Abstract
In cellular immunotherapies, natural killer (NK) cells often demonstrate potent antitumor effects in high-risk cancer patients. But Good Manufacturing Practice (GMP)-compliant manufacturing of clinical-grade NK cells in high numbers for patient treatment is still a challenge. Therefore, new protocols for isolation and expansion of NK cells are required. In order to attack resistant tumor entities, NK cell killing can be improved by genetic engineering using alpharetroviral vectors that encode for chimeric antigen receptors (CARs). The aim of this work was to demonstrate GMP-grade manufacturing of NK cells using the CliniMACS® Prodigy device (Prodigy) with implemented applicable quality controls. Additionally, the study aimed to define the best time point to transduce expanding NK cells with alpharetroviral CAR vectors. Manufacturing and clinical-scale expansion of primary human NK cells were performed with the Prodigy starting with 8-15.0 × 109 leukocytes (including 1.1–2.3 × 109 NK cells) collected by small-scale lymphapheresis (n = 3). Positive fraction after immunoselection, in-process controls (IPCs), and end product were quantified by flow cytometric no-wash, single-platform assessment, and gating strategy using positive (CD56/CD16/CD45), negative (CD14/CD19/CD3), and dead cell (7-aminoactinomycine [7-AAD]) discriminators. The three runs on the fully integrated manufacturing platform included immunomagnetic separation (CD3 depletion/CD56 enrichment) followed by NK cell expansion over 14 days. This process led to high NK cell purities (median 99.1%) and adequate NK cell viabilities (median 86.9%) and achieved a median CD3+ cell depletion of log −3.6 after CD3 depletion and log −3.7 after immunomagnetic CD3 depletion and consecutive CD56 selection. Subsequent cultivation of separated NK cells in the CentriCult® chamber of Prodigy resulted in approximately 4.2–8.5-fold NK cell expansion rates by adding of NK MACS® basal medium containing NK MACS® supplement, interleukin (IL)-2/IL-15 and initial IL-21. NK cells expanded for 14 days revealed higher expression of natural cytotoxicity receptors (NKp30, NKp44, NKp46, and NKG2D) and degranulation/apoptotic markers and stronger cytolytic properties against K562 compared to non-activated NK cells before automated cultivation. Moreover, expanded NK cells had robust growth and killing activities even after cryopreservation. As a crucial result, it was possible to determine the appropriate time period for optimal CAR transduction of cultivated NK cells between days 8 and 14, with the highest anti-CD123 CAR expression levels on day 14. The anti-CD123 CAR NK cells showed retargeted killing and degranulation properties against CD123-expressing KG1a target cells, while basal cytotoxicity of non-transduced NK cells was determined using the CD123-negative cell line K562. Time-lapse imaging to monitor redirected effector-to-target contacts between anti-CD123 CAR NK and KG1a showed long-term effector–target interaction. In conclusion, the integration of the clinical-scale expansion procedure in the automated and closed Prodigy system, including IPC samples and quality controls and optimal time frames for NK cell transduction with CAR vectors, was established on 48-well plates and resulted in a standardized GMP-compliant overall process.
Keywords: natural killer cell, CliniMACS Prodigy®, chimeric antigen receptor, transduction, alpharetroviral vector
Introduction
Natural killer (NK) cells connect the innate and adaptive immune system as the first line of defense against virally infected and malignantly transformed cells. NK cells eliminate target cells without previous activation, and cytotoxic activity is regulated by balanced signals from inhibitory and activating surface receptors.1–4 Mechanisms for early killing of target cells are based on release of immune regulatory cytokines such as interferon gamma (IFN-γ) and tumor necrosis factor alpha (TNF-α),5 death receptor ligand binding to induce caspase-dependent apoptosis,6,7 or secretion of cytotoxic granules composed of granzymes A and B, granulysin, or perforin.1,2
The main NK cell population is defined as CD56+CD3– lymphoid cells, which are divided in two subpopulations: CD56highCD16– regulatory cells and CD56dimCD16+ cytotoxic cells. Because of the advantage of preventing graft-versus-host disease (GvHD) in allogeneic settings, NK cells seem to be ideal for immunotherapies.8 However, it is still a challenge to expand high numbers of Good Manufacturing Practice (GMP)-grade NK cells for patient treatments, since several infusions of at least 1 × 107 NK cells/kg are necessary.8,9
In the past, GMP-compliant procedures relying on the CliniMACS Plus® (Plus) device were developed successfully for two-step-processes by combination of immunomagnetic CD3 T-cell depletion and CD56+ NK cell separation. However, a fully automated GMP-compliant separation and expansion of clinical-grade NK cells at sufficiently high effector cell numbers remains a great challenge.10–12 Recently, a multiple-step manufacturing process was realized by the CliniMACS Prodigy® (Prodigy) instrument for clinical grade NK cells using an automated T-cell receptor (TCR)-α/β-CD19 depletion, CD56 enrichment, followed by an automated feeder cell-based expansion period, which resulted in a mean 850-fold expansion after day 14.13
Use of NK cells as an anticancer adoptive immunotherapeutic approach is hampered by various mechanisms that tumor or leukemia cells employ to circumvent the effector cell–based immunosurveillance.14,15 To overcome these tumor defense strategies—so-called tumor immune escape mechanisms—and to enhance the specific targeting to resistant/masking malignant cells, NK cells can be transduced by engineered viral vectors to express chimeric antigen receptors (CARs). These artificial surface receptors consist of an extracellular single-chain variable fragment (scFv) domain to detect specific target molecules expressed on cancer cells. For signal transduction, the CAR molecule contains intracellular signaling domains fused to the scFv.16 Shortly after binding to CAR target molecules, activation of the CAR NK cells results in strong IFN-γ release and/or secretion of cytotoxic granules (degranulation).1
CD123 corresponds to the interleukin (IL)-3 receptor α chain that is frequently reported to be highly expressed on acute myeloid leukemia (AML) cells. Therefore, this cell surface molecule constitutes a suitable marker for AML cell detection.17,18
The objective of this study was to establish a fully integrated manufacturing platform and the appropriate quality controls (QC) for clinical-scale production of human primary NK cells. Therefore, the study connected the GMP-compliant NK cell separation by immunomagnetic CD3 depletion and CD56 separation with a NK cell expansion for 14 days in the automated closed Prodigy system. Glucose concentration and pH were monitored as parameters for NK cell growth during the whole expansion period. In-process control (IPC) samples were analyzed for a period of 14 days to assess the NK cell-based cytotoxicity against K562 target cells, levels of surface marker expression, cytokines, cytolytic granule proteins, and degranulation marker CD107a involved in NK cell expansion. NK cell viability was evaluated before cryopreservation and after thawing. The second aim was to implement optimized vector technology in the manufacturing runs during time of expansion. In order to define the best time for optimal successful alpharetroviral vector transduction of expanded NK cells with CAR vectors, IPC samples harvested at different time points in the expansion process in regard to transduction rates were analyzed. The redirected effector-mediated killing activity against the AML cell line KG1a was analyzed by flow cytometry–based cytotoxicity assays and time-lapse tracking experiments, which demonstrated improved retargeted killing that was dependent upon activation levels from expanded NK cells.
Methods
Source of cell material
Peripheral blood leukapheresis (LA) products from healthy donors were obtained from the Institute for Transfusion Medicine of Hannover Medical School following written informed consent, as approved by the ethics committee (#2159-2014) of Hannover Medical School.
Cell lines
The human cell line K562 (chronic myelogenous leukemia) was obtained from the German Collection of Microorganisms and Cell Cultures and was cultivated in Roswell Park Memorial Institute medium 1640 (Biochrom) supplemented with 10% fetal bovine serum (FBS; Biochrom). This CD123-negative cell line was used to determine basal cytotoxicities of freshly isolated, expanded, or thawed primary NK cells. The CD123-positive human cell line KG1a (acute myelogenous leukemia) was purchased from the American Type Culture Collection and was grown in Iscove's modified Dulbecco's medium (Biochrom) supplemented with 20% FBS. The KG1a cells were used in cytotoxic assays to determine the impact of the CAR on the killing capacity of CAR NK cells.
CD3 depletion, selection, and expansion of NK cells using Prodigy
Prodigy (Miltenyi Biotec) presented a closed automated GMP-compliant cell processing system that was used for enrichment of primary human NK cells out of LA, starting with a two-step procedure that consists of CD3 depletion and CD56 selection without changing the tubing set TS 310. All reagents used for manufacturing were GMP-compliant, except AB serum. For clinical manufacturing, clinical grade AB serum will be mandatory. Depletion and selection were based on CD3 CE and CD56 CE reagents (Miltenyi Biotec), respectively. The complete procedure was performed according to the manufacturer's instructions and was based on modified T-cell transduction process software v1.2.1 (Miltenyi Biotec). Different cell fractions—such as LA, positive fraction (PF) after immunoselection, IPCs from expanded cell culture, and end product (EP)—were collected and analyzed. As starting material, LA products with a median of 14 × 109 viable white blood cells (WBC; range 8–15.0 × 109 WBC) were used.
