Abstract
Objective
The purpose of this study was to investigate the HCRTR2 gene variants rs3122156, rs2653342, and rs2653349 in a large homogenous Swedish case‐control cohort in order to further evaluate the possible contribution of HCRTR2 to cluster headache.
Background
Cluster headache is a severe neurovascular disorder and the pathophysiology is not yet fully understood. Due to striking circadian and circannual patterns of this disease, the hypothalamus has been a research focus in cluster headache. Several studies with many different cohorts from Europe have investigated the hypocretin receptor 2 (HCRTR2) gene, which is expressed in the hypothalamus. In particular, one HCRTR2 single nucleotide polymorphism, rs2653349, has been subject to a number of genetic association studies on cluster headache, with conflicting results. Two other HCRTR2 gene variants, rs2653342 and rs2653349, have been reported to be linked to cluster headache in an Italian study.
Methods
We genotyped a total of 517 patients diagnosed with cluster headache and 581 controls, representing a general Swedish population, for rs3122156, rs2653342, and rs2653349 using quantitative real‐time PCR. Statistical analyses of genotype, allele, and haplotype frequencies for the 3 gene variants were performed comparing patients and controls.
Results
For rs3122156, the minor allele frequency in patients was 25.9% compared to 29.9% in controls (P = .0421). However, this significance did not hold after correction for multiple testing. The minor allele frequencies for rs2653342 (14.7% vs 14.7%) and rs2653349 (19.5% vs 18.8%) were similar for patients and controls. Furthermore, we found one haplotype that was significantly less common in patients than controls (P = .0264). This haplotype included the minor allele for rs3122156 and the major alleles for rs2653342 and rs2653349. Significance did not hold after applying a permutation test.
Conclusions
Our data show a trend for association between cluster headache and the HCRTR2 polymorphism rs3122156, where the minor allele seems to be a protective factor. However, the other 2 HCRTR2 gene variants, including the previously reported rs2653349, were not associated with cluster headache in our Swedish material. A comparison with previous studies points to variance in genotype and allele frequencies among the different populations, which most likely contributes to the opposing results regarding rs2653349. Although the results from this study do not strongly support an association, HCRTR2 remains an interesting candidate gene for involvement in the pathophysiology of cluster headache.
Keywords: polymorphism, haplotype, case‐control study
Abbreviations
- CH
cluster headache
- HCRTR2
hypocretin receptor 2
- HWE
Hardy‐Weinberg equilibrium
- SNP
single nucleotide polymorphism
Introduction
Cluster headache (CH) is a severe neurovascular disorder characterized by recurring attacks of unilateral, excruciating pain, but the pathophysiology is not yet fully understood. The activation of the trigeminal system leading to vasodilatation may be the source of pain and autonomic symptoms, but is not specific to CH.1 As opposed to other primary headache disorders, such as migraine or tension‐type headache, CH shows a distinct circadian and circannual regularity of the attacks in a majority of patients, strongly suggesting involvement of the biological clock, which is regulated in the hypothalamic region of the brain.2, 3, 4 Moreover, it has been determined that the hypothalamus is active during an attack,5 and the secretion of hormones exhibiting a circadian rhythm, such as melatonin and cortisol, is irregular in CH patients.6, 7 For this reason, the hypothalamus has been a main focus of investigations on the pathophysiology of CH.
