Abstract
Autophagy is a highly regulated, biological process that provides energy during periods of stress and starvation. This conserved process also acts as a defense mechanism and clears microbes from the host cell. Autophagy is impaired in Cystic Fibrosis (CF) patients and CF mice, as their cells exhibit low expression levels of essential autophagy molecules. The genetic disorder in CF is due to mutations in the cystic fibrosis transmembrane conductance regulator (cftr) gene that encodes for a chloride channel. CF patients are particularly prone to infection by pathogens that are otherwise cleared by autophagy in healthy immune cells including Burkholderia cenocepacia (B. cenocepacia). The objective of this study is to determine the mechanism underlying weak autophagic activity in CF macrophages and find therapeutic targets to correct it. Using reduced representation bisulfite sequencing (RRBS) to determine DNA methylation profile, we found that the promoter regions of Atg12 in CF macrophages are significantly more methylated than in the wild-type (WT) immune cells, accompanied by low protein expression. The natural product epigallocatechin-3-gallate (EGCG) significantly reduced the methylation of Atg12 promoter improving its expression. Accordingly, EGCG restricted B. cenocepacia replication within CF mice and their derived macrophages by improving autophagy and preventing dissemination. In addition, EGCG improved the function of CFTR protein. Altogether, utilizing RRBS for the first time in the CF field revealed a previously unrecognized mechanism for reduced autophagic activity in CF. Our data also offers a mechanism by which EGCG exerts its positive effects in CF.
1. Introduction
DNA methylation is the most stable, epigenetic modification controlling the transcription of the mammalian genome. DNA methylation leads to the addition of methyl groups across the entire genome, including in and around promoters [1,2]. Methylation of the promoter maintains differential gene expression patterns in a tissue-specific and developmental stage-specific manner [2]. Several technologies to measure DNA methylation exist, including a recently developed approach that achieves single-base resolution through bisulfite conversion called reduced representation bisulfite sequencing (RRBS) [2,3]. This approach has not been previously exploited in the study of cystic fibrosis (CF).
CF is the most common hereditary disease in the Caucasian population [4–9]. It is characterized by the mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) protein, a member of the ATP-binding cassette transporter family coding for a chloride channel. CFTR defect is due to many hereditary mutations, the most common mutation however is the deletion of phenylalanine at position 508 (F508del) in CFTR. Therefore, mice used in this study express global CFTR F508del protein and will be referred to as CF mice [10,11]. In epithelial cells, the CFTR channel is responsible for the transport of salt and water [12]. The disruption in the function of CFTR is accompanied by production of thick mucus in several organs. It is also accompanied by reduction of autophagic activity in epithelial [13,14]. Autophagy is a conserved pathway in all eukaryotic cells responsible for clearing nonfunctional organelles, protein aggregates and microbes [15–17]. The activation of autophagy presents as increased formation of autophagosomes that deliver their contents to lysosomes for degradation. Specific autophagy factors called ATGs (autophagy-related genes) are each required for the formation and progression of autophagosomes.
In CF epithelial cells, elegant studies by Maiuri and colleagues demonstrated that defective CFTR in epithelial cells induces the upregulation of reactive oxygen species (ROS) and tissue transglutaminase (TG2) that drives the crosslinking of autophagy molecules, leading to aggresome formation and sequestration of mutant CFTR [14].
In CF macrophages, reduced autophagy was found to be due to low expression of major autophagy molecules. Defective autophagy in CF macrophages is accompanied by increased inflammatory cytokine production and persistence of specific organisms including Burkholderia cenocepacia (B. cenocepacia). This Gram-negative bacterium is responsible for severe pneumonia and often death in CF patients [13,19]. However, B. cenocepacia is efficiently cleared by autophagy in healthy macrophages. Therefore, the clearance of B. cenocepacia is considered readout for autophagic activity.
It has been shown that green tea extract induces autophagy in lung cancer, cardiac disease [20], breast cancer [21], diabetes [22], however the mechanism was unclear. Green tea extracts from Camellia sinensis contain a number of catechins, including epigallocatechin-3-gallate (EGCG), epigallocatechin (EGC), epicatechin-gallate (ECG), and epicatechin (EC). EGCG is the most abundant polyphenol in green tea and is considered to have anti-inflammatory, anti-oxidant, and cancer preventative properties [23–25]. Several reports have also demonstrated the ability of EGCG to control intracellular infections, yet the mechanism is still unclear. EGCG is an inhibitor of DNA methyltransferases (DNMTs) by direct, inhibitory interaction with the catalytic site of DNMTs [28]. It has also been shown that EGCG reverses the methylation-mediated downregulation of specific targets. Although EGCG has been used in several reports in the CF field, no studies have investigated its effect on methylation in this disorder. Also, no studies focused on its effect on the clearance of B. cenocepacia in CF.
Determination of epigenetic alterations of autophagy genes in CF will provide valuable clues in early diagnosis and in diversifying treatment options for CF patients. The dysfunction of autophagy is a hallmark for several disease conditions including neurodegenerative disorders, autoimmune disorders, and chronic granulomatous diseases in addition to CF [26–29]. ATGs are frequently governed by epigenetic mechanisms, such as chromatin modulation, histone modification, and microRNAs [5,30–33].
In this report, using RRBS technology, we demonstrate that the promoter of autophagy gene Atg12 is significantly methylated in CF macrophages and responds to the demethylation effect of EGCG. In turn, EGCG increases the expression of this protein, improving autophagy, and clearance of intracellular B. cenocepacia. In addition, EGCG concomitantly improved CFTR function in CF macrophages, yet independently of autophagy. This is the first report investigating the DNA methylation status of autophagy genes in CF macrophages. This study also demonstrates the specific mechanism of action of EGCG in autophagy-mediated clearance of bacteria in CF. Our study provides novel therapeutic targets in CF and improves our understanding of the underlying mechanism of dysfunctional autophagy in CF and possibly other disease conditions characterized by weak autophagy.
