Abstract
The Arf and Rab/Ypt GTPases coordinately regulate membrane traffic and organelle structure by regulating vesicle formation and fusion. Ample evidence has indicated that proteins in these two families may function in parallel or complementarily; however, the manner in which Arf and Rab/Ypt proteins perform interchangeable functions remains unclear. In this study, we report that a Golgi-localized Arf, Arl1, could suppress Ypt6 dysfunction via its effector golgin, Imh1, but not via the lipid flippase Drs2. Ypt6 is critical for the retrograde transport of vesicles from endosomes to the trans-Golgi network (TGN), and its mutation leads to severe protein mislocalization and growth defects. We first overexpress the components of the Arl3-Syt1-Arl1-Imh1 cascade and show that only Arl1 and Imh1 can restore endosome-to-TGN trafficking in ypt6-deleted cells. Interestingly, increased abundance of Arl1 or Imh1 restores localization of the tethering factor Golgi associated retrograde–protein (GARP) complex to the TGN in the absence of Ypt6. We further show that the N-terminal domain of Imh1 is critical for restoring GARP localization and endosome-to-TGN transport in ypt6-deleted cells. Together, our results reveal the mechanism by which Arl1-Imh1 facilitates the recruitment of GARP to the TGN and compensates for the endosome-to-TGN trafficking defects in dysfunctional Ypt6 conditions.
INTRODUCTION
The ADP-ribosylation factors (Arfs) are a family of small GTPases that govern vesicular trafficking in eukaryotic cells (D’Souza-Schorey and Chavrier, 2006; Gillingham and Munro, 2007). Arfs have been reported to function in vesicle formation, organelle integrity, and signal transduction (Boman and Kahn, 1995; Donaldson and Jackson, 2011; Jackson and Bouvet, 2014). These small GTPases perform their functions based on their conformation, cycling between a GTP-bound active state and a GDP-bound inactive state regulated by guanine nucleotide–exchange factors (GEFs) and GTPase-activating proteins (GAPs), respectively (Donaldson and Jackson, 2000; Pasqualato et al., 2002).
The class of Arf-like proteins (Arls), which function similarly to Arfs, has been identified as having more than 20 members in mammals and only two in yeast (Lee et al., 1997; Kahn et al., 2006). Among them, Arf-like protein 1 (Arl1) is conserved from yeast to humans and is also the most studied Arl. In yeast, the activity of Arl1 is regulated by its GEF, Syt1, and GAP, Gcs1 (Liu et al., 2005; Chen et al., 2010). Arl1 is known to enforce the transport of the glycosylphosphatidylinositol (GPI)-anchored protein Gas1 to the plasma membrane to maintain cell wall integrity (Liu et al., 2006). Arl1 has also been shown to be involved in selective autophagy under normal growth conditions; this involvement occurs together with Arl3, which is necessary to activate Arl1 and its downstream effectors (Setty et al., 2003, 2004; Wang et al., 2017). Moreover, we have demonstrated that active Arl1 forms a ternary complex with the flippase Drs2 and the Arf GEF Gea2, which mediates the recruitment of the GRIP domain-containing golgin Imh1 to late Golgi (Tsai et al., 2013). Furthermore, Imh1 not only is a downstream effector of Arl1 but also acts as a feedback regulator to maintain and protect the activity of Arl1 from Gcs1-activated GTP hydrolysis (Chen et al., 2012).
Golgins are Golgi-localized proteins that contain coiled-coil regions and are required for tethering events in membrane fusion and for maintenance of the Golgi structure (Munro and Nichols, 1999; Munro, 2011; Witkos and Lowe, 2015). Some golgins target the trans-Golgi membrane via the GRIP domain at their carboxyl terminus. Four GRIP domain-containing golgins have been identified in mammalian cells, Golgin-97, Golgin-245/p230, GCC185, and GCC88, whereas Imh1 is the only one identified in yeast (Munro and Nichols, 1999; Brown et al., 2001; Gillingham and Munro, 2016). Golgin-97, Golgin-245, and Imh1 are recruited to the Golgi membrane by Arl1 and act as downstream effectors (Lu and Hong, 2003; Panic et al., 2003a; Setty et al., 2003; Wu et al., 2004). Despite extensive study of mammalian GRIP domain-containing golgins, the exact and detailed function of Imh1 in vesicular trafficking remains unclear.
Regarding the functions of Imh1, its strong genetic interaction with Ypt6 provides valuable hints, as the deletion of YPT6 along with either ARL1 or IMH1 shows synthetic lethality (Siniossoglou et al., 2000; Setty et al., 2003). Ypt6, the yeast homologue of mammalian Rab6, is associated with the Golgi complex and has been demonstrated to be required for retrograde trafficking from early endosomes to late Golgi (or the trans-Golgi network [TGN] in mammalian cells) (Tsukada et al., 1999; Siniossoglou et al., 2000; Bensen et al., 2001). However, Ypt6 does not appear to localize exclusively to the TGN, because more than 50% of GFP-Ypt6 fluorescence overlaps with the cis-Golgi marker. Recent studies suggest that mammalian Rab6 is important not only for membrane trafficking but also for the organization and maintenance of the Golgi (Starr et al., 2010; Storrie et al., 2012). Ypt6 promotes endosome-derived vesicle fusion via the recruitment of the tethering factor Golgi associated retrograde–protein (GARP) complex, which comprises four subunits, Vps51/52/53/54 (Conibear and Stevens, 2000; Conibear et al., 2003; Donaldson and Jackson, 2000; Siniossoglou and Pelham, 2001). The interaction between Ypt6 and the GARP complex occurs through its Vps52 subunit. Removal of Ypt6 or GARP blocks recycling of the late Golgi SNAREs Tlg1 and Tlg2 and the exocytic SNARE Snc1 from endosomes back to the Golgi (Siniossoglou and Pelham, 2001, 2002).
Arl1 and Ypt6 have been suggested to have overlapping functions and to be in redundant pathways (Bensen et al., 2001; Panic et al., 2003b; Tong et al., 2004; Yang and Rosenwald, 2016). Although Arl1 has also been reported to interact with Vps53 (Siniossoglou and Pelham, 2001; Panic et al., 2003b), disruption of ARL1, unlike that of YPT6, does not affect the localization of the GARP complex at the Golgi or the recycling of Snc1 from endosomes back to the Golgi (Panic et al., 2003b; Liu et al., 2006). Therefore, the role of Arl1 in Ypt6-independent endosome-to-Golgi transport pathways remains to be further elucidated. In this study, we demonstrated that Imh1 mediates recruitment of the GARP complex in the absence of Ypt6. Additionally, increased abundance of Arl1 or Imh1, but not of the N-terminally truncated Imh1, can rescue Ypt6-mediated endosome-to-TGN transport. Our findings provide new insight into the participation of the Arl1-Imh1 complex in Ypt6-mediated retrograde transport.
RESULTS
Overexpression of Arl1 or Imh1 restores endosome-to-TGN transport and suppresses temperature-sensitive growth defects in ypt6Δ cells
The Arl1 and Ypt6 pathways have been proposed to have overlapping functions in regulating vesicle-tethering events at the TGN membrane (Panic et al., 2003b; Graham, 2004). To dissect the functional redundancy between Arl1 and Ypt6, we first determined the defects that resulted from YPT6 deletion. We examined organelle protein markers in the secretory pathway, especially in the Golgi apparatus, in the absence of Ypt6. Consistent with a previous report (Siniossoglou and Pelham, 2001), we observed that the SNARE proteins Snc1 and Tlg1 mislocalized in ypt6-deleted cells from the plasma membrane and endosome/Golgi, respectively, to an intracellular, obscure pattern (Supplemental Figure S1). However, Sec7 (late-Golgi marker), Gos1 (mid-Golgi marker), Sed5 (cis-Golgi marker), FYVE (endosome marker), Vps74 (PtdIns4P marker), and GOLPH3 (PtdIns4P marker) appeared to be unaffected in ypt6Δ cells. These results suggest a defect in targeting rather than a general disruption of the Golgi, as the distribution of several Golgi-localized proteins was unaffected in the ypt6Δ strain.
Although Ypt6 is known for its function on regulating the fusion of endosome-derived vesicles to the late Golgi in yeast (Conibear and Stevens, 2000; Bensen et al., 2001; Siniossoglou and Pelham, 2001; Luo and Gallwitz, 2003; Suda et al., 2013; Makaraci et al., 2018), Rab6, the mammalian homologue of Ypt6, was shown to be involved in the secretory pathway from Golgi to the plasma membrane (Grigoriev et al., 2007; Iwanami et al., 2016). To clarify whether Snc1 mislocalization in ypt6Δ cells is a direct result of blocking the retrograde pathway from endosomes to the late Golgi or exocytic transport from the late Golgi to the plasma membrane, we examined the subcellular localization of endocytic internalization defective Snc1, Snc1-PM (Xu et al., 2017), in wild-type, ypt6Δ, arl1Δ, or imh1Δ cells (Supplemental Figure S2A). We observed that GFP-Snc1-PM was properly transported to the plasma membrane in wild-type, ypt6Δ, arl1Δ, and imh1Δ cells, suggesting that Ypt6, Arl1, and Imh1 do not directly participate in the transport of Snc1 from the late Golgi to the plasma membrane (Supplemental Figure S2A). We also examined the colocalization of GFP-Snc1 and the late Golgi marker Sec7-mRFP in wild-type, ypt6Δ, arl1Δ, and imh1Δ cells. We observed that most of the Snc1 puncta in wild-type, arl1Δ, or imh1Δ, but not ypt6Δ, cells are colocalized with the late Golgi (Supplemental Figure S2B). These data indicate that Snc1 dissociation from the plasma membrane in ypt6Δ cells is due to Snc1-containing vesicles failing to fuse to the late Golgi and confirm that Ypt6 functions in the retrograde pathway from endosomes to the late Golgi.
