Abstract
Protein arginine methyltransferases (PRMTs) are a family of enzymes that modify proteins by methylating the guanidino nitrogen atoms of arginine residues to regulate cellular processes such as chromatin remodeling, pre-mRNA splicing, and signal transduction. PRMT7 is the single type III PRMT solely capable of arginine monomethylation. To date, other than histone proteins, there are very few identified substrates of PRMT7. We therefore performed quantitative mass spectrometry experiments to identify PRMT7’s interactome and potential substrates to better characterize the enzyme’s biological function(s) in cells. These experiments revealed that PRMT7 interacts with and can methylate eukaryotic translation initiation factor 2 alpha (eIF2α), in vitro and in breast cancer cells. Furthermore, we uncovered a potential regulatory interplay between eIF2α arginine methylation by PRMT7 and stress-induced phosphorylation status of eIF2α at serine 51. Finally, we demonstrated that PRMT7 is required for eIF2α-dependent stress granule formation in the face of various cellular stresses. Altogether, our findings implicate PRMT7 as a novel mediator of eIF2α-dependent cellular stress response pathways.
INTRODUCTION
Posttranslational modifications of protein residues allow for the formation of a more intricate network of protein interactions and functions resulting in an enlarged proteomic network. This is partly due to the gain and/or loss of protein–protein, protein–DNA, and protein–RNA interactions upon the modification of specific residues within proteins. Many posttranslational modifications have been identified of which arginine methylation has shown great importance in cellular regulation.
Arginine methylation is catalyzed by protein arginine methyltransferases (PRMTs), which regulate several cellular processes such as transcriptional regulation, pre-mRNA splicing, and signal transduction (Gonsalvez et al., 2007; Litt et al., 2009; Karkhanis et al., 2012; Gayatri and Bedford, 2014; Hadjikyriacou and Yang, 2015). PRMTs are a family of enzymes that catalyze the deposition of a methyl group onto a terminal ω-guanidino nitrogen atom of an arginine residue within proteins. The methylation reaction occurs via the transfer of a methyl group from the methyl donor/cofactor, S-adenosyl-l-methionine (SAM), to the arginine substrate yielding the resultant methyl-arginine residue and S-adenosyl-l-homocysteine as a by-product (Chan et al., 2013). The family consists of nine well-characterized enzymes that are categorized into three different subtypes: type I (PRMT1, 2, 3, 4/CARM1, 6, 8), II (PRMT5, 9), and III (PRMT7). All three subtypes catalyze the formation of ω-NG-monomethylarginine (MMA). Both type I and II PRMTs can subsequently dimethylate the arginine residue forming ω-NG, NG-asymmetric dimethylarginine (aDMA) and ω-NG, NG’-symmetric dimethylarginine (sDMA), respectively (Bedford and Clarke, 2009; Yang et al., 2015). PRMT7 is unique among the nine PRMTs as it is the sole type III PRMT (Feng et al., 2013). Although PRMT7 is unable to dimethylate arginine residues, it does positively regulate dimethylation by PRMT5. Specifically, PRMT7 monomethylates arginine residues, allowing for dimethylation by PRMT5 to occur at another site on the same protein, through allosteric regulation (Jain et al., 2017).
PRMT7 has been shown to be implicated in disease progression mostly through gene regulation. For instance, PRMT7 promotes epithelial-to-mesenchymal transition (EMT) within breast cancer cells by transcriptionally repressing E-cadherin expression via the recruitment of a histone deacetylase to the promoter of the gene (Yao et al., 2014). We have additionally shown that PRMT7 promotes breast cancer cell invasion and metastasis by inducing the expression of matrix metalloproteinase 9 (Baldwin et al., 2015). Furthermore, clinical work has correlated loss-of-function mutations of PRMT7 with intellectual disability syndrome, microcephaly (abnormally small head), and brachydactyly (shortened fingers/toes; Kernohan et al., 2017; Agolini et al., 2018). Because such severe diseases are related to PRMT7 function, the enzyme most likely regulates critical cellular processes. Hence, identification of its protein interactome would aid in better understanding the biological functions of PRMT7.
Although previously misclassified due to PRMT5 contamination (Lee et al., 2005) PRMT7 is now considered as a type III PRMT (Miranda et al., 2004; Zurita-Lopez et al., 2012; Feng et al., 2013, 2014). In fact, in Caenorhabditis elegans, crystal structures demonstrate that the entrance to the substrate pocket of PRMT7 is narrower than other PRMTs due to the presence of bulky residues (Hasegawa et al., 2014). Additionally, in trypanosomal PRMT7, the bulky residues are suboptimal in forming hydrogen bonds required for appropriate SN2 geometry to produce dimethylation (Cáceres et al., 2018). The sequence of the bulky residues within the substrate pocket is analogous in mammals, implying a similar narrow pocket and substrate-binding geometry. Thus, steric constraints and geometric orientation/binding of its substrates makes it unlikely that a monomethylated arginine substrate enters the substrate-binding pocket of PRMT7 for a subsequent dimethylation reaction (Jain et al., 2016).
Although histones are known substrates of PRMT7, there are only a few nonhistone interactors/substrates that have been identified, such as CTCFL, SmD3, ASS1 (argininosuccinate synthetase) and eukaryotic elongation factor eEF2 (Jelinic et al., 2006; Gonsalvez et al., 2007; Jung et al., 2011). Yet, the nonhistone proteins identified as substrates of PRMT7 are symmetrically dimethylated and are also dependent on PRMT5 expression. Thus, it is likely that these are shared substrates and that PRMT7 may allosterically regulate the dimethylation reaction by PRMT5 as is the case with histones (Jain et al., 2017).
We report here the identification of eukaryotic translation initiation factor 2α (eIF2α) as a novel interacting protein and substrate of PRMT7. Moreover, we demonstrated a regulatory interplay between eIF2α arginine methylation by PRMT7 and eIF2α serine 51 (Ser51) phosphorylation status upon stress. Accordingly, we have shown that stress granule formation, in the face of eIF2α-dependent cellular stresses, was significantly diminished in PRMT7-knockdown cells. These results reveal eIF2α as a novel nonhistone substrate of PRMT7 and a novel functional role for PRMT7 in the cellular stress response.
RESULTS
Identification and validation of novel PRMT7 protein interactors
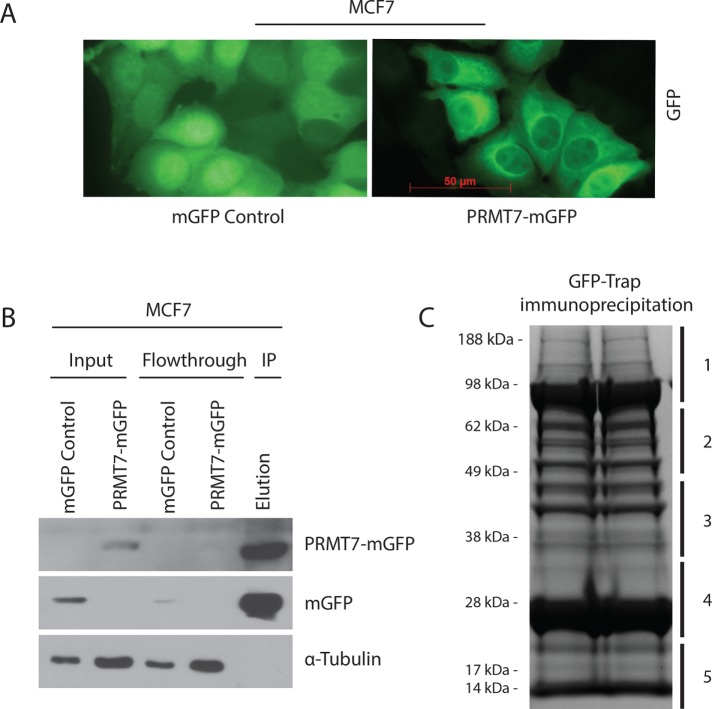
To discover unique protein interactors and/or substrates of PRMT7, SILAC (stable isotope labeling by amino acids in cell culture)-based quantitative mass spectrometry (Trinkle-Mulcahy et al., 2008; Larivière et al., 2014; Baldwin et al., 2015) was performed. MCF7 breast cancer cells were used to express either GFP as a control or PRMT7-mGFP. As previously reported (Herrmann et al., 2009; Baldwin et al., 2015), PRMT7-mGFP was found predominantly within the cytoplasm, with a weak diffused staining in the nucleus and no signal in nucleoli, in contrast to GFP, which displayed a uniformly diffused staining across the whole cell (Figure 1A). GFP-expressing cells were labeled in “light” R0K0 (L) media and our experimental condition, cells expressing PRMT7-mGFP, in “heavy” R6K4 (H) media (see Materials and Methods for details). For increased confidence, we performed a second independent experiment with reciprocal labeling. Cells were kept in culture in the isotopic-labeled media for at least five passages to ensure sufficient incorporation of the isotopes within proteins (Trinkle-Mulcahy et al., 2008; Larivière et al., 2014). Both GFP and PRMT7-mGFP were then subsequently immunopurified from cell lysates (Figure 1B). Beads from both immunoprecipitates (GFP and PRMT7-mGFP) were combined, and all bound proteins were eluted. GFP and PRMT7-mGFP were eluted in equal amounts (Figure 1B; elution lane). The eluate was resolved through SDS–PAGE (two lanes) and five equal gel pieces were used for trypsin digestion and analysis by liquid chromatography tandem mass spectrometry (LC-MS/MS; Figure 1C).
FIGURE 1:
Preparation and confirmation of immunoprecipitation for SILAC-based quantitative mass spectrometry. (A) Representative fluorescent images of MCF7 cells infected with lentiviruses expressing either mGFP control or PRMT7-mGFP showing diffused localization of PRMT7 at 40× magnification (scale bar: 50 µm). (B) Confirmation of affinity immunodepletion of PRMT7-mGFP fusion protein and mGFP using GFP-Trap beads (ChromoTek). (C) Coomassie-stained SDS–PAGE gel of the eluate (1:1 mixture of PRMT7-mGFP/mGFP) from affinity purification using GFP-Trap beads. The gel was sliced into five fragments to be sent for mass spectrometric analysis of coimmunoprecipitated proteins.
