Summary
Embryonic organizers establish gradients of diffusible signaling molecules to pattern the surrounding cells. Here, we elucidate an additional mechanism of embryonic organizers that is a secondary consequence of morphogen signaling. Using pharmacological and localized transgenic perturbations, 4D imaging of the zebrafish embryo, systematic analysis of cell motion and computational modeling, we find that the vertebrate tail organizer orchestrates morphogenesis over distances beyond the range of morphogen signaling. The organizer regulates the rate and coherence of cell motion in the elongating embryo using mechanical information that is transmitted via relay between neighboring cells. This mechanism is similar to a pressure front in granular media and other jammed systems, but in the embryo the mechanical information emerges from self-propelled cell movement and not force transfer between cells. The propagation likely relies upon local biochemical signaling that affects cell contractility, cell adhesion and/or cell polarity but is independent of transcription and translation.
Graphical Abstract
eTOC Blurb
Das, Jülich, and Schwendinger-Schreck et al. find that the zebrafish tail organizer orchestrates morphogenesis over distances beyond the range of its secreted cell signaling proteins. The organizer regulates cell migration in the elongating embryo using mechanical information that propagates via relay between neighboring cells.
One Sentence Summary:
Mechanical information expands the sphere of influence of an embryonic organizer beyond the range of morphogen signaling.
Introduction
Spemann and Mangold’s discovery of embryonic organizers and subsequent theories of morphogens and positional information, and the experimental identification of morphogen gradients are seminal breakthroughs in developmental biology. We now understand that organizers establish gradients of diffusible signaling molecules that pattern the surrounding cells in a concentration-dependent manner (Lander, 2007; Muller et al., 2013). How morphogens interlink with mechanical forces is poorly understood, but recent studies have begun to integrate morphogen patterning with morphogenesis. For example, cell rearrangement sharpens the boundaries between expression domains downstream of noisy morphogen signaling in the vertebrate neural tube (Xiong et al., 2013). In the zebrafish shield, the equivalent of the Spemann-Mangold organizer, a positive feedback loop emerges in which a morphogen increases cell adhesion which then increases reception of the morphogen signal (Barone et al., 2017). During organogenesis, folding of the vertebrate gut epithelium creates local maxima of secreted signaling molecules that then pattern the crypt-villus axis required for gut homeostasis (Shyer et al., 2015).
Much like our understanding of morphogen signaling, insights into the role of mechanical forces in development have been pioneered by studies of both Drosophila and vertebrate gastrulation (Williams and Solnica-Krezel, 2017). To generalize, these forces are generated through actomyosin contractility and transmitted to adjacent cells via cell-cell and cell-ECM adhesions that are linked to the cytoskeleton. We are just beginning to understand how coordination of these forces among cells can drive tissue morphogenesis (Heisenberg and Bellaiche, 2013; LeGoff and Lecuit, 2015). For example, the distribution of cell-ECM adhesions within a tissue is inversely correlated with the degree of cell displacement during Drosophila dorsal closure (Goodwin et al., 2016). A nice illustration of long-range organization via cellular forces is how internalization of the Drosophila endoderm generates supercellular tension that cell non-autonomously drives germband extension (Lye et al., 2015).
The vertebrate tail organizer functions within a flux of tailbud mesodermal progenitors to direct the elongation of the developing spinal column (Figure 1A) (Agathon et al., 2003; Beck and Slack, 1999; Beck et al., 2001). We previously tracked individual cell motion in the zebrafish tailbud, segmented the tailbud into four domains (excluding the notochord) and quantified collective cell behavior in these different domains (Lawton et al., 2013). The cells in the anterior dorsal medial domain of the tailbud are mostly spinal cord precursors. Here, for simplicity, we refer to this domain as the posterior neural tube (PNT). These cells migrate posteriorly towards the dorsal-medial zone (DM). The DM contains rapidly moving neural-mesodermal progenitors (Martin and Kimelman, 2012; Wilson et al., 2009). DM cells differentiating into mesoderm migrate ventrally into the Tail Organizer (TO) and then bilaterally disperse to populate the left and right presomitic mesoderm (PSM) (Das et al., 2017; Lawton et al., 2013). The TO is more fluid than the surrounding domains, and as these cell assimilate into the PSM, the tissue undergoes a fluid to solid transition in tissue mechanics (Lawton et al., 2013; McMillen and Holley, 2015; Mongera et al., 2018).
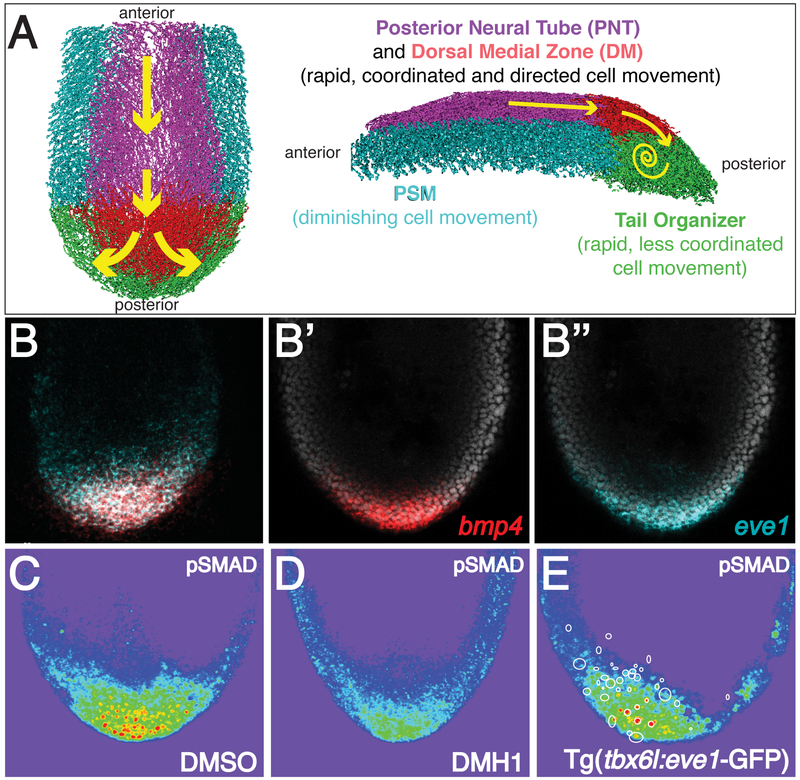
Figure 1. Bmp-eve1 signaling in the zebrafish tail organizer.
(A) A schematic of cell motion in the extending tailbud. Cell tracks are colored as indicated. The tail organizer is synonymous with the previously defined progenitor zone (Lawton et al., 2013). (B) Overlapping mRNA localization for bmp4 and eve1 in the tail organizer. pSMAD localization in control (C), DMH1 treated (D) and transgenic embryos (E). Warmer colors indicate higher levels of Bmp signaling. Circles in (E) mark the location of transgene-expressing cells.
The tail organizer is located in the posterior tailbud and both the mesoderm and epidermis express the secreted signaling proteins bmp4 and bmp2b and the transcription factor eve1 (Figure 1A-1B”) (Thisse and Thisse, 2005). Bmp signaling in the tail organizer is required for zebrafish body elongation (Connors et al., 2006; Pyati et al., 2005; Stickney et al., 2007; Yang and Thorpe, 2011). The Bmp inhibitors chordin and noggin are expressed in the posterior notochord and presomitic mesoderm (PSM), thus Bmp signaling is restricted to the posterior tailbud as indicated by phosphorylated SMAD 1/5/8 immunolocalization (Figure 1C)(O'Neill and Thorpe, 2013; Row and Kimelman, 2009). Even-skipped (eve or evx) genes are adjacent to the hox13 end of hox clusters suggesting a role in posterior animal development (Cruz et al., 2010; Li and Manley, 1998; McKay et al., 1999; Seebald and Szeto, 2011). Indeed, eve1 over-expression produces duplicated tails, and eve1 is expressed in ectopic tail organizers (Agathon et al., 2003; Barro et al., 1995; Connors et al., 2006; Cruz et al., 2010; Joly et al., 1993; Mullins et al., 1996; Row and Kimelman, 2009; Seebald and Szeto, 2011; Stickney et al., 2007; Yang and Thorpe, 2011). The patterns of bmp and eve1/evx1 expression are conserved in Xenopus and mouse tailbuds, and bmp and evx1 have tail inducing activity in Xenopus (Beck and Slack, 1999; Beck et al., 2001; Dush and Martin, 1992; Fainsod et al., 1994; Gofflot et al., 1997; Goldman et al., 2000; Ohta et al., 2007). In the mouse and chick, regulation of Bmp signaling from the ventral ectodermal ridge and mesoderm of the tailbud promotes the cessation of gastrulation and is required for subsequent tailbud elongation (Goldman et al., 2000; Ohta et al., 2007).
The consistent spatial expression pattern of zebrafish eve1, bmp4 and bmp2b in the tail organizer is maintained despite extensive cell movement (Figure 1A-1B”) (Dray et al., 2013; Fioret al., 2012; Lawton et al., 2013; Mara et al., 2007; Steventon et al., 2016). Here, we investigate how the tail organizer sustains itself and orchestrates body elongation. We find that the tail organizer regulates cell flux into the organizer downstream of Bmp signaling and non-cell autonomously via mechanical information. The in vivo data and computational modeling suggest that the mechanical information initiates a cell to cell relay that alters the movement of neighboring cells and propagates through the migrating flux of cells over distances beyond the range of Bmp signaling.
Results
Perturbation of Tail Organizer Signaling
We abrogated tail organizer function during trunk elongation using DMH1, a chemical inhibitor of Bmp receptor kinase signaling that enables temporal control of inhibition (Hao et al., 2010). The strongest reduction in pSMAD levels was observed two hours post-treatment (Figure 1D and 2A). bmp4 and eve1 transcription are sensitive to reduction of Bmp signaling while bmp2b transcription is not dependent upon Bmp signaling (Figure 2B and S1A).
