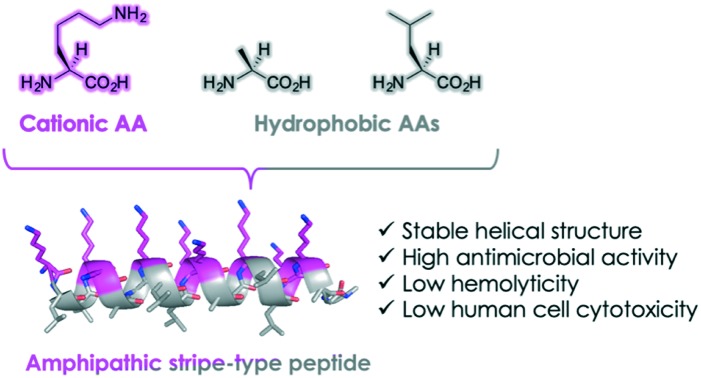
 Amphipathic helical peptide Stripe showed high antimicrobial activity, low hemolytic activity, and low human cell cytotoxicity.
Amphipathic helical peptide Stripe showed high antimicrobial activity, low hemolytic activity, and low human cell cytotoxicity.
Abstract
Antimicrobial peptides (AMPs) have garnered much attention as novel therapeutic agents against infectious diseases. They exhibit antimicrobial activity through microbial membrane disruption based on their amphipathic properties. In this study, we rationally designed and synthesized a series of novel AMPs Block, Stripe, and Random, and revealed that Stripe exhibits potent antimicrobial activity against Gram-positive and Gram-negative microbes. Moreover, we also demonstrated that Stripe disrupts both Gram-positive and Gram-negative mimetic bacterial membranes. Finally, we investigated the hemolytic activity and cytotoxicity in human blood cells and human cell lines, and found that Stripe exhibited neither. These data indicated that Stripe is a promising antimicrobial reagent that does not display significant cytotoxicity.
Introduction
Infectious disease is still a considerable problem, causing many deaths each year, despite advances in modern medicine. Currently, antimicrobial drugs that inhibit proteins expressed in microbial cells are widely used to combat these diseases.1 However, long-term administration of antimicrobial drugs can lead to the generation of multidrug-resistant bacteria (MRB), so novel antimicrobial drugs are urgently needed.2 To overcome this problem, antimicrobial peptides (AMPs), such as defensin and magainin 2 (Mag2), have garnered much attention as next-generation drugs for MRB.3Mag2, as a representative example, is an antimicrobial peptide first identified in the skin of frogs, which exerts antimicrobial activity and functions via an immuno-protective pathway.4 AMPs have amphipathic properties and function through their interaction with the microbial cell membrane, such that they insert themselves into the membrane and disrupt its structure.5 Because AMPs target the microbial membrane, the resistance against AMPs is less likely to occur than with known antimicrobial compounds.6 AMPs, therefore, represent promising reagents for MRB therapies through their actions within the microbial membrane.
Previously, we designed and synthesized a set of Lys-based amphipathic peptides and evaluated their antimicrobial activities.7 We demonstrated that the synthesized peptides form stable helical structures exhibiting potent antimicrobial activities, indicating that the helicity of peptides plays an important role in their functions. However, the antimicrobial activity of the peptides reported in the previous study7 was lowered than that of Mag2. We hypothesized that the peptide length and the amphipathic properties of these peptides were not sufficient to exert the antimicrobial activity. It also remains unclear how the distribution of cationic and hydrophobic amino acid residues in the peptides affects their anti-microbial activities. In the present study, we investigated the effect of changing the distribution of cationic amino acid residues in the helical peptide sequences and measured their antimicrobial activities as well as their hemolytic activities against human blood cells.