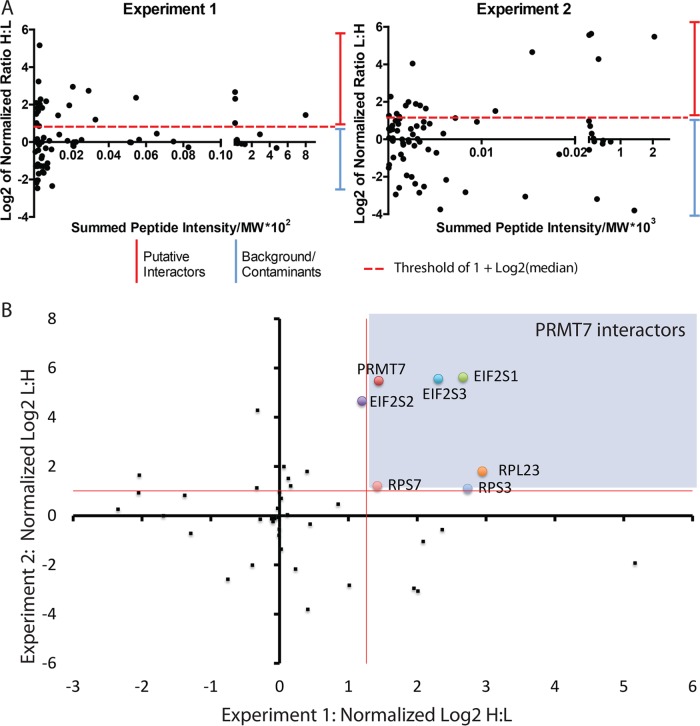
Database searching (against the human Uniprot database) and quantitation were performed using MaxQuant software v1.2.7.4, as previously described (Trinkle-Mulcahy et al., 2008; Larivière et al., 2014). ProteinCenter (Proxeon Bioinformatics; a proteomics data mining and management software) was used to eliminate redundancy, compare datasets, and convert protein IDs to gene symbols. Based on the mass differences between the R0K0 and R6K4 labeled peptides, Log2 SILAC ratios (H:L) were calculated for each identified protein (Supplemental Figure 1). For both independent mass spectrometry experiments, the normalized Log2 H:L ratio and normalized L:H ratio, for experiments 1 and 2, respectively, were plotted against normalized peptide intensity for each identified protein. We set a Log2 H:L ratio of 1 log unit above the median value as a putative interactor threshold (red line) as this indicates at least a twofold enrichment above background, nonspecific interactors (Figure 2A). A total of 72 and 76 potential protein interactors of PRMT7 were identified in experiments 1 and 2 (reverse labeling), respectively (Supplemental Table 1). Of these proteins, 41 were reproducibly pulled down in both experiments and subsequently used for our analysis. To identify specifically enriched proteins from this common set of interactors, we compared and plotted the normalized Log2 H:L ratios of each of these proteins from both experiments (Figure 2B). The thresholds are indicated on the graph as red lines (1.009 for experiment 1 and 1.26 for experiment 2). Values less than 1 Log2 unit of the median are more likely to represent either environmental contaminants or nonspecific proteins. As revealed from this analysis, the highly enriched PRMT7 interacting proteins from both experiments are present in the top right quadrant of the scatter plot. As shown in the table (Figure 3A), among the most highly enriched, reproducible protein interactors of PRMT7 from both experiments were ribosomal proteins and translation initiation factors eIF2α, β, and γ. eIF2α along with eIF2β and γ constitutes a ternary complex (eIF2αβγ-GTP-Met-tRNAMet) required for the recruitment of the 40S ribosomal subunit to mRNAs for translation initiation (Jackson et al., 2010). The interaction between PRMT7 and all three eIF2 subunits was validated via coimmunoprecipitation experiments, although the interaction was more robust with eIF2α (Figure 3B), suggesting association with 2β and γ might be indirect.
FIGURE 2:
Analysis of SILAC-based mass spectrometry results. (A) Comparison of the normalized SILAC H:L ratio of identified proteins from the two independent mass spectrometry experiments (L:H ratio for experiment 2). Thresholds for putative protein interactors are indicated by red-dashed lines. (B) Scatter plot of two independent mass spectrometry experiments comparing shared interactor proteins represented as Log2 ratios of the heavy-labeled protein to light-labeled protein. PRMT7 interactors are highlighted in purple within the graph (above red median lines).
FIGURE 3:
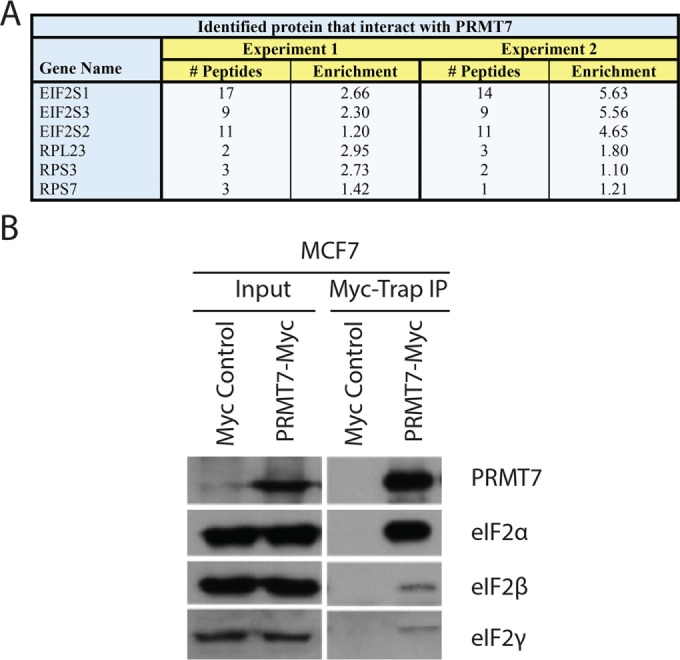
Identified PRMT7 interactors. (A) Strong interactors (high enrichment) of experiments 1 and 2 are represented in a table displaying their relative enrichment and number of identified peptides. (B) Interaction between exogenous PRMT7 and endogenous eIF2α, eIF2β, and eIF2γ was confirmed in MCF7 cells infected with either pLenti-C-MycDDK or pLenti-C-MycDDK-PRMT7 lentiviruses, using affinity pull downs with Myc-Trap beads.
PRMT7 cofractionates with the translational machinery
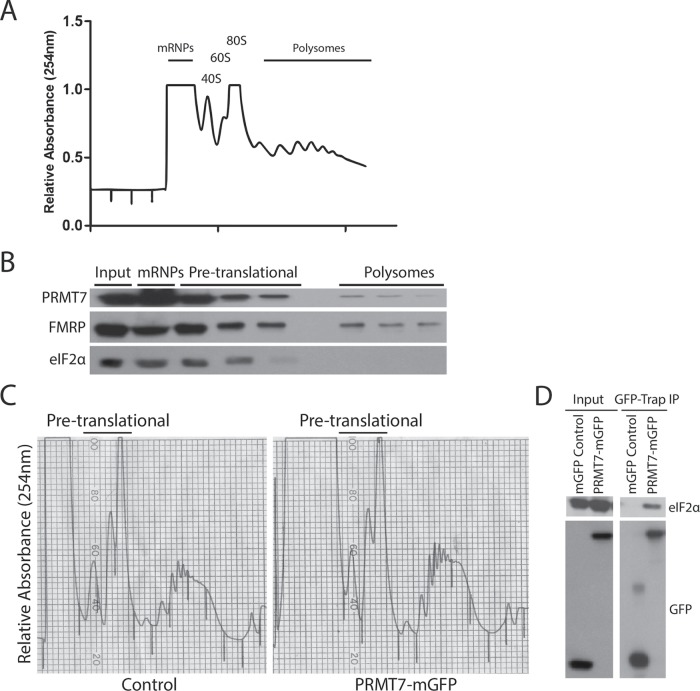
To gain a better understanding into the functional role of the PRMT7-eIF2α interaction biochemically, we explored the presence of the interaction within the translational machinery of cells using polyribosome profiling experiments. For that purpose, cytoplasmic extracts were prepared from HEK293T cells, which are highly metabolically active, and subjected to ultracentrifugation in sucrose density gradients. Because eIF2α regulates translation initiation, its protein expression is highly concentrated within the sucrose fractions containing the 40S and 60S subunits as well as monosomes (hereon referred to as “pretranslational fractions”). Similarly, we observed that PRMT7 cofractionated with eIF2α within the pretranslational fractions. Interestingly, we also observed the presence of PRMT7 within the polysomal fractions, but at a much lower level (Figure 4B). The association of PRMT7 with the translation machinery was also observed in breast cancer cell lines MCF7 and MDA-MB-231 (Supplemental Figure 2). Our mass spectrometry results support these observations as other ribosomal proteins (both small and large subunit proteins), eIF4A, and eEF1A were identified in our screens (Supplemental Table 1). Next, to determine whether PRMT7 and eIF2α interact within the pretranslational fractions, coimmunoprecipitation experiments of pooled pretranslational fractions of MCF7 cells expressing either GFP or PRMT7-mGFP were performed (Figure 4C). Those experiments confirmed that PRMT7 and eIF2α not only cofractionate within the pretranslational fractions but also interact, supporting the idea that PRMT7 may potentially regulate eIF2α’s function in translation (Figure 4D). Using a lentiviral-driven short hairpin RNA (shRNA) expression construct obtained from The RNAi Consortium (Moffat et al., 2006), we efficiently knocked down PRMT7 protein levels in both MCF7 and MDA-MB-231 (Supplemental Figure 3C). Comparing the polysome profiles of these breast cancer cells at 48 h postinfection with either control or PRMT7-targeting shRNAs, no drastic qualitative differences were observed. Nevertheless, quantification of the profiles revealed a statistically significant reduction (36% in MDA-MB-231 and 32% in MCF7 cells) in the ratio of polysomal to pretranslational fractions (Supplemental Figure 3).
FIGURE 4:
Presence of PRMT7 within the translational machinery. (A) Representative polyribosome profile of parental HEK293T cells. Relative absorbance of RNA was read at 254 nm. (B) Polyribosome fractions were run on an SDS–PAGE gel including an input of total cell lysate. Endogenous PRMT7 cofractionates with eIF2α. FMRP was used as a positive control polysome fraction. (C) Pretranslational fractions were pooled from MCF7 cells transiently expressing either mGFP or PRMT7-mGFP. (D) GFP-Trap beads were used to immunoprecipitate control mGFP and PRMT7-mGFP from pooled samples. Inputs were run alongside immunoprecipitates. PRMT7-mGFP interacts with eIF2α in pretranslational fractions.