2. Results
2.1. The promoter of Atg12 is more methylated in CF macrophages when compared to WT cells and responds to EGCG treatment
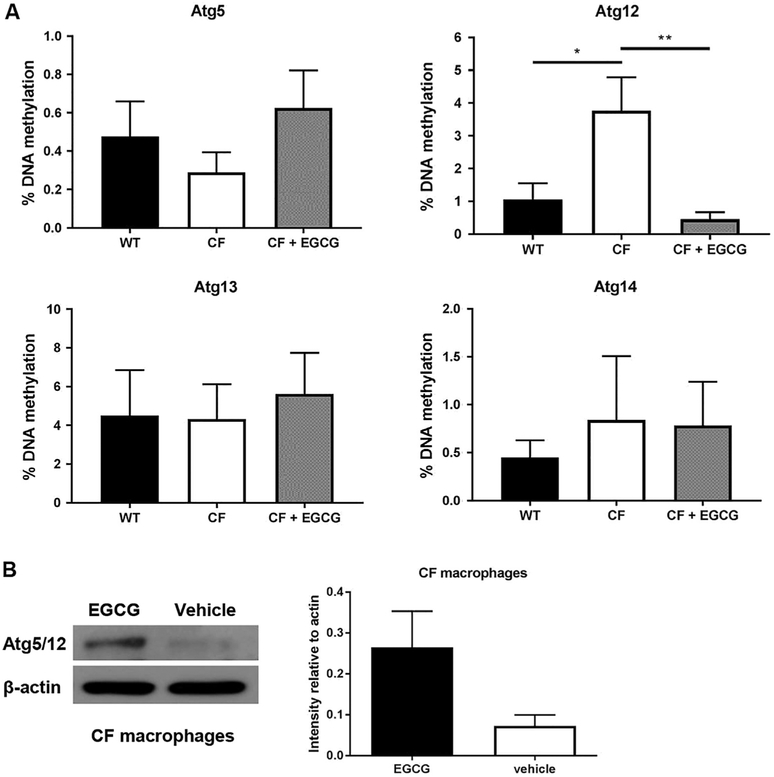
Macrophages from human subjects with CF or from the CF mouse model elicit weak autophagy activity due to low expression of autophagy proteins. ATGs are essential molecules that play specific sequential roles during the formation and maturation of autophagosomes. We and others have previously demonstrated that the expression of autophagy molecules is reduced in CF macrophages and epithelial cells [13,14,19,34–36]. We have shown that the expression of ATG5-ATG12 protein complex is low in CF macrophages, compared to that of healthy cells [13,19,35]. This complex is crucial for the early steps of autophagosome formation in all eukaryotic cells [37,38]. To better understand the mechanism behind their low expression, we studied genome-scale DNA methylation at The Ohio State University Comprehensive Cancer Center Genomics Shared Resources using Reduced Representation Bisulfite Sequencing (RRBS) technology [39,40]. Interestingly, we found that the key autophagy gene Atg12 is significantly more methylated in CF compared to WT macrophages (Fig. 1A). Notably, other autophagy genes such as Atg13 and Atg14 were not differentially methylated in CF cells compared to WT. To determine if the methylation profile reflects the level of corresponding protein expression in CF macrophages compared to WT, we analyzed cell lysates by western blot and found that the expression of ATG5-ATG12 complex is reduced in CF macrophages (Fig. 1B). The expression of ATG4 was comparable between the two (data not shown).
Fig. 1.
Methylation percentage of autophagy gene promoter Atg12 is elevated in CF (F508del) macrophages and responds to EGCG. (A) The effect of EGCG treatment on autophagy genes in CF compared to untreated CF and WT. WT (black bars), CF (white bars), and CF+EGCG (grey bars), n=3 independent experiments (B) Western blot of ATG5-ATG12 (ATG5/12) complex of wild-type (WT) and CF (F508del) macrophages treated with EGCG or vehicle control. Blot is representative of three independent experiments. Densitometry analysis of ATG5/12 complex (56 kDA) is shown as mean intensities of bands compared to housekeeping (β-actin 42 kDa). *p < .05 ** p < .01, One-way ANOVA with Holm-Sidak's correction.
Because the promoter of Atg12 is more methylated, we examined the possibility that EGCG could reduce its methylation status to WT levels and thus improve its expression. EGCG has been FDA approved as a demethylation agent for treatment for several conditions [41,42]. Studies have reported a positive effect of EGCG in CF, yet the mechanism remains unclear [6–9]. EGCG treatment was able to reduce the methylation of Atg12 promoter and restore its expression (Fig. 1A and B). Therefore, EGCG restores the expression of ATG12 by reducing the methylation of its promoter.
2.2. EGCG but not 5’AZA improves B. cenocepacia clearance in CF macrophages
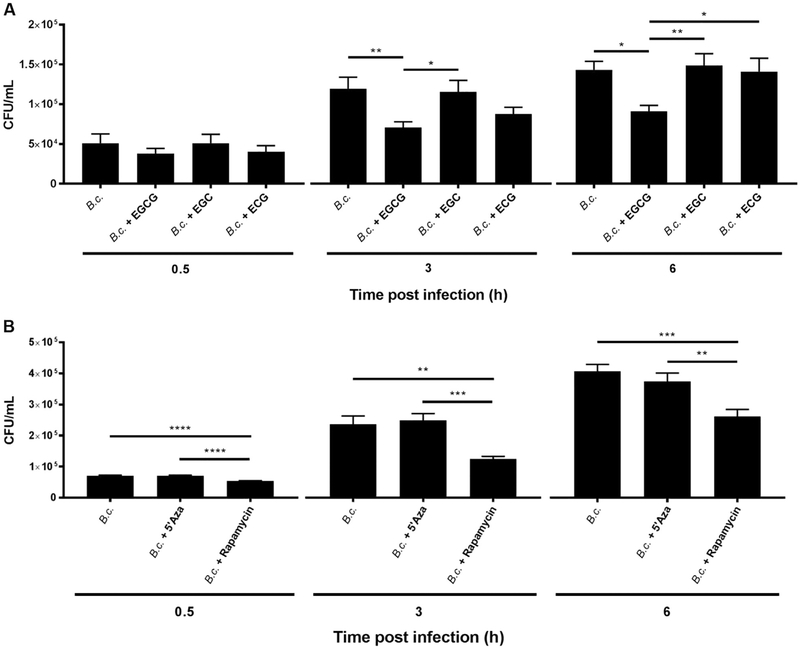
To determine if the demethylation effect of EGCG elicits a functional effect on macrophage autophagy response, we next determined if EGCG improves the clearance of B. cenocepacia in CF macrophages. B. cenocepacia is a CF-associated pathogen that persists in CF macrophages due to weak autophagy, yet is cleared in WT cells [13,19,35]. CF macrophages were pre-treated with 25 μg/mL EGCG for 24 h and then infected with B. cenocepacia and colony forming units (CFUs) were assessed (Fig. 2A). EGCG treatment of macrophages was accompanied by significant reduction of B. cenocepacia intracellular growth within 3 h of infection (Fig. 2A). Notably, EGCG was not bactericidal when added to bacterial culture in the absence of macrophages (Supplementary Fig. 1A). In addition to EGCG, green tea extract also contains a number of other catechins, including, EGC and ECG. To determine if these catechins elicit an effect similar to EGCG, CF macrophages were pre-treated with EGC or ECG then infected with B. cenocepacia. Notably, both catechins did not improve the clearance of B. cenocepacia in CF macrophages (Fig. 2A). Therefore, EGCG and not EGC or ECG improves B. cenocepacia clearance in CF macrophages. Five-azacytidine (azacitidine, 5’Aza) is a cytidine analog that is incorporated into DNA where DNMTs bind to it, thus preventing maintenance of the methylation status, promoting demethylation of various genes [43]. 5’Aza is FDA approved for treating myelodysplastic syndrome and other leukemias. Unlike EGCG, pre-treatment of CF macrophages with 5’Aza did not improve the clearance of B. cenocepacia (Fig. 2B). Rapamycin, which is a strong autophagy stimulator, was used as a positive control and it effectively improved B. cenocepacia clearance as previously published (Fig. 2B) [13]. Therefore, pre-treatment with EGCG, but not 5’Aza, improves the clearance of B. cenocepacia.
Fig. 2.
EGCG but not 5’Aza promotes the restriction of B. cenocepacia in vitro. (A) CF (F508del) macrophages were treated with 25 μg/mL EGCG, EGC, or ECG then infected with B. cenocepacia at a multiplicity of infection (MOI) of 10. Colony forming units (CFUs) were determined at 0.5, 3, and 6 h post infection. (B) CF macrophages were treated with 25 μg/mL 5’Aza or 5 μg/mL Rapamycin and infected with B. cenocepacia. CFUs were determined at 0.5, 3, and 6 h post infection. n = 3 independent experiments. *p < .05, **p < .01, ***p < .001, ****p < .0001, One-way ANOVA with Holm-Sidald's correction.
