Abstract
Across most sexually reproducing animals, centrosomes are provided to the oocyte through fertilization and must be positioned properly to establish the zygotic mitotic spindle. How centrosomes are positioned in space and time through the concerted action of key mitotic entry biochemical regulators, including protein phosphatase 2A (PP2A-B55/SUR-6), biophysical regulators, including dynein, and the nuclear lamina is unclear. Here, we uncover a role for PP2A-B55/SUR-6 in regulating centrosome separation. Mechanistically, PP2A-B55/SUR-6 regulates nuclear size before mitotic entry, in turn affecting nuclear envelope–based dynein density and motor capacity. Computational simulations predicted the requirement of PP2A-B55/SUR-6 regulation of nuclear size and nuclear-envelope dynein density for proper centrosome separation. Conversely, compromising nuclear lamina integrity led to centrosome detachment from the nuclear envelope and migration defects. Removal of PP2A-B55/SUR-6 and the nuclear lamina simultaneously further disrupted centrosome separation, leading to unseparated centrosome pairs dissociated from the nuclear envelope. Taking these combined results into consideration, we propose a model in which centrosomes migrate and are positioned through the concerted action of PP2A-B55/SUR-6–regulated nuclear envelope–based dynein pulling forces and centrosome–nuclear envelope tethering. Our results add critical precision to models of centrosome separation relative to the nucleus during spindle formation in cell division.
INTRODUCTION
Fertilization provides centrosomes to the developing embryo in many systems. Associated with the sperm pronucleus, centrosomes must grow, separate, and migrate in a coordinated manner to facilitate female and male pronuclear meeting before mitosis. In doing so, centrosomes orchestrate the microtubule cytoskeleton in the embryo such that the mitotic spindle is properly positioned, ensuring cell division occurs in a manner that facilitates further development (e.g., Caenorhabditis elegans asymmetric cell division). These events arise due to collaboration between mitotic master regulators and microtubule-based biophysical regulators.
Mechanistically, centrosomes are positioned through the concerted action of cytoplasmic flow, cytoplasmic and cortical microtubule pulling forces, and microtubule pushing forces against the cortex (Garzon-Coral et al., 2016; Nazockdast et al., 2017). On the molecular scale, it has been well established that microtubule-based motors, including dynein (Gönczy et al., 1999; Robinson et al., 1999), play important roles in centrosome migration at mitotic entry. Recently, a combination of experimental and theoretical approaches have been used to distinguish the roles of different pools of motors (i.e., nuclear and cortical), cortical flows (De Simone et al., 2016; De Simone and Gönczy, 2017), and cytoplasmic drag (De Simone et al., 2018) in regulating centrosome separation at the mesoscale. For example, in the C. elegans zygote, dynein-based microtubule pulling forces are produced at the nuclear envelope and the cortex to ensure proper centrosome separation (De Simone et al., 2016). Despite our understanding of microtubule cytoskeleton–based pulling forces, the contribution of key mitotic kinases and phosphatases to centrosome separation remains unclear.
The net effect of molecular regulators is a biophysical mechanism required for separating centrosomes. As of yet, the contributions of the nuclear envelope and its centrosome-tethering components within this model are uncertain. Centrosomes are tethered to the nuclear lamina through the linker of nucleoskeleton and cytoskeleton (LINC) complex, which is composed of SUN proteins tethered to the nuclear lamina and KASH proteins tethered to the microtubule cytoskeleton (reviewed in Chang et al., 2015). In the C. elegans embryo, both the SUN protein SUN-1 and the KASH protein ZYG-12 are required for centrosome tethering (Malone et al., 2003). Additionally, centrosome–nuclear envelope tethering is thought to be regulated by dynein, as ZYG-12 directly binds dynein’s light chain (Malone et al., 2003) and dynein is required for tethering (Gönczy et al., 1999). Dynein-based microtubule pulling forces and the centrosome–nuclear envelope tethering machinery are therefore likely to be coordinated by mitotic master regulators during centrosome separation.
Biochemically, mitotic entry is defined by the activities of master mitotic kinases and phosphatases that ensure faithful cell cycle progression (Krasinska et al., 2011; Mochida et al., 2016). As cyclin dependent kinase 1 (CDK1) activity increases at mitotic entry, its main counteracting phosphatase PP2A is also regulated. In metazoans, PP2A mainly functions as a heterotrimeric enzyme using its B55/SUR-6 adaptor subunit for substrate recognition and is the main phosphatase responsible for dephosphorylating CDK1 substrates (Mayer-Jaekel et al., 1994; Castilho et al., 2009; Mochida et al., 2009). To identify potential converse functions of the CDK1 counteracting phosphatase PP2A-B55/SUR-6, we previously used a maternal-effect genetic screen in the fly embryo (Mehsen et al., 2018), in which several components were identified as PP2A-B55/SUR-6 collaborators for proper cell cycle progression, including lamin. The biology of Drosophila melanogaster early development (e.g., the zygotic division) precluded centrosome tracking and, therefore, fundamental probing of the biophysical mechanisms at play. Thus, we examined the contributions of the genetic interaction between PP2A-B55/SUR-6 and lamin/LMN-1 to the first developmental mitosis in the C. elegans embryo (the zygote). We identify a novel role for PP2A-B55/SUR-6 in regulating centrosome separation in the embryo through the regulation of nuclear envelope–based dynein motor capacity. We propose a simple model in which dynein-based pulling forces collaborate with centrosome–nuclear envelope tethering to ensure proper centrosome separation during mitotic entry.
RESULTS
PP2A-B55/SUR-6 and LMN-1 play critical roles in centrosome separation in the Caenorhabditis elegans zygote
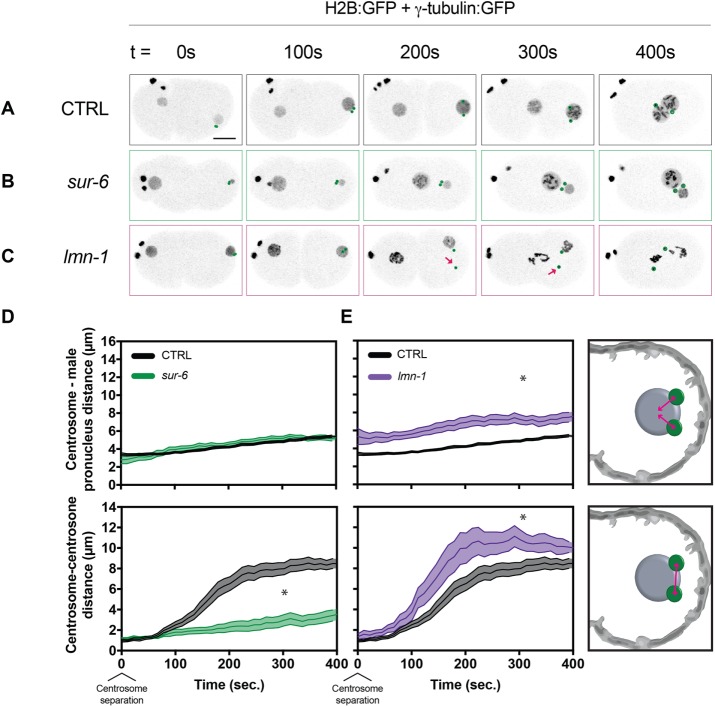
To identify potential roles for the previously discovered genetic interactions in single embryonic mitoses, we depleted individual components by RNA interference (RNAi) in the C. elegans embryo—a system in which late meiotic and first mitotic events can be readily observed by light microscopy (Supplemental Video S1 and Figure 1A). Depleting sur-6, the PP2A adapter subunit, in worms expressing H2B:green fluorescent protein (GFP) and γ-tubulin:GFP (strain TH32; see Materials and Methods) led to centrosomes that were tethered to the nuclear envelope and failed to separate from one another (Supplemental Video S2 and Figure 1, B, D, and E), while depleting LMN-1 caused centrosomes to detach from nuclear envelopes as previously reported (Meyerzon et al., 2009; Penfield et al., 2018) and increase in distance from one another (Supplemental Video S3 and Figure 1, C–E).