Results and discussion
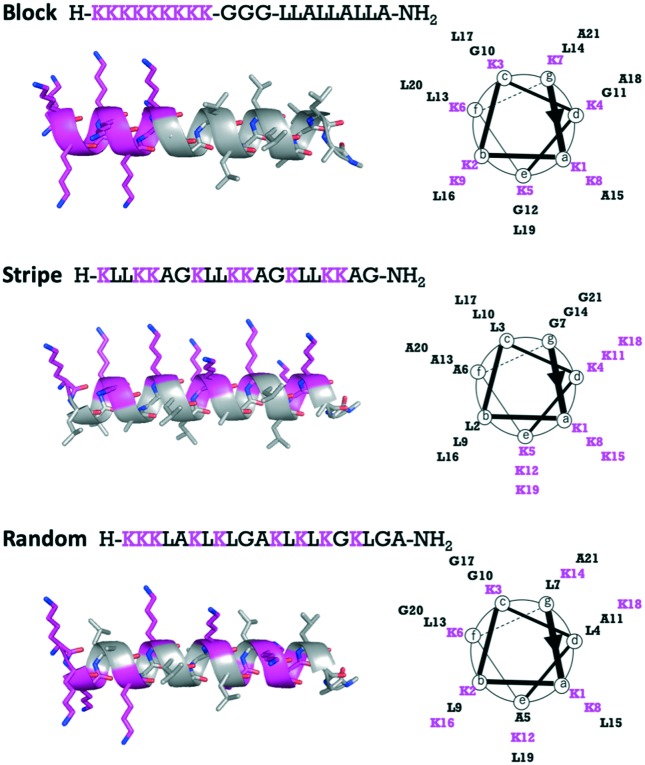
To investigate the importance of the distribution of l-Lys residues throughout the entire peptide sequences, we designed three Lys-based AMPs (Block, Stripe, and Random) containing the natural amino acids Leu and Ala found frequently in the helical motif of natural proteins8,9 and Gly as a spacer (Fig. 1). The three peptides Block, Stripe, and Random were composed of the same amino acid residues, but the distribution of the Lys residues was altered. That is, all Lys residues of Block were located at the N-terminus and hydrophobic amino acids Leu and Ala were located at the C-terminus. In Stripe, the Lys residues were located on one side of the helical structure and the Leu and Ala residues were located on the other side. In Random, all amino acid residues were scrambled and equally spaced throughout the entire peptide sequence. The designed peptides were synthesized using microwave-assisted Fmoc solid phased synthesis. Subsequently, the synthesized peptides were purified with high-performance liquid phase chromatography (HPLC) and the peptides were identified using liquid phase chromatography-mass spectrometry (LC-MS).
Fig. 1. Peptide sequences, structure (side view) and helical wheel (top view) of Block, Stripe, and Random.
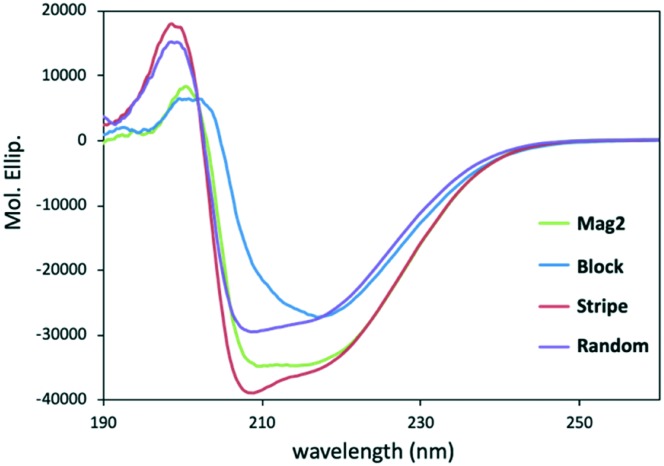
After having synthesized and purified the new peptides, we performed a secondary structural analysis of Mag2, Block, Stripe, and Random using CD spectroscopy in 20 mM phosphate buffer solution (pH = 7.4) with 1% sodium dodecyl sulfate (SDS), which has been reported to mimic the environment close to the cell membrane.10 As shown in Fig. 2, the peptides Block, Stripe, and Random showed negative maxima at around 208 and 222 nm,10,11 indicating that they formed α-helical structures similar to that of Mag2. In particular, the spectral intensity of Stripe was strong compared with those of other peptides, and this could be used as an antimicrobial agent (Fig. 2).
