Significance
The central feature of meiosis is pairing and recombination of homologous maternal and paternal chromosomes (homologs). Homolog axes become first coaligned at a certain distance; they then synapse by synaptonemal complex (SC) formation. We show that the structural and molecular pathway by which chromosomes transit from coalignment to the SC state in the fungus Sordaria macrospora involves the formation of robust interaxis bridges comprising axis component Spo76/Pds5, recombination proteins, and the evolutionary-conserved Zip2-Zip4 complex. Zip2-Zip4 mediates the recombination complex/structure interface from coalignment onward. These findings solve the conundrum of how recombination complexes move from on-axis localization at coalignment to between-axis localization on SC central regions and provoke new ideas about the molecular and mechanistic nature of SC nucleation.
Keywords: meiotic recombination, synaptonemal complex, Zip2-Zip4, chromosome structure, interaxis bridges
Abstract
A central feature of meiosis is pairing of homologous chromosomes, which occurs in two stages: coalignment of axes followed by installation of the synaptonemal complex (SC). Concomitantly, recombination complexes reposition from on-axis association to the SC central region. We show here that, in the fungus Sordaria macrospora, this critical transition is mediated by robust interaxis bridges that contain an axis component (Spo76/Pds5), DNA, plus colocalizing Mer3/Msh4 recombination proteins and the Zip2-Zip4 mediator complex. Mer3-Msh4-Zip2-Zip4 colocalizing foci are first released from their tight axis association, dependent on the SC transverse-filament protein Sme4/Zip1, before moving to bridges and thus to a between-axis position. Ensuing shortening of bridges and accompanying juxtaposition of axes to 100 nm enables installation of SC central elements at sites of between-axis Mer3-Msh4-Zip2-Zip4 complexes. We show also that the Zip2-Zip4 complex has an intrinsic affinity for chromosome axes at early leptotene, where it localizes independently of recombination, but is dependent on Mer3. Then, later, Zip2-Zip4 has an intrinsic affinity for the SC central element, where it ultimately localizes to sites of crossover complexes at the end of pachytene. These and other findings suggest that the fundamental role of Zip2-Zip4 is to mediate the recombination/structure interface at all post–double-strand break stages. We propose that Zip2-Zip4 directly mediates a molecular handoff of Mer3-Msh4 complexes, from association with axis components to association with SC central components, at the bridge stage, and then directly mediates central region installation during SC nucleation.
The central feature of meiotic prophase, which distinguishes it from the mitotic cell cycle, is a complex program of interactions between homologous maternal and paternal chromosomes (“homologs”). A major event of this program is the coming-together of homologous axes, first via coalignment and then via synapsis mediated by the tripartite protein structure called synaptonemal complex (SC).
In most organisms, coalignment and SC formation are directly mediated by recombination. Recombination initiates through DNA double-strand breaks (DSBs) that occur within axis-associated recombination complexes (reviewed in ref. 1). For coalignment, a consensus hypothesis is that one DSB end searches for a DNA partner on the homologous chromosome. A resultant nascent D-loop between the DSB end and its homologous DNA region nucleates development of a partner recombination complex, which then becomes associated with the underlying partner axis, where it mediates spatial juxtaposition of the linked two chromosome axes (reviewed in ref. 2). Occurrence of such events at many positions along the chromosomes results in coalignment of axes all along their lengths. Following coalignment, formation of the SC at some of the coalignment-mediated recombination sites brings homologous axes together by installation of SC central elements (reviewed in refs. 3–5).
The transition from coalignment to synapsis is a major event of meiotic prophase. One of the most interesting features of this transition is the fact that recombination complexes must undergo a major change in localization: from the on-axis position that occurs before and during coalignment (above) to a between-axis position on the central region of the SC. Once the SC has formed, recombination complexes are associated with the SC central element, and crossover (CO)-fated recombination complexes retain this association throughout ensuing recombination events, with mature COs finally emerging shortly before the SC is disassembled (reviewed in refs. 2 and 6).
Existing information provides some clues about the nature of this transition.
-
i)
First, EM images in Allium cepa have defined the existence of bridges between coaligned axes, including bridges with centrally localized nodules (7). Although the involved proteins remain unknown, these bridges/nodules likely represent the sites of recombinational interactions. Accordingly, in human and mouse spermatocytes, RPA (Replication Protein A) and BLM (Bloom Syndrome Protein) proteins are visible either as matching foci still attached to their coaligned axis or as coalescent foci now forming a bridge linking the two axes before onset of SC formation (8–10).
-
ii)
Second, studies in budding yeast have identified a set of molecules that coordinately mediate both SC nucleation and progression of CO-designated recombination interactions (11–13). This group, referred to as “ZMM” proteins, includes SC transverse-filament protein Zip1, the SUMO E3 ligase Zip3 that specifically marks CO sites, the XPF-domain protein Zip2, the ERCC1-like protein Spo16, the tetra-tricopeptide repeat (TPR) protein Zip4/Spo22, the 5′-3′ DNA helicase Mer3, and the interacting MutS homologs Msh4 and Msh5, which are thought to stabilize branched recombination intermediates (e.g., refs. 11 and 13–18). Recent molecular studies have further shown that Zip2 and Zip4 strongly interact and that the Zip2/Spo16 complex binds branched DNA structures in vitro (19–21). Also, similarly to budding yeast, plant and mammal orthologs of Zip2 (SHOC1) and Zip4 (TEX11) are required for the formation of COs and for WT-like SC initiation (17, 20, 22–26).
In the present study, we have exploited the power of Sordaria macrospora as a system for visualizing the program of meiotic chromosomal events to further elucidate the coalignment/SC transition, thereby allowing to more directly link cytological and molecular information. To further explore the functional roles of the Zip2-Zip4 complex, we initiated this study by identification of the Sordaria orthologs of Zip2 and Zip4. We then performed detailed chromosome localization analysis of Zip2, Zip4, Mer3, Msh4, and E3 ligase Hei10 at all prophase stages in WT and relevant mutant cases, with axes defined by Spo76/Pds5 (27) and SC visualized with Sme4/Zip1 (28). The presented results show that recombination complexes move from on-axis to between-axis localization via bridges that ultimately serve as SC nucleation sites and suggest that Zip2/Zip4 mediates the structure/recombination complex interface at all post-DSB stages, from coalignment through the end of the SC stage when crossing over is completed.
Results
Identification of Sordaria Zip2-Zip4.
Identification of Sordaria Zip2 and Zip4 by a series of phylogenomics-oriented Position-Specific Iterated BLAST protein homology searches revealed that both proteins are evolutionarily conserved. Schematic domain organization of the predicted 1,041-aa Sordaria Zip2 protein and the predicted 952-aa Sordaria Zip4 protein are shown in SI Appendix, Fig. S1A. Yeast two-hybrid analysis confirms that Sordaria Zip2 and Zip4 interact with one another, analogously to the budding yeast and mouse Zip2/SHOC1 and Zip4/TEX11 homologs (19–21). Sordaria interactions were detected between the two full-length proteins; also, the N terminus of Zip2 is sufficient for interaction with Zip4 and both the N and the C termini of Zip4 interact with Zip2, indicating that the TPR domain is not essential for the interaction (SI Appendix, Fig. S1B). Based on RT-qPCR analyses, Zip2 and Zip4 transcripts are specifically induced during the sexual cycle and remain approximately at the same level during the meiotic divisions, parallel (but at lower levels) to the temporal kinetics of transcripts for the conserved meiotic transesterase Spo11 (SI Appendix, Fig. S1C).
Zip2-Zip4 Colocalize with Mer3 and Msh4 to Leptotene Chromosome Axes.