PRMT7 methylates eIF2α within an RXR motif
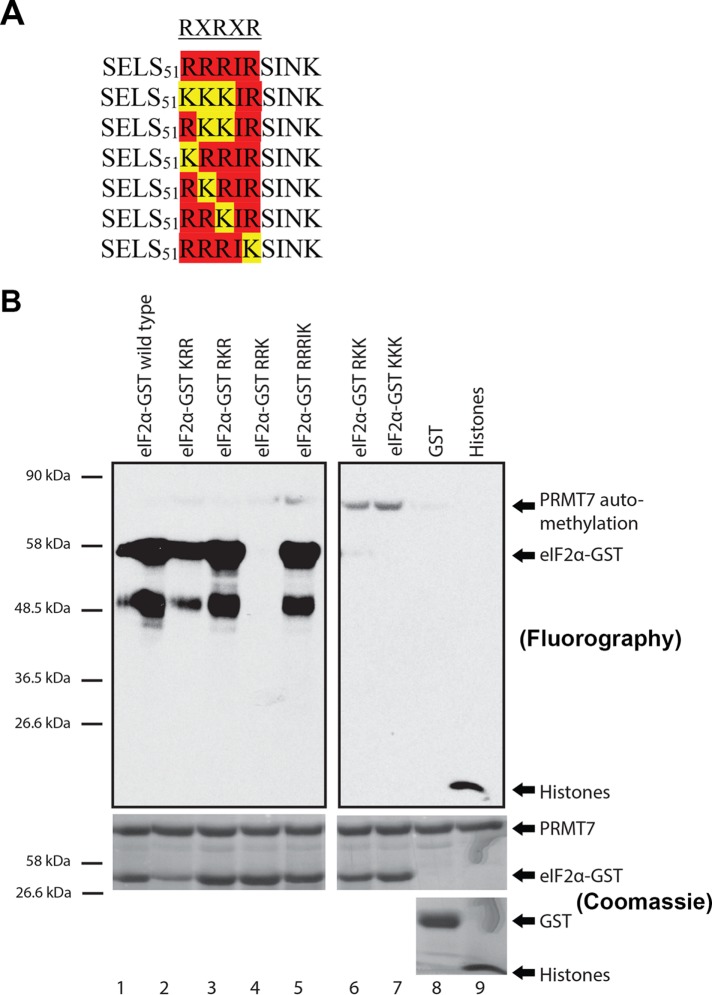
PRMT7 is known to uniquely recognize and methylate an RXR motif (Feng et al., 2013). This motif consists of a highly basic residue (X) flanked by arginine residues (R). Our in silico analysis revealed that human eIF2α contains a double RXR motif, SELS51RRRI55RSINK60, adjacent to the Ser51 residue—a site known for its crucial regulatory role within the cellular stress response upon phosphorylation by HRI, PKR, GCN2, or PERK kinases. Although I55 is not a basic residue that flanks the arginine residues within the potential RXRXR motif, the general sequence is highly basic and may be recognized by PRMT7. Using site-directed mutagenesis, the RXR sequence of eIF2α was mutated to abolish the potential PRMT7 methylation site. The arginine residues were mutated to lysine residues to preserve the positive charge. Several full-length mutants were thus generated: KRR, RKR, RRK, RKK, KKK, and RRRIK (Figure 5A). These full-length eIF2α mutant alleles were then subcloned into an expression vector to purify GST fusion proteins for in vitro methylation assays.
FIGURE 5:
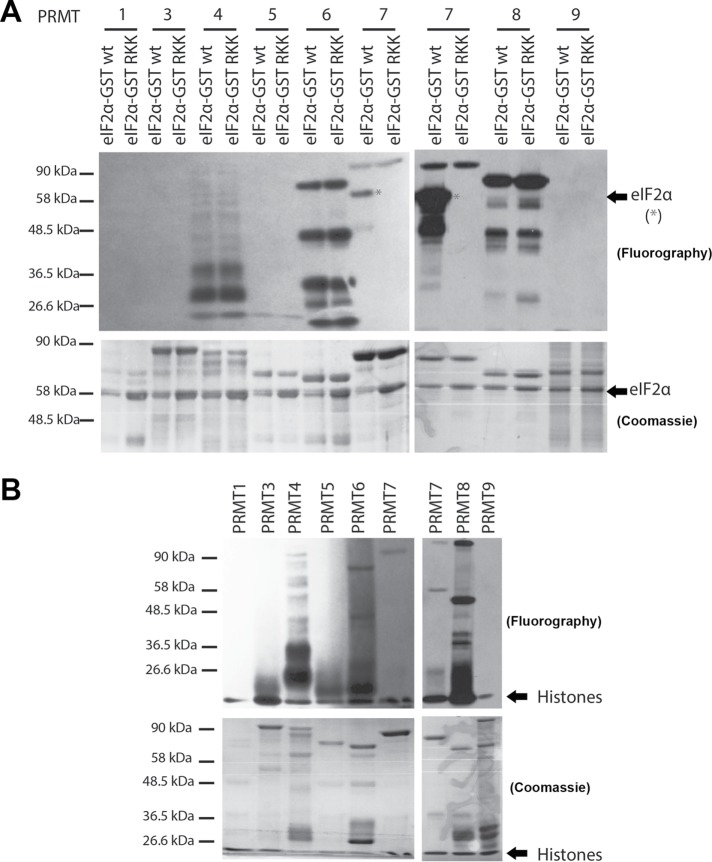
Confirmation that PRMT7 methylates eIF2α in vitro. (A) A motif recognized by PRMT7, RXRXR motif, exists adjacent to the regulatory Ser51 residue of eIF2α. Using site-directed mutagenesis, the arginine residues were mutated into lysine to create six mutants within the RXRXR motif. (B) In vitro methylation assays using 3HSAM as methyl donor, PRMT7 as the enzyme, and eIF2α as a substrate. Experiments revealed that R52R53R54I55R56 sequence motif is methylated by PRMT7—specifically, R54 is critical for its methylation. Methylation of histones was used as a positive control and GST as a negative control. Automethylation of PRMT7 was observed. Methylation assays are revealed through fluorography.
Methylation assays were performed using human PRMT7 purified from insect cells, tritium-labeled SAM (3HSAM), wild-type and mutant eIF2α-GST proteins as potential substrates, GST as a negative control, and histones as a positive control (Feng et al., 2013; Bikkavilli et al., 2014). The autoradiography revealed that wild-type, KRR, RKR, and RRRIK eIF2α-GST were methylated by PRMT7 in vitro because a strong signal was detected (Figure 5B; lanes 1, 2, 3, and 5). As expected of the positive control, methylation of histones was also prominent (lane 9). Absence of a signal was observed for the negative GST control (lane 8), eIF2α RKK mutant (lane 6), KKK mutant (lane 7), and RRK mutant (lane 4). The only signal observed in lanes 4, 6, and 7 was automethylation of PRMT7, which has been shown to be important for its activity (Geng et al., 2017). Hence, within the RXR motif, the arginine residue crucial for recognition and/or methylation was identified to be R54 as methylation was always lost whenever this residue was mutated. Furthermore, methylation assays using purified PRMT1, 3, 4, 5, 6, 8, and 9 were performed with both wild-type and R52KK mutant eIF2α-GST as substrates (Figure 6A) or histone substrates as a positive control (Figure 6B). Other than the methylation of wild-type eIF2α by PRMT7, the only signals observed were automethylation of PRMT4, 6, 7, and 8, which have been previously observed (Dillon et al., 2013; Singhroy et al., 2013; Wang et al., 2013; Feng et al., 2014; Geng et al., 2017). These results suggest that eIF2α is a PRMT7-specific substrate. PRMT2 was excluded from these studies as its activity has been shown to be very weak within cells (Lakowski and Frankel, 2009).
FIGURE 6:
eIF2α is a PRMT7-specific substrate. (A) In vitro methylation assay using 3HSAM as a methyl donor, purified PRMT 1, 3, 4, 5, 6, 7, 8, and 9 as the enzyme, and purified GST-eIF2α as a substrate (both wild type and RKK mutant). Experiments revealed that eIF2α is a PRMT7-specific substrate (asterisks). Automethylation of PRMTs 4, 6, 7, and 8 was observed. (B) Methylation of histones was used as a positive control. Automethylation was also observed for PRMTs 4, 6, 7, and 8. Methylation assays are revealed through fluorography.
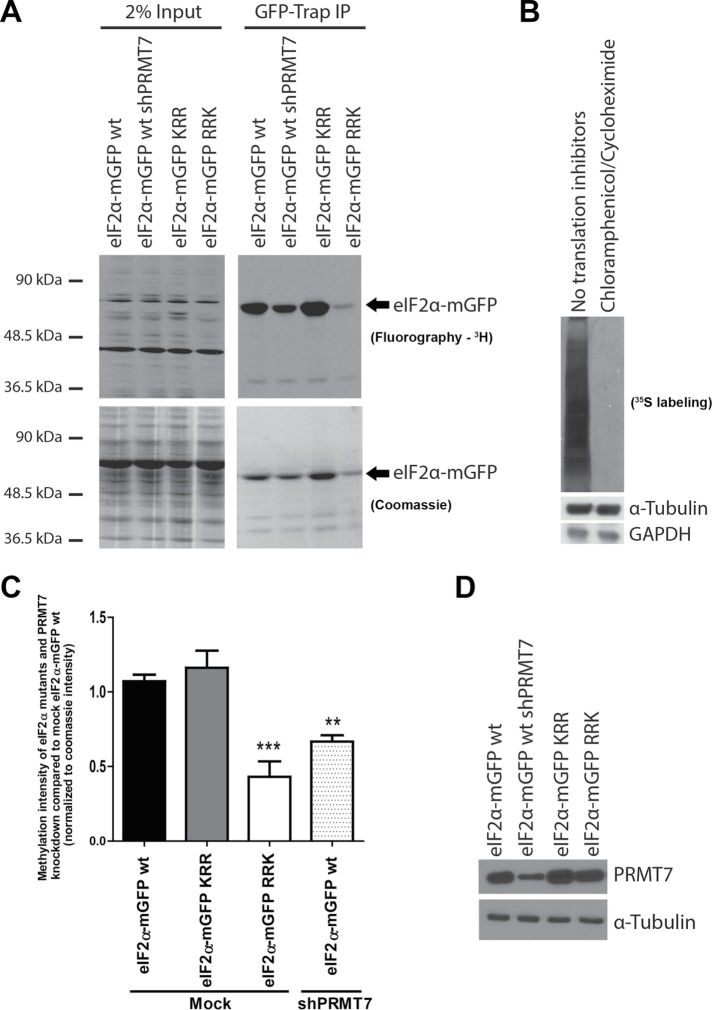
We next sought to determine whether PRMT7 was capable of methylating eIF2α in vivo. MDA-MB-231 were transduced with either wild-type eIF2α-mGFP or KRR and RRK mutants and were used for methylation assays in the presence of translation inhibitors (cycloheximide and chloramphenicol) to ensure that labeling was due to posttranslational methylation and not via direct incorporation of 3H methionine through protein synthesis (Figure 7A). To confirm that translation was indeed inhibited, metabolic labeling assays in MDA-MB-231 cells using 35S incorporation were employed with or without the use of translation inhibitors. As expected, no incorporation of 35S was observed in cells treated with the inhibitors (Figure 7B). The RRK mutant was used for these experiments because a loss of methylation was observed in vitro; KRR was used as a negative control as no changes were observed in vitro. MDA-MB-231 cells expressing wild-type eIF2α-mGFP with a knockdown in PRMT7 was also used to determine whether PRMT7 is indeed the enzyme responsible for the methylation of eIF2α in vivo. Apparent by the decrease in 3H signal, we observed a significant decrease in methylation of wild-type eIF2α-mGFP upon PRMT7 knockdown as well as for the RRK mutant when compared with wild-type eIF2α-mGFP. However, no changes in methylation signal were observed in the KRR mutant compared with wild-type eIF2α-mGFP (Figure 7A). We consistently observed lower levels of the RRK mutant in cells compared with other eIF2α alleles at the same time postinfection, suggesting this mutation might result in destabilization of the protein (see Discussion). Nevertheless, quantification of three independent experiments and normalization of eIF2α protein amounts present in the GFP-Trap pull down confirmed drastically reduced methylation for this mutant (Figure 7C), as was observed in vitro. We additionally generated knockout PRMT7 MDA-MB-231 cell lines using CRISPR/Cas9 to use for the in vivo methylation assays. Unfortunately, complete depletion of PRMT7 resulted in cell death, especially in combination with stress stimuli, preventing us from using the cells for subsequent biochemical and functional assays (unpublished data). These findings demonstrate that PRMT7 methylates eIF2α both in vitro and in vivo and that R54 within the RXR motif is important for its methylation.