Fig. 2. Secondary structures of Mag2, Block, Stripe, and Random in 20 mM phosphate buffer solution with 1% SDS solution, measured using CD spectral analysis. Peptide concentration = 200 μM.
Second, we evaluated the effects of the peptides on several microbes. These were Gram-positive Staphylococcus aureus and Staphylococcus epidermidis and Gram-negative Escherichia coli NBRC 3972, Escherichia coli DH5α, Proteus vulgaris NBRC 3045, and Pseudomonas aeruginosa NBRC 13275. As shown in Table 1, Block and Stripe showed potent and broad antimicrobial activity across the spectra of bacterial species, with the exception of P. vulgaris. In particular, Stripe exhibited 16-fold higher antimicrobial activity against S. epidermidis than Mag2 [minimum inhibitory concentration (MIC) values against S. epidermidis: Mag2, 25 μM; Stripe, 1.56 μM]. On the other hand, Random exhibited comparable antimicrobial activity against the examined microbes to Mag2. These results indicated that the distribution of cationic amino acid residues plays an important role in peptides like these to enable them to exert their antimicrobial activities, and that the strategy of placing Lys residues at the N-terminus or on one side of a helical structure can be taken advantage of, to design novel antimicrobial peptides.
Table 1. Antimicrobial activity of peptides against several microbes.
| Peptide | MIC (μM) |
|||||
| Gram(+) |
Gram(–) |
|||||
| S.a. | S.e. | DH5α | E.c. | P.v. | P.a. | |
| Mag2 | 100 | 25 | 3.125 | 3.125 | >100 | 12.5 |
| Block | 12.5 | 6.25 | 12.5 | 6.25 | >100 | 6.25 |
| Stripe | 12.5 | 1.56 | 3.125 | 3.125 | >100 | 1.56 |
| Random | 100 | 6.25 | 25 | 12.5 | >100 | 3.125 |
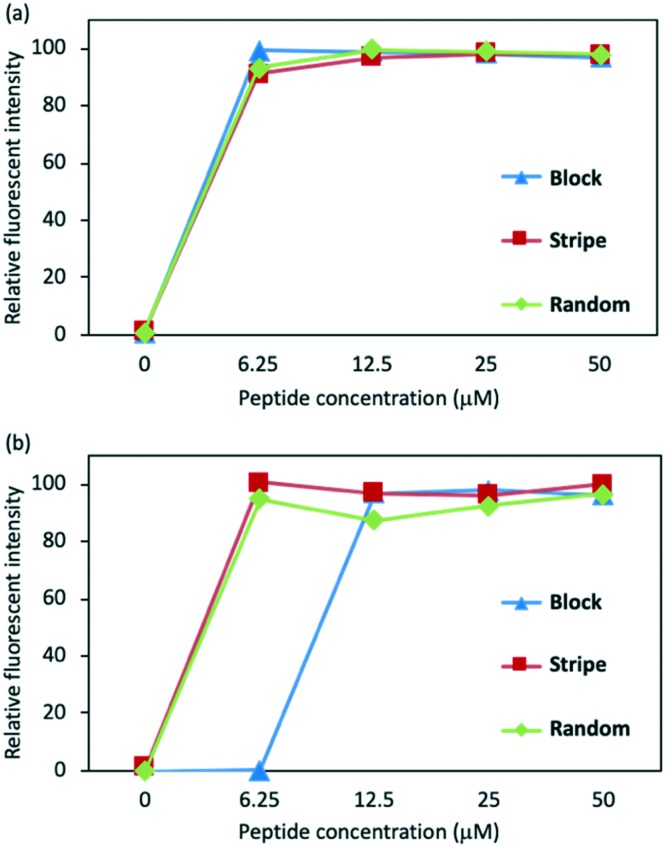
Next, we investigated whether the peptides could disrupt the microbial membrane as we hypothesized. To test this, we conducted the fluorescein leakage assay to evaluate the lytic activity against the microbial membrane. The fluorescein probe, carboxyfluorescein, encapsulated by unilamellar membrane liposomes, was prepared as previously reported,12,13 containing different proportions of lipids to mimic Gram positive or Gram negative microbial membranes. The liposomes encapsulating carboxyfluorescein were treated with Block, Stripe, and Random. The fluorescence leakage upon treatment with the peptides was estimated by measuring the fluorescence intensity. As shown in Fig. 3, the treatment of Block, Stripe, and Random at 6.25 μM induced the increase of the fluorescence intensity, indicating that these peptides may work as membrane disrupters and possibly exert their antimicrobial activities.