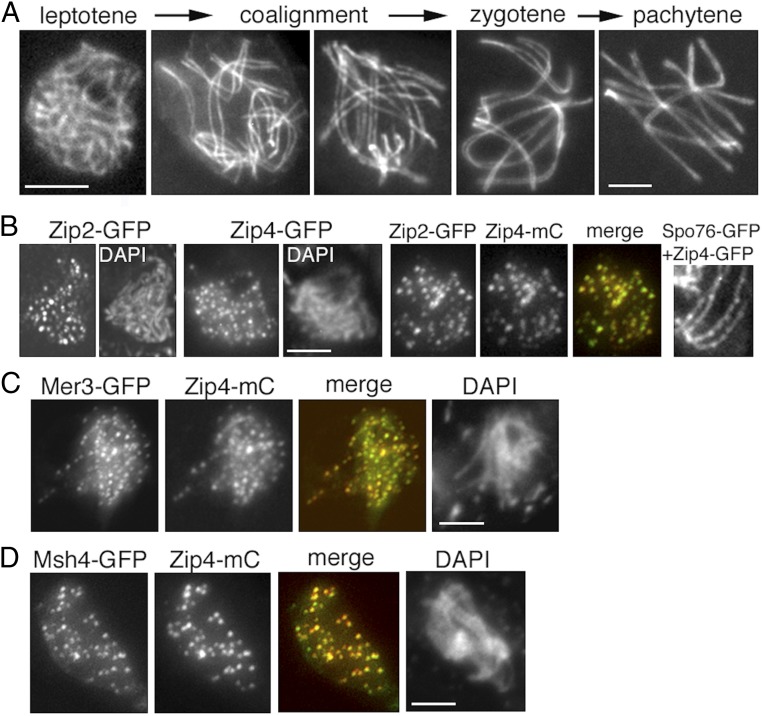
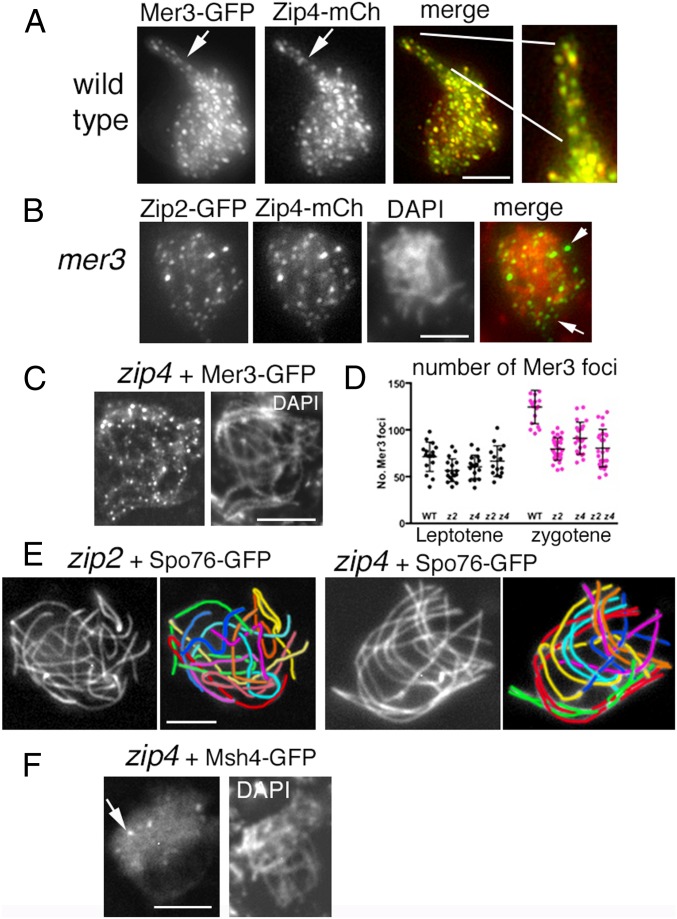
In Sordaria, chromosome axes are visible (by Spo76/Pds5-GFP) from S-phase onward, thus long before coalignment (Fig. 1A, Left). Moreover, chromosomes progress from one stage to another with a high degree of per-nucleus synchrony (Fig. 1A). This makes it possible, by wide-field or 3D-structured illumination microscopy (SIM) of single cells, to analyze in detail the very early stages of chromosome recognition and pairing, in parallel with the dynamic spatial organization of both pre-DSB and post-DSB recombination complexes by corresponding fluorescent protein tags.
Fig. 1.
Zip2 and Zip4 localizations at leptotene. (A) WT Sordaria prophase. From left to right: early leptotene, coalignment at 400 and 200 nm, partial synapsis at zygotene, and complete synapsis at pachytene. Chromosome axes are marked by Spo76/Pds5-GFP. (B) Early leptotene. (B, Left) Zip2 and Zip4 foci are regularly spaced along chromosomes with similar numbers of foci for the two molecules from early to late leptotene [respectively, for Zip2: 60 ± 9 and 73 ± 3 (n = 33 nuclei); and for Zip4: 59 ± 5 and 82 ± 11 (n = 32)]. (B, Middle) Perfect colocalization of the two proteins. (B, Right) Costaining with Spo76-GFP shows that Zip4 foci are localized on axes. (C and D) Colocalization of Zip4 foci with Mer3 foci (C) and Msh4 foci (D). (Scale bars: 2 μm.)
Analysis of early prophase stages shows that foci of Zip2 and Zip4 are already present along the chromosomes at the early leptotene stage (Fig. 1B, Left, compare their DAPI with Fig. 1A, Left). As expected from their yeast two-hybrid interaction (above), foci of the two molecules colocalize at this early stage (Fig. 1B, Middle) and then throughout coalignment and beyond, to the end of synapsis (below). Costaining with Spo76/Pds5-GFP further reveals that Zip2-Zip4 foci are localized on chromosome axes (Fig. 1B, Right). Both proteins occur as a large number of regularly spaced foci (Fig. 1 B and C) along all unpaired chromosome axes. The numbers of foci increase from early to late leptotene coordinately for the two molecules, as expected for cocomplexes (see legend of Fig. 1B). Interestingly, their foci do also colocalize with the early leptotene foci of Mer3, which mark post-DSB complexes (Fig. 1C). Msh4 appears as visible foci only at the end of coalignment (29). At that stage, Msh4 also colocalizes with Zip2 and Zip4 (Fig. 1D). Thus, by the end of the coalignment stage, chromosome axes are decorated with colocalizing complexes of Zip2, Zip4, Mer3, and Msh4.
Transitional Interaxis Bridges Link Coalignment to Synapsis.
To analyze in detail the transition from coalignment to synapsis/SC formation, the localization of Zip2, Zip4, Mer3, and Msh4 and their spatial relationships to chromosome axes, as illuminated by Spo76/Pds5-GFP, were defined by 3D-SIM analysis of nuclei progressing throughout zygotene. Early and especially mid-zygotene nuclei (SI Appendix, Fig. S2A) show all steps from coalignment to SC formation, indicating that the transition from coalignment to synapsis is very transient. To define the progression of events, we used as reference the distance between axes, which goes from the ∼400-nm early coalignment distance to the 100-nm distance seen when axes are linked by SC (Fig. 1A). Concomitantly, the analyzed proteins undergo interesting dynamic changes with diverse intermediate stages. Each of the identified morphologies (Fig. 2) is represented by 20–40 examples, as defined by analysis of over 30 zygotene nuclei.
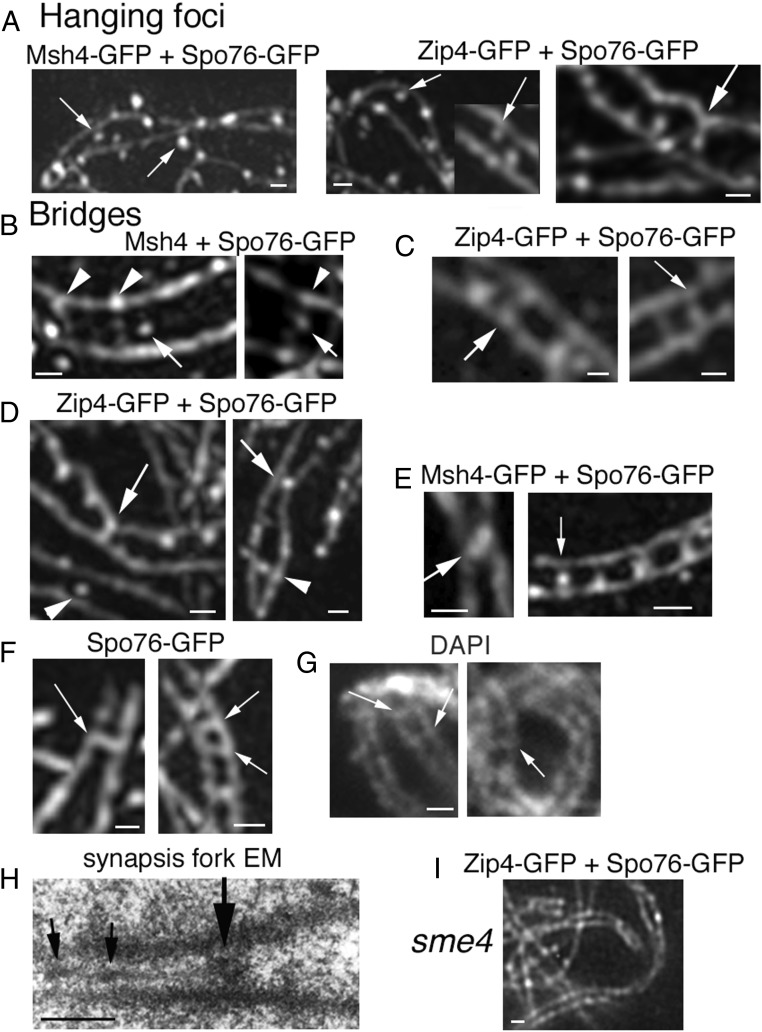
Fig. 2.