The neuropeptides hypocretin 1 and 2 (orexin A and B) are selectively synthesized in the lateral and posterior hypothalamus, and serve as ligands for the G protein‐coupled hypocretin (orexin) receptors 1 and 2 (HCRTR1 and HCRTR2).8 These neuropeptides regulate neuroendocrine and autonomic functions, such as arousal, wakefulness, and appetite, via the activation of HCRTR1 and HCRTR2.9, 10 Interestingly, Bartsch et al have demonstrated that HCRTR1/2 activation in the posterior hypothalamus affects the modulation of nociceptive input and might therefore provide a link to head pain and autonomic symptoms commonly seen in primary headaches.11 The HCRTR2 gene has been investigated in more detail, and some studies have shown an association of HCRTR2 with disorders like canine narcolepsy,12 Alzheimer’s disease,13 as well as CH.14, 15, 16
Previous studies have demonstrated a genetic contribution to CH, and 7‒20% of the patients report a first‐, second‐, or third‐degree relative who has also been diagnosed with CH.17, 18 The identification of human sequence polymorphisms that affect genes, mRNA and/or proteins is key to understanding complex genetic diseases. Several candidate genes for CH have been investigated, for example ADH4,19, 20, 21 NOS,22 CACNA1A,23 MTHFR,24 and the 2 clock genes, PER3 25 and CLOCK.26, 27, 28 In this study, we analyzed 3 genetic variants in the gene HCRTR2 that have previously been screened and reported to associate with CH in different CH cohorts in Europe. The single nucleotide polymorphism (SNP) rs2653349 in exon 5 of HCRTR2, is a non‐synonymous variant that leads to an amino acid change from isoleucine to valine in the corresponding protein, which is speculated to have an influence on the dimerization process of the receptor.16 This SNP has been screened and tested for association with CH with conflicting results,14, 15, 20, 29, 30 whereas the intronic HCRTR2 SNPs rs3122156 (intron 1) and rs2653342 (intron 4) were linked to the disease in a haplotype analysis by Rainero et al.16
Materials and Methods
Study Population and Material
The study material consists of 517 CH patients and 581 controls (Table 1) and was obtained after approval by the local ethics committee in Stockholm and informed consent of the research subjects. CH patients (mean age: 52.4 years; 68.3% men) were recruited at the Karolinska University Hospital; 439 of these patients have completed a questionnaire regarding clinical data, medication, and lifestyle. Patients were diagnosed with CH according to the International Classification of Headache Disorders (ICHD‐III) criteria.31 Control subjects were 570 anonymous blood donors with unknown age (54.9% men), as well as 11 neurologically healthy individuals not diagnosed with CH (mean age: 43.5 years; 54.5% men) recruited at Karolinska University Hospital, representing a general Swedish population (https://geblod.nu/english-engelska/). DNA was purified from whole blood samples using the Gentra Puregene Blood Kit (QIAGEN, Hilden, Germany) according to manufacturer’s instructions.
Table 1.
Demographic Characterization
| CH Patients | Controls | |
|---|---|---|
| Number of individuals | 517 | 581 |
| Age (years) | 52.4 | n/a |
| Age at onset (years) | 32.4 | – |
| Male % (n) | 68.3 (353) | 54.9 (319) |
| Heredity † % (n) | 12.5 (59) | n/a |
| Episodic CH % (n) | 89.6 (463) | – |
| Diurnal rhythmicity † % (n) | 68.3 (300) | – |
Based on a subgroup of patients for whom detailed information was available: 472 individuals (heredity) and 439 individuals (diurnal rhythmicity). CH, cluster headache; n/a, not available.
Genotyping by Quantitative Real‐Time PCR (qPCR)
The DNA samples were genotyped using predesigned TaqMan® SNP Genotyping assays on an ABI 7500 FAST Real‐Time PCR instrument (Applied Biosystems, Foster City, CA, USA). The following assays were used: C_429384_20 (rs2653342), C_1507491_10 (rs2653349), C_26549175_10 (rs3122156). Genotyping was run with the predesigned TaqMan® primer and probe (1:80), TaqMan® genotyping master mix (1:2), and 2–5 ng DNA in a total reaction volume of 10 µL using the recommended qPCR conditions for the SNP assays, except for the cycling procedure: 15 seconds at 92ºC and 90 seconds at 60ºC for 55 cycles. A post‐PCR read was performed for allelic discrimination using the 7500 software version 2.0.4 (Applied Biosystems) supplied with the instrument.