FIGURE 7:
PRMT7 methylates eIF2α in vivo. (A) In vivo methylation assay using 3H methionine in MDA-MB-231 cell lines transiently expressing wild-type eIF2α-mGFP (in the presence of control or PRMT7-targeting shRNAs), KRR, or RRK mutant alleles. (B) Representative 35S metabolic labeling demonstrating that no incorporation of isotopic amino acid is observed following treatment with translation inhibitors. (C) Quantification of the relative methylation status of eIF2α normalized to the wild-type eIF2α-mGFP mock conditions, as well as to the amount of eIF2-mGFP detected on the Coomassie stain. A significant decrease in methylation of eIF2α was observed upon PRMT7 knockdown and for the RRK mutant. No change was observed for the KRR mutant; data are presented as mean ± SEM for n = 5; **, p = 0.002; ***, p = 0.001 (ANOVA). (D) Representative Western blot depicting the efficiency of the knockdown in cells infected with the PRMT7-targeting shRNA.
Interplay between eIF2α phosphorylation and methylation
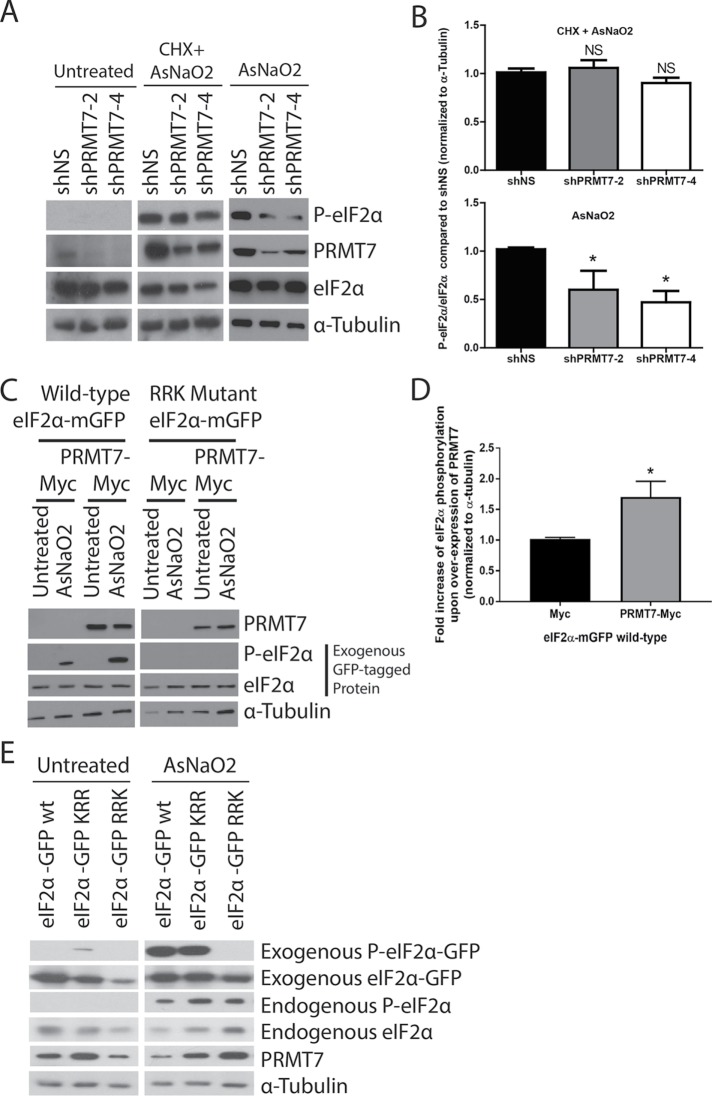
When cells are under environmental stresses such as oxidative stress, heat shock, osmotic stress, UV radiation, and/or viral infection, stress-induced kinases phosphorylate eIF2α at Ser51 leading to inhibition of bulk translation initiation and subsequent disassembly of polysomes resulting in stress granule formation (Buchan and Parker, 2009). Because the site of methylation of eIF2α was found to be adjacent to the Ser51 residue, we anticipated a potential interplay between eIF2α phosphorylation and methylation. We first looked at the effect on Ser51 phosphorylation upon knockdown of PRMT7. Using the widely used oxidative stressor, sodium arsenite (AsNaO2), we observed a significant decrease in eIF2α phosphorylation upon PRMT7 knockdown with two distinct shRNA sequences, which was apparent via Western blotting (Figure 8A) and confirmed by quantification (Figure 8B). However, we did not observe similar results when using a translation inhibitor, cycloheximide (Figure 8, A and B). In the context of a reciprocal experiment, an increase in wild-type exogenous eIF2α-mGFP phosphorylation was observed when overexpressing PRMT7 under stressed conditions (Figure 8, C and D). Importantly, an eIF2α mutant (RRK) that cannot get methylated by PRMT7 did not get phosphorylated at Ser51 upon stress, and consequently, overexpression of PRMT7 failed to induce Ser51 phosphorylation. Finally, we looked at the phosphorylation status of the wild-type, KRR, and RRK eIF2α-mGFP mutants upon exposure to oxidative stress (sodium arsenite). As shown in the Western blots, only wild-type and KRR eIF2α-mGFP were phosphorylated upon cellular stress (Figure 8E). Taken together, these results indicate that arginine methylation of eIF2α by PRMT7 (likely at R54) can influence the stress-induced phosphorylation status at Ser51.
FIGURE 8:
Interplay between eIF2α methylation and phosphorylation. Phosphorylation of eIF2α decreased upon transient knockdown of PRMT7 in MDA-MB-231 cells as shown in the (A) Western blot and (B) quantification (data are presented as mean ± SEM for n = 5; **, p = 0.01, ANOVA). This effect is not seen when experiments were performed in the presence of cycloheximide (CHX). (C) Representative Western blot depicts an increase in eIF2α phosphorylation upon transient overexpression of PRMT7-Myc in MDA-MB-231 cells and (D) its quantification (data are presented as mean ± SEM for n = 5; *, p = 0.05; two-tailed t test). (E) Western blots depicting phosphorylation of transiently expressed wild-type and KRR mutant eIF2α upon treatment with sodium arsenite for 30 min at 500 µM in MDA-MB-231 cells. Phosphorylation is absent in the RRK mutant.
We further looked at endogenous monomethylation levels of eIF2α under stressed conditions. For these experiments, we immunoprecipitated GFP and eIF2α-mGFP from untreated and AsNaO2-treated (500 µM for 30 min) HEK293T cells and subsequently probed for monomethylation using a previously validated antibody specific for arginine monomethylation (Guo et al., 2014). A significant increase in eIF2α monomethylation, correlating with high levels of Ser51 phosphorylation, was observed in AsNaO2-treated cells compared with untreated cells (Figure 9). Furthermore, in AsNaO2-treated cells, where high levels of eIF2α monomethylation and Ser51 phosphorylation were detected, the association between PRMT7 and eIF2α was drastically reduced. These findings suggest that PRMT7 actively methylates eIF2α under stressed conditions, leading to an increase in monomethylated and Ser51 phosphorylated eIF2α, which in turn seems to trigger release of PRMT7.
FIGURE 9:
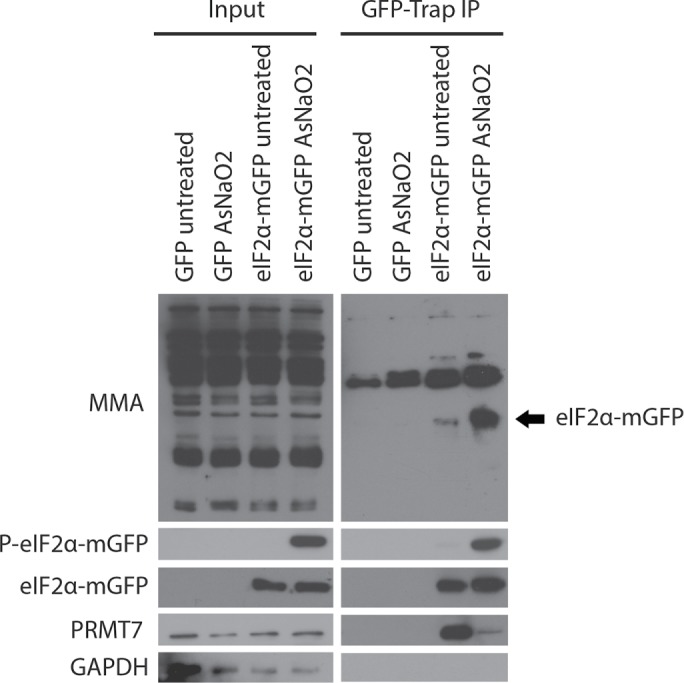
PRMT7 methylates eIF2α under stressed conditions. Transiently expressed eIF2α-mGFP and GFP (negative control) were immunoprecipitated from untreated and AsNaO2-treated (500 µM for 30 min) HEK293T cells, followed by immunoblotting using a monomethylation-specific antibody. Western blot depicts a significant increase in eIF2α monomethylation within AsNaO2-treated cells. Furthermore, immunoblotting of the same membrane for PRMT7 reveals a drastic reduction in the interaction of PRMT7 with eIF2α-mGFP under stress conditions.
PRMT7 activity is induced as part of the stress response
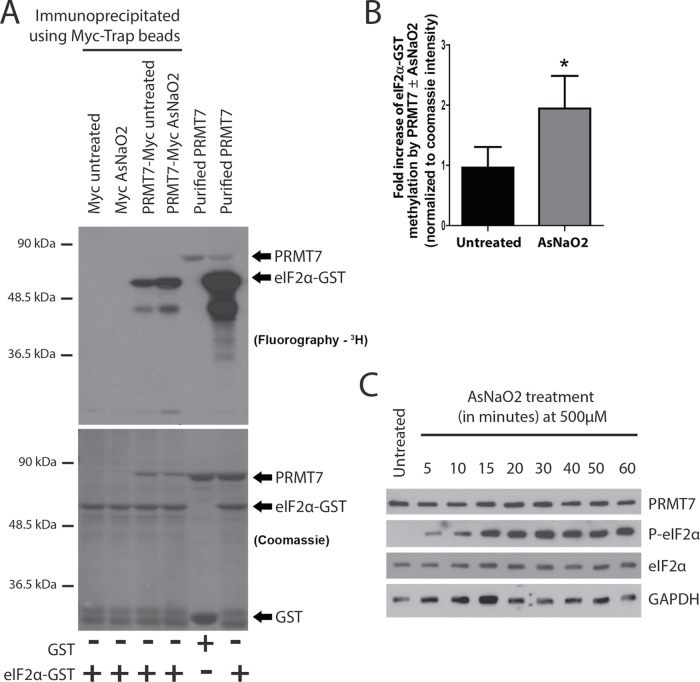
These results suggest that PRMT7 may somehow get induced following stress. To test this idea, an in vitro methylation assay was performed using immunoprecipitated PRMT7-Myc (or Myc as a negative control) from untreated and AsNaO2-treated MDA-MB-231 cells as the enzyme for the reaction and wild-type eIF2α-GST as a substrate. Purified PRMT7 with either GST or eIF2α-GST as substrates were utilized as negative and positive controls, respectively, for the methylation assays. Strikingly, PRMT7-Myc immunoprecipitated from AsNaO2 cells more actively methylated eIF2α-GST by approximately twofold (Figure 10, A and B). Importantly, these changes were not due to alterations in PRMT7 expression upon exposure to AsNaO2 (Figure 10C). To ensure that our observations were not specific to AsNaO2 treatment, we also repeated some of those experiments in the context of ER stress induced by thapsigargin. As with AsNaO2, knockdown of PRMT7 led to reduced levels of eIF2α Ser51 phosphorylation upon thapsigargin treatment (Supplemental Figure 4A). An induction of PRMT7 activity, albeit less dramatic, was also observed following thapsigargin treatment (Supplemental Figure 4, B and C). Intriguingly, however, the ER stress-dependent induction of ATF4 was not affected upon PRMT7 knockdown (Supplemental Figure 4A). Altogether, these findings suggest that PRMT7 activity is stimulated upon stress, potentially to regulate eIF2α phosphorylation, and may thus represent a novel player in the cellular stress response pathways.