FIGURE 1:
sur-6 and lmn-1 RNAi affect centrosome separation through distinct mechanisms. (A–C) Representative maximum-intensity projections of embryonic confocal stacks. All RNAi treatments were performed with L4-stage worms expressing H2B:GFP and γ-tubulin:GFP for 24 h via feeding unless otherwise noted. Circles highlight individual centrosomes and arrows denote detached centrosomes. Scale bar: 10 µm. (D, E) Centrosome–centrosome distance and centrosome–male pronucleus distance were measured in three dimensions between respective centroids. Error bars indicate SEM and are represented as shaded areas around means. Embryos analyzed per condition: n > 10. Statistics: t test where *p < 0.05. Time t = 0 s corresponds to centrosome separation.
Movie S1.
Control C. elegans zygotes expressing H2B:GFP and γ-tubulin:GFP. Maximum intensity projection of confocal stacks. Related to Figure 1.
Movie S2.
sur-6 RNAi in C. elegans zygotes expressing H2B:GFP and γ-tubulin:GFP. Maximum intensity projection of confocal stacks. Related to Figure 1.
Movie S3.
lmn-1 RNAi in C. elegans zygotes expressing H2B:GFP and γ-tubulin:GFP. Maximum intensity projection of confocal stacks. Related to Figure 1.
We hypothesized that PP2A-B55/SUR-6 may regulate centrosome migration through biochemical regulation of known centrosome separation components, including dynein. Partial RNAi-mediated depletion of the dynein motor heavy-chain dhc-1 led to centrosome migration defects, while centrosomes remained tethered to the nuclear envelope (Supplemental Video S4 and Supplemental Figure S1, B, F, and E). Similarly, partially depleting the dynein regulator lis-1, which has been shown to regulate dynein-dependent centrosome separation (Cockell et al., 2004), led to centrosome-migration phenotypes, while centrosome tethering was maintained (Supplemental Video S5 and Supplemental Figure S1, C, F, and E). Taken together, these data suggest partial depletion of dynein’s heavy chain or of its regulator lis-1 phenocopied sur-6 centrosome separation phenotypes, supporting a potential role for PP2A-B55/SUR-6 in regulating centrosome separation through dynein activity independently of centrosome–nuclear envelope tethering regulation.
Movie S4.
16h dhc-1 RNAi in C. elegans zygotes expressing H2B:GFP and γ-tubulin:GFP. Maximum intensity projection of confocal stacks. Related to Figure 1.
Movie S5.
lis-1 RNAi in C. elegans zygotes expressing H2B:GFP and γ-tubulin:GFP. Maximum intensity projection of confocal stacks. Related to Figure S1.
Given the role of LMN-1 as the principal structural component of the nuclear lamina, we hypothesized that lmn-1 centrosome migration phenotypes may be the result of failed centrosome–nuclear envelope tethering. Consistent with previous reports (Malone et al., 2003), depletion of the outer nuclear envelope LINC complex component zyg-12 led to the loss of centrosome–nuclear envelope tethering and increased centrosome–centrosome distance (confirmed in Supplemental Video S6; Supplemental Figure S1, D, F, and G). Similarly, depleting the inner nuclear envelope LINC complex component sun-1 compromised centrosome–nuclear envelope tethering (Supplemental Video S7 and Supplemental Figure S1, E–G), as described previously (Malone et al., 2003; De Simone et al., 2016). Collectively, lmn-1, zyg-12, and sun-1 led to centrosome detachment from the nuclear envelope and an increase in centrosome–centrosome distance, supporting a role for the nuclear lamina in promoting centrosome–nuclear envelope cohesion during centrosome separation. Taken together, these data suggest PP2A-B55/SUR-6 regulates centrosome separation independently of centrosome–nuclear envelope tethering, while the nuclear lamina and the LINC complex provide a congruent centrosome-tethering mechanism.
Movie S6.
zyg-12 RNAi in C. elegans zygotes expressing H2B:GFP and γ-tubulin:GFP. Maximum intensity projection of confocal stacks. Related to Figure 1.
Movie S7.
sun-1 RNAi in C. elegans zygotes expressing H2B:GFP and γ-tubulin:GFP. Maximum intensity projection of confocal stacks. Related to Figure S1.
Nuclear envelope–based dynein motor activity is regulated by PP2A-B55/SUR-6
How biochemical regulators such as PP2A-B55/SUR-6 and components of the LINC complex ensure centrosome separation in the embryo was unclear. To gain mechanistic insight into centrosome separation regulation, we measured nuclear envelope–based dynein motor capability by imaging EBP2:GFP foci velocities via a previously described approach (Srayko et al., 2005). Caenorhabditis elegans embryos expressing the microtubule plus-end binding protein EBP2:GFP were imaged before pronuclear migration, and microtubule growth velocities were measured (Supplemental Video S8). Importantly, regulation of global microtubule polymerization rates in RNAi-compromised conditions was measured by tracking cytoplasmic microtubules and was found to be unperturbed (Supplemental Figure S2). On the basis of cytoplasmic microtubule growth velocities, we estimated that microtubules with measured mean velocities above 1.5 µm/s are likely to be accelerated by external forces, as few microtubules exhibited effective polymerization rates >1.5 µm/s in the cytoplasm.
Movie S8.
Control C. elegans zygotes expressing EBP2:GFP. Single confocal plane. Related to Figure 2 and Figure S2.
Taking advantage of the untethered nature of growing microtubules at this mitotic stage (Supplemental Video S8), we measured microtubule growth velocities for microtubules interacting with the nuclear envelope of the female pronucleus exclusively, as centrosome-organized microtubules are exclusively present in the vicinity of the male pronucleus. Measuring microtubule growth velocities on the female pronucleus revealed that >70% of microtubules were accelerated relative to cytoplasmic microtubules (Figure 2A). In control embryos, median cytoplasmic microtubule growth velocity was 1.09 µm/s, or ∼0.4 µm/s slower than nuclear associated microtubules (Supplemental Figure S2A). Reconstituted mammalian dynein complexes have been found to exhibit average velocities of just below 0.6 µm/s in vitro (Gutierrez et al., 2017). Therefore, our observed 0.4 µm/s average increase in apparent microtubule growth velocities in the vicinity of the nuclear envelope are within dynein’s ability to accelerate microtubules. Consistent with previous reports, microtubule acceleration in the vicinity of the nuclear envelope was dependent on dynein, as depletion of dhc-1 by RNAi led to a reduction in the population of accelerated microtubules in a titratable manner between 16-h (Figure 2B) and 24-h (Figure 2C) RNAi treatments (Gusnowski and Srayko, 2011). Depleting the dynein regulator lis-1 also led to a reduction in the fraction of accelerated microtubules, supporting dynein’s nuclear envelope–based motor activity (Figure 2D).