Interaxis bridges. (A–E) 3D-SIM pictures with costaining of Spo76-GFP (axes) and Zip4-GFP or Msh4-GFP. (A) First step: Msh4 foci (Left; arrows) and Zip4 foci (Middle; arrows) partially “detach” from their coaligned axes. (A, Right) Axis indentation with attached focus located in-between axes (arrow). (B) Examples of early bridges when axes are at 400-nm distance with Msh4 foci located in-between axes (arrows) sometimes (Right) linked by bridge-like structures (arrowheads) or nearby such structures (Left). (C) At the 200-nm coalignment stage, Zip4 foci are located either at matching sites (arrow, Left) or on bridges (arrow, Right). (D) Two examples of single Zip4-GFP foci located in-between axis. At 100-nm distance (arrows), axes are “constricted” by foci that are either fusing (Left) or are single (Right). At 200-nm distance, foci are in the middle between two straight axes (arrowheads). (E, Left) Two fusing Msh4 foci (arrow) with close axes. (E, Right) Row of four bridges with single Zip4 foci (arrow). (F) Spo76-GFP is visible on bridges (arrows) when axes are at 200-nm (Left) and almost 100-nm (Right) distance (3D-SIM). (G) Examples of DAPI bridges visible when homolog axes are at 200-nm (arrows) distance (classical fluorescent microscope). (H) EM section of a pairing fork with a bridge-like structure (large arrow). Note that SC initiates at one side of this structure and exhibits two small recombination nodules (small arrows). (I) In the absence of Sme4, homologs coalign but Zip4 foci remain on axes. (Scale bars: 200 nm.)
First, contrary to their axis association seen during early leptotene (above), as coalignment is achieved, Zip2, Zip4, and Msh4 foci progress from their on-axis position to a “hanging” position on the inner (and outer) edge of the chromosome axes (arrows in Fig. 2A). This configuration implies that the corresponding proteins have been released from their previous tight axis association. Second, while the existence of coaligned axes strongly implies the presence of some type of interaxis linkage, no cytologically visible links are observable by staining of Spo76 and/or costaining with Zip4 or Msh4 in chromosomes with axes coaligned at ∼400 nm (SI Appendix, Fig. S2B). However, as axes begin to progress from ∼400- to ∼200-nm distance, visible interaxis connections begin to emerge and then further evolve as axes approach the ∼100-nm distance. Intermediates can be grouped into three stages.
The earliest interaxis links, seen at ∼400-nm axis distance, are rare (less than 5% of all bridge examples) and are detected either as foci still attached to axis but located between axes and accompanied by an indentation (arrow in Fig. 2A, Right) or as foci located between the axes (Fig. 2B, arrows) accompanied by a bridge-like structure (Fig. 2B, arrowheads). By the time axes are at ∼200-nm distance, links are more obvious. Two types of morphologies are now prominently observed: bridges that exhibit Zip4 or Msh4 foci located at matching sites on the two axes in a “bidentate” structure (Fig. 2C, Left, arrow) or bridges that lack clear foci but tend to have a focus-like structure at one bridge/axis junction (Fig. 2C, Right). Both morphologies are seen with equal frequency in the 32 segments analyzed. There is, therefore, no clear indication of the temporal order with which these two morphologies may occur.
Finally, single foci are seen when axes are at either 200- or 100-nm distance (Fig. 2D; arrowheads and arrows, respectively). However, in all cases when axes are at 100-nm distance (n = 35), links between axes are seen either (i) as “fusing” foci (Fig. 2D, Left, arrow; Fig. 2E, Left), (ii) as single foci not localized on a visible bridge (Fig. 2D, Right, arrow), or (iii) as single foci localized on the bridge in the middle of the space between the two axes with an underlying signal (presumptively reflecting Spo76 localization) still visible (Fig. 2E, Right). The strong correlation between 100-nm interaxis distance and the associated focus morphologies points to very tight temporal, and thus functional, linkage among (i) progression of pairs of well-separated foci to fusing or single forms; (ii) shortening of the associated Spo76 bridge signal; and (iii) local axis juxtaposition from 200- to 100-nm distance. Also, while multiple bridges in a row are not observed when axes are at ∼200-nm distance, they are more frequent at mid-late zygotene when longer segments are close to the 100-nm distance (e.g., Fig. 2E, Right), pointing to a tendency for bridges to accumulate at this stage.
Bridges at 200- and 100-nm distance also contain Spo76/Pds5, a prominent axis component, as shown by staining of Spo76/Pds5-GFP alone (Fig. 2F, Left and Right, respectively) as well as in combination with Zip2-Zip4 or Msh4 (Fig. 2 A–E). In favorable cases, DAPI staining shows that bridges can also contain DNA (Fig. 2G). Interestingly, bridge-like links are also visible in EM sections at synaptic forks (large arrow in Fig. 2H; small arrows indicate early recombination nodules).
Taken together, these findings suggest a four-step pathway for the coalignment to synapsis transition: (i) release of single Zip2-Zip4-Msh4 foci from their previous tight axis location; (ii) movement of this ensemble of foci to a between-axis position on bridges that also contain Spo76/Pds5 and/or DNA, with different transitional morphologies; (iii) reduction in the distance between axes, implying a coordinate process involving focus fusion and bridge shortening until, finally, (iv) only a single Zip2-Zip4-Msh4 focus remains between the two axes that now have locally converged at the 100-nm distance characteristic of SC.
Finally, we find that the very first step in bridge formation, release of Msh4/Zip2-Zip4 foci from axes, requires the main SC central-region transverse-filament protein Sme4/Zip1 (Fig. 2I). While this molecule is not yet cytologically visible at this stage, these findings imply that it plays a critical functional role in anticipation of its role in SC nucleation per se.
Zip2-Zip4-Mer3-Msh4 Localization Patterns Define SC Nucleation.
Previous analysis of EM serial sections of Sordaria zygotene nuclei (n = 60) (30) showed that SC nucleates at sites of early recombination nodules and then spreads out on one side of the nodule or bridge-like structure, as in Fig. 2H. Shorter SC segments have a nodule at one end, while somewhat longer SC segments have long and very short segments emanating outward in opposite directions from the associated nodule, thus suggesting that the nodule is the nucleation site (30). This asymmetry in spreading has been proposed to reflect underlying asymmetry of the DNA recombination intermediates at nucleation sites (30). We can now identify the Zip2-Zip4-Mer3-Msh4 localization patterns that correspond to the EM-defined SC nucleation.
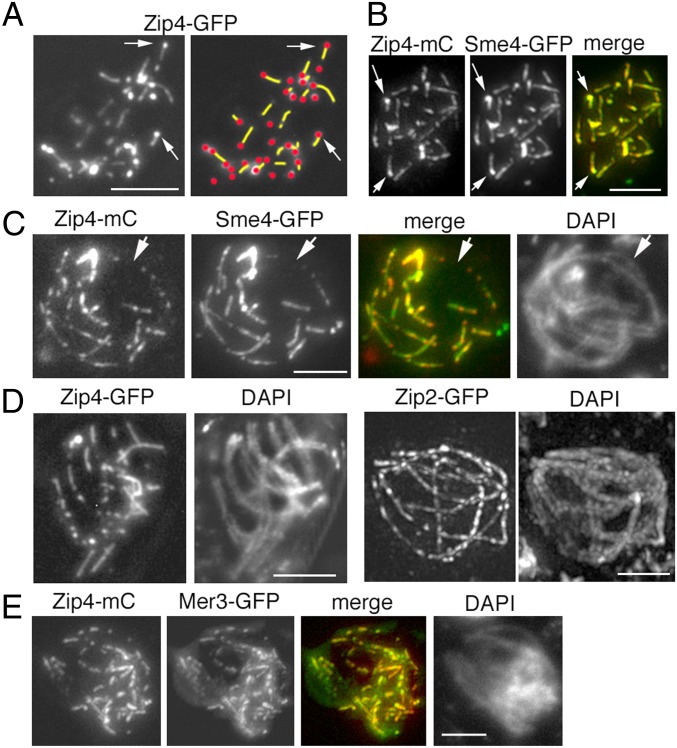
At early zygotene (defined by small amounts of Sme4/Zip1 loading), Zip2-Zip4 occur either as single foci, as foci with small lines extending in one direction from each focus, or as small lines with one to two foci (Fig. 3A and SI Appendix, Fig. S2 C and D). The number of lines adjacent to single foci increases until mid-zygotene (8% and 59% of the 82 ± 11 foci, respectively; n = 40 nuclei), as expected for sites of SC nucleation. Furthermore, both single foci and the early short tracks of Zip2-Zip4 are perfectly superimposable on early foci and tracks of Sme4/Zip1 (Fig. 3B, arrows), implying that they do in fact correspond to initial segments of SC formation. At mid-late zygotene, the number of Zip2-Zip4 lines with two or three foci increases (Fig. 3 C and D, Left), until, eventually, continuous lines with multiple foci are observed (Fig. 3D, Right), presumptively by extension and eventual fusion of initiated segments. Also, at this stage, as during early zygotene, remaining single foci and tracks of Zip2-Zip4 are perfectly superimposable on foci and tracks of Sme4/Zip1 (Fig. 3C).
