Abstract
Many DNA-processing enzymes have been shown to contain a [4Fe4S] cluster, a common redox cofactor in biology. We find using DNA electrochemistry that binding of the DNA polyanion promotes a negative shift in [4Fe4S] cluster potential, which corresponds thermodynamically to ~ 500-fold increase in DNA binding affinity for the oxidized [4Fe4S]3+ cluster versus the reduced [4Fe4S]2+ cluster. This redox switch can be activated from a distance using DNA charge transport chemistry. DNA-processing proteins containing the [4Fe4S] cluster are enumerated with possible roles for the redox switch, highlighted. A model is described where repair proteins may signal one another using DNA-mediated charge transport as a first step in their search for lesions. The redox switch in eukaryotic DNA primases appears to regulate polymerase handoff, and in DNA polymerase δ, the redox switch provides a means to modulate replication in response to oxidative stress. Thus we describe redox signaling interactions of DNA-processing [4Fe4S] enzymes as well as the most interesting potential players to consider in delineating new DNA-mediated redox signaling networks.
Keywords: DNA charge transport, iron sulfur clusters, base excision repair, DNA primase, DNA polymerase, redox signaling, oxidative stress
I. Introduction
Iron sulfur clusters are modular, tunable metal cofactors found in all domains of life that serve as one-electron carriers operating over a wide range of physiological potentials, from approximately −500 mV vs. NHE to 300 mV vs. NHE (1–3). Cytosolic and membrane-bound proteins have been found to coordinate a cubane [4Fe4S] cluster at a range of redox potentials that vary depending on the local protein environment and solvent exposure (Figure 1) (4, 5). Found in the [4Fe4S]2+ resting state, high potential [4Fe4S] clusters, like those in high potential iron (HiPIP) proteins, can be oxidized to the [4Fe4S]3+ state and lower potential clusters, like those in ferredoxins, can be reduced to the [4Fe4S]+ state (6). These cofactors commonly mediate redox reactions in nitrogen fixation, photosynthesis, and respiration (7–9), often acting in a chain of metal cofactors within an otherwise insulating protein matrix (10, 11).
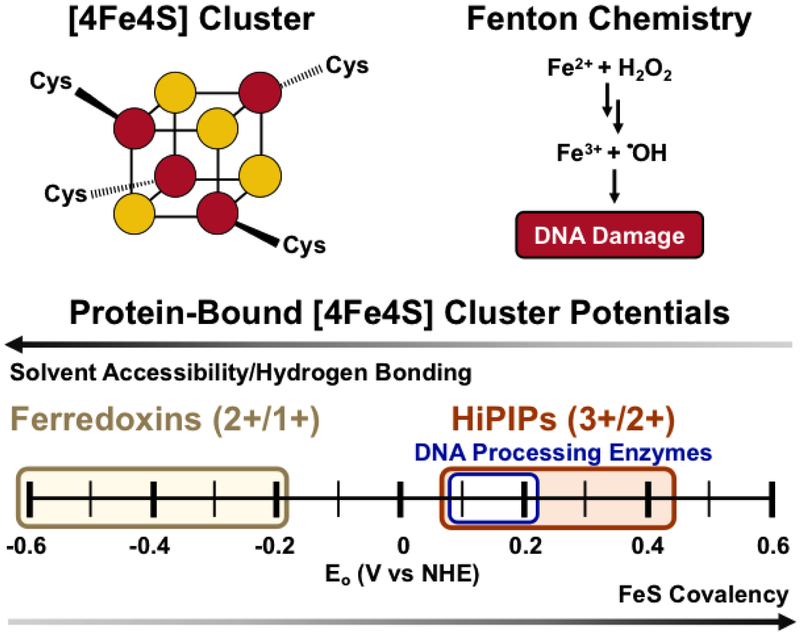
Figure 1. [4Fe4S] Clusters.
(Above, left) Schematic of the cubane [4Fe4S] cluster, with Fe in red and S in yellow. (Above, right) Cofactors containing iron can react with cellular oxidants leading to Fenton chemistry and DNA damage by the hydroxyl radical. (Below) The potential of the [4Fe4S] cluster cofactor is tunable over a wide range of physiological redox potential values. Ferredoxins access the [4Fe4S]2+/1+ couple, upon reduction from the resting [4Fe4S]2+ state. (yellow). High potential iron proteins (HiPIPs) access the [4Fe4S]3+/2+ couple upon oxidation to the [4Fe4S]3+ state from the resting [4Fe4S]2+ state (red). DNA-processing enzymes with [4Fe4S] cofactors have DNA-bound redox potentials which fall within the HiPIP [4Fe4S]3+/2+ potential range, at approximately ~65-150mV vs. NHE (blue). The solvent accessibility and hydrogen bonding/electrostatic environment of the cluster all contribute to tuning the redox potential of the cofactor (4,5).
Thirty years ago, these [4Fe4S] clusters were found also to be associated with a protein involved in DNA repair (12), and over time, more and more proteins involved in DNA processing were found to contain [4Fe4S] clusters. What were their roles? Were they structural factors or perhaps ancestral relics? As described here, we are finding that these [4Fe4S] clusters carry out redox reactions in DNA-processing proteins, serving as redox switches to regulate binding to the DNA polyanion.
[4Fe4S] Cluster Biogenesis and Loading to Target Proteins
After decades of progress, it is now understood that the incorporation of [4Fe4S] cofactors occurs through a highly regulated and coordinated series of metabolically expensive steps by a network of iron sulfur cluster biogenesis proteins (13–19). General biogenesis is carried out by the ISC pathway (Figure 2), where free iron and reduced sulfide (S2-, derived from free cysteine) are scaffolded onto components of biogenesis machinery, delivered, and loaded to apo-protein targets in a process facilitated by chaperone proteins and driven by ATP hydrolysis. In prokaryotes, this entire process occurs within the cytosol. In eukaryotes, biogenesis begins in the mitochondria, and for cytoplasmic and nuclear-bound cluster proteins (which include repair and replication enzymes), is completed in the cytoplasm by the cytosolic iron sulfur assembly (CIA) machinery (16). At present, specialized biogenesis components have not been identified for [4Fe4S] repair proteins in prokaryotes. However, prokaryotic biogenesis has been linked to pathogenesis and antibiotic resistance (20). Future studies will likely be quite informative for uncovering new regulatory roles for FeS biogenesis in all domains of life.
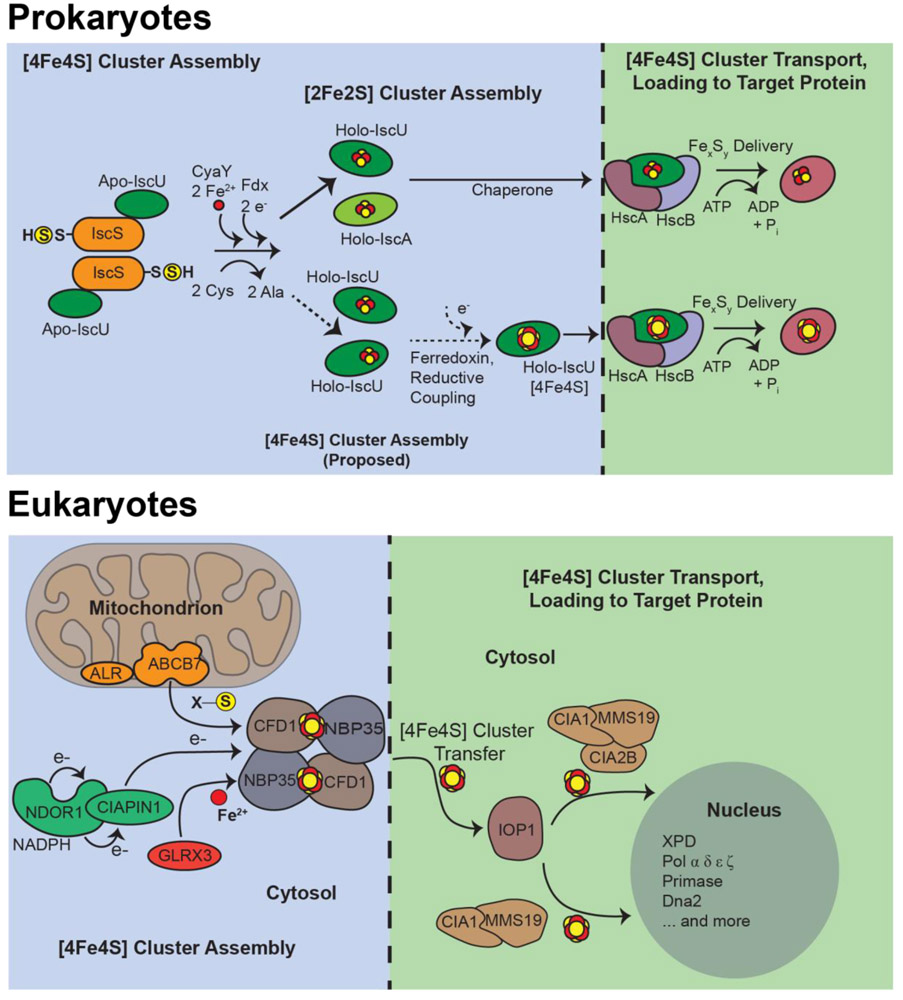
Figure 2. [4Fe4S] cluster biogenesis and loading into prokaryotic (above) and eukaryotic (below) target proteins.
Scaffold proteins, together with cysteine desulfurases and ferrous iron sources, assemble the cluster and bind chaperone machinery to then transport the cofactor first to machinery responsible for cluster delivery, then finally to target proteins. This process is not identical in bacteria and eukaryotes. However each process requires the concerted action of several specialized proteins and requires significant metabolic expense for the cell (13-19).
Mechanisms of protein target recognition by biogenesis machinery have been brought to the forefront recently. Bioinformatic signatures of the coordinating cysteines in repair and replication proteins are surprisingly weak (21); however, newly discovered target sequences recognized by biogenesis machinery, which include an LYR motif and a KKX6–10KK sequence, have been found to be essential for association with an ISC co-chaperone in yeast (13). Continued investigation of recognition motifs will be important for understanding cluster biogenesis, unraveling new facets in iron metabolism, and identifying new cluster proteins, such as those involved in DNA processes.
DNA-Processing [4Fe4S] Enzymes
The surprising discovery of a [4Fe4S] cluster in the base excision repair (BER) glycosylase Endonuclease III (EndoIII) from Escherichia coli (E. coli) in 1989 (12) soon led to the discovery of [4Fe4S] clusters in MutY (an EndoIII paralog) and Uracil DNA Glycosylase in Archaeoglobus fulgidus (AfUDG) (22, 23). Over the following decades, nucleic acid processing enzymes across several pathways were shown to contain [4Fe4S] cofactors (Table 1) (24–30). In most cases, discovery of the [4Fe4S] cluster occurred years after the first isolation of the gene products (27). As predictive tools and protein isolation methods become more and more sophisticated, we and others expect that even more [4Fe4S] clusters will be observed in essential DNA processing enzymes (27, 31).
Table 1. DNA-processing, [4Fe4S] enzymes are found in prokaryotes, archaea, and eukaryotes.
