Abstract
Acute myeloid leukemia (AML) is a heterogeneous disease with respect to its genetic and molecular basis and to patients´ outcome. Clinical, cytogenetic, and mutational data are used to classify patients into risk groups with different survival, however, within-group heterogeneity is still an issue. Here, we used a robust likelihood-based survival modeling approach and publicly available gene expression data to identify a minimal number of genes whose combined expression values were prognostic of overall survival. The resulting gene expression signature (4-GES) consisted of 4 genes (SOCS2, IL2RA, NPDC1, PHGDH), predicted patient survival as an independent prognostic parameter in several cohorts of AML patients (total, 1272 patients), and further refined prognostication based on the European Leukemia Net classification. An oncogenic role of the top scoring gene in this signature, SOCS2, was investigated using MLL-AF9 and Flt3-ITD/NPM1c driven mouse models of AML. SOCS2 promoted leukemogenesis as well as the abundance, quiescence, and activity of AML stem cells. Overall, the 4-GES represents a highly discriminating prognostic parameter in AML, whose clinical applicability is greatly enhanced by its small number of genes. The newly established role of SOCS2 in leukemia aggressiveness and stemness raises the possibility that the signature might even be exploitable therapeutically.
Subject terms: Acute myeloid leukaemia, Cancer stem cells
Introduction
Acute myeloid leukemia (AML) is a highly heterogeneous disease in terms of clinical course and outcome: some patients do not tolerate, or are refractory to induction chemotherapy, many achieve a remission but go on to relapse with frequently therapy resistant disease, while yet others attain a permanent remission and long-term survival. AML is organized in a hierarchic manner: leukemic stem cells (LSCs) at the apex of this hierarchy give rise to the bulk of the leukemic cells (LCs), exhibit higher resistance to chemotherapy than the latter, and represent the cellular source of relapse1. The clinical heterogeneity of AML is related to a number of parameters, of which patient age as well as genetic and molecular alterations present in the malignant cells are of particular importance2–5. Numerous recurrent chromosome aberrations and point mutations have been identified in AML, and shown to act as drivers of the disease as well as to have prognostic significance2–6. In addition, AML is associated with extensive alterations of the transcriptome, and, like genetic aberrations, changes in gene expression contribute to leukemogenesis and may represent therapeutic targets3,7–10. With respect to prognostication, transcriptional patterns may even outperform the clinical and genetic parameters commonly used for this purpose11. Accordingly, a number of studies have identified gene expression signatures that were associated with survival, and in several cases could be cross-validated in additional data sets and even used to refine the European Leukemia Net (ELN) score7,12–20, which integrates cytogenetic and mutational information and is the current gold standard for prognostication in AML21. However, some of these signatures were established by applying a prior gene selection process that may impede their general applicability, and most contain too many genes to be realistically implemented in clinical routine.
Suppressor Of Cytokine Signalling 2 (SOCS2) is transcriptionally activated by JAK-STAT signalling and encodes a negative feedback regulator of this pathway, which is aberrantly activated in many cancers including AML22. SOCS2 would therefore be predicted to act as a tumor suppressor, and was indeed down-regulated in several types of solid tumors22. On the other hand, oncogenic roles of SOCS2 were reported, e.g., in colon and prostate cancer23–25. In the healthy murine hematopoietic system, Socs2 was expressed at high levels in stem cells (HSCs) and down-regulated during differentiation26,27. A requirement for Socs2 function in HSCs, however, became apparent only in specific experimental settings like 5-Fluorouracil treatment or a longer series of consecutive transplantations26,27. As for hematopoietic malignancies, SOCS2 expression was significantly increased in patients with chronic myeloid leukemia (CML) in blast crisis as compared to chronic phase patients and healthy controls28, but its absence did not alter the latency or histopathologic features of CML like disease in mice transplanted with BCR-ABL1 transduced bone marrow cells26. A crucial role of JAK-STAT signalling in AML, including in LSCs, is well documented29, but so far, little is known about the role of SOCS2 in this disease.
Here, we report the establishment of a gene expression signature that was composed of 4 genes and consistently associated with survival in 7 cohorts of AML patients with publicly available gene expression and survival data. The top gene in this signature was SOCS2. Experiments using mouse models of AML as well as malignant human myeloid cell lines demonstrated a role of SOCS2 in disease aggressiveness and stemness.