Fig. 3.
SC nucleation. (A) At early zygotene, Zip2-Zip4 occur either as single foci or as foci with small lines extending in one direction from a focus (arrows) or as lines with no obvious focus. (Right) Corresponding drawing with indications of lines (yellow) and foci (red). (B) Zip4 foci and foci with lines (arrows), colocalize perfectly with corresponding Sme4/Zip1 morphologies (arrows). (C) Zip4 and Sme4/Zip1 morphologies colocalize also throughout zygotene (arrow points to an unsynapsed region, clearly visible in DAPI; Right). (D, Left) At late-zygotene, Zip4-GFP elongates further. (Right) At early pachytene, Zip2-GFP makes continuous foci/lines along all homologs. (E) Colocalization of Mer3 foci with Zip4 foci during zygotene. (Scale bars: 2 μm.)
Additionally, Mer3 and Msh4 foci perfectly colocalize with the Zip2 and Zip4 foci/tracks throughout zygotene (Fig. 3E and SI Appendix, Fig. S2E), clearly indicating that assemblies of all of these molecules follow the same progression, as a single morphological unit, from coalignment, through the bridge stage, to SC nucleation.
Localization of Zip2-Zip4, Mer3, and Msh4 to SC nucleation sites directly matches the fact that, in budding-yeast meiosis, these same molecules have been specifically identified as localizing to, and being required for, SC nucleation (the Introduction and refs. 12, 13, and 31). Additionally, budding-yeast Spo16 (which interacts with Zip2) is also frequently located at ends of partially elongated Zip1 lines (11), indicating a conservation of this asymmetric loading process between the two organisms.
To further explore the role of Zip2-Zip4, we carried out detailed analysis of Zip2-Zip4 dynamics, in relation to structure and recombination complex dynamics, throughout prophase, as described in the following sections.
Zip2-Zip4 Localizes to Leptotene Axes but with No Detectable Role for Formation of Axis or DSBs.
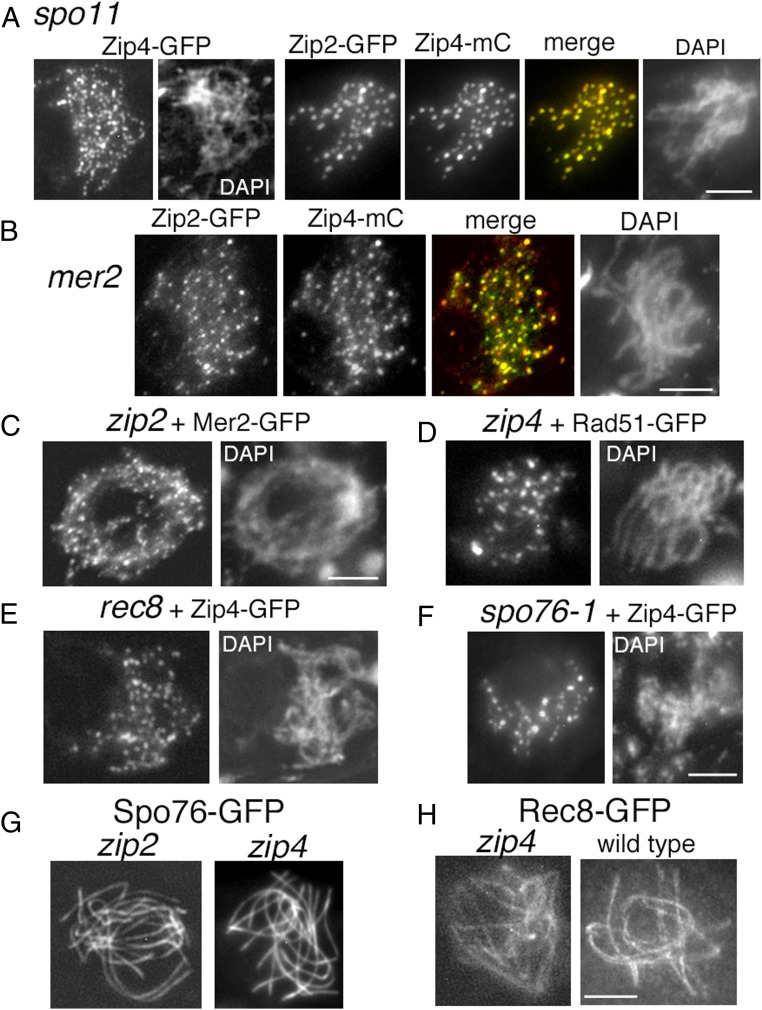
As shown above, Zip2 and Zip4 occur at leptotene along unpaired chromosome axes as regularly spaced foci (Fig. 1B). This early leptotene localization is independent of proteins involved in DSB formation and thus DSBs. Single foci (Fig. 4A, Left) and Zip2-Zip4 cofoci (Fig. 4A, Right) are detected in a mutant lacking Spo11, which promotes recombination-initiating DSBs. Zip2-Zip4 cofoci are also present in the absence of Mer2 (Fig. 4B), which is required for axis-associated pre-DSB complexes (32). Conversely, Zip2 and Zip4 are not required either to (i) load Mer2 (Fig. 4C) or (ii) promote DSB formation per se, because zip2 and zip4 null mutants show WT-like localization (Fig. 4D) and numbers of Rad51 foci that mark DSBs: 47 ± 10 and 53 ± 7 versus 52 ± 8 in WT (n = 25, 20, and 30 nuclei, respectively).
Fig. 4.
Localization of Zip2-Zip4 in the absence of DSBs and cohesins, plus Mer2, Rad51, Spo76, and Rec8 localizations in the absence of Zip2 and/or Zip4. (A) In spo11-null mutant, Zip4 makes regularly spaced foci along all chromosomes (Left) that colocalize with Zip2 foci (Right). (B) Zip2 and Zip4 foci colocalize also in the absence of Mer2. (C and D) WT-like loading of Mer2-GFP (Left) and Rad51-GFP (Right) in zip2 and zip4 null mutants. (E) Zip4 loads normally on axes in the absence of Rec8. (F) Reduced number of Zip4 foci in the nonnull spo76-1 mutant, likely due to the mutant’s defective sister cohesion and/or abnormal chromatin diffuseness (as seen in the corresponding DAPI picture, compared with the DAPI of rec8). (G and H) Spo76 (G) and Rec8 (H) loading is WT-like along all axes in the absence of Zip2 or Zip4. Note that Rec8 lines (Right) are more irregular than the Spo76 lines (Left) in both mutant and WT backgrounds (WT Spo76-GFP in Fig. 1A). (Scale bars: 2 μm.)
Leptotene axis localization of Zip2 and Zip4 is also independent of axis cohesin-component Rec8 (Fig. 4E). However, the number of foci is reduced (over 50%) in a nonnull mutant of cohesin-associated protein Spo76/Pds5 (Fig. 4F), which shows a tendency for split axes from late leptotene onward (27), implying sensitivity to axis structure. Conversely, Zip2 and Zip4 are not required for WT-like loading of Spo76 (Fig. 4G and below) or Rec8 along axes (Fig. 4H). Moreover, axis lengths are WT-like in the corresponding null mutants (respectively, 54 ± 7 and 55 ± 6 μm compared with 55 ± 4 μm in WT; n = 25, 20, and 120 nuclei).
These findings suggest that at early leptotene, Zip2-Zip4 is essentially a passive “passenger protein complex,” with an intrinsic affinity for leptotene axes that does not depend on recombination initiation or axis-associated pre-DSB complexes and has no discernible role for DSBs, post-DSB Rad51 loading or axis formation.
Zip2-Zip4 Is Recruited to Mer3-Marked Recombination Complexes and Promotes Coalignment.
Previous studies have directly implicated Mer3 and Msh4 as important players in the coalignment process (29). In the absence of Mer3, some coalignment occurs but different chromosomes are interwoven, indicating that Mer3 is required to achieve topologically regular side-by-side alignment. In the absence of Msh4, later stages in coalignment are defective (29).
As shown above (Fig. 1 C and D), Zip2 and Zip4 colocalize with Mer3 and Msh4 at leptotene. This colocalization includes also dual localization to both members of the matching focus pairs described previously for Mer3 alone (Fig. 5A, Right). Strikingly, in the absence of Mer3, Zip2-Zip4 foci no longer localize exclusively to the aberrantly coaligned chromosomes but are instead mostly delocalized to the nucleoplasm (arrows in Fig. 5B and SI Appendix, Fig. S2F). In contrast, in the absence of Zip4 (thus with WT Mer3 present), Mer3 still localizes exclusively on axes (Fig. 5C) and with WT-like numbers of foci at early/mid-leptotene (Fig. 5D, Left) when Mer3 foci initially form only on the DSB “donor” chromosome (29). Zip2-Zip4 complexes could become localized to the sites of Mer3-marked recombination complexes either by binding to Mer3 directly and/or by binding to Mer3-stabilized DNA structures.