FIGURE 2:
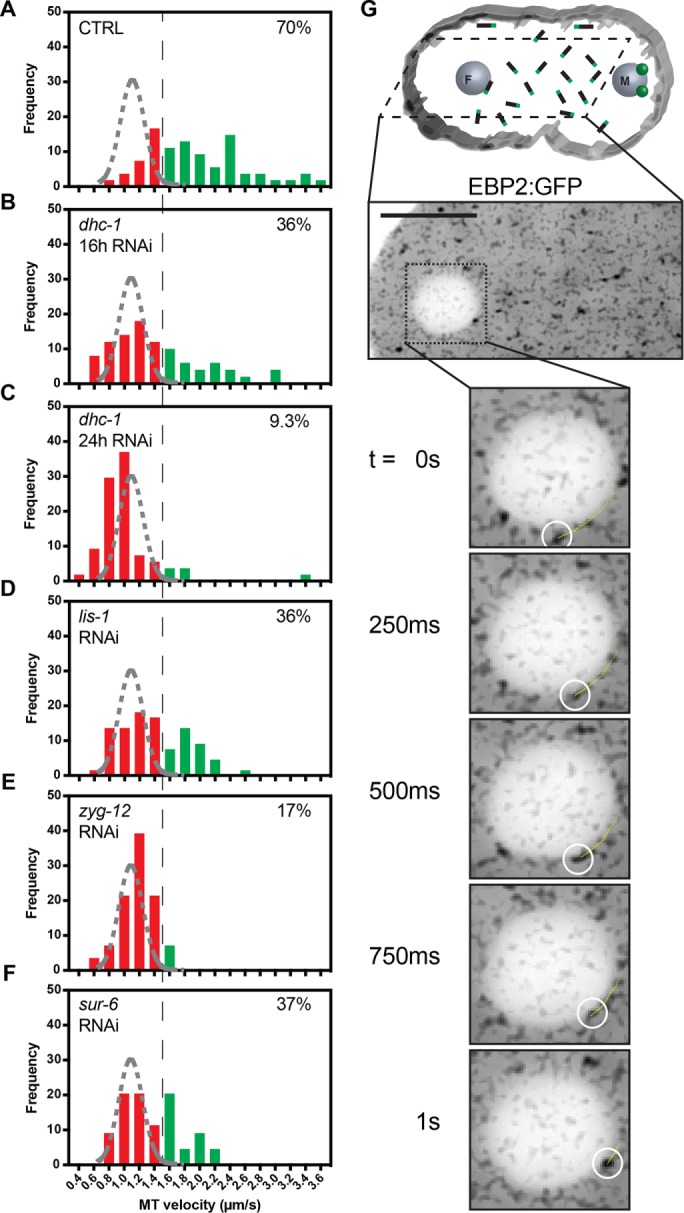
PP2A-B55/SUR-6 regulates dynein-dependent microtubule growth velocity acceleration at the outer nuclear envelope. (A–F) Frequency distributions of EBP2:GFP foci velocities for microtubules contacting the nuclear envelope of the female pronucleus before pronuclear migration. The black dashed line represents a threshold over which microtubules are assumed to be accelerated by nuclear envelope–tethered dynein. The gray dashed curve represents a Gaussian distribution fit to control unaccelerated cytoplasmic microtubule growth velocities. Microtubules analyzed per condition: n > 50 across > 3 embryos. Percentage inlays represent the proportion of microtubules growing at velocities above the 1.5 µm/s threshold in each condition. (G) Drawing illustrating the imaging plane, cytoplasmic microtubules in black with green plus ends representing EBP2, and representative single-plane confocal image of embryos expressing EBP2:GFP. The yellow trajectory line represents a track for up to 10 frames (2.5 s) following the displayed frame using TrackMate. Scale bar: 10 µm.
For determination of how components known to affect centrosome separation may affect dynein-mediated pulling forces, growth velocities of microtubules interacting with the nuclear envelope were measured in zyg-12– (Figure 2E) and sur-6–depleted (Figure 2F) embryos. Depleting zyg-12, which has previously been shown to be required for DHC-1 localization to the nuclear envelope in the germ line and in embryos (Malone et al., 2003), decreased the proportion of accelerated microtubules at the female pronucleus (Figure 2E). Depleting sur-6 also led to a decrease in the proportion of accelerated microtubules at the female pronucleus (Figure 2F), supporting a role for PP2A-B55/SUR-6 in regulating centrosome separation through microtubule-based dynein motor capacity.
PP2A-B55/SUR-6 regulates nuclear envelope–based dynein density
Biochemical and biophysical regulation of centrosome separation through dynein could occur through several mechanisms such as dynein affinity for the nuclear envelope (mean amount) or dynein density on the nuclear envelope (amount per surface area). On the basis of the results reported earlier, we asked how nuclear envelope–based dynein motor capacity could be regulated by PP2A-B55/SUR-6. To distinguish between potential dynein regulatory mechanisms, we measured amounts of endogenous DHC-1 (Heppert et al., 2018) on the nuclear envelope during mitotic entry (Supplemental Video S9).
Movie S9.
Control C. elegans zygotes expressing DHC-1:mNeonGreen. Single confocal plane. Related to Figure 3 and Figure S3.
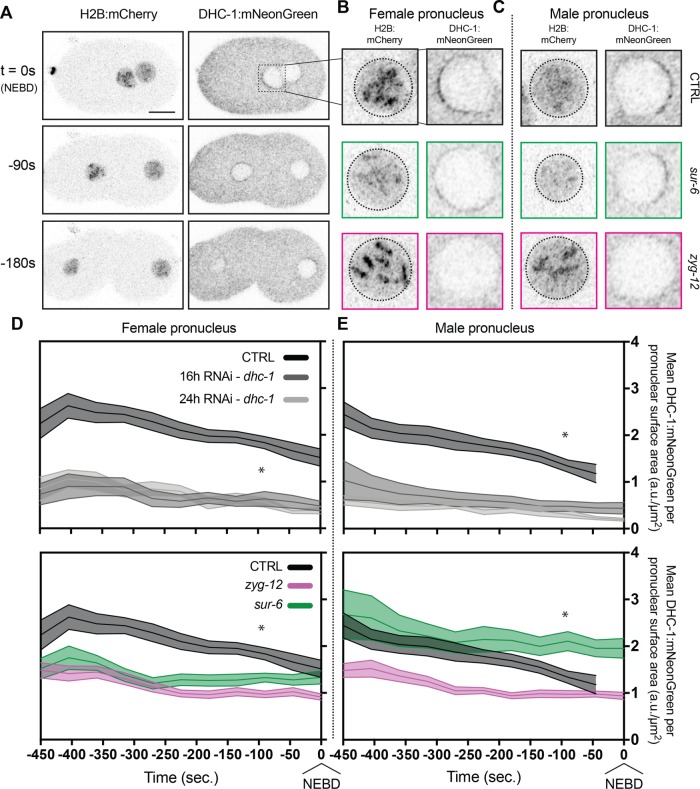
Targeting dhc-1 via RNAi in embryos expressing DHC-1:mNeonGreen significantly depleted the amount of nuclear envelope DHC-1:mNeonGreen (Supplemental Figure S3A). Importantly, the amount of DHC-1 on the nuclear envelope could be depleted in a titratable manner between 16-h and 24-h dhc-1 depletions. The loss of DHC-1:mNeonGreen in dhc-1 depletions at the nuclear envelope could also be quantified in terms of DHC-1 density on the nuclear envelope based on nuclear surface area (Figure 3, D and E).