Fig. 3. Fluorescence leakage assay with lipid bilayer mimicking (a) Gram-positive and (b) negative microbial membranes. Composition of the lipid bilayer mimicking Gram-positive, phosphatidylglycerol (PG) : cardiolipin (CL) = 3 : 1; Gram-negative, PG : CL: phosphatidylethanolamine (PE) = 2 : 7 : 1.14 Fluorescence intensity at 6.25 μM treatment of Triton X-100 is defined as 100%.
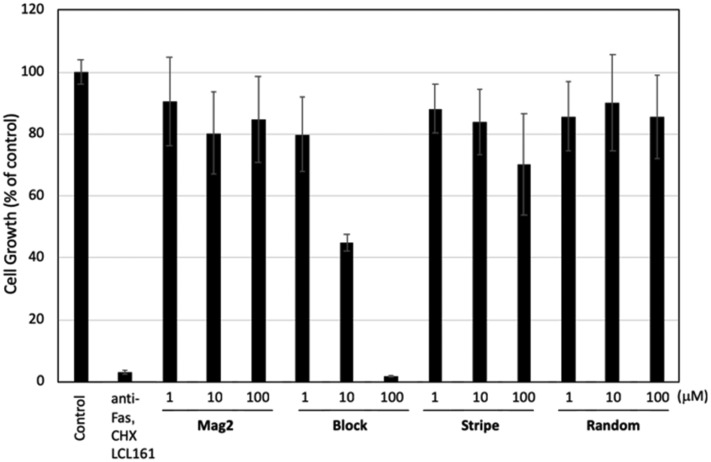
As described above, the peptides target the microbial membrane and exhibit potential antimicrobial activities. On the other hand, there is a possibility that the peptides disrupt not only the microbial membrane but also human cells, resulting in induction of adverse effects. Therefore, we performed a hemolysis assay on human blood cells and a water-soluble tetrazolium salt (WST) assay on cell lines to evaluate the cytotoxicity of the peptides. Human blood cells were treated with the peptides at indicated concentration, and absorption at 550 nm was detected. Spider toxin (M-lycotoxin), which is a representative lytic peptide, was used as a positive control. As shown in Table 2, M-lycotoxin showed potent hemolytic activity with an IC50 value of 0.78 μM, while Mag2 showed no hemolytic activity. In contrast, Block showed a higher hemolytic activity than Mag2 with an IC50 value of 25 μM. Stripe and Random showed no significant hemolytic activity. Furthermore, we performed the WST assay in the presence of the peptides in TIG-3 cells to evaluate their cytotoxicity. As shown in Fig. 4, Block showed significant cytotoxicity at 100 μM concentration. On the other hand, Stripe and Random exhibited zero or very slight cytotoxicity at all concentrations up to and including 100 μM treatment. Based on the hemolytic assay, the anti-human cell activity of Block was likely dependent on its cell membrane disrupting activity, whereas Stripe could only exert its membrane disrupting activity against microbes. These results indicated that the localization of Lys throughout the entire peptide plays a pivotal role in regulating their antimicrobial and hemolytic activities.