Fig. 5.
Zip2-Zip4, Mer3, and Msh4 localizations and zip2-zip4 pairing defects. (A) Zip4 localizes to the matching pairs of Mer3 foci in WT. (A, Right) Enlarged segment of the coaligned region. (B) Zip2 and Zip4 colocalize in the absence of Mer3, but foci are no longer exclusively located on chromosomes, as shown by their merge with DAPI (Right; arrows point to foci which do not overlap with DAPI staining). (C) Mer3 foci are regularly spaced along all zip4 chromosomes. (D) Number of Mer3 foci in single and double zip2 zip4 mutants (z2, z4, z2z4) compared with WT at leptotene (Left, black dots) and at early zygotene (Right, pink dots) nuclei (determined by ascus sizes, which grow from 20 μm at early leptotene to 60 μm at zygotene and 100 μm at pachytene, in mutants and WT). (E) zip2 and zip4 mutant nuclei show either no coalignment at all (Left) or partial coalignment (Right). The seven homologs are distinguishable by their lengths and color in the corresponding drawings. In the zip4 nucleus (Right), red, green, cyan, and orange pairs are coaligned while purple pair is partially coaligned and yellow plus blue pairs are not coaligned. Chromosome axes are marked by Spo76-GFP. (F) Only few Msh4 foci (arrow) are visible in the absence of Zip4 (compare with Mer3 foci in C). (Scale bars: 2 μm.)
Zip2-Zip4 also plays a central role for homologous coalignment. In most zip2 and zip4 mutant nuclei, there is virtually no complete coalignment at the 400- or 200-nm distance (Fig. 5E, Left). However, coalignment at larger distances may sometimes occur (Fig. 5E, Right; more examples in SI Appendix, Fig. S3A). Overall, coalignment is slightly less defective in zip4 [34% of homologs show partial 400-nm coalignment and 24% show complete coalignment at 200 nm (231 pairs examined in 33 nuclei)] than in zip2 [26% and 17.6% partial and complete coalignment, respectively (91 pairs tested in 13 nuclei)]. Although not rigorously demonstrated, the role of Zip2-Zip4 in coalignment is likely mediated via its localization to Mer3-marked recombination complexes.
In WT, the number of Mer3 foci doubles at late leptotene/early zygotene (Fig. 5D, Right) when the “searching” DSB end/partner complex becomes associated with the underlying partner axis (29). In zip2, zip4, and zip2 zip4 double mutants, the number of Mer3 foci is reduced below this “double” WT number (Fig. 5D, Right) but is higher than the number seen at leptotene (Fig. 5D, Left). This modest reduction contrasts with the severe reduction in coalignment in these mutants (Fig. 5E). This disparity could be explained if, in the zip2 and zip4 null mutants, homolog partner recognition and partner axis association are relatively efficient but that the next step, the actual process of spatial juxtaposition into the coaligned configuration, is defective.
WT homolog coalignment also requires Msh4, which acts at a later stage of the process than Mer3 (above). In zip2- and zip4-null mutants, Msh4 foci still appear (Fig. 5F), but in very small numbers [respectively, 1–5 in zip2 and 3–10 in zip4 (n = 30 nuclei each), compared with 81 ± 8 in WT (n = 50)], roughly in proportion to the extent of full coalignment in the mutants. Therefore, the defect in Msh4 focus number could potentially be an indirect consequence of the mutants’ alignment defects rather than resulting from absence of a direct Msh4-Zip2-Zip4 interaction.
Zip2-Zip4 Localizes to the SC Central Element Throughout Pachytene and Colocalizes with Hei10 CO Sites at Late Pachytene.
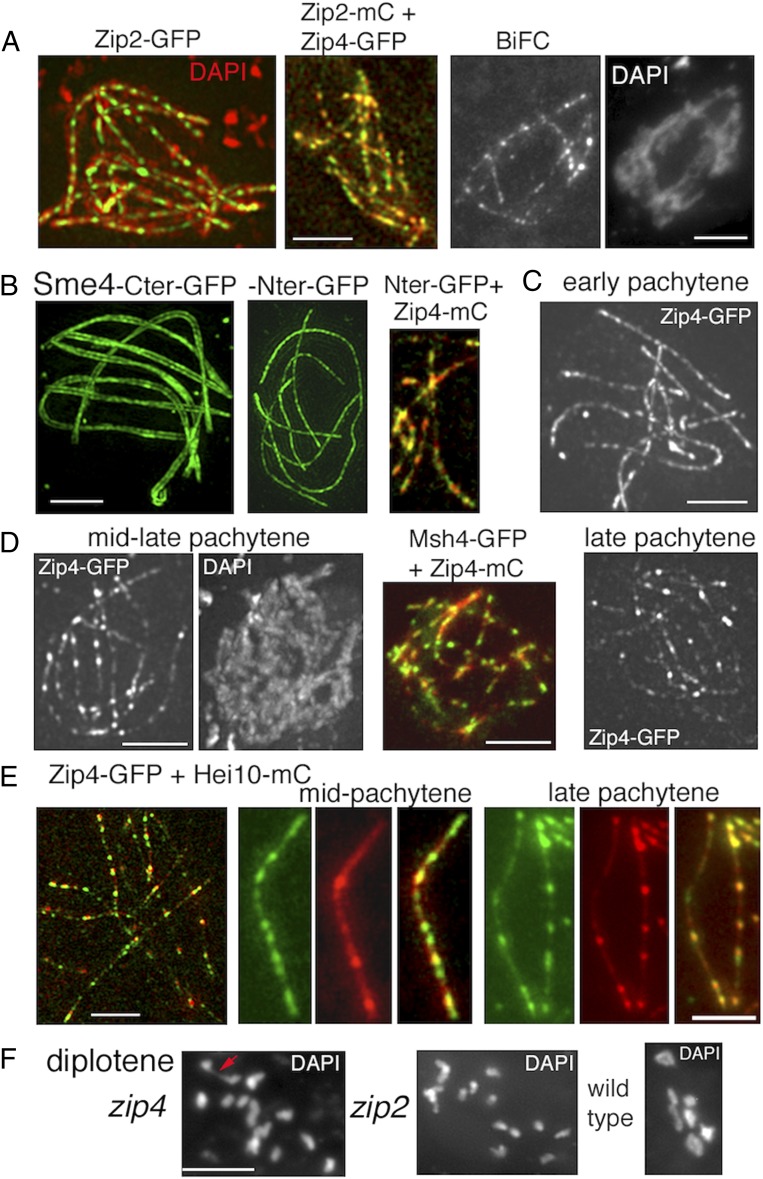
At early pachytene, Zip2 and Zip4 localize along the SC central region as discrete, regularly spaced foci (Fig. 6A, Left). Their perfect colocalization with one another along the SC (Fig. 6A, Middle) is confirmed by bimolecular fluorescence complementation (BiFC) analysis (33). When the N terminus of GFP is attached to the N terminus of Zip4 and the C terminus of GFP is attached to the C terminus of Zip2, the two proteins are close enough to reconstitute a single GFP signal along the SC central region (Fig. 6A, Right and SI Appendix, Fig. S3B). Also, like in budding yeast (11, 12), Zip2 and Zip4 are dependent upon one another for axes and SC localization (SI Appendix, Fig. S3C).
Fig. 6.
Pachytene localization of Zip2-Zip4 versus Sme4/Zip1 and Hei10. (A) All nuclei are at Pachytene. (A, Left) Costaining of Zip2-GFP with DAPI (red). (A, Middle) Colocalization of Zip2-mCherry and Zip4-GFP. (A, Right) Reconstitution of a single GFP signal by Zip2-Zip4 BiFC analysis. (B) 3D-SIM pictures of Sme4 C terminus–GFP define two lines (Left), while the N terminus–GFP is seen as a single line (second picture from left). (B, Middle) Colocalization of Sme4 N terminus–GFP with Zip4-mCherry. (C) Early pachytene nucleus with regularly spaced Zip4 foci (as Zip2 foci in A). (D) During mid-late pachytene, some Zip4 foci increase in volume and brightness (Left, compare with C). They colocalize with Msh4 foci (Middle). (D, Right) At late pachytene, the nonbright Zip4 foci are even more dim. (E) Costaining of Zip4-GFP and Hei10-mCherry during mid and late pachytene. (E, Left) 3D-SIM picture of a late pachytene nucleus. (E, Middle and Right) Foci along one homolog. At mid-pachytene (Middle), Zip4 foci (green) are more numerous and thus only partially colocalize with Hei10 foci (red), while at late pachytene (Right), the two types of foci colocalize mostly. (F) Diplotene nuclei. (F, Left) zip4 with 1 chiasma (red arrow) and zip2 with 14 univalents (Middle) versus WT nucleus with 7 chiasmate bivalents (Right). (Scale bars: 2 μm.)