FIGURE 3:
Quantification of endogenous DHC-1:mNeonGreen reveals nuclear envelope density regulation by PP2A-B55/SUR-6. (A) Representative confocal images of embryos expressing mCherry:HIS-58 and endogenous DHC-1:mNeonGreen. mCherry:HIS-58 images represent maximum-intensity projections of confocal stacks, and DHC-1:mNeonGreen images are single planes. Scale bar: 10 µm. (B, C) Inlays of female pronuclei (B) and male (C) pronuclei one frame before nuclear envelope breakdown (NEBD). (D, E) Density measurements of DHC-1:mNeonGreen (a.u., arbitrary units) per surface area of nuclear envelope (µm2) on female pronuclei (D) and male pronuclei (E). Embryos analyzed per condition: n > 14. Statistics: t test where *p < 0.05. Time t = 0 s corresponds to NEBD.
Depletion of zyg-12 in the embryo caused a significant loss in DHC-1:mNeonGreen density before nuclear envelope breakdown on both the female and male pronuclei (Figure 3, D and E). ZYG-12–dependent localization of DHC-1 is reflected both in DHC-1 density on the surface of the nuclear envelope and in the average amount of DHC-1 on the envelope (Supplemental Figure S3B).
Depletion of sur-6 led to insignificant changes in DHC-1:mNeonGreen on female and male pronuclei (Supplemental Figure S3B), suggesting that PP2A-B55/SUR-6 does not regulate dynein’s affinity for the nuclear envelope. However, density measurements of DHC-1 with respect to nuclear envelope surface area revealed female pronuclei had significantly less DHC-1 per unit area compared with their control counterparts, whereas male pronuclei had higher DHC-1 densities (Figure 3, D and E). Interestingly, depleting sur-6 led to significant pronuclear size changes in which female pronuclei were 66% larger and male pronuclei were 18% smaller in volume (Supplemental Figure S4), consistent with observations using a temperature-sensitive SUR-6 mutant in embryos (O’Rourke et al., 2011). Taken together, measurements of DHC-1 amounts on pronuclei revealed that ZYG-12 regulates dynein motor activity throughout mitotic entry by regulating DHC-1 amounts on the outer nuclear envelope, as expected. In contrast to regulating dynein amounts, PP2A-B55/SUR-6 affects dynein motor activity through the regulation of pronuclear size, affecting pronuclear DHC-1 densities.
PP2A-B55/SUR-6 regulates cortical dynein motor activity
In addition to nuclear envelope–based dynein, cortex-associated dynein has also been shown to be critical for centrosome separation in the C. elegans embryo (De Simone et al., 2016). Microtubules interacting with dynein tethered to the cortex have been shown to be accelerated in a dynein-dependent manner during metaphase (Gusnowski and Srayko, 2011). For determination of whether PP2A-B55/SUR-6 could be regulating centrosome separation through the regulation of cortical dynein in addition to nuclear envelope–associated dynein, untethered microtubules growing in the vicinity of the cortex were imaged using EBP2:GFP foci as proxies before pronuclear migration using total internal reflection fluorescence (TIRF) microscopy (Supplemental Video S10).
Movie S10.
Control C. elegans zygotes expressing EBP2:GFP. Single TIRF illuminated plane. Related to Figure 4.
At the cortex, most microtubules grew at rates above the 1.5 µm/s threshold (Figure 4A), indicating prevalent microtubule accelerating forces. The measured polymerization rates were dependent on dynein in a titratable manner, as expected (Figure 4, B and C), as well as on the dynein regulator LIS-1 (Figure 4D). Interestingly, depleting zyg-12 led to a reduction in the proportion of dynein-accelerated microtubules (Figure 4E) to the same extent as a 24-h dhc-1 depletion (Figure 4C). How ZYG-12 could be regulating cortical dynein activity is unclear and should be the subject of future work.
FIGURE 4:
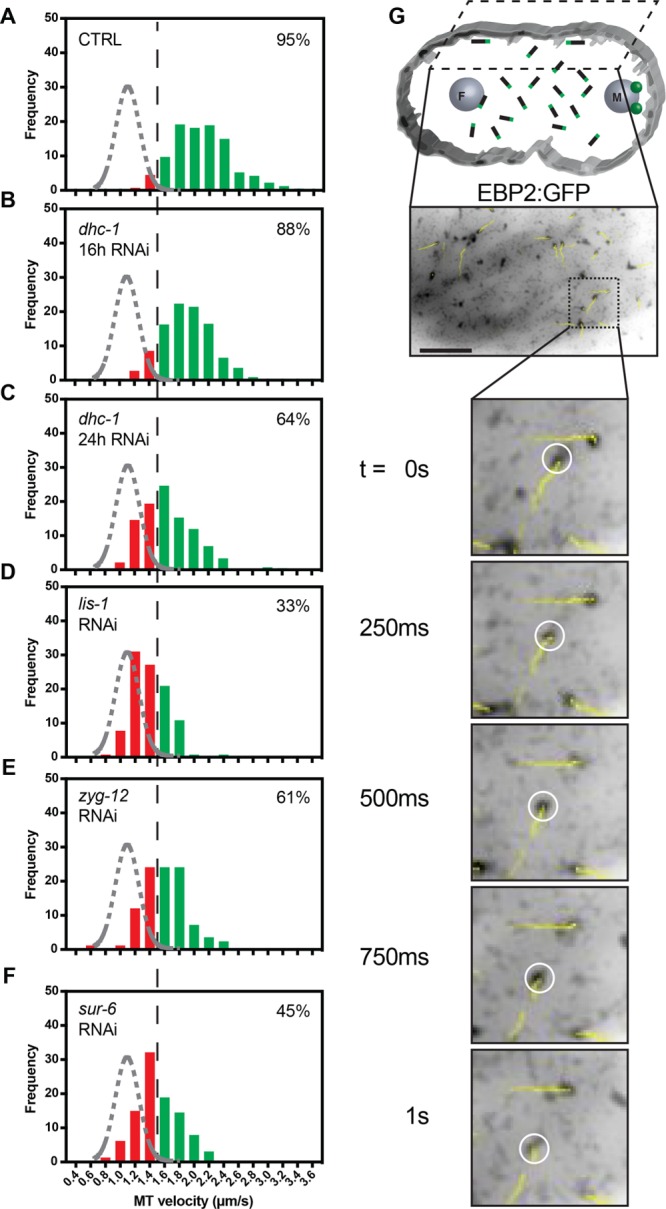
PP2A-B55/SUR-6 regulates dynein-dependent microtubule growth velocity acceleration at the cortex. (A–F) Frequency distributions of EBP2:GFP foci velocities for microtubules polymerizing along the cortex of the embryo before pronuclear migration as visualized by TIRF microscopy. The black dashed line represents a threshold over which microtubules are assumed to be accelerated by cortical dynein. The gray dashed curve represents a Gaussian distribution fit to control unaccelerated cytoplasmic microtubule growth velocities. Microtubules analyzed per condition: n > 80 across > 3 embryos. Percentage inlays represent the proportion of microtubules growing at velocities above the 1.5 µm/s threshold in each condition. (G) Drawing illustrating the imaging plane, cytoplasmic microtubules in black with green plus ends representing EBP2, and representative single-plane confocal image of embryos expressing EBP2:GFP. Yellow trajectory lines represent tracks for up to 10 frames (2.5 s) following the displayed frame using TrackMate. Scale bar: 10 µm.
For determination of whether PP2A-B55/SUR-6 regulates cortical dynein in addition to nuclear envelope–based dynein, sur-6 was depleted via RNAi. sur-6 depletions led to a reduction in more than 50% of the proportion of microtubules being accelerated by dynein (Figure 4E), suggesting that PP2A-B55/SUR-6 regulates cortical dynein in addition to nuclear envelope–tethered dynein.