2.3. EGCG improves the clearance of B. cenocepacia by improving autophagy activity and CF macrophage survival
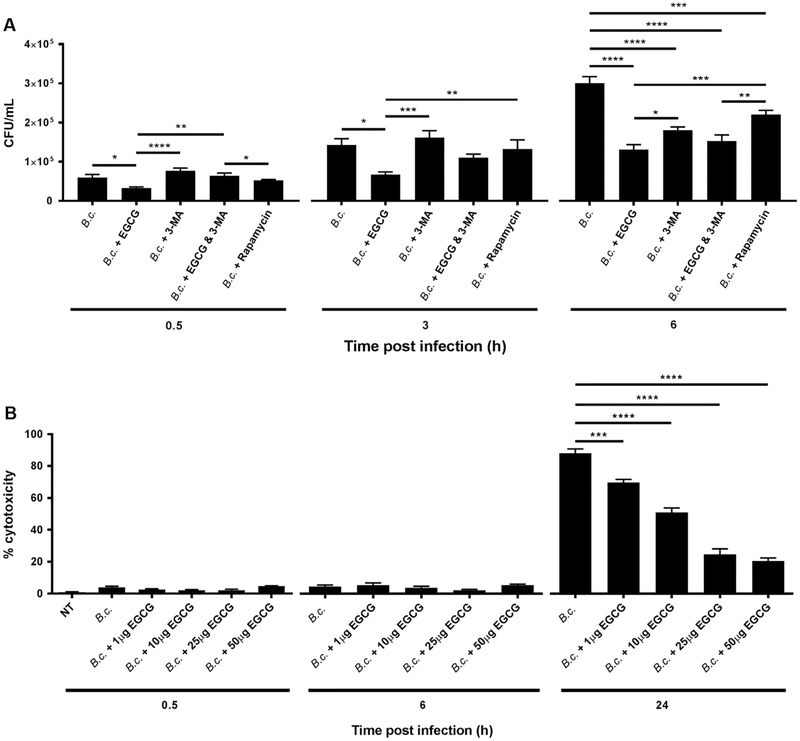
We and others have previously demonstrated that the expression of autophagy molecules is reduced in CF macrophages and epithelial cells [13,14,19,36]. Low Atg expression in CF immune cells is accompanied by persistence of B. cenocepacia infection [13,19,35]. To determine if the ability of EGCG to improve B. cenocepacia clearance in CF macrophages is mediated by improved autophagic activity, we examined the bacterial burdens in CF macrophages pre-treated with EGCG in the presence or absence of the autophagy inhibitor 3-methyl-adenine (3-MA) (Fig. 3A). Notably, EGCG reduced bacterial burdens, yet this effect was diminished in the presence of 3-MA at 0.5 and 3 h (Fig. 3A). Therefore, EGCG improves the clearance of B. cenocepacia via improving autophagy. Bacterial infection is often associated with the demise of the host cell allowing the dissemination of the infectious agents to other cells. To determine if EGCG can prevent B. cenocepacia-associated macrophage death, CF macrophages were treated with varying concentrations of EGCG (1 −50 μg/mL) for 24 h then infected with B. cenocepacia. Cytotoxicity was determined by assaying supernatants for release of lactate dehydrogenase (LDH). EGCG significantly reduced macrophage death in a dose dependent manner starting at 1 μg/mL (Fig. 3B). We also examined if 5’Aza was able to reduce CF macrophage death during B. cenocepacia infection. Notably, 5’Aza failed to reduce cell death at concentrations such as 1 and 10 μg/mL, but was able to significantly reduce CF macrophage death starting at 25 μg/mL (Supplementary Fig. 2A). Similarly, the autophagy stimulating drug Rapamycin also reduced CF macrophage death during B. cenocepacia infection at the dose of 5 μg/mL (Supplementary Fig. 2A). In addition, the other catechins EGC and ECG did not promote cell survival of the host cell post-infection (Supplementary Fig. 2B). Therefore, EGCG is more effective in reducing macrophage death during B. cenocepacia infection than 5’Aza, EGC, or ECG. Together, EGCG improves autophagy-mediated restriction and prevents cell death in response to B. cenocepacia.
Fig. 3.
EGCG promotes the clearance of B. cenocepacia in CF (F508del) macrophages via autophagy and reduces cell death. (A) CF (F508del) macrophages were treated with 25 μg/mL EGCG and/or 1 mM 3-MA, or 5 μg/mL rapamycin, then infected with B. cenocepacia at an multiplicity of infection (MOI) of 10. Colony forming units (CFUs) were then assessed 0.5, 3, and 6 h. (B) CF macrophages were treated with increasing concentration (1-50 μg/mL) of EGCG, and LDH release from cells was assessed 0.5, 6, and 24 h post infection. n = 3 independent experiments. *p < .05, **p < .01, ***p < .001, ****p < .0001, One-way ANOVA with Holm-Sidak correction.
2.4. EGCG prevents the growth and dissemination of B. cenocepacia and accompanying inflammation in live CF mice
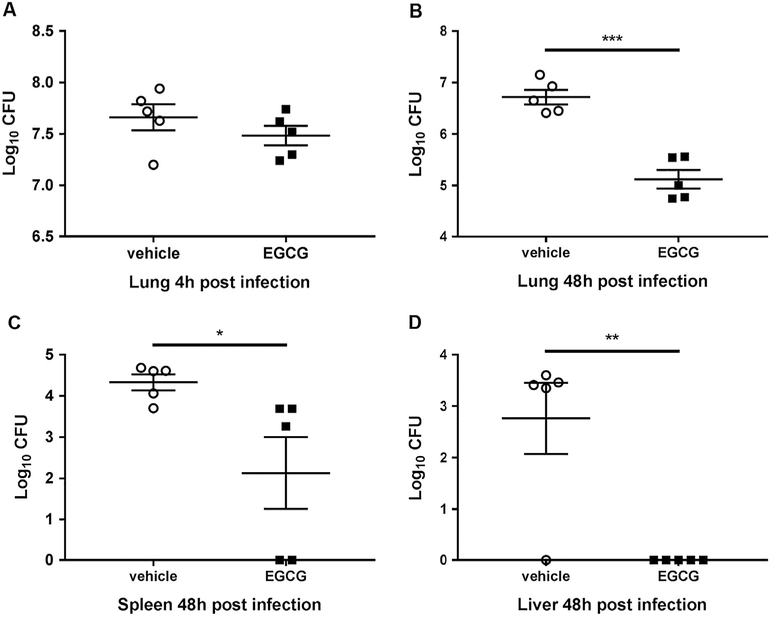
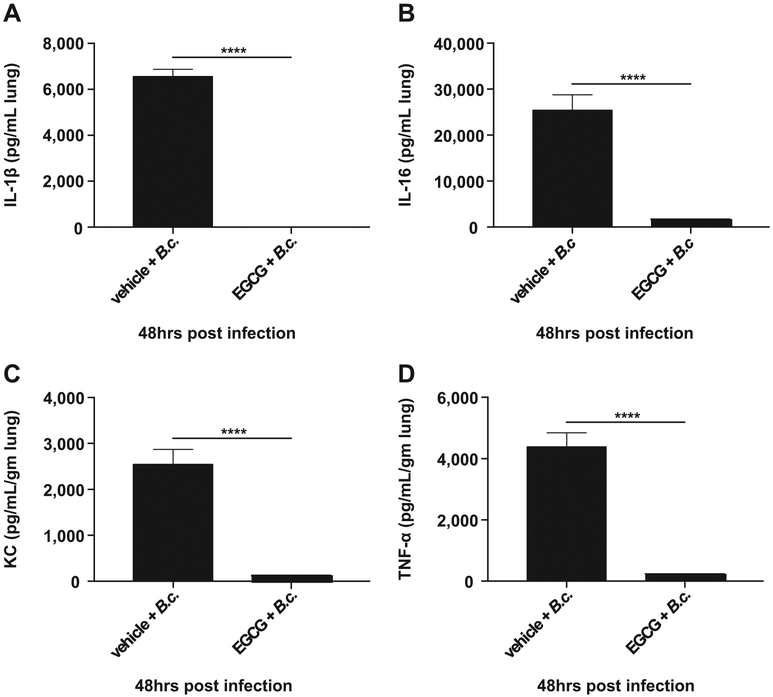