Table 2. Hemolysis assay of the peptides against human blood cells. M-Lycotoxin, a spider toxin,15 was used as a positive control.
| Peptide | Hemolysis (μM) |
| M-Lycotoxin | 0.78 |
| Mag2 | >100 |
| Block | 25 |
| Stripe | >100 |
| Random | 100 |
Fig. 4. The effect of the antimicrobial peptides Mag2, Block, Stripe, and Random on the cell growth in fibroblasts. TIG-3 cells were incubated with the indicated concentration of the compounds for 72 h and subsequently were subjected to a WST assay. Data in the graphs are means ± standard deviation (n = 4). A mixture of anti-Fas antibodies (100 ng mL–1), cycloheximide (CHX) (10 μg mL–1), and LCL-161 (1 μM) was used as an inducer for apoptosis. Although activation of Fas is a well-known inducer of apoptotic cell death, fibroblasts are resistant to Fas-mediated apoptosis.17,18 Anti-apoptotic proteins, such as IAP (inhibitor of apoptosis protein) and FLIP (FLICE-like inhibitor protein), are thought to be involved in this resistance. LCL-161 is an inhibitor for IAP. A protein translation inhibitor CHX reduces the levels of FLIP, because FLIP is a short-lived protein. Therefore, we use the treatment with anti-Fas mAb in combination with LCL-161 and CHX as a control.
Conclusions
In summary, we rationally designed and synthesized a set of novel antimicrobial peptides Block, Stripe, and Random, and investigated their mechanism of action using carboxyfluorescein filled unilamellar membranes. We also performed hemolysis and WST assays to assess their potential cytotoxicities. Here, we showed that Stripe may exhibit potent antimicrobial activity against both Gram-positive and Gram-negative microbes based on its lytic activity at the microbial membrane. Moreover, we demonstrated that Stripe showed neither hemolytic activity nor cytotoxicity in the effective concentration range against human blood cells and a well-characterized cell line. These data suggest that the distribution of cationic and hydrophobic residues throughout an entire peptide significantly affects not only its antimicrobial activity but also its hemolytic activity. We predict that Stripe may be a promising reagent for combatting infectious diseases, and that our design strategy could be broadly applied to develop novel AMPs. Further structural development of peptides that display anti-microbial activity against MRBs is in progress.
Experimental
General information
Chemicals were purchased from Sigma-Aldrich Co. LLC, Kanto Chemicals Co. Inc., Tokyo Chemical Industry Co. Ltd., Wako Pure Chemical Industries Ltd., and Watanabe Chemical Industries, Ltd., and used without further purification. Mass spectra were obtained on a Shimadzu IT-TOF MS equipped with an electrospray ionization source.
Peptide synthesis
Peptides Mag2, Block, Stripe, and Random, and M-lycotoxin were synthesized using Fmoc-based solid-phase methods. A representative coupling and deprotection cycle are described as follows. NovaPEG Rink amide resin was soaked for 30 min in CH2Cl2. After the resin had been washed with DMF, Fmoc-amino acid (4 equiv.) and HBTU (4 equiv.) dissolved in N-methyl-2-pyrrolidone (NMP) were added to the resin. DIPEA (4 equiv.) and 0.1 M HOBt in NMP were added for the coupling reaction. Fmoc protective groups were deprotected using 20% piperidine in DMF. The resin was suspended in cleavage cocktail (95% TFA, 2.5% water, 2.5% triisopropylsilane) at room temperature for 3 h. TFA was evaporated to a small volume under a stream of N2 and dripped into cold ether to precipitate the peptide. The peptides were dissolved in DMSO and purified by reverse-phase high performance liquid chromatography using a Discovery® BIO Wide Pore C18 column (25 cm × 21.2 mm, solvent A: 0.1% TFA/water, solvent B: 0.1% TFA/MeCN, flow rate: 10.0 mL mL–1, gradient: 10–90% gradient of solvent B over 30 min). After being purified, the peptide solutions were lyophilized. The peptide purity was assessed using an analytical HPLC and a Discovery® BIO Wide Pore C18 column (25 cm × 4.6 mm, solvent A: 0.1% TFA/water, solvent B: 0.1% TFA/MeCN, flow rate: 1.0 mL mL–1, gradient: 10–100% gradient of solvent B over 30 min).