Computational simulations of centrosome migration partially recapitulate PP2A-B55/SUR-6’s role in regulating centrosome separation through pronuclear size and nuclear envelope dynein density
Given the pleotropic nature of PP2A-B55/SUR-6–mediated dynein regulation, we turned to computational simulations to gain insight into phenotypes relevant to centrosome separation. As a mitotic master regulator, PP2A-B55/SUR-6 is known to regulate several aspects of mitotic entry and exit, including mitotic entry timing (Mochida et al., 2009), chromosome decondensation and nuclear envelope reformation (Afonso et al., 2014; Mehsen et al., 2018), and cytokinesis initiation (Cundell et al., 2013). For determination of whether the roles uncovered for PP2A-B55/SUR-6 in affecting pronuclear size (Supplemental Figure S4), nuclear envelope dynein density (Figure 3, B–E), and cortical dynein activity (Figure 4F) are sufficient to affect centrosome separation, a previously described Cytosim-based computational model was used (Supplemental Video S11 and Figure 5A) (De Simone et al., 2016).
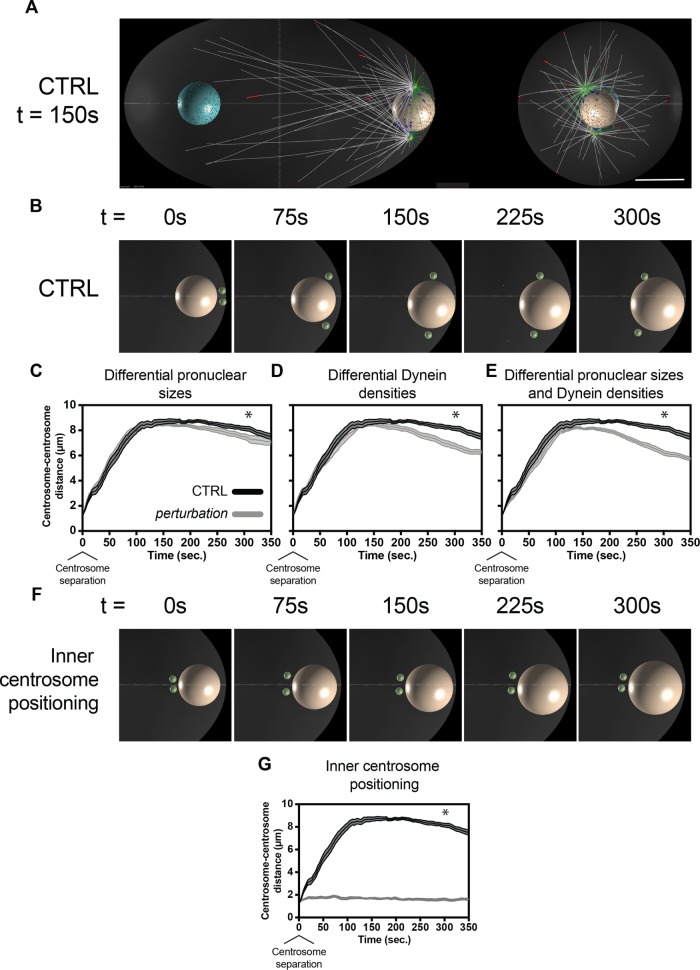
FIGURE 5:
Cytosim-based computational simulations predict that differences in pronuclear size and nuclear envelope–tethered dynein density affect centrosome separation, while initial inner centrosome separation is dominant. (A) Representative image of Cytosim-based centrosome separation simulations at 150 s after centrosome separation. Red and blue dots represent dynein–microtubule binding at the cortex and at the nuclear envelope, respectively. White microtubules represent inwardly directed microtubules, and green microtubules represent rearward-directed microtubules with respect to centrosomes (green). The rightmost image represents an end-on view of the posterior of the embryo. Scale bar: 10 µm. (B) Representative images of embryo inlays through early centrosome migration. Male pronuclei are displayed in beige and centrosomes in green. (C) Predicted centrosome–centrosome distances in control embryos and in embryos with 18% smaller male pronuclei and 66% larger female pronuclei (µm3). (D) Predicted centrosome–centrosome distances in control embryos and in embryos with 40% denser dynein on male pronuclei and 25% sparser dynein on female pronuclei (a.u./µm2). (E) Predicted centrosome–centrosome distances in control embryos and in embryos with both differentially sized pronuclei and dynein densities, as in C and D. (F) Representative images of simulated centrosome separation with inner initial centrosome separation. (G) Predicted centrosome–centrosome distances in control embryos and in embryos with inner initial centrosome separation. All simulated perturbations reflect experimental measurements of sur-6 RNAi phenotypes. Error bars indicate SEM and are represented as shaded areas around means of 10 simulation runs per condition. Statistics: t test where *p < 0.05. Time t = 0 s corresponds to centrosome separation.
Movie S11.
Cytosim-based simulation of centrosome separation displaying pronuclei (female - teal and male - beige), inward (white) and outward (green) microtubules, centrosomes (green), cortical Dynein-microtubule binding (red) and nuclear envelope-associated Dynein-microtubule binding (blue). Related to Figure 5 and Figure S5.
Separating sur-6 effects, we simulated differential pronuclear sizes, dynein densities on pronuclear envelopes, and cortical dynein motor activity during centrosome separation. First, pronuclear sizes were modified as to reflect sur-6 effects while maintaining dynein densities. Female pronuclei were simulated to have initial volumes 66% larger (µm3) than control, while male pronuclei were 18% smaller, as measured experimentally. Interestingly, modifying only pronuclear volumes had a subtle yet significant effect on centrosome separation (Figure 5C). Second, pronuclear dynein densities were modified to reflect experimental values independently of pronuclear size differences. Decreasing dynein surface density on the female pronucleus by 25% (arbitrary units per surface area of nuclear envelope [a.u./µm2]) while increasing dynein surface density on the male pronucleus by 40% also led to significant differences in centrosome separation (Figure 5D). Third, cortical dynein motor activity was simulated as 50% slower than control to reflect experimental values. Cortical dynein on its own did not have significant effects on centrosome separation (Supplemental Figure S5), nor did it have synergistic effects when combined with pronuclear size and dynein density perturbations (unpublished data), suggesting PP2A-B55/SUR-6 regulation of cortical dynein is not a significant contributor to centrosome separation.
To predict whether pronuclear size and nuclear envelope dynein density collaborate in ensuring centrosome separation, we simulated perturbations to pronuclear size and dynein density simultaneously (Figure 5E). Simulating sur-6 based phenotypes together reduced centrosome–centrosome distance additively. These data support a role for PP2A-B55/SUR-6 in regulating centrosome separation through nuclear size regulation and its effect on dynein density on the nuclear envelope.
Evidently, the sur-6 phenotype could only be partially recapitulated through its regulation of nuclear size and nuclear envelope dynein densities, indicating that sur-6 likely induces other processes relevant to centrosome separation that have yet to be uncovered. Another clear centrosome-based sur-6 phenotype was the initial position of centrosome pairs before centrosome separation (Figure 1B). In sur-6 depletions, centrosome pairs were positioned internally to the embryo as opposed to being proximal to the cortex, potentially disrupting the orientation of microtubule pulling or pushing forces against the cortex or the female pronucleus. Simulating centrosome migration in embryos in which centrosomes were internal to the embryo (Figure 5F) abolished centrosome separation, suggesting initial centrosome separation with respect to the cortex is required for centrosome migration (Figure 5G). Taken together, these data suggest that PP2A-B55/SUR-6–mediated regulation of pronuclear size and nuclear envelope–based DHC-1 density are important for proper centrosome separation, while early centrosome positioning with respect to the embryo’s cortex is dominant in regulating centrosome separation.