Genotyping by Pyrosequencing
For the HCRTR2 polymorphism rs2653342, randomly selected samples were additionally genotyped by pyrosequencing to confirm the TaqMan® assay results, where the distinction between the 3 genotype clusters was consistently poor. Primers (forward: 5′‐CTCGCTGTCATCTTT GTATCCC‐3′; reverse: 5′‐TCGGAGTAACTGGGCAATAGA‐3′) for the preceding PCR and the pyrosequencing primer (5′‐CCTATAAATAGCAC‐3′) were designed using the web‐based softwares Primer332 and mfold.33 Primer sequences were screened on the NCBI webpage (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to confirm specificity. The PCR was carried out using a non‐biotinylated forward and a biotinylated reverse primer as well as Taq polymerase (Thermo Scientific, Ulm, Germany) to amplify fragments. The biotinylated PCR products were immobilized onto streptavidin‐coated sepharose beads (GE Healthcare, Uppsala, Sweden) using a PyroMark® vacuum prep tool (QIAGEN), denatured and purified in 70% ethanol, 0.2 M NaOH, and washing buffer according to manufacturer’s instructions, and finally annealed to the sequencing primer for 2 minutes at 80ºC. The SNP was analyzed using a PyroMark® Reagent Kit and Q96 ID system (QIAGEN). Samples rerun with pyrosequencing corresponded 100% with the TaqMan® assay results for the 3 different genotypes, and the original TaqMan® results were therefore considered reliable.
Statistical Analysis
Power calculations were made with the free power and sample size calculation software PS v3.0.34 With this sample size (n = 1098) and an estimated minor allele frequency of 0.15 in individuals with European descent (http://www.ncbi.nlm.nih.gov/SNP), we have 80% power to detect true odds ratios for CH below 0.59 or above 1.55, respectively. Deviation from the Hardy‐Weinberg equilibrium (HWE) was tested for using the web‐based Online Encyclopedia for Genetic Epidemiology studies software.35 Genotype association was evaluated with chi‐square (χ2) test and allele association was analyzed with Fisher’s exact test or χ2 test (for N > 2000) using GraphPad Prism v5.04 (GraphPad Software Inc, La Jolla, CA, USA). Both tests were run with Bonferroni correction for multiple testing when applicable. For logistic regression analysis with sex as covariate, PLINK whole genome association analysis toolset v1.0736 was used. Haplotype analysis and permutation test with 10,000 permutations were performed in Haploview v4.2.37
In Silico Analysis
To evaluate the possible effect of the non‐synonymous SNP rs2653349 on the mRNA structure of HCRTR2, the secondary structure was predicted using the publicly available software mfold.33 Partial HCRTR2 mRNA sequences, including flanking sequences (70 nucleotides) on each side of the SNP, were analyzed and compared to the wild‐type sequence.
Results
We analyzed 3 SNPs in HCRTR2 (rs3122156, rs2653342, and rs2653349), as shown in Table 2. For each SNP we found all 3 genotypes present in both CH patients and controls. The observed frequencies of patients and controls were in agreement with the HWE for rs2653342 and rs2653349 (data not shown). For rs3122156, only patients were in agreement with HWE (χ2 = 0.27, P = .60), and in the controls we detected an excess of homozygotes (data not shown). Further genetic analysis showed there was a significant difference in occurrence of the less common allele (G) between CH patients and controls (P = .0421) for rs3122156 (Table 2). The G allele was more prevalent in controls (29.9%) than in patients (25.9%), indicating that rs3122156 might constitute a protective factor for CH. However, this significance did not remain after correction for multiple testing. No association with CH was found in genotype or allele frequencies for rs2653342 and rs2653349. Additionally, logistic regression analysis was performed with sex as covariate, in order to exclude any bias introduced by the skewed sex ratio in the patient group (68.3% males in the patient group vs 54.9% males in the control group). The analysis was run under a full genotypic as well as dominant or additive model. Under the dominant and the full genotypic model no association between any of the 3 SNPs and CH was found (data not shown). Using the additive model, there was a significant association with CH for rs3122156, even after correction for multiple testing (Pcorr = 0.0492). In summary, using 2 different statistical methods, we could determine that there is no significant difference on the genotypic level, but a weak association with CH on the allelic level.
