome III loss using the A-like faker (ALF) assay (Novoa et al., 2018) in the single and double mutants at permissive temperature of 25°C, we found a dramatic synergy between disruption of SNU114 or HSH155 and loss of the SAC through MAD1 deletion (Figure 4D). The ALF assay measures the stability of the MAT locus on chromosome III and loss of this locus leads to quantifiable aberrant mating events (see Materials and Methods). Overall, these data are consistent with a mitotic defect in splicing mutants that requires the activity of the SAC for genome maintenance.
FIGURE 4:
Mitotic defects in spliceosome mutants. (A) Proportion of G2/M cells determined by budding relative to nuclear state determined by Hta2-mCherry fluorescence (left panels, scale bar = 2 μm); Fisher’s exact test. (B) Western blot of Clb2 protein levels in the indicated strains and temperatures. (C) Spot dilution assay of benomyl sensitivity (15 μg/ml benomyl). (D) Frequency of MAT loss in single (black bars) and mad1Δ double mutants (gray bars), grown at 25°C. Right, representative images of WT, mad1Δ and double mutant spots. Student’s t test was used to calculate significance. (A, D) Mean values and SEM are shown, n = 3.
Tubulin levels control genome integrity in splicing mutants
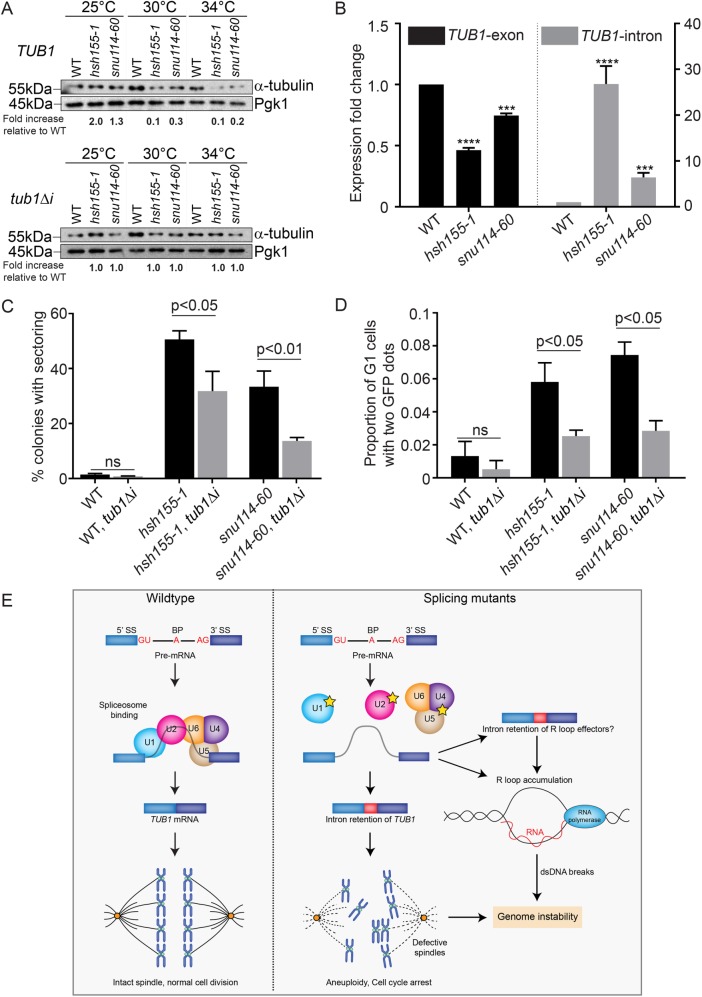
Only ∼5% of yeast genes are spliced, and the bulk of splicing flux is driven by ribosomal protein transcripts (Parenteau et al., 2008). Splicing of the TUB1 transcript, encoding α-tubulin, has previously been implicated in cell-cycle delays in other splicing mutants (Burns et al., 2002; Dahan and Kupiec, 2002). Moreover, mutant alleles of TUB1 such as the cold-sensitive tub1-1 have been shown to increase chromosome missegration at low temperatures (Hoyt et al., 1990) and lead to decreased Tub1 protein (Supplemental Figure S1E). Tub1 protein levels decreased in both hsh155-1 and snu114-60 mutants as temperature increased (Figure 5A). RT-qPCR of TUB1 mRNA under the same conditions indicated a mild decrease in TUB1 mRNA expression in hsh155-1 and snu114-60, which was accompanied by considerable intron retention (Figure 5B), supporting the idea that defective TUB1 mRNA splicing drives loss of protein. Indeed, we observed Tub1 protein-level decreases of variable penetrance in a panel of splicing factor mutants (i.e., alleles of YHC1, SNU13, SYF1, CWC2, PRP4, PRP31, and PRP6) (Supplemental Figure S1F), supporting that this is a general phenomenon. To directly connect defective TUB1 splicing to genome maintenance, we retested CIN in hsh155-1 and snu114-60 encoding an intronless TUB1 gene, tub1Δi. As expected, tub1Δi increased the amount of Tub1 protein expressed in each splicing mutant relative to WT (Figure 5A). More importantly, intronless TUB1 partially suppressed the CTF phenotype observed in snu114-60 and hsh155-1 (Figure 5C). We also tested the effect of tub1Δi on the rate of endogenous chromosome III missegregation using the LacO-LacI-GFP system and found suppression of aneuploidy in both splicing mutants (Figure 5D). As expected, tub1Δi has no effect on R-loop levels or recombination in snu114-60, demonstrating that there are multiple mechanisms, operating concurrently, that