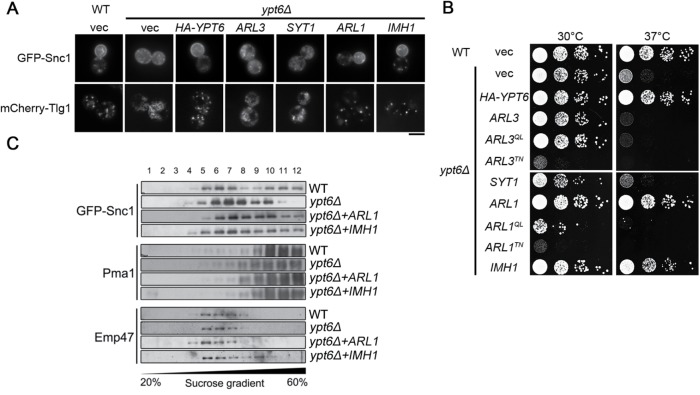
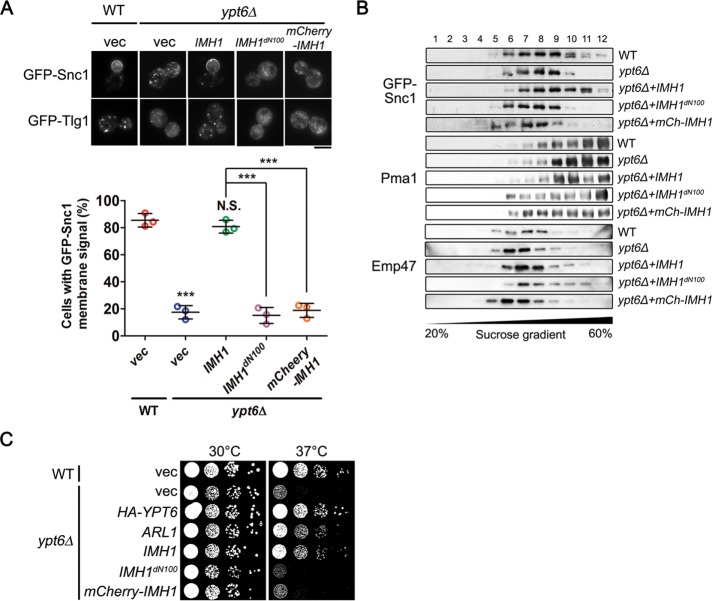
We next examined whether Arl1 or Imh1 could restore the mislocalization of Snc1 and Tlg1 in the absence of Ypt6. While the deletion of ARL1 or IMH1 did not affect the localization of Snc1, Tlg1, and other markers in the secretory pathway (Supplemental Figure S3A), the overexpression of Arl1 or Imh1 restored the plasma membrane distribution of Snc1 and the late Golgi localization of Tlg1 and also suppressed the temperature sensitivity of ypt6Δ cells (Figure 1, A and B, and Supplemental Figure S4A). Interestingly, Arl3 and Syt1, upstream of the Arl1-Imh1 cascade, failed to rescue these phenotypes in ypt6Δ cells. These findings were further verified using sucrose–density gradient centrifugation by observing the distribution of Snc1. Under the disruption of YPT6, Snc1 mislocalized from the higher-density plasma membrane fractions to the lower-density fractions, whereas overexpressing Arl1 or Imh1 restored its distribution (Figure 1C). Our results indicate that Arl1 and Imh1 are able to maintain the retrograde transport of Snc1 and Tlg1 in the absence of Ypt6.
FIGURE 1:
Overexpression of Arl1 or Imh1 restores endosome-to-TGN transport and suppresses temperature-sensitive growth defects in ypt6Δ cells. (A) Overexpression of Arl1 or Imh1, but not Syt1 or Arl3, restores Snc1 and Tlg1 transport defects in ypt6Δ cells. Fluorescently tagged Snc1 or Tlg1 was coexpressed with Arl3, Syt1, Arl1, or Imh1 in ypt6Δ cells. Live cells were observed in mid–log phase using fluorescence microscopy (scale bar, 5 μm). (B) Overexpression of Arl1 or Imh1 suppresses the temperature-sensitive growth defects in ypt6Δ cells. ypt6Δ cells transformed with Ypt6, different forms (wild-type [WT], constitutively active [QL], and constitutively inactive [TN]) of Arl1 or Arl3, and Imh1 were serially diluted 10-fold as indicated, spotted on plates, and incubated at 30 or 37°C. (C) Arl1 and Imh1 restore the distribution of GFP-Snc1 in ypt6Δ cells. WT and ypt6Δ cells were transformed with Arl1 and Imh1. Lysates of spheroplasts from cells were fractionated by sucrose gradient (20–60%) centrifugation. Proteins in the fractions were precipitated, separated by SDS–PAGE, and analyzed by immunoblotting. Pma1 (plasma membrane marker), GFP-Snc1, and Emp47 (early Golgi marker) were identified with specific antibodies and detected using an ECL system. Gradient fractions were numbered from the top.
We further tested whether other proteins in the Arl3-Syt1-Arl1-Imh1 cascade could suppress temperature-sensitive growth defects in the ypt6Δ strain (Supplemental Figure S3B). We found that the malfunction of any gene in the Arl3-Syt1-Arl1-Imh1 cascade would not exhibit temperature-sensitive growth defects (Supplemental Figure S3B). Similarly to the results above, only Arl1 and Imh1, not Arl3 or Syt1, suppressed temperature-sensitive growth defects in ypt6Δ cells. Moreover, both the constitutively active Arl1Q72L (Arl1QL) and the constitutively inactive Arl1T32N (Arl1TN) failed to suppress defective growth at 37°C in ypt6Δ cells, suggesting that Arl1 required proper cycling between the GTP-bound active state and the GDP-bound inactive state to perform its functions (Figure 1B). These findings reveal the functional redundancy between Arl1-Imh1 and Ypt6, suggesting that Arl1 and Imh1 regulate trafficking and compensate for the absence of Ypt6.
Arl1 and Imh1 function interdependently in suppressing defects of endosome-to-TGN transport and temperature-sensitive growth in ypt6Δ cells
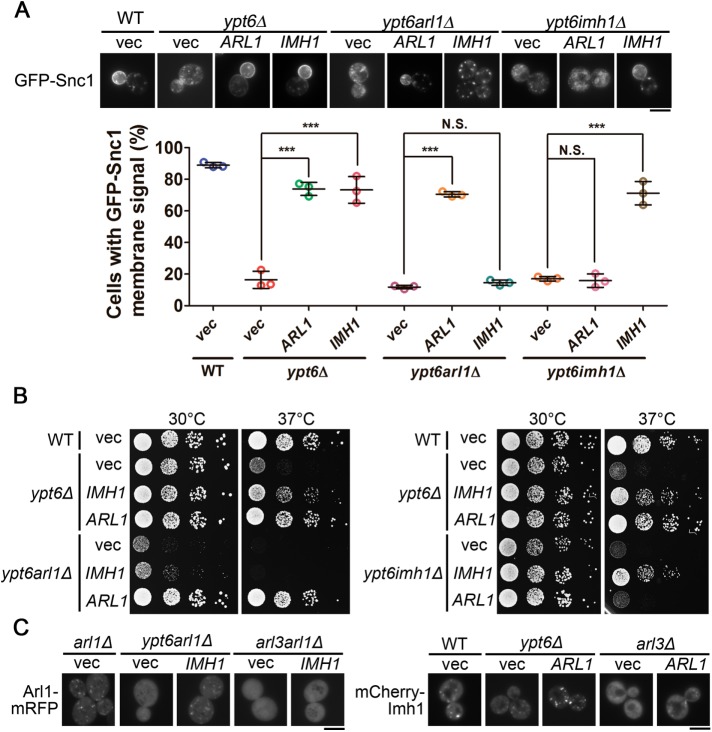
To determine whether Arl1 and Imh1 function in concert to suppress defects in the absence of Ypt6, we first generated a conditional knockout strain by replacing the native promotor of ARL1 with the GAL1 promotor (hereafter termed ypt6arl1Δ). This manipulation mimics ARL1 and YPT6 double deletion in glucose-containing culture medium and avoids the synthetic lethality between ARL1 and YPT6. Compared with ypt6Δ cells, ypt6arl1Δ cells exhibit similar defects in the localization of Snc1 and Tlg1. We observed that overexpression of Arl1, but not of Imh1, restored the subcellular localization of Snc1 and Tlg1 in ypt6arl1Δ cells, suggesting that Imh1 depends on Arl1 for its suppression abilities (Figure 2A and Supplemental Figure S4B). Similarly, overexpression of Imh1, but not of Arl1, restored the trafficking defects of Snc1 and Tlg1 in ypt6imh1Δ cells (Figure 2A and Supplemental Figure S4B). Additionally, Arl1 and Imh1 exhibited interdependence in suppressing temperature-sensitive growth (Figure 2B). These results indicate that Arl1 and Imh1 function coordinately to suppress endosome-to-Golgi trafficking and high temperature–sensitive growth in the absence of Ypt6.