These enzymes are found in different pathways, such as repair, replication, and transcription, and perform specialized, distinct tasks in cells.
| [4Fe4S] Protein | Pathway | Function | Reference | ||
|---|---|---|---|---|---|
| Bacteria | Archaea | Eukarya | |||
| EndonucleaseIII | NTHL1, Ntg2 | Base Excision Repair | Bifunctional glycosylase, removal of oxidized pyrimidine bases | 12,24 | |
| MutY | MUTYH | Base Excision Repair | Glycosylase, removes adenines mispaired with 8-oxo-guanine | 22,24,58 | |
| XPD | XPD, Rad3 | NucleotideExcision Repair | 5'-3' helicase, unwinds DNA surrounding TT dimers | 65,97 | |
| DinG | Replication-Coupled Repair | Helicase, unwinds R-loop structures | 64 | ||
| FANCJ | Replication-Coupled Repair | 5'-3' helicase, generates ssDNA overhangs for homologous recombination | 65 | ||
| Rtel1, Chl1 | Telomere processing, meiotic crossover | 5'-3' helicase, unwinds specialized DNA structures | 83,89 | ||
| AddAB | Replication-Coupled Repair | Helicase-nuclease with 5'-3' nuclease activity, generates 3'-ssDNA overhangs | 90 | ||
| Dna2 | Replication-Coupled Repair | Helicase-nuclease, ssDNA-dependent ATPase, Okazaki fragment processing/double-strand break repair | 107 | ||
| DNA Primase | Replication | DNA-dependent RNA polymerase, synthesizes 8-14nt RNA primer on ssDNA | 62,63 | ||
| DNAPolymerase α | Replication | Extends RNA primer by 10-20nt | 106 | ||
| DNAPolymerase δ | Replication | Lagging Strand DNA polymerase, 3'-5' exonuclease | 106 | ||
| DNA Polymerase ε | Replication | Leading Strand DNA polymerase, 3'-5' exonuclease | 106, 124 | ||
| DNAPolymerase ζ | Replication | Translesion synthesis polymerase | 106, 124, 134 | ||
| RNA Polymerase | Elp3 | Transcription | Template-directed RNA synthesis | 24,29 | |
| Cas4 | CRISPR adaptive immunity | 5'- 3' exonuclease | 26 | ||
| PhrB | Photoreactivation DNA repair | Repair of UV-induced cyclopyrimidine dimers | 25 | ||
| UvrC | Nucleotide Excision Repair | 5'-, 3'- endonuclease | 28 | ||
| Spore Photoproduct Lyase | DNA Repair (UV-induced lesions) | (5R)-5-(α-thyminyl)-5,6-dihydrothymidine lesion repair | 30 | ||
The question of what role the [4Fe4S] clusters played, however, was less straightforward to answer, as early studies demonstrated that the clusters were isolated in the electron paramagnetic resonance (EPR) silent [4Fe4S]2+ state and resistant to powerful chemical oxidants and reductants (12, 23, 32). Moreover, the spectroscopic signature of coordinating cysteines was unchanged upon binding a damaged substrate, leading to the initial conclusion that the cluster had a structural role (12, 33, 34). MutY, however, could be denatured and refolded in the apo form, challenging this early conclusion (35). A substrate-sensing role was proposed for the cluster in light of this result, but a general, chemical function for the cofactor eluded observation.
II. Characterizing the Fundamental Properties of DNA-Mediated Charge Transport
In addition to the metabolic expense undertaken by cells to load a cluster into a target protein, placing an iron-containing cofactor in a DNA-binding enzyme can put the bound nucleic acid at risk of damage. A labile ferrous iron from the cofactor can undergo Fenton chemistry, creating reactive oxygen species and damaging nearby DNA bases (Figure 1) (36). Why then does Nature spend the requisite energy incorporating a redox-active inorganic cofactor into a DNA-processing enzyme?
At the same time that these proteins involved in DNA processing were being found to contain [4Fe4S] clusters, experiments were being conducted to characterize DNA charge transport chemistry (DNA CT), where electrons rapidly migrate through well stacked duplex DNA (37). The native substrate of these [4Fe4S] enzymes, double stranded DNA (dsDNA), was initially predicted to conduct charge in the dry, solid state (38), as the π -stacked DNA bases resemble the structure of graphite, a conductive material (Figure 3). To assess whether DNA conducted charge in biologically relevant aqueous conditions, new platforms were developed to examine this chemistry. Two important characteristics of this chemistry emerged: (i) DNA CT can occur over long molecular distances with shallow distance dependence, and (ii) DNA CT is exquisitely sensitive to perturbations in π-stacking of the bases.
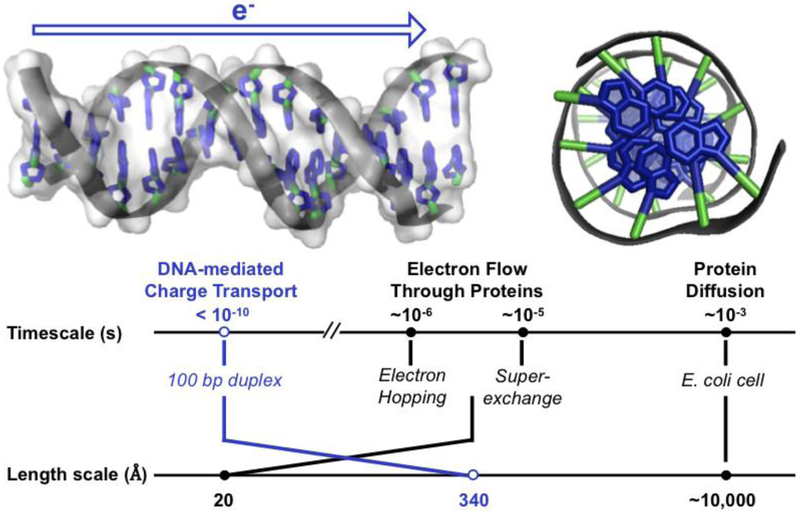
Figure 3. The structure of DNA facilitates long-range, rapid electron transfer.
(Top, left) Side view of DNA. The aromatic bases at the center of the DNA helix are oriented so that the π orbitals of adjacent bases overlap with one another in the duplex. This structural property suggests that charge could pass through the π-stacked base pairs of DNA. (Top, right) View down the helical axis of aromatic base pairs (blue) stacked in 3.4 Å layers at the center of DNA. (Bottom) Time scale and length scale of various electron transfer pathways through biomolecules (10, 11,41,43). PDB ID 3BSE.
A range of studies using DNA-bound electron donors and acceptors were used to characterize DNA CT chemistry. In an early photophysical study, a DNA oligomer was prepared containing a tethered luminescent ruthenium intercalator at one end and an intercalating rhodium oxidant at the other. While the tethered, DNA-bound ruthenium complex luminesced in the absence of the rhodium complex, in its presence, the luminescence of the ruthenium complex was quenched by electron transfer, remarkably over a distance > 40 Å (39). In a subsequent experiment using ethidium as the luminescent donor, electron transfer quenching was also evident but was attenuated in the presence of a single base mismatch intervening between the bound ethidium and rhodium (40). Long-range CT through a 63 bp duplex DNA substrate has been observed with DNA-intercalating photooxidants, where the DNA-bound photooxidant can promote oxidative damage at guanine residues from a distance. A covalent rhodium photooxidant at one end of a DNA duplex, for example, oxidizes guanine bases at the 5′- guanine of a guanine doublet, the site of low oxidation potential, through DNA CT, generating 7,8-dihydro-8-oxo-2′-deoxyguanosine lesions 200 Å from the site of intercalation (41). Experiments monitoring base-base CT utilizing 2-aminopurine furthermore showed that these DNA CT reactions can occur on the picosecond timescale, gated by the motions of the bases (42), and moving 10 times the single-step tunneling distance through protein in a miniscule fraction of the time (10, 11, 37, 43, 44)!
Again, however, perturbations in base stacking, as occur with base mismatches or kinks in the DNA, turn off this long range CT chemistry. In fact, proteins that bind and kink the DNA, such as TATA-binding protein, can be used to turn off DNA CT. In contrast, proteins that bind DNA without affecting DNA base stacking, as with histones, do not alter DNA CT (45–47).
We also explored DNA CT in the ground state using DNA electrochemistry. Here, as with the photophysical studies, we observe long range CT as long as the DNA is well stacked. Indeed, ground state CT through DNA was observed over 100 base pairs to a tethered, intercalating redox probe, methylene blue, using DNA-modified gold electrodes, but a single base mismatch in the 100-mer was sufficient to attenuate the redox signal severely (48).
III. Measuring Redox Potentials of [4Fe4S] Enzymes Bound to DNA
For our early electrochemistry studies, we had used proteins to modulate DNA CT to a small DNA-bound redox probe (46), but we considered that DNA electrochemistry could be used also to examine DNA CT to a redox cofactor within a DNA-bound protein. Could a DNA-binding protein containing a redox cofactor carry out DNA CT chemistry? If so, DNA CT experiments could be used to characterize the redox centers of DNA-binding proteins and to determine their DNA-bound potentials.
DNA-Mediated Electrochemistry
DNA-modified Au electrodes have become a useful platform for assessing whether a DNA-processing, [4Fe4S] enzyme is redox-active in solution under physiologically relevant conditions (Figure 4). Gold surfaces are functionalized with alkanethiol-modified DNA duplexes through formation of a thiol-gold bond. The Au can be used as the working electrode in a three-electrode cell after surface washing and passivation (48–50). DNA-bound redox potentials of [4Fe4S] proteins can be measured with this method, where charge flows from the electrode through the DNA to the cluster. In this platform, the DNA, functionalized onto an electrode surface, is biologically accessible; restriction enzymes, for example, can cut the DNA on the electrode with sequence specificity, as in solution. Electrochemical studies on these platforms have been central to the prediction and discovery of redox signaling between DNA-bound [4Fe4S] enzymes across different repair pathways (51–53).
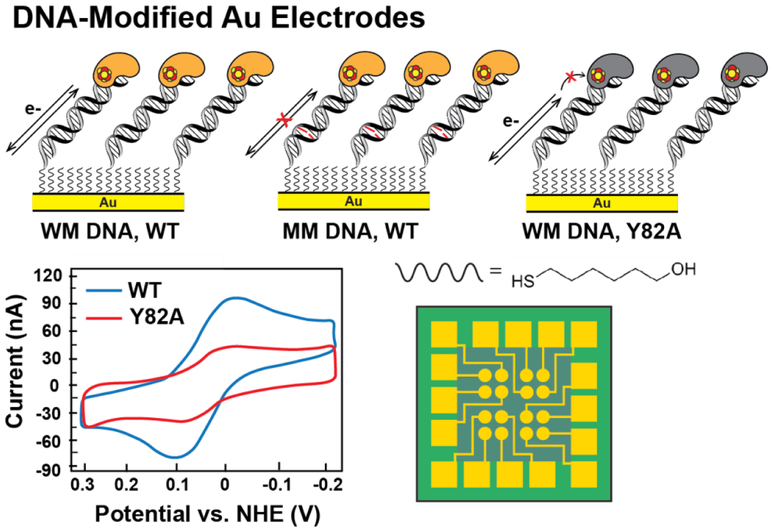
Figure 4. Measuring the redox potential of DNA-bound [4Fe4S] enzymes.
(Top) DNA electrochemistry on Au electrodes, where DNA duplexes containing a tethered alkanethiol are attached to a gold electrode passivated with β-mercaptohexanol, can be used to assess redox signals from DNA-bound, [4Fe4S] proteins (yellow, left). Signals are attenuated when base lesions (red, center) in the duplex are present, or when the redox pathway within the [4Fe4S] protein, as for Y82A (gray, right) is deficient in CT. Cyclic voltammetry (below, left) scanning can be used to measure the DNA-mediated redox signal from CT-proficient proteins (WT EndoIII, blue) and CT-deficient proteins (EndoIII Y82A, red). A multiplexed chip platform (below, right) has now been adapted to measure [4Fe4S] protein signals on 16 separate DNA-modified electrodes, with replicates on a single surface (32, 48-50).
We first examined MutY, EndoIII, and AfUDG, the three base excision repair proteins found to contain [4Fe4S] clusters (32). Earlier studies using strong chemical oxidants and reductants had suggested that the clusters were redox-inactive at physiological potentials, but these studies had been conducted in the absence of DNA. It was reasonable to consider that binding the DNA polyanion might change the potential of the cluster within the protein. Our studies showed first that a reversible signal was detectable at ~80 mV versus NHE, within the physiological window, for each of these proteins bound to the DNA-modified electrode. The potentials were consistent with those found for the clusters in HiPIP proteins and were at equal potentials for all of the repair proteins examined. Moreover, the presence of an abasic site on the DNA intervening between the bound protein and the electrode surface served to attenuate the signal from the cluster. This result established two important points: (i) we were measuring the DNA-bound potential of the protein and (ii) we were observing DNA-mediated CT between the electrode and the cluster within the protein. Many repair and replication proteins have now been studied using this DNA electrochemistry platform, and in each case we have observed reversible, redox signals in the physiological potential range, near ~80 mV vs. NHE, corresponding to cycling between the [4Fe4S]2+ and [4Fe4S]3+ oxidation states (28, 32, 51, 52, 54–57).
It is noteworthy that the most recent generation of DNA-modified electrodes utilizes a multiplexed chip so multiple experimental conditions can be examined in parallel (Figure 4) (48–50). Patterned, silicon chips with sixteen independently addressable Au electrodes uniform in area can be physically divided into four quadrants and used to monitor the redox activity of a single protein on as many as four different DNA substrates on the same surface. The platform can also be used to compare CT efficiency of WT and mutant protein on the same chip. All of the experiments described can moreover be carried out under strictly anaerobic conditions.