create genome instability in this strain (Supplemental Figures S1G and S1H). Importantly, the partial suppression of CIN phenotypes in both hsh155-1 and snu114-60 by tub1Δi support the idea that TUB1-independent mechanisms of genome instability must also be at play. We feel that it is likely that minor contributions from defects in multiple intron containing genes may combine with the effects of TUB1 dysregulation to promote the overall increase in genome instability seen in splicing mutants.
FIGURE 5:
Tubulin stability contributes to genome maintenance in splicing mutants. (A) Western blot of relative α-tubulin protein levels. Top, TUB1; bottom, intronless TUB1. (B) Quantification of TUB1 mRNA transcript levels from exon region (left) and TUB1 intron region (right) by RT-qPCR. Asterisks show p values of ΔΔCt: ***p = 0.0002; ****p < 0.0001. (C) CTF phenotypes in TUB1 or tub1Δi strains. (D) Endogenous Chr III stability in hsh155-1 and snu114-60 with intronless TUB1 (gray bars). (C, D) Fisher’s exact test. (E) Model of defective splicing-induced genome instability in yeast. (B, C, D) Mean values with SEM are shown, n = 3.
Tubulin levels must be tightly regulated, and maintaining an equimolar ratio of α- to β-tubulin is known to be critical for functional spindles (Katz et al., 1990). Our data support a model (Figure 5E) where intron retention and associated decreases in α-tubulin lead to sporadic chromosome missegregation, buffered by the SAC (Figure 4). If this is true, then simple reduction of TUB1 transcription should also cause genome instability even with normal splicing. To test this, we engineered strains where the TUB1 gene was under the control of a galactose-regulated promoter in a WT or tub1Δ background. Shifting the strains to dextrose repressed Tub1 expression in the tub1Δ background (Supplemental Figure S1I) and led to a significant increase in CIN by the ALF assay (Supplemental Figure S1J). Thus, altered α-tubulin levels cause CIN, whether through splicing defects or decreased transcription.
Perspective
Determining sources of genome instability is important to understand the accumulation of mutations during cellular adaptation and in human disease. While much is known about DNA replication, repair, and mitosis, considerably less is known about the effects of other cellular pathways on the genome. Nonetheless, these noncanonical pathways account for a large proportion of reported genome maintenance factors (Stirling et al., 2011). RNA processing has emerged as a major contributor to genome maintenance and various mechanisms have been described. Direct roles for some RNA processing factors have been found in DNA repair, such as the moonlighting function of Prp19 in ATR activation (Marechal et al., 2014), or the role of the spliceosome in R-loop mediated ATM activation (Tresini et al., 2015). More recently, dominant cancer-associated splicing mutations have been linked to RNA polymerase pausing and R-loop accumulation (Chen et al., 2018). Still other studies have suggested a role for the aberrant gene expression landscapes produced in RNA processing mutants as drivers of genome instability (Blazek et al., 2011; Vohhodina et al., 2017).