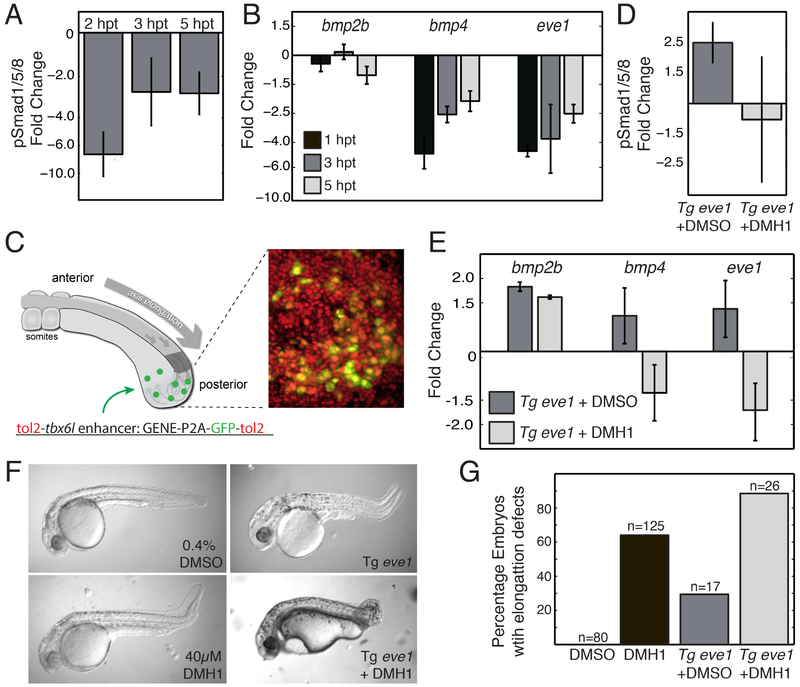
Figure 2. Temporally and spatially controlled perturbation of the tail organizer.
Western blot for pSmad (A) and qRT-PCR for nascent transcription (B) of pooled, dissected tailbuds from embryos treated with DMH1 or DMSO at the 5-somite stage. pSmad levels (A) and bmp4 and eve1 transcription are reduced 1-5 hours post DMH1 treatment (hpt) relative to DMSO treated controls. n=3 replicates for each timepoint. (C) A local perturbation of the tail organizer is introduced in transient transgenics that express eve1 along with GFP using the tbx6l enhancer. On the right, an experimental image of a tail organizer is shown in which nuclei of all cells are labeled with nls-RFP (red) and transgenic cells are labeled with GFP (green). Transgenic expression of eve1 increases pSmad levels (D) as well as bmp2b, bmp4 and eve1 transcription (E). Blocking the Bmp receptor with DMH1 eliminates the increase in pSmad (D) and bmp4 and eve1 transcription, but not bmp2b transcription (E). Measurements were made 3 hours post DMH1 treatment. n=3 experimental replicates for each condition. (F) Representative images of body elongation phenotypes following perturbation of the tail organizer. (G) A summary of the percent of embryos with body elongation defects. Total number of embryos from at least three experimental replicates are indicated. Error bars denote the standard error. See also Figure S1.
We performed a targeted spatial perturbation of organizer signaling by mosaically co-expressing eve1 and GFP under the control of the tbx6l enhancer (Figure 2C) (Dray et al., 2013; Szeto and Kimelman, 2004). tbx6l expression largely overlaps eve1 expression, thus the transgene mosaically disrupts the levels and regulation of eve1 within its normal expression domain in the posterior tailbud. Mosaic over-expression of eve1 elevated total pSMAD levels and increased pSMAD in both transgenic and non-transgenic cells within the tail organizer, producing a more heterogeneous pattern (n=12) (Figure 1E and 2D). The eve1 transgene also increased transcription of bmp2b, bmp4 and endogenous eve1 (Figure 2E). Transgenic embryos treated with DMH1 showed wild-type levels of pSMAD but reduced transcription of both eve1 and bmp4 (Figure 2D and 2E). These data indicate that the tail organizer is sustained within the flux of cells transiting the organizer via positive feedback between eve1, bmp4 and bmp2b.
DMH1 treatment, transgenic expression of eve1 and transgenic expression of Tg eve1 + DMH1 treatment beget defects in body elongation with the latter producing the most frequent and strongest defects (Figure 2F and 2G). Given that Tg eve1 + DMH1 have normal levels of Bmp signaling in the tail organizer, these data indicate that wild-type body elongation requires the regulated spatiotemporal pattern of Bmp signaling produced by the bmp1-eve1 circuit. To determine the kinematic basis of the elongation phenotypes, we systematically analyzed cell motion in the tailbud after perturbation of the tail organizer. We acquired 3D confocal timelapses of nuclear localized RFP in four DMSO-treated control embryos, four DMH1-treated embryos, three transgenic DMSO-treated embryos and three transgenic DMH1-treated embryos.
Cell motion in the tail organizer is aberrant in both DMH1 and Tg eve1 + DMH1 embryos. Cell track straightness is reduced in both conditions, and the mean coefficient of variation (C.V.) of cell track speed is increased in DMH1 embryos (Figure 3A and 3B). We examined the mean square displacement (MSD), and modeled the data using a diffusion coefficient and directional velocity (Figure 3C and 3D) (Dray et al., 2013; Monnier et al., 2012). The directional velocity is reduced in the tail organizer of DMH1 embryos (Figure 3D). Cell flux through the tail organizer includes a medial domain comprised of both a dorsal to ventral flow and a medial to lateral flow that are segregated from flows on the lateral periphery that are predominantly posterior to anterior and ventral to dorsal (Figure 3G and S2). These flows intermix after perturbation of TO signaling, and the disruption of the dorsal to ventral flow likely obstructs cell flow into and through the tail organizer, particularly in Tg eve1 + DMH1 embryos.
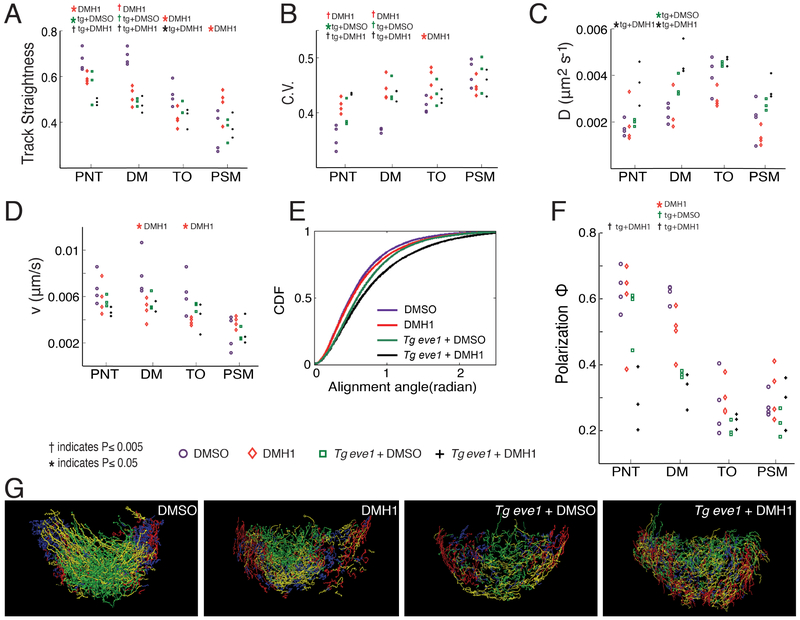
Figure 3. Perturbation of the tail organizer has long-range effects on tailbud cell motion.
Treatments which gave a significant change from wild type in a given tailbud domain are indicated at the top of each panel. (A) Cell track straightness. (B) Mean coefficient of variation of cell track speed. (C) Diffusion coefficient and (D) directional velocity obtained by fitting the data to a MSD model. (E) Local order as measured by the CDF (cumulative distribution function) of the local alignment angle. The CDFs for DMSO-treated controls and Tg eve1 + DMH1 embryos differ (p<0.05, t-test). A steeper curve indicates more ordered cell motion. (F) Global order as measured by Polarization Φ. In panels A-D and F, the metrics are shown for four domains of the tail (PNT, DM, TO and PSM). In panel E, local order is shown only for the PNT. (G) Cell flow within the TO visualized by displaying only cell tracks with the greatest displacement from dorsal to ventral (green), medial to lateral (yellow), posterior to anterior (red) and ventral to dorsal (blue). In DMSO control embryos, the dorsal to ventral flow is concentrated medially while the posterior to anterior and ventral to dorsal flows are concentrated laterally. This represents the predominant pattern of cell flow through the tail organizer from the DM and into the PSM. In experimental embryos, these flows are less well segregated indicating a more disordered flux through the tail organizer, particularly in Tg eve1 + DMH1 treated embryos. See also Figures S2 and S3.
Outside of the tail organizer in the PNT and DM, cell track straightness is reduced and the mean C.V. of cell track speed was increased in all experimental conditions (Figure 3A and 3B). The diffusion coefficient normally spikes in the tail organizer, but in both Tg eve1 and Tg eve1 + DMH1 embryos, the DM diffusion coefficient is increased, and this increase extends into the PNT in Tg eve1 + DMH1 embryos (Figure 3C). Reduction in directional velocity was also observed in the DM of DMH1 embryos (Figure 3D). These data indicate that perturbation of TO signaling affects cell motion outside of the organizer. To measure the effect these perturbations have on collective cell behavior, we examined global order of cell motion. Global order within each domain is quantified via the Polarization (Φ) which provides a normalized measure of the coherence of cell velocities. DM global order is reduced in each experimental condition while PNT global order is reduced in Tg eve1 + DMH1 embryos (Figure 3F). Local order quantifies the alignment of instantaneous velocities within a 20 micron radius of each cell and provides the statistics of the alignment angles expressed in a cumulative distribution function (CDF). While the global and local order typically correlate, there are experimental perturbations that cause them to differ (Das et al., 2017). We were particularly interested in the apparent long-range effects of the perturbations observed in the PNT and quantified local order in this domain. We found that the local order of cell motion is reduced in the PNT of Tg eve1 + DMH1 embryos (Figure 3E and S3A).
Long Range Effects of Organizer Perturbation
The observations that transgenic expression of eve1 in the tail organizer and inhibition of Bmp signaling, which is restricted to the tail organizer, are sufficient to affect cell motion in the posterior neural tube suggest a long-range organizing function for the tail organizer beyond the range of Bmp signaling. We hypothesized that the long-range orchestration of cell motion by the tail organizer is mechanical and mediated by a relay between migrating cells. In the embryo, there is unlikely to be force transmitted from cell to cell due to viscous drag. Instead, physical contact between cells could induce a biochemical response such as cell contact mediated repulsion or changes in cell contractility, adhesion or polarity that alter the active migration of the cell. This relay would pass posterior to anterior from cell to cell until it dissipates due to both viscous drag and resistance by the predominant anterior to posterior flow of cells in the neural tube.