FIGURE 2:
Arl1 and Imh1 act interdependently to suppress defects in endosome-to-TGN transport and temperature-sensitive growth in ypt6Δ cells. (A) Arl1 and Imh1 require each other to suppress the transport defects in ypt6Δ cells. Arl1 or Imh1 was coexpressed with GFP-Snc1 in the indicated cells. Live cells were observed in mid–log phase using fluorescence microscopy. Cells showing the GFP-Snc1 plasma membrane signal were quantified (n = 100). The data are reported as the mean ± SD of three independent experiments (***p < 0.001; N.S. not significant; one-way ANOVA; scale bar, 5 μm). (B) Both Arl1 and Imh1 are required to suppress temperature-sensitive growth defects in ypt6arl1Δ and ypt6imh1Δ cells. Yeast strains transformed with Arl1 or Imh1 were serially diluted 10-fold as indicated, spotted on plates, and incubated at 30 or 37°C. (C) Cytosolic distribution of Arl1 and Imh1 in ypt6Δ cells can be restored by overexpressing Imh1 or Arl1, respectively, in an Arl3-dependent manner. Arl1-mRFP or mCherry-Imh1 was coexpressed with Imh1 or Arl1, respectively, in the indicated cells. Live cells were observed in mid–log phase using fluorescence microscopy (scale bar, 5 μm).
Previous studies have shown that Ypt6 is essential for the Golgi localization of Arl1 and Imh1 (Benjamin et al., 2011). Because we demonstrated the strong interdependence of Arl1 and Imh1 in suppressing the dysfunction of Ypt6, we speculated that the overexpression of either Arl1 or Imh1 would be capable of restoring their localization in the absence of Ypt6. In YPT6-deletion cells, Arl1-mRFP and mCherry-Imh1 were diffuse throughout the cytosol. The dispersed patterns of Arl1-mRFP and mCherry-Imh1 were restored by overexpressing Imh1 and Arl1, respectively (Figure 2C). However, the overexpression of Arl1 or Imh1 did not restore their localization in the absence of Arl3 (Figure 2C), which excluded a potential dominant off-target effect of overexpression. These results again demonstrated the positive-feedback regulation between Arl1 and Imh1, suggesting that they depend on each other for their Golgi localization in ypt6Δ cells.
The Arl1- and Imh1-mediated suppression of Ypt6 dysfunction is independent of the Arf GEF Syt1
Our previous studies have demonstrated that the Arf GEF Syt1 is the only characterized GEF of Arl1. Syt1 is the GEF that is specifically responsible for activating Arl1 to promote the recruitment of the golgin Imh1 to the TGN (Chen et al., 2010). To our surprise, Arl1 and Imh1 were still capable of exerting their suppression effects in the absence of Syt1 (Supplemental Figure S5). In contrast, the ability of Arl1 and Imh1 to suppress high-temperature growth defects in ypt6arl3Δ cells was attenuated. Moreover, we demonstrated that the overexpression of Syt1 was incapable of suppressing defects in endosome-to-Golgi trafficking of ypt6Δ cells (Figure 1A). Together, these data imply that Arl1 is potentially regulated by another unknown GEF and thus can bypass Syt1-mediated activation to support trafficking in ypt6Δ cells.
The Arl1-Drs2-Gea2 ternary complex is not involved in the suppression of defects in endosome-to-TGN transport and temperature-sensitive growth in ypt6Δ cells
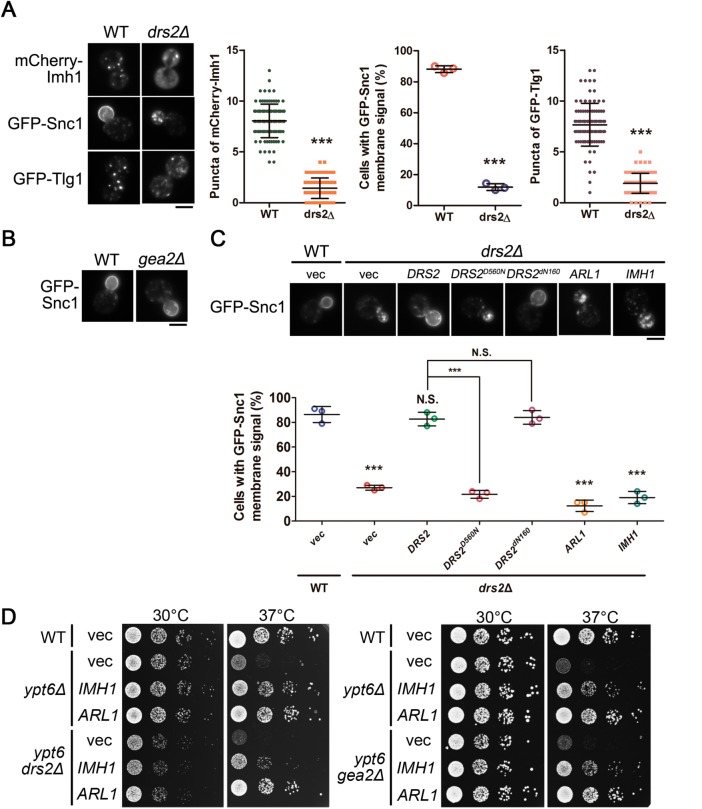
Our previous findings showed that Arl1 forms a complex with Drs2 and the Arf GEF Gea2 at the TGN that mediates the recruitment of the golgin Imh1 (Tsai et al., 2013). To test whether this ternary complex is involved in Arl1- and Imh1-mediated suppression events under conditions of Ypt6 dysfunction, we examined the retrograde transport of Snc1 with DRS2 or GEA2 disruption. Consistent with the previous findings (Hua et al., 2002; Furuta et al., 2007), GFP-Snc1 and GFP-Tlg1 failed to maintain their proper localization in drs2Δ cells (Figure 3A). Interestingly, unlike Drs2, Gea2 did not seem to be involved in the transport of Snc1 (Figure 3B). Therefore, we speculate that instead of functioning as a Drs2-Gea2 complex, Drs2 might perform its own independent function in mediating the retrograde transport of Snc1. To test this hypothesis, we rescued drs2Δ cells with wild-type and different mutant forms of Drs2 and found that wild-type Drs2, but not the flippase activity–defective Drs2D560N, was able to restore the transport of Snc1 in drs2Δ cells. Intriguingly, Drs2dN160, which cannot interact with Arl1 (Tsai et al., 2013), was able to rescue Snc1 retrograde transport in drs2Δ cells (Figure 3C). Moreover, overexpression of Arl1 or Imh1 failed to suppress Snc1 mislocalization in drs2Δ cells (Figure 3C). These results indicate that Drs2 functions in regulating Snc1 in an Arl1-independent manner. We further showed that overexpression of Arl1 or Imh1 restored Snc1 and Tlg1 localization in ypt6gea2Δ cells but not in ypt6drs2Δ cells (Supplemental Figure S6). Moreover, overexpression of Arl1 or Imh1 was still capable of restoring temperature-sensitive growth defects in ypt6drs2Δ and ypt6gea2Δ cells (Figure 3D). Conclusively, Arl1 mediates retrograde transport via an Imh1-dependent, but Drs2-Gea2 complex-independent, pathway in ypt6Δ cells.
FIGURE 3:
The Arl1-Drs2-Gea2 ternary complex is not involved in the suppression of defects in endosome-to-TGN transport and temperature-sensitive growth in ypt6Δ cells. (A) Drs2 mediates the transport of Snc1 and Tlg1. Fluorescently tagged Imh1, Snc1, or Tlg1 was transformed into WT or drs2Δ cells (scale bar, 5 μm). Cells showing the GFP-Snc1 plasma membrane signal were quantified (n = 100) and the data are reported as the mean ± SD of three independent experiments (***p < 0.001; one-way ANOVA). Cells showing the mCherry-Imh1 or GFP-Tlg1 puncta were quantified (n = 100), and the data are reported as the mean ± SD of three independent experiments (***p < 0.001; paired t test). (B) Gea2 is not required for the proper transport of Snc1. GFP-Snc1 was transformed into WT or gea2Δ cells and examined by fluorescence microscopy (scale bar, 5 μm). (C) Arl1 is not involved in Drs2-mediated Snc1 transport. GFP-Snc1 was expressed in wild-type or coexpressed with Arl1, Imh1, or different forms of Drs2 in drs2Δ cells. Live cells were observed in mid–log phase by fluorescence microscopy. Cells showing the GFP-Snc1 plasma membrane signal were quantified (n = 100). The data are reported as the mean ± SD of three independent experiments (***p < 0.001; N.S. not significant; one-way ANOVA; scale bar, 5 μm). (D) The ternary complex of Arl1-Drs2-Gea2 is not involved in Arl1 and Imh1 suppressing temperature-sensitive growth defects in ypt6Δ cells. The indicated strains (wild-type, ypt6Δ, ypt6drs2Δ, ypt6gea2Δ) transformed with Arl1 or Imh1 were serially diluted 10-fold as indicated, spotted on plates, and incubated at 30 or 37°C.