Table 2.
Genotype and Allele Frequencies for 3 HCRTR2 SNPs in Controls (n = 581) and CH Patients (n = 517)
| Controls % (n) | CH Patients % (n) | χ2 (df) | OR (95% CI) | P Value | P corr Value | |||
|---|---|---|---|---|---|---|---|---|
| rs3122156 | Genotype | TT | 51.4 (298) | 55.3 (286) | 5.620 (2) | .0602 | ||
| TG | 37.4 (217) | 37.5 (194) | ||||||
| GG | 11.2 (65) | 7.2 (37) | ||||||
| Allele | T | 70.1 (813) | 74.1 (766) | 4.129 (1) | 0.82 (0.68–0.99) | .0421 | .1263 | |
| G | 29.9 (347) | 25.9 (268) | ||||||
| rs2653342 | Genotype | GG | 73.2 (424) | 72.1 (373) | 1.951 (2) | .3770 | ||
| GA | 24.2 (140) | 26.3 (136) | ||||||
| AA | 2.6 (15) | 1.5 (8) | ||||||
| Allele | G | 85.3 (988) | 85.3 (882) | 0.002 (1) | 1.00 (0.79–1.27) | .9622 | ||
| A | 14.7 (170) | 14.7 (152) | ||||||
| rs2653349 | Genotype | GG | 66.3 (385) | 64.2 (332) | 0.854 (2) | .6525 | ||
| GA | 29.9 (174) | 32.5 (168) | ||||||
| AA | 3.8 (22) | 3.3 (17) | ||||||
| Allele | G | 81.2 (944) | 80.5 (832) | 0.165 (1) | 1.05 (0.85–1.30) | .6843 | ||
| A | 18.8 (218) | 19.5 (202) |
χ2, chi‐square; CH, cluster headache; CI, confidence interval; df, degrees of freedom; OR, odds ratio; P corr value, P value corrected for multiple testing.
Since about 68% of the patients who filled out the questionnaire reported that they experience a temporal pattern in attack recurrence, a stratified analysis was performed. In this additional analysis, the genotype and allele frequencies from CH patients with diurnal rhythmicity were compared to controls for the 3 SNPs in HCRTR2. The stratified analysis did not yield a significant association of CH with any of the 3 HCRTR2 SNPs (data not shown).
A haplotype analysis was carried out for the 3 HCRTR2 polymorphisms, and only subjects with a known genotype for all SNPs (n = 1095) were included (Table 3). The position of each SNP in the HCRTR2 haplotypes is as follows: (1) rs3122156, (2) rs2653342, and (3) rs2653349. The haplotype G‐G‐G with the minor allele for rs3122156 and the major alleles for rs2653342 and rs2653349 had a higher frequency in control subjects than CH patients, which suggests that this haplotype is a protective factor (P = .0264). However, the significance was lost when performing a permutation test (Ppermut = .1160).
Table 3.
Haplotype Analysis for 3 HCRTR2 SNPs in CH Patients and Controls
| Haplotype | Control | CH Patients | χ2 (df = 2) | P Value | Ppermut Value | ||
|---|---|---|---|---|---|---|---|
| n | % | n | % | ||||
| T‐G‐G † | 640 | 55.4 | 606 | 58.6 | 2.367 | .1239 | |
| G‐G‐G | 297 | 25.7 | 224 | 21.6 | 4.932 | .0264 | .1160 |
| T‐A‐A | 158 | 13.7 | 148 | 14.3 | 0.161 | .6881 | |
| G‐G‐A | 41 | 3.5 | 42 | 4.1 | 0.482 | .4873 | |
| T‐G‐A | 9 | 0.8 | 10 | 0.9 | 0.116 | .7334 | |
| G‐A‐A | 8 | 0.6 | 2 | 0.2 | 2.545 | .1107 | |
| T‐A‐G | 3 | 0.3 | 2 | 0.2 | 0.101 | .7506 | |
χ2, chi‐square; CH, cluster headache; df, degrees of freedom; Ppermut value, P value after test with 10,000 permutations.