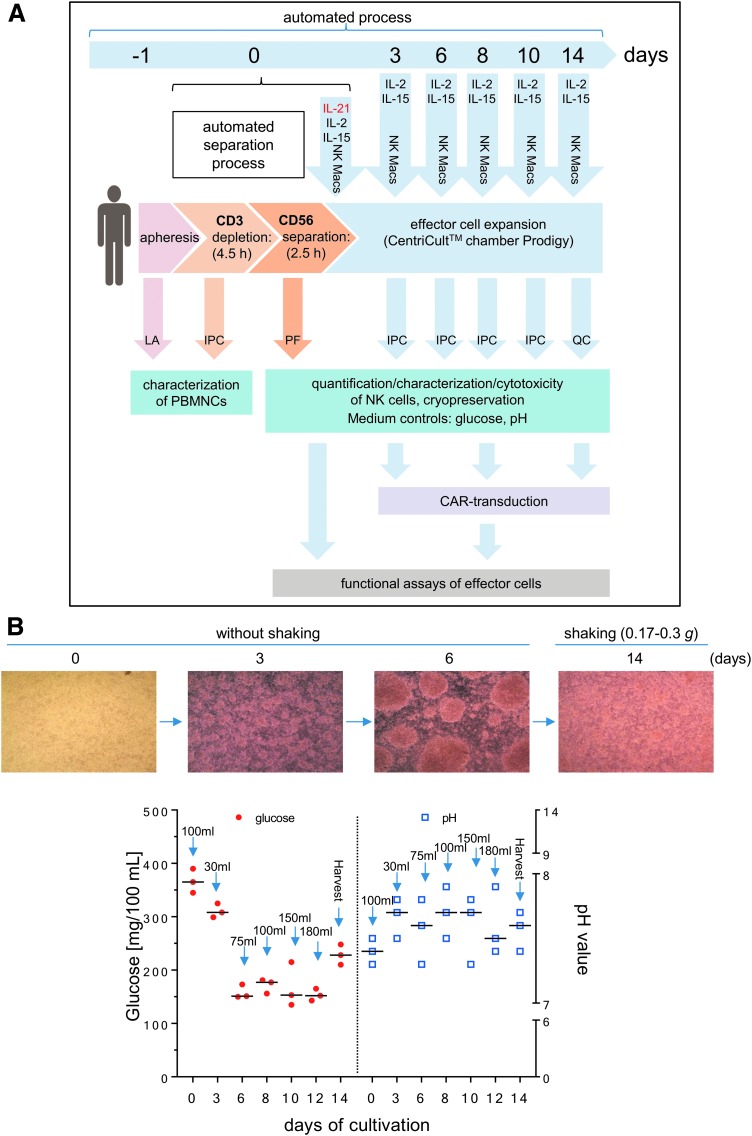
As part of the preassembled tubing set, the CentriCult® unit (Miltenyi Biotec) allowed a maximum volume of 250 mL for cell cultivation. The starting concentration of primary human NK cells was 2–4 × 106 NK cells/mL while expanding the cells for 14 days at 37°C and 5% CO2 in NK MACS® expansion medium supplemented with 5% human type AB serum (Biochrom), IL-21 (1 IU/mL; initial; Miltenyi Biotec), IL-2 (500 IU/mL; Proleukin S; Novartis Pharma GmbH), and IL-15 (140 IU/mL; Miltenyi Biotec), and contained 2% NK MACS® supplement (Miltenyi Biotec). Growth medium was exchanged every 2–3 days, as shown in Fig. 1. During the first 6 days, NK cells grew without shaking, while a gentle shaking mode (0.17–0.3 g) was activated for the CentriCult® unit on day 7.
Figure 1.
Manufacturing of Good Manufacturing Practice (GMP) grade natural killer (NK) cells. (A) Schematic overview of the automated process steps for the separation and expansion of donor NK cells using CliniMACS Prodigy® (Prodigy). Unstimulated leukapheresis (LA) products were used for depletion of CD3+ cells followed by immunomagnetic enrichment of CD56+ cells. Subsequently, expansion of isolated CD56+/CD3– NK cells (positive fraction [PF]) were performed in NK MACS growth medium supplemented initially with interleukin (IL)-21, IL-2, and IL-15 on day 0. Medium replenishment followed on days 3, 6, 8, and 10 with NK MACS medium supplemented with IL-2 and IL-15. Samples were collected every 2–3 days (in-process control [IPC] samples; quality control [QC] samples) for NK cell quantification and functional effector cell analysis, surface marker expression levels, and growth parameters (glucose, pH), and to test suitability of cryopreservation. To determine the adequate time period for transduction, small samples of expanded NK cells were collected on expansion days 2 or 3, 8, and 14. Moreover, cell samples were collected for analysis of basal NK cell cytotoxicity, degranulation/cytokine secretion, as well as chimeric antigen receptor (CAR) detection, as indicated in the schematic procedure overview. (B) Culture conditions for NK cells expanded in the Prodigy CentriCult® chamber. NK cells were cultured in NK MACS growth medium for the first 6 days without shaking to maintain effector cell cluster formations. Shaking mode was initiated on day 7 to ensure optimal nutrient supply for the NK cells. Glucose (red circles) and pH (blue squares) values were determined over 14 days (last day of expansion indicated as “harvest”), and individual feeding steps are marked by arrows and volume of growth medium added.
Alpharetroviral vectors and transduction
Alpharetroviral self-inactivating (SIN) vectors harbored large deletions of transcriptional elements in the U3 of the long terminal repeat regions and included the strong myeloproliferative sarcoma virus promoter as an internal promoter to direct expression of the third-generation CAR transgene (anti-CD123 CAR) followed by an internal ribosomal entry site for enhanced green fluorescent protein (eGFP) expression (Fig. 5A).19,20 The anti-CD123 CAR construct has been described previously21 and contained a signal peptide derived from granulocyte–macrophage colony-stimulating factor receptor α-chain, anti-CD123 scFv,22 CD28, CD137 (4-1BB), and CD3ζ. A vector that solely expressed eGFP was used as a control.
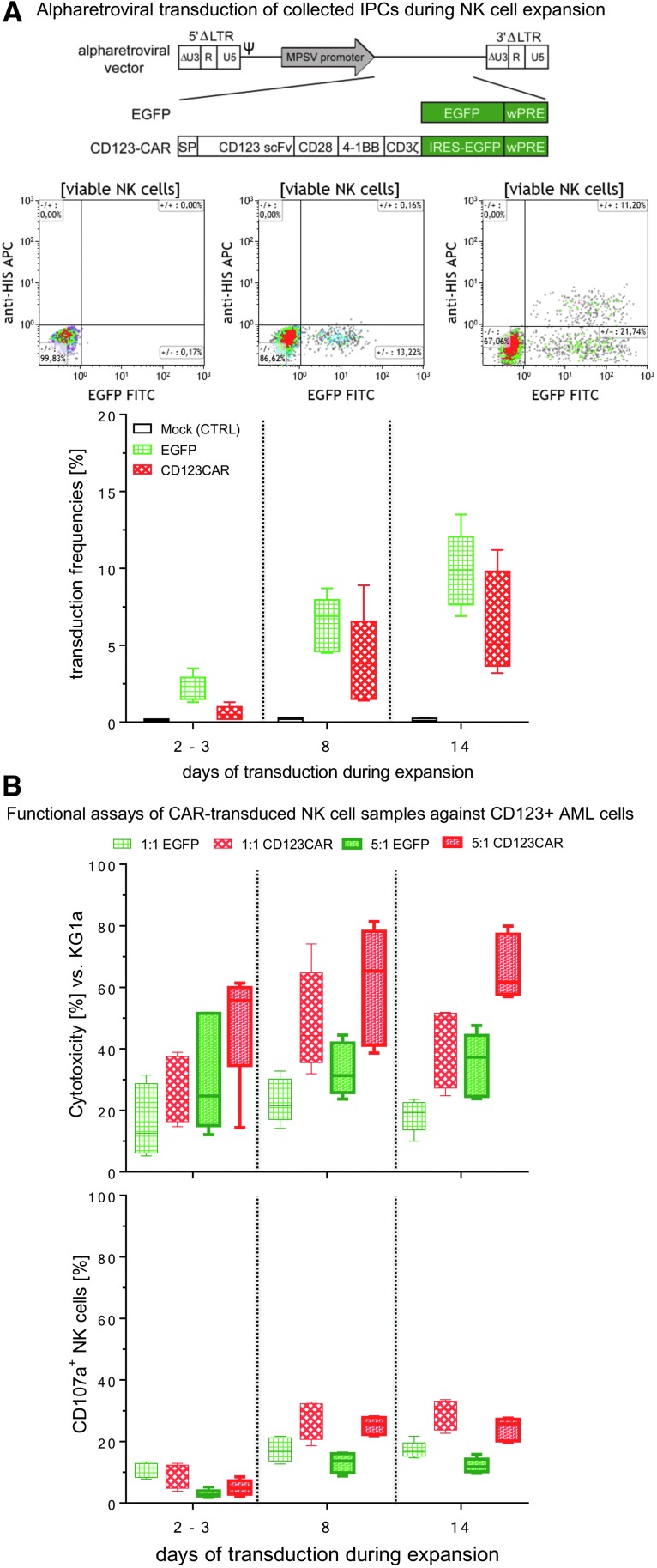
Figure 5.
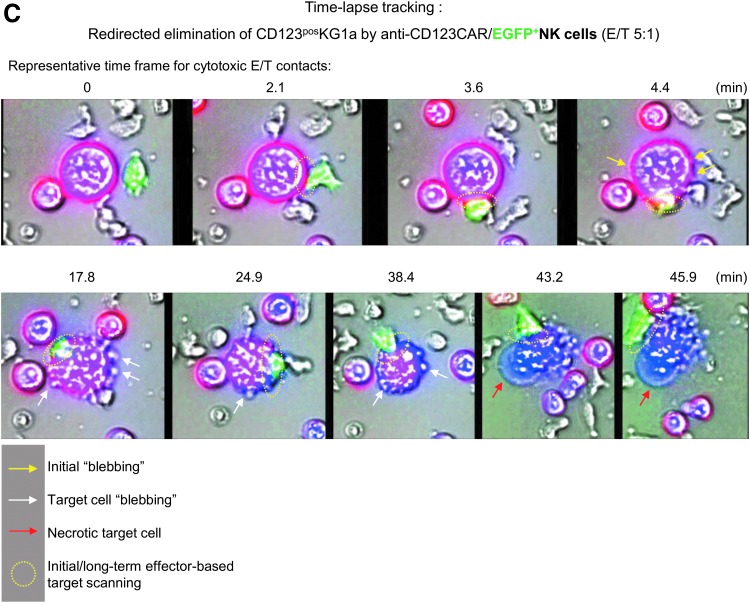
Transduction efficacy and functional characterization of anti-CD123 CAR NK cells. In order to find the appropriate time point for genetic engineering of cultured NK cells, several IPCs were collected during the expansion period (2–3, 8, and 14 days in Prodigy). NK cells were modified with RD114/TR-pseudotyped alpharetroviral vector encoding anti-CD123 CAR/enhanced green fluorescent protein (eGFP) or only eGFP at a multiplicity of infection (MOI) of 1 or 3 (A, upper illustration). For mock control (CTRL), the transduction procedure was performed with NK cells but without any vector constructs. Transduction frequencies of anti-CD123 CAR NK cells were quantified by incubation with recombinant CD123 peptides (to bind the single-chain variable fragment) conjugated with C-terminal HIS-Tag, followed by incubation with anti-HIS mABs conjugated with APC (A, flow cytometric graphs). Quantification of anti-CD123 CAR surface levels and intracellular eGFP signals in CD123 CAR/eGFP- and eGFP-modified NK cells were compared to non-modified mock controls (A, lower graph). (B) Retargeted killing properties and CD107a degranulation levels were analyzed after 24 h of co-cultivation against CD123-positive KG1a cells at indicated E/T ratios (1:1 and 5:1). Data are shown as median values with ranges given from five independent experiments performed in duplicate. (C) Tracking experiments were performed to monitor redirected killing activity of primary CAR NK cells. KG1a targets were intracellularly stained by eFluor® 450 and anti-CD34 mAB PE (blue/violet cells), while positive transduced effector cells are green due to eGFP expression. Anti-CD123/eGFP CAR NK cells were co-incubated with KG1a cells at an E/T ratio of 5:1, and redirected E/T cell interactions (representative time frame for cytotoxic E/T contacts) were tracked overnight by time-lapse fluorescent microscopy via scanR automated image and data analysis software (Olympus). Yellow (initial) and white arrows indicate blebbing of target cells due to apoptosis and followed by necrosis (red arrows) of KG1a cells caused by anti-CD123/eGFP CAR NK cells (green). The dotted yellow circles indicate specific, long-term E/T contacts between NK cells and KG1a cells.