The recruitment of the GARP complex to the TGN by Arl1 and Imh1 is critical for suppressing defects of endosome-to-TGN transport in ypt6Δ cells
The GARP complex, the effector of Ypt6, is a multisubunit tethering complex (MTC) that is responsible for tethering vesicles in endosome-to-Golgi transport at late Golgi (Bonifacino and Hierro, 2011). Ypt6, but not Arl1 or Imh1, is known to regulate the recruitment of the GARP complex under normal conditions (Panic et al., 2003b), and malfunction of the GARP complex also leads to mislocalization of Snc1 and Tlg1, which is similar to the situation in ypt6Δ cells (Conibear et al., 2003; Supplemental Figures S1 and S7). Therefore, we speculated that the GARP complex might be involved in the Arl1- and Imh1-mediated suppression of endosome-to-TGN transport defects in ypt6Δ cells. To test our hypothesis, we first examined whether overexpression of Arl1 or Imh1 restores endosome-to-Golgi trafficking defects in the absence of the GARP complex. We observed that increased abundance of Arl1 or Imh1 failed to suppress the mislocalization of Snc1 and Tlg1 in vps52Δ and vps53Δ cells (Figure 4A), indicating that the GARP complex is strictly required for proper endosome-to-Golgi trafficking. Likewise, ypt6vps53Δ cells still displayed defective transport of Snc1 and Tlg1 upon overexpression of Arl1 or Imh1, again showing that the suppression ability of Arl1 and Imh1 required the presence of the GARP complex (Figure 4B).
FIGURE 4:
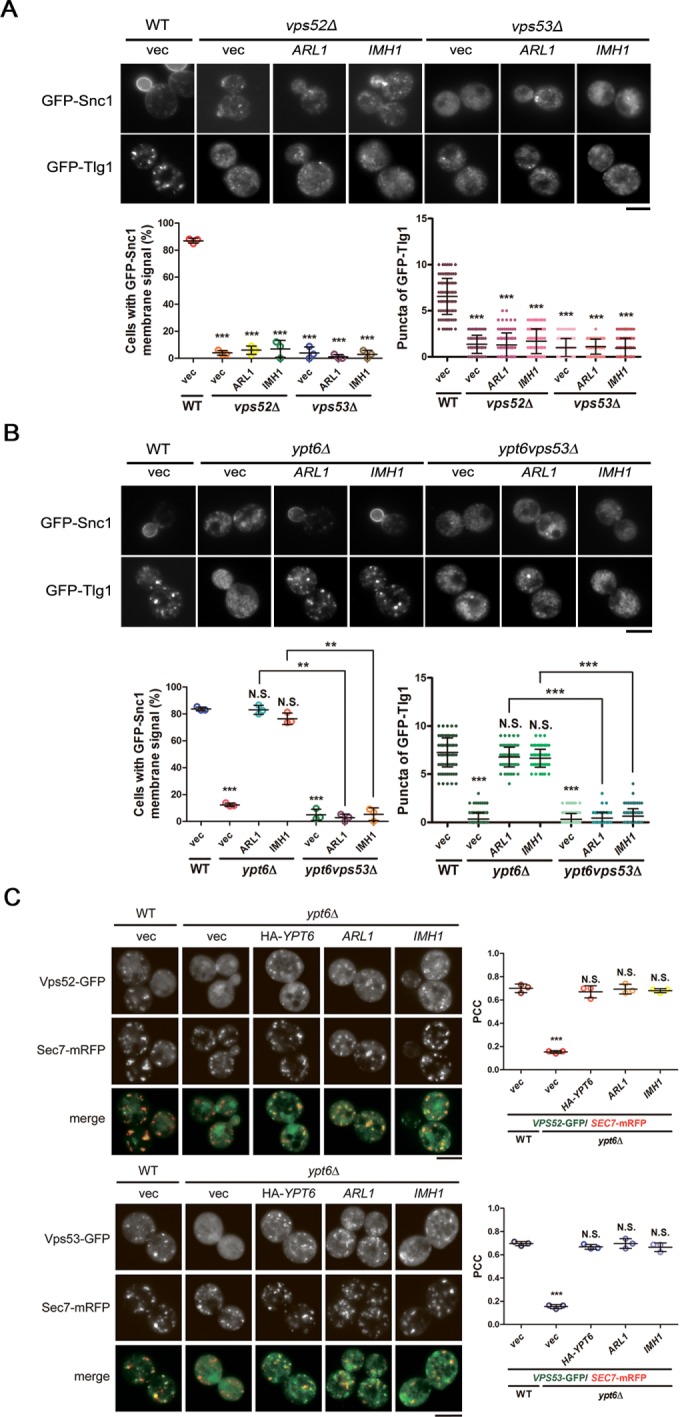
The recruitment of the GARP complex to the TGN by Arl1 and Imh1 is critical for suppressing defects in endosome-to-TGN transport in ypt6Δ cells. (A) Overexpression of Imh1 or Arl1 is unable to suppress GFP-Snc1 and GFP-Tlg1 mislocalization in GARP-complex subunit-deletion strains. Arl1 or Imh1 was coexpressed with GFP-Snc1 or GFP-Tlg1 in vps52Δ and vps53Δ cells. Live cells were observed in mid–log phase by fluorescence microscopy. Cells showing the GFP-Snc1 plasma membrane signal or the GFP-Tlg1 puncta were quantified (n = 100). The data are reported as the mean ± SD of three independent experiments (***p < 0.001; one-way ANOVA; scale bar, 5 μm). (B) The GARP complex is required for Arl1 and Imh1 to suppress Snc1 and Tlg1 transport defects in ypt6Δ cells. Fluorescently tagged Snc1 or Tlg1 was coexpressed with Arl1 or Imh1 in ypt6Δ or ypt6vps53Δ cells. Live cells were observed in mid–log phase using fluorescence microscopy. Cells showing the GFP-Snc1 plasma membrane signal or the GFP-Tlg1 puncta were quantified (n = 100). The data are reported as the mean ± SD of three independent experiments (**p < 0.005, ***p < 0.001; N.S. not significant; one-way ANOVA; scale bar, 5 μm). (C) Overexpression of Arl1 or Imh1 restores the late Golgi puncta of the GARP complex in ypt6Δ cells. The Arl1-, Imh1-, or HA-Ypt6-overexpressed ypt6Δ cells were coexpressed with fluorescently tagged Vps52 or Vps53 and Sec7-mRFP (late Golgi marker). Live cells were observed in mid–log phase using fluorescence microscopy (scale bar, 5 μm). At least 100 cells were included in the experiments from three independent biological repeats. The ratios of colocalization between Vps52/53-GFP and the late Golgi indicator (Sec7-mRFP) were represented as the Pearson correlation coefficient (PCC) using the ImageJ plug-in Just Another Colocalization Plugin with Costes Automatic Thresholding as described in Materials and Methods. The data are presented as the mean ± SD, and the p values were determined applying one-way ANOVA (***p < 0.001; N.S. not significant).
We thus hypothesized that the overexpression of Arl1 and Imh1 in ypt6Δ cells might act to restore proper recruitment of the GARP complex, thus facilitating endosome-to-Golgi trafficking. Therefore, we examined the localization of GARP complex subunits in ypt6Δ cells overexpressing Arl1 or Imh1. We found that increased abundance of Arl1 or Imh1 indeed restored the dispersed pattern of GARP complex subunits into a late Golgi distribution in ypt6Δ cells (Figure 4C). Altogether, our finding indicates that increased abundance of Arl1 or Imh1 is able to recruit the GARP complex back to the TGN in the absence of Ypt6 and further restore the defects of endosome-to-TGN transport in the absence of Ypt6.