Reference haplotype (T‐G‐G) corresponds to wild‐type allele of each HCRTR2 single nucleotide polymorphism (SNP) where rs3122156 is in position 1, rs2653342 in position 2, and rs2653349 in position 3.
Our in silico analysis of mRNA folding shows that rs2653349 leads to changes in secondary structure and mRNA folding energy (ΔG), 34.34 kcal/mol (A allele) vs 36.70 kcal/mol (G allele) (Fig. 1).
Figure 1.
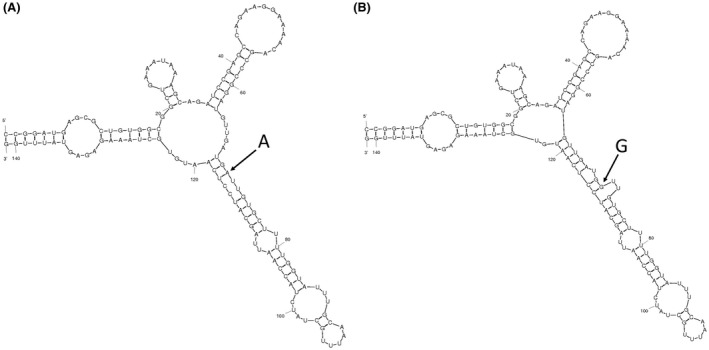
Comparison of mRNA folding with the 2 different rs2653349 alleles. Structures were generated using the mfold program. (A) The mRNA structure with the A allele has an initial folding energy of ΔG = −34.34 kcal/mol. (B) The mRNA structure with the G allele has a mean folding energy of ΔG = −36.70 kcal/mol. The G allele leads to a small additional loop in the mRNA structure.
Discussion
Genotypic and allelic analysis of the investigated HCRTR2 polymorphisms in CH patients suggests that there is no association between rs2653342 or rs2653349 and CH in Sweden. However, a trend for a protective effect of the rs3122156 minor allele G on the HCRTR2 gene could be seen, which is in agreement with an earlier work by Rainero et al.16 Moreover, haplotype analysis of the 3 HCRTR2 polymorphisms revealed a significantly associated protective haplotype, which included the minor allele for rs3122156 and the major alleles for rs2653342 and rs2653349. However, this association is most likely due to presence of the rs3122156 minor allele. Interestingly, the observed frequencies of controls, but not patients, deviated from the HWE for rs3122156. This could possibly be explained by that the locus is under selection, which leads to a false observation of excess homozygotes. When comparing genotype and allele frequencies of patients and controls with data from the 1000 Genomes Project (http://www.ensembl.org/Homo_sapiens/Info/Index) and previous reports for this SNP, it becomes obvious that our control population, as well as controls from Italy (Rainero et al), has a higher frequency for the less frequent homozygous genotype GG (11.2% and 12.8% vs 7.4%) and the G allele (29.9% and 32.0% vs 27.6%) compared to 1000 Genomes (Table 4).16 This might be caused by a positive selection for this SNP in these 2 control populations, but could also depend on genotyping errors or the existence of population stratification. Taken together, these results suggest a protective role for the less common G allele of rs3122156 on the HCRTR2 gene in CH in the Swedish population. Additionally, an in silico analysis of mRNA folding shows that rs2653349 can lead to minor changes in the mRNA structure, which could affect, for example, mRNA stability of HCRTR2.
Table 4.