Results
Establishment of a 4-gene expression signature with prognostic value in AML
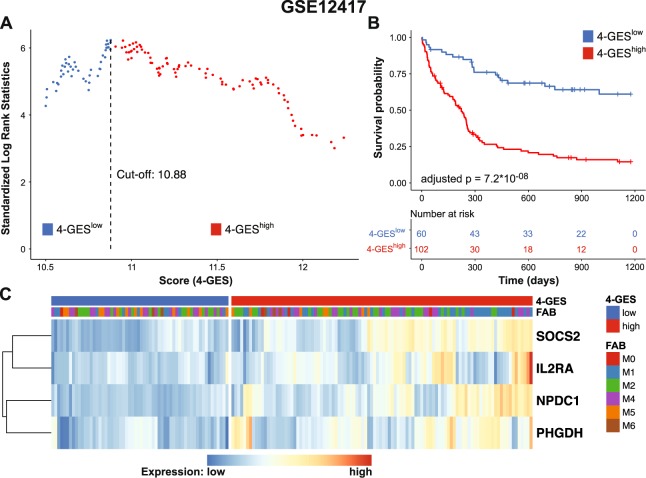
Cohort 1 of data set GSE12417 was used as training set, because it includes patients of all age groups, but is restricted to AML with a normal karyotype, which is the prognostically most heterogeneous of the cytogenetically defined subgroups of AML (Table 1)13. After removal of an MDS sample, gene expression data of 162 cytogenetically normal AML patients remained for model calculation. A forward gene selection was employed and the optimal prognostic model was selected by using the criterion of minimal AIC, an approach to minimize model complexity while maintaining maximum fit of the model to the data (Table 2). This approach resulted in the identification of 4 genes (SOCS2, IL2RA, NPDC1 and PHGDH), whose combined expression values were prognostic of OS in this cohort of patients. A 4-gene expression score (4-GES) was calculated using the expression values and the β-coefficients from a multivariable Cox regression analysis of each of the 4 genes. Based on the resulting score, patients were classified into low risk (N = 60) and high risk (N = 102) groups using maximally selected rank statistics (Fig. 1A). Kaplan Meier analysis and log rank testing revealed that AML patients with a high 4-GES had significantly shorter OS than patients with a low 4-GES (median survival: 223 days vs. not reached, adjusted p = 7.2 * 10−08; Fig. 1B). GSE12417 data were generated using Affymetrix arrays, and on these, many genes are represented by more than one probe set. The forward selection process used to establish the 4-GES picked only one of these per gene. However, all other probe sets for the 4 genes were also prognostic of OS when analysed individually (adjusted p < 0.05 in all cases; Supplementary Fig. S1).
Table 1.
Summary of AML gene expression data sets retrieved from GEO and TCGA/Cancer Browser databases.
| Accession number | Platform | Patients (N)a | Ageb | Cytogenetics |
|---|---|---|---|---|
| GSE12417, cohort 1 | Affymetrix, HG-U133A | 162 | 58 (17–83) | Normal |
| GSE12417, cohort 2 | Affymetrix, HG-U133_Plus_2 | 78 | 62 (18–85) | Normal |
| GSE6891, cohort 1 | Affymetrix, HG-U133_Plus_2 | 222 | 43 (15–60) | Heterogeneous |
| GSE6891, cohort 2 | Affymetrix, HG-U133_Plus_2 | 185 | 45 (17–60) | Heterogeneous |
| GSE37642 | Affymetrix, HG-U133A | 379 | 57 (18–83) | Heterogeneous |
| GSE71014 | Illumina HumanHT-12 V4.0 | 104 | na | Normal |
| TCGA_LAML | RNA-sequencing | 142 | 60 (18–88) | Heterogeneous |
aNumber of patients after exclusion of samples with FAB M3/unknown FAB type or with MDS. bAge in years, median (range). na, not available.
Table 2.
Survival associated gene expression model identified by forward gene selection using GSE12417 cohort 1.
| Probe ID | Gene Symbol | nloglik | AIC | Selected |
|---|---|---|---|---|
| 203373_at | SOCS2 | 452.01 | 910.02 | * |
| 218086_at | NPDC1 | 449.26 | 906.52 | * |
| 211269_s_at | IL2RA | 447.91 | 905.81 | * |
| 201397_at | PHGDH | 444.23 | 900.46 | * |
| 218966_at | MYO5C | 444.23 | 902.45 | |
| 209386_at | TM4SF1 | 443.2 | 902.39 | |
| 203372_s_at | SOCS2 | 443.17 | 904.33 | |
| 201540_at | FHL1 | 441.85 | 903.71 | |
| 211597_s_at | HOPX | 440.17 | 902.33 |
Nloglik, negative log likelihood; AIC, Akaike information criterion; AIC is provided for the prognostic model including the respective gene and all genes above that gene in the list.
Figure 1.
Prognostic value of the 4-GES in 162 cytogenetically normal AML patients (GSE12417, cohort 1). (A) Cut-off value for stratification of AML patients into 4-GESlow (blue) and 4-GEShigh (red) was calculated by maximally selected rank statistics. (B) Kaplan Meier curves for overall survival of 162 AML patients classified as 4-GESlow or 4-GEShigh. Log rank tests were calculated and p-values were adjusted for multiple testing according to Altman et al.58. (C) Heatmap summarizing expression values of SOCS2, IL2RA, NPDC1 and PHGDH in 4-GESlow (blue) and 4-GEShigh (red) AML patients. Blue, low expression; red, high expression.