FIGURE 10:
PRMT7 activity toward eIF2α is increased under stressed conditions. (A) PRMT7-Myc was transiently expressed in MDA-MB-231 cells and then affinity-purified using Myc-Trap beads. Affinity-purified PRMT7-Myc, from Mock- or AsNaO2-treated (500 µM for 30 min) was then used as a source of enzyme to perform in vitro methylation assays using 3HSAM as methyl-donor wild-type eIF2α-GST as a substrate. Cells transfected with a Myc empty vector were used as a negative control. As a positive control, purified PRMT7 with either GST or wild-type eIF2α-GST was used. (B) Quantification of the methylation status of eIF2α was calculated for the AsNaO2-treated condition and normalized to the amount of PRMT7-Myc detected in the Coomassie stain. A significant increase in methylation of eIF2α was observed in AsNaO2-treated cells; data are presented as mean ± SEM for n = 5; *, p = 0.02; two-tailed t test. (C) Western blot depicting unchanged endogenous PRMT7 expression AsNaO2 treatment (500 µM for the indicated times) in parental MDA-MB-231 cells.
PRMT7 regulates eIF2α-dependent stress granule formation
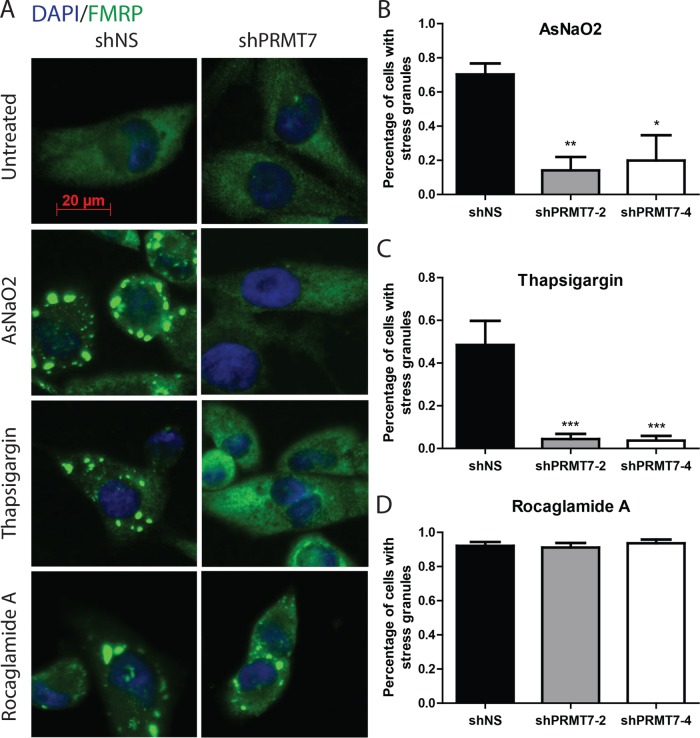
Stress-induced eIF2α Ser51 phosphorylation blocks translation initiation, reduces the formation of polysomes, and ultimately leads to the formation of so-called stress granules. These cytoplasmic foci are nonmembranous ribonucleoprotein structures consisting of nontranslating mRNAs, translation initiation factors, the 40S ribosomal subunit, RNA-binding proteins, and signaling molecules such as proapoptotic proteins (Balagopal and Parker, 2009; Farny et al., 2009; Fournier et al., 2010). To determine whether PRMT7 affects stress granule formation, we compared MDA-MB-231 cells expressing either control or PRMT7-targeting shRNAs, treated with two eIF2α-dependent stressors (AsNaO2 and thapsigargin), or with an eIF2α-independent stressor (rocaglamide A; Sadlish et al., 2013; Kedersha et al., 2016; Aulas et al., 2017). We then evaluated the percentage of cells harboring stress granules (number of cells with stress granules/total number of cells per field image). Strikingly, we observed a 50–90% reduction in the percentage of cells exhibiting stress granules upon knock down of PRMT7 compared with control MDA-MB-231 cells when using eIF2α-dependent stressors (Figure 11, A–C, and Supplemental Figure 5). In contrast, PRMT7 knockdown had no effect on stress granule formation when cells were treated with the eIF2α-independent stressor rocaglamide A (Figure 11, A and D). Altogether, these findings demonstrate that PRMT7 is required for the proper formation of eIF2α-dependent stress granules most likely through its ability to methylate eIF2α and subsequently regulate Ser51 phosphorylation.
FIGURE 11:
Stress granule formation is abrogated upon PRMT7 knockdown. (A) Representative immunofluorescent images of MDA-MB-231 cells depicting decreased stress granule formation upon transient knockdown of PRMT7 when exposed to AsNaO2 (500 µM for 30 min) and thapsigargin (10 µM for 2 h). No change was observed when cells were treated with rocaglamide A (2 µM for 2 h). FMRP (green) was used as a stress granule marker (scale bar: 20 µm). (B) Quantitation of “percentage of cells with at least five stress granules” for n = 5 independent experiments in triplicate when treated with AsNaO2; data are presented as mean ± SEM; *, p = 0.04; **, p = 0.005 (ANOVA). (C) Similar quantitation was performed when treating with thapsigargin; ***, p = 0.0001 (ANOVA), and (D) rocaglamide A (not significant).
DISCUSSION
PRMTs have been emerging as crucial regulatory enzymes in various cellular pathways because aberrant expression of these enzymes has been correlated with aggressive human diseases such as cancers, cardiovascular diseases, and neurodegenerative diseases (Bedford and Clarke, 2009; Yang and Bedford, 2013; Blanc and Richard, 2017). PRMT7, the only type III PRMT, has been shown to promote breast cancer aggressiveness and metastasis (Thomassen et al., 2009; Yao et al., 2014; Baldwin et al., 2015). However, PRMT7 is still not well characterized functionally; thus, further studies are necessary to better elucidate its role. The objective of the current study was to gain insights into the biological functions of PRMT7 by identifying its protein interactors and potential substrates. Hence, we performed SILAC-based quantitative mass spectrometry. We identified and validated many protein interactors of PRMT7 including the eukaryotic translation initiation factor eIF2α. Specifically, we demonstrated that PRMT7 can methylate eIF2α and regulate its stress-induced phosphorylation at Ser51. We have also shown that PRMT7 is required for eIF2α-dependent stress granule formation, thus altogether identifying PRMT7 as a novel key player in the cellular response to stress.
eIF2α is a novel substrate of PRMT7
Our SILAC quantitative proteomic experiments have identified the eIF2 complex as specific interactors of PRMT7. While we were able to validate the interaction of PRMT7 with all three subunits using coimmunoprecipitation experiments, the association with eIF2α was significantly more robust than with the β and γ subunits (see Figure 3B). We reason that this may reflect the fact that eIF2α is a substrate for PRMT7 and thus, potentially interacts directly, in its free monomeric form as well as within the ternary complex, with the enzyme; the β and γ subunits being indirect interactions in this context. Using mutagenesis, we have pinpointed the PRMT7 methylation site within eIF2α to be within a duplicated RXR motif (SELS51RRRIRSINK60) adjacent to the Ser51 residue, a site important for eIF2α’s role in the cellular stress response. We noticed in those experiments that the nonmethylated RRK eIF2α mutant was found expressed at consistently lower levels compared with other alleles at the same time postinfection, suggesting that preventing methylation by PRMT7 may result in an unstable protein. We wondered whether this apparent instability might be due to the inability of the unmethylatable RRK mutant to incorporate in the eIF2 ternary complex. To test this possibility, we performed GFP–pull-down experiments using either wild-type, KRR, or RRK mutants, but no difference in the ability to interact with the eIF2β or γ subunits was seen between the different alleles (Supplemental Figure 6), suggesting that methylation does not regulate assembly of the ternary complex and a distinct mechanism is likely responsible for the apparent instability of the RRK mutant. As previously mentioned, PRMT7 is a type III PRMT capable of forming MMA final products, whereas other PRMTs are involved in dimethylation. Generally, it is presumed that the MMA product is a transient modification that is subsequently dimethylated by either the same PRMT or another family member, suggesting that the MMA mark acts as a priming posttranslational modification (PTM) (Yang and Bedford, 2013; Jain et al., 2017). In our in vitro methylation assays, we noted that the only PRMT capable of methylating purified eIF2α was PRMT7. Consistent with this result, mining the supplementary data from a high-throughput LC-MS/MS study revealed eIF2α to be monomethylated and not asymmetrically dimethylated in mouse brain and embryo (Guo et al., 2014). However, symmetric dimethylation of arginine residues was not explored. Similarly, this was not explored in our study and we cannot rule out that once monomethylated by PRMT7, eIF2α could become a substrate for other PRMTs. Furthermore, the Guo et al. mass spectrometry study identified the R54 site as monomethylated in mice, but also the R53 position, within the same RXR motif described here, which is in fact conserved from yeast, fruit fly, mouse to humans (Hendrick et al., 2016). However, these methylation sites were not identified in their screen using mammalian colorectal carcinoma cells (Guo et al., 2014), suggesting that this methylation event might not be constitutive. This also raises the question as to whether MMA can behave as a final product under certain conditions and/or species.
Notably, in our in vivo methylation assays, we observed a decrease in methylation status of eIF2α by knocking down PRMT7. These findings suggest that PRMT7 is responsible for a major methylation site on eIF2α. A more drastic decrease in methylation status was observed in the RRK mutant compared with the knockdown cell line suggesting that there is residual PRMT7 still capable of methylating eIF2α in the knockdown cell line. The use of a CRISPR-Cas9 knockout cell line was not possible because the addition of translation inhibitors, as required by the protocol, resulted in massive cell stress and death in the complete absence of PRMT7. Nevertheless, an incomplete loss of methylation in the RRK mutant suggests that there are potentially other sites of methylation within eIF2α to be further identified. Moreover, methylation by other PRMTs through scavenging could occur. Even though we have shown that the other PRMTs do not methylate eIF2α in vitro, this does not eliminate the possibility of methylation occurring in vivo. Thus, future experiments should include looking into whether eIF2α is a shared substrate in vivo among the PRMT family of enzymes and whether substrate scavenging occurs with PRMT7.