Graphite Electrochemistry to Compare DNA-bound and DNA-free [4Fe4S] Cluster Potentials
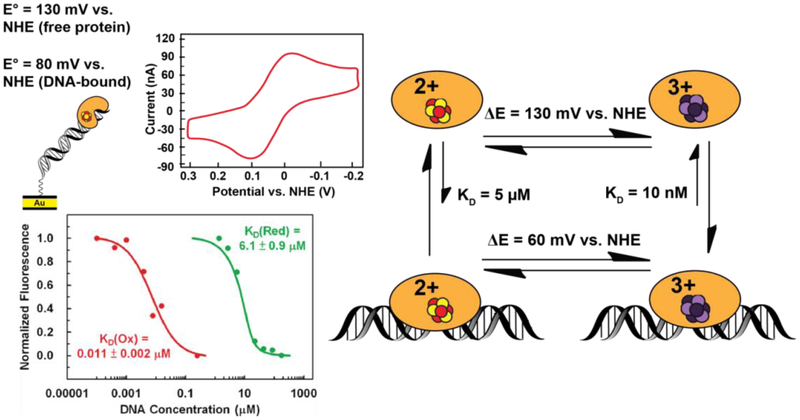
We became interested in monitoring directly the shift in potential for [4Fe4S] repair proteins associated with binding DNA, and that required measuring the protein potential both in the absence and presence of DNA. DNA electrochemistry on Au electrode surfaces is amenable to measuring physiological potentials ranging from −200 mV to +500 mV vs. NHE (28, 50–59). However, scanning beyond this range is necessary to observe a redox signal from a protein in the absence of DNA, which has a higher (more reductive) midpoint potential due to the absence of the DNA polyanion (51, 53, 56, 60, 61). In order to measure the DNA-free and DNA-bound redox potentials of EndoIII, highly oriented pyrolytic graphite (HOPG) electrodes were used (60). Bare surfaces or surfaces modified with pyrene-functionalized duplex DNA (which creates a DNA monolayer through π stacking between pyrene and graphite) facilitated direct comparison of DNA-free and DNA-bound EndoIII [4Fe4S] cluster redox potentials. A negative shift of approximately 200 mV was observed for the DNA-bound EndoIII relative to DNA-free EndoIII. Given that protein binding importantly does not lead to a large conformational change in the protein or DNA, this shift in redox potential for the [4Fe4S] cluster thermodynamically reflects a roughly 500-fold tighter DNA binding affinity for the oxidized [4Fe4S]3+ state, relative to the reduced [4Fe4S]2+ state, based on the Nernst equation (Figure 5). Oxidation of the [4Fe4S]2+ cluster to the [4Fe4S]3+ state thus should serve as a redox switch for DNA binding.
Figure 5. DNA binding shifts the potential of [4Fe4S] cluster enzymes.
Endonuclease III has a redox potential of approximately 80mV vs. NHE for the [4Fe4S]3+/2+ couple measured on a DNA-modified Au electrode. (Above, left) This potential is a negative shift from the ~130mV vs. NHE potential for this couple when the protein is not bound to DNA. This shift corresponds thermodynamically to a stabilization of the oxidized [4Fe4S]3+ state upon binding the DNA polyanion (right). Microscale thermophoresis on electrochemically oxidized and native reduced Endonuclease III (below, left) indicates that a ~550-fold increase in DNA binding affinity is associated with the conversion from [4Fe4S]2+ to the [4Fe4S]3+ state, (53) consistent with this negative shift in potential.
Spectroscopic Observation of [4Fe4S] Cluster Redox Activity in DNA-Processing Proteins
Electrochemical observations of the [4Fe4S] clusters in DNA-processing enzymes was also complemented by spectroscopic analysis. EPR spectroscopy requires chemical or electrochemical conversion of the resting, diamagnetic [4Fe4S]2+ enzymes to the paramagnetic oxidized [4Fe4S]3+ or reduced [4Fe4S]+ redox states, and thus was used to establish the resting state for the DNA-bound repair protein as [4Fe4S]2+ (28, 62–65). We also demonstrated that the DNA-bound protein can be oxidized photochemically, from a distance, using DNA CT from a distantly tethered intercalating photooxidant (32, 51). Importantly, we were also able to demonstrate spectroscopically that the cluster could be oxidized by guanine radicals, generated using flash-quench experiments monitored by transient absorption spectroscopy (66). Indeed, these studies highlight how the oxidized [4Fe4S]3+ cluster could be generated within the cell, from a distance using DNA CT from guanine radicals under conditions of oxidative stress, and in so doing, activating the DNA repair machinery.
IV. A Shift in Cluster Potential Reflects a Redox Switch in DNA Binding
While we had seen several examples of DNA binding yielding a shift in redox potential for the clusters within the repair proteins, from which one can infer a difference in DNA binding affinity for the protein with a [4Fe4S]3+ cluster versus a [4Fe4S]2+ cluster, we were not at first able to measure this difference directly. In the absence of DNA, the [4Fe4S]3+ cluster degrades further to a [3Fe4S]+ cluster, which affects protein binding. As a result, binding affinities for the [4Fe4S]3+/2+ clusters needed to be determined anaerobically.
Recently we found that microscale thermophoresis could be used under anaerobic conditions to carry out DNA binding experiments for the [4Fe4S] proteins in the two oxidation states. EndoIII containing the [4Fe4S]3+ cluster was first generated on DNA-modified electrodes and then, under strictly anaerobic conditions, the thermophoresis experiments were conducted (53). Consistent with the shift in potential associated with DNA binding, oxidized EndoIII with the [4Fe4S]3+ cluster was indeed found to bind >500 times more tightly to dsDNA than the reduced EndoIII with the [4Fe4S]2+ cluster (Figure 5). This difference in binding affinity is understandable based simply on electrostatic considerations. In fact, calculations based upon the distance of the cluster to the DNA polyanion and the intervening protein dielectric well reflect the change in binding affinity that we see. It is interesting in that context that we find similar shifts in potential for all of the DNA repair proteins examined, and the clusters generally appear to be ~ 20 Å from the polyanionic DNA backbone. Based upon these results, then, we can consider that binding of these repair proteins to the DNA polyanion serves to tune the potential of the cluster by altering the electrostatic environment, activating the cluster toward oxidation and lowering the [4Fe4S]3+/2+ couple into a physiologically accessible potential range.
V. [4Fe4S] Proteins in Nucleic Acid Processing and Repair
Thousands of DNA damage sites are generated by endogenous and exogenous agents in each cell daily (67, 68). An arsenal of DNA repair pathways have evolved to address the structurally and chemically diverse lesions, though a comprehensive understanding of how repair pathways efficiently identify and remove damaged bases has remained elusive (69, 70). In the case of the repair proteins that contain [4Fe4S] clusters, what they share is a low copy number within the cell and a moderate specificity in binding their target lesion versus unmodified DNA. But with few players and not a high specificity in targeting, how do they effectively find all the lesions within the cell on a timescale appropriate to the organism?
We have considered that DNA charge transport chemistry might provide a first step in localizing repair proteins that contain these clusters in the vicinity of lesions, essentially redistributing the proteins to regions of damage, irrespective of the actual lesion characteristics as long as the damage interferes with DNA CT. We had actually found earlier that many common base lesions interfere with DNA CT (71). Moreover, the fact that these proteins share a similar redox potential means that essentially they can work together, transferring electrons from one to another to carry out a first scan of the genome.
A Model for Scanning the Genome Utilizing DNA CT Chemistry
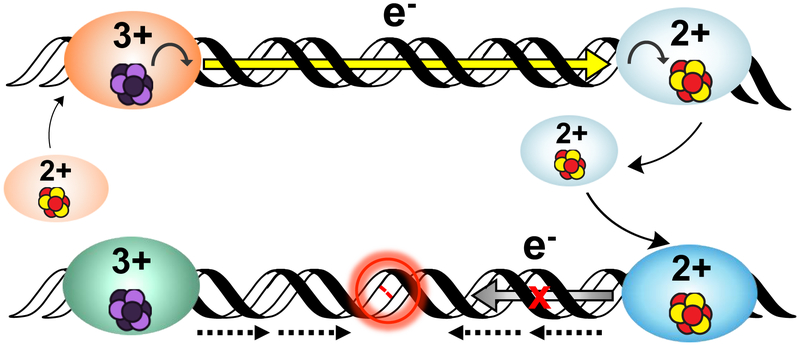
To describe how DNA-mediated CT chemistry could be utilized for more efficient lesion detection, we developed a model for redox signaling among a network of [4Fe4S] repair proteins (Figure 6) (32). Upon association of freely-diffusing protein in the [4Fe4S]2+ state onto duplex DNA, the protein is activated toward oxidation and can reduce a distally bound protein from the [4Fe4S]3+ state to the more weakly binding [4Fe4S]2+ state (Figure 5, top). Alternatively, guanine radicals (G.+), a product of oxidative stress, can generate the [4Fe4S]3+ species by DNA CT over long molecular distances (41, 66, 72). Iterations of this scanning can occur throughout the cell. However, if there is an intervening region of dsDNA containing a lesion that attenuates CT, redox signaling between the cluster proteins is interrupted. Both proteins in the tightly-binding [4Fe4S]3+ state persist on the DNA and can then localize to the precise site of damage (Figure 5, bottom).
Figure 6. A model for DNA-mediated redox signaling between repair proteins.
Enzymes with the cluster in the native [4Fe4S]2+ state first bind DNA, causing the cluster to become activated toward oxidation. Oxidative stress initiates the damage search when highly reactive species such as the guanine radical cation are formed; these can oxidize DNA-bound proteins in their vicinity. Oxidation of the cluster to the [4Fe4S]3+ state leads to a > 500-fold increase in DNA binding affinity, so oxidized proteins remain bound and diffuse along the DNA. Another [4Fe4S]2+ protein bound at a distant site can reduce the oxidized protein, effectively scanning the intervening DNA for lesions through DNA-mediated CT. At this point, on damage-free DNA (above) the reduced protein binds less tightly to DNA and can diffuse away, while the newly oxidized protein continues the damage search. This process of redox exchange continues until a segment of DNA containing a lesion is approached. Since even subtle lesions can disrupt base stacking (below), CT is attenuated and any nearby oxidized proteins remain bound. Thus, DNA CT allows repair proteins to scan large sections of the genome and redistribute to areas containing damage (24, 37).
We have, by now, found many types of DNA repair proteins containing [4Fe4S] clusters that are involved in redox signaling on DNA, and we have probed how these redox signaling networks work cooperatively utilizing DNA CT through their [4Fe4S] clusters. We have also found CT deficiencies in mutated proteins strongly linked to disease. In the context of redox signaling, included below is an overview of the major repair protein families that coordinate a [4Fe4S] cluster, and illustrations of how they may utilize redox signaling.
Glycosylases
Glycosylases are key players in BER, a highly conserved pathway responsible for recognizing and removing single-base lesions generated by spontaneous deamination, alkylating agents, and oxidative stress, among other sources of damage (Figure 7) (73, 74). For EndoIII, MutY, and AfUDG, biochemical studies have very elegantly demonstrated that mutations at coordinating cysteines or in the cluster binding domain can affect protein expression, enzymatic function, and DNA binding, even though the cluster is located remotely from the glycosylase active site and there is not a large conformational change associated with the binding to DNA (34, 75–77). Specific to mammalian BER, a connection has been established between the glycosylases, NTHL1 and MUTYH, and multiple cancers, most notably MUTYH-associated polyposis (MAP). These syndromes are characterized by increased risk of aggressive, early-onset colorectal cancer (68, 74, 78).
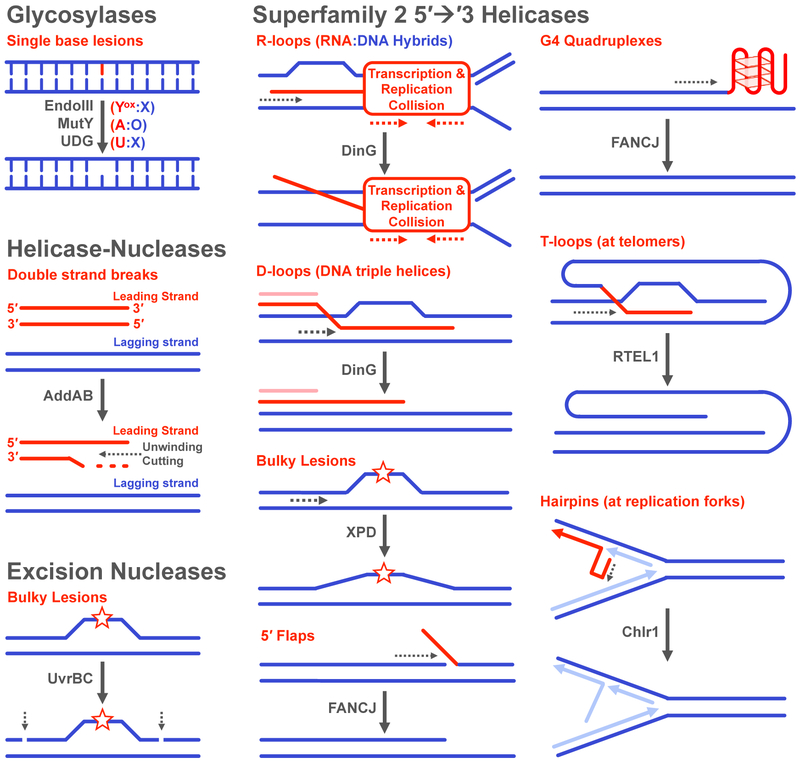
Figure 7. Reactions and Substrates for DNA repair enzymes containing [4Fe4S] clusters.