HT1080 fibroblasts were used to estimate titers of alpharetroviral vector supernatants using standard transduction protocols. Briefly, 5 × 104 HT1080 cells were seeded in each well of 12-well plates (Sarstedt) the day before transduction. To transduce HT1080 cells, culture medium was replaced with 500 μL of fresh culture medium containing protamine sulfate (4 μg/mL; Sigma–Aldrich). Various volumes of supernatants containing alpharetroviral vector particles were then added to different wells, and the cells were centrifuged for 1 h at 400 g at ∼35°C). Four to six days later, cells were harvested and analyzed by flow cytometry for eGFP expression. To minimize underestimation of viral vector titers due to multiple infectious events, viral vector titers were calculated as transducing units (or infectious units) from samples exhibiting transduction efficiencies between 3% and 30%.23
A detailed description of the alpharetroviral vector system (pAlpha.SIN.noTATA) and production of alpharetroviral vector supernatants has been published previously.19,20,24 Briefly, 5 × 106 293T cells were seeded onto 10 cm culture dishes the day before transfection. The next day, 293T cells were transfected with a mixture of the appropriate alpharetroviral vector (5 μg), a codon-optimized alpharetroviral gag/pol helper plasmid (2.5 μg; GenBank accession no. HM130053) and RD114-TR envelope plasmid25 (2 μg) using the calcium phosphate method. After approximately 6 h, the medium was exchanged for 9 mL fresh DMEM containing 10 mM HEPES. To harvest viral particles, supernatants were collected 24 and 48 h after transfection, filtered through 0.22 μm pore-size filters (Millipore), pooled, concentrated by ultracentrifugation, and stored at −80°C until further use.
Transduction of NK cells with alpharetroviral vectors (at a multiplicity of infection [MOI] of 1 or 3) was accomplished with a RetroNectin-based method on 48-well plates, as essentially described by Suerth et al.,19,20 and was performed separately from the Prodigy device to determine the optimal time period for alpharetroviral NK cell transduction. Briefly, NK cells were transduced on a 48-well plate, and the wells were coated with RetroNectin (Takara) overnight at 4°C. RetroNectin was removed the next day, the wells were blocked with sterile-filtered phosphate-buffered saline (PBS) containing 2% bovine serum albumin (30 min, room temperature), washed with HBSS/HEPES immediately prior to adding alpharetroviral vector supernatants onto the RetroNectin pre-coated 48-well plates, which were then centrifuged for 30 min at 400 g at 4°C. After loading the alpharetroviral vector supernatants, the supernatant was removed, and the NK cells were added, incubated for 24 h, and then manually transferred to uncoated wells for an additional 6 days of expansion.
Definition of appropriate time periods for successful CAR transduction in expanded NK cells
NK cell samples were collected at different days of expansion (day 2 or 3, day 8, and day 14) for RetroNectin-based transduction on 48-well-plates.19,20 For transduction, alpharetroviral SIN vectors included an expression cassette containing only eGFP, or a combination of an anti-CD123 CAR and eGFP were used. After transduction at a MOI of 1 or 3, NK cells were expanded for 6 days on 48-well-plates containing NK MACS® basal medium supplemented with 5% human type AB serum, IL-2 (500 IU/mL), IL-15 (140 IU/mL), and 2% NK MACS® supplement. Transduction frequency, cytotoxicity, and NK cell degranulation (after 4 h co-culture with KG1a at effector-to-target [E/T] cell ratios of 1:1 and 5:1) were determined by flow cytometry.
Monitoring of glucose and pH
Determinations of glucose concentration and pH value of the cell-free supernatant were performed in parallel every 2–3 days using blood glucose meter Accu Check Aviva and pH indicator strips for a pH range between 6.5 and 10 (MColorpHast™; Merck).
Flow cytometric analysis of surface markers
Purified NK cells and other peripheral blood cells collected in LA, PF, IPC samples, and EP were analyzed for specific surface markers on a Navios flow cytometer (Beckman Coulter) in a no-wash single-platform procedure,26,27 adding Flow-Count fluorospheres™ (Beckman Coulter) to calculate cell concentrations. Cell staining was done using monoclonal antibodies (mABs) such as NK group 2 member D (NKG2D; CD314)-phycoerythrin (PE), NKp30 (CD337)-PE, NKp44 (CD336)-PE, NKp46 (CD335)-phycoerythrin-cyanin-7 (PC7), CD19-fluorescein isothiocyanate (FITC) or allophycocyanin-Alexa Fluor 750 (APC750), CD14-ECD (phycoerythrin-TexasRed®-X), CD69-ECD, CD16-allophycocyanin (APC) or -pacific blue (PB), CD56-PC7, CD3-PB or -allophycocyanin-Alexa Fluor 700 (APC700), and CD45-krome orange (KO) purchased from Beckman Coulter. Additional antibodies (CD137-APC, CD178-APC, and CD253-APC) were manufactured by BD Biosciences. The proportion of living and dead cells in the samples was quantified by 7-AAD (Beckman Coulter) staining. The quality control strategy followed ISHAGE guidelines27 and is illustrated in Fig. 2.
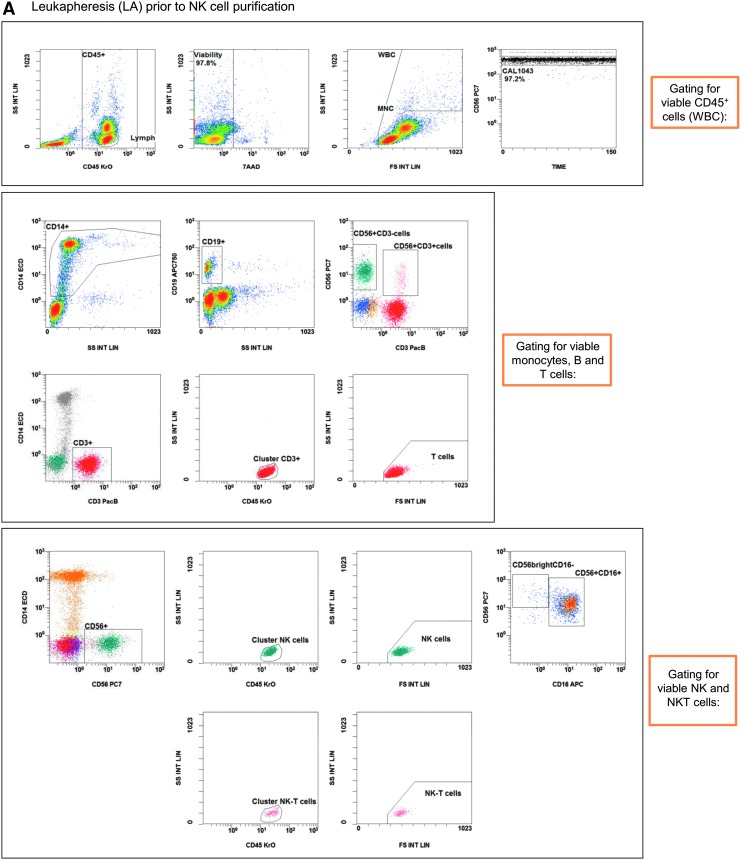
Figure 2.
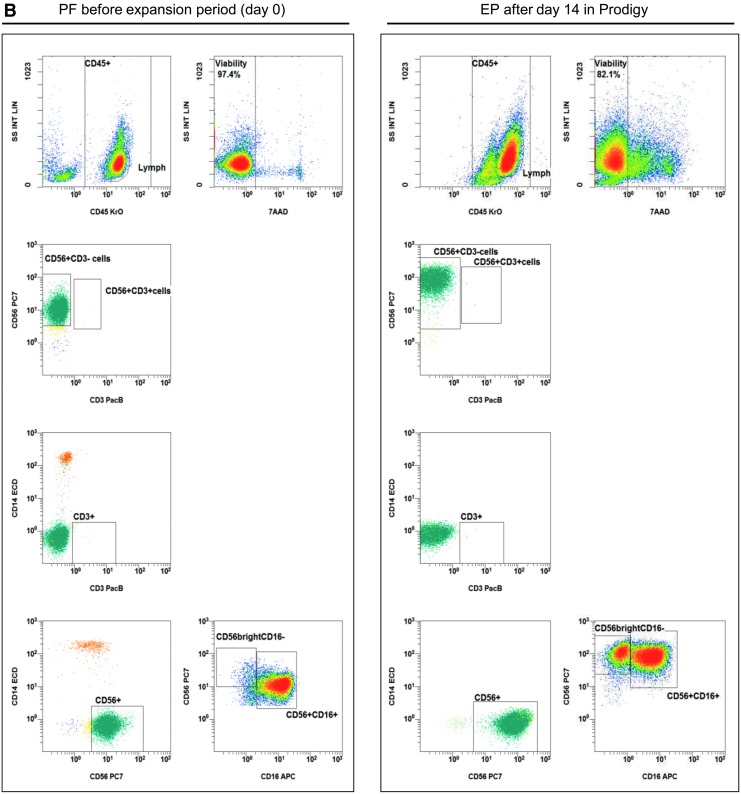
Gating strategy: flow cytometric quality controls for the quantification of CD3–CD56+ NK cells. IPCs from different process fractions (LA, PF, and EP) were stained by monoclonal antibodies (mABs) to visualize CD3–CD56+ NK cells and analyzed by a no-wash, single-platform method based on an eight-color flow cytometric panel. (A) Initial, CD45−, 7-AAD+, and non-specifically stained debris discriminated by low forward and side scatter (FS/SS) signals were excluded from viable CD45+ cells (WBC). The CAL histogram describes event number of the region “beads” along the time frame to calculate the events for cells/μL and to detect consistent sample flow (upper row, right plot). To identify monocytes and T and B cells, pre-analyzed viable CD45+ cells were used to detect CD14+ monocytes and lymphocyte subpopulations based on CD3 or CD19 surface levels. FS/SS dot plots were gated on the region “MNC” to discriminate nonspecific cell debris and outline the corresponding lymphocyte regions. Gating for viable NK and NKT cells were performed by discrimination (second line: CD3 vs. CD56) of CD3+CD56+ NKT cells to identify CD3−CD56+ NK cells (LA, PF, and EP). (B) One NK manufacturing and expansion process is exemplarily presented. Shown are analyses of PF and EP and the gating strategy for the initial immunomagnetic separation (day 0) and expansion of activated NK cells (day 14).