In a multivariable setting, the 4-GES remained significantly associated with OS after adjusting for patient age (p = 8.8 * 10−08, HR = 3.8; Table 3). The expression pattern of SOCS2, IL2RA, NPDC1, and PHGDH in GSE12417 cohort 1 is shown in Fig. 1C. Application of the 4-GES to GSE12417 cohort 2, which contains samples from 78 patients with AML, yielded similar results as described for cohort 1 (p = 0.035, HR = 4.58 after adjusting for age; Table 3). Overall, these findings demonstrate that high expression of SOCS2, IL2RA, NPDC1 and PHGDH may be of prognostic relevance for AML patients. Thus, we proceeded to validate the model in 5 additional patient cohorts.
Table 3.
Multivariable Cox regression analysis for overall survival of AML patients.
| Data set/cohort | Variable | 4-GES | L-24 | M-7 | W-3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | p-value | HR | 95% CI | p-value | HR | 95% CI | p-value | HR | 95% CI | p-value | ||
| GSE12417/1 | GE score, high vs. low | 3.8 | 2.3–6.2 | 8.8 * 10−08 | 2.25 | 1.5–3.4 | 7.5 * 10−05 | na | Na | na | 1.51 | 1.07–2.14 | 0.021 |
| Age (years) | 1.02 | 1.0–1.0 | 0.006 | 1.03 | 1.0–1.04 | 0.0003 | na | Na | na | 1.03 | 1.01–1.04 | 0.0004 | |
| GSE12417/2 | GE score, high vs. low | 4.58 | 1.1–18.9 | 0.035 | ns | ns | ns | 2.68 | 1.5–4.9 | 0.0016 | 2.01 | 1.1–3.7 | 0.022 |
| Age (years) | 1.04 | 1.0–1.1 | 0.008 | ns | ns | ns | 1.03 | 1.0–1.06 | 0.0063 | 1.03 | 1.0–1.1 | 0.0079 | |
| GSE6891/1 | GE score, high vs. low | 1.69 | 1.2–2.5 | 0.008 | 1.1 | 0.8–1.6 | 0.56 | 2.0 | 1.4–2.8 | 5.3 * 10−05 | ns | ns | ns |
| Cytogenetic riska | 1.91 | 1.5–2.5 | 1.4 * 10−06 | 1.97 | 1.5–2.6 | 3.7 * 10−07 | 1.98 | 1.6–2.5 | 2.9 * 10−08 | ns | ns | ns | |
| CEBPAm (w/m/b) | 0.79 | 0.1–6.4 | 0.828 | 0.84 | 0.1–8.1 | 0.878 | 0.95 | 0.1–6.4 | 0.957 | ns | ns | ns | |
| CEBPAm (yes/no) | 0.94 | 0.0–57.1 | 0.976 | 0.71 | 0.0–61.2 | 0.878 | 0.66 | 0.01–27.5 | 0.83 | ns | ns | ns | |
| EVI1 expression (+/−) | 0.82 | 0.5–1.5 | 0.511 | 0.95 | 0.5–1.7 | 0.854 | 0.89 | 0.5–1.6 | 0.681 | ns | ns | ns | |
| FLT3-ITD | 1.38 | 0.9–2.0 | 0.106 | 1.69 | 1.2–2.5 | 0.006 | 1.82 | 1.3–2.6 | 0.0008 | ns | ns | ns | |
| GSE6891/2 | GE score, high vs. low | 2.24 | 1.5–3.4 | 0.0001 | 1.33 | 0.9–2.0 | 0.173 | 1.7 | 1.1–2.5 | 0.0086 | ns | ns | ns |
| Age (years) | 1.01 | 1.0–1.03 | 0.281 | 1.01 | 1.0–1.03 | 0.128 | 1.01 | 1.0–1.03 | 0.102 | ns | ns | ns | |
| Cytogenetic riska | 1.13 | 0.8–1.6 | 0.528 | 1.22 | 0.9–1.75 | 0.27 | 1.35 | 1.0–1.92 | 0.092 | ns | ns | ns | |
| EVI1 expression (+/−) | 3.73 | 1.8–7.6 | 0.0002 | 2.95 | 1.5–6.0 | 0.0025 | 2.9 | 1.4–5.8 | 0.0028 | ns | ns | ns | |
| GSE37642 | GE score, high vs. low | 2.13 | 1.6–2.8 | 1.8 * 10−07 | 1.49 | 1.2–1.9 | 0.0016 | na | Na | na | ns | ns | ns |
| Age (years) | 1.03 | 1.0–1.04 | 5.6 * 10−12 | 1.03 | 1.0–1.04 | 1.5 * 10−13 | na | Na | na | ns | ns | ns | |
| ELN scoreb | 1.26 | 1.1–1.4 | 7.1 * 10−05 | 1.33 | 1.2–1.5 | 4.1 * 10−07 | na | Na | na | ns | ns | ns | |
| FAB, M1 vs. others | 1.06 | 0.8–1.4 | 0.718 | 0.95 | 0.7–1.3 | 0.745 | na | Na | na | ns | ns | ns | |
| FAB, M4 vs. others | 1.32 | 1.0–1.8 | 0.074 | 1.45 | 1.1–2.0 | 0.017 | na | Na | na | ns | ns | ns | |
| TCGA_LAML | GE score, high vs. low | 1.52 | 1.0–2.3 | 0.051 | 1.3 | 0.8–2.1 | 0.249 | ns | Ns | ns | 2.45 | 1.4–4.3 | 0.002 |