Overall, our data reveal how changes in information flux due to allele-specific effects on RNA maturation can influence specific mechanisms of genome instability. This is important as we have shown previously that >150 yeast genes with functions in transcription, RNA processing and translation can be mutated to cause a genome instability phenotype (Stirling et al., 2011). In principle, our data suggest that many of these mutants with broad impacts on gene expression could selectively impair a specific aspect of genome maintenance that can be identified. Indeed, cancer-associated mutations in core splicing factors like SF3B1, the orthologue of yeast Hsh155, are invariably nonsynonymous coding variants suggesting that subtle but specific changes in the transcriptome are required for cancer formation and maintenance (Darman et al., 2015). It will be important to employ sensitized genetic backgrounds and sensitive assays to enable direct study of cancer-associated SF3B1/Hsh155 alleles in yeast genome maintenance. Mechanistic studies of how specific mutations in the spliceosome alter the proteome in a specific cellular context could elucidate roles for spliceosomal mutations in cancer.
MATERIALS AND METHODS
Yeast strains, growth, and CIN assays
All yeast strains were in the s288c background (Supplemental Table S3) and were grown under standard conditions in the indicated media and growth temperature. To assess benomyl sensitivity, strains were compared between yeast extract peptone dextrose (YPD) + 0.2% dimethyl sulfoxide (DMSO) (control) or YPD + 15 μg/ml benomyl (Sigma-Aldrich cat. no.45339). Growth curves were conducted in YPD using a Tecan M200 plate reader and were compared using the area under the curve as previously described (Chang et al., 2017). Briefly, logarithmic phase cultures were diluted to OD 0.05 in a 96-well plate and grown for 24 h, with OD600 readings taken every 25 min. The chromosome transmission fidelity (CTF) and ALF assays were performed as described (Stirling et al., 2011; Novoa et al., 2018). In the ALF assay involving tub1∆, tub1∆ mutant strain carrying a galactose-inducible TUB1 plasmid was grown in either synthetic complete (SC) + 2% galactose or 2% dextrose media for 24 h, mixed with an excess of mating tester and plated onto synthetic defined (SD) media + 2% galactose or 2% dextrose at 30°C. The CTF assay measures inheritance of an artificial chromosome fragment as monitored by colony color to indicate chromosome instability–an induced ade2-101 mutation pigments colonies red, and when supplemented with a chromosome fragment carrying suppressor mutation SUP11, the chromosome instability read-out was white (indicating no loss of fragment) or red (loss of fragment). The ALF assay measures the frequency of MATα locus loss in α-mating type haploid yeast, which indicates dedifferentiation to a-mating type haploid yeast. Cells that have lost this locus were detected by selection for mated diploid yeast. For plasmid loss, strains carrying pRS313::HIS3 were grown overnight in SC-histidine and then plated on YPD and allowed to form colonies without selection before replica plating onto SC-histidine. The frequency of colonies that could not grow on SC-histidine is reported as the plasmid loss rate. For all experiments, significance of the differences was determined using Prism7 (GraphPad Software). For all experiments, sample means were compared with Fisher’s exact test, Student’s t tests or analysis of variance (ANOVA) for multiple comparisons as indicated.
Synthetic genetic array and validation
SGA screening was performed as described previously for URA3-marked query strains (Stirling et al., 2011; Costanzo et al., 2016) and scored using the Balony software package (Young and Loewen, 2013). From raw colony scores, we chose a cutoff of p < 0.05 and an experiment-control score of ≥|0.5| for list analysis and GO term enrichment using the Princeton generic GO term-finder (http://go.princeton.edu/cgi-bin/GOTermFinder) and visualized with REVIGO (Supek et al., 2011). snu114-60 and yhc1-1 interactions are available at http://thecellmap.org (Usaj et al., 2017). For hit validation, fresh double mutant strains were made by tetrad dissection and tested in quantitative growth curves or by spot dilution assays. Observed area under the curve for double mutants was compared with a multiplicative model of the predicted fitness based on the fitness of the two single mutants.
Image and cell-cycle analysis
Imaging of budding index by differential interference contrast (DIC) or fluorescence of Rad52-GFP and GFP labeled chromosome III was conducted on a Leica DMi8 microscope using an HCX plan apochromat 1.4 NA oil immersion 100× lens. The images were captured at room temperature by an ORCA Flash 4.0 V2 camera (Hamamatsu Photonics), using MetaMorph Premier acquisition software (Molecular Devices). Scoring was done in ImageJ (National Institutes of Health). For Rad52 foci, all cells were scored as negative or positive for a focus. Imaging of LacO-LacI-GFP labeled chromosome III was performed as described (Woodruff et al., 2009). For scoring, first unbudded cells were selected in the DIC channel and then scored for the presence of 1 or ≥2 LacI-GFP dots. Chromosome spreads were performed exactly as described; primary DNA-RNA Hybrid [S9.6] (Kerafast cat. no. ENH001); secondary Alexa Fluor 568 goat anti-mouse immunoglobulin G (Invitrogen cat. no. A-11004) (Wahba et al., 2011).