Flow cytometry–based cytotoxicity assay and CD107a degranulation assay
To assess the cytotoxicity of the NK cells, effector cells were co-cultured with target cell lines at various E/T ratios in NK MACS containing 5% HSA (Octapharma). NK cells were incubated with the K562 cell line for 4 h at E/T ratios of 0.5:1, 1:1, and 5:1 to measure basal NK cytotoxicity, while co-cultivation of CAR-modified NK cells and CD123-positive KG1a cells was accomplished for 24 h at E/T ratios of 1:1 and 5:1 to detect specific anti-CD123 CAR-dependent killing. For detection of viable cells, samples were stained with 7-AAD, and specific mABs (Beckman Coulter) against surface markers were used (NK cells: CD16-APC or -PB, CD56-PC7 or -APC, CD45-KO; K562: CD15-FITC; KG1a: CD34-PC7) in no-wash, single-platform flow cytometric measurements (Navios; Beckman Coulter), as illustrated in Supplementary Fig. S1.
To evaluate NK cell-mediated cytolytic activity based on decline of viable target cells, the following equation was used:
Cytotoxicity (%) = [1 – concentration (co-cultured target cells/μL)/concentration (control target cells/μL] × 100%.
Degranulation of NK cells was detected in the presence of mAB CD107a-PE (Beckman Coulter) after 1 h of co-incubation with target cells at E/T ratios described previously for cytotoxicity assessments. After this 1 h staining with CD107a, Monensin and Brefeldin were added (each diluted 1:1,000; eBioscience), and cells were incubated for an additional 3 h to determine basal NK degranulation using CD123-negative K562 cells or for an additional 4 h to determine CAR-specific NK cell degranulation using CD123-positive KG1a cells. Cells were then washed with PBS (Biochrom). Cell staining and flow cytometric analyses were accomplished as described above for cytotoxic assays.
CAR detection
Anti-CD123 CAR-modified NK cells were incubated with the recombinant and polyhistidine-labeled protein CD123 (1 μg/mL final concentration; Sino Biological, Inc.) for 30 min at 4°C. Unbound CD123 was removed by washing with PBS before transduced NK cells were stained with 7-AAD, CD45-KO, and anti-HIS-APC (R&D Systems) and analyzed by flow cytometric analysis.
Cytokine analysis
Measurements of soluble cytokines, pro-apoptotic markers (TNF-α and IFN-γ), and cytotoxic granule proteins (perforin, granzyme A, granzyme B, and granulysin) were performed in supernatants of cytotoxicity assays using a bead-based immunoassay (LEGENDplex™, human CD8 panel, 13-plex, BioLegend®) according to the manufacturer's instructions.
Cryopreservation and thawing
IPC samples were collected between days 0 and 14 of the NK expansion period (Fig. 1A) in a CentriCult® unit (Prodigy). To freeze the unmodified NK cells, cells were re-suspended in Composol PS® (Fresenius Kabi) containing 2.86% HSA (20% Octalbin; Octapharma). Cells were then mixed with a Composol–dimethyl sulfoxide (DMSO; Sigma–Aldrich) solution (20% DMSO) for cell protection. Freezing was performed in an automated cryo freezer Biofreeze BV50 (Consarctic) before cells were stored in the vapor phase of liquid nitrogen. After 3 months, cells were quickly thawed at 37°C and re-suspended in fresh pre-warmed NK MACS expansion medium (5% AB serum, 2% supplement, 500 IU/mL IL-2, 140 IU/mL IL-15, initial 1 IU/mL IL-21). Cell concentrations were initially set to 2 × 106 NK cells/mL. Samples were measured by flow cytometry after 24, 48, and 72 h to determine recovery, expansion, and surface markers (NKG2D, NKp30, CD178, and CD253).
Time-lapse microscopy
Cell–cell interactions between transduced primary human NK cells and target cells (KG1a) were monitored by fluorescence tracking for 8 h under standard incubation conditions (37°C, 5% CO2, 95% relative humidity) using the IX81 microscope (Olympus). All transduced NK cells expressed eGFP, while KG1a cells were labeled with the intracellular dye eFluor™ 450 (Affymetrix eBioscience™), and extracellular staining was performed with mAB CD34-PE (Beckman Coulter). For co-incubation, effector and target cells were mixed at an E/T ratio of 5:1 on chamber slides. In time-lapse recordings, images of selected slide sections were generated every 30 s with Olympus scanR data software.
Statistical analyses
For statistical analysis and generation of graphs, GraphPad Prism v6.05 (GraphPad Software, Inc.) was used to analyze data sets from three Prodigy runs individually for NK cell fractions (LA, PF, EP, and IPC samples), NK cell expansion rates, recovery, viability, effector cell–based properties, and transduction frequencies. Data are indicated as the median with range (minimum to maximum) within the individual text sections.
Results
Automated separation and expansion of primary NK cells
Three different NK cell manufacturing runs on the Prodigy device were performed by starting the immunomagnetic separations via CD3 depletion followed by CD56 enrichment (Fig. 1A). After removal of CD3+ cells, CD56+ NK cell separation was immediately performed by positive selection using CliniMACS CD56 CE reagent and the same tubing set (TS310). CD3+ cells were depleted in a median frequency of log −3.58 (range log −3.6 to log −3.24) after the depletion step. Following CD56 selection, the overall separation process was increased to a median log −3.66 (range log −3.68 to log −3.6; Table 1). Medians of purity and viability of selected NK cells prior to expansion were >90% (median purity 93.2%, range 78.3–94.9%; median viability 96.5%, range 96.2–97.4%). The median of total NK cell recovery was 65.9% (range 63.2–67.5%), with an absolute number of 818.4 × 103 contaminating CD3+ cells (median, range 574.0–1035.0 × 103) and 543.4 × 103 CD3+ CD56+ cells (median, range 201.6–575.0 × 103; Table 1).
Table 1.
Results of the automated separation and expansion process of CD3-CD56+ NK cells using lymphapheresis on Prodigy
| Prodigy manufacturing runs | Run 1 | Run 2 | Run 3 | Median | Min. | Max. |
|---|---|---|---|---|---|---|
| WBC in starting material ( × 109 abs., LA) | 14 | 8 | 15 | 14 | 8 | 15 |
| NK cells in starting material ( × 109 abs., LA) | 2.2 | 1.1 | 2.3 | 2.2 | 1.1 | 2.3 |
| Decrease of CD3+ cells (CD3 depletion; log) | –3.60 | –3.24 | –3.58 | –3.58 | –3.60 | –3.24 |
| Depletion of CD3+ cells after overall process (log) | –3.68 | –3.66 | –3.60 | –3.66 | –3.68 | –3.60 |
| Purity (%) of NK cells (CCB2) | 93.2 | 78.3 | 94.9 | 93.2 | 78.3 | 94.9 |
| Viability (%) of NK cells after CD56 separation | 97.4 | 96.2 | 96.5 | 96.5 | 96.2 | 97.4 |
| Recovery (%) of NK cells after CD3+ depletion (reapplication bag) | 81.8 | 82.6 | 93.0 | 82.6 | 81.8 | 93.0 |
| Recovery (%) of NK cells after CD56+ separation | 82.5 | 79.8 | 68.0 | 79.8 | 68.0 | 82.5 |
| Total recovery (%) of NK cells (CCB2) | 67.5 | 65.9 | 63.2 | 65.9 | 63.2 | 67.5 |
| Contamination of CD3+ T cells (*103 abs., CCB2) | 818.4 | 574.0 | 1035.0 | 818.4 | 574.0 | 1035.0 |
| Contamination of CD3+CD56+ cells ( × 103 abs., CCB2) | 543.4 | 201.6 | 575.0 | 543.4 | 201.6 | 575.0 |
| IL-21/IL-2/IL-15 expansion | ||||||
| NK cell expansion rates (day 14; x-fold) | 5.85 | 4.18 | 8.45 | 5.85 | 4.18 | 8.45 |
| NK cell expansion rates (after day 3; x-fold) | 11.31 | 12.46 | 17.94 | 12.46 | 11.31 | 17.94 |
| Purity (%) of NK cells (day 14) | 99.2 | 98.8 | 99.1 | 99.1 | 98.8 | 99.2 |
| Viability (%) of NK cells (day 14) | 86.9 | 86.0 | 89.2 | 86.9 | 86.0 | 89.2 |
| Ratio CD56+CD16–/CD56+CD16+ NK cells (day 14) | 2.3 | 0.9 | 0.2 | 0.9 | 0.2 | 2.3 |
| Contamination of CD3+ T cells ( × 103 abs., day 14) | 135.0 | 140.0 | 635.3 | 140.0 | 135.0 | 635.3 |
| Contamination of CD3+CD56+ cells ( × 103 abs., day 14) | 156.3 | 97.5 | 231.0 | 156.3 | 97.5 | 231.0 |
CD3–CD56+ NK cells were finally collected from three independent manufacturing runs in cell collection bags 2 (CCB2). For process evaluation, purity (%), recovery (%), NK subset ratios and cell viability (%) were determined, as well as CD3+ cell depletion and CD56+ cell expansion. Cell viability was assessed by 7-AAD staining and subsequent flow cytometric analyses.
NK, natural killer; LA, lymphapheresis; IL, interleukin.
Subsequent NK cell cultivation steps within the same manufacturing process (Prodigy) were performed by effector cell activation with repeated addition of IL-2 and IL-15 (every 2–3 days), and initial IL-21 (day 0) to reach median expansion rates of 5.85-fold (range 4.18–8.45) after 14 days. However, the often-described loss of freshly separated NK cells between expansion day 0 and 328–30 was detected for all three manufacturing runs and resulted in a lower increase in expansion rates between days 0 and 14. Consequently, it would be more useful to determine NK expansion rates after the initial loss of NK cells, for example between expansion days 3 and 14, as indicated in Table 1 where a median expansion rate of 12.46-fold (range 11.31–17.94) was observed. The NK cell purity (median 99.1%, range 98.8–99.2%) increased with expansion, and viability (median 86.9%, range 86.0–89.2%) slightly decreased. After 14 days of expansion, the T-cell contamination decreased to a median of 140.0 × 103 CD3+ cells (range 135.0–635.3 × 103) and 156.3 × 103 CD3+/CD56+ cells in total (range 97.5–231.0 × 103; Table 1). The ratio of NK cell subpopulations (CD56bright/CD16– cells vs. CD56dim/CD16+ cells) was also examined and displayed a wide range between 0.2 and 2.3 (median 0.9; Table 1).
During the first 6 days of initial NK cell expansion, the cells were grown without any shaking of the cultivation chamber to promote effector cell contact and cell clustering. After 6 days, activated NK cells showed strongly increased cell numbers and much larger cluster formations (Fig. 1B). Due to higher glucose consumption and increased pH, the nutrient/medium supply for effector cells were increased (every 2 days), and the suspension was shaken slightly (shake type 1: every 30 s unidirectional shaking for 2 s) to ensure better mixing of the medium and cells. During the expansion phase, glucose concentrations varied (range 150–390 mg/100 mL), while pH values ranged between 7.3 and 7.9 (Fig. 1B).