CD spectrometry
CD spectra were recorded using a 1.0 mm path length cell. The data are expressed in terms of [θ]; i.e., total molar ellipticity (deg cm2 dmol–1). 20 mM phosphate buffer solution (pH = 7.4) with 1% sodium dodecyl sulphate (SDS) was used as a solvent. Peptide concentration; 200 μM.
Biology
Reagents
Tissue culture plastics were purchased from Greiner Bio-One (Frickenhausen, Germany). Cycloheximide (CHX) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Anti-Fas monoclonal antibodies (mAb) were purchased from Medical & Biological Laboratories Co., Ltd. (Aichi, Japan). LCL-161 was purchased from Active Biochem (Kowloon, Hong Kong). TIG-3 cells were obtained from the Japanese Cancer Research Resources Bank (Tokyo, Japan).
Antimicrobial activity
Selected bacterial strains were obtained from Biological Resource Center, NITE (NBRC; Tokyo, Japan). Escherichia coli DH5α was purchased from BioDynamics Laboratory Inc. (Tokyo, Japan). The antimicrobial activities of the peptides against the bacteria, including two Gram-positive bacteria and four Gram-negative bacteria: Staphylococcus aureus, Staphylococcus epidermidis, E. coli DH5α, E. coli NBRC 3972, Proteus vulgaris NBRC 3045, and Pseudomonas aeruginosa NBRC 13275, were measured using the standard broth micro-dilution method as previously described. Briefly, the bacteria were inoculated and grown to mid-log phase in fresh LB medium at 37 °C. Each peptide was 2-fold serially diluted with an initial concentration of 200 μM to 1.56 μM for use. Then, 50 μL per well of the peptide solution was added to each well of a sterile 96-well plate. Subsequently, 50 μL per well of inoculation with 106 CFUs (colony forming units) per mL were added to each well, and the plate was incubated for 18 h at 35 °C. The minimal inhibitory concentration (MIC) was defined as the lowest concentration of the peptide which completely inhibited the growth of bacteria by visual inspection at 535 nm.
Hemolysis activity
Human red blood cells (RBCs) were kindly supplied by the Japanese Red Cross Society (Tokyo, Japan), which collects from volunteers under informed consent. The hemolytic activity of the peptides was assayed by a previously reported standard procedure.16 Briefly, the RBCs were washed three times and resuspended in PBS buffer, and 50 μL of RBC solution were incubated with 50 μL of each diluted peptide for 3 h at 37 °C. Then, intact RBC solutions were centrifuged and the absorbance of the supernatant was measured at 535 nm. Triton X-100 and M-lycotoxin13 served as the positive controls.
Liposome preparation
Lipids, including phosphatidylglycerol (PG), phosphatidylethanolamine (PE) and cardiolipin (CL), were obtained from Sigma-Aldrich. Two types of liposomes with different lipid ratios were prepared as follows: PG/CL (3 : 1, w/w) to mimic the S. aureus membrane and PG/CL/PE (2 : 1 : 7, w/w) to mimic the E. coli membrane. Lipids at each of the aforementioned ratios were dissolved in chloroform, the organic solvent was dried by rotary evaporation to form a thin film on the side of the round bottomed flask, and then placed under a stream of nitrogen overnight, and the lipids were resuspended in 50 mM carboxyfluorescein dye buffer solution (1 N NaOH, 20 mM Tris-HCl, pH 7.4). Each suspension was incubated for 30 min at 70 °C, vortexed for 1 min, and centrifuged at 10 000 rpm for 30 min, at 10 °C. Untrapped dye was removed by gel filtration on a Sephadex G-50 column.