The N-terminus of Imh1 is critical for recruiting the GARP complex to the TGN in the absence of Ypt6
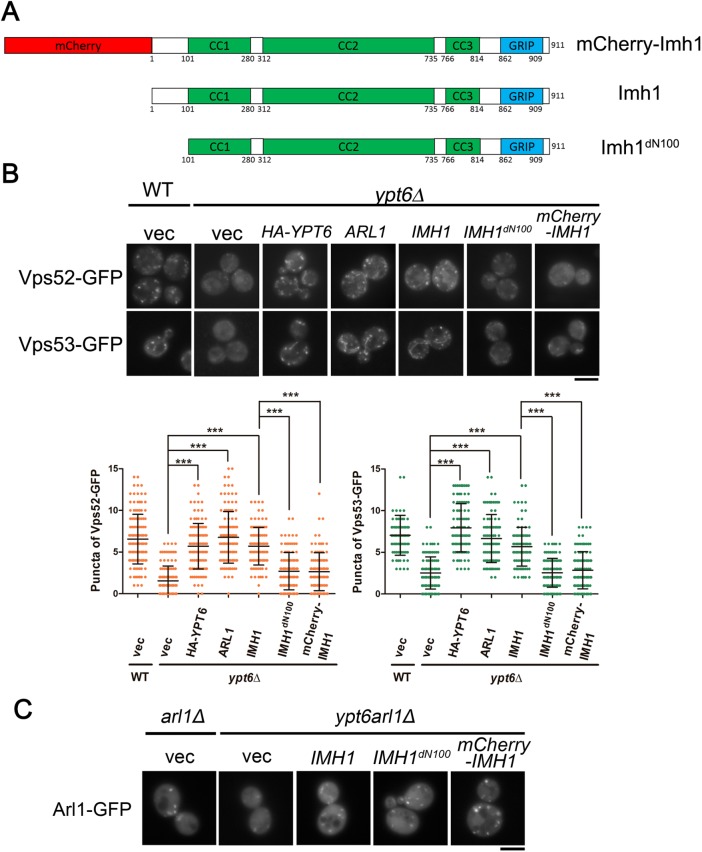
Golgins play important roles in vesicle trafficking and are required for tethering specific cargoes before membrane fusion (Witkos and Lowe, 2015). For most of the golgins, the N-terminal motif provides the ability to recognize and capture specific cargoes (Wong et al., 2017). Accordingly, we speculated that Imh1 also utilizes its N-terminal motif to restore the dispersed GARP complex in ypt6Δ cells. Imh1 contains three coiled-coil domains and one GRIP domain, which have been reported to be involved in dimerization and Arl1 interaction, respectively (Wu et al., 2004). We therefore tested the function of the uncharacterized domain before the first coiled-coil domain at the N-terminal region, deleting the first 100 amino acids of Imh1 (Imh1dN100). On the other hand, the fluorescent protein mCherry was fused to the N-terminus of Imh1 (mCherry-Imh1) to examine whether the tag would mask or obstruct the cargo recognition of Imh1 (Figure 5A). In ypt6Δ cells, the GARP complex preserved its TGN localization with Arl1 or Imh1 overexpression. However, cells overexpressing Imh1dN100 and mCherry-Imh1 could not recruit the GARP complex to the Golgi (Figure 5B), suggesting that the intact N-terminal domain of Imh1 must be exposed for Imh1 to perform its function.
FIGURE 5:
The N-terminal region of Imh1 is critical for recruiting the GARP complex to the TGN in the absence of Ypt6. (A) Schematic diagram of Imh1dN100 and mCherry-Imh1 constructs. (B) The N-terminal region of Imh1 is required for restoring GARP complex localization in ypt6Δ cells. GFP-tagged Vps52 or Vps53 was coexpressed with wild-type Imh1, Imh1dN100, or mCherry-Imh1. Cells containing Vps52-GFP and Vps53-GFP puncta were quantified (n = 150). The data are reported as the mean ± SD of three biological repeats (***p < 0.001; N.S. not significant; one-way ANOVA; scale bar, 5 μm). (C) The N-terminal region of Imh1 is not involved in the localization of Arl1-GFP. Different forms of Imh1 were coexpressed with Arl1-GFP in ypt6arl1Δ cells to examine the localization of Arl1. Live cells were observed in mid–log phase using fluorescence microscopy (scale bar, 5 μm).
Early findings provide evidence that Arl1 associates with the GARP complex through Vps53 (Panic et al., 2003b); therefore, it is possible that Arl1 makes major contributions to GARP complex recruitment in the absence of Ypt6. However, we have demonstrated that the GRIP domain of Imh1 binds to Arl1 to preserve its activity (Chen et al., 2012). The N-terminal mutants of Imh1 in this case, showing defective GARP complex recruitment ability, should hypothetically be functional in regulating Arl1 activity. To confirm our speculation, we examined Arl1 localization in ypt6arl1Δ cells overexpressing Imh1dN100 or mCherry-Imh1. Interestingly, full-length Imh1, Imh1dN100, and mCherry-Imh1 all restored the proper localization of Arl1 (Figure 5C). These results suggested that without the additional function of the N-terminus of Imh1, Arl1 is insufficient for recruiting the GARP complex in the absence of Ypt6.
The N-terminus of Imh1 is essential for suppressing defects of Ypt6 dysfunction
We next asked whether the N-terminus of Imh1 is also involved in suppressing endosome-to-Golgi trafficking defects and temperature-sensitive growth in the absence of Ypt6. We first examined the localization of Snc1 and Tlg1 in ypt6Δ cells overexpressing full-length Imh1, Imh1dN100, or mCherry-Imh1. We observed that full-length Imh1, but not Imh1dN100 or mCherry-Imh1, restored the trafficking defects of these cargoes in ypt6Δ cells (Figure 6A). Consistently, the findings were supported by sucrose-density gradient centrifugation; only full-length Imh1, not Imh1dN100 or mCherry-Imh1, suppressed the improper distribution of GFP-Snc1 in ypt6Δ cells (Figure 6B). We also examined the importance of the N-terminus of Imh1 in suppressing temperature-sensitive growth defects in ypt6Δ cells. In contrast to overexpression of full-length Imh1, overexpression of Imh1dN100 or mCherry-Imh1 failed to suppress temperature-sensitive growth defects in ypt6Δ cells (Figure 6C). In conclusion, we demonstrated that the ability of Imh1 to suppress endosome-to-TGN transport and to suppress temperature-sensitive growth defects depends on the N-terminus of Imh1, as it plays a role in recruiting the GARP complex back to the TGN.
FIGURE 6:
The N-terminal region of Imh1 is essential for restoring defects in endosome-to-Golgi transport and temperature-sensitive growth in ypt6Δ cells. (A) Imh1dN100 and mCherry-Imh1 failed to suppress the Snc1 and Tlg1 transport defects in ypt6Δ cells. Fluorescently tagged Snc1 or Tlg1 was coexpressed with different forms of Imh1 in ypt6Δ cells. Live cells were observed in the mid–log phase, and the percentages of cells with GFP-Snc1 membrane signals were quantified by fluorescence microscopy (n = 100). The data are reported as the mean ± SD of three biological repeats (***p < 0.001; N.S. not significant; one-way ANOVA; scale bar, 5 μm). (B) The N-terminal region is critical for Imh1 to restore the proper GFP-Snc1 cellular distribution in ypt6Δ cells. WT and ypt6Δ cells transformed with different forms of Imh1 were lysed and subjected to centrifugation in a 20–60% sucrose gradient as described in Materials and Methods. Fractions were collected and analyzed by Western blotting using antibodies against Pma1 (plasma membrane marker) and GFP-Snc1 and Emp47 (early Golgi marker). (C) Imh1dN100 and mCherry-Imh1 are unable to suppress temperature-sensitive growth defects in ypt6Δ cells. ypt6Δ cells transformed with different forms of Imh1 were serially diluted 10-fold as indicated, spotted on plates, and incubated at 30 or 37°C.
The Arl1-independent association between Imh1 and the GARP complex does not require the N-terminus of Imh1 but is insufficient for GARP complex recruitment
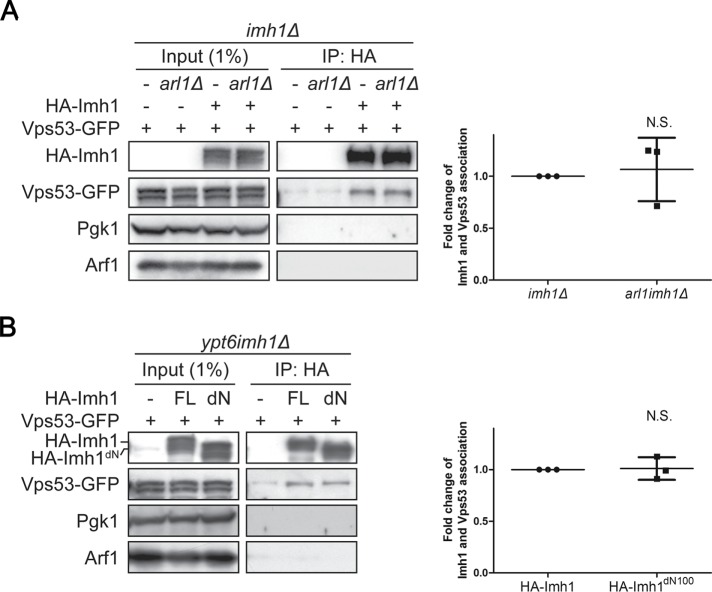
Given that the N-terminal mutants of Imh1 did not alter the localization of Arl1 (Figure 5C), we speculated that the recruitment of the GARP complex might rely mainly on the association with Imh1. To test this hypothesis, we carried out a coimmunoprecipitation assay and showed that Vps53-GFP coprecipitated with HA-Imh1 and that the depletion of Arl1 did not affect their association (Figure 7A). Strikingly, we found that in contrast to full-length Imh1, Imh1dN100 showed no significant difference in Vps53 association (Figure 7B). These results suggest that the N-terminal 100 amino acids of Imh1 have additional functions in restoring trafficking defects in ypt6Δ cells other than associating with the GARP complex.