(Top, left) Glycosylases remove a number of single-base lesions caused by endogenous and exogenous agents. (Middle, left) The helicase-nuclease AddAB processes double strand breaks. (Bottom, left) The UvrBC complex cleaves the phosphodiester backbone around the damaged strand of DNA. (Right) Superfamily 2 5′→3′ helicases participate in a number of pathways and are involved in unwinding very diverse substrates. The substrate specificity is overlapping for many of the helicases (ex. FANCJ and RTEL1), though genome location (ex. telomeres) and cell cycle phase (ex. replication in the S phase) appear to be factors in activity (73, 77, 81, 82, 87, 91,93).
As described above, several BER glycosylases were demonstrated to participate in DNA-mediated CT chemistry (32). Further examination of the available structures of EndoIII in the free and DNA-bound forms revealed a pathway of aromatic residues between the cluster and the DNA, separated by 15-20 Å, and predicted to provide a CT pathway (10, 11, 79, 80). Informed by these crystal structures, several key mutants of EndoIII have been generated and characterized, including a CT deficient/enzymatically proficient mutant Y82A, a complementary CT proficient but enzymatically deficient mutant D138A, and charge-flipped mutants to explore the electrostatics near the cluster (61, 79, 80). Unique to the HiPIP-like [4Fe4S] repair proteins, the DNA polyanion is the governing factor that shifts the potential and stabilizes the cluster in the [4Fe4S]3+ state (5, 61). Furthermore, redox sensing of base stacking perturbations on DNA-modified gold electrodes containing a lesion has been observed, even when that lesion is not a substrate for the glycosylase (32, 80).
Superfamily 2 (SF2) 5' → 3' Helicases
SF2 helicases were the second family of enzymes discovered to coordinate a [4Fe4S] cluster (65). SF2 helicases are NTP-dependent proteins that directionally translocate and unwind duplexes (81). Distinct from the glycosylase family, the FeS helicases are involved in several repair pathways (Figure 7). These pathways respond to stress from multiple sources, and there are many examples of extensive crosstalk and cooperativity among a complex network of repair pathways that include SF2 helicases with [4Fe4S] clusters (27). As might be expected, mutations in SF2 helicases containing [4Fe4S] clusters are associated with a host of genetic disorders and cancers and are being targeted for cancer therapies (82, 83).
A common theme with SF2 helicases is the multifunctional nature of their activities within the cell (Figure 7). In bacteria, DinG resolves R-loops, RNA:DNA hybrids formed at collisions between replication and transcription machinery; DinG has also been shown to be active on D-loops (displacement loops, triple stranded DNA) (64, 84). Notably, cysteine mutants of DinG are more susceptible to proteolytic degradation in vitro (64). A new, well-conserved bacterial protein YoaA, was identified in a genetics screen to be involved in coordinating repair and replication machinery at blocked replication forks. Based on sequence similarity to DinG, YoaA was predicted to be a [4Fe4S] protein (85).
The first archaeal/eukaryotic [4Fe4S] SF2 helicase discovered, XPD, is part of the transcription factor II H (TFIIH) complex and is involved in both nucleotide excision repair (NER) and transcription initiation (65). In NER, helicase activity is critical for removing damaged oligomers, which can be disrupted by mutating coordinating cysteines. In contrast, only the association of XPD with TFIIH is needed to initiate transcription, which is thought to aid assembly of other proteins with TFIIH. Many other facets of the XPD have also been studied, including regulation of XPD by other proteins, the timing of cluster insertion relative to XPD incorporation with TFIIH, how XPD is involved in cell cycle control, and a role for XPD in preventing oxidative damage in the mitochondria (86).
Both XPD and DinG can participate in DNA CT chemistry with a shared DNA-bound redox potential of approximately 80 mV vs. NHE and sense signal attenuating lesions (54, 59). Further probing of the CT activity found that upon addition of ATP, the signal for XPD and DinG quite stunningly increases in a concentration-dependent manner without any shift in the midpoint potential. Helicase activity thus increases the electrochemical signal through better coupling of the cluster to the π-stacked DNA bases, essentially signaling the helicase activity through DNA CT. The increased coupling upon cofactor binding appears to be an important feature of the SF2 helicase family that helps coordinate repair and replication, and signaling from a distance that the helicase is active.
Another eukaryotic SF2 helicase, FANCJ has a role in several pathways; compromised FANCJ function, which can result from mutations in the cluster binding domain, has been linked to several cancers, and FANCJ upregulation has been found in many tumor types (87). FANCJ is known to act on many substrates, including duplex substrates, D-loops and G4 quadruplexes (87). FANCJ association with numerous other proteins, including BRCA1, can depend on the timing of the damage response and the type of lesions generated. Furthermore, FANCJ activity has been observed to alleviate replication stress through resolution of stalled replication forks, and particularly at telomeres rich in G4 quadruplexes. Two other SF2 helicases that coordinate a [4Fe4S] cluster, RTEL1 and Chlr1, resolve several different types of structures in the process of facilitating replication (83, 88). Both RTEL1 and Chlr1 have been also found to associate with various replication proteins. Mutations in these enzymes are similarly associated with an array of diseases (88, 89). Electrochemical studies of these proteins have not yet been conducted, but studying these proteins in the context of redox signaling will likely illuminate how these multifunctional enzymes may coordinate their activities.
Helicase-Nucleases
In 2009, the first helicase-nuclease containing a [4Fe4S] cluster identified was AddB, part of the AddAB heterodimer found in gram positive bacteria and some proteobacteria (90, 91). Helicase-nuclease activity is involved in double strand break (DSB) repair, which can be caused by several factors, including collapsed replication forks. Located in the C-terminal nuclease domain of AddB, the cluster was found to be essential for binding of DNA substrates, but not essential for complexation with AddA or for maintaining structure, though a stabilizing role of the cluster was suggested. The crystal structure of the AddAB in complex with a DNA substrate revealed a DNA binding loop supported by the cluster domain, providing explanation of why mutating coordinating cysteines abolished substrate binding (92). A homologous [4Fe4S] helicase-nuclease, Dna2, was later found in eukaryotes (vide infra).
Excision Nucleases
UvrC, an excision nuclease from E. coli, was reported in 2018 to be a [4Fe4S] protein (28). Only found in bacterial and some archaeal NER pathways, UvrC (in complex with another NER protein, UvrB) uniquely catalyzes incisions in the phosphodiester backbone both 5′- and 3′- to damaged substrates (93). Distinct from the other known [4Fe4S] repair proteins in bacteria, UvrC was found to coordinate an O2-sensitive [4Fe4S] cluster, causing protein aggregation upon oxidative degradation. Mutation of coordinating cysteines to alanine led to aggregation or severely reduced overexpression, the latter of which has been reported before for MutY (94). Additionally, UvrC in its holo form binds to duplex DNA without lesions, enabling demonstration that UvrC participates in DNA CT chemistry. UvrC shares a DNA-bound midpoint potential with EndoIII, MutY, and DinG. UvrC has been noted in several reports to be unstable in vitro and the scarce in vivo (~10 copies/cell), so continued investigation should provide insights as to how chemically and structurally diverse lesions are detected by UvrC in its holo form.
Monitoring Redox Signaling Between Repair Proteins and Repair Pathways
Given the multitude of these clusters, their similarity in redox potential and their diversity in lesions targeted, we became interested in examining how the proteins might cooperate in searching for DNA lesions. To study cooperative, redox communication between [4Fe4S] proteins, a series of in vitro and in vivo assays were developed to monitor signaling networks. In vitro, atomic force microscopy (AFM) has facilitated visualization of [4Fe4S] repair protein distribution on well-matched (WM) DNA versus duplex DNA containing a single base mismatch (MM) (53, 55, 59, 79, 80). Though strands containing a single, engineered C:A mismatch are not native substrates for any of these repair enzymes studied, preferential binding to the 3.8 kb long MM strands, expressed as a binding density ratio of 1.5 (Figure 8), has been seen for all CT-proficient repair proteins. Mixtures of [4Fe4S] proteins from different repair pathways, and, remarkably, mixtures of cluster proteins from distinct organisms also signal cooperatively one to another, localizing to damaged strands.
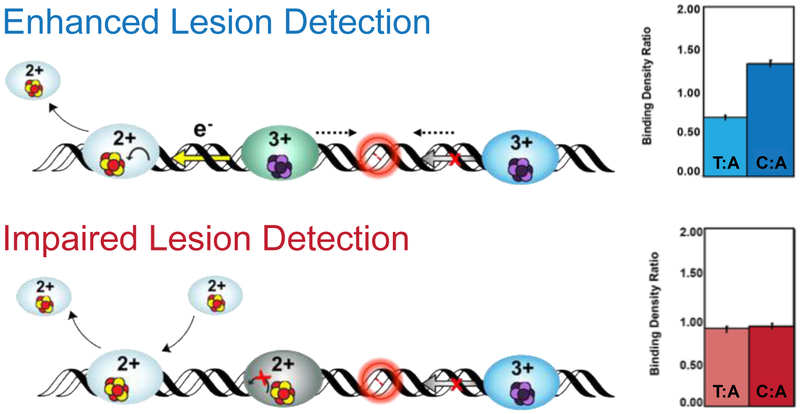
Figure 8. Visualization of Protein Localization on Damaged DNA by Atomic Force Microscopy (AFM).
In the AFM redistribution assay, [4Fe4S] protein or protein mixtures are incubated with DNA substrates which contain either long, well matched DNA strands (3.8 kb) or strands containing a single C:A mismatch in the 3.8 kb duplex along with short, undamaged DNA strands (1.6 and 2.2 kb). Images are collected and proteins bound to long strands of DNA are counted, normalized to the proteins bound to the short strands, and expressed as a binding density ratio (right). As shown at right (top), a greater density of proteins is found on the strands containing a mismatch (C:A) compared to the well-matched (T:A) strand, even though the repair proteins do not bind the C:A mismatch as a substrate. If the protein is defective in carrying out DNA CT, however, the binding density is the same on the mismatched and matched strands (bottom right) (55, 59, 79, 80).
Two factors have been found to affect the efficiency of the damage search: (i) the CT proficiency of a protein and (ii) the extent to which the protein population is oxidized. Mixing CT-proficient and CT-deficient proteins (eg. EndoIII and Y82A) impairs localization to MM strands; the binding density ratio is 1. Protein samples that are 99% oxidized generated anaerobically using DNA electrochemistry also do not redistribute to MM strands, likely due to the >500-fold increase in binding affinity. Redistribution of oxidized EndoIII, however, can be restored with addition of reduced DinG. The same phenomenon was seen in the reciprocal experiment with oxidized DinG and reduced EndoIII. These data underscore that long-range redox signaling between [4Fe4S] enzymes is dependent on the shared DNA-bound redox potential and CT proficiency of the protein. Furthermore, these experiments highlight the ability of the proteins to signal one another over kilobase distances.
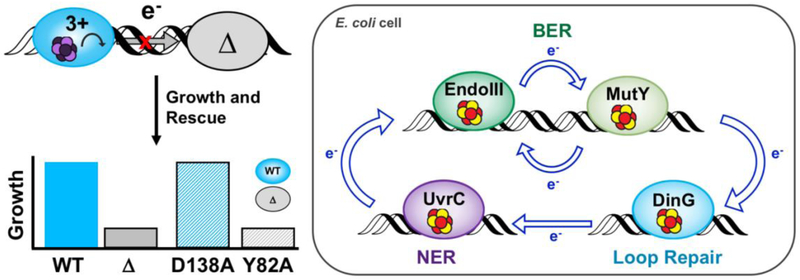
Complementary genetics assays were developed using strains of E. coli that depend on the repair activity of MutY, DinG, or UvrC for growth. Putative redox signaling networks can be disrupted by genetically knocking out a signaling partner (57, 59, 79). Limited growth in the knocked out strains therefore points to diminished repair activity occurring, owing to limited availability of signaling partners (Figure 9). Complementation plasmids for EndoIII, which include WT enzyme, Y82A, and D138A have been used to monitor how rescue with wild type and EndoIII mutants can restore survival. With this genetic approach, evidence of signaling among BER enzymes, DinG, and UvrC has been found to be necessary for repair activity and growth of cells (57, 59, 79). Rescue with the CT competent but enzymatically deficient D138A but not the CT deficient but enzymatically competent Y82A restores growth in EndoIII knockouts, demonstrating that CT proficiency and not enzymatic activity is needed for redox signaling and efficient repair activity in other pathways.