PP2A-B55/SUR-6 collaborates with LMN-1 for proper centrosome separation
Given the role uncovered for PP2A-B55/SUR-6 in regulating dynein-based pulling forces and the previously identified genetic interaction between PP2A-B55/SUR-6 and the nuclear lamina (Mehsen et al., 2018), we predicted that pulling forces collaborate with centrosome–nuclear envelope tethering during centrosome separation. To determine how PP2A-B55/SUR-6 regulation of centrosome separation is coordinated with LINC complex function, we depleted components of both pathways simultaneously via RNAi.
As described previously, DHC-1 outer nuclear envelope localization is dependent on the hook protein ZYG-12 (Malone et al., 2003; Figure 3, D and E), and both DHC-1 (Gönczy et al., 1999) and ZYG-12 are required for centrosome–nuclear envelope cohesion (Malone et al., 2003; De Simone et al., 2016; Supplemental Figure S1D). Here, we titrated the amount of DHC-1 required for centrosome–nuclear envelope cohesion while affecting centrosome separation (Supplemental Figure S1B) and motor ability at the nuclear envelope (Figure 2B) and the cortex (Figure 4B). Compromising lis-1 via partial RNAi also created a condition under which centrosomes remained tethered to the nuclear envelope (Supplemental Figure S1C) while compromising dynein motor ability at the nuclear envelope (Figure 2D) and at the cortex (Figure 4D). Partial RNAi depletions of dhc-1 and lis-1 therefore allow us to test the contribution of dynein-mediated pulling forces in addition to centrosome–nuclear envelope tethering in centrosome separation in the C. elegans embryo.
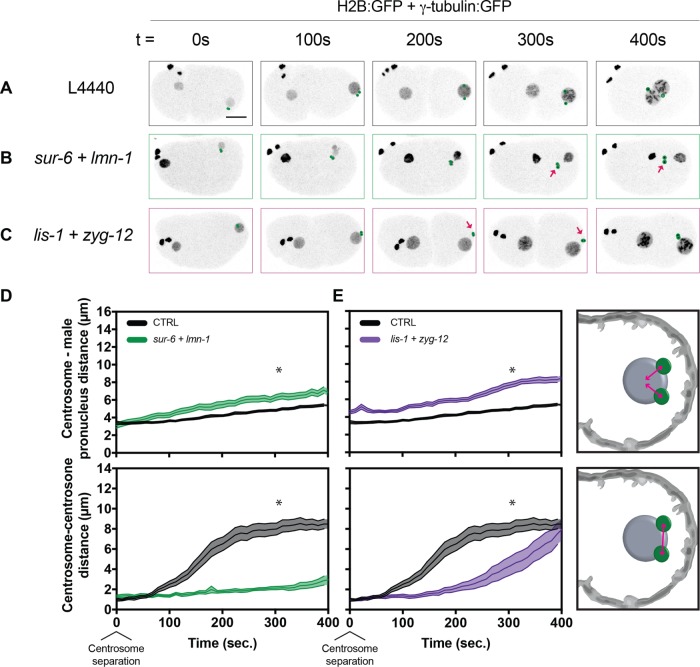
To test whether PP2A-B55/SUR-6 and the nuclear lamina collaborate in separating centrosomes, we codepleted sur-6 and lmn-1 in embryos (Supplemental Video S12). Centrosomes migrated to the center of embryos detached from the male pronuclear envelope and with compromised centrosome–centrosome distances (Figure 6, B and D). The measured centrosome separation phenotype in sur-6 + lmn-1 (Figure 6D) was only slightly more severe than in sur-6 alone (Figure 1B), although a combination of centrosome separation and nuclear envelope–tethering phenotypes were observed. Taken together, results from codepletion of sur-6 and lmn-1 suggest that centrosome separation is achieved through the complementary regulation of dynein-mediated pulling forces and centrosome–nuclear envelope tethering.
FIGURE 6:
Codepleting sur-6 and lmn-1 or lis-1 and zyg-12 leads to defects in both centrosome attachment and separation. (A–C) Representative maximum-intensity projections of embryonic confocal stacks. All RNAi treatments were performed with L4-stage worms expressing H2B:GFP and γ-tubulin:GFP for 24 h via feeding unless otherwise noted. Circles highlight individual centrosomes, and arrows denote detached pairs of centrosomes. Scale bar: 10 µm. (D, E) Centrosome–centrosome distance and centrosome–male pronucleus distance were measured in three dimensions between respective centroids. Error bars indicate SEM and are represented as shaded areas around means. Embryos analyzed per condition: n > 15. Statistics: t test where *p < 0.05. Time t = 0 s corresponds to centrosome separation.
Movie S12.
sur-6 and lmn-1 co-depletion via RNAi in C. elegans zygotes expressing H2B:GFP and γ-tubulin:GFP. Maximum intensity projection of confocal stacks. Related to Figure 6.
To gain mechanistic insight, we then tested whether collaboration of dynein-based pulling forces with centrosome–nuclear envelope tethering to position centrosomes occurs independently of sur-6 effects on pronuclear size and initial centrosome positioning. We reasoned that simultaneously compromising other components of the dynein motor machinery and of the LINC complex would similarly abrogate centrosome separation, as in sur-6 and lmn-1 codepletions. To test this, we codepleted lis-1 and zyg-12 using partial RNAi depletions (Supplemental Video S13 and Figure 6C). Centrosomes became detached from the male pronuclear envelope, and centrosome separation was severely impaired (Figure 6E). We conclude that dynein-mediated pulling forces regulated by LIS-1 and centrosome–nuclear envelope tethering regulated by ZYG-12 collaborate for proper centrosome separation during mitotic entry.
Movie S13.
lis-1 and zyg-12 co-depletion via RNAi in C. elegans zygotes expressing H2B:GFP and γ-tubulin:GFP. Maximum intensity projection of confocal stacks. Related to Figure 6.
Taken together, sur-6 and lmn-1 or lis-1, and zyg-12 codepletions led to additive phenotypes in which centrosomes are detached from the male pronucleus during centrosome separation and fail to separate from one another. In both conditions, centrosome separation is severely compromised, suggesting that dynein-based pulling forces and centrosome–nuclear envelope tethering collaborate through parallel pathways during the early stages of centrosome separation.
DISCUSSION
By combining quantitative live cell-imaging, particle tracking, and computational simulations, we determined that PP2A-B55/SUR-6 contributes to centrosome separation though a pathway that collaborates with the nuclear lamina. sur-6 also affects nuclear size via an unclear mechanism; however, PP2A-B55/SUR-6 regulation through the nuclear export of its negative regulator during mitotic entry in Drosophila has been described previously (Wang et al., 2013) and may prove to be important in the worm embryo. Importantly, nucleo-cytoplasmic trafficking has been shown to regulate nuclear and chromosome size scaling in C. elegans (Ladouceur et al., 2015) as well as nuclear expansion rates (Boudreau et al., 2018), making nucleo-cytoplasmic trafficking a likely regulatory candidate in ensuring proper PP2A-B55/SUR-6 function. In addition to the sur-6 phenotypes considered here, PP2A-B55/SUR-6 may also affect centrosome separation through the regulation of centriole duplication (Song et al., 2011) and centrosome disassembly (Enos et al., 2018), as has been shown for mitotic exit of the first embryonic mitosis.