Flow cytometry was done using the BD FACSCalibur platform. Cells were arrested in G1 by α-factor arrest using 10 µg/ml α-factor (Cedarlane, cat. no. Y1001) for 2.5 h, and samples were collected at 30, 90, and 150 min after wash-out. Cells were fixed with 70% ethanol and stained with 16 μg/ml propidium iodide (Sigma-Aldrich cat. no. 287075). The proportion of cells in G1, S, and G2/M cell cycles was quantified using FlowJo.
Splicing efficiency assay
Splicing assay protocol was performed as described (Galy et al., 2004). All measurements were taken with individual transformants in triplicate. Cells were struck as a patch on SC-leucine and then replica plated to glycerol-lactate–containing SC medium without leucine (GGL-leu). Cells from each patch were inoculated in liquid GGL-leu media for 2 h at 30°C and then were induced with final 2% galactose for 4 h. Cells carrying reporters were lysed and assayed for β-galactosidase assay using a Gal-Screen β-galactosidase reporter gene assay system for yeast or mammalian cells (Applied Biosystems) as per the manufacturer’s instructions and read with a SpectraMax i3 (Molecular Devices). Relative light units were normalized to cell concentration as estimated by measuring OD600.
Recombination assays
To construct the recombination plasmids, pRS314GLB (de la Loza et al., 2009) was linearized with BglII and ligated with URA3 sequences with BglII sites (Aksenova et al., 2013). Following transformation and plasmid isolation, the presence of URA3 was confirmed by Sanger sequencing. Direct repeat recombination assays were performed as described (Chang et al., 2017).
Western blot
Whole-cell extracts were prepared by 100% trichloroacetic acid extraction (2 × 106 cells). Lysates were separated on a 10% or 15% SDS–PAGE gel, transferred to nitrocellulose, and probed with the following antibodies: anti-Tub1 (Invitrogen cat. no. 32-2500) (1:500 dilution), anti-Pgk1 (Santa Cruz cat. no. sc-130335) (1:1000 dilution) as a loading control, Yra1 antibody (a gift from David Bentley, University of Colorado, Denver) (1:10,000 dilution), and Clb2 antibody (Santa Cruz Biotechnology cat. no. sc-6697, discontinued) (1:1000 dilution). ImageJ software was used to quantify protein bands (Schneider et al., 2012).
RNA isolation, cDNA preparation, and reverse transcription–quantitative PCR analysis
Total RNA was isolated from 0.5–1 OD cell cultures shifted to 34°C for 3.5 h, using the yeast RiboPure RNA Purification kit (Ambion). cDNA (1 µg) was reverse transcribed using anchored-oligo(dT)18 primer and Transcriptor Reverse transcription (Roche). Reverse transcription–quantitative PCRs were performed and analyzed using SYBR green PCR Master Mix and a StepOnePlus Real-Time PCR system (Applied Biosystems). RQ values were normalized to an unspliced SPT15 transcript and expressed relative to WT.
Supplementary Material
Acknowledgments
We thank A. Aguilera, H. Klein, D. G. Drubin, B. Palancade, S. A. Elela, D. Koshland, and P. Hieter for strains and plasmids and D. Bentley for the gift of Yra1 antibody. A.S.T. is a supported by a Cordula and Gunter Paetzold Fellowship. P.C.S. is a Canadian Institutes of Health Research (CIHR) New Investigator and Michael Smith Foundation for Health Research scholar. This work was supported by CIHR (MOP-136982) and the Natural Sciences and Engineering Research Council of Canada (RGPIN 2014-04490).
Abbreviations used:
- CIN
chromosomal instability
- GFP
green fluorescent protein
- GO
gene ontology
- snRNP
small nuclear ribonucleoprotein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-07-0439) on November 21, 2018.