Our data above demonstrates that EGCG improves autophagy-mediated B. cenocepacia clearance in vitro. To determine if EGCG improves the clearance of B. cenocepacia in live CF mice, EGCG was administrated intra-tracheally at 25mg/kg (300 μg) to lightly anesthetized mice (Supplementary Fig. 3). Then, 10 × 106 bacterial CFUs were delivered to the lungs of treated mice via intra-tracheal administration as we previously described [13,19,35]. Following infection, mice were treated with two additional doses of EGCG and then sacrificed 48 h after infection. The lungs were collected and homogenized and plated for CFUs (Supplementary Fig. 3). Treatment of CF mice with EGCG significantly reduced B. cenocepacia infection in the lungs after 48 h of infection (Fig. 4A and B) without affecting the initial infectious dose as reflected by 4 h data (Fig. 4A). Because B. cenocepacia infection is often associated with dissemination to other organs, bacterial loads were evaluated in the liver and spleen of infected mice. EGCG treatment significantly reduced the spread of B. cenocepacia to other organs (Fig. 4C and D). Together, these results demonstrate that EGCG reduces bacterial infection in CF lungs and prevents dissemination to liver and spleen (Fig. 4A-D). To determine if EGCG reduces the inflammation that accompanies B. cenocepacia infection in vivo, cytokines were measured in the lungs of mice treated with EGCG or vehicle control. The administration of EGCG prior to B. cenocepacia infection significantly reduced the production of inflammatory mediators IL-1β, IL-6, KC, and TNFα 48 h post infection (Fig. 5 A-D). Together, these results demonstrate that EGCG reduces bacterial infection, dissemination, and the accompanying inflammation in vivo (Figs. 4 and 5).
Fig. 4.
EGCG contributes to the restriction of B. cenocepacia in vivo. B. cenocepacia colony forming units (CFUs) from CF (F508del) mice treated with vehicle (PBS) or 25 mg/kg EGCG obtained from lung homogenates at (A) 4 h and (B) 48 h post-infection. Disseminated bacterial CFU recovered from (C) spleen and (D) liver 48 h post-infection. Vehicle (PBS) and EGCG, n = 5 mice. *p < .05, **p < .01, unpaired t-test with Welch's correction.
Fig. 5.
EGCG reduces inflammation after infection with B. cenocepacia in vivo. MSD V-PLEX Proinflammatory Panel 1 (mouse) cytokine analysis from lung homogenate supernatants analyzed for (A) IL-1B, (B) IL-6, (C) KC, and (D) TNF-α. Vehicle, n = 3 mice, Vehicle + B.c., EGCG, and EGCG + B.c. treated groups, n = 5 mice. *p < .05, **p < .01, ****p < .0001, unpaired t-test with Welch's correction.
2.5. EGCG increases the expression of autophagy proteins in the lungs of CF mice
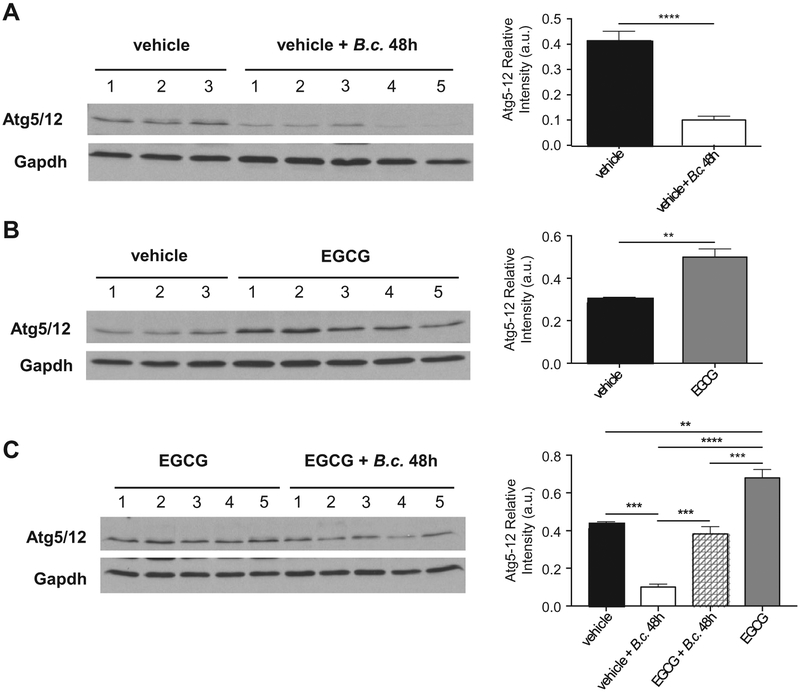
We and others have previously demonstrated that the expression of autophagy molecules is reduced in CF macrophages and epithelial cells [13,14,19,34–36]. We next determined if B. cenocepacia exerts an effect on the expression of autophagy molecules in vivo (Fig. 6). CF mice were infected with B. cenocepacia intratracheally, then, lungs were isolated and homogenized. The expression of ATG5-ATG12 complex was characterized. Notably, infection with B. cenocepacia significantly reduces the expression of autophagy molecules (Fig. 6A). Interestingly, EGCG alone improves the expression of ATG5-ATG12 complex (Fig. 6B) and when added before infection, it prevents the B. cenocepacia-mediated reduction of expression (Fig. 6C). Therefore, EGCG effectively improves the expression of ATG5-ATG12 complex in vivo preventing B. cenocepacia-mediated reduction of ATG5-ATG12 expression.
Fig. 6.
EGCG promotes expression of autophagy members during B. cenocepacia infection in vivo. Western blot of ATG5-ATG12 (ATG5/12) complex from lung homogenates of CF (F508del) mice treated with (A) vehicle or vehicle + B.c. (B) vehicle or EGCG (C) EGCG or EGCG + B.c. Densitometry analysis of ATG5/12 (56 kDa) are shown as mean intensities of bands compared to housekeeping protein (GAPDH36 kDa) ± SEM. Vehicle n=3 mice and treatment groups n=5 mice. **p < .01, ***p < .001, ****p < .0001, (A&B) unpaired t-test with Welch's correction (C) One-way ANOVA with Holm-Sidak correction.