IPCs were evaluated to quantify cellular compositions of LA prior purification, purified NK cell product (PF, day 0), and subsequent expanded NK cells after 14 days (EP, Fig. 2A and B). Prior to NK cell separation, LA contained populations of CD14+ monocytes, CD19+ B cells, and CD3+ T cells (including CD3+/CD56+ NKT cells), as well as CD3–/CD56+ NK cells. After depletion of CD3+ cells and separation of CD56+ cells, the population of CD3–/CD56+ NK cells was highly purified, demonstrating nearly no residual CD3+/CD56+ NKT cells and a minimal population of CD14+ monocytes (Fig. 2B, day 0). After 14 days, the residual CD14+ cells and CD3+ T cells (including CD3+/CD56+ NKT cells) were largely removed due to specific cell culture conditions (Fig. 2B, day 14), as the NK MACS® basal medium was specially optimized for NK cell growth. The depletion of monocytes and T cells prevents the development of GvHD, since both immune cell populations are able to produce inflammatory cytokines (e.g., TNF-α).
Comparison of surface markers, cytotoxicity, and degranulation before and after expansion of NK cells
The surface expression of NCRs of freshly isolated NK cells was analyzed and compared to the NCR levels detected after 14 days of expansion (Fig. 3A). Total NK cells expressed initially low to moderate levels of NKG2D (median 23%, range 3.3–53.4%), NKp30 (median 0.5%, range 0.1–1.82%), NKp44 (median 1.0%, range 0.3–4.5%), and NKp46 (median 43.3%, range 7.1–44.9%). The surface markers increased during the 14-day expansion in the NK MACS medium supplemented with cytokines IL-21, IL-2, and IL15 (NKG2D: median 97.3%, range 95.1–99.2%; NKp30: median 72.8%, range 36.0–93.1%; NKp44: median 95.5%, range 69.4–95.8%; NKp46: median 71.4%, range 50.8–89.6%). Death receptors (CD178 and CD253) and activation markers (CD69 and CD137) were also expressed at higher levels after cell expansion compared to freshly separated NK cells (Fig. 3B). In particular, the expression of the early activation marker CD69 increased from 2.21% (range 0.2–17.1%) to 95.1% (range 80.8–90.4%). The expression of the apoptotic marker CD178 (FasL) displayed a similar rate of increase starting at 24.8% (range 0.4–39.7%) and reached 89.7% (range 62.2–99.4%) on day 14. The expression of CD137 (day 0: median 0.2%, range 0.2–1.1%; day: 14 median 3.5%, range 1.7–4.23%) and CD253 (day 0: median 33.4%, range 12.2–47.9%; day 14: median 82.9%, range 81.6–100%) increased as well.
Figure 3.
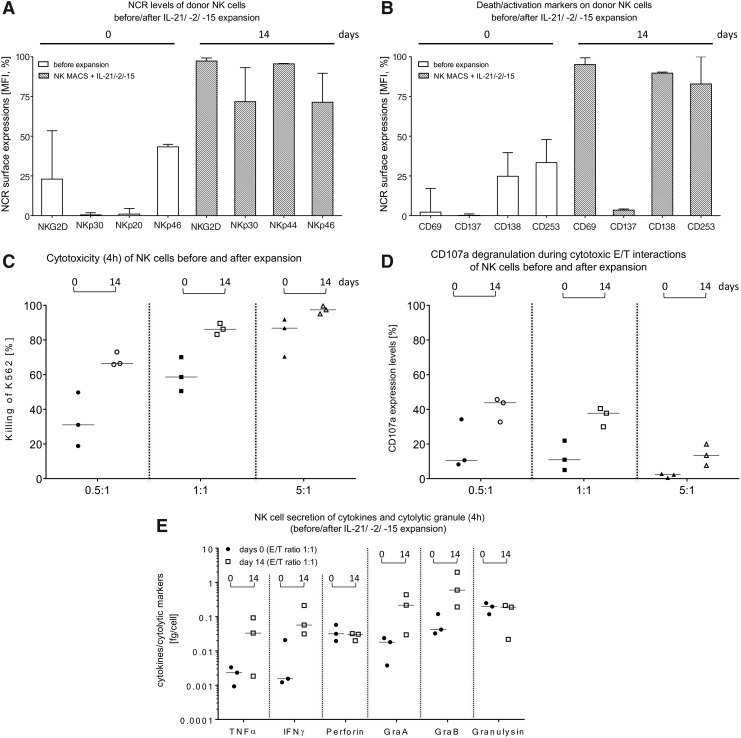
Comparison of cytotoxic properties and activation markers on primary NK cells before and after the expansion process in Prodigy. IPCs (approximately 2.0–2.5 mL) were collected as indicated in Fig. 1A for analysis of NK cell-dependent cytotoxicity, CD107a degranulation, surface marker expression levels, and cytokine secretion. NK cell phenotyping of (A) NCRs (NKG2D, NKp30, NKp44, and NKp46), (B) activation markers (CD69 and CD137), and death receptors (CD178 and CD253) were analyzed by flow cytometry for 2-week-cultured NK cells compared to highly purified NK cells before the expansion period. Correspondingly, effector cell-mediated cytotoxicity (C) and CD107a degranulation rates (D) were analyzed for expanded NK cells against K562 cells at indicated effector-to-target (E/T) ratios and compared to freshly separated NK cells before starting the expansion period (day 0). (E) Secretion of tumor necrosis factor alpha, interferon gamma, perforin, granzyme A and B, and granulysin were determined after cytolytic reactions in supernatants of co- and mono-cultured NK and K562 cells. Medians with ranges are shown from three independent experiments performed in duplicate.
Killing activities of non-expanded, freshly separated NK cells were compared to activated NK cells after 14 days using K562 target cells at the indicated E/T ratios (Fig. 3C). In general, the NK cell cytotoxicity increased after expansion and highest killing rates (%) derived at an E/T ratio of 5:1 (Fig. 3C). The strongest increase in NK cell cytotoxicity was observed at an E/T ratio of 0.5:1, with an initial median cytotoxicity of 31.0% (range 18.8–49.8%, day 0), which increased to a median cytotoxicity of 66.5% (range 65.9–73.1%) on day 14. Degranulation assays detected CD107a as a marker for stimulation-induced granule exocytosis. Analyses demonstrated enhanced CD107a expression for expanded (day 14) NK cells compared to non-activated NK cells (day 0; Fig. 3D). It is notable that an E/T ratio of 0.5:1 showed the highest degranulation effect (freshly isolated NK cells: median 10.6%, range 8.2–34.2% vs. expanded NK cells: median 43.8%, range 32.7–45.7%) compared to an E/T ratio of 1:1 (non-activated NK cells: median 10.9%, range 5.0–21.9% vs. expanded NK cells: median 37.7%, range 29.9–40.4%) and 5:1 (freshly isolated NK cells: median 2.3%, range 0.5–2.8% vs. expanded NK cells: median 13.4%, range 7.6–19.9%). Supernatants of cytotoxic experiments with NK cells before and after 14 days of expansion were collected, and pro-inflammatory cytokines (TNF-α and IFN-γ) and cytotoxic granule proteins (perforin, granzyme A, granzyme B, and granulysin) were analyzed (Fig. 3E). Concentrations of TNF-α, IFN-γ, granzyme A, and granzyme B increased during expansion (TNF-α: day 0 median 0.002 fg/cell, range 0.0009–0.003 vs. day 14 median 0.033 fg/cell, range 0.0018–0.092; IFN-γ: day 0 median 0.002 fg/cell, range 0.001–0.02 vs. day 14 median 0.057 fg/cell, range 0.03–0.2; granzyme A: day 0 median 0.018 fg/cell, range 0.004–0.02 vs. day 14 median 0.216 fg/cell, range 0.03–0.43; granzyme B: day 0 median 0.042 fg/cell, range 0.032–0.12 vs. day 14 median 0.598 fg/cell, range 0.19–1.98), while no changes were detected for perforin (freshly isolated NK cells: day 0 median 0.031 fg/cell, range 0.019–0.058 fg/cell vs. expanded NK cells: day 14 median 0.031 fg/cell, range 0.02–0.031 fg/cell) or granulysin (freshly isolated NK cells: day 0 median 0.178 fg/cell, range 0.118–0.248 fg/cell vs. expanded NK cells: day 14 median 0.190 fg/cell, range 0.022–0.212 fg/cell).
Expansion of NK cells after cryopreservation
To assess the stability of expanded NK cells after cryopreservation, IPC samples were frozen directly after separation (day 0) or on day 3, 6, 8, 10, or 14 after isolation during expansion. After 3 months, cryopreserved NK cells were thawed and expanded for 24, 48, and 72 h to determine killing activity and surface marker expression (NKG2D, NKp30, CD178, and CD253; Fig. 4). Due to cell loss on day 3 prior to the cell expansion phase, all NK cells harvested on day 3 were used solely for functional assays (Fig. 4G). NK cells stored in liquid nitrogen on day 10 after separation showed the best ability to expand, especially 72 h after re-cultivation (72 h median 1.1-fold, range 1.1–5.7-fold), while moderate cell revival ability was found on cells harvested on day 6 (72 h median 0.9-fold, range 0.88–1.7-fold), day 8 (72 h median 1.7-fold, range 1.7–3.9-fold), and day 14 (72 h median 2.2-fold, range 1.6–3.36-fold). The amount of NK cells that were frozen immediately after isolation (day 0) decreased during the next 72 h (median 0.4-fold, range 0.4–0.5-fold) due to the well-known loss of NK cells between days 0 and day 6 of expansion (Fig. 4A).28-30 After 24 h of re-cultivation, NK cells demonstrated the lowest viability (medians <80%) for all collected IPC samples and increased during the expansion period after 72 h (medians >80%; Fig. 4B). Activating NKp30 was highly expressed (median >80%; range 95.5–100%) in all NK cells harvested between day 0 and day 14 and re-cultivated after thawing between 24 and 72 h (Fig. 4D). Activating cell surface receptor NKG2D showed similar expression, but NK cells harvested directly after isolation (day 0) demonstrated a lower expression level during expansion for 24 and 48 h (Fig. 4C), and only after 72 h of expansion did approximately 80% (median, range 77.0–99.9%) of total NK cells stain positive for NKG2D. The death receptor ligands FasL (CD178) and TRAIL (CD253) were highly expressed on NK cells harvested between days 6 and 14 (CD178: median >60%, range 46.3–99.5%; CD253: median >70%, range 57.8–99.8%) and cultivated after thawing for 24–72 h (Fig. 4E and F), while CD178 and CD253 were expressed to a lower degree on NK cells harvested immediately after separation (day 0) in all re-cultivation experiments. There was a slight loss of CD178-positive and CD253-positive NK cells after 48 h of expansion, which was overcome after 72 h of expansion.