This relay mechanism has similarities to a pressure front traveling through granular media and jammed matter such as glassy and gel-like materials (Boudet et al., 2009; Rericha et al., 2002). However, this phenomenon is not well studied in active matter or biological systems. To explore this mechanism theoretically, we developed a 3D computational model of the elongating tailbud that represents cells as self-propelled elastic spheres (Figure 4A and S4A)(Gönci et al., 2008; Szabó et al., 2006). As in our prior 2D models, there are adhesive and repulsive interactions between cells and a propensity for neighboring cells to align their instantaneous velocities (Das et al., 2017; Lawton et al., 2013)(See SI for details). Note that repulsion in the simulations may represent a number of in vivo mechanisms such as volume exclusion between cells, as two cells cannot occupy the same space, as well as biochemical processes such as contact mediated inhibition.
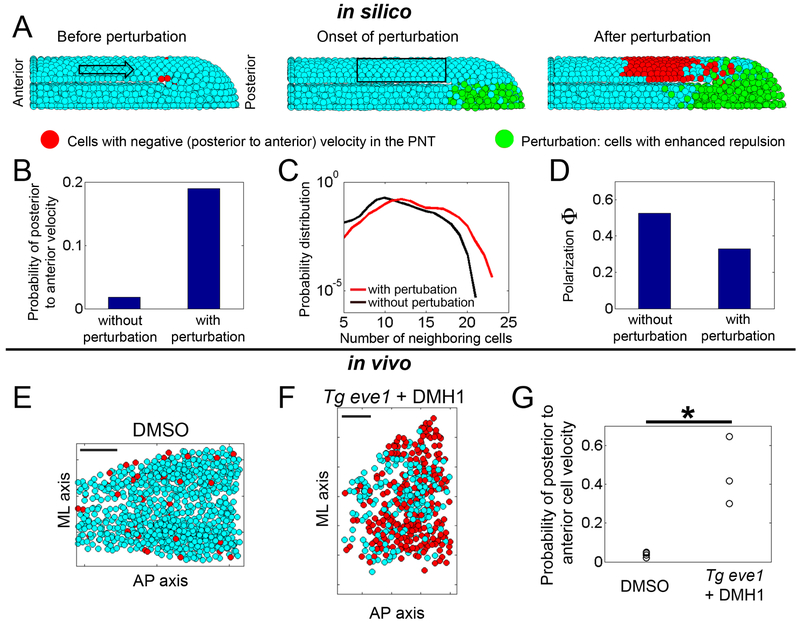
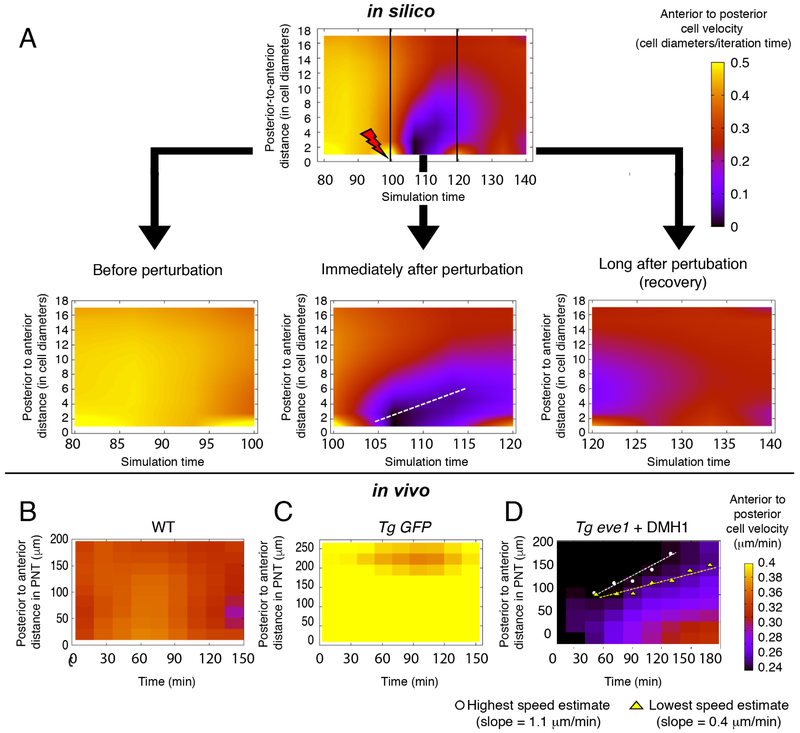
Figure 4. Analysis of mechanical information propagation following a perturbation in the tail organizer using 3D computer simulations and in vivo data.
Simulations: (A) Before the perturbation (left panel), the overall flow of PNT cells is from anterior to posterior (arrow), though a few cells may stochastically move posterior to anterior at any instant (3 red cells). The perturbation is then introduced (middle panel) by stochastically increasing cell-cell repulsion in tail organizer cells (green). After the onset of the perturbation (right panel), a large group of cells in the PNT display posterior to anterior velocities (red cells), and the front of this disturbance propagates posterior to anterior over time (see Movie S1). (B) The probability of posterior to anterior cell velocities is higher after the perturbation (p<0.05, t-test). (C) Cell density is quantified via the probability distribution of the number of neighboring cells. These two probability distributions (before and after perturbation) show that the number of neighboring cells is higher after perturbation (p<0.05, t-test). (D) Global order in cell motion, polarization (Φ), is reduced after the perturbation (p<0.05, t-test). All quantities in panels B-D are calculated by sampling within a fixed volume (black rectangle in A) over 30 simulations with and 30 simulations without perturbation. In B and D, the standard deviations are < 6 %. Experiments: Snapshots of cells of the PNT in (E) a DMSO-treated control embryo (see Movie S2) and (F) a DMH1-treated tbx6l:eve1-transgenic (See Movie S3). Red dots indicate cells with a posterior to anterior velocity. Scale bars in E and F represent 50 microns. (G) Tg eve1 + DMH1 embryos (n=3) have an increased probability of PNT cells having a posterior to anterior velocity relative to DMSO control embryos (n=4) (p<0.05, t-test).
Analysis of pressure fronts in granular matter suggests that a disturbance created by a local perturbation can travel through the material (Boudet et al., 2009; Rericha et al., 2002). Thus, to probe the transmission of mechanical information in our model, we introduced a local perturbation by increasing the repulsion between cells in the organizer. The perturbation is switched on stochastically in individual cells within the tail organizer to mimic transgene expression in experiments (Figure 4A). Consistent with the in vivo phenotype of Tg eve1 + DMH1 embryos, the perturbation decreases cell track straightness and the mean C.V. of cell track speed in both the PNT and TO (Figure S4B-S4C).
If mechanical information is being transmitted from the tail organizer to the PNT, the perturbation would mostly disturb the anterior-posterior component of cell velocities, and this disturbance should propagate posterior to anterior through the PNT. Therefore, in the presence and absence of the perturbation, we examined the probability of posterior to anterior instantaneous velocities which are directed oppositely to the general anterior to posterior flow of cells in the PNT. We found that the perturbation increased the probability of posterior to anterior cell velocities in the PNT (Figure 4A, 4B, S4D and Movie S1). The perturbation also increased cell density within the PNT as reflected by a shift in the probability distribution of the number of cell neighbors (Figure 4C and S4E). This theoretical result is consistent with local jamming of cells, similar to a traffic jam on a highway. A traffic jam starts from the site of an accident and then travels against the flow of traffic as a disturbance. The presence of competing cells moving forward and backward in the PNT following the perturbation decreases global order (Figure 4D and S4F). This lower polarization (Φ) in silico is again consistent with the observed in vivo phenotype of Tg eve1 + DMH1 embryos.
We reexamined the in vivo data for the signatures of mechanical information suggested by this theory. Indeed, Tg eve1 + DMH1 embryos exhibit an increased probability of posterior to anterior cell velocities in the PNT (Figure 4E-4G and Figure S3B). Thus, the computer simulations provide a theoretical mechanism for the propagation of mechanical information in the tailbud. The information is transmitted as disturbance, which travels posterior to anterior, and then damps out due to the combined influence of viscosity and the competing active cell migration from anterior to posterior (Figure S4G-S4I).
Wnt and Fgf signaling in the tail organizer
Wnt and Fgf signaling are both prominent in the zebrafish tail organizer and abrogation of either signaling pathway causes body elongation defects (Das et al., 2017; Furthauer et al., 2002; Lawton et al., 2013; Row and Kimelman, 2009; Sawada et al., 2001; Steventon et al., 2016). Thus, it is possible that Bmp signaling could induce long-range effects indirectly by modulating Fgf or Wnt signaling. We examined expression of Wnt and Fgf target genes after DMH1 treatment and in Tg eve1 + DMSO and Tg eve1 + DMH1 embryos. DMH1 treatment alters axin2, dkk1, sef and sprouty transcription (Figure S1A). However, Tg eve1 and Tg eve1 + DMH1 embryos exhibit similar levels of Wnt and Fgf target gene expression meaning that alteration of Wnt and Fgf signaling does not explain the stronger long-range effects observed in Tg eve1 + DMH1 embryos (Figure S1B).
Previously, we pharmacologically inhibited Fgf receptor signaling using SU5402 and reduced Wnt signaling by over-expression of notumn1a (Lawton et al., 2013). The mean C.V. of cell track speed is increased and track straightness is reduced after either Fgf or Bmp inhibition, but the directional velocity is only reduced in DMH1 and Tg eve1 + DMH1 embryos. notum1a overexpression did not change any of these metrics (Das et al., 2017; Lawton et al., 2013). Thus, DMH1 embryos and Tg eve1 + DMH1 embryos exhibit stronger tail organizer cell motion defects than reduction of Wnt or Fgf signaling. Accordingly, we do not observe an increase in posterior to anterior cell velocities in the PNT of embryos with reduced Wnt or Fgf signaling (Figure S3C). All together, these data indicate that the long-range effects observed in Tg eve1 + DMH1 embryos are unlikely to be mediated by changes in Wnt or Fgf signaling.