2.6. EGCG improves the function of the CFTR channel independently of autophagy
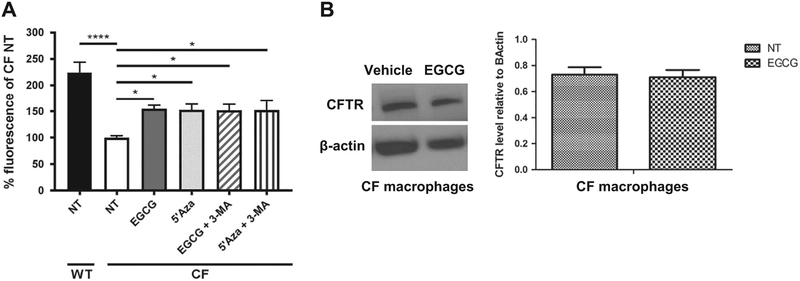
The CF cells expressing the F508del mutant protein are characterized by weak CFTR channel function which is implicated in many phenotypes in CF patients and CF mouse model. To determine if EGCG improves CFTR function, CF macrophages were treated with EGCG and CFTR activity was determined by SPQ assay, as previously described [35,44]. The SPQ assay is commonly used to evaluate CFTR-mediated iodide efflux (reflecting CFTR function) in epithelial cells and we have successfully modified it to use it to measure CFTR function in mouse macrophages [35]. Macrophages were loaded with SPQ by incubation with a hypotonic solution then incubated with an iodide containing buffer to quench SPQ fluorescence. The CFTR channel was then activated by adding forskolin to increase intracellular cAMP (since CFTR is a cAMP-activated channel) [45]. Efflux of iodide was reflected by increase in SPQ fluorescence intensity. We found that EGCG and 5’Aza significantly improved CFTR function in CF macrophages (Fig. 7A). However, the addition of autophagy inhibitor 3-MA did not prevent the increase in SPQ activity mediated by EGCG (Fig. 7A). To determine if improved CFTR function in macrophages by EGCF is due to increased CFTR expression, we analyzed CF macrophages treated with EGCG by western blot using specific antibody to mouse CFTR. We found that the expression of mouse CFTR in CF macrophages did not increase in response to EGCG (Fig. 7B). Therefore, EGCG improves CFTR function in macrophages independently of autophagic activity and expression.
Fig. 7.
EGCG increases CFTR activity in CF (F508del) macrophages. (A) SPQ assay was performed on WT and CF (F508del) macrophages treated or not with 25 μg/mL EGCG or 5’Aza with or without 1mM3-MA overnight. Values depicted as mean±SEM, are normalized to CF no treatment as 100% functionality. n=4 independent experiments, *p < .05, **p < .01. (B) Western blot of (CFTR) protein of WT and CF macrophages treated with EGCG or vehicle control. Blot is representative of three independent experiments. Densitometry analysis of CFTR (165 kDA) is shown as mean intensities of bands compared to housekeeping (β-actin 42 kDa).
3. Discussion
The pathobiology of CF is multi-factorial, including impairment of autophagy in epithelial cells and macrophages. Macrophages from cystic fibrosis patients and mice allow the persistence and growth of B. cenocepacia, Pseudomonas, Staphylococcus, and non-tuberculosis mycobacteria [46,47]. This permissiveness is mainly due to weak autophagic activity as reports from our lab and other groups clearly demonstrated that autophagy is compromised in CF immune cells and epithelial cells [13,14,48]. We also showed that the expression of ATGs such as ATG5, ATG12, and ATG7 is significantly reduced in CF macrophages when compared to non-CF counterparts [13,35]. Despite the rapidly growing literature describing defective autophagy in CF, the underlying mechanisms remain enigmatic. Autophagy activity can be promoted by several drugs including rapamycin in live CF mice and their macrophages as well as human CF macrophages. However, B. cenocepacia infection blocks the effect of any autophagy stimulating drug even rapamycin [13,35]. This finding suggests that stimulation of autophagy in CF should be used to prevent but not treat infections such as B. cenocepacia. Nonetheless, rapamycin exerts severe side effects making it unsuitable for patients with CF or any other chronic disease [49,50]. Similarly, EGCG treatment after B. cenocepacia infection failed to restore autophagic activity (data not shown). Therefore, correcting autophagy in CF has to be achieved before the onset of infection with a vicious autophagy suppressing organism such as B. cenocepacia. Therefore, safer autophagy-stimulating drugs are desperately needed for chronic conditions where autophagy is down-regulated.
To better understand the mechanism behind low expression of autophagy molecules in CF, we analyzed epigenetic regulations of Atg genes. Epigenetic modification of DNA causes changes in gene expression without altering the DNA sequence. Our previous studies demonstrated that the microRNA cluster Mir-17-92 is over-expressed in CF macrophages. Notably, this cluster repetitively targets autophagy genes, reducing their transcription. Lowering the expression of specific Mirs within the cluster in CF macrophages improved the expression of ATG7 and ATG16l1, but not other ATGs including Atg12 [35].
In this report, we studied epigenetic regulation of autophagy by DNA methylation. The role of methylation in disease states such as cancer is becoming increasingly important leading to an increase in FDA-approved demethylation agents [25,42]. The four main sequencing technologies used for exploring genome-wide DNA methylation include methylated DNA binding domain sequencing, methylated DNA immunoprecipitation sequencing, whole genome bisulfite sequencing (WGBS), and reduced representation bisulfite sequencing (RRBS) [39,40]. The former two use the enrichment of methylated DNA to acquire a maximum resolution of 150 bp while the latter two achieve single-base resolution through the bisulfite conversion [51]. Compared with WGBS, RRBS is a cost-effective approach for studying genomewide patterns of DNA methylation. Using RRBS for the first time in the CF field, we demonstrate that the promoter of Atg12 is more methylated in CF macrophages when compared to WT cells. The higher methylation of this gene correlated with its low expression in CF cells. Whether, this finding will translate into a viable marker to follow CF progression and exacerbations remains to be elucidated.
On the other hand, several studies have reported the hypermethylation of the CFTR gene in human and murine CF cells [4,5]. However, our analysis using RRBS did not reveal significant methylation of the promoter region of the cftr gene. Additionally, the treatment of CF macrophages with EGCG did not increase the total expression of CFTR. Nevertheless, EGCG improved CFTR function yet independently of autophagy. Thus, it is possible that EGCG increases the stability of mature plasma membrane bound CFTR without increasing the total protein level. Notably, several reports demonstrated that EGCG improves CFTR function and expression of human CFTR mature form in epithelial cells from CF patients. EGCG was used in combination with cysteamine [6–9] that increases the expression of Beclin 1 and re-establishes the autophagic pathway which improved the clearance of Pseudomonas [52]. Thus, EGCG improves CFTR function through multiple mechanisms that may differ between epithelial cells and macrophages.
EGCG has been extensively studied primarily because of its anticarcinogenic effects [53,54]. It has been used in other studies demonstrating its ability to improve microbial clearance [42]. Several reports suggested that EGCG improves autophagy [6–9], yet, the mechanism was unknown. Here we found that the treatment of CF macrophages with EGCG significantly reduces the methylation of Atg12 promoter and is accompanied by improvement of expression of the corresponding protein. This led to improved autophagic activity and in turn, increased B. cenocepacia clearance in vitro and in vivo. The impairment of autophagy has been implicated in other infectious disease conditions, cancer, and neurodegenerative disorders.
Our study herein offers mechanistic insight into CF pathobiology and additional disease conditions characterized by weak autophagic activity.
4. Materials and methods
4.1. Bacterial strains and reagents
B. cenocepacia K56-2 is a clinical isolate obtained from a CF patient. MH1K is a gentamicin-sensitive derivate of K56-2. All bacteria were grown overnight in LB media at 37 °C and 200 rpm.