| Age (years) | 1.03 | 1.0–1.1 | 0.00013 | 1.03 | 1.0–1.1 | 8.8 * 10−05 | ns | Ns | ns | 1.04 | 1.0–1.1 | 2.3 * 10−05 | |
| Cytogenetic riska | 1.29 | 0.9–1.9 | 0.179 | 1.25 | 0.8–1.9 | 0.265 | ns | Ns | ns | 1.49 | 1.0–2.1 | 0.033 | |
Parameters provided with the respective data sets were first tested in univariable analyses (Supplementary Table S1); those that resulted as significant were included in the multivariable models, whose results are summarized in this table. 4-GES, 4-gene expression score; L-24, 24-gene expression signature by Li et al.16; M-7, 7-gene expression signature by Marcucci et al.19; W-3, 3-gene expression signature by Wilop et al.20; HR, hazard ratio; CI, confidence interval; GE score, gene expression score; CEBPAm, CEBPA mutation; w, wild type; m, monoallelic; b, biallelic. aAssignment to cytogenetic risk groups were included in the respective GEO entries. bAssignment to ELN risk groups was provided by T. Herold, University of Munich, Department of Internal Medicine III, Munich, Germany. No relevant patient data were provided in GSE71014; therefore, multivariable analyses could not be performed. Significant p-values and corresponding HRs and Cis are indicated in bold letters. na, score could not be calculated because 2 signature genes were not represented on HG-U133A microarrays; ns, no statistical significance found in univariable analyses, thus, no multivariable analyses were performed.
Validation of the 4-GES in patients with cytogenetically heterogeneous AML
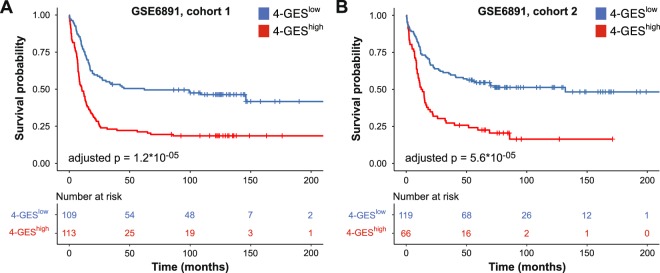
To determine whether the 4-GES has prognostic value also in cytogenetically heterogeneous AML, survival analyses were performed using data set GSE6891, which consists of 2 cohorts of AML patients with variable karyotypes30. Because treatment of patients with AML M3, characterized by the translocation t(15;17), differs substantially from that of all other AML subgroups, the corresponding samples were removed from this and all other cytogenetically heterogeneous data sets. The 4-GES again predicted OS in a highly significant manner (adjusted p = 1.2 * 10−05 and adjusted p = 5.6 * 10−05 for cohorts 1 and 2, respectively; Fig. 2A,B). In both cohorts, the 4-GES was also significant in multivariable analyses including several other prognostic parameters as shown in Table 3. GSE6891 contains information about cytogenetic risk groups. To obtain larger groups, we merged cohorts 1 and 2 and repeated survival analyses separately for the three cytogenetic subgroups. A highly significant prognostic value of the 4-GES was seen for the cytogenetically intermediate subgroup (n = 242, adjusted p = 5.1 * 10−06), but not for the substantially smaller cytogenetically favourable (n = 74) and poor (n = 80) subgroups (Supplementary Fig. S2).
Figure 2.
Validation of the 4-GES in gene expression data set GSE6891, containing cytogenetically heterogeneous AML patients. (A) Cohort 1 (N = 222), (B) cohort 2 (N = 185). Kaplan Meier curves for overall survival of 4-GESlow (blue) and 4-GEShigh (red) patients are shown. Statistical significance was calculated using the log rank test and p-values were adjusted for multiple testing according to Altman et al.58.