The respective roles of nuclear envelope and cortical dynein in pulling on centrosomes have recently become clearer (De Simone et al., 2016, 2018; De Simone and Gönczy, 2017). How dynein-based pulling forces are transmitted through the LINC complex to allow for centrosome separation and pronuclear migration have yet to be uncovered. Compromising the LINC complex components zyg-12 or sun-1 caused similar centrosome-separation phenotypes, which were also phenocopied by compromising the nuclear lamina as a whole. Although LMN-1 and SUN-1 are likely required for dynein localization on the nuclear envelope in addition to ZYG-12, models in which the LINC complex functions in tethering centrosomes to the nuclear envelope and in promoting dynein motor activity on the nuclear envelope surface are not mutually exclusive. Partial depletions of dhc-1 and lis-1 led to reduced dynein motor capacity at the nuclear envelope (Figure 2, B and D) and at the cortex (Figure 4, B and D). Importantly, centrosome separation was compromised, while centrosome–nuclear envelope cohesion was maintained (Figure 1C and Supplemental Figure S1). In fact, quantification of endogenous nuclear envelope–associated DHC-1 suggests that DHC-1 amounts are lower in 16-h dhc-1 RNAi treatments compared with zyg-12–compromised embryos (Figure 3). These data suggest that centrosomes are tethered to the nuclear envelope through more than dynein itself. Centrosome–nuclear envelope tethering has also been suggested to occur through ZYG-12 dimerization between outer nuclear envelope–based and centrosome-based pools of the hook protein (Malone et al., 2003). Alternative tethering mechanisms include ZYG-12–microtubule binding, which is supported by the presence of microtubule binding domains in most mammalian hook proteins (Walenta et al., 2001), and through nuclear pore–microtubule binding, as identified in several metazoans (reviewed in Goldberg, 2017).
Several of PP2A-B55/SUR-6’s uncovered roles were evaluated separately through computational simulations (Figure 5). Perturbing pronuclear size (Figure 5C) and nuclear envelope–based dynein density (Figure 5D) led to defects late in centrosome separation, potentially indicating defects in centrosome centration rather than centrosome separation. Similarly, defects in initial centrosome positioning relative to the cortex (Figure 5, F and G) reveal early centrosome separation requirements consistent with previous observations in the initial separation of the male pronucleus (De Simone and Gönczy, 2017). Although the molecular mechanisms by which PP2A-B55/SUR-6 may regulate initial centrosome separation remain elusive, differential dynein densities on female and male pronuclei in sur-6 (Figure 3E) in addition to differential pronuclear sizes (Supplemental Figure S4) may contribute to premature centrosome migration toward the interior of the embryo. Interestingly, centrosome distance in lis-1 + zyg-12 recovered ∼400 s following centrosome separation (Figure 6E), indicating that a collaboration between dynein-based pulling forces and centrosome–nuclear envelope cohesion may only be required early in centrosome separation. This is supported by the finding that embryos without a male pronucleus and compromised cortical pulling forces fail to separate centrosomes until pronuclear meeting (De Simone et al., 2018). On the other hand, initial centrosome positioning in sur-6 (Figure 1B) may play a dominant role beyond centrosome separation (Figure 1F), revealing potential roles of cortical dynein, cytoplasmic dynein, and other regulatory mechanisms at later stages that could not be captured by our computational simulations. Importantly, despite centrosome distance recovery in lis-1 + zyg-12 at later time points, severe chromosome congression and segregation phenotypes were prevalent (Supplemental Video S12), indicating that early centrosome separation and migration are likely critical for subsequent mitotic events.
In conclusion, we propose a model in which PP2A-B55/SUR-6 regulates centrosome separation through dynein-mediated force generation. This force-generation pathway is coordinated with centrosome–nuclear envelope cohesion during mitotic entry to ensure proper centrosome migration and separation. The collaboration between these pathways broadly and between PP2A-B55/SUR-6 and lamin/LMN-1 specifically are critical for ensuring mitotic fidelity and are likely to occur across tissues and organisms.
MATERIALS AND METHODS
Computational simulations
Computational modeling of centrosome dynamics was performed using Cytosim under parameters described previously (De Simone et al., 2016). Brownian motion of elastic fibers and solids in a constant homogenous viscous medium are described by overdamped Langevin equations. The embryo is simulated as an ellipsoid (50 × 30 × 30 μm) confining all embryonic elements. Centrosomes serve as microtubule nucleation sites. Microtubules undergo dynamic instability and can interact with dynein motors that are evenly distributed on pronuclear surfaces and the cell cortex. The female and male pronuclei are at their respective anterior and posterior locations of the embryo at t = 0. Centrosomes are 1.2 μm apart and are located between the male pronucleus and the posterior cortex.
Each condition was simulated 10 times. The position of centrosomes (solids) was exported from Cytosim, and centrosome distance was measured using a script written in Matlab (MathWorks). All changes in simulation parameters, reflecting different perturbations, were performed using experimentally obtained values.
Caenorhabditis elegans use, RNAi, and microscopy
Worm strains were grown and maintained at 20°C using standard procedures. Bacterial strains containing a vector expressing double-stranded RNA under the isopropyl β-d-1-thiogalactopyranoside promoter were obtained from the Ahringer library (Bob Goldstein’s laboratory, University of North Carolina at Chapel Hill, Chapel Hill, NC). Targets were confirmed by sequencing.
Worm strains used were TH32 (pie-1::bg-1::GFP + unc-119(+), pie-1::GFP::H2B + unc-119(+)) and MAS37 (pie-1p::ebp-2::GFP + unc-119(+)). DHC-1:mNeonGreen quantification experiments were performed in a DHC-1:mNeonGreen and mCherry:HIS-58 strain made by crossing LP560 (cp268[dhc::mNG-C1^3×Flag]) I with (pie-1p::mCherry::his-58 + unc-119(+)) IV obtained through a backcross from OD426.
For centrosome distance and DHC-1:mNeonGreen density measurements, worm embryos were mounted in egg buffer (118 mmol/l NaCl, 48 mmol/l KCl, 2 mmol/l CaCl2, 2 mmol/l MgCl2, 25 mmol/l HEPES, pH 7.3) between a 1.5 coverslip and a microscope slide spaced by 22.81-µm glass beads (Whitehouse Scientific) and sealed with Valap (1:1:1 lanolin, petroleum jelly, and parafilm wax). Embryos were then imaged on a Nikon A1r resonant scanning confocal microscope using a 60× Apo water-immersion objective (Nikon), GaASP PMT detectors, and NIS-Elements (Nikon) at 22°C.
Cytoplasmic and cortical microtubule polymerization dynamics were measured using embryos mounted in egg buffer between a 1.5 coverslip and a 4% agarose pad in egg buffer and a microscope slide and sealed with Valap. For cytoplasmic microtubule experiments, embryos were then imaged on a Nikon A1r resonant scanning confocal microscope using the Galvano scanner, a 60× Apo TIRF oil-immersion objective (Nikon), GaASP PMT detectors, and NIS-Elements (Nikon) at 22°C. For cortical microtubule experiments, embryos were then imaged on a Nikon TIRF microscope using a 100× Apo TIRF oil-immersion objective (Nikon), an Andor iXon3 EMCCD, camera and NIS-Elements (Nikon) at 22°C.
Image analysis
All image analysis was performed using Fiji (Schindelin et al., 2012). For centrosome tracking, TrackMate (Tinevez et al., 2017) was used with a DoG detection and the Simple LAP tracker. For microtubule plus-tip tracking, TrackMate was used with a DOG detection and the linear motion LAP tracker. Pronuclear size, DHC-1:mNeonGreen fluorescence intensity measurements, and image processing were performed using custom Fiji plug-ins available upon request.
Supplemental information
The Supplemental Material includes five figures and can be found with this article. Videos are hyperlinked within the article’s text and are available here: https://github.com/viboud12/Centrosome-positioningseparation-bioRxiv.