4.2. Mice
C57BL/6 wild-type (WT) mice were obtained from The Jackson Laboratory (Bar Harbor, ME, USA). F508del (CF), C57BL6 background mice were purchased from the Cystic Fibrosis Mouse Models Core at Case Western Reserve University (CWRU). All mice were housed in a pathogen-free facility and experiments were conducted with approval from the Animal Care and Use Committee at The Ohio State University (Columbus, OH, USA).
4.3. Cell culture and reagents
Murine bone marrow-derived macrophages (BMDMs) were isolated from C57BL/6 (WT) mice, as previously described [13,55–57]. Prior to infection, macrophages were treated overnight with 25 μg/mL EGCG (Sigma Aldrich #E4143), EGC (Sigma Aldrich #E3768), ECG (Sigma Aldrich #E3893), or 5’Aza-deoxycytidine (5’Aza, Sigma Aldrich, #A3656), in combination or not with 1 mM 3-MA (Sigma Aldrich, M9281) in IMDM media supplemented with 10% FBS. All in vitro infections were performed with MOI 10:1. Infections were synchronized by centrifugation 400 rpm for 5 min. For the Gentamicin Protection Assay macrophages were infected for 0.5 h followed by 0.5 h incubation with 50 μg/mL gentamicin to avoid extracellular bacterial replication. EGCG, or 5’Aza, with or without 3-MA treatments were added to macrophages for specified times. To stimulate autophagy, cells were incubated with 5 μg/mL rapamycin (cat # R0395, Sigma Aldrich) 1 h prior to infection and then continuously until the end of the experiment.
4.4. Reduced representation bisulfite sequencing (RRBS)
Genome-wide methylation analysis was performed on WT or CF-derived macrophages treated with either vehicle or EGCG (3 replicates per treatment group) using RRBS. This approach utilizes the methylation-insensitive restriction enzyme MspI to cut DNA at CCGG sites in the genome. This effectively enriches for fragments derived from CpG-rich regions. The RRBS libraries were prepared using an input amount of 200 ng DNA following the protocol described by Gu et al. with modifications [51]. Briefly, 200 ng genomic DNA was digested with MspI (New England Biolabs, #R0106L) followed by AMPure XP beads (A63882, Beckman Coulter, Inc.) cleanup. Digested DNA (75 ng) was end-repaired, adenylated and ligated with methylated adapters (BIOO NEXTFlex Bisulfite-seq kit (NOVA-5119-02, PerkinElmer, Inc.). This was followed by bead-based cleanup and fragment size selection. Bisulfite conversion was performed using EZ DNA Methylation-Gold Kit (Zymo Research Corp, #D5005). Converted DNA was amplified for 18 cycles followed by beads cleanup. Quantity and quality of RRBS libraries were assayed by Qubit dsDNA HS kit (Invitrogen, #Q32854) and Bioanalyzer HS 12000 DNA chip (Agilent Technologies, #5067–1509,), respectively. Sequencing was performed using Illumina HiSeq 4000 Sequencer (Illumina, San Diego, CA) to ~40 million pass filter paired-end 150 bp clusters per sample.
4.5. Bioinformatics analysis
RRBS data in fastq format was preprocessed with Trim Galore (v0.4.0) [58]. It employs Cutadapt to perform both adapter and quality trim (bases with Phred score < 20) on paired-end 150 bp RRBS reads. Trimmed reads were aligned to the bisulfite converted human genome using the Bismark (v0.17.0) aligner [59]. FastQC and Bismark output files were parsed through a custom python script that generated RRBS post-alignment quality control statistics including parameters such as trimming percentage, alignment rates, and incomplete conversion rate. BAM files were sorted using SAMtools [60] and then passed to methylKit to create text files containing % methylation values for each RRBS read. Differential methylation analysis was performed using methylKit [61] for extraction of CpG methylation values. Only CpGs with coverage of at least ≥80 reads across all three replicates were considered for analysis. Gene promoter regions were defined to be ±1 kb of the transcription start site (TSS) according to NCBI's mm 10 RefSeq annotation. However, when repetitive elements are present in gene promoters, the multiply-mapped nature of sequencing reads derived from these regions will make it difficult to call methylation value accurately. In these scenarios, the proximal promoter was then defined to be ±1 kb or the longest region uninterrupted by repetitive elements on either side of TSS.
4.6. In vivo infection
F508del mice were treated intratracheally for 3 days with 100 μL EGCG (25 mg/kg) or vehicle (PBS). On the fourth day, mice were infected with 10 × 106 K56–2. The four hour mouse group was sacrificed and lungs were isolated for RNA, protein, and initial bacterial CFU counts. EGCG or PBS treatments continued for the second group and 48 h later, this group was sacrificed. Again, lung, but also liver, and spleen were obtained for RNA, protein, and CFUs to determine bacterial load and dissemination (Supplemental Fig. 2). Data are presented as colony forming units (CFU) per gram of organ.
4.7. Immunoblotting
Protein, RNA and DNA were extracted from macrophages using Qiagen AllPrep DNA RNA protein mini kit according to the manufacturer's instructions. (Qiagen #80004). Lung tissue was homogenized using TRIzol (Thermo Fisher Scientific #15596026) followed by protein isolation. Briefly, after phase separation using chloroform, 100% ethanol was added to the interphase/phenol-chloroform layer to precipitate genomic DNA. Subsequently, the phenol-ethanol supernatant was mixed with isopropanol to isolate proteins. The Bradford method was used to determine protein concentrations. Equal amounts of protein were separated by 12% SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane. Membranes were incubated overnight with antibodies against ATG5-ATG12 complex (Sigma Aldrich #A0856), CFTR (Santa Cruz #sc376683) and GAPDH (Cell Signaling Technologies #2118) or Actin (Abcam #ab8226). Corresponding secondary antibodies conjugated with horseradish peroxidase and in combination with enhanced chemiluminescence reagent (Amersham #RPN2209) were used to visualize protein bands. Densitometry analyses were performed by normalizing target protein bands to their respective GAPDH/β-actin loading control using ImageJ software as we previously described [30,34,35,48].
4.8. CFTR channel activity assay
The CFTR channel transports chloride as well as iodide. Therefore, CFTR function was assessed by measuring iodide efflux using the fluorescent and halide-sensitive dye 6-Methoxy-N-(3-sulfopropyl) quinolinium (SPQ) as previously described by our group [35]. Briefly, macrophages were plated in a 96 well-plate to confluency. Cells were loaded with SPQ using hypotonic shock and were incubated with 10 mM SPQ in Opti-MEM/water (1:1) for 15 min at 37 °C. Cells were then washed and incubated twice for 10 min with fluorescence quenching NaI buffer [130 mM NaI, 5 mM KNO3, 2.5 mM Ca(NO3)2, 2.5 mM Mg(NO3)2, 10 mM d-glucose, 10 mM N-(2-hydroxyethyl) piperazine-N′-(2-ethanesulfonic) acid (HEPES, pH 7.4)]. Subsequently, cells were switched to a dequenching isotonic NaNO3 buffer (identical to NaI buffer except that 130 mM NaI was replaced with 130 mM NaNO3) in the presence of 20 μM forskolin and 100 μM 8-(4-chlorophenylthio)-c-AMP to activate CFTR. Non-specific increase in fluorescence was measured by incubating the cells with the activation cocktail and the specific CFTR inhibitor GlyH101 (10 μM). Fluorescence was measured using the plate reader VICTOR X3 (Perkin Elmer) with excitation wavelength at 350 nm and DAPI emission filter. Results are expressed as % increase of fluorescence and represent CFTR-mediated Iodide efflux that was calculated by subtracting total fluorescence (measured in presence of CFTR activators) – fluorescence CFTR-independent (measured in presence of CFTR activators plus inhibitor) as we [35] and others [62] previously published.