Estimating the speed of mechanical information
To estimate the rate at which mechanical information propagates, we displayed the mean anterior to posterior cell velocity as a heat map plotted as a function of position along the anterior-posterior axis and time (Figure 5). In computer simulations, a disturbance of mean velocity propagates anteriorly after an increase in cell repulsion in the tail organizer. This disturbance appears as the purple flare (Figure 5A). We divided the plot into three time-intervals representing the states before perturbation, immediately after perturbation and long after perturbation. These three plots reflect the outcomes of the in vivo experiments in which we have less precise control of the perturbation. The propagation rate of the front of the disturbance is given by the slope of the flare (Figure 5A, lower middle plot). This propagation damps out due to viscosity and resistance from the predominant anterior to posterior cell flow, and the velocity field slowly returns to a normal state (Figure 5A, lower right plot).
Figure 5. Estimating the propagation speed of mechanical information.
(A) Computer simulations were performed with the perturbation triggered at time-point 100 as described in Figure 4. Mean anterior to posterior cell velocity was plotted as heat map as a function of posterior to anterior distance along the PNT and time. The mechanical information is revealed by the propagation of a disturbance in mean cell velocity from posterior to anterior (the purple flare). The plot was subdivided into three time-intervals to increase the temporal resolution: before perturbation, immediately after perturbation and long after perturbation. The speed of the posterior to anterior propagation of the mechanical information is estimated from the slope of the purple flair representing the disturbance over time (dashed line). In simulations, time is measured in units of 30 in silico iterations and the length is in units of cell diameters. (B) The heat maps for wild-type embryos are relatively uniform much like the simulations prior to or long after perturbation (see also Figure S5A). (C) Tg tbx6l:GFP transgenic control embryos produce heat maps that are relatively uniform (see also Figure S5C). (D) Tg tbx6l:eve1-GFP embryos treated with DMH1 at the onset of imaging exhibit heterogenous heat maps. Four of nine embryos exhibited the posterior to anterior propagation of a disturbance in mean anterior to posterior cell velocity. We estimated the maximum and minimum slope of the propagation for each of these embryos. (See also Figure S5E-H).
We sought to replicate the in silico experiment in vivo by delaying the addition of DMH1 to Tg eve1 embryos. In prior experiments, DMH1 was added two hours before the start of the timelapse (and before the embryos were mounted for imaging) to ensure that Bmp inhibition had been established. Here, we added DMH1 to Tg eve1 embryos after embryos had been mounted for imaging and just before the start of the timelapse. The objective was to image the onset of the Bmp inhibition and visualize any posterior to anterior propagation of disturbance in the cell velocities. Analysis of wild-type embryos and Tg GFP embryos produced velocity heat maps similar to those observed in simulations before and long after perturbation (Figures 5B, 5C, S5A and S5C). Plots of global order of cell motion (polarization Φ) in the PNT of these control embryos were relatively stable over time (Figure S5B and S5D). Tg eve1 + DMH1 embryos exhibited two classes of phenotypes: Group 1 (5 of 9 embryos) and Group 2 (4 of 9 embryos). Group 1 had velocity heat maps and polarization Φ values resembling controls (Figure S5E and S5F). By contrast, Group 2 exhibited heat maps with propagating posterior to anterior disturbances in mean velocity and highly variable polarization Φ values (Figure 5D, S5E and S5G). Group 2 Tg eve1 + DMH1 embryos also displayed evidence of mechanical information in the form of an increase in the probability of posterior to anterior cell velocities compared to Group 1 Tg eve1 + DMH1 embryos and Tg GFP controls (Figure S5H). Thus, experimental variability produces two classes of embryos with Group 1 indistinguishable from controls and Group 2 exhibiting evidence of propagation of mechanical information after perturbation. This range of phenotypes likely arises from the stochasticity of transgene expression and variation in DMH1 absorption in embryos mounted in low melt agarose for imaging. Using the heat maps, we estimated the maximum and minimum rates of posterior to anterior propagation of the disturbance (Figure 5D and Figure S5E). The average propagation speed is estimated at 1 μm per minute.
Increasing cell contractility in the tail organizer induces retrograde motion in the PNT
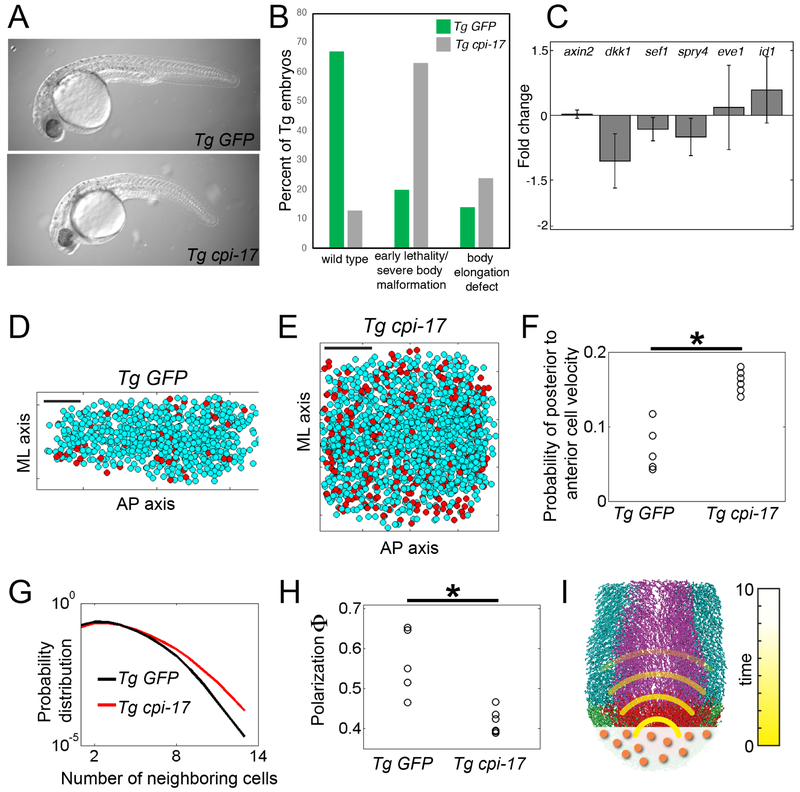
To more directly examine mechanical information transfer, we generated a targeted mechanical disruption of the tail organizer and quantified cell motion in the PNT. We mosaically increased actomyosin contractility in the tail organizer via expression of an inhibitor of myosin phosphatase (cpi-17) under the control of the tbx6l enhancer, cpi-17 has been shown to modulate cell migration during zebrafish gastrulation (Weiser et al., 2009). Moreover, an increase in actomyosin contractility mimics the increase in cell repulsion in our simulations. Tg cpi-17 embryos exhibit a higher incidence of early developmental and body elongation defects compared to Tg GFP controls (Figure 6A and 6B). Embryos with severe body malformations are not suitable for our analysis of cell motion. Therefore, we used embryos that exhibited relatively mild phenotypes, typically retarded body elongation.
Figure 6. Mechanical information propagation after increasing cell contractility in the tail organizer.
(A) Transient tbx6l transgenics expressing either GFP (control) or cpi-17 and GFP in the tail organizer. (B) Tg tbx6l:GFP transgenics produce mostly normal embryos (3 experiments; n=363) whereas Tg tbx6l:cpi-17 embryos (4 experiments; n=416) exhibit a high frequency of early developmental defects and embryos with abnormal body elongation. (C) Nascent transcription of Wnt, Fgf and Bmp target genes was examined by qRT-PCR on pooled dissected tailbuds. Expression in Tg tbx6l:cpi-17 tailbuds was normalized to expression in Tg tbx6l:GFP controls. Data are averages from 3 independent experiments and display standard error. Snapshots of cells of the PNT in (D) a Tg tbx6l:GFP control (see Movie S4) and (E) a Tg tbx6l:cpi-17 embryo (see Movie S5). Red dots indicate cells with a negative (i.e. posterior to anterior) velocity. Scale bars in D and E represent 50 microns. (F) The probability of posterior to anterior cell velocities the PNT in five Tg tbx6l:GFP controls and six Tg tbx6l:cpi-17 embryos. (G) Cell density in the PNT is displayed via the probability distribution of the number of neighboring cells. Each probability distribution is obtained by pooling the data from each experimental condition. These two probability distributions differ (p<0.05, t-test), and show that cell density is higher in Tg tbx6l:cpi-17 embryos. (H) Global order of PNT cell velocities as measured by the polarization Φ. (I) Mechanical information (yellow) extends the sphere of influence of the tail organizer beyond that of a secreted signaling molecule (orange). Asterisks denote p<0.05 (t-test).
We examined tailbud expression of Bmp, Wnt and Fgf target genes in Tg cpi-17 tailbuds (Figure 6C). Bmp and Fgf target gene expression were not altered relative to Tg GFP controls. One Wnt target, dkk1, exhibited a small decrease while the other Wnt target, axin2, was unchanged. Since we do not observe evidence of mechanical information after stronger perturbation of Wnt signaling (Figure S3C), any alteration in PNT cell motion in Tg tbx6l:cpi-17 embryos should not be a consequence of altered tail organizer signaling.
Mosaic expression of cpi-17 in the tail organizer reduces global order (polarization, Φ) as well as local order in the PNT revealing a long-range effect of a local mechanical perturbation of the organizer (Figure 6H and Figure S3D and S3E). As predicted by theory, the mechanical information correlates with an increase in the probability of posterior to anterior cell velocity in the PNT as well as an increase in cell density (Figure 6D-6G). These data reveal a long-range effect of mechanical perturbation of the vertebrate tail organizer.
Discussion
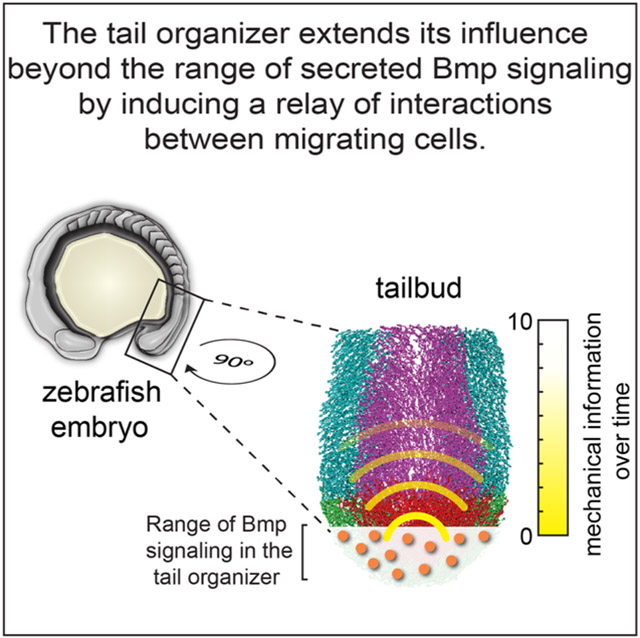
Here, we find that the zebrafish tail organizer orchestrates morphogenesis over distances beyond the range of Bmp signaling. This long-range organization is a secondary effect downstream of canonical organizer cell signaling and likely propagates through local, adhesive and repulsive interactions between neighboring cells (Figure 6I). In other words, there is no transfer of force from one cell to another since viscous drag leads to overdamping. Thus, in contrast to pressure front propagation in granular media and other jammed materials which is induced by an external force, mechanical information emerges from a relay of cell intrinsic forces. The propagation is likely mediated by biochemical signaling that affects cell contractility, cell adhesion and/or cell polarity but is independent of transcription and translation.