Figure 4.
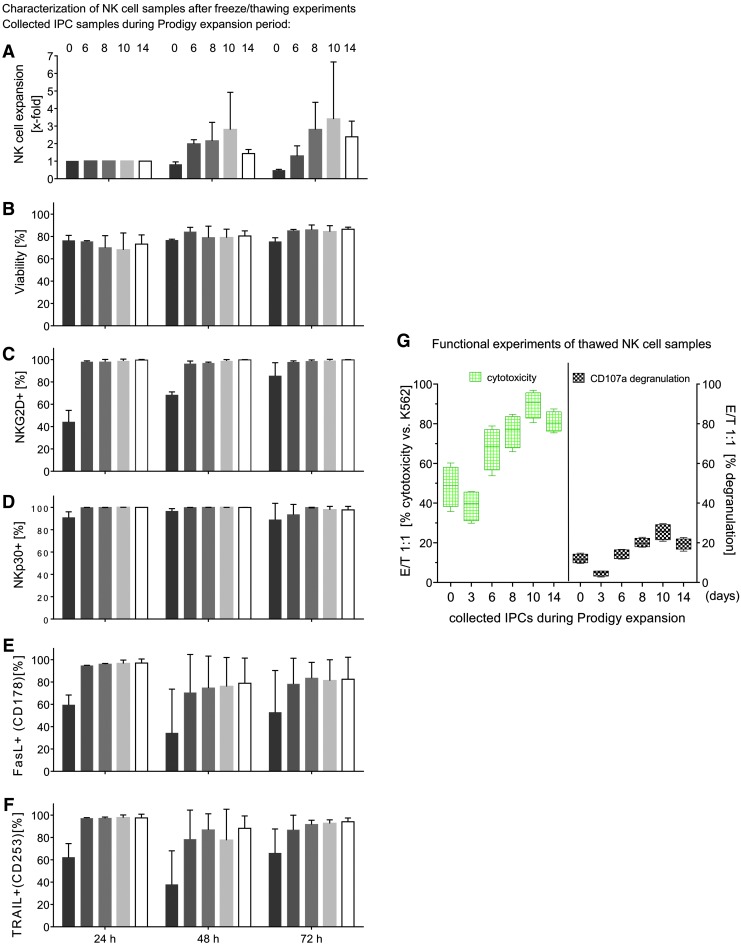
Freeze and thawing experiments from collected IPCs before, during, and after NK expansion in Prodigy. IPCs of isolated and expanded NK cells were harvested/collected directly after immunomagnetic separation (Prodigy) and during NK cell cultivation at different time points (3, 6, 8, 10, and 14 days) and cryopreserved in liquid nitrogen for 3 months before thawing and re-cultivation was started for 24, 48, and 72 h. NK cell expansion properties (A) and viability (B) during re-cultivation, and percentages of NK cells with expression of NKG2D, NKp30, FasL, and TRAIL (C–F) were determined by flow cytometry. Medians with ranges are shown from two to three independent experiments performed in duplicate. (G) Functional experiments (killing activity/CD107a degranulation) with thawed NK cell samples derived from frozen IPCs were performed against K562 at an E/T ratio of 1:1. Median with ranges is presented from four independent experiments accomplished in duplicates.
Functional assays after cryopreservation
NK cells harvested on various days during the expansion period were analyzed for killing activity and their ability to release cytotoxic granules after 3 months of freezing. Therefore, NK cells from different IPC samples (0–14 days) were re-cultured for 72 h and subsequently co-cultured with K562 target cells (E/T ratio 1:1) to examine their cytotoxicity and degranulation potential. NK cells harvested on day 10 displayed the highest lytic activity (median 90%, range 80.5–96.9%), while the same NK cells collected on day 3 exhibited the lowest cytotoxicity (median 39%, range 29.9–45.8 %). Basal killing activity peaked for NK cells harvested after day 10 (median 90%, range 80.5–96.9%) and declined after day 10 (day 14 median 81%, range 75.4–87.4%; Fig. 4G, left side).
NK cell samples that were assessed in cytotoxic assays were also analyzed for CD107a expression to estimate degranulation. In accordance with the cytotoxic data, the lowest degranulation was detected for cells harvested on day 3 (median 4.7%, range 2.8–5.8%), while peak expression of CD107a was found for NK cells harvested on day 10 (median 25.5%, range 20.8–29.7%). For NK cells harvested on day 14, CD107a levels decreased (median 19.8%, range 15.8–22.7%; Fig. 4G, right side).
Retargeted anti-CD123 CAR NK cells and their ability for cytotoxic killing and degranulation
NK cells were transduced at several time points during the expansion period in order to determine the best time point for transduction efficiency. Alpharetroviral vectors that encoded either an anti-CD123 CAR combined with eGFP (Fig. 5A) or solely eGFP as a control were used to transduce NK cells. Transduction frequency of CAR-engineered NK cells was quantified by flow cytometry to measure the number of eGFP-positive cells. Surface expression of CAR molecules was detected with a polyhistidine-labeled CD123 antigen and anti-HIS mABs conjugated with APC. As shown in Fig. 5A, eGFP signals were higher (21.7%) than signals for CD123 antigen binding (11.2%), which implies a different expression of eGFP and functional CARs, although both genes were part of the same vector (Fig. 5A). The highest transduction efficiencies of alpharetroviral CAR constructs were achieved with NK cells harvested on day 14 (median 5.2%, range 3.2–11.2%), while the lowest transduction rate was determined for NK cells that were modified 2–3 days after NK cell separation (median 0.5%, range 0.2–1.3%). Control experiments with the eGFP vector confirmed these results (day 2–3: median 2.3%, range 1.3–3.5%; day 14: median 9.9%, range 6.9–13.5%), implying that NK cells in expansion are more receptive for alpharetroviral vectors at later time points (Fig. 5A).
Retargeted killing activity of CAR-engineered NK cells was examined with CD123-positive KG1a cells in potency assays for cytotoxicity and CD107a degranulation using different E/T ratios (1:1 and 5:1). After 24 h of co-culturing, a decrease of the KG1a target cell population was determined by single-platform flow cytometric assays. Anti-CD123 CAR/eGFP-modified NK cells were compared to eGFP-positive NK cells (without CAR expression), which served as controls for redirected elimination of KG1a. Anti-CD123/eGFP transduced NK cells showed a higher killing activity against KG1a cells compared to eGFP-modified NK cells at both examined E/T ratios (Fig. 5B). The highest cytotoxic rates were determined from anti-CD123 CAR/eGFP NK cells transduced on days 8 and 14 of expansion, suggesting that this interval is a suitable time point for gene modification of expanded NK cells. Comparison of killed target cells for an E/T ratio of 5:1 (day 8: anti-CD123 CAR/eGFP: median 65.4%, range 38.6–81.4% vs. eGFP: median 31.3%, range 23.7–39.3%) and for an E/T ratio of 1:1 (day 8: anti-CD123/eGFP: median 44.3%, range 31.9–74.1%; eGFP: median 21.5%, range 14.1–32.8%) showed a higher killing rate for an E/T ratio of 5:1.
In general, anti-CD123 CAR/eGFP NK cells exhibited higher CD107a surface levels compared to eGFP-expressing NK cells (Fig. 5B). As also observed for NK cell cytotoxicity, degranulation was enhanced for NK cells modified on days 8 and 14 of expansion, while NK cells transduced on days 2–3 of expansion showed lower CD107a surface levels for both E/T ratios. NK cells modified on day 8 or 14 and subsequently co-cultured with KG1a at an E/T ratio of 1:1 exhibited the highest CD107a degranulation (day 8: anti-CD123/eGFP: median 29.7%, range 18.7–32.8%; eGFP: median: 16.8%, range 12.7–21.7%). In contrast, an E/T ratio of 5:1 displayed lower extracellular CD107a release (day 8: anti-CD123/eGFP: median 25.7%, range 21.9–28.0%; eGFP: median: 13.7%, range 8.8–16.2%). In all examined NK cell samples, an E/T ratio of 1:1 showed higher 107a degranulation rates compared to an E/T ratio of 5:1.
In order to track specific retargeting of E/T cell contacts, anti-CD123/eGFP-transduced NK cells were co-incubated for 8 h with labeled CD123-positive KG1a cells (E/T ratio of 5:1). The scanR automated image and data analysis software (Olympus) was used for time-lapse imaging of redirected cell contacts of CAR NK and KG1a cells that resulted in specific, long-term E/T interactions. Shortly after making contact with target cells, anti-CD123 CAR NK cells initiated membrane protrusions (“blebbing” of target cells) on the KG1a that resulted in apoptotic and/or necrotic cell reactions, while effector cells stayed vital, as indicated in the time-lapse photographs shown in Fig. 5C and Supplementary Video S1. Imaging analysis showed that anti-CD123 CAR-expressing NK cells acted specifically against CD123-positive target cells.
Discussion
In the last decade, several clinical studies (NCT 00995137, NCT 01974479, NCT 03056339, NCT 03415100, NCT 03579927, NCT 03692767, NCT 03690310, NCT03692663, NCT 03692637, and NCT 03824964) have demonstrated the growing interest in CAR-modified primary NK cells as a gene therapy medicinal product against different tumor entities. However, GMP-compliant manufacturing of cells on a clinical scale still has to overcome various hurdles before the CAR NK cell strategy can be made broadly available. This work examined the potential of the closed Prodigy system to separate highly purified primary NK cells in combination with clinical-scale expansion. Although only three Prodigy runs are reported here, all three runs demonstrated consistent results regarding cellular composition, viability, purity, expansion rates, and functionality of the produced NK cells. This implies that a robust standardized manufacturing process can be implemented. However, it will also be important to perform the transduction procedure on Prodigy to validate the optimal transduction time point that was determined in experiments outside of the Prodigy system. Since the NK cell content of the peripheral blood shows individual variations and comprises up to 10–20% of all lymphocytes,31,32 optimal donor NK cell concentrations should be high. For clinical treatment in a Phase II study, Stern et al. demonstrated that an NK cell dose of 1 × 107 NK cells/kg could be isolated from one unstimulated LA in the majority of all cases,8 but NK cells have to be expanded for multiple dose treatments. The present results are consistent with this observation. NK cells from a standard LA will be sufficient for one to two doses for an adult patient (expected dose of 1 × 107 NK cells/kg; expected body weight ranges between 70 and 100 kg), while freshly expanded NK cells would probably be available for >10 doses. Another important point was to identify the time period for optimal NK cell transduction during the NK cell expansion, since it is well known that NK cells undergo strong cell number reduction during the early stages of culture after immunomagnetic separation. Moreover, effector stability and potency experiments demonstrated that cryopreserved NK cells can also be employed to eradicate target cells.