4.9. Statistical analyses
Data were analyzed using GraphPad Prism 7. All figures display the mean value and standard error of the mean (SEM) from at least three biologically independent samples and/or experiments. Comparisons between groups were conducted with Student's t-test, one-way, or two-way ANOVA. P-values <.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Daniel M. Layman for his technical assistance. Studies in the Amer laboratory are supported by Ohio State University (OSU) Bridge funds, Public Health Preparedness for Infectious Disease (PHPID), Center for Clinical and Translational Science (CCTS), R21 Al113477, R01 Al24121, and R01 HL127651-01A1 and OSU Comprehensive Cancer Center Support Grant P30 CA016058. Cytokine analysis, using the V-PLEX Proinflammatory Panel 1 (mouse) kit from MSD (MesoScale Diagnostics, LLC), was supported by Award Number Grant UL1TR001070 from the National Center For Advancing Translational Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center For Advancing Translational Sciences of the National Institutes of Health. KC is supported by a Cystic Fibrosis Postdoctoral Research Fellowship. KK is supported by Deutsche Forschungsgemeinschaft (DFG – German Research Foundation). AB is supported by funding from the Egyptian Government. The authors declare they have no conflicts of interest.
Funding
This work was supported by the NIH, National Institute of Allergy and Infectious Diseases (NIAID), [R01AI24121]; National Heart, Lung, and Blood Institute (NHBLI), [R01HL127651-01A1].
Footnotes
Disclosure statement
No potential conflict of interest was reported by the authors.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jcf.2019.01.011.
References
- [1].Ravichandran M, Jurkowska RZ, Jurkowski TP. Target specificity of mammalian DNA methylation and demethylation machinery. Org Biomol Chem 2018;16:1419–35. [DOI] [PubMed] [Google Scholar]
- [2].Tirado-Magallanes R, Rebbani K, Lim R, Pradhan S, Benoukraf T. Whole genome DNA methylation: beyond genes silencing. Oncotarget 2017;8:5629–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res 2005;33:5868–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Magalhaes M, Tost J, Pineau F, Rivals I, Busato F, Alary N, et al. Dynamic changes of DNA methylation and lung disease in cystic fibrosis: lessons from a monogenic disease. Epigenomics 2018;10:1131–45. [DOI] [PubMed] [Google Scholar]
- [5].Magalhaes M, Rivals I, Claustres M, Varilh J, Thomasset M, Bergougnoux A, et al. DNA methylation at modifier genes of lung disease severity is altered in cystic fibrosis. Clin Epigenetics 2017;9:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang S, Stoll G, Pedro J, Sica V, Sauvat A, Obrist F, et al. Evaluation of autophagy inducers in epithelial cells carrying the DeltaF508 mutation of the cystic fibrosis transmembrane conductance regulator CFTR. Cell Death Dis 2018;9:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Izzo V, Pietrocola F, Sica V, Durand S, Lachkar S, Enot D, et al. Metabolic interactions between cysteamine and epigallocatechin gallate. Cell Cycle 2017;16:271–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tosco A, De Gregorio F, Esposito S, De Stefano D, Sana I, Ferrari E, et al. A novel treatment of cystic fibrosis acting on-target: cysteamine plus epigallocatechin gallate for the autophagy-dependent rescue of class II-mutated CFTR. Cell Death Differ 2016; 23:1380–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].De Stefano D, Villella VR, Esposito S, Tosco A, Sepe A, De Gregorio F, et al. Restoration of CFTR function in patients with cystic fibrosis carrying the F508del-CFTR mutation. Autophagy 2014;10:2053–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Donaldson SH, Pilewski JM, Griese M, Cooke J, Viswanathan L, Tullis E, et al. Tezacaftor/Ivacaftor in Subjects with Cystic Fibrosis and F508del/F508del-CFTR or F508del/G551D-CFTR. Am J Respir Crit Care Med 2018;197:214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Welsh MJ, Denning GM, Ostedgaard LS, Anderson MP. Dysfunction of CFTR bearing the delta F508 mutation.J Cell Sci 1993(Suppl. 17):235–9. [PubMed] [Google Scholar]
- [12].Bradbury NA. Intracellular CFTR: localization and function. Physiol Rev 1999;79:S175–91. [DOI] [PubMed] [Google Scholar]
- [13].Abdulrahman BA, Khweek AA, Akhter A, Caution K, Kotrange S, Abdelaziz DH, et al. Autophagy stimulation by rapamycin suppresses lung inflammation and infection by Burkholderia cenocepacia in a model of cystic fibrosis. Autophagy 2011;7:1359–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Luciani A, Villella VR, Esposito S, Brunetti-Pierri N, Medina D, Settembre C, et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat Cell Biol 2010;12:863–75. [DOI] [PubMed] [Google Scholar]
- [15].Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature 2011;469:323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Takahama M, Akira S, Saitoh T. Autophagy limits activation of the inflammasomes. Immunol Rev 2018;281:62–73. [DOI] [PubMed] [Google Scholar]
- [17].Seveau S, Turner J, Gavrilin MA, Torrelles JB, Hall-Stoodley L, Yount JS, et al. Checks and Balances between Autophagy and Inflammasomes during Infection. J Mol Biol 2017;430:174–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Abdulrahman BA, Khweek AA, Akhter A, Caution K, Tazi M, Hassan H, et al. Depletion of the ubiquitin-binding adaptor molecule SQSTM1/p62 from macrophages harboring cftr DeltaF508 mutation improves the delivery of Burkholderia cenocepacia to the autophagic machinery. J Biol Chem 2013;288:2049–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yanagi S, Matsumura K, Marui A, Morishima M, Hyon SH, Ikeda T, et al. Oral pretreatment with a green tea polyphenol for cardioprotection against ischemia-reperfusion injury in an isolated rat heart model.J Thorac Cardiovasc Surg 2011;141:511–7. [DOI] [PubMed] [Google Scholar]
- [21].Liu SM, Ou SY, Huang HH. Green tea polyphenols induce cell death in breast cancer MCF-7 cells through induction of cell cycle arrest and mitochondrial-mediated apoptosis. J Zhejiang Univ Sci B 2017;18:89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhou H, Chen Y, Huang SW, Hu PF, Tang LJ. Regulation of autophagy by tea polyphenols in diabetic cardiomyopathy.J Zhejiang Univ Sci B 2018;19:333–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sergent T, Piront N, Meurice J, Toussaint O, Schneider YJ. Anti-inflammatory effects of dietary phenolic compounds in an in vitro model of inflamed human intestinal epithelium. Chem Biol Interact 2010;188:659–67. [DOI] [PubMed] [Google Scholar]
- [24].Elbling L, Herbacek I, Weiss RM,Jantschitsch C, Micksche M, Gerner C, et al. Hydrogen peroxide mediates EGCG-induced antioxidant protection in human keratinocytes. Free Radic Biol Med 2010;49:1444–52. [DOI] [PubMed] [Google Scholar]