Figure 9. Genetic Assays for Detection of DNACT Signaling Among E. coli [4Fe4S] Repair Proteins.
(Top left) E. coli parent (blue) and EndoIII knockout (KO, gray) strains that report on the repair protein activity have been used to monitor DNA-mediated communication between putative signaling partners in vivo. Complementation plasmids expressing CT-proficient (D138A) or CT-deficient (Y82A) versions of EndoIII are introduced to EndoIII KO strains to evaluate if the parent phenotype can be rescued (bottom left). Rescue can only be achieved with a CT-proficient enzyme (blue bar, gray outline), strongly indicating that DNA-mediated redox signaling is necessary for efficient repair. (Right) The redox signaling network in E. coli includes BER, loop repair, and NER (57, 59, 79, 80).
Diseases and Cancers Related to Mutations in [4Fe4S] Proteins
Mutations linking cancer with compromised repair activity by human [4Fe4S] proteins have been reported (24, 58). Investigations into the redox chemistry of mutant proteins has helped illuminate how impaired redox signaling capacity could contribute to disease. For example, the CT-deficient EndoIII Y82A mutant corresponds the Y166 of the human homolog, MUTYH. This Y166S mutation has been known to be associated with MAP colorectal cancers (95). Within the context of DNA CT chemistry, a mutation which compromises the CT proficiency of one repair protein could understandably lead to compromised crosstalk of [4Fe4S] repair pathways across the genome.
A novel MUTYH germline variant, C306W, was recently found in a patient with colonic polyposis and a family history of early-onset colon cancer (58). Mutation of C306 does not fully abolish cluster loading or DNA-bound redox activity, but is destabilizing, leading to rapid oxidative degradation of the [4Fe4S] cluster to the [3Fe-4S]+ species on a DNA electrode. Degradation of the cluster results in diminished binding to DNA substrates and enzymatic function. The C306W mutant is a powerful example of how cluster degradation disrupts enzymatic activity and redox signaling, emphasizing the critical role of the cluster in preventing the onset of disease. Moreover this degradation occurs only with oxidation, underscoring the important role of the cluster in carrying out redox chemistry.
Mutations in XPD have also been linked to disease, in particular, to three distinct but related disorders with extreme photosensitivity: trichothiodystrophy (TTD), Cockayne syndrome (CS), and xeroderma pigmentosum (XP) (27, 86). Crystal structures of XPD have helped illuminate how common mutations in different domains of XPD lead to specific disorders (96, 97). For example, the L325V mutation (L461 in human XPD), specifically associated with XP and TTD, is CT-deficient relative to the WT enzyme, with diminished ability to distinguish well-matched versus mismatched strands in our AFM assay (55). Another mutation, G34R (G47R in human XPD), is associated with loss of ATP binding and helicase activity. Consistent with this data, the G34R mutant did not exhibit enhanced electronic coupling upon addition of ATP on DNA-modified electrodes (54). Thus, these disease-relevant mutations not only impair enzymatic activity, but also diminish redox signaling capacity.
The study of the redox chemistry and cooperative signaling between disease-relevant mutants has exemplified intimate connections between cluster stability, enzymatic activity, redox activity, and CT proficiency (27). Mutations in other [4Fe4S] proteins may also affect enzymatic or redox signaling activity, and we expect that many heretofore uncharacterized clinically relevant mutations will be linked to compromised redox signaling. The cluster within these proteins thus represents an intriguing new therapeutic target.
Redox-Mediated Catalysis by [4Fe4S] Radical SAM Enzymes
For all of the proteins described thus far, the [4Fe4S] cluster has been involved in redox chemistry but not directly in the enzymatic reaction carried out by the protein. This, however, is not the case for more than 13,500 known radical S-adenosyl-L-methionine (SAM) enzymes (98, 99). Radical SAM enzymes employ the tunable, versatile [4Fe4S] cluster cofactor to catalyze essential biochemical reactions, acting on a variety of substrates in multiple metabolic pathways, including repair of UV-induced DNA damage and modification of tRNAs (30, 98, 100–102). These repair proteins, unlike those that carry out redox signaling, contain ferredoxin-like clusters coordinated by three cysteines, which cycle between the [4Fe4S]+ and [4Fe4S]2+ states during activity. The primary reaction catalyzed by the redox function of the cluster is the generation of a 5’-deoxyadenosyl radical, a powerful, aliphatic oxidant which then catalyzes further biochemical reactions.
One example of repair by a radical SAM enzyme is cleavage of a methylene bridge in a UV-induced DNA lesion by the spore photoproduct lyase (SP lyase), which is responsible for repairing (5R)-5-(α-thyminyl)-5,6-dihydrothymidine, or the spore photoproduct, (30). SP lyase has been structurally and biochemically characterized, though chemical properties of the DNA-bound protein are less understood. SP lyase binds a polyanionic substrate that could electrostatically alter the environment and potential of the cluster (103). Comparison of the redox potentials for the [4Fe4S]2+/+ couple in the presence and absence of a nucleic acid substrate has not yet been explored but will be interesting to consider regarding the chemistry driving catalysis.
VI. [4Fe4S] Enzymes in Eukaryotic DNA Replication
Diverse, specialized replication enzymes, many of which contain [4Fe4S] clusters, duplicate large eukaryotic genomes with high fidelity (104, 105). DNA primase, B-family polymerases α, δ, and ε, the helicase-nuclease Dna2, and the translesion DNA polymerase ζ all contain the [4Fe4S] cofactor (62, 63, 106, 107). These multisubunit enzymes, along with the replicative helicase (CMG), processivity factors such as proliferating cell nuclear antigen (PCNA), replication factor C (RFC), and single-stranded binding protein Replication Protein A (RPA), work together to coordinate replication (108–113).
Before polymerases can synthesize new DNA, replication origin sites in the genome must be recognized and licensed through binding of the hexameric origin recognition complex (ORC), followed by Cdc6, Cdt4, and the Mcm2-7 helicase (114–116). Loading of additional proteins, including DNA polymerase ε, subsequently forms the pre-initiation complex (113, 114). GINS and Cdc45 associate with Mcm 2-7 and form the active CMG helicase (117–119). The replicative helicase anneals AT-rich DNA at origin sites, beginning bidirectional replication on the two parent strands of genomic DNA. Phosphorylation by cyclin dependent kinase and the Dbf4-dependent kinase spatiotemporally regulates these steps. After annealing and origin firing, polymerases begin DNA synthesis.
DNA Polymerase-α-Primase Begins Replication through Coordinated Binding and Dissociation Events
DNA polymerase-α-primase (pol-prim) is the heterotetrameric complex responsible for synthesizing an RNA-DNA primer which begins DNA synthesis on a template (120). Primase consists of an RNA polymerase subunit p48 and a regulatory subunit p58, and synthesizes an 8-14nt RNA primer on ssDNA (121–123). Polymerase α consists of a catalytic subunit p180 and an auxiliary subunit p70 and synthesizes a ~10-30nt DNA segment downstream of the RNA primer. The C-terminal domain of the primase auxiliary subunit (p58C), and putatively the C-terminal domain of the polymerase α catalytic subunit (p180), coordinate [4Fe4S] cofactors (62, 63, 124). To synthesize RNA/DNA primers, primase first binds ssDNA and two nucleotide triphosphates (NTPs). After substrate binding and synthesis of a phosphodiester bond between NTPs, primase is converted to the active form. Primase then rapidly elongates the primer, and finally truncates synthesis, handing off the template to polymerase α (121–123, 125). After this transfer step, polymerase α binds the RNA/DNA template and deoxynucleotide triphosphates (dNTPs), polymerizing ~10-30 dNTPs downstream of the RNA primer. After this sequence is completed, DNA polymerases ε and δ can take over replication.
Primase and polymerase α contain [4Fe4S] clusters but are otherwise structurally and functionally distinct from polymerases δ and ε (120, 122). Primase and polymerase α are heterodimers containing catalytic and regulatory subunits. These low-fidelity enzymes lack a proofreading exonuclease domain and have error rates of ~10−2 (121–123) and ~10−4-10−5 (120) respectively, and unlike polymerases ε and δ, polymerase α and primase also do not interact with PCNA. Primase [4Fe4S] cluster assembly and fidelity are, moreover, negatively affected by oxidative stress conditions in the cell (126), suggesting cluster sensitivity to the redox environment.
Primase has a Redox Switch
The [4Fe4S] cluster of DNA primase was recently discovered to function as a redox switch, regulating DNA binding and signaling (52). The [4Fe4S] domain of primase, p58C, can bind DNA, independently of the primase enzyme (126, 127). On a DNA-modified electrode, this protein was anaerobically oxidized or reduced using bulk electrolysis. Subsequent CV scans showed that the oxidized [4Fe4S]3+ protein was redox-active on DNA, while the reduced [4Fe4S]2+ form could not be detected (52). The oxidized protein was bound to the DNA-modified electrode, while the reduced form was only loosely associated. This electrochemically controlled switch is mediated by conserved tyrosines between the cluster and the DNA binding interface (52, 126). Mutation of these tyrosines to phenylalanine or leucine attenuates redox activity on DNA electrodes and compromises primase initiation on ssDNA but not catalytic activity. Moreover, primase truncation is gated by DNA CT in vitro; a single base mismatch in a nascent primer abrogates truncation.
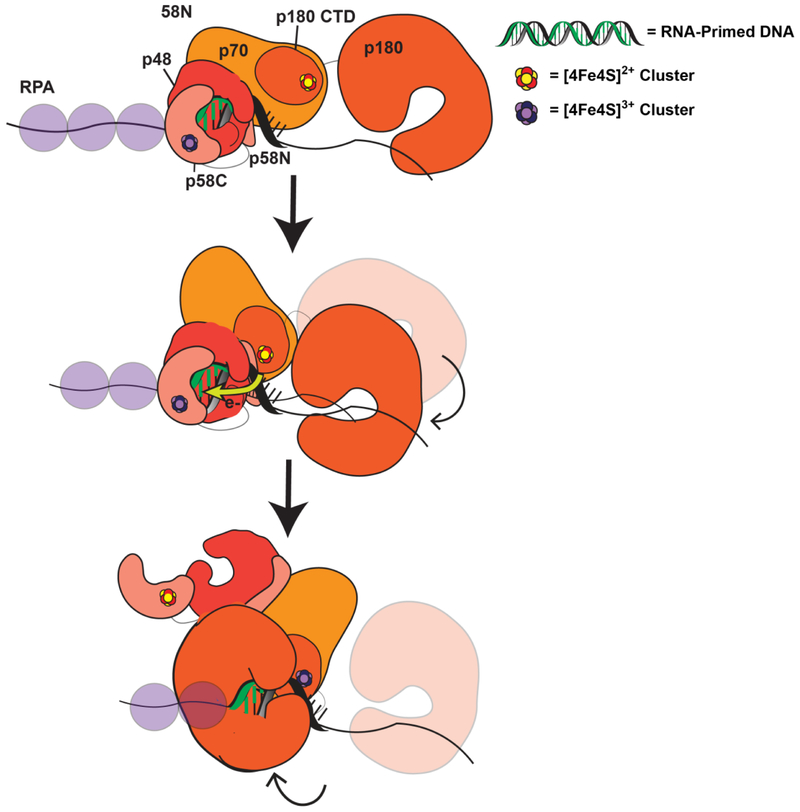
In a proposed model of primase-polymerase α handoff (Figure 10), oxidized, tightly bound [4Fe4S]3+ primase, coupled into the RNA/DNA duplex, synthesizes the RNA primer. When a reduced [4Fe4S]2+ polymerase α cluster contacts the RNA/DNA duplex, polymerase α is oxidized by primase through DNA CT. Our results with human DNA primase indicated that a mismatch formed in the growing DNA/RNA hybrid inhibited this handoff, consistent with the idea that DNA CT facilitates this rapid, long range signaling. Through CT, polymerase α becomes tightly bound in the [4Fe4S]3+ form, and primase is reduced to the [4Fe4S]2+ form. Primase dissociates from the substrate, allowing polymerase α to bind and synthesize DNA on the template (Figure 10). Redox switching driven by the [4Fe4S] cofactor thus may coordinate these binding and dissociation events.