Comparison of Minor Allele Frequencies of rs3122156, rs2653342, and rs265334
| Study | Minor Allele Frequency | |||||
|---|---|---|---|---|---|---|
| rs3122156 | rs2653342 | rs2653349 | ||||
| CH (n) | Controls (n) | CH (n) | Controls (n) | CH (n) | Controls (n) | |
| Fourier et al | 0.259 (268) | 0.299 (347) | 0.147 (152) | 0.147 (170) | 0.195 (202) | 0.188 (218) |
| 1000 Genomes (EUR) | – | 0.276 (278) | – | 0.147 (148) | – | 0.184 (185) |
| Rainero et al14, 16 | 0.216 (47) | 0.320 (135) | 0.257 (56) | 0.166 (70) | 0.037 (8) | 0.126 (53) |
| Zarilli et al20 | – | – | – | – | 0.120 (13) | 0.078 (31) |
| Schürks et al15 | – | – | – | – | 0.133 (60) | 0.201 (107) |
| Baumber et al29 (British) | – | – | – | – | 0.190 (24) | 0.208 (37) |
| Baumber et al29 (Swedish) | – | – | – | – | 0.173 (34) | 0.220 (46) |
| Baumber et al29 (Danish) | – | – | – | – | 0.219 (42) | 0.271 (39) |
| Weller et al30 | – | – | – | – | 0.210 (242) | 0.227 (397) |
| Fan et al28 | 0.290 (65) | 0.255 (98) | 0.058 (13) | 0.044 (17) | 0.067 (15) | 0.042 (16) |
CH, cluster headache.
Rainero et al16 performed a haplotype analysis of 6 HCRTR2 polymorphisms including the 3 SNPs analyzed in this project. They found a haplotype associated with CH that contains the wild‐type alleles for rs3122156 and rs2653349, and the mutant allele for rs2653342. This allele combination was only found at very low frequencies in our material with no difference between cases and controls. The finding of this significant risk haplotype might be influenced by the strong association found for rs2653349 in Rainero’s cohort. Interestingly, all haplotypes in Rainero’s study containing the allele combination from the protective haplotype G‐G‐G in our study were slightly more common in controls than cases, although not significantly. However, despite the differences in haplotype frequencies and results, this points to a role of the haplotype G‐G‐G in protecting from developing CH across different populations.
An association of rs2653349 with CH was reported in an Italian14 and a German15 cohort, but could not be confirmed in a second Italian cohort,20 a Chinese Han cohort,28 a mixed Northern European (British, Danish, and Swedish) cohort,29 and a Dutch cohort (Table 4).30 The genotype and allele frequencies at rs2653349 observed in our material correspond to those reported for controls in previous studies from Germany, the Netherlands, and in the British and Swedish cohorts from the study by Baumber et al. However, they differ substantially from the 2 Italian studies and Baumber’s Danish cohort. Since our Swedish cohort, with the second largest sample size after the Dutch study, did not show an association with CH, this study adds to the conflicting results of previous ones and no definite conclusion can be drawn regarding rs2653349 and CH. The differences observed in genotype and allele frequencies of the control groups hint that the effect might be population‐dependent.
One possible limitation of our study is the lack of clinical information for our control material since we cannot exclude the occurrence of CH patients in the control group. However, statistically the occurrence of CH should be extremely low, or non‐existent, corresponding to the prevalence of CH in the Swedish population (0.05‐0.1).38
Conclusion
To conclude, in our Swedish cohort we found a trend of association between the protective variant rs3122156 in HCRTR2 and CH. We could not confirm the association between the 2 HCRTR2 SNPs rs2653342 and rs2653349 conferring increased risk for CH as was suggested in previous studies performed by other groups.
Statement of Authorship
Category 1
-
Conception and Design
Carmen Fourier, Caroline Ran, Andrea Carmine Belin
-
Acquisition of Data
Carmen Fourier, Caroline Ran, Anna Steinberg, Christina Sjöstrand, Elisabet Waldenlind
-
Analysis and Interpretation of Data
Carmen Fourier, Caroline Ran, Andrea Carmine Belin
Category 2
-
Drafting the Manuscript
Carmen Fourier
-
Revising It for Intellectual Content
Carmen Fourier, Caroline Ran, Andrea Carmine Belin
Category 3
-
Final Approval of the Completed Manuscript
Carmen Fourier, Caroline Ran, Anna Steinberg, Christina Sjöstrand, Elisabet Waldenlind, Andrea Carmine Belin
Acknowledgments
We thank Fengqing Xiang for excellent technical assistance and Ann‐Christin Karlsson for help with recruitment of patients.