REFERENCES
- Aguilera A, Garcia-Muse T. (2012). R loops: from transcription byproducts to threats to genome stability. Mol Cell , 115–124. [DOI] [PubMed] [Google Scholar]
- Aksenova AY, Greenwell PW, Dominska M, Shishkin AA, Kim JC, Petes TD, Mirkin SM. (2013). Genome rearrangements caused by interstitial telomeric sequences in yeast. Proc Natl Acad Sci USA , 19866–19871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon A, Tyers M, Futcher B, Nasmyth K. (1993). Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell , 993–1007. [DOI] [PubMed] [Google Scholar]
- Barta I, Iggo R. (1995). Autoregulation of expression of the yeast Dbp2p ’DEAD-box’ protein is mediated by sequences in the conserved DBP2 intron. EMBO J , 3800–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek D, Kohoutek J, Bartholomeeusen K, Johansen E, Hulinkova P, Luo Z, Cimermancic P, Ule J, Peterlin BM. (2011). The cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev , 2158–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet A, Grosso AR, Elkaoutari A, Coleno E, Presle A, Sridhara SC, Janbon G, Geli V, de Almeida SF, Palancade B. (2017). Introns protect eukaryotic genomes from transcription-associated genetic instability. Mol Cell , 608–621. e606. [DOI] [PubMed] [Google Scholar]
- Burns CG, Ohi R, Mehta S, O’Toole ET, Winey M, Clark TA, Sugnet CW, Ares M, Jr, Gould KL. (2002). Removal of a single alpha-tubulin gene intron suppresses cell cycle arrest phenotypes of splicing factor mutations in Saccharomyces cerevisiae. Mol Cell Biol , 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrocci TJ, Zoerner DM, Paulson JC, Hoskins AA. (2017). SF3b1 mutations associated with myelodysplastic syndromes alter the fidelity of branchsite selection in yeast. Nucleic Acids Res , 4837–4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YA, Aristizabal MJ, Lu PY, Luo Z, Hamza A, Kobor MS, Stirling PC, Hieter P. (2014a). Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet , e1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YA, Hieter P, Stirling PC. (2014b). Mechanisms of genome instability induced by RNA-processing defects. Trends Genet , 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EY, Novoa CA, Aristizabal MJ, Coulombe Y, Segovia R, Chaturvedi R, Shen Y, Keong C, Tam AS, Jones SJM, et al. (2017). RECQ-like helicases Sgs1 and BLM regulate R-loop-associated genome instability. J Cell Biol , 3991–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chen JY, Huang YJ, Gu Y, Qiu J, Qian H, Shao C, Zhang X, Hu J, Li H, et al. (2018). The augmented R-loop is a unifying mechanism for myelodysplastic syndromes induced by high-risk splicing factor mutations. Mol Cell , 412–425. e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, et al. (2016). A global genetic interaction network maps a wiring diagram of cellular function. Science , aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan O, Kupiec M. (2002). Mutations in genes of Saccharomyces cerevisiae encoding pre-mRNA splicing factors cause cell cycle arrest through activation of the spindle checkpoint. Nucleic Acids Res , 4361–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D, Bailey SL, Bhavsar EB, Chan B, Colla S, et al. (2015). Cancer-associated SF3B1 hotspot mutations induce cryptic 3? splice site selection through use of a different branch point. Cell Rep , 1033–1045. [DOI] [PubMed] [Google Scholar]
- Davis CA, Grate L, Spingola M, Ares M., Jr (2000). Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res , 1700–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Loza MC, Wellinger RE, Aguilera A. (2009). Stimulation of direct-repeat recombination by RNA polymerase III transcription. DNA Repair (Amst) , 620–626. [DOI] [PubMed] [Google Scholar]
- Dolatshad H, Pellagatti A, Fernandez-Mercado M, Yip BH, Malcovati L, Attwood M, Przychodzen B, Sahgal N, Kanapin AA, Lockstone H, et al. (2015). Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia , 1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. (2004). Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell , 63–73. [DOI] [PubMed] [Google Scholar]
- Garcia-Rubio M, Aguilera P, Lafuente-Barquero J, Ruiz JF, Simon MN, Geli V, Rondon AG, Aguilera A. (2018). Yra1-bound RNA-DNA hybrids cause orientation-independent transcription-replication collisions and telomere instability. Genes Dev , 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavalda S, Santos-Pereira JM, Garcia-Rubio ML, Luna R, Aguilera A. (2016). Excess of Yra1 RNA-binding factor causes transcription-dependent genome instability, replication impairment and telomere shortening. PLoS Genet , e1005966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, Garcia-Rubio M, Bermejo R, Gaillard H, Shirahige K, Marin A, Foiani M, Aguilera A. (2011). Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J , 3106–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. (2011). Hallmarks of cancer: the next generation. Cell , 646–674. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Stearns T, Botstein D. (1990). Chromosome instability mutants of Saccharomyces cerevisiae that are defective in microtubule-mediated processes. Mol Cell Biol , 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno S, Rondon AG, Luna R, Aguilera A. (2002). The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J , 3526–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi P, Halene S, Abdel-Wahab O. (2017). How do messenger RNA splicing alterations drive myelodysplasia? Blood , 2465–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz W, Weinstein B, Solomon F. (1990). Regulation of tubulin levels and microtubule assembly in Saccharomyces cerevisiae: consequences of altered tubulin gene copy number. Mol Cell Biol , 5286–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Manley JL. (2005). Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell , 365–378. [DOI] [PubMed] [Google Scholar]