Further analysis shows that the Zip2-Zip4 complex localizes to the SC central element. Observation of pachytene nuclei by 3D-SIM indicates that a Sme4/Zip1 C terminus–GFP tag defines two lines along the homolog axes (Fig. 6B, Left), while an N terminus–GFP tag gives a thin smooth line that defines the SC central element (Fig. 6B, second from left). In relation to this Sme4 organization, Zip2 and Zip4 single dotted lines colocalize with the Sme4 thin N-terminal lines that define the central element (Fig. 6B, Middle). Thus, at early pachytene, Zip2-Zip4 has an intrinsic affinity for SC central elements, irrespective of its localization to recombination sites.
Interestingly, while at early pachytene Zip2 and Zip4 foci show similar sizes and brightness (Fig. 6 A and C), from mid to late pachytene, they exhibit new morphologies: some foci are now dimmer, while others are enlarged and much brighter (Fig. 6D, Left and SI Appendix, Fig. S4A for detailed staging by DAPI staining). Msh4 foci still colocalize with Zip2-Zip4 foci at this stage (Fig. 6D, Middle). In Sordaria, SC formation nucleates at all CO sites and a subset of other recombination sites whose interactions are presumably resolved as noncrossovers (NCOs) (30). We speculate that the subset of foci that are dimmer are those that were originally localized to the subset of SC-nucleating NCO-fated interactions.
At late pachytene, even more foci get dimmer (Fig. 6D, Right). Most of the bright Zip2-Zip4 foci (∼60%; n = 100) now either colocalize or lie side-by-side with late Hei10 foci, which mark the sites of COs (Fig. 6E, Left: entire nucleus; Middle and Right: single homologs), pointing to an involvement of Zip2-Zip4 in the maturation of CO formation in the context of the SC. Overall, these patterns, as well as the continuous colocalization of Zip2-Zip4 foci with Msh4 foci through pachytene, suggest that Zip2-Zip4 may continue to play roles at the SC/recombination complex interface throughout these later stages of the recombination process.
Direct assessment of this possibility is precluded by the fact that zip2 and zip4 mutants are already strongly defective in coalignment (above). As expected from earlier pairing defects, almost no Hei10 foci occur in zip2 and zip4: respectively, 0–1 and 1–3 (SI Appendix, Fig. S4B; n = 30 nuclei, respectively) compared with the 22 ± 3 foci seen in WT (34). The number of chiasmata is correspondingly reduced (Fig. 6F, Left), with 70% of diplotene nuclei showing only univalents (Fig. 6F, Middle; n = 30) compared with the seven chiasmate bivalents seen in WT (Fig. 6F, Right). Notably, however, there is no evidence of broken chromosomes at metaphase I, which indicates that all DSBs are converted to intact duplex products, either as NCOs or via interactions between sister chromatids. Thus, while Zip2 and Zip4 are important to ensure normal maturation of CO-fated recombination intermediates, they are not required for the biochemical events of NCO and/or sister chromatid recombination.
Discussion
The progression from chromosome coalignment to synapsis is a universal step in the basic meiotic program. During this transition, two key logistical problems must be solved. First, chromosome axes must move from ∼400- to 200-nm separation as seen at the coalignment stage to ∼100-nm separation as seen along the SC. Second, recombination complexes must change their association with underlying structures: from tight linkage to individual homolog axes during coalignment to tight linkage with SC central components.
We have now provided further information about how this transition occurs. We previously showed that homolog axes are coaligned but invisibly connected, presumably by the corresponding DNA recombination intermediates (29). We now show that these invisible connections develop into robust interaxis bridges that contain chromosome structure molecule(s) like Spo76/Pds5, DNA, Mer3-Msh4 recombination complexes, and the evolutionarily conserved Zip2-Zip4 complex. The geometry and dynamics of these bridges suggest solutions to the two basic logistical challenges described above. (i) Movement of recombination complexes from axes to bridges is the event by which these complexes relocate from on-axis to between-axis positions. (ii) Evolution of bridges from a ∼200-nm form to the 100-nm form provides the spatial juxtaposition required for installation of SC central components between homologous axes. Interestingly, the main SC transverse-filament protein Sme4/Zip1 has an essential but “cytologically invisible” role in this transition, at the release step and thus long before any tripartite SC is formed. Existence of this anticipatory role at the very onset of bridge formation raises the possibility that Sme4/Zip1 might play additional (invisible) roles throughout the bridge stage (below).
The Coalignment to Synapsis Transition.
Overall, positioning of colocalizing Zip2-Zip4-Mer3-Msh4 foci between the juxtaposed 100-nm axes allows recombination complexes to be already positioned in the SC central region as it is installed. Moreover, the single cofocus Zip2-Zip4-Mer3-Msh4 ensemble appears at one end of each asymmetrically spreading segment of nucleated SC. This pattern corresponds to limited asymmetric spreading of SC outward from recombination nodules seen in Sordaria EM serial sections (above and ref. 30) or from bridge-like structures visible at pairing forks in different organisms (e.g., refs. 35 and 36). Plus these early segments colocalize with Sme4/Zip1 early segments.
The underlying mechanisms of the observed progression from coalignment to SC nucleation remain to be determined. However, it is notable that there is very tight linkage between morphological changes in bridges and axis juxtaposition, from 400 to 200 nm and from 200 to 100 nm, which suggests that bridge shortening with accompanying Zip2-Zip4-Msh4 focus changes, finally including fusion of bidentate foci, underlies juxtaposition. It is also interesting that interaxis bridges contain cohesin-associated protein Spo76/Pds5 and DNA. Since Spo76/Pds5 is a major component of the chromosome axes, bridges might be thought of as “miniature axes.” This analogy is supported by the fact that in mouse spermatocytes, meiosis-specific cohesin RAD21L does also form bridges between aligned axes (37).
The presented findings allow linkage between previously disparate findings concerning bridges. For example, the morphologies described here correspond directly to those previously elucidated by EM studies of Allium cepa chromosomes (7), namely, hanging single foci, the two types of 200-nm morphologies that show asymmetric bridge/axis bulges and bidentate structures respectively and, finally, local juxtaposition of axes at 100 nm with an accompanying nodule as a prelude to SC nucleation. We now show the direct involvement of Zip2-Zip4, Mer3, and Msh4 in these morphological changes. Other findings provide further evidence that the processes described here are widely conserved. (i) Release of RPA and RAD51/DMC1 from axes, plus their presynaptic localization between axes, is evident in EM spreads of mouse and rat spermatocytes (reviewed in ref. 8). (ii) In human and mouse spermatocytes, at early zygotene, RPA and BLM proteins are visible either as matching foci still attached to their axis or as coalescent foci linking the two axes (8–10). (iii) Analysis of zygotene in six plants shows that their synaptic forks contain “early recombination nodules” labeled with Rad51 antibodies (38). (iv) When Locusta migratoria homologs are coaligned at 300 nm, they are connected by transverse filaments at synaptic forks (35).
We also note that Sordaria Mer2 and budding yeast Zip3 show repositioning from axis to SC central region (32, 39), suggesting that, in their respective organisms, they move as part of the process defined here by analysis of Sordaria Mer3, Msh4, and Zip2-Zip4.
Zip2-Zip4: Mediator of the Recombination Complex/Structure Interface at All Post-DSB Stages.
Our analysis suggests that Zip2-Zip4 exhibits two types of affinities: one for chromosome structures (axes and SC) and another for recombination complexes (marked by Mer3, Msh4, and Hei10). Moreover, the specific substrates for these affinities change, independently, over time.
-
i)
An intrinsic affinity for chromosome structure is revealed by localization of Zip2-Zip4 foci. At early leptotene, foci localize all along the lengths of the chromosome axes, independently of pre-DSB recombination complex assembly (in the absence of Mer2), and DSB formation (in the absence of Spo11). At early pachytene, Zip2-Zip4 foci localize continuously along the length of the SC central element, irrespective of focal localization to recombination complexes. The specific molecular partners of Zip2-Zip4 at both stages are not known. With respect to chromosome axes, Spo76/Pds5 and Rad21L (above in mouse) are possible candidates, as are meiosis-specific axis components, which have not yet been analyzed in Sordaria. With respect to SC central components, the N terminus of Sme4/Zip1 is an attractive candidate because of its colocalization with Zip2-Zip4, but dedicated central element proteins could also/alternatively be involved [e.g., Ecm11 and Gmc2 (40)].