Peptide-induced dye leakage assay
The leakage of carboxyfluorescein was detected by measuring the fluorescence intensity at an excitation wavelength of 535 nm (emission). 100% dye release was achieved from the liposomes with the addition of 10% (v/v) Triton X-100 in 20 mM Tris-HCl (pH 7.4). The percentage of carboxyfluorescein release was calculated using the equation: dye release (%) = (Fobs – F0)/(F100 – F0) × 100%, where is the fluorescence intensity of a peptide released from liposome/carboxyfluorescein solution.
Cell culture
Human normal fibroblast TIG-3 cells were cultured in Dulbecco's modified Eagle's medium (Sigma-Aldrich) containing 10% FBS and 100 μg ml–1 kanamycin.
Cell viability assay
Cell viability was determined using water-soluble tetrazolium WST-8 {4-[3-(2-methoxy-4-nitrophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate} for the spectrophotometric assay and conducted according to the manufacturer's instructions (Dojindo, Tokyo, Japan). Cells were seeded at a concentration of 5 × 103 cells per well in a 96-well culture plate. After 24 h, the cells were treated with the indicated compounds for 72 h. The WST-8 reagent was added, and the cells were incubated for 0.5 h at 37 °C in a humidified atmosphere of 5% CO2. The absorbance at 450 nm of the medium was measured using the EnVision multilabel plate reader (Perkin Elmer, Waltham, MA, USA).
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This study was supported in part by grants from AMED under Grant Number JP18mk0101120, the Japan Society for the Promotion of Science (KAKENHI, Grants 18K14880 to T. M., 17k08385 to Y. D.), Terumo Foundation for Life Sciences and Arts (to T. M. and Y. D.), Takeda Science Foundation (to T. M. and Y. D.), Shorai Foundation for Science and Technology (to T. M.), The Tokyo Biochemical Foundation (to T. M.), The Naito Foundation (to Y. D.), and the NOVARTIS Foundation (Japan) for the Promotion of Science (to Y. D.).
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c9md00166b
References
- (a) Song J. H. Int. J. Infect. Dis. 2003;1:S1–S4. doi: 10.1016/s1201-9712(03)90064-6. [DOI] [PubMed] [Google Scholar]; (b) Suga T., Yamaguchi K. Asian Med. J. 2009;52:103. [Google Scholar]
- (a) Nikaido H. Annu. Rev. Biochem. 2009;78:119. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chang H. H., Cohen T., Grad Y. H., Hanage W. P., O'Brien T. F., Lipstich M. Microbiol. Mol. Biol. Rev. 2015;79:101. doi: 10.1128/MMBR.00039-14. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Munita J. M., Arias C. A. Microbiol. Spectrum. 2016;4:10. doi: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Ageitos J. M., Sanchez-Perez A., Calo-Meta P., Villa T. G. Biochem. Pharmacol. 2017;133:117. doi: 10.1016/j.bcp.2016.09.018. [DOI] [PubMed] [Google Scholar]; (b) Savjani J. K., Gajjar A. K., Savjani K. T. Mini Rev. Med. Chem. 2009;9:194. doi: 10.2174/138955709787316038. [DOI] [PubMed] [Google Scholar]