FIGURE 7:
The Arl1-independent association between Imh1 and the GARP complex does not require the N-terminus of Imh1 but is insufficient for GARP complex recruitment. (A) The association of Imh1 and Vps53 is unaltered in the absence of Arl1. Yeast cells were lysed, centrifuged to collect clear lysate, and incubated with monoclonal anti–HA-agarose antibody. The bound proteins were analyzed by Western blotting. The intensities of bound HA-Imh1, bound Vps53-GFP, and input Vps53-GFP were measured by ImageJ Fiji software. The bound Vps53-GFP signals were normalized to the bound HA-Imh1 and input Vps53-GFP and then the readouts of arl1imh1Δ cells were compared with those of imh1Δ cells. The data are reported as the mean ± SD of three independent experiments (N.S. not significant; paired t test). (B) The N-terminal truncated Imh1 does not disrupt the association between Imh1 and Vps53. Yeast cells were lysed, centrifuged to collect clear lysate, and incubated with monoclonal anti–HA-Agarose antibody. The bound proteins were analyzed by Western blotting. The intensities of bound HA-Imh1, bound Vps53-GFP, and input Vps53-GFP were measured by ImageJ Fiji software. The bound Vps53-GFP signals were normalized to the bound HA-Imh1 and input Vps53-GFP and then the readouts of HA-Imh1dN100-expressing cells were compared with those of HA-Imh1-expressing cells. The data are reported as the mean ± SD of three independent experiments (N.S. not significant; paired t test).
DISCUSSION
The Arf and Ypt/Rab small GTPases are key regulators of membrane trafficking via their mediation of vesicle formation and membrane tethering versus vesicle fusion, respectively. Many reports have indicated that complementary function and cross-talk between these two families are important for membrane trafficking coordination in cells (Jones et al., 1999; Deretic, 2013; McDonold and Fromme, 2014). In this study, we discovered that increased abundance of Arl1 and its effector Imh1 interdependently recruits the GARP complex to the TGN in the absence of Ypt6, and this recruitment subsequently tethers endosome-derived vesicles and suppresses the growth defect. Our findings reveal the molecular mechanism of the functional redundancy between Arl1 and Ypt6 and provide further mechanistic insight into the cross-talk between the Arf and Ypt families.
Genetic interactions between components of the Arl1 and Ypt6 pathways have suggested overlapping functions (Benjamin et al., 2011), while our data show that Arl1 and its effector Imh1 act interdependently to suppress defects of endosome-to-TGN transport and temperature-sensitive growth in ypt6Δ cells and that increased abundance of Arl1 must be activated by a GEF other than Syt1. It will be interesting to identify the unknown Arl1 GEF by screening genes involved in the Arl1-mediated suppression of Ypt6 dysfunction. In addition, our results show that both constitutively active Arl1Q72L and constitutively inactive Arl1T32N failed to suppress defective growth at 37°C in ypt6Δ cells, indicating that Arl1 required proper cycling between the GTP-bound active state and the GDP-bound inactive state to perform its functions.
Membrane asymmetry, curvature, and dynamics are known to have major roles in vesicle transport (McMahon and Boucrot, 2015). Lipid translocases (flippases) have been implicated in vesicle formation through the generation of membrane curvature (Chen et al., 1999). The Graham group previously reported that the Arf GEF Gea2 binds to and stimulates the activity of the flippase Drs2 (Chantalat et al., 2004). Our previous study further demonstrated that activated Arl1 interacts with both Gea2 and Drs2, forming a ternary complex, and that each interaction is necessary for the stability of the complex and is critical for flippase activity at the TGN (Tsai et al., 2013). This Arl1-Drs2-Gea2 complex is specifically required for recruiting the golgin Imh1 to the TGN. Consistent with a previous report that Drs2 is essential for supporting protein transport between endosomes and the TGN (Hua et al., 2002), we show that GFP-Snc1 and GFP-Tlg1 failed to maintain their proper localization in drs2Δ cells (Figure 3A). However, we observed that unlike Drs2, Gea2 is not involved in the transport of Snc1 (Figure 3B), and that Drs2 functions in regulating Snc1 in an Arl1-independent manner. Moreover, the findings from Tanaka’s group have identified that Rcy1, an F-box protein that was reported to be involved in recycling plasma membrane proteins, would bind directly to the Drs2 C-terminal regulatory domain, implying that the role of Rcy1 is to act on Drs2 flippase activity, thus providing another putative Arl1-independent pathway in which Drs2 participates in endosome–to–-late Golgi protein transport (Hanamatsu et al., 2014). On the basis of our results and the report from Tanaka’s group, we speculate that instead of functioning in an Arl1-Drs2-Gea2 complex at the TGN, Drs2 might regulate the retrograde transport of Snc1, which is not based on an Arl1-related mechanism. Therefore, we conclude that the ternary complex formed by Arl1-Drs2-Gea2 is not involved in Arl1- and Imh1-mediated suppression events in ypt6Δ cells.
Cross-talk between the Arl1 and Ypt6 pathways has been suggested by the fact that Arl1 and Ypt6 share the ability to bind GARP through Vps53 and Vps52, respectively (Panic et al., 2003b). The GARP complex functions to tether and facilitate retrograde transport of cargoes, such as Snc1 or Tlg1, to the TGN (Conibear et al., 2003). However, the deletion of YPT6, but not of ARL1, affects GARP complex localization at the TGN; thus, the major recruitment of the GARP complex to the TGN is via Ypt6 (Siniossoglou and Pelham, 2001). Our findings show that neither Imh1dN100 nor mCherry-Imh1 could restore GARP localization and endosome-to-TGN transport in ypt6-deleted cells, although both Imh1dN100 and mCherry-Imh1 can stabilize Arl1 on the Golgi. These results suggest that the interaction between Arl1 and the GARP complex is not sufficient to recruit GARP to the TGN, thus implying that Imh1 may interact with the GARP complex through its N-terminal region. Our results indeed show that Imh1 can associate with GARP in vivo in the absence of Arl1. However, the N-terminal region of Imh1 is not required for its association with the GARP complex, indicating that the association of the GARP complex and Imh1 is not sufficient to restore GARP complex recruitment. Therefore, we hypothesize that the N-terminus of Imh1 may recruit other factors that either interact with the GARP complex or sustain the integrity of the membrane environment that the GARP complex favors. This complex, in turn, performs its appropriate functions to facilitate retrograde transport with its vesicle-tethering ability.
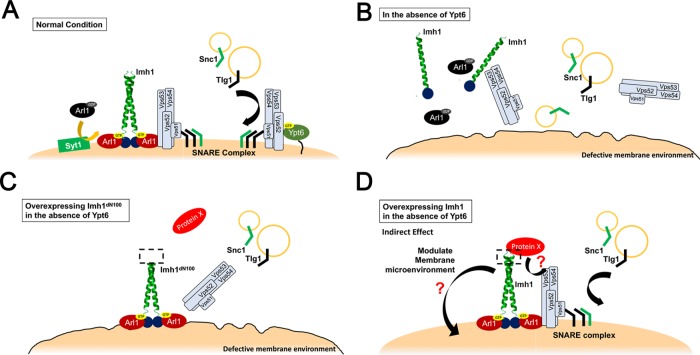
We propose a model (Figure 8) in which, under normal conditions, endosome-to-TGN retrograde transport is mainly mediated by the Ypt6-dependent GARP complex. Dysfunction of YPT6 may lead to a defective Golgi membrane environment, which impairs the TGN localization of Arl1, Imh1, and the GARP complex and leads to defects in endosome-to-TGN transport. Increased abundance of Arl1 is activated in a Syt1-independent manner and subsequently recruits the golgin Imh1 to help restore the GARP complex to the TGN in the absence of Ypt6. Then, Imh1 might recruit a protein X and either associate with the GARP complex through protein X or together with it contribute to restoring the defective membrane environment resulting from YPT6 deletion.
FIGURE 8:
Model of Arl1-Imh1 acting on the Ypt6-independent endosome-to-TGN retrograde transport. (A) Under normal conditions, endosome-to-TGN retrograde transport is mainly mediated by the Ypt6-dependent GARP complex. (B) Dysfunction of YPT6 leads to a defective Golgi membrane environment, which impairs TGN localization of the GARP complex and endosome-to-TGN transport. (C) Overexpression of the N-terminal mutant Imh1 can stabilize active Arl1 at the TGN but cannot recruit the GARP complex to the TGN to promote proper targeting of endosome-derived vesicles. (D) We hypothesize that Imh1 might direct its function through indirect mechanisms, as the association of Imh1dN100 with Vps53 is insufficient to restore GARP localization. Imh1 might recruit a protein X and either associate with the GARP complex through protein X or together with it contribute to restoring the defective membrane environment resulting from YPT6 deletion.
In conclusion, we reveal a novel action of Arl1-Imh1 in compensating for the defects of dysfunctional Ypt6 on endosome-to-TGN traffic by facilitating the recruitment of GARP to the TGN. Our findings identify an unexpected function of Imh1 in addition to fine-tuning Arl1 activation in the absence of Ypt6 and provide a link to Syt1-independent Arl1 activation. The N-terminal domain of Imh1 is critical for restoring GARP localization in ypt6-deleted cells. Previous findings indicate that GARP orchestrates retrograde transport from endosomes to the TGN by promoting vesicle tethering and the assembly of SNARE complexes in consecutive, independent steps (Perez-Victoria and Bonifacino, 2009), yet the molecular mechanism of the spatial and temporal control of GARP recruitment is still unknown. Imh1 is a multiphosphorylated protein and has been reported to potentially interact with many cellular proteins. Further analysis of the regulation of Imh1 will improve our understanding of the mechanism of Arl1 activity and Imh1 function at the late Golgi.