Looking more closely at the RXR motif, methylation and phosphorylation of eIF2α was lost when R54 was mutated to a lysine residue (RRK), abolishing the RXR sequence motif. However, introduction of the KRR and RRRIK mutants had no impact on methylation of eIF2α. One proposition is that within the duplicate RXR motif, SELS51RRRIRSINK60, only R54 is important for recognition because if mutated, both RXR motifs are lost. For instance, the KRR mutant still maintains an RXR motif: KRRIR as does the RRRIK mutant. Conversely, the RRK mutant no longer contains any RXR motif: RRKIR. Moreover, as the KRRIR mutant is still methylated, this suggests that neutral residues may still be recognized within an RXR motif by PRMT7 if the general region is extremely basic. We also cannot rule out methylation at R52 and R56, as mutation of R54 could prevent modification at these sites due to loss of the RXR motif. In fact, evidence was previously provided for methylation by PRMT7 of all three arginine residues in the RKRSR sequence of Histone H2B (Feng et al., 2013).
Interplay between eIF2α arginine methylation and serine phosphorylation
As there are many instances where PTMs can be either mutually exclusive or synergistic, we wanted to determine whether phosphorylation of eIF2α at Ser51 was affected by nearby methylation by PRMT7. First, we observed that the absence of PRMT7 when cells were under oxidative stress resulted in decreased eIF2α phosphorylated at Ser51, whereas overexpression of PRMT7 resulted in an increase in phospho-Ser51 eIF2α. Positive cross-talk between arginine methylation and serine phosphorylation has been observed previously for other PRMTs. For instance, methylation of apoptosis signal-regulating kinase 1 (ASK1) by PRMT5 at R89 enhances the ASK1-Akt interaction resulting in phosphorylation of ASK1 at S83 to inhibit endothelial cell apoptosis (Chen et al., 2016). Furthermore, a mutant allele of eIF2α (R54-K) that does not get methylated was incapable of being phosphorylated upon exposure to oxidative stress, suggesting that methylation may actually be a prerequisite for stress-induced phosphorylation of Ser51 to take place. However, when experiments were performed in the presence of the translation elongation inhibitor cycloheximide, eIF2α phosphorylation at Ser51 was still observed upon PRMT7 knockdown (see Figure 8A). Thus, it is possible that the change in amino acid (from arginine to lysine) at position 54 may have contributed to the effect observed on phosphorylation. This result also implies that methylation of eIF2α by PRMT7 may regulate the phosphorylation status of eIF2α at a later stage during the stress response. For example, methylation of eIF2α could positively impact its recognition by GADD43 to promote dephosphorylation of eIF2α (Rojas et al., 2015).
PRMT7 activity is induced as part of the cellular response to stress
We have shown that there is an increase in eIF2α methylation upon cellular stress, correlating with an increase in PRMT7 activity under stressed conditions. These results suggest that eIF2α is not constitutively methylated and that methylation is induced as part of the cellular response to stress. We have observed this phenomenon in response to oxidative stress (sodium arsenite) as well as ER stress (thapsigargin), but it will be important to confirm whether this can also be generalized to other cellular stresses. Further experimentation is also required in order to determine the precise mechanism(s) by which PRMT7 activity is stimulated under stress conditions.
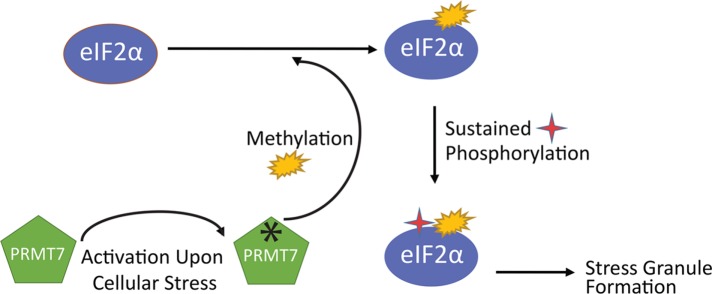
A well-known feature of PRMTs is that they tend to interact less strongly with their substrates once methylated. Consistent with this notion, we have observed that the association between PRMT7 and eIF2α is greatly reduced following stress (see Figure 9). Altogether, our results support a novel role for PRMT7, through its methylation of eIF2α, as an integral player in the cellular stress response regulatory pathway(s) (see model in Figure 12).
FIGURE 12:
Working model depicting how PRMT7 participates in the eIF2α-dependent cellular response to stress. Upon exposure to stress, such as oxidative or ER stress, PRMT7 activity is stimulated, which leads to increased methylation of eIF2α within the RXR motif. Methylation of eIF2α by PRMT7 then promotes stable phosphorylation levels of eIF2α at Ser51, ultimately inducing stress granule formation.
Novel role for PRMT7 in eIF2α-dependent stress granule formation
Because PRMT7 did not seem to affect translation as noted with unchanged polyribosomal profiles upon knockdown of PRMT7, we further explored the role of PRMT7 in a downstream eIF2α readout: stress granule formation. Interestingly, we observed a drastic decrease in stress granule formation within cells treated with stressors that are known to induce stress granules through an eIF2α-dependent mechanism. Importantly, no impact on stress granules was observed upon PRMT7 knockdown when an eIF2α-independent stressor (rocaglamide A) was used. Thus, these results support a direct role for PRMT7, likely through its methylation of eIF2α, in promoting stress granule formation. These observations are also consistent with our results showing that no effect on eIF2α phosphorylation upon PRMT7 knockdown was seen when experiments were performed in the presence of cycloheximide (Figure 8A), which is known to prevent induction of stress granules.
Implications of the eIF2α/PRMT7 interaction in breast cancer
eIF2α protein levels are up-regulated in malignant melanocytic and colonic epithelial neoplasms, thyroid carcinomas, and non-Hodgkin’s lymphomas (Wang et al., 1999, 2001; Rosenwald et al., 2003). Phosphorylated eIF2α is similarly up-regulated in triple negative breast cancer and could represent a novel prognostic factor (Guo et al., 2017). Furthermore, P-eIF2α is implicated in promoting cell survival of tumor cells when under stressed conditions (Rajesh et al., 2015; Tenkerian et al., 2015). Phosphorylation of eIF2α at Ser51 by the stress-activated eIF2α kinases also leads to selective translation of mRNAs harboring upstream open reading frames such as ATF4 (Harding et al., 2003). Intriguingly, we have not seen any impact on ATF4 induction following ER stress upon PRMT7 knockdown (see Supplemental Figure 4A). Because this role of eIF2α is at the level of translation initiation, it is conceivable that transient phosphorylation at Ser51 is sufficient to promote this activity whereas a more sustained phosphorylation level may be required to support formation of stress granules. It is also possible that such alterations were not observed as residual eIF2α phosphorylation in our PRMT7-knockdown lines may be sufficient to promote ATF4 induction.
Stress granules have been shown to exist in breast tumors (potentially induced by the hypoxic core microenvironment), promoting cell survival (Moeller and Dewhirst, 2006; Baguet et al., 2007). Furthermore, several chemotherapeutic drugs (bortezomib, 5-fluorouracil, 6-thioguanine, and 5-azacytidine) have been shown to induce stress granule formation thereby resulting in the cells resisting drug-induced apoptosis (Fournier et al., 2010; Kaehler et al., 2014; Anderson et al., 2015). PRMT7 has also been implicated in breast cancer. For example, the chromosomal region of PRMT7 has been shown in a gene expression meta-analysis study to be positively correlated with breast cancer aggression and metastasis (Thomassen et al., 2009). Moreover, as previously mentioned, PRMT7 promotes breast cancer cell invasion, metastasis, and EMT (Yao et al., 2014; Baldwin et al., 2015; Geng et al., 2017). Interestingly, PRMT7 was originally identified in a screen of genetic suppressor elements to discover genes conferring susceptibility to chemotherapeutic cytotoxicity in hamster cells (Gros et al., 2003). Because it is currently unknown, mechanistically, how PRMT7 confers resistance toward chemotherapeutic drugs, it is possible that PRMT7 may promote resistance, at least in part, via its role in eIF2α-dependent stress granule formation. Thus, targeting PRMT7 in breast cancer via the use of small molecule inhibitors to prevent its methyltransferase activity could prove to be a beneficial therapeutic approach.
MATERIALS AND METHODS
Cell lines
MCF7, MDA-MB-231, and HEK293T cells were purchased from the American Type Culture Collection. The cells were cultured in complete DMEM supplemented with 10% fetal bovine serum (FBS), 1% sodium pyruvate, 1% 50 IU/ml penicillin, and 50 mg/ml streptomycin. MCF7 cells were additionally supplemented with 2.75 µg/ml insulin. Mycoplasma testing was performed monthly via immunostaining against 4′,6-diamidino-2-phenylindole (DAPI). Three different types of cell lines were used throughout the paper to ensure the observed effects could be broadly applicable and not limited to one specific cell line.
Antibodies
PRMT7 (#14762S), eIF2α (#5324S), P- eIF2α (#3597S), ATF4 (#11815), and monomethyl-arginine (#8015S) antibodies were purchased from Cell Signaling Technology; FMRP antibody (#MAB2160) and puromycin (#MABE343) were purchased from EMD Millipore; GFP antibody (#sc-8334) and TIAR antibody (#sc-1749) were purchased from Santa Cruz Biotechnology; eIF2β (#A301-743A) and eIF4G (#A301-7741) were purchased from Bethyl Laboratories; eIF2γ antibody (#11162-1-AP) was purchased from Proteintech; and α-tubulin (#T6199) was purchased from Sigma Aldrich. All antibodies were used at a 1:1000 dilution overnight at 4°C.
Plasmids
To overexpress either C-terminal Myc-tagged or GFP-tagged PRMT7, pLenti-C-MycDDK or pLenti-C-mGFP, respectively, were purchased from Origene. A control vector expressing only GFP or Myc was created via blunt-end digestion and subsequent ligation of the pLenti-C-mGFP-PRMT7 and pLenti-C-MycDDK-PRMT7 vectors, respectively (characterized in Baldwin et al., 2015). Mutant eIF2α-GST protein from pGEX-4T2 vector was designed: R52R53R54 mutated to KKK, RKK, KRR, RKR, and RRK. Primers were designed using QuickChange Primer Design by Agilent Technologies. These constructs were then subcloned into the pLenti-C-mGFP backbone as described above. To knock down PRMT7, RNA interference was performed using pLKO.1 vectors obtained from The RNAi Consortium containing either an shRNA with a luciferase sequence for control (5′-CAAATCACAGAATCGTCGTAT-3′) or two independent PRMT7 sequences (5′-GCTAACCACTTGGAAGATAAA-3′ and 5′-CGATGACTACTGCGTATGGTA-3′).
Production of lentivirus and cell transduction
HEK293T cells were transiently transfected with packaging plasmids, pMD2.G and psPAX2 along with the expression plasmid. After 48 h incubation, lentivirus was harvested from the media by filtering it through a 0.45 μm syringe filter. Media containing virus with 8 µg/ml polybrene was placed on target cells (seeded at 250,000 per 10-cm plate 24 h before) and allowed to infect for 48 h. Media was then changed and target cells were used for experimentation at the 48 h time point postinfection. For SILAC mass spectrometry experiments, infected cells were maintained in culture for more than 1 month to ensure stable but low levels of expression (comparable to endogenous levels). No selection was used because no selectable markers are present within the pLenti-C-mGFP vectors used for those experiments.