The posterior tailbud is a complex cell signaling domain with active Fgf, Wnt and Bmp signaling all of which have distinct effects on cell movement and cell fate. Fgf and Wnt are in gene regulatory networks that govern segmental patterning of the paraxial mesoderm and differentiation of bipotential neural-mesodermal progenitors (Gouti et al., 2017; Pourquie, 2011). Inhibition of Fgf decreases the speed of cell motion in the posterior tailbud of the chick, and in zebrafish, Fgf inhibition increases variability in cell speed and decreases track straightness in the posterior tailbud and reduces convergence in the PSM (Benazeraf et al., 2010; Lawton et al., 2013; Steventon et al., 2016). Partial inhibition of Wnt signaling via injection of moderate levels of notum1a mRNA increases ordered cell motion in the DM which in turn leads to loss of symmetric elongation (Das et al., 2017; Lawton et al., 2013). The increased order is likely due to inhibition of a two-step epithelial to mesenchymal transition (EMT) in neural-mesodermal progenitors (Manning and Kimelman, 2015). The first step in the EMT is regulated by Wnt and the second step is regulated by Fgf (Goto et al., 2017). This integration of Wnt and Fgf signaling is observed in the extensive crosstalk between the two pathways in the zebrafish tailbud (Stulberg et al., 2012). Data presented here indicate that Bmp signaling exhibits some cross-regulation with Fgf and Wnt. Indeed, prior studies found that Bmp signaling opposes cell fate specification by Wnt and Fgf. Tailbud Bmp signaling promotes vasculature and tail fin cell fates in zebrafish (Connors et al., 2006; Pyati et al., 2005; Row et al., 2018). Bmp signaling promotes vascular fates by increasing expression of id genes which then inhibit the function of basic helix loop helix transcription factors that specify paraxial mesoderm cell fates downstream of Wnt and Fgf signaling (Row et al., 2018).
Within the tail organizer, Bmp signaling increases the directional velocity and promotes ordered cell flow, yet the flux of cells through the organizer is constrained by the requirement to maintain Bmp signaling. Cells must secrete sufficient Bmp protein while transiting the organizer to activate the bmp-eve1 positive feedback in newly arrived cells. If this positive feedback is weakened, then the flux through the organizer must be reduced in order to maintain the organizer signaling domain. Cell flux through the tail organizer is characterized by a medial domain of both dorsal to ventral flow and medial to lateral flow that is segregated from flows moving posterior to anterior and ventral to dorsal along the lateral sides of the organizer. In Tg eve1 + DMH1 embryos, these flows are intermixed, and the loss of the dorsal to ventral channel likely creates congestion that leads to the local decrease in cell velocities that propagates from posterior to anterior in the form of a disturbance. This decrease in cell speed may help maintain tail organizer signaling domain albeit at the cost of retarding tail elongation.
In contrast to the positive feedback between bmp4 and eve in the tail organizer, eve1 represses bmp2b, bmp4 and eve1 transcription on the ventral side of the zebrafish gastrula (Figure S1C)(Cruz et al., 2010). This inversion in gene regulation is also observed in the Fgf regulation of brachyury/ntla in which Fgf promotes brachyury/ntla expression during gastrulation but inhibits brachyury/ntla expression in the tailbud during body elongation (Goto et al., 2017). These switches in regulatory feedback may underlie the transition from gastrulation to tailbud elongation.
We estimate that the mechanical information in the posterior neural tube travels at roughly 1 micron per minute when measured at 18°C, which is a similar order of magnitude to the 0.39 micron per minute average anterior to posterior velocity of posterior neural tube cells in wild-type embryos when measured at 18°C (Das et al., 2017). Zebrafish are ectothermic, and their rate of development increases dramatically with the temperature at which they are raised. Thus, one would expect the propagation of mechanical information to also scale with temperature. To aid our cell tracking, we image embryos in a chamber at 18°C whereas the standard temperature for raising and staging zebrafish is 28.5°C (Kimmel et al., 1995). The effect of temperature on the rate of development during somitogenesis has been quantified and using that information, we estimate that mechanical information would travel at a rate of 3 to 4 microns per minute at 28.5°C (Schroter et al., 2008). With an average cell diameter of ~10 microns, the mechanical information passes from cell to cell in roughly 10 minutes at 18°C and in 2 to 3 minutes at 28.5°C. These rates are too rapid to be mediated via transcription and translation. Furthermore, the transcription factors most linked to mechanotransduction, Yap/Wwtr, function in the notochord and epidermis but not in the spinal cord or paraxial mesoderm progenitors during zebrafish body elongation (Kimelman et al., 2017).
The mechanical information does not appear to propagate faster than morphogen diffusion (Muller et al., 2013). However, mechanical information could have a larger effective range as it may dissipate more slowly than the rate at which a morphogen concentration drops below a minimum threshold. Mechanical information may also more easily propagate anisotropically. For example, mechanical information may travel most effectively through a migrating population of cells. Since we only observe mechanical information as a disturbance in cell migration, we are tautologically unable to address this hypothesis. We are also not able to determine whether mechanical information travels downstream of the tail organizer into the presomitic mesoderm because the population of the presomitic mesoderm with eve1 transgenic cells and other tail organizer cells makes it impossible to rule out cell autonomous effects.
In summary, we find that an embryonic organizer orchestrates morphogenesis over distances beyond the range of morphogen signaling. This mechanical information is a secondary effect of canonical organizer cell signaling, and it is unclear whether it affects either gene expression or cell fate. Mechanical information is likely mediated by biochemical responses to cell-cell contact and not via force transfer between cells. Improved experimental methods for simultaneous spatiotemporal control of gene function and live imaging may reveal that mechanical information orchestrates morphogenesis in other contexts from embryonic development to organogenesis and regeneration.
Star Methods
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Scott Holley (scott.holley@yale.edu).
Experimental Model and Subject Details
Zebrafish care and breeding followed standard protocols (Nüsslein-Volhard and Dahm, 2002) as approved by Yale IACUC. Briefly, zebrafish are housed in a recirculating tank system. Embryos are attained from adults placed pairwise in mating tanks for natural spawning. Adults are returned to their regular tanks after mating and given at least two weeks before the next mating. Wild-type strains used for morphological and qPCR analyses were Tubingen and for imaging and western blots were TL and TLAB. For all assays, embryos were raised at 28°C for two hours, followed by 22°C for eleven hours and then 20°C until treatment. All experiments are performed on embryos within the first 48 hours of development well before sex determination in zebrafish (Wilson et al., 2014).
Quantitative Real-Time PCR
Embryos were treated with 40 μM DMH1 or 0.4% DMSO vehicle control at approximately the 5-somite stage (5ss) for one, three or five hours. Embryos were manually dissected to isolate the presomitic mesoderm in Mark’s Modified Ringer buffer with 10mM HEPES. Approximately 50 tailbuds were pooled per experimental replicate. RNA was extracted using the RNeasy Micro Kit (Qiagen) and cDNA was reverse transcribed using the High-Capacity cDNA RT kit (Applied Biosystems/Thermofisher). qRT-PCR was performed as previously described (Stulberg et al., 2012). cDNA was mixed with primers, buffer, and power SYBR green (Applied Biosystems) and loaded in a 7900 HT Applied Biosystems machine. Cycle parameters were 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 58°C for 1 min. Primer concentrations were adjusted to attain primer efficiencies between 90–120%. The Tg cpi-17 experiments were performed on a QuantStudio3 (Applied Biosystems). Three replicates were performed for all measurements. Primer sequences are provided in Table S1. Most primers were designed to include one exonic and one intronic sequence, so only nascent transcripts were measured. When intronic primers were unavailable, both primers were in exonic regions. Fold change was calculated as 2−ΔΔCt, where expression was normalized to β-actin and compared to a control, and converted to log10. Statistical comparisons were made using the Student’s unpaired t test.
Western Blot
Embryos were treated at the 5-somite stage with 40 μM DMH1 or 0.4% DMSO for three hours. Embryos were manually dissected in Mark’s Modified Ringer with 10 mM HEPES, and tissue was deyolked and protein extracted as in (Link et al., 2006). Protein samples were run on a 12% SDS-PAGE gel at 200V for 90 minutes. After blocking with 5% milk in TBST, blots were incubated two nights in 1:500 α-pSmad1/5/8 (Cell Signaling Technologies) at 4°C in block. Next, blots were rinsed twice in TBST, washed 1× 10 minutes and 3× 5 minutes before incubating for 90 minutes in 1:10,000 α-H2B (Abcam) in block. Blot was washed in TBST as before and incubated 1:50,000 in goat α-rabbit HRP (Sigma-Aldrich) for two hours before a third wash procedure and detection with Pierce ECL 2 (Thermo Scientific). Western blots were visualized using a Typhoon FLA 9500 Phosphorimager (GE) and quantified using ImageQuant software.
In Situ Hybridization and Immunohistochemistry
For immunohistochemistry, embryos were stained using anti-pSMAD1/5/8 primary antibody (Cell Signaling Technologies) at a concentration of 1:100, and the endogenous GFP signal was used to identify transgenic cells. Shown are heat maps of a single representative 1.2μm optical slice of pSMAD1/5/8 staining with GFP-positive cells circled in white. Double fluorescent in situ hybridization for eve1 and bmp4 was performed as previously described (Brend and Holley, 2009; Jülich et al., 2005). Probes were amplified by PCR from wild-type cDNA. Primers used were eve1F: tggcttggaagagaaacagtgacg, eve1R+T7: aatacgactcactatagtcaggtctggaatgacacaggagt, bmp4F: atgattcctggtaatcgaatg and bmp4R+T7: cgtaatacgactcactatagggttagcggcagccacacccctc. Shown is projection view of z-stack from a representative embryo. Images were taken with a Zeiss 510 LSM confocal microscope. Three experimental replicates were performed for each assay with a total of twelve embryos imaged for GFP/pSMAD1 antibody stains, and ten embryos for eve1/bmp4 in situs.