Figure 10. Model for primase-polymerase α handoff through redox switching.
(52) Oxidized [4Fe4S]3+ primase is bound to the RNA/DNA primer during primer synthesis. Polymerase α is DNA-dissociated and reduced but flexibly tethered to primase (Top). When the RNA primer reaches appropriate length, polymerase α orients in a manner coupling the [4Fe4S] cluster into the RNA/DNA substrate and can be oxidized by DNA CT through this segment, sending an electron through the primed template to reduce DNA primase (Middle). Reduced primase dissociates from the RNA-primed DNA, and oxidized polymerase α can then synthesize DNA downstream of the primase product (Bottom).
We recently observed that the redox-driven DNA binding switch is conserved in yeast as well as human primase (128). On DNA electrodes, oxidized [4Fe4S]3+ p58C is tightly bound and redox-active, whereas reduced [4Fe4S]2+ p58C is loosely associated with DNA and redox-inert in yeast and human systems. Yeast and human redox switches, moreover, are mediated by conserved tyrosines positioned between the p58C DNA binding interface and [4Fe4S] cluster. Remarkably the tyrosines are positioned differently but mediate the same chemistry in yeast and human primase. Mutations at Y397 in yeast primase moreover cause partial loss of redox signaling and lead to oxidative degradation to a [3Fe4S]+ species on DNA. This effect is severe enough to cause lethality in yeast for the p58C Y397L mutation, underscoring the importance of this redox signaling chemistry in essential replication processes.
DNA Polymerases ε and δ Divide Labor between Leading and Lagging Strands
Polymerases ε and δ are the larger, high-fidelity polymerases responsible for DNA synthesis on the leading and lagging strands, with proofreading 3’→5’ exonuclease domains and with mutation rates of ~105-10−6 and ~10−4-10−6, respectively (120, 129). PCNA binding moreover enhances the intrinsic processivity of both enzymes. The high intrinsic fidelity is necessary for these enzymes, as their products remain in the daughter DNA copy. Pol-prim products, on the other hand, are removed during Okazaki fragment maturation.
The division of replicative labor has been extensively debated and investigated (130–133). Error-prone polymerase studies monitoring ribonucleotide or mispaired base incorporation in an RnaseH knockout cell line by error-prone polymerase ε or δ mutants has illuminated the distribution of DNA synthesis. A polymerase ε variant, pol2M644G, caused ribonucleotide incorporation on the leading strand (130, 131). A low-fidelity polymerase δ variant, pol3L612M, conversely caused an increase in replication errors, localized on the lagging strand (132). Although polymerase δ can replicate the leading strand templates under certain conditions, polymerase ε putatively synthesizes the leading strand and polymerase δ, the lagging strand under normal conditions (133).
The four-subunit polymerase ε holoenzyme has a large polymerase subunit, Pol2, which contains two [4Fe4S] clusters. The first cluster, located within the active polymerase domain, is essential for polymerase activity but dispensable for exonuclease activity (134). The second [4Fe4S] cluster is ligated in the Pol2 C-terminal domain, further from the active polymerase site, and is stabilized by coordination with Dpb2 (124). The Dpb2 subunit of polymerase ε is associated with the C-terminus of Pol2 and is essential for replisome assembly and checkpoint activation (120). The noncatalytic Dpb3 and Dpb4 subunits likely enhance polymerase ε processivity. Characterizing the putative [4Fe4S] clusters in polymerase ε will illuminate new roles for DNA-mediated redox signaling in replication in the context of a complex, multisubunit enzyme.
The three-subunit polymerase δ coordinates a [4Fe4S] cluster in the Pol3 subunit catalytic domain. Polymerase δ auxiliary subunit Pol31 associates with Pol3 to stabilize the cluster (124) and the subunit, Pol32 (120, 129). After replication factor C loads PCNA onto DNA, polymerase δ coordinates with PCNA to extend the Okazaki fragments begun by pol-prim (135, 136). PCNA binding greatly enhances polymerase δ processivity, and biochemical evidence suggests that a conformational switch may occur during polymerase δ PCNA binding and activity. (129). Polymerase δ moreover is uniquely capable of strand displacement synthesis (120), consistent with its role in Okazaki fragment maturation, interacting with protein partners like Dna2. DNA polymerase δ is additionally stabilized in the presence of stalled forks during replication stress and may play a role in response to changes in the environment as with oxidative stress (137).
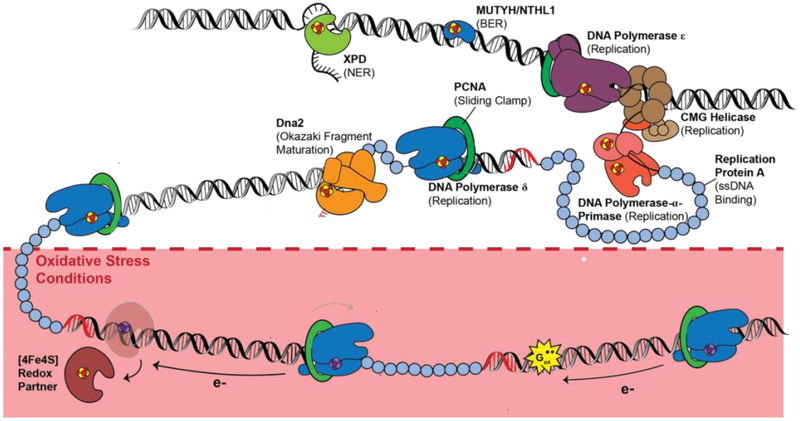
Polymerase δ has been demonstrated electrochemically to be redox-active on DNA, with a midpoint potential near 100 mV vs. NHE in the presence of PCNA (51). With PCNA bound, polymerase δ already binds very tightly to DNA, so it is difficult to consider the cluster oxidation to function as a redox switch for binding. Instead what we observe is that oxidation to the [4Fe4S]3+ state slows polymerase activity in vitro, an effect which can be electrochemically reversed by reducing the protein back to the [4Fe4S]2+ state. These results inspired a model where PCNA-bound, reduced [4Fe4S]2+ polymerase δ processively synthesizes DNA. Under oxidizing conditions, the cluster is converted to the [4Fe4S]3+ form, inducing tighter DNA binding, so tight as to stall activity. Polymerase δ then remains stalled in the oxidized form, consistent with the stalling of polymerase δ found under conditions of oxidative stress. While repair proteins then may be activated with cluster oxidation under conditions of oxidative stress, replication would be inhibited. However, the polymerase can be reduced and restored to the processive form after DNA damage resolution, potentially through long range signaling using DNA CT from repair proteins (Figure 11). Clearly, this signaling needs still to be established, though the electrochemistry and biochemistry show that this long range signaling is possible.
Figure 11. [4Fe4S] enzymes in eukaryotic repair and replication.
B-family polymerases, DNA primase, Dna2 helicase-nuclease, BER/NER enzymes such as MUTYH, NTHL1, and XPD, all coordinate a [4Fe4S] cluster cofactor. Several of these proteins have been demonstrated to participate in DNA-mediated redox signaling; characterization of their redox roles is ongoing. (Below) Under oxidative stress conditions, polymerase δ may be converted to the [4Fe4S]3+ state as a means to stall synthesis under poor cellular conditions. Polymerase δ can be reversibly oxidized and reduced through DNA CT, which may regulate polymerase activity on the lagging strand.
More [4Fe4S] Proteins in DNA Processing
In addition to the primary replicative eukaryotic DNA polymerases, several other enzymes have now been discovered to contain [4Fe4S] cofactors (29, 107, 124). Dna2 is a [4Fe4S] helicase-nuclease enzyme important in double strand break repair, Okazaki fragment maturation, and processing stalled replication forks (138). The cluster in Dna2 is located in the nuclease domain, approximately 10 A from the bound ssDNA substrate. Dna2 5′ → 3′ endonuclease activity is primarily associated with long flap processing during Okazaki fragment maturation (120). Dna2 also plays a role in preventing regression of stalled replication forks, however, and the enzyme has weak ATP-dependent helicase activity (139, 140). Dna2 function, especially helicase and nuclease coordination, is still unclear; [4Fe4S] redox signaling may be important for regulating the multiple cellular roles for Dna2 and its interaction with other [4Fe4S] enzymes, such as polymerase δ.
The translesion polymerase ζ also contains a [4Fe4S] cluster, coordinated in the Rev3 catalytic subunit (112, 124), which is homologous to other B-family polymerase catalytic subunits. This enzyme also contains a subunit Rev7, and two subunits found in polymerase δ, Pol31 and Pol32, but lacks an exonuclease domain. Interacting with PCNA and many other replication factors, polymerase ζ catalyzes mutagenic polymerase activity in the presence of lesions that stall replication fork progression (112). Finally RNA polymerase II, which synthesizes RNA during transcription, has been demonstrated to contain a [4Fe4S] cluster. (29, 112, 141).These proteins may use redox signaling on DNA to coordinate activity in cells, and investigation of their [4Fe4S] redox properties will illuminate important biochemical features of the proteins and their pathways.
VII. Summary and Perspectives
Long-range, DNA-mediated redox signaling provides a means to coordinate replication and repair activity across the nucleus. The redox switching driven by a change in cluster oxidation state regulates DNA association and dissociation with the protein and this redox switch can function rapidly and from a distance without a change in structure of the intervening DNA. This capacity is integral to the DNA CT-driven lesion search in [4Fe4S] repair proteins, in polymerase handoff and in the oxidative stress response. The change in cluster redox potential upon binding the DNA polyanion, the capacity of these DNA-bound proteins to cycle reversibly between [4Fe4S]2+ and [4Fe4S]3+ redox states, and the ability to promote reactions on DNA from a distance using DNA CT offer powerful chemistry of which to take advantage. DNA CT thus offers a means of rapid communication across the genome among DNA-processing proteins to activate a response, and the [4Fe4S] cluster provides the essential cofactor to carry out this chemistry. As more DNA processing proteins are discovered to contain [4Fe4S] clusters, questions to explore include: (i) which proteins are signaling partners with one another; (ii) how are their redox properties modulating protein activities; and (iii) when protein mutations lead to defects in DNA CT, can we begin to target these defects therapeutically? Certainly, continued investigation of DNA-processing [4Fe4S] enzymes will illuminate the rich and important functional roles of this redox chemistry in the genome.
Acknowledgements.
We are grateful to the NIH (GM126904) for their continued financial support and to our many coworkers and collaborators in elucidating this chemistry.