- (a) Zasloff M., Martin B., Chen H. C. Proc. Natl. Acad. Sci. U. S. A. 1988;85:910. doi: 10.1073/pnas.85.3.910. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Matsuzaki K. Biochim. Biophys. Acta. 1998;1376:391. doi: 10.1016/s0304-4157(98)00014-8. [DOI] [PubMed] [Google Scholar]
- (a) Tamba Y., Yamazaki M. J. Phys. Chem. B. 2009;113:4846. doi: 10.1021/jp8109622. [DOI] [PubMed] [Google Scholar]; (b) Karai M. A., Alam L. M., Takahashi T., Levandy V., Yamazaki M. Langmuir. 2015;31:3391. doi: 10.1021/la503318z. [DOI] [PubMed] [Google Scholar]; (c) Kamakar S., Maity P., Halder A. ACS Omega. 2017;2:8859. doi: 10.1021/acsomega.7b01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki K. Biochim. Biophys. Acta. 2009;1788:1687. doi: 10.1016/j.bbamem.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Misawa T., Imamura M., Ozawa Y., Haishima K., Kurihara M., Kikuchi Y., Demizu Y. Bioorg. Med. Chem. Lett. 2017;27:3950. doi: 10.1016/j.bmcl.2017.07.074. [DOI] [PubMed] [Google Scholar]
- Karle L. L. Acta Crystallogr., Sect. B: Struct. Sci. 1992;48:341. doi: 10.1107/s0108768192000673. [DOI] [PubMed] [Google Scholar]
- Oku H., Yamada K., Katakai R. Biopolymers. 2008;89:270. doi: 10.1002/bip.20904. [DOI] [PubMed] [Google Scholar]
- (a) Yamashita H., Kato T., Oba M., Misawa T., Hattori T., Ohoka N., Tanaka M., Naito M., Demizu Y. Sci. Rep. 2016;6:33003. doi: 10.1038/srep33003. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kobayashi H., Misawa T., Oba M., Hirata N., Kanda Y., Tanaka M., Matsuno K., Demizu Y. Chem. Pharm. Bull. 2018;66:575. doi: 10.1248/cpb.c18-00079. [DOI] [PubMed] [Google Scholar]
- Toniolo C., Polese A., Formaggio F., Crisma M., Kamphuis J. J. Am. Chem. Soc. 1996;118:2744. [Google Scholar]
- Dong W., Mao X., Guan Y., Kang Y., Shang D. Sci. Rep. 2017;7:40228. doi: 10.1038/srep40228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsu T., Imamura T., Komagome K., Masuda K., Mizushima T. Anal. Sci. 2007;23:517. doi: 10.2116/analsci.23.517. [DOI] [PubMed] [Google Scholar]
- (a) Murzyn K., Rog T., Pasenkiewicz-Gierula M. Biophys. J. 2005;88:1091. doi: 10.1529/biophysj.104.048835. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) McGeachy A. C., Caudill E. R., Liang D., Cui Q., Pederson J. A., Geiger F. M. Chem. Sci. 2018;9:4285. doi: 10.1039/c8sc00804c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akishiba M., Takeuchi T., Kawaguchi Y., Sakamoto K., Yu H. H., Nakase I., Takatani-Nakase T., Madni F., Graslund A., Futaki S. Nat. Chem. 2017;9:751. doi: 10.1038/nchem.2779. [DOI] [PubMed] [Google Scholar]
- Strandberg E., Zerweck J., Horn D., Pritz G., Berditsch M., Burck J., Wadhwani P., Ulrich A. Pept. Sci. 2015;21:436. doi: 10.1002/psc.2780. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Yoshimi M., Maeyama T., Hagimoto N., Kuwano K., Nara N. Eur. Respir. J. 2002;20:359. doi: 10.1183/09031936.02.00252602. [DOI] [PubMed] [Google Scholar]
- Ajayi I. O., Sisson T. H., Higgins P. D., Booth A. J., Sagana R. L., Huang S. K., White E. S., King J. E., Moore B. B., Horowitz J. C. Am. J. Respir. Cell Mol. Biol. 2013;49:86. doi: 10.1165/rcmb.2012-0224OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.