-
ii)
Affinity of Zip2-Zip4 for recombination complexes is first apparent by their localization to sites of Mer3-marked post-DSB recombination complexes, strictly dependent on Mer3. Although not demonstrated, this association is likely of functional significance since the absence of either Mer3 or Zip2 or Zip4 results in a coalignment defect. Msh4 foci colocalize also with these complexes as coalignment is completed. The Zip2-Zip4-Mer3-Msh4 ensemble remains intact throughout the transition from coalignment to synapsis, i.e., from the end of coalignment before release from axes, during that release, on bridges, and at SC nucleation sites, even as the complex changes its structural partner from axes to SC central elements. At early pachytene, Mer3 foci disappear; however, Msh4 foci remain through mid-late pachytene, where they generally colocalize with brighter Zip2-Zip4 foci. Furthermore, thereafter, CO-correlated foci of Hei10 colocalize with Zip2-Zip4 bright foci at late pachytene.
As far as we can discern, the global associations of Zip2-Zip4 with axes and SC central elements are not important for the structures per se. Zip2-Zip4 plays no role for leptotene axis morphogenesis. Furthermore, since a subset of Zip2-Zip4 foci is changed/lost along the central element as pachytene progresses, Zip2-Zip4 is likely also not an essential component of the SC. We thus infer that when Zip2-Zip4 is present at structure-associated recombination sites, these localizations are manifestations of roles of the complex as a mediator of the recombination complex/structure interface. By this interpretation, the Zip2-Zip4 “couple” mediates the recombination complex/structure interface at all post-DSB stages throughout dynamic changes in the composition of associated recombination complexes and the molecular nature of the underlying structural component. By maintaining its association with Mer3-Msh4 while changing its affinity for structure from axes to SC central elements, Zip2-Zip4 serves as a platform that switches its Mer3/Msh4 cargo from one structural substrate to the other.
The multistage recombination complex/structure mediator roles of Zip2-Zip4 defined for Sordaria in the present study can begin to explain the functional significance of the several biochemical properties defined by molecular studies of their counterparts in budding yeast and mouse. These include interactions of Zip4 with Msh5 (partner of Msh4) and axis/SC lateral component Red1 and of Zip2/Spo16 with branched DNA structures (19–21). Analogously, in mouse, SHOC1/ZIP2 and TEX11/ZIP4 colocalize substantially with MSH4 (20, 22) and TEX11 interacts with SC lateral-component SYCP2 (22). Importantly, the current results suggest that all of these activities may come into play, directly or indirectly, at multiple stages.
A Model for the Coalignment-to-Synapsis Transition as Mediated by Zip2-Zip4 and Sme4/Zip1 (Fig. 7).
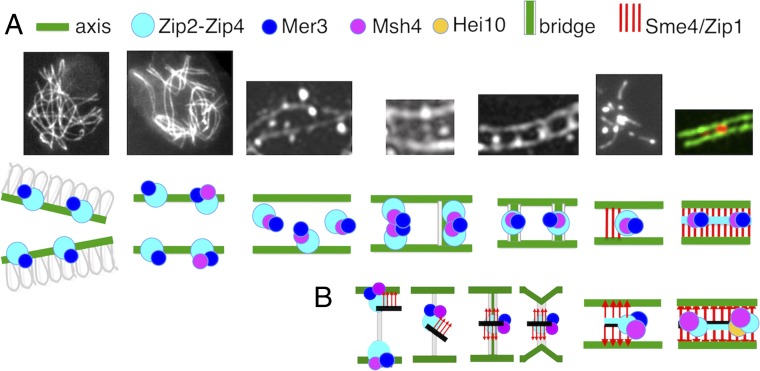
Fig. 7.
From early leptotene to SC formation. (A, Upper) From left to right: pictures corresponding to the different stages described in the paper. (A, Lower) Cartoon of the experimentally observed proteins (indicated above) involved in the corresponding transitions. (B) Proposed model for Zip2-Zip4-mediated coalignment-to-synapsis transition. From left to right: Zip2-Zip4-Mer3-Msh4–mediated recruitment of SC central component Sme4/Zip1 (red arrows) and SC central components (black line). The resulting ensemble, bound to axes components, is released from the axes through bridge formation before relocalization halfway between axes. A signal for SC nucleation then occurs which disassembles the bridges and brings axes closer together, allowing SC initiation. During this process, the Zip2-Zip4-Mer3-Msh4 ensemble now gets associated only with SC central components. SC is thus nucleated by centrally located Zip2-Zip4-Mer3-Msh4, and SC initiation occurs at sites of recombination specifically because of their association with Zip2-Zip4. Recombination complexes are thereby automatically localized on SC central elements. Instead of recombination complexes being “moved” to the SC, the SC comes to the recombination complex.
As discussed above, Zip2-Zip4 has likely an intrinsic affinity for the SC central element, irrespective of association with recombination complexes. If SC central elements can recruit Zip2-Zip4 during elongation of the SC, then, conversely, Zip2-Zip4 should be able to recruit SC central element components to sites of Zip2-Zip4-Mer3-Msh4 complexes at earlier stages, i.e., during the bridge transition. We also show that Sme4/Zip1 is required to release Zip2-Zip4 and Mer3/Msh4 from association with chromosome axes at the onset of bridge formation. Furthermore, the N terminus of Sme4/Zip1 colocalizes with Zip2-Zip4 to the central element. Thus, Zip2-Zip4 might recruit Sme4/Zip1, via its N terminus, to Zip2-Zip4-Mer3-Msh4 complexes on coaligned axes, thereby enabling their Sme4/Zip1-mediated release (Fig. 7B). Support of this hypothesis is given by zygotene EM pictures of pairing forks where central region “material” extends out from a formed SC segment toward one of the still diverging homolog lateral element suggesting a pre-SC formation/localization of SC central element proteins along the lateral element (e.g., ref. 36).
Since Sme4/Zip1 has an important function at the onset of bridge formation, this association might remain (invisibly) throughout the bridge transition. Such an association might, for example, allow Sme4/Zip1 to mediate bridge contraction and, in any case, being in place, to then directly nucleate installation of the SC. The joint occurrence on bridges of both axis components and this SC central-element component could also facilitate the transfer of Mer3/Msh4 complexes from axes to SCs. Given that Zip2-Zip4 can interact with both types of components (above), bridges might include a multivalent complex in which Zip2-Zip4 interacts not only with Mer3-Msh4 but also, simultaneously, with both an axis component and an SC central element component on evolving bridges. Such a complex could include a transitional intermediate in a direct handoff of recombination complexes from axes to SC central regions. Overall this model would imply that, as mediated by the critical role of Zip2-Zip4, SC components are brought to the sites of recombination complexes, rather than the reverse, and provides a specific molecular basis for the fact that, in most organisms, SC formation nucleates at sites of recombination complexes (reviewed in ref. 30).
Materials and Methods
Cloning, Plasmids, and Transformation of Sordaria.
Null zip2 and zip4 mutants were obtained by single-step gene replacement: a hygromycin-resistance cassette replaces the entire ORF. Transformants carrying a null allele were constructed as described previously (32). Transformants carrying a null allele were selected for hygromycin resistance and confirmed by DNA sequencing. Transformations were performed in a ku70-null mutant background, which increases the homologous integration events. Further crosses with a KU70 WT strain eliminated the ku70 allele. Introduction of an ectopic WT ZIP2 or ZIP4 gene into the null mutant restored a WT phenotype in both cases.
In all GFP or mCherry fusions, their coding sequences [p-EGFP-1 (Clontech); pRsetB-mCherry] were fused just after the initiating methionine codon for ZIP4 and just after the last C-terminal amino acid predicted from the ORF for ZIP2. The GFP/mCherry-tagged versions of the genes, under control of their own promoters, were introduced in the WT strain by cotransformation with a plasmid encoding the nourseothricine (pbc-nour, for GFP versions) or the hygromycin-resistance cassette (Pan7.1, GenBank accession no. Z32698, for mCherry versions) at ectopic locations and by further crosses into the different mutant strains. Zip4-GFP/-mCherry and Zip2-GFP/-mCherry proteins are fully functional: they complement all meiotic and sporulation defects of their cognate null mutants.
In all plasmids generated for the BiFC assay, the two halves of the GFP (NGFP or CGFP) were generated by PCR and used to replace the GFP in the two plasmids containing the two GFP fusions. The two fusions with the two halves of the GFP were then introduced at ectopic locations into a WT strain by cotransformation with a plasmid encoding the hygromycin-resistance cassette.
Yeast Two-Hybrid Experiment.
The strains PJ69-4A/a (MATa/α ade4 trp1-901 leu2-3,112 ura3-52 his3-200 Gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ) were used for all two-hybrid experiments. Yeast media (yeast peptone dextrose adenine, synthetic dextrose-Leu –Leu; –Trp; –Leu/–Trp; –Ade/–His/–Leu/–Trp with or without agar) were prepared using the pouches according to the manufacturer (Yeast Media Set 2; Clontech, Ozyme).