The 4-GES is able to refine the ELN risk classification
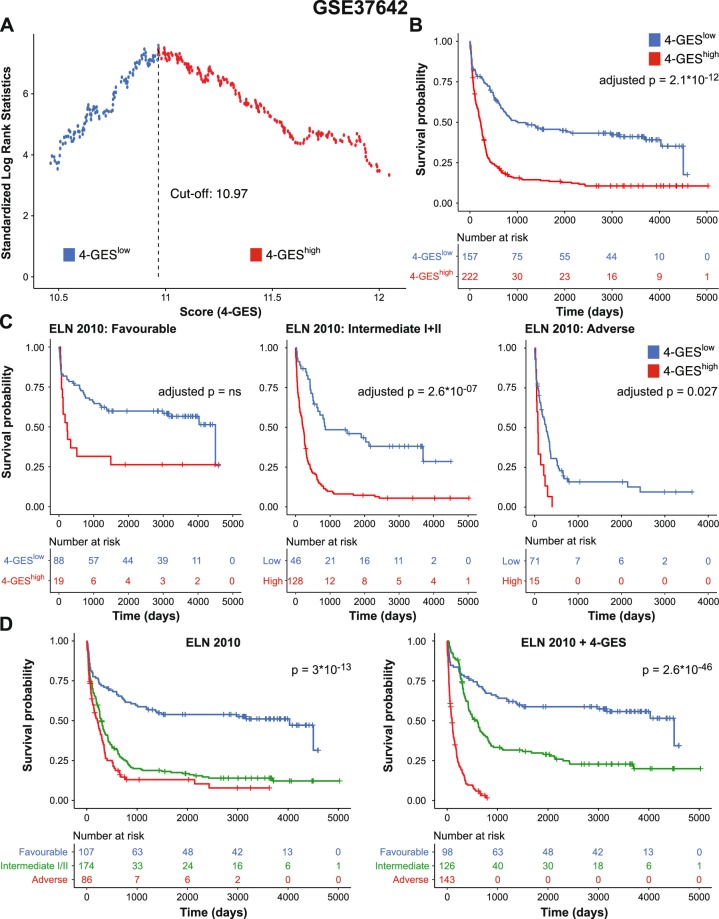
Like GSE6891, GSE37642 contains data from cytogenetically heterogeneous AML16; in addition, ELN risk classification information was available. In a first step, the entire study population was divided into a 4-GEShigh (N = 222) and a 4-GESlow (N = 157) group (Fig. 3A). Patients in the 4-GEShigh group had significantly shorter OS than patients in the 4-GESlow group (adjusted p = 2.1 * 10−12; Fig. 3B). ELN classification was available for 367 patients: 107 were assigned to the favourable risk group, 100 to the intermediate I, 74 to the intermediate II, and 86 to the adverse risk group. Multivariable Cox regression analysis revealed that the 4-GES remained significantly associated with OS after adjusting for patient age and ELN score (adjusted p = 1.8 * 10−07, HR = 2.13; Table 3). Next, we investigated whether the 4-GES was prognostic also within the ELN risk groups. Stratification of the patients within the favourable, intermediate I/II, and adverse groups into 4-GEShigh and 4-GESlow subgroups resulted in a clear identification of patients with shorter OS in the ELN intermediate I/II (adjusted p = 2.6 * 10−07) and adverse (adjusted p = 0.027) groups (Fig. 3C). Among the ELN favourable patients, the difference did not reach statistical significance after adjustment for multiple testing. To ask whether the 4-GES was able to refine the ELN classification, patients from the six groups generated in the previous analysis were combined into three new groups based on median survival: ELN favourable/4-GEShigh patients with low median OS were re-assigned to ELN intermediate risk group, ELN intermediate/4-GEShigh patients with low median OS were re-assigned to ELN adverse risk group, and ELN adverse/4-GESlow patients with high median OS were re-assigned to ELN intermediate risk group. The resulting ELN + 4-GES classification indeed substantially refined the ELN score (p = 2.6 * 10−46 vs. p = 3 * 10−13 for the ELN + 4-GES and the ELN scores, respectively; Fig. 3D and Supplementary Fig. S3A). Similar results were obtained when these analyses were done separately for patients younger or older than 60 years of age (Supplementary Fig. S3B).
Figure 3.
Validation of the 4-GES in gene expression data set GSE37642, consisting of 379 cytogenetically heterogeneous AML patients. (A) Cut-off value for stratification of AML patients into 4-GESlow (blue) and 4-GEShigh (red) was calculated by maximally selected rank statistics. (B) Kaplan Meier curves for overall survival of 379 AML patients classified as 4-GESlow (blue) and 4-GEShigh (red). (C) Kaplan Meier curves for overall survival of 107 ELN favourable, 174 ELN intermediate I/II and 86 ELN adverse risk AML patients stratified into 4-GESlow (blue) and 4-GEShigh (red). Statistical significance was calculated using the log rank test and p-values were adjusted for multiple testing according to Altman et al.58. (D) Kaplan Meier curves for overall survival of AML patients stratified into favourable, intermediate, and adverse based on ELN 2010 classification (left) and ELN 2010 + 4-GES classification (right). ELN, European Leukemia Net.