- [25].Du GJ, Zhang Z, Wen XD, Yu C, Calway T, Yuan CS, et al. Epigallocatechin Gallate (EGCG) is the most effective cancer chemopreventive polyphenol in green tea. Nutrients 2012;4:1679–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Xie W, Zhou J. Aberrant regulation of autophagy in mammalian diseases. Biol Lett 2018;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hua Y, Shen M, McDonald C, Yao Q. Autophagy dysfunction in autoinflammatory diseases. J Autoimmun 2018;88:11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fujikake N, Shin M, Shimizu S. Association between Autophagy and Neurodegenerative Diseases. Front Neurosci 2018;12:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tschurtschenthaler M, Adolph TE. The Selective Autophagy Receptor Optineurin in Crohn's Disease. Front Immunol 2018;9:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Khalil H, Tazi M, Caution K, Ahmed A, Kanneganti A, Assani K, et al. Aging is associated with hypermethylation of autophagy genes in macrophages. Epigenetics 2016;11:381–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Collins PL, Oltz EM. Histone methylation keeps the brakes on autophagy. Mol Cell Biol 2013;33:3974–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Baek SH, Kim KI. Epigenetic Control of Autophagy: Nuclear events gain more attention. Mol Cell 2017;65:781–5. [DOI] [PubMed] [Google Scholar]
- [33].Shin HR, Kim H, Kim KI, Baek SH. Epigenetic and transcriptional regulation of autophagy. Autophagy 2016;12:2248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Krause K, Kopp BT, Tazi MF, Caution K, Hamilton K, Badr A, et al. The expression of Mirc1/Mir17-92 cluster in sputum samples correlates with pulmonary exacerbations in cystic fibrosis patients. J Cyst Fibros 2017;17:454–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tazi MF, Dakhlallah DA, Caution K, Gerber MM, Chang SW, Khalil H, et al. Elevated Mirc1/Mir17-92 cluster expression negatively regulates autophagy and CFTR (cystic fibrosis transmembrane conductance regulator) function in CF macrophages. Autophagy 2016;12:2026–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Luciani A, Villella VR, Esposito S, Gavina M, Russo I, Silano M, et al. Targeting autophagy as a novel strategy for facilitating the therapeutic action of potentiators on DeltaF508 cystic fibrosis transmembrane conductance regulator. Autophagy 2012;8:1657–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Walczak M, Martens S. Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation. Autophagy 2013;9:424–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 2007;282:37298–302. [DOI] [PubMed] [Google Scholar]
- [39].Hahn MA, Li AX, Wu X, Pfeifer GP. Single base resolution analysis of 5-methylcytosine and 5-hydroxymethylcytosine by RRBS and TAB-RRBS. Methods Mol Biol 2015;1238:273–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang T, Liu Q, Li X, Wang X, Li J, Zhu X, et al. RRBS-analyser: a comprehensive web server for reduced representation bisulfite sequencing data analysis. Hum Mutat 2013;34:1606–10. [DOI] [PubMed] [Google Scholar]
- [41].Nihal M, Roelke CT, Wood GS. Anti-melanoma effects of vorinostat in combination with polyphenolic antioxidant (−)-epigallocatechin-3-gallate (EGCG). Pharm Res 2010;27:1103–14. [DOI] [PubMed] [Google Scholar]
- [42].Steinmann J, Buer J, Pietschmann T, Steinmann E. Anti-infective properties of epigallocatechin-3-gallate (EGCG), a component of green tea. Br J Pharmacol 2013;168:1059–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bender CM, Pao MM, Jones PA. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res 1998; 58:95–101. [PubMed] [Google Scholar]
- [44].Cheng SH, Rich DP, Marshall J, Gregory RJ, Welsh MJ, Smith AE. Phosphorylation of the R domain by cAMP-dependent protein kinase regulates the CFTR chloride channel. Cell 1991;66:1027–36. [DOI] [PubMed] [Google Scholar]
- [45].Jurkuvenaite A, Varga K, Nowotarski K, Kirk KL, Sorscher EJ, Li Y, et al. Mutations in the amino terminus of the cystic fibrosis transmembrane conductance regulator enhance endocytosis. J Biol Chem 2006;281:3329–34. [DOI] [PubMed] [Google Scholar]
- [46].Armstrong DS, Grimwood K, Carzino R, Carlin JB, Olinsky A, Phelan PD. Lower respiratory infection and inflammation in infants with newly diagnosed cystic fibrosis. BMJ 1995;310:1571–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nixon GM, Armstrong DS, Carzino R, Carlin JB, Olinsky A, Robertson CF, et al. Early airway infection, inflammation, and lung function in cystic fibrosis. Arch Dis Child 2002;87:306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Krause K, Caution K, Badr A, Hamilton K, Saleh A, Patel K, et al. CASP4/caspase-11 promotes autophagosome formation in response to bacterial infection. Autophagy 2018:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tarasewicz A, Debska-Slizien A, Rutkowska B, Szurowska E, Matuszewski M. Efficacy and safety of mammalian target of rapamycin inhibitor use-long-term follow-up of first Tuberous Sclerosis complex patient treated de novo with sirolimus after kidney transplantation: a case report. Transplant Proc 2018;50:1904–9. [DOI] [PubMed] [Google Scholar]
- [50].Easton JB, Houghton PJ. Therapeutic potential of target of rapamycin inhibitors. Expert Opin Ther Targets 2004;8:551–64. [DOI] [PubMed] [Google Scholar]
- [51].Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc 2011;6:468–81. [DOI] [PubMed] [Google Scholar]
- [52].Ferrari E, Monzani R, Villella VR, Esposito S, Saluzzo F, Rossin F, et al. Cysteamine reestablishes the clearance of Pseudomonas aeruginosa by macrophages bearing the cystic fibrosis-relevant F508del-CFTR mutation. Cell Death Dis 2017;8:e2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lu JW, Hsieh PS, Lin CC, Hu MK, Huang SM, Wang YM, et al. Synergistic effects of combination treatment using EGCG and suramin against the chikungunya virus. Biochem Biophys Res Commun 2017;491:595–602. [DOI] [PubMed] [Google Scholar]
- [54].Moyers SB, Kumar NB. Green tea polyphenols and cancer chemoprevention: multiple mechanisms and endpoints for phase II trials. Nutr Rev 2004;62:204–11. [DOI] [PubMed] [Google Scholar]
- [55].Akhter A, Gavrilin MA, Frantz L, Washington S, Ditty C, Limoli D, et al. Caspase-7 activation by the Nlrc4/Ipaf inflammasome restricts Legionella pneumophila infection. PLoS Pathog 2009;5:e1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Abdelaziz DH, Gavrilin MA, Akhter A, Caution K, Kotrange S, Khweek AA, et al. Apoptosis-associatedspeck-like protein (ASC) controls Legionella pneumophila infection in human monocytes. J Biol Chem 2011;286:3203–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Akhter A, Caution K, Abu Khweek A, Tazi M, Abdulrahman BA, Abdelaziz DH, et al. Caspase-11 promotes the fusion of phagosomes harboring pathogenic bacteria with lysosomes by modulating actin polymerization. Immunity 2012;37:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Krueger T, Fisher PL, Becker S, Pontasch S, Dove S, Hoegh-Guldberg O, et al. Transcriptomic characterization of the enzymatic antioxidants FeSOD, MnSOD, APX and KatG in the dinoflagellate genus Symbiodinium. BMC Evol Biol 2015;15:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011;27:1571–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol 2012;13:R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Clancy JP, Bebok Z, Ruiz F, King C, Jones J, Walker L, et al. Evidence that systemic gentamicin suppresses premature stop mutations in patients with cystic fibrosis. Am J Respir Crit Care Med 2001;163:1683–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.