Fluorescence microscopy
MCF7 cells stably expressing either mGFP or PRMT7-mGFP cells were grown over glass coverslips in complete DMEM at 250,000 cells per six-well plate and incubated at 5% carbon dioxide atmosphere at 37°C overnight to allow cells to adhere. Cells were then washed three times with 1X PBS (phosphate-buffered saline), fixed with 4% paraformaldehyde in 1X PBS for 10 min, and washed again three times with 1X PBS. Coverslips were mounted onto glass slides using Vectashield mounting media with DAPI purchased from Vector Laboratories and allowed to dry overnight. Fluorescent images were taken at 40× magnification using a Zeiss LSM 5 Pascal/Axiovert 200 epifluorescence microscope.
SILAC-based affinity purification and mass spectrometry
MCF7 cells stably expressing either mGFP or PRMT-mGFP were grown in either light media (R0K0; 12C6-arginine/12C6-lysine; Sigma) or heavy media (R6K4; 13C6-arginine/4,4,5,5-D4-lysine; Cambridge Isotope Laboratories) supplemented with dialyzed fetal bovine serum, penicillin/streptomycin, and insulin. After at least 5–10 cell doublings in the labeling media, cells were trypsinized and pellets were flash-frozen in liquid nitrogen. Pellets were lysed using freshly prepared lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, and protease inhibitors). For complete cell lysis, cell extracts were sonicated on ice (6 × 10 s at full power). Protein concentrations were obtained using the Bradford assay. Equal amounts of total cell extract were used for each condition and were precleared with protein A/G agarose beads for 1 h at 4°C while nutating. Cleared extracts were then incubated with GFP-Trap_A agarose beads (Chromotek) and nutated for 1 h at 4°C. Beads were then washed with lysis buffer and the differentially labeled samples combined. After several more washes, bound proteins were eluted with 1% SDS, reduced using DTT (dithiothreitol), and alkylated using iodoacetamide. The eluate was separated on an SDS–PAGE gel, stained with Simply Blue Safestain (Thermo-Fisher/Invitrogen), and cut into five equal fragments. Each fragment was further cut into small pieces for in-gel digestion using Trypsin Gold (Promega) as previously described (refer to Trinkle-Mulcahy et al., 2008; Larivière et al., 2014). An aliquot of each tryptic digest was analyzed by LC-MS/MS on an LTQ Orbitrap XL hybrid MS with nanospray source (Thermo Scientific) coupled to an UltiMate 3000 RSLC nano HPLC (Dionex). Statistical and bioinformatic analyses were performed using MaxQuant software v1.2.7.4. For a more detailed protocol, refer to Trinkle-Mulcahy et al. (2008) and Larivière et al. (2014).
Coimmunoprecipitation
Coimmunoprecipitation experiments were carried out in a similar manner to that described above. Briefly, HEK293T, MCF7, or MDA-MB-231 cells were lysed in freshly prepared lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, and protease inhibitors). Lysate was precleared with protein A/G agarose beads for 1 h at 4°C with end-to-end rotation. Cleared extract was then nutated at 4°C with GFP-Trap_A beads (Chromotek) for 1 h at 4°C. Beads were then washed with lysis buffer. Proteins were eluted using 1% SDS and samples were prepared to run on an SDS–PAGE gel. Basic Western blotting was performed and antibodies mentioned above were used to identify protein.
Polyribosome profiling
HEK293T, MCF7, or MDA-MB-231 cells were seeded at 80% confluency for experiments. Cells were lysed on ice in freshly prepared buffer (20 mM Tris-HCl, 150 mM NaCl, 1.25 mM MgCl2, 1% NP-40, 1 mM DTT, 8 U/ml RNase Out, and protease inhibitors, pH 7.4). Lysates were centrifuged and supernatant quantified for RNA content at 260 nm using a NanoDrop 2000 spectrophotometer (Thermo Scientific). Supernatants were loaded onto a 15–50% sucrose gradient, and ultracentrifugation was performed using an Optima L-90K ultracentrifuge and SW 41Ti rotor (Beckman) at 234,326 × g for 2.5 h at 4°C. Samples were then fractionated using a Foxy R1 Fraction Collector (Brandel) and UA-6 Absorbance Detector (ISCO). For analysis, proteins were precipitated from fractions using chloroform/methanol and dried in vacuo. Pellets were resuspended in water and analyzed by Western blotting. For coimmunoprecipitation using pooled pretranslational fractions, samples were diluted 1:5 in lysis buffer and the aforementioned coimmunoprecipitation protocol was followed. For quantification of polysomal to pretranslational fractions, profiles were first traced above background using Adobe Illustrator. For comparison, pixels for each section were quantified via histogram densitometry (Adobe Photoshop).
Protein purification
PRMT7 was purified as previously described (Feng et al., 2013). Purified PRMT5 was a generous gift from Derrick Gibbings (University of Ottawa). GST fusion proteins (PRMT1, 3, 4, 6, 8, 9, and both wild-type and mutant eIF2α) were expressed using pGEX-4T2 vectors and purified from Escherichia coli BL-21 (DES) cells (New England Biolabs). BL-21 cells were induced with a final concentration of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Pelleted cells were lysed in 1X PBS supplemented with protease inhibitors and sonicated on ice (5 × 30 s at full power). The GST fusion proteins were purified using glutathione-agarose beads (Sigma Aldrich) and washed with 1X PBS. Proteins were eluted with 20 mg/ml glutathione in 1X PBS. Purified proteins were then dialyzed overnight at 4°C in 1X PBS. The dialyzed proteins were concentrated using centrifugal filter units (EMD Millipore) and quantified using Bradford assay. Purified recombinant histones were purchased from Sigma Aldrich.
Methylation assays
In vitro: 10 µg of purified PRMT enzyme was combined with 5 µg of the substrate protein (histones or eIF2α) and 1 µCi µM S-adenosyl-l-[methyl-3H] methionine (Perkin Elmer) in a reaction buffer (50 mM potassium HEPES, 10 mM NaCl, and 1 mM DTT, pH 7.5) for 22 h at room temperature. Methylation reaction was quenched using Laemmli reducing buffer (25% glycerol, 125 mM Tris-HCl, pH 6.8, 4% SDS, 700 mM β-mercaptoethanol, 0.1% bromophenol blue) and subsequently run on an SDS–PAGE gel. The gel was then Coomassie stained and processed for fluorography by soaking in En3Hance reagent (Perkin Elmer) for 30 min at room temperature. The gel was washed three times in water for 30 min and dried in vacuo. The 3H-labeled proteins were visualized after a 2-wk exposure at −80°C.
In vivo: MDA-MB-231 cells were treated with 100 µg/ml cycloheximide and 40 µg/ml chloramphenicol in DMEM without methionine. Cells were incubated for 1 h at 37°C. l-[methyl-3H]-methionine (33 µCi/ml) (Perkin Elmer) was added to cells in fresh methionine-free DMEM with cycloheximide and chloramphenicol. Cells were incubated for 3 h and then subsequently lysed and resolved on an SDS–PAGE gel. The gel was then Coomassie stained and soaked in En3Hance buffer (Perkin Elmer) for 30 min at room temperature. The gel was washed three times in water for 30 min and dried in vacuo. The 3H-labeled proteins were visualized by fluorography after a 2-wk exposure at −80°C.
Immunofluorescence: stress granules
MDA-MB-231 cells were seeded at 70% confluency onto glass coverslips in a six-well plate and incubated overnight to allow cells to adhere. Cells were washed twice with 1X PBS before fixation in 1% formaldehyde (10 min) at room temperature followed by cold methanol fixation for 10 min. Cells were then washed three times with 1X PBS and blocked for 1 h with 5% bovine serum albumin in 1X PBS. Cells were then washed once with PBS and incubated with primary antibody (1:50 dilution) at room temperature for 1 h. Following the incubation, cells were washed twice with 1X PBS and incubated with secondary antibody (1:300 dilution) for 1 h at room temperature in the dark. Washes were performed twice with 1X PBS, and coverslips were mounted onto glass slides using Vectashield mounting media with DAPI (Vector Laboratories). Images were taken at 40× magnification using a Zeiss LSM 5 Pascal/Axiovert 200 epifluorescence microscope. Within randomly imaged fields (five fields per sample), the cells were counted according to DAPI staining in an unbiased manner—fields were chosen randomly without looking at the field. Subsequently, cells with at least five stress granules were counted as cells containing granules. The number of cells with stress granules was then divided by the total number of cells, giving the percentage of cells with stress granules. As controls, untreated cells and cells treated with both stressor and cycloheximide (a suppressor of stress granule formation) were used.
Statistical analysis
Data are presented as mean ± SE for the number of experiments indicated in the figure legends. Statistical analysis was performed using a two-tailed Student’s t test or by one-way or two-way analysis of variance (ANOVA) using a Bonferroni posttest using GraphPad Prism software where applicable. p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by the Canadian Breast Cancer Foundation and the Canadian Research Society funding agencies granted to J.C. N.H. was supported by the Canadian Institutes of Health Research Doctoral Award. We thank Bernard Jasmin for his valuable feedback throughout the editing process and Sabina Sarvan and Jean-François Couture for their assistance.
Abbreviations used:
- AsNaO 2
sodium arsenite
- eIF2α
eukaryotic translation initiation factor 2α
- FMRP
fragile X mental retardation protein
- G3BP1
GTPase-activating protein-binding protein 1
- PRMT
protein arginine methyltransferase
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-05-0330) on January 30, 2019.