Validation of the 4-GES in gene expression data sets generated with alternative technologies
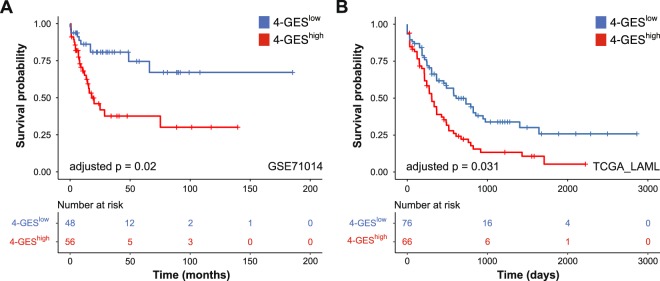
To investigate whether the 4-GES retains prognostic value in AML patients whose samples were analysed on platforms other than Affymetrix microarrays, we performed survival analyses on data sets GSE71014 and TCGA_LAML2,31. GSE71014 consists of 104 cytogenetically normal AML samples, which were analysed using the Illumina BeadArray platform. Forty-eight patients were 4-GESlow and 56 were 4-GEShigh, and the latter had significantly shorter OS (adjusted p = 0.02; Fig. 4A). Multivariable analysis was not possible because no additional potentially prognostic parameters are provided in this data set.
Figure 4.
Validation of the 4-GES in the gene expression data sets (A) GSE71014 and (B) TCGA_LAML. Kaplan Meier curves for overall survival of 4-GESlow (blue) and 4-GEShigh (red) patients are shown. Statistical significance was calculated using the log rank test and p-values were adjusted for multiple testing according to Altman et al.58.
Data set TCGA_LAML contains RNA-sequencing data from 142 AML samples (76 4-GESlow and 66 4-GEShigh), and a high 4-GES was significantly associated with shorter OS (adjusted p = 0.031; Fig. 4B). Multivariable analyses of TCGA_LAML, including the parameters age, cytogenetic risk score, and 4-GES, revealed a HR of 1.52 for the 4-GES, however, statistical significance was just not reached (p = 0.051, Table 3).
Overall, these findings demonstrate that the 4-GES is a highly reliable independent prognostic factor in AML.
Comparison of the 4-GES to other prognostic gene expression signatures
As outlined above, several gene expression based scores have been proposed for the prognostication of AML. To further validate the 4-GES, we therefore compared it to a selected subset of these signatures: the 24-gene signature reported by Li et al.16 (L-24), the 7-gene signature of Marcucci et al.19 (M-7), and Wilop’s 3-gene signature20 (W-3). The L-24 was chosen because it was the first signature that was rigorously validated in several additional data sets and shown to be able to refine the ELN score16. The other two signatures were selected because, like the 4-GES, they contain <10 genes, making them particularly attractive for potential clinical use. In addition, the L-24 and the M-7 were included in a recent comparison of several prognostic scores32 and yielded favourable results. The L-24 and the M-7 divide patients into two prognostic groups, while the W-3 is based on three groups. There is no overlap of genes between these signatures and the 4-GES (Supplementary Table S3). In univariable analyses, the 4-GES was significant in all seven patient cohorts (Supplementary Table S4). The L-24 reached significance in 5/7 cohorts, and the M-7 in 4/5 cohorts (in the remaining two cohorts, it was not applicable because not all signature genes were represented on the array type used). The W-3 performed well in three cohorts, two of which were used to establish it (Supplementary Table S4). In multivariable analyses that included each signature together with all other available prognostic parameters, the 4-GES yielded higher hazard ratios than the other signatures in 4/6 data sets (GSE71014 could not be analyzed because no additional clinical data were provided with it), performed similar to the M-7 in one data set, and was superseded by the W-3 in the TCGA data set, which was part of the training set used to establish this signature (Table 3). In summary, therefore, the 4-GES compared favourably to other prognostic gene expression scores.