Drug Treatment
Embryos were treated at the 5-somite stage with 40 μM DMH1 or 0.4% DMSO. DMH1 was dissolved in DMSO to a stock concentration of 10 mM, and further diluted in E2 for treatment. Embryos were dechorionated with pronase and kept at 28°C after treatment. Morphological analysis was performed using a dissecting microscope at both 5 and 24 hours post treatment.
Transgenic Embryos
Embryos were injected with 30 ng/μl pT2 tbx6l-βglobin:eve1-P2A-emGFP DNA and 150 ng/μl transposase mRNA, 150 ng/μl pT2 tbx6l-βglobin:cpi17-P2A-emGFP DNA and 50 ng/μl transposase mRNA or 150 ng/μl pT2 tbx6l-βglobin:emGFP DNA and 50 ng/μl transposase mRNA at the 1-cell stage and raised as described above. The tbx6l enhancer drives transcription in the posterior tailbud (Dray et al., 2013; Jülich et al., 2015; Szeto and Kimelman, 2004). The 5’ UTR of human beta globin was used to increase translation. The P2A sequence enabled expression of two peptides from a single mRNA (Kim et al., 2011). Embryos were manually sorted for strong GFP fluorescence in the tail between the tailbud and 5-somite stage using a fluorescent dissecting microscope.
Confocal Imaging
Embryos were injected with 100 ng/μl nls-RFP mRNA at the 1-cell stage and raised as above. Embryos were mounted on their tail in 1.5% low-melt agarose on a 24×50mm coverslip (FisherScientific), and chambers were backfilled with DMH1 or DMSO solution. Movies were taken at 18°C using a Linkam Scientific PE100 cooling stage on a Zeiss LSM 510 confocal microscope for 1.5 to 3 hours. Cell movement was tracked and manually sorted based on tailbud region (PNT, DM, TO/PZ, PSM) using Imaris software (Bitplane) as previously described (Lawton et al., 2013).
Image Analysis
Distributions for cell track straightness, mean coefficient of variation (C.V.) of cell track speed and track mean speed for all cells within each region (PNT, DM, TO/PZ, PSM) were exported directly from Imaris. Average values for each region are shown, but statistical tests were based on the full distribution using ANOVA analysis with bootstrapping. The data extracted from Imaris were analyzed to calculate the polarization, MSD, local order, distribution of nearest neighbors, and probability of negative Vy. For the calculations of these quantities, we used the center of mass of the anterior 50 μm of the PSM as our reference frame to correct for global growth of the embryo as described previously (Lawton et al., 2013). The individual cell positions and velocities were then corrected as below:
Here and are the instantaneous individual positions and velocities respectively extracted from Imaris (raw data); and are the corrected instantaneous positions and velocities respectively; is the velocity of the center of mass of the anterior 50 μm of the PSM; and Δt is the time step (Δt is about 3 min in the experimental movies). All quantities were calculated using custom codes written in Matlab, and are defined below.
Polarization (Quantification of global order):
The polarization is defined as:
Here, the summation indicates an average taken over the total number of cells (N) at each time point, and the outer brackets indicate an average over all time points. For high global order, Φ is close to 1, and for global disorder Φ is close to 0.
Mean square displacement (MSD):
The MSD for each individual track is defined by , where τ is the lag-time. Diffusion and velocity coefficients for each tailbud region were calculated as before (Dray et al., 2013) by utilizing a Bayesian approach to select the most appropriate model (Monnier et al., 2012). The best-fitted model was a drift-diffusive motion for all the experimental movies.
Alignment angle (Quantification of local order):
For each i-th cell, we defined the 'alignment angle' as . Here, denotes a local mean velocity averaged over the cells (including cell i) that lie within a sphere of 20 μm radius, centering on cell i. For high local order, there is a greater probability of θi to be close to zero.
Top 10% displacement images:
We previously found that patterns of cell flow in the tail organizer can be visualized by displaying only the top 10% of cell tracks with the greatest displacement in a given direction (Lawton et al., 2013). To create the top 10% displacement images, the top 10% of cell tracks displacing ventrally within the PZ were selected and subsequently deleted from the region before selecting and deleting the top 10% of tracks that displaced laterally, then anteriorly, and finally in the dorsal direction so as to ensure no tracks were double-counted. Selected tracks were color-coded and grouped together for visualization.
Simulation Methods
Based on our previous 2D model of tailbud cell migration (Das et al., 2017), we developed a more realistic 3D model of zebrafish tail elongation. Each cell, i, is modeled as a self-propelled soft particle with a radius ai and instantaneous position , which moves with a constant self-propelling speed v0 in a well-defined direction . In the overdamped limit (i.e. when cell inertia is negligible), the dynamics of the i-th cell is given by
| (1) |
Here, μ is the mobility parameter, and is an intercellular force between the i-th and j-th cells. The intercellular force is summed over all the nearest neighbors. Thus, the instantaneous velocity of each individual cell is determined by two distinct factors: one is the cellular self-propelling velocity () stemming from an active force, and the other is the net intercellular force exerted by its neighbors.
To model the emergence of collective order, we combined the essential feature of local velocity alignment of individual cells from a 2D model (Szabo et al., 2006) with a 3D Vicsek model (Gönci et al., 2008). The direction of cellular self-propulsion attempts to align itself to the direction of the instantaneous velocity , but with some uncertainty. The self-propelling directions of the cells were updated following the rule
| (2) |
Here Δt is the time step, set at Δt = 0.005 in the simulations. is a rotation matrix that represents a random rotation of the instantaneous direction of individual velocity about a random axis (defined by the unit vector ), and by a random angle, ξ. The unit vector is a random vector perpendicular to , and chosen from a uniform distribution. Thus, defines a random axis for the rotation. The random angle, ξ represents the 'angular noise', which is a random number chosen uniformly in the interval η[−π, π], where η is the strength of the angular noise. The value of η varies from 0 to 1. When the angular noise strength is zero (η = 0), ordered motion quickly emerges. At maximum noise (η = 1), the motion is completely random.
The intercellular force is modeled as a short-ranged and piece-wise linear function of intercellular distance . This force is repulsive for distances smaller than Req, while it is attractive for distances Req ≤rij ≤R0, and zero if cells are farther apart than R0:
| (3) |
Here, frep and fadh are the maximum repulsive and adhesive forces, and . Note that repulsive forces in the simulation may represent a number of in vivo mechanisms including volume exclusion between cells, as two cells cannot occupy the same space as well as biochemical processes such as contact mediated repulsion. The adhesive forces result from cell-cell attractive interactions mediated by proteins like Cadherins. We set frep >> fadh to avoid any clustering (or crystallization) of cells, which was never observed experimentally. The equilibrium distance between two cells (i and j) is taken to be Req = (ai + aj), i.e. the sum of the corresponding radii. We assigned the individual cellular radii within the range ai ⌊ [a0 − δ,a0 + δ], i. e. uniformly distributed about the average radius a0 with root-mean-square fluctuations (δ = 0.05).
We simulated the cell motion by numerically integrating Eqs. (1) and (2) using the 'explicit Euler’ method with a time step Δt = 0.005, and assuming a perfectly reflecting boundary condition (rigid boundaries). We start with random initial positions and self-propulsion directions of the cells. The cells are initialized at a high packing fraction (0.95), as cells are closely packed inside the tailbud. The shape of the tailbud is modeled as a half-cylindrical structure, and its medial-lateral (ML) dimension matches that of the experimental datasets (see Figure S4A). The ML dimension of the tailbud is roughly L0 / 2a0 ∪12 cells wide, while the width of the PSM is about half of the PNT (Figure S4A). Here L0 is the ML width of the tailbud and a0 is the average cell radius. We set L0 = 10 and 2a0 = 5/6 in the simulations.
To make the tailbud grow in time, cells were introduced in the PNT at a constant rate from the anterior boundary (Figure S4A). This approximates the posterior flow of cells in the embryo partly due to convergent extension (Roszko et al., 2009; Steventon et al., 2016). We introduce a cell in every γ iteration steps, where γ is a parameter that controls the cell influx. In response to the increase in cell number in the PNT, we simultaneously slide the curved posterior boundary and the boundary separating left and right PSMs in the posterior direction (see the bold red arrows in Figure S4A). The barrier between PNT and PSM represents the extra-cellular matrix (ECM) that progressively forms over time and binds the cells to the epidermis. To estimate the increase in the anterio-posterior (AP) length of the half-cylindrical structure, we assumed that the global cell density is constant, i.e.
| (4) |
Here, N(t) is the total cell number inside the tailbud, and V(t) is the net volume of the half-cylindrical tailbud at an instant, t. Thus, the increase in volume ΔV = V(t + Δt)−V(t) can be calculated from the increase in cell number ΔN = N(t + Δt)−N(t). From geometrical consideration, the total volume of the tailbud at any instant is given by
| (5) |
Here, LPSM is the instantaneous AP length of the PSM (Note that the length of PNT is the same as the length of PSM in our model, see Figure S2A). So the overall AP length of the tailbud is Ltot = LpsM + LTO. The length of the tail-organizer, LTO is kept fixed over time; while the length of the PSM (LPSM) and consequently the total length (Ltot) increase over time as below
| (6) |
Here, ΔL is the length increment at each time step, by which amount the boundaries need to be moved posteriorly, preserving the shape of the half-cylindrical structure (see Figure S2A). The length increment ΔL is calculated by simultaneously solving Equations 4 and 5 at each time step.
Parameters values
To simulate the wild-type cell motion, we used v0 = 1, μ = 1, γ = 5, η = 0.7, fadh = 1, frep = 30, and R0 = 1.2Req, consistent with our earlier 2D model (Das et al., 2017). This set of parameters mimics the observed wild-type cell motion in the PNT, and correctly reproduces the wild-type value of polarization.