MATERIALS AND METHODS
Strains, plasmids, antibodies, and reagents
The yeast strains used in this study are listed in Supplemental Table S1. The plasmids used in this study are listed in Supplemental Table S2. The pRS416-GFP-SNC1 and pRS416-GFP-SNC1-PM plasmids were generously provided by Todd R. Graham, Vanderbilt University, Nashville, TN. The pRS313-VPS52-GFP and pRS313-VPS53-GFP plasmids were kindly provided by Li-Ting, Jang, National Taiwan University, Taipei, Taiwan.
The following antibodies were used for Western blotting: Emp47 (1:5000), HA (1:5000; Covance, Princeton, NJ), Pma1 (1:5000), GFP (1:5000; Molecular Probes), and Arl1 (1:3000). The anti-Emp47, Pma1, and Arl1 antibodies were generated in our laboratory. The procedures of generating rabbit anti-Emp47 and anti-Arl1 antibodies were referred to in our previous studies (Li et al., 2007; Chiang et al., 2019). Oryctolagus cuniculus (female New Zealand white rabbits) were immunized with purified recombinant Emp47N200 protein (N-terminal 200 residues of Emp47) or Arl1 protein, respectively. Rabbit anti-Pma1 antibodies were prepared against a synthetic peptide corresponding to residues 905–918 of Pma1 (AAMQRVSTQHEKET). All the animal experiments were conducted obeying the institutional guidelines that were approved by the Institutional Animal Care and Use Committee (IACUC), National Taiwan University.
Plating assay for high temperature sensitivity
Mid–log phase yeast cells were spotted with 10-fold serial dilution onto synthetic medium plates. The plates were incubated at 30°C or 37°C for 2 d before imaging.
Microscopy and fluorescent images analysis
Images of live cells containing fluorescent fusion proteins were obtained after overnight culture in synthetic medium with 2% (wt/vol) glucose. The mid–log phase cells were examined, and images were captured using a Zeiss Axioskop microscope equipped with a Cool Snap FX-camera. For all microscopic examinations, the exposure times and image processing procedures were identical for each sample within an experiment. The ratio of cells with membrane signal was determined following the previous reports (Hankins et al., 2015; Xu et al., 2017). The intensities of plasma membrane GFP-Snc1 were detected by ImageJ Fiji software and compared with total fluorescence. At least 100 cells were included in each experiment with three biological repeats. The cells containing Vps52-GFP or Vps53-GFP punctate signals were measured using ImageJ Fiji software (n = 150, three biological repeats). To quantify the colocalization between mCherry-Tlg1 and Sec7-GFP or GFP-Sed5, the Pearson correlation coefficient was assessed by the ImageJ plug-in Just Another Colocalization Plugin with Costes Automatic Thresholding (Bolte and Cordelières, 2006; Xu et al., 2017).
Sucrose density–gradient centrifugation
Wild-type or ypt6∆ yeast cells exogenously expressing full-length Imh1 or Arl1 were cultured in 50 ml of minimal medium to mid–log phase (A600 ∼ 1). Approximately 30 A600 units of yeast cells was resuspended in 1 ml of 20 mM NaN3 and NaF and then incubated with a Zymolyase-100T mixture in 1 ml of K-Pi buffer (190 mM K2HPO4, 310 mM KH2PO4, and 1.2 M sorbitol) containing β-mercaptoethanol to form spheroplasts at 30°C. Spheroplasts were lysed in 5 ml of cold lysis buffer (20 mM HEPES [pH 6.8], 1 mM EDTA [pH 8.0], 50 mM KOAc, 100 mM sorbitol, 1 mM dithiothreitol [DTT], and a protease inhibitor cocktail) with a nitrogen bomb. After 600 × g centrifugation for 10 min, equal amounts of clarified lysates were layered on top of a manually generated sucrose gradient (stacked with a 20–60% sucrose mixture with 500 µl of each layer in a 5% interval) diluted in dilution buffer (40 mM HEPES [pH 6.8], 2 mM EDTA [pH 8.0], 100 mM KOAc), which was then subjected to centrifugation (179,640 × g for 16 h at 4°C in a Beckman SW55-Ti rotor). Fractions were collected manually from the top, precipitated with 10% trichloroacetic acid, mixed with an SDS sample buffer, and analyzed by Western blot analysis.
Protein interaction analysis
To analyze the interaction between Vps53 and Imh1, cells were exogenously transformed with GFP-tagged Vps53 and full-length Imh1 or N-terminal truncated Imh1. Cells were cultured in minimal medium containing 2% glucose lacking amino acids according to the selection of the plasmids. After overnight culture to mid–log phase, yeast cells (100 A600 units) were harvested and washed with 1 ml of 20 mM NaN3 and NaF, lysed with glass beads in 1 ml of lysis buffer (50 mM Tris-Cl [pH 7.5], 50 mM NaCl, 10 mM MgCl2, 1 mM EDTA [pH 8.5], 5% glycerol, 0.15% NP-40, and a protease inhibitor cocktail) at 4°C and subsequently centrifuged at 3500 × g for 10 min to collect clear cell lysate. The clarified cell lysates were then incubated with prewashed monoclonal anti–HA-agarose antibody (Sigma-Aldrich) for 2 h at 4°C and washed three times with lysis buffer. The bound proteins were then analyzed by Western blotting for the presence of GFP-tagged Vps53.
Statistical analysis
All statistical data were analyzed by either paired t test or one-way analysis of variance (ANOVA) followed by Dunnett’s post hoc multiple comparison tests using GraphPad Prism (GraphPad Software) with mean ± SD. Significant statistical differences (***p < 0.001) are indicated.
Supplementary Material
Acknowledgments
We thank Randy Haun, Chia-Jung Yu, and Ya-Wen Liu for their critical review of the manuscript. We also express our deep gratitude to Todd R. Graham and Li-Ting Jang for providing the Snc1 and Vps52/53 expression plasmids. This work was supported by grants from the Ministry of Science and Technology in Taiwan (106-2320-B-002-055-MY3 and 107-3017-F-002-002) and the Center of Precision Medicine from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education in Taiwan to F.S.L.
Abbreviations used:
- Arf
ADP-ribosylation factor
- Arl
Arf-like protein
- GAP
GTPase-activating protein
- GARP
Golgi-associated retrograde protein
- GEF
guanine nucleotide exchange factor
- TGN
trans-Golgi network
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-09-0579) on February 6, 2019.