- Luna R, Jimeno S, Marin M, Huertas P, Garcia-Rubio M, Aguilera A. (2005). Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol Cell , 711–722. [DOI] [PubMed] [Google Scholar]
- Marechal A, Li JM, Ji XY, Wu CS, Yazinski SA, Nguyen HD, Liu S, Jimenez AE, Jin J, Zou L. (2014). PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol Cell , 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday V, Stirling PC. (2016). Navigating yeast genome maintenance with functional genomics. Brief Funct Genomics , 119–129. [DOI] [PubMed] [Google Scholar]
- Novoa CA, Ang JS, Stirling PC. (2018). The A-like faker assay for measuring yeast chromosome III stability. Methods Mol Biol , 1–9. [DOI] [PubMed] [Google Scholar]
- Parenteau J, Durand M, Veronneau S, Lacombe AA, Morin G, Guerin V, Cecez B, Gervais-Bird J, Koh CS, Brunelle D, et al. (2008). Deletion of many yeast introns reveals a minority of genes that require splicing for function. Mol Biol Cell , 1932–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. (2009). A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell , 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Manley JL. (2017). R loops and links to human disease. J Mol Biol , 3168–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Strasser K, Hurt E. (2002). An intron in the YRA1 gene is required to control Yra1 protein expression and mRNA export in yeast. EMBO Rep , 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods , 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. (2011). Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell , 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling PC, Bloom MS, Solanki-Patil T, Smith S, Sipahimalani P, Li Z, Kofoed M, Ben-Aroya S, Myung K, Hieter P. (2011). The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genet , e1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling PC, Chan YA, Minaker SW, Aristizabal MJ, Barrett I, Sipahimalani P, Kobor MS, Hieter P. (2012). R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev , 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, Smuc T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One , e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Rodriguez-Santiago S, Wang J, Pu J, Yuste A, Gupta V, Moldon A, Xu YZ, Query CC. (2016). SF3B1/Hsh155 HEAT motif mutations affect interaction with the spliceosomal ATPase Prp5, resulting in altered branch site selectivity in pre-mRNA splicing. Genes Dev , 2710–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresini M, Warmerdam DO, Kolovos P, Snijder L, Vrouwe MG, Demmers JA, van IWF, Grosveld FG, Medema RH, Hoeijmakers JH, et al. (2015). The core spliceosome as target and effector of non-canonical ATM signalling. Nature , 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usaj M, Tan Y, Wang W, VanderSluis B, Zou A, Myers CL, Costanzo M, Andrews B, Boone C. (2017). TheCellMap.org: a web-accessible database for visualizing and mining the global yeast genetic interaction network. G3 , 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen J, Pons C, Mellor JC, Yamaguchi TN, Friesen H, Koschwanez J, Usaj MM, Pechlaner M, Takar M, Usaj M, et al. (2016). Exploring genetic suppression interactions on a global scale. Science , aag0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vohhodina J, Barros EM, Savage AL, Liberante FG, Manti L, Bankhead P, Cosgrove N, Madden AF, Harkin DP, Savage KI. (2017). The RNA processing factors THRAP3 and BCLAF1 promote the DNA damage response through selective mRNA splicing and nuclear export. Nucleic Acids Res , 12816–12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Amon JD, Koshland D, Vuica-Ross M. (2011). RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell , 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Zheng X, Chen H, Guo Y, Jiang H, He X, Zhu X, Zheng Y. (2015). Splicing function of mitotic regulators links R-loop-mediated DNA damage to tumor cell killing. J Cell Biol , 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Drubin DG, Barnes G. (2009). Dynein-driven mitotic spindle positioning restricted to anaphase by She1p inhibition of dynactin recruitment. Mol Biol Cell , 3003–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young BP, Loewen CJ. (2013). Balony: a software package for analysis of data generated by synthetic genetic array experiments. BMC Bioinformatics , 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.