Numerical rules to introduce perturbation in the tail organizer
To introduce a local perturbation in the tail-organizer (TO), we stochastically increased the cell repulsion of a group of cells that migrate into the TO after an onset of the perturbation. Each cell that migrates into the TO from the DM has a given probability of 80% to increase its repulsion parameter value, and any cell that comes into contact with this 'perturbed' cell, experiences a higher cell-cell repulsion. Once a cell switches to a higher value of the repulsion parameter, it cannot return to the 'wild-type' state again. Thus, the perturbation creates a continuous pool of cells with high repulsion in the TO. The enhanced value of the repulsion is 7 times the wild-type value (fpurt = 7 frep).
Analysis of the simulation data
The codes for the model simulations were written in FORTRAN 90. The outputs of the program were averaged over 30 simulations of the tailbud in perturbed and in unperturbed conditions for data shown in Figure 4.
Analysis of front propagation in the simulations
In the simulations, we quantified the response in cell motion in the PNT following a perturbation in the TO (Figure S4D-S4F). We quantified the net AP velocity in the PNT, Vy (defined as: PNT, where is the instantaneous y-component of the i-th cell, and N is the instantaneous number of cells in a box of fixed volume in the PNT). This AP velocity frequently becomes negative after the perturbation (Figure S4D). We also found that the cell density in the PNT increases after the perturbation (Figure S4E). Finally, the polarization sharply drops once the perturbation is switched on (Figure S4F).
To quantify the propagation of the disturbance created by the perturbation, we partitioned the PNT into a number of boxes with equal size starting from the posterior end of the PNT, and measured the spatial average of the net AP velocity 〈Vy〉 in each box, and plotted 〈Vy〉 as a function of the distance from the posterior end of PNT. In this velocity profile, the average AP velocity has both positive and negative parts (Figure S2E) and the zero-crossing point (black arrow heads in Figure S4G) progressively shifts towards the anterior. The zero-crossing point of 〈Vy〉 represents the front position at any instant (Cartoon below Figure S4G). A similar analysis for the density profile showed that the density has a peak near the posterior end, and this peak position slowly shifts towards the anterior with time (Figure S4H). To see how quickly the front propagates, we plotted the front position as a function of the elapsed time from the onset of the perturbation (Figure S4I). After a sharp initial rise, the front propagates almost linearly in time, and then damps out.
Significance test
To establish if there is any significant difference between samples for a given quantity, we used a two-sample T-test provided by the Matlab function 'ttest2'. Significance was defined as p< 0.05.
Supplementary Material
Movie S1. A 3D computer simulation of the tailbud demonstrating the mechanical information propagation. Related to Figure 4. Red circles denote the cells in the PNT with instantaneous velocities directed posterior to anterior (negative AP velocity). Green circles denote the perturbed cells with high repulsion. The perturbation is switched on at time-point 100. Each time units equal 30 in silico iterations, and the length scale is in units of cell diameters. The parameters are specified in Supplemental Information.
Movie S2. Cell motion in the PNT of DMSO-treated control embryos. Related to Figure 4. An experimental timelapse is projected on a dorsal plane. All circles denote the nuclei. Red circles denote the nuclei with instantaneous velocities directed posterior to anterior (negative AP velocity).
Movie S3. Cell motion in the PNT of DMH1-treated tbx6l:eve1 transgenics. Related to Figure 4. An experimental timelapse is projected on a dorsal plane. All circles denote the nuclei. Red circles denote the nuclei with instantaneous velocities directed posterior to anterior (negative AP velocity).
Movie S4. Cell motion in the PNT of tbx6l:GFP transgenic embryos. Related to Figure 6. An experimental timelapse is projected on a dorsal plane. All circles denote the nuclei. Red circles denote the nuclei with instantaneous velocities directed posterior to anterior (negative AP velocity).
Movie S5. Cell motion in the PNT of tbx6l:cpi-17 transgenic embryos. Related to Figure 6. An experimental timelapse is projected on a dorsal plane. All circles denote the nuclei. Red circles denote the nuclei with instantaneous velocities directed posterior to anterior (negative AP velocity).
Highlights.
Positive feedback between Bmp signaling and eve1 functions in the tail organizer.
Perturbation of organizer signaling has long-range effects on cell motion.
These long-range effects are beyond the range organizer signaling.
These long-range effects are mediated by mechanical information.
Acknowledgements
We thank Miriam Genuth and Abdel-Rahman Hassan for comments on the manuscript and Josien van Wolfswinkel’s lab for assistance with the qRT-PCR experiments. Research support provided by award R33GM114257 from the NIH Common Fund Single Cell Analysis Program to (S.A.H, T.E. and C.S.O.) and award 1R01GM129149-01 from NIGMS to (S.A.H).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Agathon A, Thisse C, and Thisse B (2003). The molecular nature of the zebrafish tail organizer. Nature 424, 448–452. [DOI] [PubMed] [Google Scholar]
- Barone V, Lang M, Krens SFG, Pradhan SJ, Shamipour S, Sako K, Sikora M, Guet CC, and Heisenberg CP (2017). An Effective Feedback Loop between Cell-Cell Contact Duration and Morphogen Signaling Determines Cell Fate. Dev Cell 43, 198–211.e112. [DOI] [PubMed] [Google Scholar]
- Barro O, Vriz S, Joly JS, Joly C, Condamine H, and Boulekbache H (1995). Widespread expression of the eve1 gene in zebrafish embryos affects the anterior-posterior axis pattern. Developmental genetics 17, 117–128. [DOI] [PubMed] [Google Scholar]
- Beck CW, and Slack JM (1999). A developmental pathway controlling outgrowth of the Xenopus tail bud. Development 126, 1611–1620. [DOI] [PubMed] [Google Scholar]
- Beck CW, Whitman M, and Slack JM (2001). The role of BMP signaling in outgrowth and patterning of the Xenopus tail bud. Dev Biol 238, 303–314. [DOI] [PubMed] [Google Scholar]
- Benazeraf B, Francois P, Baker RE, Denans N, Little CD, and Pourquie O (2010). A random cell motility gradient downstream of FGF controls elongation of an amniote embryo. Nature 466, 248–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet JF, Cassagne J, and Kellay H (2009). Blast shocks in quasi-two-dimensional supersonic granular flows. Phys Rev Lett 103, 224501. [DOI] [PubMed] [Google Scholar]
- Brend T, and Holley SA (2009). Zebrafish whole mount high-resolution double fluorescent in situ hybridization. J Vis Exp, 1229. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors SA, Tucker JA, and Mullins MC (2006). Temporal and spatial action of tolloid (mini fin) and chordin to pattern tail tissues. Dev Biol 293, 191–202. [DOI] [PubMed] [Google Scholar]
- Cruz C, Maegawa S, Weinberg ES, Wilson SW, Dawid IB, and Kudoh T (2010). Induction and patterning of trunk and tail neural ectoderm by the homeobox gene eve1 in zebrafish embryos. Proc Natl Acad Sci U S A 107, 3564–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D, Chatti V, Emonet T, and Holley SA (2017). Patterned disordered cell motion ensures vertebral column symmetry. Dev Cell 42, 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray N, Lawton AK, Nandi A, Jülich D, Emonet T, and Holley SA (2013). Cell-Fibronectin interactions propel vertebrate trunk elongation via tissue mechanics. Curr Biol 23, 1335–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dush MK, and Martin GR (1992). Analysis of mouse Evx genes: Evx-1 displays graded expression in the primitive streak. Dev Biol 151, 273–287. [DOI] [PubMed] [Google Scholar]
- Fainsod A, Steinbeisser H, and De Robertis EM (1994). On the function of BMP-4 in patterning the marginal zone of the Xenopus embryo. Embo J 13, 5015–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fior R, Maxwell AA, Ma TP, Vezzaro A, Moens CB, Amacher SL, Lewis J, and Saude L (2012). The differentiation and movement of presomitic mesoderm progenitor cells are controlled by Mesogenin 1. Development 139, 4656–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furthauer M, Lin W, Ang SL, Thisse B, and Thisse C (2002). Sef is a feedback-induced antagonist of Ras/MAPK-mediated FGF signalling. Nat Cell Biol 4, 170–174. [DOI] [PubMed] [Google Scholar]
- Gofflot F, Hall M, and Morriss-Kay GM (1997). Genetic patterning of the developing mouse tail at the time of posterior neuropore closure. Dev Dyn 210, 431–445. [DOI] [PubMed] [Google Scholar]
- Goldman DC, Martin GR, and Tam PP (2000). Fate and function of the ventral ectodermal ridge during mouse tail development. Development 127, 2113–2123. [DOI] [PubMed] [Google Scholar]
- Gönci B, Nagy M, and Vicsek T (2008). Phase transition in the scalar noise model of collective motion in three dimensions. The European Physical Journal Special Topics 157, 53–59. [Google Scholar]
- Goodwin K, Ellis SJ, Lostchuck E, Zulueta-Coarasa T, Fernandez-Gonzalez R, and Tanentzapf G (2016). Basal Cell-Extracellular Matrix Adhesion Regulates Force Transmission during Tissue Morphogenesis. Dev Cell 39, 611–625. [DOI] [PubMed] [Google Scholar]
- Goto H, Kimmey SC, Row RH, Matus DQ, and Martin BL (2017). FGF and canonical Wnt signaling cooperate to induce paraxial mesoderm from tailbud neuromesodermal progenitors through regulation of a two-step EMT. Development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouti M, Delile J, Stamataki D, Wymeersch FJ, Huang Y, Kleinjung J, Wilson V, and Briscoe J (2017). A Gene Regulatory Network Balances Neural and Mesoderm Specification during Vertebrate Trunk Development. Dev Cell 41, 243–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Ho JN, Lewis JA, Karim KA, Daniels RN, Gentry PR, Hopkins CR, Lindsley CW, and Hong CC (2010). In vivo structure-activity relationship study of dorsomorphin analogues identifies selective VEGF and BMP inhibitors. ACS chemical biology 5, 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg CP, and Bellaiche Y (2013). Forces in tissue morphogenesis and patterning. Cell 153, 948–962. [DOI] [PubMed] [Google Scholar]
- Joly JS, Joly C, Schulte-Merker S, Boulekbache H, and Condamine H (1993). The ventral and posterior expression of the zebrafish homeobox gene eve1 is perturbed in dorsalized and mutant embryos. Development 119, 1261–1275. [DOI] [PubMed] [Google Scholar]