Literature Cited
- 1.Beinert H, Holm RH, Munck E. 1997. Iron-sulfur clusters: Nature’s modular, multipurpose structures. Science 277:653–59 [DOI] [PubMed] [Google Scholar]
- 2.Rees DC, Howard JB. 2003. The interface between the biological and inorganic worlds: Iron-sulfur metalloclusters. Science 300:929–31 [DOI] [PubMed] [Google Scholar]
- 3.Meyer J 2008. Iron-sulfur protein folds, iron-sulfur chemistry, and evolution. J. Biol. Inorg. Chem 13:157–70 [DOI] [PubMed] [Google Scholar]
- 4.Dey A, Jenney FE, Adams MWW, Babini E, Takahashi Y, et al. 2007. Solvent tuning of electrochemical potentials in the active sites of HiPIP versus ferredoxin. Science 318:1464–68 [DOI] [PubMed] [Google Scholar]
- 5.Ha Y, Arnold AR, Nuñez NN, Bartels PL, Zhou A, et al. 2017. Sulfur K-Edge XAS Studies of the Effect of DNA Binding on the [Fe4S4] Site in EndoIII and MutY. J. Am. Chem. Soc 139:11434–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertini I, Gray HB, Lippard SJ, Valentine JS. 1994. Bioinorganic Chemistry University Books [Google Scholar]
- 7.Mortenson LE, Valentine RC, Carnahan JE. 1962. An electron transport factor from Clostridium pasteurianum. Biochem. Biophys. Res. Commun 7:448–52 [DOI] [PubMed] [Google Scholar]
- 8.Sands RH, Beinert H. 1960. Studies on mitochondria and submitochondrial particles by paramagnetic resonance (EPR) spectroscopy. Biochem. Biophys. Res. Commun 3:47–52 [Google Scholar]
- 9.Arnon DI, Whatley FR, Allen MB. 1957. Triphosphopyridine nucleotide as a catalyst of photosynthetic phosphorylation. Nature 180:182–85 [DOI] [PubMed] [Google Scholar]
- 10.Gray HB, Winkler JR. 2010. Electron flow through metalloproteins. Biochim. Biophys. Acta - Bioenerg. 1797:1563–72 [DOI] [PubMed] [Google Scholar]
- 11.Winkler JR, Gray HB. 2014. Electron Flow through Metalloproteins. Chem. Rev 114:3369–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunningham RP, Asahara H, Bank JF, Scholes CP, Salerno JC, et al. 1989. Endonuclease III Is an Iron-Sulfur Protein. Biochemistry 28:4450–55 [DOI] [PubMed] [Google Scholar]
- 13.Rouault TA. 2015. Mammalian iron-sulphur proteins: novel insights into biogenesis and function. Nat Rev Mol Cell Biol 16:45–55 [DOI] [PubMed] [Google Scholar]
- 14.Johnson DC, Dean DR, Smith AD, Johnson MK. 2005. Structure, Function, and Formation of Biological Iron-Sulfur Clusters. Annu. Rev. Biochem 74:247–81 [DOI] [PubMed] [Google Scholar]
- 15.Braymer JJ, Lill R. 2017. Iron-sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem 292:12754–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paul VD, Lill R. 2015. Biogenesis of cytosolic and nuclear iron-sulfur proteins and their role in genome stability. Biochim. Biophys. Acta - Mol. Cell Res 1853:1528–39 [DOI] [PubMed] [Google Scholar]
- 17.Rouault TA, Maio N. 2017. Biogenesis and functions of mammalian iron-sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. J. Biol. Chem 292:12744–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubio LM, Ludden PW. 2008. Biosynthesis of the Cofactor of Nitrogenase. Annu. Rev. Microbiol 62:93–111 [DOI] [PubMed] [Google Scholar]
- 19.Wayne Outten F 2015. Recent advances in the Suf Fe-S cluster biogenesis pathway: Beyond the Proteobacteria. Biochim. Biophys. Acta - Mol. Cell Res 1853:1464–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ezraty B, Vergnes A, Banzhaf M, Duverger Y, Huguenot A, et al. 2013. Fe-S cluster biosynthesis controls uptake of aminoglycosides in a ROS-less death pathway. Science 340:1583–87 [DOI] [PubMed] [Google Scholar]
- 21.White MF, Dillingham MS. 2012. Iron-sulphur clusters in nucleic acid processing enzymes. Curr. Opin. Struct. Biol 22:94–100 [DOI] [PubMed] [Google Scholar]
- 22.Guan Y, Manuel RC, Arvai AS, Parikh SS, Mol CD, et al. 1998. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat. Struct. Biol 5:1058–64 [DOI] [PubMed] [Google Scholar]
- 23.Hinks JA, Evans MCW, De Miguel Y, Sartori AA, Jiricny J, Pearl LH. 2002. An iron-sulfur cluster in the Family 4 uracil-DNA glycosylases. J. Biol. Chem 277:16936–40 [DOI] [PubMed] [Google Scholar]
- 24.Bartels PL, O’Brien E, Barton JK. 2017. DNA signaling by iron-sulfur cluster proteins In Biochemistry, Biosynthesis and Human Diseases: Biochemistry, Biosynthesis, and Human Diseases, ed TA Rouault. 2:405–23. Berlin, Boston: De Gruyter [Google Scholar]
- 25.Zhang F, Scheerer P, Oberpichler I, Lamparter T, Krauss N. 2013. Crystal structure of a prokaryotic (6-4) photolyase with an Fe-S cluster and a 6,7-dimethyl-8-ribityllumazine antenna chromophore. PNAS 110:7217–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Kasciukovic T, White MF. 2012. The CRISPR Associated Protein Cas4 Is a 5′ to 3′ DNA Exonuclease with an Iron-Sulfur Cluster. PLoS One. 7:e47232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuss JO, Tsai CL, Ishida JP, Tainer JA. 2015. Emerging critical roles of Fe-S clusters in DNA replication and repair. Biochim. Biophys. Acta - Mol. Cell Res 1853:1253–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva RMB, Grodick MA, Barton JK. 2018. in preparation [Google Scholar]
- 29.Hirata A, Klein BJ, Murakami KS. 2008. The X-ray crystal structure of RNA polymerase from Archaea. Nature 451:851–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjdia A, Heil K, Barends TRM, Carell T, Schlichting I. 2012. Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme. Nucleic Acids Res 40:9308–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rouault TA. 2015. Iron-sulfur proteins hiding in plain sight. Nat. Chem. Biol 11:442–45 [DOI] [PubMed] [Google Scholar]
- 32.Boal AK, Yavin E, Lukianova OA, O’Shea VL, David SS, Barton JK. 2005. DNA-bound redox activity of DNA repair glycosylases containing [4Fe-4S] clusters. Biochemistry 44:8397–8407 [DOI] [PubMed] [Google Scholar]
- 33.Fu W, O’Handley S, Cunningham RP, Johnson MK. 1992. The role of the iron-sulfur cluster in Escherichia coli endonuclease III. A resonance Raman study. J. Biol. Chem 267:16135–37 [PubMed] [Google Scholar]
- 34.Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer J a. 1995. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 14:4108–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muty G, Porello SL, Cannon MJ, David SS. 1998. A Substrate Recognition Role for the [4Fe-4S] 2 + Cluster of the DNA Repair. Society. 2960:6465–75 [DOI] [PubMed] [Google Scholar]
- 36.Imlay JA. 2013. The molecular mechanisms andphysiological consequences of oxidativestress: lessons from a model bacterium. Nat. Rev. Microbiol 11:443–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Genereux JC, Barton JK. 2010. Mechanisms for DNA charge transport. Chem. Rev 110:1642–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eley DD, Spivey DI. 1962. Semiconductivity of organic substances. IX. Nucleic acid in the dry state. Trans. Faraday Soc 58:411–15 [Google Scholar]
- 39.Murphy CJ, Arkin MR, Jenkins Y, Ghatlia ND, Bossmann SH, et al. 1993. Long-range photoinduced electron transfer through a DNA helix. Science 262:1025–29 [DOI] [PubMed] [Google Scholar]
- 40.Kelley SO, Barton JK. 1999. Electron transfer between bases in double helical DNA. Science 283:375–81 [DOI] [PubMed] [Google Scholar]
- 41.Nunez ME, Hall DB, Barton JK. 1999. Long- range oxidative damange to DNA: effects of distance and sequence. Chem. Biol 6:85–97 [DOI] [PubMed] [Google Scholar]
- 42.O’Neill MA, Becker HC, Wan C, Barton JK, Zewail AH. 2003. Ultrafast Dynamics in DNA-Mediated Electron Transfer: Base Gating and the Role of Temperature. Angew. Chemie - Int. Ed 42:5896–5900 [DOI] [PubMed] [Google Scholar]
- 43.Giese B, Graber M, Cordes M. 2008. Electron transfer in peptides and proteins. Curr. Opin. Chem. Biol 12:755–59 [DOI] [PubMed] [Google Scholar]
- 44.Phillips R, Kondev J, Theriot J. 2008. Physical Biology of the Cell London: Garland Science, Taylor and Francis Group; 1st ed. [Google Scholar]
- 45.Rajski SR, Barton JK. 2001. How different DNA-binding proteins affect long-range oxidative damage to DNA. Biochemistry 40:5556–64 [DOI] [PubMed] [Google Scholar]
- 46.Boon EM, Salas JE, Barton JK. 2002. An electrical probe of protein-DNA interactions on DNA-modified surfaces. Nat. Biotechnol 20:282–86 [DOI] [PubMed] [Google Scholar]
- 47.Nunez ME, Noyes KT, Barton JK. 2002. Oxidative charge transport through DNA in nucleosome core particles. Chem. Biol 9:403–15 [DOI] [PubMed] [Google Scholar]
- 48.Slinker JD, Muren NB, Renfrew SE, Barton JK. 2011. DNA charge transport over 34 nm. Nat. Chem 3:228–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slinker JD, Muren NB, Gorodetsky AA, Barton JK. 2010. Multiplexed DNA-modified electrodes. J. Am. Chem. Soc 132:2769–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pheeney CG, Arnold AR, Grodick MA, Barton JK. 2013. Multiplexed electrochemistry of DNA-bound metalloproteins. J. Am. Chem. Soc 135:11869–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bartels PL, Stodola JL, Burgers PMJ, Barton JK. 2017. A Redox Role for the [4Fe4S] Cluster of Yeast DNA Polymerase δ. J. Am. Chem. Soc 139:18339–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Brien E, Holt ME, Thompson MK, Salay LE, Ehlinger AC, et al. 2017. The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport. Science 355:eaag1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tse ECM, Zwang TJ, Barton JK. 2017. The Oxidation State of [4Fe4S] Clusters Modulates the DNA-Binding Affinity of DNA Repair Proteins. J. Am. Chem. Soc 139:12784–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mui TP, Fuss JO, Ishida JP, Tainer JA, Barton JK. 2011. ATP-stimulated, DNA-mediated redox signaling by XPD, a DNA repair and transcription helicase. J. Am. Chem. Soc 133:16378–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sontz PA, Mui TP, Fuss JO, Tainer JA, Barton JK. 2012. DNA charge transport as a first step in coordinating the detection of lesions by repair proteins. PNAS 109:1856–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ekanger L, Oyala PH, Moradian A, Sweredoski MJ, Barton JK. 2018. Submitted [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou A, Ph.D thesis, California Institute of Technology, 2018. [Google Scholar]
- 58.McDonnell KJ, Chemler JA, Bartels PL, O’Brien E, Marvin ML, et al. A human MUTYH variant linking colonic polyposis to redox degradation of the [4Fe4S]2+ cluster. Nat. Chem [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grodick MA, Segal HM, Zwang TJ, Barton JK. 2014. DNA-mediated signaling by proteins with 4Fe-4S clusters is necessary for genomic integrity. J. Am. Chem. Soc 136:6470–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gorodetsky Alon A., Amie K. Boal A, Barton JK. 2006. Direct Electrochemistry of Endonuclease III in the Presence and Absence of DNA. J. Am. Chem. Soc 128:12082–83 [DOI] [PubMed] [Google Scholar]
- 61.Bartels PL, Zhou A, Arnold AR, Nunez NN, Crespilho FN, et al. 2017. Electrochemistry of the [4Fe4S] Cluster in Base Excision Repair Proteins: Tuning the Redox Potential with DNA. Langmuir 33:2523–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klinge S, Hirst J, Maman JD, Krude T, Pellegrini L. 2007. An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat. Struct. Mol. Biol 14:875–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weiner BE, Huang H, Dattilo BM, Nilges MJ, Fanning E, Chazin WJ. 2007. An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J. Biol. Chem 282:33444–51 [DOI] [PubMed] [Google Scholar]
- 64.Ren B, Duan X, Ding H. 2009. Redox control of the DNA damage-inducible protein DinG helicase activity via its iron-sulfur cluster. J. Biol. Chem 284:4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rudolf J, Makrantoni V, Ingledew WJ, Stark MJR, White MF. 2006. The DNA Repair Helicases XPD and FancJ Have Essential Iron-Sulfur Domains. Mol. Cell. 23:801–8 [DOI] [PubMed] [Google Scholar]
- 66.Yavin E, Boal AK, Stemp EDA, Boon EM, Livingston AL, et al. 2005. Protein-DNA charge transport: Redox activation of a DNA repair protein by guanine radical. PNAS 102:3546–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ciccia A, Elledge SJ. 2010. The DNA Damage Response: Making It Safe to Play with Knives. Mol. Cell 40:179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marsden CG, Dragon JA, Wallace SS, Sweasy JB. 2017. Base Excision Repair Variants in Cancer. Methods Enzymol 591:119–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gorman J, Greene EC. 2008. Visualizing one-dimensional diffusion of proteins along DNA. Nat. Struct. Mol. Biol 15:768–74 [DOI] [PubMed] [Google Scholar]
- 70.Christmann M, Kaina B. 2013. Transcriptional regulation of human DNA repair genes following genotoxic stress: Trigger mechanisms, inducible responses and genotoxic adaptation. Nucleic Acids Res 41:8403–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boal AK, Barton JK. 2005. Electrochemical detection of lesions in DNA. Bioconjug. Chem 16:312–21 [DOI] [PubMed] [Google Scholar]