Financial Support: This work was supported by the Swedish Brain Foundation, Swedish Research Council, Swedish Headache Society, and Karolinska Institutet.
Conflict of Interest: None.
References
- 1. May A, Bahra A, Büchel C, et al. PET and MRA findings in cluster headache and MRA in experimental pain. Neurology. 2000;55:1328‐1335. [DOI] [PubMed] [Google Scholar]
- 2. Barloese M, Lund N, Petersen A, et al. Sleep and chronobiology in cluster headache. Cephalalgia. 2015;35:969‐978. [DOI] [PubMed] [Google Scholar]
- 3. Tousson E, Meissl H. Suprachiasmatic nuclei grafts restore the circadian rhythm in the paraventricular nucleus of the hypothalamus. J Neurosci. 2004;24:2983‐2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Steinberg A, Fourier C, Ran C, et al. Cluster headache – clinical pattern and a new severity scale in a Swedish cohort. Cephalalgia. 2018;38:1286‐1295. [DOI] [PubMed] [Google Scholar]
- 5. May A, Bahra A, Büchel C, et al. Hypothalamic activation in cluster headache attacks. Lancet. 1998;352:275‐278. [DOI] [PubMed] [Google Scholar]
- 6. Waldenlind E, Gustafsson SA, Ekbom K, et al. Circadian secretion of cortisol and melatonin in cluster headache during active cluster periods and remission. J Neurol Neurosurg Psychiatry. 1987;50:207‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leone M, Bussone G. A Review of Hormonal findings in cluster headache. Evidence for hypothalamic involvement. Cephalalgia. 1993;13:309‐317. [DOI] [PubMed] [Google Scholar]
- 8. Sakurai T, Amemiya A, Ishii M, et al. Orexins and orexin receptors: A family of hypothalamic neuropeptides and G protein‐coupled receptors that regulate feeding behavior. Cell. 1998;92:573‐585. [DOI] [PubMed] [Google Scholar]
- 9. Date Y, Ueta Y, Yamashita H, et al. Orexins, orexigenic hypothalamic peptides, interact with autonomic, neuroendocrine and neuroregulatory systems. Proc Natl Acad Sci U S A. 1999;96:748‐753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Willie JT, Chemelli RM, Sinton CM, et al. To eat or to sleep? Orexin in the regulation of feeding and wakefulness. Annu Rev Physiol. 2001;24:429‐458. [DOI] [PubMed] [Google Scholar]
- 11. Bartsch T, Levy MJ, Knight YE, et al. Differential modulation of nociceptive dural input to [hypocretin] orexin A and B receptor activation in the posterior hypothalamic area. Pain. 2004;109:367‐378. [DOI] [PubMed] [Google Scholar]
- 12. Lin L, Faraco J, Li R, et al. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell. 1999;98:365‐376. [DOI] [PubMed] [Google Scholar]
- 13. Gallone S, Boschi S, Rubino E, et al. Is HCRTR2 a genetic risk factor for Alzheimer’s disease? Dement Geriatr Cogn Disord. 2014;38:245‐253. [DOI] [PubMed] [Google Scholar]
- 14. Rainero I, Gallone S, Valfrè W, et al. A polymorphism of the hypocretin receptor 2 gene is associated with cluster headache. Neurology. 2004;63:1286‐1288. [DOI] [PubMed] [Google Scholar]
- 15. Schürks M, Kurth T, Geissler I, et al. Cluster headache is associated with the G1246A polymorphism in the hypocretin receptor 2 gene. Neurology. 2006;66:1917‐1919. [DOI] [PubMed] [Google Scholar]
- 16. Rainero I, Gallone S, Rubino E, et al. Haplotype analysis confirms the association between the HCRTR2 gene and cluster headache. Headache. 2008;48:1108‐1114. [DOI] [PubMed] [Google Scholar]
- 17. Russell MB, Andersson PG, Thomsen LL. Familial occurrence of cluster headache. J Neurol Neurosurg Psychiatry. 1995;157:341‐343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leone M, Russell MB, Rigamonti A, et al. Increased familial risk of cluster headache. Neurology. 2001;56:1233‐1236. [DOI] [PubMed] [Google Scholar]