- Jülich D, Cobb G, Melo AM, McMillen P, Lawton AK, Mochrie SGJ, Rhoades E, and Holley SA (2015). Cross-scale Integrin regulation organizes ECM and tissue topology. Dev Cell 34, 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jülich D, Lim C-H, Round J, Nicolaije C, Davies A, Schroeder J, Geisler R, Consortium TS, Lewis J, Jiang Y-J, et al. (2005). beamter/deltaC and the role of Notch ligands in the zebrafish somite segmentation, hindbrain neurogenesis and hypochord differentiation. Dev Biol 286, 391–404. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA, and Choi SY (2011). High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6, e18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimelman D, Smith NL, Lai JKH, and Stainier DY (2017). Regulation of posterior body and epidermal morphogenesis in zebrafish by localized Yap1 and Wwtr1. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WM, R KS, Ullmann B, and Schilling TF. (1995). Stages of Embryonic Development of the Zebrafish. Dev Dynam 203, 253–310. [DOI] [PubMed] [Google Scholar]
- Lander AD (2007). Morpheus unbound: reimagining the morphogen gradient. Cell 128, 245–256. [DOI] [PubMed] [Google Scholar]
- Lawton AK, Nandi A, Stulberg MJ, Dray N, Sneddon MW, Pontius W, Emonet T, and Holley SA (2013). Regulated tissue fluidity steers zebrafish body elongation. Development 140, 573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeGoff L, and Lecuit T (2015). Mechanical Forces and Growth in Animal Tissues. Cold Spring Harbor perspectives in biology 8, a019232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, and Manley JL (1998). Even-skipped represses transcription by binding TATA binding protein and blocking the TFIID-TATA box interaction. Mol Cell Biol 18, 3771–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link V, Shevchenko A, and Heisenberg CP (2006). Proteomics of early zebrafish embryos. BMC Dev Biol 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lye CM, Blanchard GB, Naylor HW, Muresan L, Huisken J, Adams RJ, and Sanson B (2015). Mechanical Coupling between Endoderm Invagination and Axis Extension in Drosophila. PLoS Biol 13, e1002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AJ, and Kimelman D (2015). Tbx16 and Msgn1 are required to establish directional cell migration of zebrafish mesodermal progenitors. Dev Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mara A, Schroeder J, Chalouni C, and Holley SA (2007). Priming, Initiation and Synchronization of the Segmentation Clock by deltaD and deltaC. Nat Cell Biol 9, 523–530. [DOI] [PubMed] [Google Scholar]
- Martin BL, and Kimelman D (2012). Canonical Wnt signaling dynamically controls multiple stem cell fate decisions during vertebrate body formation. Dev Cell 22, 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay LM, Carpenter B, and Roberts SG (1999). Evolutionary conserved mechanism of transcriptional repression by even-skipped. Nucleic Acids Res 27, 3064–3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillen P, and Holley SA (2015). The tissue mechanics of vertebrate body elongation and segmentation. Curr Opin Genet Dev 32, 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongera A, Rowghanian P, Gustafson HJ, Shelton E, Kealhofer DA, Carn EK, Serwane F, Lucio AA, Giammona J, and Campas O (2018). A fluid-to-solid jamming transition underlies vertebrate body axis elongation. Nature 561, 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier N, Guo SM, Mori M, He J, Lenart P, and Bathe M (2012). Bayesian approach to MSD-based analysis of particle motion in live cells. Biophys J 103, 616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P, Rogers KW, Yu SR, Brand M, and Schier AF (2013). Morphogen transport. Development 140, 1621–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, Brand M, van Eeden FJM, Furutani-Seiki M, Granato M, Haffter P, Heisenberg C-P, et al. (1996). Genes establishing dorsoventral pattern formation in the zebrafish embryo: the ventral specifying genes. Development 123, 81–93. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, and Dahm R, eds. (2002). Zebrafish (Oxford, UK: Oxford University Press; ). [Google Scholar]
- O’Neill K, and Thorpe C (2013). BMP signaling and spadetail regulate exit of muscle precursors from the zebrafish tailbud. Dev Biol 375, 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S, Suzuki K, Tachibana K, Tanaka H, and Yamada G (2007). Cessation of gastrulation is mediated by suppression of epithelial-mesenchymal transition at the ventral ectodermal ridge. Development 134, 4315–4324. [DOI] [PubMed] [Google Scholar]
- Pourquie O (2011). Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell 145, 650–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyati UJ, Webb AE, and Kimelman D (2005). Transgenic zebrafish reveal stage-specific roles for Bmp signaling in ventral and posterior mesoderm development. Development 132, 2333–2343. [DOI] [PubMed] [Google Scholar]
- Rericha EC, Bizon C, Shattuck MD, and Swinney HL (2002). Shocks in supersonic sand. Phys Rev Lett 88, 014302. [DOI] [PubMed] [Google Scholar]
- Roszko I, Sawada A, and Solnica-Krezel L (2009). Regulation of convergence and extension movements during vertebrate gastrulation by the Wnt/PCP pathway. Semin Cell Dev Biol 20, 986–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row RH, and Kimelman D (2009). Bmp inhibition is necessary for post-gastrulation patterning and morphogenesis of the zebrafish tailbud. Dev Biol 329, 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row RH, Pegg A, Kinney BA, Farr GH 3rd, Maves L, Lowell S, Wilson V, and Martin BL (2018). BMP and FGF signaling interact to pattern mesoderm by controlling basic helix-loop-helix transcription factor activity. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada A, Shinya M, Jiang YJ, Kawakami A, Kuroiwa A, and Takeda H (2001). Fgf/MAPK signalling is a crucial positional cue in somite boundary formation. Development 128, 4873–4880. [DOI] [PubMed] [Google Scholar]
- Schroter C, Herrgen L, Cardona A, Brouhard GJ, Feldman B, and Oates AC (2008). Dynamics of zebrafish somitogenesis. Dev Dyn 237, 545–553. [DOI] [PubMed] [Google Scholar]
- Seebald JL, and Szeto DP (2011). Zebrafish evel regulates the lateral and ventral fates of mesodermal progenitor cells at the onset of gastrulation. Dev Biol 349, 78–89. [DOI] [PubMed] [Google Scholar]
- Shyer AE, Huycke TR, Lee C, Mahadevan L, and Tabin CJ (2015). Bending gradients: how the intestinal stem cell gets its home. Cell 161, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steventon B, Duarte F, Lagadec R, Mazan S, Nicolas JF, and Hirsinger E (2016). Species tailoured contribution of volumetric growth and tissue convergence to posterior body elongation in vertebrates. Development 143, 1732–1741. [DOI] [PubMed] [Google Scholar]
- Stickney HL, Imai Y, Draper B, Moens C, and Talbot WS (2007). Zebrafish bmp4 functions during late gastrulation to specify ventroposterior cell fates. Dev Biol 310, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stulberg MJ, Lin A, Zhao H, and Holley SA (2012). Crosstalk between Fgf and Wnt Signaling in the Zebrafish Tailbud. Dev Biol 369, 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo B, Szollosi GJ, Gonci B, Juranyi Z, Selmeczi D, and Vicsek T (2006). Phase transition in the collective migration of tissue cells: experiment and model. Physical review E, Statistical, nonlinear, and soft matter physics 74, 061908. [DOI] [PubMed] [Google Scholar]
- Szabó B, Szöllösi GJ, Gönci B, Jurányi Z, Selmeczi D, and Vicsek T (2006). Phase transition in the collective migration of tissue cells: Experiment and model. Physical Review E 74, 061908. [DOI] [PubMed] [Google Scholar]
- Szeto DP, and Kimelman D (2004). Combinatorial gene regulation by Bmp and Wnt in zebrafish posterior mesoderm formation. Development 131, 3751–3760. [DOI] [PubMed] [Google Scholar]
- Thisse C, and Thisse B (2005). High Throughput Expression Analysis of ZF-Models Consortium Clones. ZFIN Direct Data Submission. [Google Scholar]
- Weiser DC, Row RH, and Kimelman D (2009). Rho-regulated myosin phosphatase establishes the level of protrusive activity required for cell movements during zebrafish gastrulation. Development 136, 2375–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ML, and Solnica-Krezel L (2017). Regulation of gastrulation movements by emergent cell and tissue interactions. Curr Opin Cell Biol 48, 33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CA, High SK, McCluskey BM, Amores A, Yan YL, Titus TA, Anderson JL, Batzel P, Carvan MJ 3rd, Schartl M, et al. (2014). Wild sex in zebrafish: loss of the natural sex determinant in domesticated strains. Genetics 198, 1291–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson V, Olivera-Martinez I, and Storey KG (2009). Stem cells, signals and vertebrate body axis extension. Development 136, 1591–1604. [DOI] [PubMed] [Google Scholar]
- Xiong F, Tentner AR, Huang P, Gelas A, Mosaliganti KR, Souhait L, Rannou N, Swinburne IA, Obholzer ND, Cowgill PD, et al. (2013). Specified neural progenitors sort to form sharp domains after noisy Shh signaling. Cell 153, 550–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, and Thorpe C (2011). BMP and non-canonical Wnt signaling are required for inhibition of secondary tail formation in zebrafish. Development 138, 2601–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. A 3D computer simulation of the tailbud demonstrating the mechanical information propagation. Related to Figure 4. Red circles denote the cells in the PNT with instantaneous velocities directed posterior to anterior (negative AP velocity). Green circles denote the perturbed cells with high repulsion. The perturbation is switched on at time-point 100. Each time units equal 30 in silico iterations, and the length scale is in units of cell diameters. The parameters are specified in Supplemental Information.
Movie S2. Cell motion in the PNT of DMSO-treated control embryos. Related to Figure 4. An experimental timelapse is projected on a dorsal plane. All circles denote the nuclei. Red circles denote the nuclei with instantaneous velocities directed posterior to anterior (negative AP velocity).
Movie S3. Cell motion in the PNT of DMH1-treated tbx6l:eve1 transgenics. Related to Figure 4. An experimental timelapse is projected on a dorsal plane. All circles denote the nuclei. Red circles denote the nuclei with instantaneous velocities directed posterior to anterior (negative AP velocity).
Movie S4. Cell motion in the PNT of tbx6l:GFP transgenic embryos. Related to Figure 6. An experimental timelapse is projected on a dorsal plane. All circles denote the nuclei. Red circles denote the nuclei with instantaneous velocities directed posterior to anterior (negative AP velocity).
Movie S5. Cell motion in the PNT of tbx6l:cpi-17 transgenic embryos. Related to Figure 6. An experimental timelapse is projected on a dorsal plane. All circles denote the nuclei. Red circles denote the nuclei with instantaneous velocities directed posterior to anterior (negative AP velocity).