- 72.Hall DB, Holmlin RE, Barton JK. 1996. Oxidative DNA damage through long-range electron transfer. Nature 382:731–35 [DOI] [PubMed] [Google Scholar]
- 73.David SS, Williams SD. 1998. Chemistry of Glycosylases and Endonucleases Involved in Base-Excision Repair. Chem. Rev 98:1221–62 [DOI] [PubMed] [Google Scholar]
- 74.David SS, O’Shea VL, Kundu S. 2007. Base-excision repair of oxidative DNA damage. Nature. 447:941–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fromme JC, Verdine GL. 2003. Structure of a trapped endonuclease III-DNA covalent intermediate. EMBO J. 22:3461–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Engstrom LM, Partington OA, David SS. 2012. An iron-sulfur cluster loop motif in the archaeoglobus fulgidus uracil-DNA glycosylase mediates efficient uracil recognition and removal. Biochemistry 51:5187–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lukianova OA, David SS. 2005. A role for iron-sulfur clusters in DNA repair. 9(2):145–51 [DOI] [PubMed] [Google Scholar]
- 78.Weren RDA, Ligtenberg MJL, Geurts van Kessel A, De Voer RM, Hoogerbrugge N, Kuiper RP. 2018. NTHL1 and MUTYH polyposis syndromes: two sides of the same coin? J. Pathol 244:135–42 [DOI] [PubMed] [Google Scholar]
- 79.Boal AK, Genereux JC, Sontz PA, Gralnick JA, Newman DK, Barton JK. 2009. Redox signaling between DNA repair proteins for efficient lesion detection. PNAS 106: 15237–15242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Romano CA, Sontz PA, Barton JK. 2011. Mutants of the base excision repair glycosylase, endonuclease III: DNA charge transport as a first step in lesion detection. Biochemistry 50:6133–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu Y, Brosh RM. 2012. DNA helicase and helicase-nuclease enzymes with a conserved iron-sulfur cluster. Nucleic Acids Res 40:4247–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brosh RM. 2013. DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer 13:542–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vannier JB, Sarek G, Boulton SJ. 2014. RTEL1: Functions of a disease-associated helicase. Trends Cell Biol 24:416–25 [DOI] [PubMed] [Google Scholar]
- 84.Voloshin ON, Camerini-Otero RD. 2007. The DinG protein from Escherichia coli is a structure-specific helicase. J. Biol. Chem 282:18437–47 [DOI] [PubMed] [Google Scholar]
- 85.Brown LT, Sutera VA, Zhou S, Weitzel CS, Cheng Y, Lovett ST. 2015. Connecting Replication and Repair: YoaA, a Helicase-Related Protein, Promotes Azidothymidine Tolerance through Association with Chi, an Accessory Clamp Loader Protein. PLoS Genet 11:e1005651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Houten B Van, Kuper J, Kisker C. 2016. Role of XPD in cellular functions: To TFIIH and beyond. DNA Repair 44:136–42 [DOI] [PubMed] [Google Scholar]
- 87.Brosh RM, Cantor SB. 2014. Molecular and cellular functions of the FANCJ DNA helicase defective in cancer and in Fanconi anemia. Front. Genet 5:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grodick MA, Muren NB, Barton JK. 2015. DNA charge transport within the cell. Biochemistry 54:962–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guo M, Hundseth K, Ding H, Vidhyasagar V, Inoue A, et al. 2015. A distinct triplex DNA unwinding activity of ChlR1 helicase. J. Biol. Chem 290:5174–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yeeles JTP, Cammack R, Dillingham MS. 2009. An iron-sulfur cluster is essential for the binding of broken DNA by AddAB-type helicase-nucleases. J. Biol. Chem 284:7746–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lenhart JS, Schroeder JW, Walsh BW, Simmons LA. 2012. DNA Repair and Genome Maintenance in Bacillus subtilis. Microbiol. Mol. Biol. Rev 76:530–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saikrishnan K, Yeeles JT, Gilhooly NS, Krajewski WW, Dillingham MS, Wigley DB. 2012. Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex. EMBO J. 31:1568–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kisker C, Kuper J, Houten B Van, Houten B Van. 2013. Prokaryotic nucleotide excision repair. Cold Spring Harb. Perspect. Biol 5:1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Golinelli MP, Chmiel NH, David SS. 1999. Site-directed mutagenesis of the cysteine ligands to the [4Fe-4S] cluster of Escherichia coli MutY. Biochemistry. 38:6997–7007 [DOI] [PubMed] [Google Scholar]
- 95.Cheadle JP, Sampson JR. 2007. MUTYH-associated polyposis-From defect in base excision repair to clinical genetic testing. DNA Repair 6:274–79 [DOI] [PubMed] [Google Scholar]
- 96.Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, et al. 2008. XPD Helicase Structures and Activities: Insights into the Cancer and Aging Phenotypes from XPD Mutations. Cell 133:789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wolski SC, Kuper J, Hanzelmann P, Truglio JJ, Croteau DL, et al. 2008. Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol 6:1332–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lanz ND, Booker SJ. 2015. Auxiliary iron-sulfur cofactors in radical SAM enzymes. Biochim. Biophys. Acta - Mol. Cell Res 1853:1316–34 [DOI] [PubMed] [Google Scholar]
- 99.Maiocco SJ, Grove TL, Booker SJ, Elliott SJ. 2015. Electrochemical Resolution of the [4Fe-4S] Centers of the AdoMet Radical Enzyme BtrN: Evidence of Proton Coupling and an Unusual, Low-Potential Auxiliary Cluster. J. Am. Chem. Soc 137:8664–67 [DOI] [PubMed] [Google Scholar]
- 100.Marquet A, Florentin D, Ploux O, Bui BTS. 1998. In vivo formation of C-S bonds in biotin. An example of radical chemistry under reducing conditions. J. Phys. Org. Chem 11:529–35 [Google Scholar]
- 101.Anton BP, Saleh L, Benner JS, Raleigh EA, Kasif S, Roberts RJ. 2008. RimO, a MiaB-like enzyme, methylthiolates the universally conserved Asp88 residue of ribosomal protein S12 in Escherichia coli. PNAS 105:1826–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schwalm EL, Grove TL, Booker SJ, Boal AK. 2016. Crystallographic capture of a radical S-adenosylmethionine enzyme in the act of modifying tRNA. Science 352:309–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kneuttinger AC, Heil K, Kashiwazaki G, Carell T. 2013. The radical SAM enzyme spore photoproduct lyase employs a tyrosyl radical for DNA repair. Chem. Commun 49:722–24 [DOI] [PubMed] [Google Scholar]
- 104.Moran U, Phillips R, Milo R. 2010. SnapShot: Key numbers in biology. Cell 141:1262. [DOI] [PubMed] [Google Scholar]
- 105.Ganai RA, Johansson E. 2016. DNA Replication - a Matter of Fidelity. Mol. Cell 62:745–55 [DOI] [PubMed] [Google Scholar]
- 106.Netz DJA, Stith CM, Stumpfig M, Köpf G, Vogel D, et al. 2011. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol 8:125–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pokharel S, Campbell JL. 2012. Cross talk between the nuclease and helicase activities of Dna2: Role of an essential iron-sulfur cluster domain. Nucleic Acids Res 40:7821–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Baranovskiy AG, Babayeva ND, Zhang Y, Gu J, Suwa Y, et al. 2016. Mechanism of Concerted RNA-DNA Primer Synthesis by the Human Primosome. J. Biol. Chem 291:10006–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Johansson E, Majka J, Burgers PMJ. 2001. Structure of DNA Polymerase δ from Saccharomyces cerevisiae. J. Biol. Chem 276:43824–28 [DOI] [PubMed] [Google Scholar]
- 110.Chilkova O, Jonsson B-H, Johansson E. 2003. The Quaternary Structure of DNA Polymerase ε from Saccharomyces cerevisiae. J. Biol. Chem 278:14082–86 [DOI] [PubMed] [Google Scholar]
- 111.Nelson JR, Lawrence CW, Hinkle DC. 1996. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science 272:1646–49 [DOI] [PubMed] [Google Scholar]
- 112.Makarova AV, Burgers PM. 2015. Eukaryotic DNA polymerase ζ. DNA Repair 29:47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yeeles JTP, Deegan TD, Janska A, Early A, Diffley JFX. 2015. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519:431–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fragkos M, Ganier O, Coulombe P, Mechali M. 2015. DNA replication origin activation in space and time. 16(6):360–74 [DOI] [PubMed] [Google Scholar]
- 115.Iyer LM, Leipe DD, Koonin EV., Aravind L. 2004. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol 146:11–31 [DOI] [PubMed] [Google Scholar]
- 116.Duncker BP, Chesnokov IN, McConkey BJ. 2009. The origin recognition complex protein family. Genome Biol 10:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Moyer SE, Lewis PW, Botchan MR. 2006. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. PNAS. 103:10236–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pacek M, Tutter AV., Kubota Y, Takisawa H, Walter JC. 2006. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell 21:581–87 [DOI] [PubMed] [Google Scholar]
- 119.Labib K, Gambus A. 2007. A key role for the GINS complex at DNA replication forks. Trends Cell Biol 17:271–78 [DOI] [PubMed] [Google Scholar]
- 120.Burgers PMJ, Kunkel TA. 2017. Eukaryotic DNA Replication Fork. Annu. Rev. Biochem 86:417–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Arezi B, Kuchta RD. 2000. Eukaryotic DNA primase. Trends Biochem. Sci 25:572–76 [DOI] [PubMed] [Google Scholar]
- 122.Kuchta RD, Stengel G. 2010. Mechanism and evolution of DNA primases. Biochim. Biophys. Acta - Proteins Proteomics. 1804:1180–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Frick DN, Richardson CC. 2001. DNA primases. Annu. Rev. Biochem 70:39–80 [DOI] [PubMed] [Google Scholar]
- 124.Netz DJA, Mascarenhas J, Stehling O, Pierik AJ, Lill R. 2014. Maturation of cytosolic and nuclear iron-sulfur proteins. Trends Cell Biol 24:303–12 [DOI] [PubMed] [Google Scholar]
- 125.Núñez-Ramírez R, Klinge S, Sauguet L, Melero R, Recuero-Checa MA, et al. 2011. Flexible tethering of primase and DNA Pol α in the eukaryotic primosome. Nucleic Acids Res 39:8187–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu L, Huang M. 2015. Essential role of the iron-sulfur cluster binding domain of the primase regulatory subunit Pri2 in DNA replication initiation. Protein Cell 6:194–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vaithiyalingam S, Warren EM, Eichman BF, Chazin WJ. 2010. Insights into eukaryotic DNA priming from the structure and functional interactions of the 4Fe-4S cluster domain of human DNA primase. PNAS 107:13684–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.O’Brien E, Salay LE., Epum EA, Friedman KL, Chazin WJ, Barton JK. 2018. Submitted [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Garg P, Burgers PMJ. 2005. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol 40:115–28 [DOI] [PubMed] [Google Scholar]
- 130.McElhinny SAN, Kumar D, Clark AB, Watt DL, Watts BE, et al. 2010. Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol 6:774–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA. 2013. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol. Cell 50:437–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McElhinny SAN, Stith CM, Burgers PMJ, Kunkel TA. 2007. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase. J. Biol. Chem 282:2324–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Johnson RE, Klassen R, Prakash L, Prakash S. 2015. A Major Role of DNA Polymerase δ in Replication of Both the Leading and Lagging DNA Strands. Mol. Cell 59:163–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jain R, Vanamee ES, Dzikovski BG, Buku A, Johnson RE, et al. 2014. An iron-sulfur cluster in the polymerase domain of yeast DNA polymerase ε. J. Mol. Biol 426:301–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tsurimoto T, Stillman B. 1991. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J. Biol. Chem 266:1950–60 [PubMed] [Google Scholar]
- 136.Yuzhakov A, Kelman Z, Hurwitz J, O’Donnell M. 1999. Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J. 18:6189–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.De Piccoli G, Katou Y, Itoh T, Nakato R, Shirahige K, Labib K. 2012. Replisome Stability at Defective DNA Replication Forks Is Independent of S Phase Checkpoint Kinases. Mol. Cell 45:696–704 [DOI] [PubMed] [Google Scholar]
- 138.Zhou C, Pourmal S, Pavletich NP. 2015. Dna2 nuclease-helicase structure, mechanism and regulation by Rpa. Elife 4:e09832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hu J, Sun L, Shen F, Chen Y, Hua Y, et al. 2012. The intra-S phase checkpoint targets Dna2 to prevent stalled replication forks from reversing. Cell 149:1221–32 [DOI] [PubMed] [Google Scholar]
- 140.Budd ME, Choe WC, Campbell JL. 2000. The nuclease activity of the yeast Dna2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem 275:16518–29 [DOI] [PubMed] [Google Scholar]
- 141.Jennings ME, Lessner FH, Karr EA, Lessner DJ. 2017. The [4Fe-4S] clusters of Rpo3 are key determinants in the post Rpo3/Rpo11 heterodimer formation of RNA polymerase in Methanosarcina acetivorans. Microbiologyopen. 6:e00399. [DOI] [PMC free article] [PubMed] [Google Scholar]