Transformations were performed using the high efficiency method of Schiestl and Gietz (41). Specific steps were performed as described previously (32): cDNA sequences were amplified from RT-PCR at day 4 (meiotic-prophase stage) with appropriate primers and Phusion (ThermoFisher Scientific) according to the manufacturer’s indications; amplified fragments were cloned directly into pJET1.2 (ThermoFisher Scientific); after plasmid amplification in Escherichia coli, all inserts were sequenced to check for the absence of adventitious mutations before further digestion and cloning into pGBT9 (GenBank accession no. U07646) and pGAD424 (GenBank accession no. U07647). Full ZIP2 or ZIP4 cDNA were used to construct ZIP2N, ZIP2C, ZIP4N, and ZIP4C subdomain fragments by amplification using primers containing restriction enzyme sites. Restriction sites were selected for fusing the coding sequence of the amplification fragment with GAL4 binding domain (pGBT9) or activating domain (pGAD424). Recombinant pGBT9 and pGAD424 plasmids were amplified in E. coli and transformed in PJ69-4A (for pGAD424) or PJ69-4a (for pGBT9) yeast strains.
RT-qPCR Experiment.
cDNA and RT-qPCR were performed as described in ref. 32.
Cytology.
GFP, mCherry, and DAPI (0.5 μg/mL) signals were observed, either on living material or after fixation in 4% paraformaldehyde, with a Zeiss Axioplan microscope with a CCD Princeton camera, a Leica DMIRE2 microscope (Leica) with a CoolSNAPHQ CCD camera (Roper Scientific), or a Delta Vision OMXTM platform (3D-SIM; Applied Precision). MetaMorph software (Universal Imaging Corp.) and public domain software ImageJ (https://imagej.nih.gov/ij/) were used to deconvolute Z-series and treat the images.
Supplementary Material
Acknowledgments
We thank Edith Heard (Institut Curie, Paris) for providing access to her 3D-SIM microscope. This work, E.D., A.D.M., J.L.S., K.B., M.L., R.D., D.Z., and E.E. were supported by grants from the CNRS (UMR 9198, I2BC) and by a subcontract collaboration with N.K. supported by NIH Grant R01 GM044794.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901237116/-/DCSupplemental.
References
- 1.Lam I, Keeney S (2014) Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol 7:a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter N. (2015) Meiotic recombination: The essence of heredity. Cold Spring Harb Perspect Biol 7:a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Page SL, Hawley RS (2004) The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol 20:525–558. [DOI] [PubMed] [Google Scholar]
- 4.Fraune J, et al. (2016) Evolutionary history of the mammalian synaptonemal complex. Chromosoma 125:355–360. [DOI] [PubMed] [Google Scholar]
- 5.Gao J, Colaiácovo MP (2018) Zipping and unzipping: Protein modifications regulating synaptonemal complex dynamics. Trends Genet 34:232–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zickler D, Kleckner N (2015) Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol 7:a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albini SM, Jones GH (1987) Synaptonemal complex spreading in Allium cepa and A. fistulosum. I. The initiation and sequence of pairing. Chromosoma 95:324–338. [Google Scholar]
- 8.Moens PB, Marcon E, Shore JS, Kochakpour N, Spyropoulos B (2007) Initiation and resolution of interhomolog connections: Crossover and non-crossover sites along mouse synaptonemal complexes. J Cell Sci 120:1017–1027. [DOI] [PubMed] [Google Scholar]
- 9.Oliver-Bonet M, Campillo M, Turek PJ, Ko E, Martin RH (2007) Analysis of replication protein A (RPA) in human spermatogenesis. Mol Hum Reprod 13:837–844. [DOI] [PubMed] [Google Scholar]
- 10.Holloway JK, Morelli MA, Borst PL, Cohen PE (2010) Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J Cell Biol 188:779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shinohara M, Oh SD, Hunter N, Shinohara A (2008) Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat Genet 40:299–309. [DOI] [PubMed] [Google Scholar]
- 12.Tsubouchi T, Zhao H, Roeder GS (2006) The meiosis-specific Zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with Zip2. Dev Cell 10:809–819. [DOI] [PubMed] [Google Scholar]
- 13.Börner GV, Kleckner N, Hunter N (2004) Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117:29–45. [DOI] [PubMed] [Google Scholar]
- 14.Mazina OM, Mazin AV, Nakagawa T, Kolodner RD, Kowalczykowski SC (2004) Saccharomyces cerevisiae Mer3 helicase stimulates 3′-5′ heteroduplex extension by Rad51; implications for crossover control in meiotic recombination. Cell 117:47–56. [DOI] [PubMed] [Google Scholar]
- 15.Snowden T, Acharya S, Butz C, Berardini M, Fishel R (2004) hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell 15:437–451. [DOI] [PubMed] [Google Scholar]
- 16.Lynn A, Soucek R, Börner GV (2007) ZMM proteins during meiosis: Crossover artists at work. Chromosome Res 15:591–605. [DOI] [PubMed] [Google Scholar]
- 17.Macaisne N, et al. (2008) SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Curr Biol 18:1432–1437. [DOI] [PubMed] [Google Scholar]
- 18.Voelkel-Meiman K, Cheng S-Y, Morehouse SJ, MacQueen AJ (2016) Synaptonemal complex proteins of budding yeast define reciprocal roles in MutSγ-mediated crossover formation. Genetics 203:1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Muyt A, et al. (2018) A meiotic XPF-ERCC1-like complex recognizes joint molecule recombination intermediates to promote crossover formation. Genes Dev 32:283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guiraldelli MF, et al. (2018) SHOC1 is a ERCC4-(HhH)2-like protein, integral to the formation of crossover recombination intermediates during mammalian meiosis. PLoS Genet 14:e1007381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arora K, Corbett KD (2019) The conserved XPF:ERCC1-like Zip2:Spo16 complex controls meiotic crossover formation through structure-specific DNA binding. Nucleic Acids Res 47:2365–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang F, et al. (2008) Meiotic failure in male mice lacking an X-linked factor. Genes Dev 22:682–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chelysheva L, et al. (2007) Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet 3:e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen Y, et al. (2012) ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. J Cell Sci 125:2581–2591. [DOI] [PubMed] [Google Scholar]
- 25.Adelman CA, Petrini JHJ (2008) ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS Genet 4:e1000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q, Shao J, Fan H-Y, Yu C (2018) Evolutionarily-conserved MZIP2 is essential for crossover formation in mammalian meiosis. Commun Biol 1:147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Heemst D, James F, Pöggeler S, Berteaux-Lecellier V, Zickler D (1999) Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs. Cell 98:261–271. [DOI] [PubMed] [Google Scholar]
- 28.Espagne E, et al. (2011) Sme4 coiled-coil protein mediates synaptonemal complex assembly, recombinosome relocalization, and spindle pole body morphogenesis. Proc Natl Acad Sci USA 111:10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storlazzi A, et al. (2010) Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell 141:94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Espagne E, de Muyt A, Zickler D, Kleckner NE (2014) Interference-mediated synaptonemal complex formation with embedded crossover designation. Proc. Natl. Acad. Sci. U.S.A. 111:E5059–E5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chua PR, Roeder GS (1998) Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell 93:349–359. [DOI] [PubMed] [Google Scholar]
- 32.Tessé S, et al. (2017) Asy2/Mer2: An evolutionarily conserved mediator of meiotic recombination, pairing, and global chromosome compaction. Genes Dev 31:1880–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghosh I, Hamilton AD, Regan L (2000) Antiparallel leucine zipper-directed protein reassembly: Application to the green fluorescent protein. J Am Chem Soc 122:5658–5659. [Google Scholar]
- 34.De Muyt A, et al. (2014) E3 ligase Hei10: A multifaceted structure-based signaling molecule with roles within and beyond meiosis. Genes Dev 28:1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moens PB. (1969) The fine structure of meiotic chromosome polarization and pairing in Locusta migratoria spermatocytes. Chromosoma 28:1–25. [DOI] [PubMed] [Google Scholar]
- 36.Holm PB. (1977) Three-dimensional reconstruction of chromosome pairing during the zygotene stage of meiosis in Lilium longiflorum (thunb.). Carlsberg Res Commun 42:103–151. [Google Scholar]
- 37.Rong M, Matsuda A, Hiraoka Y, Lee J (2016) Meiotic cohesin subunits RAD21L and REC8 are positioned at distinct regions between lateral elements and transverse filaments in the synaptonemal complex of mouse spermatocytes. J Reprod Dev 62:623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson LK, Hooker KD, Stack SM (2001) The distribution of early recombination nodules on zygotene bivalents from plants. Genetics 159:1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serrentino M-E, Chaplais E, Sommermeyer V, Borde V (2013) Differential association of the conserved SUMO ligase Zip3 with meiotic double-strand break sites reveals regional variations in the outcome of meiotic recombination. PLoS Genet 9:e1003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Humphryes N, et al. (2013) The Ecm11-Gmc2 complex promotes synaptonemal complex formation through assembly of transverse filaments in budding yeast. PLoS Genet 9:e1003194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schiestl RH, Gietz RD (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16:339–346. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.