Supplementary Material
Acknowledgments
We greatly thank Jennifer Heppert (Goldstein lab, UNC–Chapel Hill) for reagents and Tony Perdue (Microscopy Core, Biology Department, UNC–Chapel Hill) for support. We thank Kevin Cannon, Carlos Patiño-Descovich, and Michael Werner for critical reading of the article. V.B. was supported by predoctoral fellowships from the Fonds de Recherche Santé–Québec. This study was also supported by National Science Foundation CAREER Award 1652512 to P.S.M.
Abbreviations used:
- CDK1
cyclin-dependent kinase 1
- GFP
green fluorescent protein
- LINC
linker of nucleoskeleton and cytoskeleton
- NEBD
nuclear envelope breakdown
- PP2A
protein phosphatase 2A
- RNAi
RNA interference
- TIRF
total internal reflection fluorescence.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-10-0631) on March 6, 2019.
REFERENCES
- Afonso O, Matos I, Pereira AJ, Aguiar P, Lampson MA, Maiato H. (2014). Feedback control of chromosome separation by a midzone Aurora B gradient. Science , 332–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau V, Hazel J, Sellinger JK, Chen P, Manakova K, Radzyminski R, Garcia HG, Allard J, Gatlin J, Maddox PS. (2018). Nucleo-cytoplasmic trafficking regulates nuclear surface area during nuclear organogenesis. Biorxiv 326140. [Google Scholar]
- Castilho PV, Williams BC, Mochida S, Zhao Y, Goldberg ML. (2009). The M phase kinase greatwall (Gwl) promotes inactivation of PP2A/B55δ, a phosphatase directed against CDK phosphosites. Mol Biol Cell , 4777–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Worman HJ, Gundersen GG. (2015). Accessorizing and anchoring the LINC complex for multifunctionality. J Cell Biol , 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell MM, Baumer K, Gönczy P. (2004). lis-1 is required for dynein-dependent cell division processes in C. elegans embryos. J Cell Sci , 4571–4582. [DOI] [PubMed] [Google Scholar]
- Cundell MJ, Bastos R, Zhang T, Holder J, Gruneberg U, Novak B, Barr FA. (2013). The BEG (PP2A-B55/ENSA/Greatwall) pathway ensures cytokinesis follows chromosome separation. Mol Cell , 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone A, Gönczy P. (2017). Computer simulations reveal mechanisms that organize nuclear dynein forces to separate centrosomes. Mol Biol Cell , 3165–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone A, Nédélec F, Gönczy P. (2016). Dynein transmits polarized actomyosin cortical flows to promote centrosome separation. Cell Rep , 2250–2262. [DOI] [PubMed] [Google Scholar]
- De Simone A, Spahr A, Busso C, Gönczy P. (2018). Uncovering the balance of forces driving microtubule aster migration in C. elegans zygotes. Nat Commun , 938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enos SJ, Dressler M, Gomes B, Hyman AA, Woodruff JB. (2018). Phosphatase PP2A and microtubule-mediated pulling forces disassemble centrosomes during mitotic exit. Biol Open , bio029777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon-Coral C, Fantana HA, Howard J. (2016). A force-generating machinery maintains the spindle at the cell center during mitosis. Science , 1124–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MW. (2017). Nuclear pore complex tethers to the cytoskeleton. Semin Cell Dev Biol , 52–58. [DOI] [PubMed] [Google Scholar]
- Gönczy P, Pichler S, Kirkham M, Hyman AA. (1999). Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol , 135–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusnowski EM, Srayko M. (2011). Visualization of dynein-dependent microtubule gliding at the cell cortex: implications for spindle positioning. J Cell Biol , 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez PA, Ackermann BE, Vershinin M, McKenney RJ. (2017). Differential effects of the dynein-regulatory factor Lissencephaly-1 on processive dynein-dynactin motility. J Biol Chem , 12245–12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppert JK, Pani AM, Roberts AM, Dickinson DJ, Goldstein B. (2018). A CRISPR tagging-based screen reveals localized players in Wnt-directed asymmetric cell division. Genetics , genetics.300487.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasinska L, Domingo-Sananes M, Kapuy O, Parisis N, Harker B, Moorhead G, Rossignol M, Novák B, Fisher D. (2011). Protein phosphatase 2A controls the order and dynamics of cell-cycle transitions. Mol Cell , 437–450. [DOI] [PubMed] [Google Scholar]
- Ladouceur A-M, Dorn JF, Maddox PS. (2015). Mitotic chromosome length scales in response to both cell and nuclear size. J Cell Biol , 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Bot N, Tsai M-C, Campbell JM, Ahringer J, White JG. (2003). The C. elegans Hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell , 825–836. [DOI] [PubMed] [Google Scholar]
- Mayer-Jaekel R, Ohkura H, Ferrigno P, Andjelkovic N, Shiomi K, Uemura T, Glover D, Hemmings BA. (1994). Drosophila mutants in the 55 kDa regulatory subunit of protein phosphatase 2A show strongly reduced ability to dephosphorylate substrates of p34cdc2. J Cell Sci , 2609–2616. [DOI] [PubMed] [Google Scholar]
- Mehsen H, Boudreau V, Garrido D, Bourouh M, Larouche M, Maddox PS, Swan A, Archambault V. (2018). PP2A-B55 promotes nuclear envelope reformation after mitosis in Drosophila. J Cell Biol , jcb.201804018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerzon M, Gao Z, Liu J, Wu J-C, Malone CJ, Starr DA. (2009). Centrosome attachment to the C. elegans male pronucleus is dependent on the surface area of the nuclear envelope. Dev Biol , 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida S, Ikeo S, Gannon J, Hunt T. (2009). Regulated activity of PP2A–B55δ is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J , 2777–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida S, Rata S, Hino H, Nagai T, Novák B. (2016). Two bistable switches govern M phase entry. Curr Biol , 3361–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazockdast E, Rahimian A, Needleman D, Shelley M. (2017). Cytoplasmic flows as signatures for the mechanics of mitotic positioning. Mol Biol Cell , 3261–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke SM, Carter C, Carter L, Christensen SN, Jones MP, Nash B, Price MH, Turnbull DW, Garner AR, Hamill DR, et al. (2011). A survey of new temperature-sensitive, embryonic-lethal mutations in C. elegans: 24 alleles of thirteen genes. PLoS One , e16644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield L, Wysolmerski B, Mauro M, Farhadifar R, Martinez MA, Biggs R, Wu H-Y, Broberg C, Needleman D, Bahmanyar S. (2018). Dynein-pulling forces counteract lamin-mediated nuclear stability during nuclear envelope repair. Mol Biol Cell , 852–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Wojcik EJ, Sanders MA, McGrail M, Hays TS. (1999). Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J Cell Biol , 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Liu Y, Anderson ED, Jahng W, O’Connell KF. (2011). Protein phosphatase 2A-SUR-6/B55 regulates centriole duplication in C. elegans by controlling the levels of centriole assembly factors. Dev Cell , 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srayko M, Kaya A, Stamford J, Hyman AA. (2005). Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev Cell , 223–236. [DOI] [PubMed] [Google Scholar]
- Tinevez J-Y, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW. (2017). TrackMate: an open and extensible platform for single-particle tracking. Methods , 80–90. [DOI] [PubMed] [Google Scholar]
- Walenta JH, Didier AJ, Liu X, Krämer H. (2001). The Golgi-associated Hook3 protein is a member of a novel family of microtubule-binding proteins. J Cell Biol , 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Galan JA, Normandin K, É Bonneil, Hickson GR, Roux PP, Thibault P, Archambault V. (2013). Cell cycle regulation of Greatwall kinase nuclear localization facilitates mitotic progression. J Cell Biol , 277–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.