REFERENCES
- Benjamin JJR, Poon PP, Drysdale JD, Wang X, Singer RA, Johnston GC. (2011). Dysregulated Arl1, a regulator of post-Golgi vesicle tethering, can inhibit endosomal transport and cell proliferation in yeast. Mol Biol Cell , 2337–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen ES, Yeung BG, Payne GS. (2001). Ric1p and the Ypt6p GTPase function in a common pathway required for localization of trans-Golgi network membrane proteins. Mol Biol Cell , 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Cordelières FP. (2006). A guided tour into subcellular colocalization analysis in light microscopy. J Microsc , 213–232. [DOI] [PubMed] [Google Scholar]
- Boman AL, Kahn RA. (1995). Arf proteins: the membrane traffic police? Trends Biochem Sci , 147–150. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Hierro A. (2011). Transport according to GARP: receiving retrograde cargo at the trans-Golgi network. Trends Cell Biol , 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DL, Heimann K, Lock J, Kjer-Nielsen L, van Vliet C, Stow JL, Gleeson PA. (2001). The GRIP domain is a specific targeting sequence for a population of trans-Golgi network derived tubulo-vesicular carriers. Traffic , 336–344. [DOI] [PubMed] [Google Scholar]
- Chantalat S, Park SK, Hua Z, Liu K, Gobin R, Peyroche A, Rambourg A, Graham TR, Jackson CL. (2004). The Arf activator Gea2p and the P-type ATPase Drs2p interact at the Golgi in Saccharomyces cerevisiae. J Cell Sci , 711–722. [DOI] [PubMed] [Google Scholar]
- Chen CY, Ingram MF, Rosal PH, Graham TR. (1999). Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J Cell Biol , 1223–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K-Y, Tsai P-C, Hsu J-W, Hsu H-C, Fang C-Y, Chang L-C, Tsai Y-T, Yu C-J, Lee F-JS. (2010). Syt1p promotes activation of Arl1p at the late Golgi to recruit Imh1p. J Cell Sci , 3478–3489. [DOI] [PubMed] [Google Scholar]
- Chen KY, Tsai PC, Liu YW, Lee F-JS. (2012). Competition between the golgin Imh1p and the GAP Gcs1p stabilizes activated Arl1p at the late-Golgi. J Cell Sci , 4586–4596. [DOI] [PubMed] [Google Scholar]
- Chiang TS, Lin MC, Tsai MC, Chen CH, Jang LT, Lee F-JS. (2019). ADP-ribosylation factor-like 4A interacts with Robo1 to promote cell migration by regulating Cdc42 activation. Mol Biol Cell , 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Cleck JN, Stevens TH. (2003). Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol Biol Cell , 1610–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. (2000). Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell , 305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Chavrier P. (2006). ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol , 347–358. [DOI] [PubMed] [Google Scholar]
- Deretic D. (2013). Crosstalk of Arf and Rab GTPases en route to cilia. Small GTPases , 70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. (2000). Regulators and effectors of the ARF GTPases. Curr Opin Cell Biol , 475–482. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. (2011). ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol , 362–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta N, Fujimura-Kamada K, Saito K, Yamamoto T, Tanaka K. (2007). Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol Biol Cell , 295–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. (2007). The small G proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol , 579–611. [DOI] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. (2016). Finding the Golgi: golgin coiled-coil proteins show the way. Trends Cell Biol , 399–408. [DOI] [PubMed] [Google Scholar]
- Graham TR. (2004). Membrane targeting: getting Arl to the Golgi. Curr Biol , R483–R485. [DOI] [PubMed] [Google Scholar]
- Grigoriev I, Splinter D, Keijzer N, Wulf PS, Demmers J, Ohtsuka T, Modesti M, Maly IV, Grosveld F, Hoogenraad CC, Akhmanova A. (2007). Rab6 regulates transport and targeting of exocytotic carriers. Dev Cell , 305–314. [DOI] [PubMed] [Google Scholar]
- Hanamatsu H, Fujimura-Kamada K, Yamamoto T, Furuta N, Tanaka K. (2014). Interaction of the phospholipid flippase Drs2p with the F-box protein Rcy1p plays an important role in early endosome to trans-Golgi network vesicle transport in yeast. J Biochemistry , 51–62. [DOI] [PubMed] [Google Scholar]
- Hankins HM, Sere YY, Diab NS, Menon AK, Graham TR. (2015). Phosphatidylserine translocation at the yeast trans-Golgi network regulates protein sorting into exocytic vesicles. Mol Biol Cell , 4674–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Fatheddin P, Graham TR. (2002). An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol Biol Cell , 3162–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanami N, Nakamura Y, Satoh T, Liu Z, Satoh AK. (2016). Rab6 is required for multiple apical transport pathways but not the basolateral transport pathway in Drosophila photoreceptors. PLoS Genet , e1005828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CL, Bouvet S. (2014). Arfs at a glance. J Cell Sci , 4103–4109. [DOI] [PubMed] [Google Scholar]
- Jones S, Jedd G, Kahn RA, Franzusoff A, Bartolini F, Segev N. (1999). Genetic interactions in yeast between Ypt GTPases and Arf guanine nucleotide exchangers. Genetics , 1543–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Cherfils J, Elias M, Lovering RC, Munro S, Schurmann A. (2006). Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J Cell Biol , 645–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FJ, Huang CF, Yu WL, Buu LM, Lin CY, Huang MC, Moss J, Vaughan M. (1997). Characterization of an ADP-ribosylation factor-like 1 protein in Saccharomyces cerevisiae. J Biol Chem , 30998–31005. [DOI] [PubMed] [Google Scholar]
- Li CC, Chiang TC, Wu TS, Pacheco-Rodriguez G, Moss J, Lee F-JS. (2007). ARL4D recruits cytohesin-2/ARNO to modulate actin remodeling. Mol Biol Cell , 4420–4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-W, Huang C-F, Huang K-B, Lee F-JS. (2005). Role for Gcs1p in regulation of Arl1p at trans-Golgi compartments. Mol Biol Cell , 4024–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-W, Lee S-W, Lee F-JS. (2006). Arl1p is involved in transport of the GPI-anchored protein Gas1p from the late Golgi to the plasma membrane. J Cell Sci , 3845–3855. [DOI] [PubMed] [Google Scholar]
- Lu L, Hong W. (2003). Interaction of Arl1-GTP with GRIP domains recruits autoantigens Golgin-97 and Golgin-245/p230 onto the Golgi. Mol Biol Cell , 3767–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Gallwitz D. (2003). Biochemical and genetic evidence for the involvement of yeast Ypt6-GTPase in protein retrieval to different Golgi compartments. J Biol Chem , 791–799. [DOI] [PubMed] [Google Scholar]
- Makaraci P, Cruz MD, McDermott H, Nguyen V, Highfill C, Kim K. (2018). Yeast dynamin and Ypt6 function in parallel for the endosome-to-Golgi retrieval of Snc1. Cell Biol Int, 10.1002/cbin.11042. [DOI] [PubMed] [Google Scholar]
- McDonold CM, Fromme JC. (2014). Four GTPases differentially regulate the Sec7 Arf-GEF to direct traffic at the trans-golgi network. Dev Cell , 759–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. (2015). Membrane curvature at a glance. J Cell Sci , 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. (2011). The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb Perspect Biol , a005256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S, Nichols BJ. (1999). The GRIP domain—a novel Golgi-targeting domain found in several coiled-coil proteins. Curr Biol , 377–380. [DOI] [PubMed] [Google Scholar]
- Panic B, Perisic O, Veprintsev DB, Williams RL, Munro S. (2003a). Structural basis for Arl1-dependent targeting of homodimeric GRIP domains to the Golgi apparatus. Mol Cell , 863–874. [DOI] [PubMed] [Google Scholar]
- Panic B, Whyte JR, Munro S. (2003b). The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr Biol , 405–410. [DOI] [PubMed] [Google Scholar]
- Pasqualato S, Renault L, Cherfils J. (2002). Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for “front–back” communication. EMBO Rep , 1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Victoria FJ, Bonifacino JS. (2009). Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans-Golgi network. Mol Cell Biol , 5251–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty SR, Shin ME, Yoshino A, Marks MS, Burd CG. (2003). Golgi recruitment of GRIP domain proteins by Arf-like GTPase 1 is regulated by Arf-like GTPase 3. Curr Biol , 401–404. [DOI] [PubMed] [Google Scholar]
- Setty SR, Strochlic TI, Tong AH, Boone C, Burd CG. (2004). Golgi targeting of ARF-like GTPase Arl3p requires its N alpha-acetylation and the integral membrane protein Sys1p. Nat Cell Biol , 414–419. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Peak-Chew SY, Pelham HRB. (2000). Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. EMBO J , 4885–4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Pelham HR. (2001). An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes. EMBO J , 5991–5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Pelham HR. (2002). Vps51p links the VFT complex to the SNARE Tlg1p. J Biol Chem , 48318–48324. [DOI] [PubMed] [Google Scholar]
- Starr T, Sun Y, Wilkins N, Storrie B. (2010). Rab33b and Rab6 are functionally overlapping regulators of Golgi homeostasis and trafficking. Traffic , 626–636. [DOI] [PubMed] [Google Scholar]
- Storrie B, Micaroni M, Morgan GP, Jones N, Kamykowski JA, Wilkins N, Pan TH, Marsh BJ. (2012). Electron tomography reveals Rab6 is essential to the trafficking of trans-Golgi clathrin and COPI-coated vesicles and the maintenance of Golgi cisternal number. Traffic , 727–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda Y, Kurokawa K, Hirata R, Nakano A. (2013). Rab GAP cascade regulates dynamics of Ypt6 in the Golgi traffic. Proc Natl Acad Sci USA , 18976–18981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. (2004). Global mapping of the yeast genetic interaction network. Science , 808–813. [DOI] [PubMed] [Google Scholar]
- Tsai P-C, Hsu J-W, Liu Y-W, Chen K-Y, Lee F-JS. (2013). Arl1p regulates spatial membrane organization at the trans-Golgi network through interaction with Arf-GEF Gea2p and flippase Drs2p. Proc Natl Acad Sci USA , E668–E677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Will E, Gallwitz D. (1999). Structural and functional analysis of a novel coiled-coil protein involved in Ypt6 GTPase-regulated protein transport in yeast. Mol Biol Cell , 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IH, Chen YJ, Hsu JW, Lee FJ. (2017). The Arl3 and Arl1 GTPases co-operate with Cog8 to regulate selective autophagy via Atg9 trafficking. Traffic , 580–589. [DOI] [PubMed] [Google Scholar]
- Witkos TM, Lowe M. (2015). The golgin family of coiled-coil tethering proteins. Front Cell Dev Biol , 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M, Gillingham AK, Munro S. (2017). The golgin coiled-coil proteins capture different types of transport carriers via distinct N-terminal motifs. BMC Biol , 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Lu L, Hong W, Song H. (2004). Structural basis for recruitment of GRIP domain golgin-245 by small GTPase Arl1. Nat Struct Mol Biol , 86–94. [DOI] [PubMed] [Google Scholar]
- Xu P, Hankins HM, MacDonald C, Erlinger SJ, Frazier MN, Diab NS, Piper RC, Jackson LP, MacGurn JA, Graham TR. (2017). COPI mediates recycling of an exocytic SNARE by recognition of a ubiquitin sorting signal. eLife , e28342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Rosenwald AG. (2016). Autophagy in Saccharomyces cerevisiae requires the monomeric GTP-binding proteins, Arl1 and Ypt6. Autophagy , 1721–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.