SOCS2 promotes AML aggressiveness and stemness
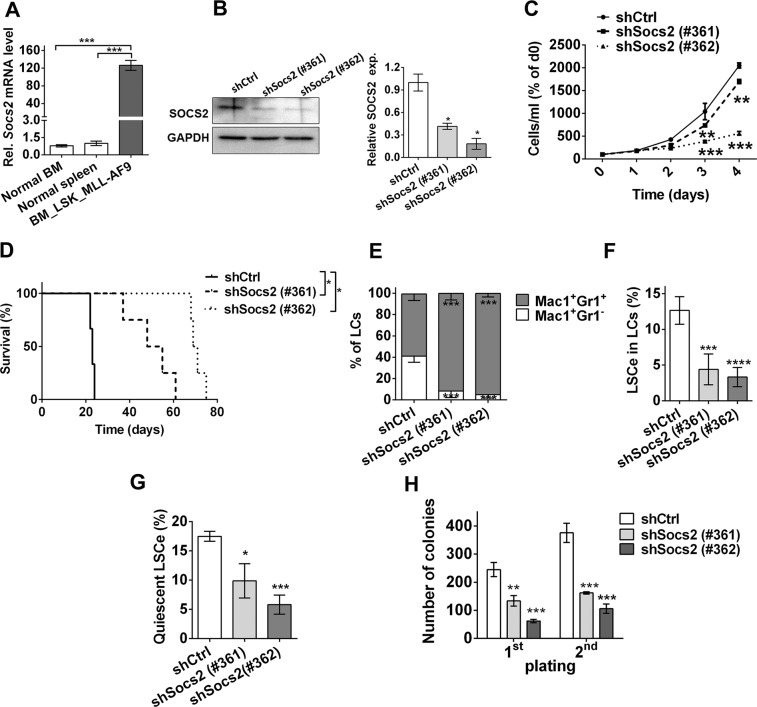
Because the expression of SOCS2 was found to be most strongly associated with OS according to the robust likelihood-based approach, and expression differences between 4-GESlow and 4-GEShigh AML samples were higher for SOCS2 than for the other genes of the model, this gene was subjected to functional analysis. In normal hematopoietic cells SOCS2 levels were high in hematopoietic stem cells and substantially decreased during cell differentiation (Supplementary Fig. S4). To investigate the role of Socs2 in leukemogenesis, a well-established mouse model of human AML driven by the fusion oncogene MLL-AF9 was used33. Transplantation of Lin− Sca-1+ c-Kit+ (LSK) cells transduced with pMSCV_MLL-AF9_IRES_Venus34 into sublethally irradiated C57BL/6 recipient mice led to a rapid-onset AML-like disease (Supplementary Fig. S5) as reported35. Socs2 mRNA expression was highly up-regulated in leukemic cells (LCs) from terminally ill mice compared to normal BM or spleen cells (Fig. 5A). To knock down Socs2, LCs were transduced with two different shRNAs (shSocs2 #361 and #362), expressed in the lentiviral vector pLKO.1_puro_CMV_TagRFP, or with non-target shRNA (shCtrl) as a control. Venus+ RFP+ cells were isolated by flow cytometry, and down-regulation of SOCS2 by the gene specific shRNAs was confirmed by immunoblot analysis (Fig. 5B). Knock-down of Socs2 significantly inhibited growth of primary murine LCs in vitro (Fig. 5C), and delayed disease onset upon transplantation into C57BL/6 recipient mice (Fig. 5D). In LCs from terminally ill mice, it effected a shift from immature (Mac-1+ Gr-1−) to more mature (Mac-1+ Gr-1+) myeloid cells (Fig. 5E). Moreover, experimental down-regulation of Socs2 negatively affected the abundance and functional properties of LSCs. In the MLL-AF9 model, LSCs are strongly enriched in the Lin− Sca1− c-Kit+ CD34+ CD16/CD32hi population35, henceforth termed LSCe. Knock-down of Socs2 caused a decrease of immunophenotypically defined LSCe among Venus+ RFP+ LCs (Fig. 5F), lessened the number of quiescent LSCe (Fig. 5G), and reduced clonogenicity in a serial replating assay (Fig. 5H), indicating that Socs2 enhanced LSC abundance and function in the MLL-AF9 mouse model of AML.
Figure 5.
Effects of Socs2 knock-down in an MLL-AF9 driven mouse model of AML. (A) Socs2 mRNA levels in MLL-AF9+ BM LCs and in BM and spleen cells of healthy mice were determined by qRT-PCR and normalised to those of the housekeeping gene ß-2-microglobulin using the ∆∆CT method. Mean ± SEM of three independent experiments; ***p < 0.001 (Student’s two-tailed t-test). (B) Left panel, immunoblot analysis for SOCS2 expression in shCtrl or shSocs2 transduced MLL-AF9+ LCs. Right panel, quantification of immunoblot results; mean ± SEM of two independent experiments; *p < 0.05 (Student’s two-tailed t-test). (C) shCtrl or shSocs2 transduced MLL-AF9+ LCs were maintained in suspension culture and counted on the indicated days. Mean ± SEM of three independent experiments. **p < 0.01; ***p < 0.001 (2-way ANOVA followed by Bonferroni’s post-hoc test). (D) Kaplan-Meier plot of mice transplanted with shCtrl or shSocs2 transduced MLL-AF9+ LCs (300.000 Venus+ RFP+ cells per mouse, n = 4). **p < 0.01 (log-rank test). (E–H) Analyses of LCs from mice terminally ill after transplantation with shCtrl or shSocs2 transduced MLL-AF9+ LCs. Mean ± SEM of 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 (2-way ANOVA followed by Bonferroni’s post-hoc test). (E) Myeloid differentiation of spleen LCs. Mac-1+ Gr-1−, immature LCs; Mac-1+ Gr-1+, mature LCs. (F) Abundance of LSC enriched cells (LSCe; Lin− cKit+ Sca-1− CD34+ CD16/CD32hi cells) among Venus+ RFP+ spleen LCs. (G) Cell cycle distribution of spleen LSCe was determined by Ki67 and DAPI staining. Quiescent cells were defined as Ki67− cells with a 2n DNA content. (H) Colony formation by BM LCs serially plated into methyl cellulose.