- 19. Rainero I, Rubino E, Gallone S, et al. Cluster headache is associated with the alcohol dehydrogenase 4 (ADH4) gene. Headache. 2010;50:92‐98. [DOI] [PubMed] [Google Scholar]
- 20. Zarrilli F, Tomaiuolo R, Ceglia C, et al. Molecular analysis of cluster headache. Clin J Pain. 2015;31:52‐57. [DOI] [PubMed] [Google Scholar]
- 21. Fourier C, Ran C, Steinberg A, et al. Screening of two ADH4 variations in a Swedish cluster headache case‐control material. Headache. 2016;56:835‐840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sjöstrand C, Modin H, Masterman T, et al. Analysis of nitric oxide synthase genes in cluster headache. Cephalalgia. 2002;22:758‐764. [DOI] [PubMed] [Google Scholar]
- 23. Sjöstrand C, Giedratis V, Ekbom K, et al. CACNA1A gene polymorphisms in cluster headache. Cephalalgia. 2001;21:953‐958. [DOI] [PubMed] [Google Scholar]
- 24. Schürks M, Neumann FA, Kessler C, et al. MTHFR 677C>T polymorphism and cluster headache. Headache. 2011;51:201‐207. [DOI] [PubMed] [Google Scholar]
- 25. Ofte HK, Tronvik E, Alstadhaug KB. Lack of association between cluster headache and PER3 clock gene polymorphism. J Headache Pain. 2016;17:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cevoli S, Mochi M, Pierangeli G, et al. Investigation of the T3111C CLOCK gene polymorphism in cluster headache. J Neurol. 2008;255:299‐300. [DOI] [PubMed] [Google Scholar]
- 27. Fourier C, Ran C, Zinnegger M, et al. A genetic CLOCK variant associated with cluster headache causing increased mRNA levels. Cephalagia. 2018;38:496‐502. [DOI] [PubMed] [Google Scholar]
- 28. Fan Z, Hou L, Wan D, et al. Genetic association of HCRTR2, ADH4 and CLOCK genes with cluster headache: A Chinese population‐based case‐control study. J Headache Pain. 2018;19:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baumber L, Sjöstrand C, Leone M, et al. A genome‐wide scan and HCRTR2 candidate gene analysis in a European cluster headache cohort. Neurology. 2006;66:1888‐1893. [DOI] [PubMed] [Google Scholar]
- 30. Weller CM, Wilbrink LA, Houwing‐Duistermaat JJ, et al. Cluster headache and the hypocretin receptor 2 reconsidered: A genetic association study and meta‐analysis. Cephalalgia. 2015;35:741‐747. [DOI] [PubMed] [Google Scholar]
- 31. Headache Classification Committee of the International Headache Society . The International Classification of Headache Disorders, 3rd edition (beta version). Cephalalgia. 2013;33:629‐808. [DOI] [PubMed] [Google Scholar]
- 32. Untergasser A, Cutcutache I, Koressaar T, et al. Primer3 – new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406‐3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dupont WD, Plummer WD. Power and sample size calculations: A review and computer program. Control Clin Trials. 1990;11:116‐128. [DOI] [PubMed] [Google Scholar]
- 35. Rodriguez S, Gaunt TR, Day INM. Hardy‐Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol. 2009;169:505‐514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Purcell S, Neale B, Todd‐Brown K, et al. PLINK: A tool set for whole‐genome association and population‐based linkage analyses. Am J Hum Genet. 2007;81:559‐575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barrett JC, Fry B, Maller J, et al. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263‐265. [DOI] [PubMed] [Google Scholar]
- 38. Ekbom K, Svensson DA, Pedersen NL, et al. Lifetime prevalence and concordance risk of cluster headache in the Swedish twin population. Neurology. 2006;67:798‐803. [DOI] [PubMed] [Google Scholar]