The depletion of CD3+ cells and the separation of CD56+ cells from LA were done in consecutive steps within the same tubing set. The purity of NK cells is critical because of the T cells characteristic to trigger GvHD. It has been demonstrated that patients developed an acute GvHD ≥grade II when receiving ≥0.5 × 105 T cells/kg body weight, while NK cell doses are very well tolerated when T-cell contamination is 0.03 × 105 T cells/kg body weight.8 The depletion median rate of log −3.6 and a median purity of 93.2% in the final product were very similar to former published data (CD3+ depletion: log −3.5; CD56+ purity: 95.4%)11 using the Prodigy instrument. The median total recovery (65.9%) and the median viability (96.5%) of the CD56+ cells were also comparable to previously collected data (total recovery: 60.4%; viability: 96.3%), suggesting the robustness of these protocols.11
Moreover, the translation of the optimized in vitro expansion protocol11 to the CentriCult® chamber (Prodigy) to generate a single-process procedure for clinical-scale NK cell manufacturing were realized by using GMP-compliant NK MACS® basal medium supplemented with GMP-compliant cytokines (initial dose of IL-21, and repeated doses of IL-2/IL-15 for 14 days). The essential impact of IL-21 on NK cells has been described in detail previously,33,34 as well as the combination of IL-21 and IL-2 as important factors for NK cell viability, expansion, and activity.11,35–37 Granzin et al. demonstrated an expansion rate of 22-fold for NK cells co-cultured with feeder cells, and that expansion could be increased to 53-fold with the addition of IL-21 to the feeder cell co-cultures.38 Kloess et al. reported similar results with feeder cell co-culture, leading to an increased expansion rate from 50.9- to 84.8-fold compared to IL-21 alone.11 Because contamination of insufficiently irradiated feeder cells could cause severe side effects when co-infused with NK cells for clinical purposes, feeder cells were omitted in the experiments described here and instead a constant dose of IL-2 and IL-15 was used. The supply of repeated IL-2 and IL-15 application and initial IL-21 in the present experiments demonstrated a median NK cell expansion of 5.85-fold, which is clearly lower when compared to growth rates based on feeder cells plus IL-2 that ranged from 22- to 51-fold.11,38 This expansion rate rises to 12.46-fold when calculation based on the lowest NK cell concentration, which was on day 3. It was reported that the combination of IL-2 and IL-15 or IL-2 and IL-21 enhances the cytolytic properties of NK cells,39 which corroborates the results of the functional assays in the present study.
This study demonstrated increasing levels of NCRs (NKG2D, NKp30, NKp44, and NKp46) and death/activation markers (CD69, CD137, CD178, CD253), as well as a strong increase in cytokines (TNF-α and IFN-γ), cytolytic proteins (granzymes A and B), and enhanced CD107a expression levels on NK cells that were expanded 14 days. The upregulation of the surface markers indicated that NK cells were highly activated after expansion, which was also reflected by functional assays (cytotoxicity, CD107a degranulation) and confirmed by other publications.11,13 The current degranulation experiments showed higher CD107a expression levels for low E/T ratios such as 0.5:1 or 1:1 compared to lower surface CD107a on NK cells at an E/T ratio of 5:1, indicating different predominant killing mechanisms. Factors such as secretory lysosome contents, adhesion promoting structures on target cells, or sensitivity of triggering death pathways may mediate this enhanced NK cell cytotoxicity.40 The observation of high CD107a degranulation for low E/T ratios was also reported by others.41,42
Monitoring glucose consumption and pH reflect NK cell density and metabolism rates during expansion in the CentriCult® chamber. High NK cell metabolism was associated with increased glucose reduction in the growth medium. To ensure an optimal nutrition supply, NK cells were fed every 2 days during the linear growth phase, which stabilized the glucose concentration in the range of 150–228 mg/mL between day 6 and day 14 of NK cell expansion. During the first 6 days, NK cells are fragile, as demonstrated by the decline of the cell concentration, and accompanied by a low glucose consumption and little exchange of growth medium in the first 3–4 days. For this reason, gentle cell shaking was not applied until day 7. Gentle cell mixing started on day 7 allowed high cell densities.13 Medium exchange every 2 days limited the pH fluctuation to within a physiologically tolerable range between pH 7.3 and 7.9.
In order to assure NK cell quality after cell cryopreservation, IPC samples were harvested at various time points during the cultivation process for control experiments followed by storing in vapor phase of liquid nitrogen for 3 months. After thawing, NK cells were expanded for an additional 3 days to restore NK cell functions. The highest revitalization was found for NK cells collected on day 10 of expansion, while less growth only occurred for NK cells collected immediately after the separation process. This was also reflected by the lowest expression of NKG2D, NKp30, CD178, and CD253, indicating that NK cells collected directly after separation demonstrated low killing activity and degranulation. Similar results were shown by Childs and Berg43 who then recommended re-culturing thawed NK cells for several days before administration to patients. Indeed, this study observed increased killing activity of NK cells and degranulation by NK cells collected up to day 10, while day 14-collected EP demonstrated slightly lowered cytotoxicity and degranulation (compared to day 10 NK cells) after re-cultivation. This may be partially explained by the finding that chronic stimulation with IL-2 and IL-15 induced NK cell exhaustion.44,45 On the other hand, cytokines are necessary for in vitro cultivation of NK cells that would demonstrate decreased cell numbers within 72 h without cytokines.
Customized immune therapies with a particular focus on CAR-modified immune cells have emerged in recent years because of the advantage of attacking cancer cells even after development of resistance to conventional agents. CAR NK cells seem to be a potential source for “off-the-shelf” therapies,46 since they do not cause GvHD while mediating antitumor and antileukemic reactions.47,48 Thus, clinical-grade manufacturing of CAR NK cells has to be developed and optimized. In order to detect adequate time periods for optimal transduction rates by alpharetroviral vector constructs encoding either anti-CD123/eGFP or only eGFP, IPC samples were collected on days 2–3, 8, and 14 after selection. Transduction rates (measured on day 6 after transduction) rose continuously until day 14 after NK selection. However, functional assays against KG1a demonstrated similar killing activity and CD107a degranulation for NK cells transduced on day 8 or day 14 of expansion, suggesting that cytolytic properties of CAR-transduced NK cells might be optimized on day 8 without any opportunity for substantial increase. This implies that NK cells should be transduced on day 8 (with subsequent expansion for 6 days) without any disadvantages in generating highly activated NK cells. Determination of the optimal time period to transduce primary NK cells is required to integrate the transduction procedure into future Prodigy processes. The described RetroNectin-based transduction method seems to be inappropriate for routine Prodigy processes, since the 48-well-plate procedure is an open system and therefore it is not GMP-compliant. In addition, it is difficult to combine the RetroNectin-based method with the closed Prodigy system. An alternative method might be transduction based on vectofusin-1, a synthetic nontoxic histidine-rich peptide that promotes T-cell transduction with lentiviral vectors.49 This soluble GMP-compliant enhancer could be easily added in advanced Prodigy settings for NK cell transduction. Time-lapse tracking experiments performed by fluorescent microscopy confirmed the results of expanded CAR NK cell cytotoxicity and demonstrated redirected E/T cell contacts of anti-CD123 CAR NK cells toward CD123-positive AML cells and subsequent apoptotic and necrotic reactions in the attacked AML cells.
In the present experiments, the used vectors were SIN vectors that are replication incompetent, demonstrate a singular infectiousness, and decrease the risk of insertional mutagenesis after transduction.21,24 Nevertheless, the freedom of infectious particles in the final product should be achieved. Usually this is accomplished via manifold washing of the cell suspension during the post-transduction cultivation.50,51 Such washing procedures can be integrated into the activity matrix of the Prodigy device. In this context, methods should be implemented to check residual vector concentration in supernatants (Jang et al. described a PCR-based technique to quantify residual vectors).52
In addition to CAR NK cells, CAR T cells present an additional immunotherapy option to treat AML. The development of T cells expressing CD123-specific CARs demonstrated high cytolytic activities in in vitro experiments and in vivo mouse models.53,54 However, serious side effects such as impaired normal hematopoiesis were reported in mice.55 Nevertheless, several Phase I/II studies in China and the United States (NCT02159495, NCT03203369, NCT03190278, NCT03585517, NCT03672851, and NCT03631576) using anti-CD123 CAR T cells recently began patient recruitment. Future preclinical studies with primary CAR NK cells should examine the benefits for clinical treatment, comparing both cytotoxic and regulatory NK cell populations. Moreover, primary CAR-transduced NK cells could be included in current therapeutic protocols containing the application of checkpoint inhibitors, especially for NK cell relevant checkpoints (e.g., TIM-3, LAG-3) on malignant target cells.
Supplementary Material
Acknowledgments
The authors thank D. Lenz for excellent technical support. This project was funded by grants of the DFG (Collaborative Research Center 738, CRC738, and Cluster of Excellence REBIRTH), the Integrated Research and Treatment Center Transplantation (IFB-Tx, Ref. No. 01EO0802 and 01EO1302), and the European Union (Horizon 2020 program SCIDNET).
Author Disclosure
J.K. and V.H. were employees of Miltenyi Biotec GmbH, although V.H. left Miltenyi Biotec shortly after the experimental phase. A.S. is co-inventor on a patent application describing alpharetroviral SIN vectors. The remaining authors declare no financial or commercial conflicts of interest.