REFERENCES
- Agolini E, Dentici ML, Bellacchio E, Alesi V, Radio FC, Torella A, Musacchia F, Tartaglia M, Dallapiccola B, Nigro V, et al. (2018). Expanding the clinical and molecular spectrum of PRMT7 mutations: 3 additional patients and review. Clin Genet , 675–681. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, Ivanov P. (2015). Stress granules, P-bodies and cancer. Biochim Biophys Acta , 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas A, Fay MM, Lyons SM, Achorn CA, Kedersha N, Anderson P, Ivanov P. (2017). Methods to classify cytoplasmic foci as mammalian stress granules. J Vis Exp , 10.3791/55656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguet A, Degot S, Cougot N, Bertrand E, Chenard M-P, Wendling C, Kessler P, Le Hir H, Rio MC, Tomasetto C. (2007). The exon-junction-complex-component metastatic lymph node 51 functions in stress-granule assembly. J Cell Sci , 2774. [DOI] [PubMed] [Google Scholar]
- Balagopal V, Parker R. (2009). Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol , 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin RM, Bejide M, Trinkle-Mulcahy L, Cote J. (2015). Identification of the PRMT1v1 and PRMT1v2 specific interactomes by quantitative mass spectrometry in breast cancer cells. Proteomics , 2187–2197. [DOI] [PubMed] [Google Scholar]
- Baldwin RM, Haghandish N, Daneshmand M, Amin S, Paris G, Falls TJ, Bell JC, Islam S, Côté J. (2015). Protein arginine methyltransferase 7 promotes breast cancer cell invasion through the induction of MMP9 expression. Oncotarget , 3013–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG. (2009). Protein arginine methylation in mammals: who, what, and why. Mol Cell , 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikkavilli RK, Avasarala S, Van Scoyk M, Karuppusamy Rathinam MK, Tauler J, Borowicz S, Winn RA. (2014). In vitro methylation assay to study protein arginine methylation. J Vis Exp 2014, e51997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc RS, Richard S. (2017). Arginine methylation: the coming of age. Mol Cell , 8–24. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Parker R. (2009). Eukaryotic stress granules: the ins and outs of translation. Mol Cell , 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres TB, Thakur A, Price OM, Ippolito N, Li J, Qu J, Acevedo O, Hevel JM. (2018). Phe71 in type III trypanosomal protein arginine methyltransferase 7 (TbPRMT7) restricts the enzyme to monomethylation. Biochemistry , 1349–1359. [DOI] [PubMed] [Google Scholar]
- Chan KK, Thompson S, O’Hagan D. (2013). The mechanisms of radical SAM/cobalamin methylations: an evolving working hypothesis. Chembiochem , 675–677. [DOI] [PubMed] [Google Scholar]
- Chen M, Qu X, Zhang Z, Wu H, Qin X, Li F, Liu Z, Tian L, Miao J, Shu W. (2016). Cross-talk between Arg methylation and Ser phosphorylation modulates apoptosis signal–regulating kinase 1 activation in endothelial cells. Mol Biol Cell , 1358–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon MB, Rust HL, Thompson PR, Mowen KA. (2013). Automethylation of protein arginine methyltransferase 8 (PRMT8) regulates activity by impeding S-adenosylmethionine sensitivity. J Biol Chem , 27872–27880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farny NG, Kedersha NL, Silver PA. (2009). Metazoan stress granule assembly is mediated by P-eIF2α-dependent and -independent mechanisms. RNA , 1814–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Hadjikyriacou A, Clarke SG. (2014). Substrate specificity of human protein arginine methyltransferase 7 (PRMT7): the importance of acidic residues in the double E loop. J Biol Chem , 32604–32616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Maity R, Whitelegge JP, Hadjikyriacou A, Li Z, Zurita-Lopez C, Al-Hadid Q, Clark AT, Bedford MT, Masson JY, Clarke SG. (2013). Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine- and arginine-rich regions. J Biol Chem , 37010–37025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier MJ, Gareau C, Mazroui R. (2010). The chemotherapeutic agent bortezomib induces the formation of stress granules. Cancer Cell Int , 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayatri S, Bedford MT. (2014). Readers of histone methylarginine marks. Biochim Biophys Acta , 702–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng P, Zhang Y, Liu XX, Zhang N, Liu Y, Liu XX, Lin C, Yan X, Li Z, Wang G.et al. (2017). Automethylation of protein arginine methyltransferase 7 and its impact on breast cancer progression. FASEB J , 2287–2300. [DOI] [PubMed] [Google Scholar]
- Gonsalvez GB, Tian L, Ospina JK, Boisvert FM, Lamond AI, Matera AG. (2007). Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J Cell Biol , 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros L, Delaporte C, Frey S, Decesse J, de Saint-Vincent BR, Cavarec L, Dubart A, Gudkov AV, Jacquemin-Sablon A. (2003). Identification of new drug sensitivity genes using genetic suppressor elements: protein arginine N-methyltransferase mediates cell sensitivity to DNA-damaging agents. Cancer Res , 164–171. [PubMed] [Google Scholar]
- Guo L, Chi Y, Xue J, Ma L, Shao Z, Wu J. (2017). Phosphorylated eIF2α predicts disease-free survival in triple-negative breast cancer patients. Sci Rep , 44674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A, Gu H, Zhou J, Mulhern D, Wang Y, Lee KA, Yang V, Aguiar M, Kornhauser J, Jia X, et al. (2014). Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol Cell Proteomics , 372–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjikyriacou A, Yang Y. (2015). Unique features of human protein arginine methyltransferase 9 (PRMT9) and its substrate RNA splicing factor SF3B2. J Biol Chem , 16723–16743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. (2003). An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell , 619–633. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Toma-Fukai S, Kim J-DD, Fukamizu A, Shimizu T. (2014). Protein arginine methyltransferase 7 has a novel homodimer-like structure formed by tandem repeats. FEBS Lett , 1942–1948. [DOI] [PubMed] [Google Scholar]
- Hendrick HM, Welter BH, Hapstack MA, Sykes SE, Sullivan WJ, Temesvari LA, Temesvari LA. (2016). Phosphorylation of eukaryotic initiation factor-2α during stress and encystation in Entamoeba species. PLoS Pathog , e1006085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann F, Pably P, Eckerich C, Bedford MT, Fackelmayer FO. (2009). Human protein arginine methyltransferases in vivo—distinct properties of eight canonical members of the PRMT family. J Cell Sci , 667–677. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol , 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain K, Jin CY, Clarke SG. (2017). Epigenetic control via allosteric regulation of mammalian protein arginine methyltransferases. Proc Natl Acad Sci USA , 10101–10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain K, Warmack RA, Debler EW, Hadjikyriacou A, Stavropoulos P, Clarke SG. (2016). Protein arginine methyltransferase product specificity is mediated by distinct active-site architectures. J Biol Chem , 18299–18308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinic P, Stehle JC, Shaw P. (2006). The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol , e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung GA, Shin BS, Jang YS, Sohn JB, Woo SR, Kim JE, Choi G, Lee KM, Min BH, Lee KH, et al. (2011). Methylation of eukaryotic elongation factor 2 induced by basic fibroblast growth factor via mitogen-activated protein kinase. Exp Mol Med , 550–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaehler C, Isensee J, Hucho T, Lehrach H, Krobitsch S. (2014). 5-Fluorouracil affects assembly of stress granules based on RNA incorporation. Nucleic Acids Res , 6436–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkhanis V, Wang L, Tae S, Hu YJ, Imbalzano AN, Sif S. (2012). Protein arginine methyltransferase 7 regulates cellular response to DNA damage by methylating promoter histones H2A and H4 of the polymerase delta catalytic subunit gene, POLD1. J Biol Chem , 29801–29814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, Anderson P. (2016). G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol , 845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernohan KD, McBride A, Xi Y, Martin N, Schwartzentruber J, Dyment DA, Majewski J, Blaser S, Care4Rare Canada Consortium, Boycott KM, et al. (2017). Loss of the arginine methyltranserase PRMT7 causes syndromic intellectual disability with microcephaly and brachydactyly. Clin Genet , 708–716. [DOI] [PubMed] [Google Scholar]
- Lakowski TM, Frankel A. (2009). Kinetic analysis of human protein arginine N-methyltransferase 2: formation of monomethyl- and asymmetric dimethyl-arginine residues on histone H4. Biochem J , 253–261. [DOI] [PubMed] [Google Scholar]
- Larivière N, Law J, Trinkle-Mulcahy L. (2014). Dissection of a novel autocrine signaling pPathway via quantitative secretome and interactome mapping. J Proteome Res , 3432–3443. [DOI] [PubMed] [Google Scholar]
- Lee JH, Cook JR, Yang ZH, Mirochnitchenko O, Gunderson SI, Felix AM, Herth N, Hoffmann R, Pestka S. (2005). PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J Biol Chem , 3656–3664. [DOI] [PubMed] [Google Scholar]
- Litt M, Qiu Y, Huang S. (2009). Histone arginine methylations: their roles in chromatin dynamics and transcriptional regulation. Biosci Rep , 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda TB, Miranda M, Frankel A, Clarke S. (2004). PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J Biol Chem , 22902–22907. [DOI] [PubMed] [Google Scholar]
- Moeller BJ, Dewhirst MW. (2006). HIF-1 and tumour radiosensitivity. Br J Cancer , 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. (2006). A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell , 1283–1298. [DOI] [PubMed] [Google Scholar]
- Rajesh K, Krishnamoorthy J, Kazimierczak U, Tenkerian C, Papadakis AI, Wang S, Huang S, Koromilas AE. (2015). Phosphorylation of the translation initiation factor eIF2α at serine 51 determines the cell fate decisions of Akt in response to oxidative stress. Cell Death Dis , e1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M, Vasconcelos G, Dever TE. (2015). An eIF2α-binding motif in protein phosphatase 1 subunit GADD34 and its viral orthologs is required to promote dephosphorylation of eIF2α. Proc Natl Acad Sci USA , E3466–E3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald IB, Wang S, Savas L, Woda B, Pullman J. (2003). Expression of translation initiation factor eIF-2α is increased in benign and malignant melanocytic and colonic epithelial neoplasms. Cancer , 1080–1088. [DOI] [PubMed] [Google Scholar]
- Sadlish H, Galicia-Vazquez G, Paris CG, Aust T, Bhullar B, Chang L, Helliwell SB, Hoepfner D, Knapp B, Riedl R, et al. (2013). Evidence for a functionally relevant rocaglamide binding site on the eIF4A-RNA complex. ACS Chem Biol , 1519–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhroy DN, Mesplede T, Sabbah A, Quashie PK, Falgueyret JP, Wainberg MA. (2013). Automethylation of protein arginine methyltransferase 6 (PRMT6) regulates its stability and its anti-HIV-1 activity. Retrovirology , 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenkerian C, Krishnamoorthy J, Mounir Z, Kazimierczak U, Khoutorsky A, Staschke KA, Kristof AS, Wang S, Hatzoglou M, Koromilas AE. (2015). mTORC2 balances AKT activation and eIF2α serine 51 phosphorylation to promote survival under stress. Mol Cancer Res , 1377–1388. [DOI] [PubMed] [Google Scholar]
- Thomassen M, Tan Q, Kruse TA. (2009). Gene expression meta-analysis identifies chromosomal regions and candidate genes involved in breast cancer metastasis. Breast Cancer Res Treat , 239–249. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia R, Boisvert F-M, Vandermoere F, Morrice NA, Swift S, Rothbauer U, Leonhardt H, et al. (2008). Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J Cell Biol , 223–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Charoensuksai P, Watson NJ, Wang X, Zhao Z, Coriano CG, Kerr LR, Xu W. (2013). CARM1 automethylation is controlled at the level of alternative splicing. Nucleic Acids Res , 6870–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Lloyd RV, Hutzler MJ, Rosenwald IB, Safran MS, Patwardhan NA, Khan A. (2001). Expression of eukaryotic translation initiation factors 4E and 2α correlates with the progression of thyroid carcinoma. Thyroid , 1101–1107. [DOI] [PubMed] [Google Scholar]
- Wang S, Rosenwald IB, Hutzler MJ, Pihan GA, Savas L, Chen JJ, Woda BA. (1999). Expression of the eukaryotic translation initiation factors 4E and 2α in non-Hodgkin’s lymphomas. Am J Pathol , 247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Bedford MT. (2013). Protein arginine methyltransferases and cancer. Nat Rev Cancer , 37–50. [DOI] [PubMed] [Google Scholar]
- Yang Y, Hadjikyriacou A, Xia Z. (2015). PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145. Nat Commun , 6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Jiang H, Ma Y, Wang L, Wang L, Du J, Hou P, Gao Y, Zhao L, Wang G, et al. (2014). PRMT7 induces epithelial-to-mesenchymal transition and promotes metastasis in breast cancer. Cancer Res , 5656–5667. [DOI] [PubMed] [Google Scholar]
- Zurita-Lopez CI, Sandberg T, Kelly R, Clarke SG. (2012). Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming ω-NG-monomethylated arginine residues. J Biol Chem , 7859–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.