Supplementary Material
References
- 1. Smyth MJ, Cretney E, Kelly JM, et al. . Activation of NK cell cytotoxicity. Mol Immunol 2005;42:501–510 [DOI] [PubMed] [Google Scholar]
- 2. Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol 2008;9:495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luetke-Eversloh M, Killig M, Romagnani C. Signatures of human NK cell development and terminal differentiation. Front Immunol 2013;4:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moretta L, Pietra G, Vacca P, et al. . Human NK cells: From surface receptors to clinical applications. Immunol Lett 2016;178:15–19 [DOI] [PubMed] [Google Scholar]
- 5. Vivier E, Raulet DH, Moretta A, et al. . Innate or adaptive immunity? The example of natural killer cells. Science 2011;331:44–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Srivastava S, Lundqvist A, Childs RW. Natural killer cell immunotherapy for cancer: a new hope. Cytotherapy 2008;10:775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer 2016;16:7–19 [DOI] [PubMed] [Google Scholar]
- 8. Stern M, Passweg JR, Meyer-Monard S, et al. . Pre-emptive immunotherapy with purified natural killer cells after haploidentical SCT: a prospective Phase II study in two centers. Bone Marrow Transplant 2013;48:433–438 [DOI] [PubMed] [Google Scholar]
- 9. Passweg JR, Koehl U, Uharek L, et al. . Natural-killer-cell-based treatment in haematopoietic stem-cell transplantation. Best Pract Res Clin Haematol 2006;19:811–824 [DOI] [PubMed] [Google Scholar]
- 10. Koehl U, Brehm C, Huenecke S, et al. . Clinical grade purification and expansion of NK cell products for an optimized manufacturing protocol. Front Oncol 2013;3:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kloss S, Oberschmidt O, Morgan M, et al. . Optimization of human NK cell manufacturing: fully automated separation, improved ex vivo expansion using IL-21 with autologous feeder cells, and generation of anti-CD123-CAR-expressing effector cells. Hum Gene Ther 2017;28:897–913 [DOI] [PubMed] [Google Scholar]
- 12. Koehl U, Kalberer C, Spanholtz J, et al. . Advances in clinical NK cell studies: donor selection, manufacturing and quality control. Oncoimmunology 2016;5:e1115178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Granzin M, Soltenborn S, Muller S, et al. . Fully automated expansion and activation of clinical-grade natural killer cells for adoptive immunotherapy. Cytotherapy 2015;17:621–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kloess S, Huenecke S, Piechulek D, et al. . IL-2-activated haploidentical NK cells restore NKG2D-mediated NK-cell cytotoxicity in neuroblastoma patients by scavenging of plasma MICA. Eur J Immunol 2010;40:3255–3267 [DOI] [PubMed] [Google Scholar]
- 15. Raneros AB, Puras AM, Rodriguez RM, et al. . Increasing TIMP3 expression by hypomethylating agents diminishes soluble MICA, MICB and ULBP2 shedding in acute myeloid leukemia, facilitating NK cell-mediated immune recognition. Oncotarget 2017;8:31959–31976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oberschmidt O, Kloess S, Koehl U. Redirected primary human chimeric antigen receptor natural killer cells as an “off-the-shelf immunotherapy” for improvement in cancer treatment. Front Immunol 2017;8:654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Munoz L, Nomdedeu JF, Lopez O, et al. . Interleukin-3 receptor alpha chain (CD123) is widely expressed in hematologic malignancies. Haematologica 2001;86:1261–1269 [PubMed] [Google Scholar]
- 18. Pizzitola I, Anjos-Afonso F, Rouault-Pierre K, et al. . Chimeric antigen receptors against CD33/CD123 antigens efficiently target primary acute myeloid leukemia cells in vivo. Leukemia 2014;28:1596–1605 [DOI] [PubMed] [Google Scholar]
- 19. Suerth JD, Maetzig T, Galla M, et al. . Self-inactivating alpharetroviral vectors with a split-packaging design. J Virol 2010;84:6626–6635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suerth JD, Morgan MA, Kloess S, et al. . Efficient generation of gene-modified human natural killer cells via alpharetroviral vectors. J Mol Med (Berl) 2016;94:83–93 [DOI] [PubMed] [Google Scholar]
- 21. Hubner J, Hoseini SS, Suerth JD, et al. . Generation of genetically engineered precursor T-cells from human umbilical cord blood using an optimized alpharetroviral vector platform. Mol Ther 2016;24:1216–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stein C, Kellner C, Kugler M, et al. . Novel conjugates of single-chain Fv antibody fragments specific for stem cell antigen CD123 mediate potent death of acute myeloid leukaemia cells. Br J Haematol 2010;148:879–889 [DOI] [PubMed] [Google Scholar]
- 23. Fehse B, Kustikova OS, Bubenheim M, et al. . Pois(s)on—it's a question of dose. Gene Ther 2004;11:879–881 [DOI] [PubMed] [Google Scholar]
- 24. Suerth JD, Maetzig T, Brugman MH, et al. . Alpharetroviral self-inactivating vectors: long-term transgene expression in murine hematopoietic cells and low genotoxicity. Mol Ther 2012;20:1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sandrin V, Boson B, Salmon P, et al. . Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood 2002;100:823–832 [DOI] [PubMed] [Google Scholar]
- 26. Zimmermann SY, Esser R, Rohrbach E, et al. . A novel four-colour flow cytometric assay to determine natural killer cell or T-cell-mediated cellular cytotoxicity against leukaemic cells in peripheral or bone marrow specimens containing greater than 20% of normal cells. J Immunol Methods 2005;296:63–76 [DOI] [PubMed] [Google Scholar]
- 27. Koehl U, Bochennek K, Esser R, et al. . ISHAGE-based single-platform flowcytometric analysis for measurement of absolute viable T cells in fresh or cryopreserved products: CD34/CD133 selected or CD3/CD19 depleted stem cells, DLI and purified CD56+CD3- NK cells. Int J Hematol 2008;87:98–105 [DOI] [PubMed] [Google Scholar]
- 28. Kim EK, Ahn YO, Kim S, et al. . Ex vivo activation and expansion of natural killer cells from patients with advanced cancer with feeder cells from healthy volunteers. Cytotherapy 2013;15:231–241.e1. [DOI] [PubMed] [Google Scholar]
- 29. Siegler U, Meyer-Monard S, Jorger S, et al. . Good manufacturing practice-compliant cell sorting and large-scale expansion of single KIR-positive alloreactive human natural killer cells for multiple infusions to leukemia patients. Cytotherapy 2010;12:750–763 [DOI] [PubMed] [Google Scholar]
- 30. Parkhurst MR, Riley JP, Dudley ME, et al. . Adoptive transfer of autologous natural killer cells leads to high levels of circulating natural killer cells but does not mediate tumor regression. Clin Cancer Res 2011;17:6287–6297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cerwenka A, Lanier LL. Natural killer cells, viruses and cancer. Nat Rev Immunol 2001;1:41–49 [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y, Wallace DL, de Lara CM, et al. . In vivo kinetics of human natural killer cells: the effects of ageing and acute and chronic viral infection. Immunology 2007;121:258–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Davis MR, Zhu Z, Hansen DM, et al. . The role of IL-21 in immunity and cancer. Cancer Lett 2015;358:107–114 [DOI] [PubMed] [Google Scholar]
- 34. Wagner J, Pfannenstiel V, Waldmann A, et al. . A two-phase expansion protocol combining interleukin (IL)-15 and IL-21 improves natural killer cell proliferation and cytotoxicity against rhabdomyosarcoma. Front Immunol 2017;8:676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wendt K, Wilk E, Buyny S, et al. . Interleukin-21 differentially affects human natural killer cell subsets. Immunology 2007;122:486–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Q, Ye LJ, Ren HL, et al. . Multiple effects of IL-21 on human NK cells in ex vivo expansion. Immunobiology 2015;220:876–888 [DOI] [PubMed] [Google Scholar]
- 37. Granzin M, Wagner J, Kohl U, et al. . Shaping of natural killer cell antitumor activity by ex vivo cultivation. Front Immunol 2017;8:458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Granzin M, Stojanovic A, Miller M, et al. . Highly efficient IL-21 and feeder cell-driven ex vivo expansion of human NK cells with therapeutic activity in a xenograft mouse model of melanoma. Oncoimmunology 2016;5:e1219007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang W, Feng H, Chen Q, et al. . The functional potency of natural killer cells in response to IL-2/IL-15/IL-21 stimulation is limited by a concurrent upregulation of Tim-3 in bladder cancer. Exp Cell Res 2018;372:92–98 [DOI] [PubMed] [Google Scholar]
- 40. Bryceson YT, Fauriat C, Nunes JM, et al. . Functional analysis of human NK cells by flow cytometry. Methods Mol Biol 2010;612:335–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Penack O, Gentilini C, Fischer L, et al. . CD56dimCD16neg cells are responsible for natural cytotoxicity against tumor targets. Leukemia 2005;19:835–840 [DOI] [PubMed] [Google Scholar]
- 42. Fischer L, Penack O, Gentilini C, et al. . The anti-lymphoma effect of antibody-mediated immunotherapy is based on an increased degranulation of peripheral blood natural killer (NK) cells. Exp Hematol 2006;34:753–759 [DOI] [PubMed] [Google Scholar]
- 43. Childs RW, Berg M. Bringing natural killer cells to the clinic: ex vivo manipulation. Hematology Am Soc Hematol Educ Program 2013;2013:234–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huenecke S, Zimmermann SY, Kloess S, et al. . IL-2-driven regulation of NK cell receptors with regard to the distribution of CD16+ and CD16- subpopulations and in vivo influence after haploidentical NK cell infusion. J Immunother 2010;33:200–210 [DOI] [PubMed] [Google Scholar]
- 45. Elpek KG, Rubinstein MP, Bellemare-Pelletier A, et al. . Mature natural killer cells with phenotypic and functional alterations accumulate upon sustained stimulation with IL-15/IL-15Ralpha complexes. Proc Natl Acad Sci U S A 2010;107:21647–21652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Iyer RK, Bowles PA, Kim H, et al. . Industrializing autologous adoptive immunotherapies: manufacturing advances and challenges. Front Med (Lausanne) 2018;5:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Glienke W, Esser R, Priesner C, et al. . Advantages and applications of CAR-expressing natural killer cells. Front Pharmacol 2015;6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mehta RS, Rezvani K. Chimeric antigen receptor expressing natural killer cells for the immunotherapy of cancer. Front Immunol 2018;9:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Piovan C, Marin V, Scavullo C, et al. . Vectofusin-1 promotes RD114-TR-pseudotyped lentiviral vector transduction of human HSPCs and T lymphocytes. Mol Ther Methods Clin Dev 2017;5:22–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cesani M, Plati T, Lorioli L, et al. . Shedding of clinical-grade lentiviral vectors is not detected in a gene therapy setting. Gene Ther 2015;22:496–502 [DOI] [PubMed] [Google Scholar]
- 51. Bagutti C, Schmidlin M, Mueller M, et al. . Washout kinetics of viral vectors from cultured mammalian cells. Appl Biosaf 2012;17:188–197 [Google Scholar]
- 52. Jang YH, Song HI, Yang Y, et al. . Reliable RT-qPCR-based titration of retroviral and lentiviral vectors via quantification of residual vector plasmid DNA in samples. Biotechnol Lett 2016;38:1285–1291 [DOI] [PubMed] [Google Scholar]
- 53. Mardiros A, Dos Santos C, McDonald T, et al. . T cells expressing CD123-specific chimeric antigen receptors exhibit specific cytolytic effector functions and antitumor effects against human acute myeloid leukemia. Blood 2013;122:3138–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mardiros A, Forman SJ, Budde LE. T cells expressing CD123 chimeric antigen receptors for treatment of acute myeloid leukemia. Curr Opin Hematol 2015;22:484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gill S, Tasian SK, Ruella M, et al. . Preclinical targeting of human acute myeloid leukemia and myeloablation using chimeric antigen receptor-modified T cells. Blood 2014;123:2343–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.