To confirm these observations, a second AML mouse model driven by the combined action of a Flt3 gene with an activating internal tandem duplication (Flt3-ITD) and a mutated nucleophosmin gene (NPM1c) was employed36. As with the MLL-AF9 model, spleen cells from terminally ill Flt3-ITD/NPM1c mice expressed highly elevated levels of Socs2 mRNA compared to normal spleen cells (Supplementary Fig. S6A). Knock-down of Socs2 in Flt3-ITD/NPM1c LCs led to a rapid and complete loss of RFP+ cells in culture (Supplementary Fig. S6B). While this prevented the use of these cells for transplantation experiments, it provided strong evidence for an essential role of Socs2 in the proliferation and/or survival of Flt3-ITD/NPM1c driven LCs. Noteworthy, analyses of expression microarray data from sorafenib or DMSO treated murine pro-B cells with stable FLT3-ITD expression revealed significantly lower expression of Socs2, Il2ra and Phgdh in sorafenib treated cells compared to DMSO controls (Supplementary Fig. S7). Further confirming the role of SOCS2 in leukemic cell proliferation, retroviral expression of this gene in the malignant human myeloid cell lines U937 and HL60 caused significant increases in cell numbers compared to empty vector transduced cells (Supplementary Fig. S8).
Discussion
Several prognostic gene expression signatures for AML patients have been reported, but were established based on a priori assumptions about the identity of the potentially relevant genes, and/or contained relatively large numbers of genes, making them too complex for potential clinical applications7,12–20. We therefore aimed to establish a signature composed of a small number of genes, both whose identity and number were determined through unbiased approaches. As a training set, a cohort of cytogenetically normal patients representing all age groups was used. The resulting signature, 4-GES (consisting of SOCS2, IL2RA, NPDC1, and PHGDH) was confirmed as an independent prognostic parameter in five of six additional data sets, including some that contained cytogenetically heterogeneous patient groups and/or employed different gene expression profiling technologies, and just fell short of significance in the sixth data set. When the 4-GES was applied to distinct cytogenetic subgroups of AML a highly significant prognostic value was observed for the cytogenetically intermediate subgroup, however, for the cytogenetically poor subgroup statistical significance was not reached after correction for multiple testing. Because the cytogenetically poor subgroup was rather small, the 4-GES should be evaluated in a much broader cohort of adverse cytogenetic risk AML patients in future studies. The 4-GES performed as well or better than other rigorously tested prognostic signatures, and was able to substantially refine the ELN classification both in younger (<60 years) and older (≥60 years) patients with AML. Further studies are necessary to develop a standardized, fast and easy to use (multiplex) real-time PCR approach for the 4 genes and housekeepers to stratify AML patients into low and high risk groups prospectively.
At present, risk stratification in AML is based on recurrent genetic alterations. In addition to their prognostic value, these contribute to various aspects of disease pathology, and some of them have been developed as therapeutic targets2–6,37. Some genes whose individual aberrant expression had prognostic significance in AML were also shown to contribute to leukemogenesis9,10, and in a recent report, a gene expression signature associated with LSCs and poor therapy response was used to identify targeted drugs potentially useful for the treatment of AML8. The 4-GES comprises only four genes, making it appear likely that most or all of these also make a functional contribution to AML pathogenesis. To begin to address this question, the role of SOCS2 in AML was investigated using suitable experimental models. SOCS2 was selected because it resulted as the top gene from the survival modeling approach; in addition, it was strongly and significantly up-regulated at relapse of AML, a disease stage that is often refractory to therapy and thus can be considered as inherently aggressive38. SOCS genes are transcriptional targets of activated JAK-STAT signalling22, a pathway that plays a major pro-leukemogenic role in AML, contributes to the growth and maintenance of AML LSCs29, and is being explored as target for rationally designed therapeutics39. SOCS proteins act as substrate recruiting components of E3-ubiquitin ligase complexes and initiate the degradation of cytokine receptors and signaling proteins, thus acting as negative feedback regulators of the cytokine activated pathways leading to their induction22. They would therefore be expected to serve as tumor suppressors, and SOCS2 was indeed down-regulated in several cancer types22. In contrast, SOCS2 was up-regulated in AML compared to healthy controls, and particularly in samples with activating ITD mutations of the tyrosine kinase receptor FLT340. SOCS2 promoted the degradation of activated FLT3, and decreased proliferation and colony formation of cells experimentally expressing FLT3-ITD40; however, these experiments were performed using the murine pro-B cell line Ba/F3, which may not be an ideal model for human AML. In contrast, the data reported here show that experimental expression of SOCS2 had a small but significant growth promoting effect in malignant human myeloid cell lines (Supplementary Fig. S8). Correspondingly, its knock-down in primary cells from the Flt3-ITD/NPM1c driven model of cytogenetically normal AML retarded proliferation and/or promoted cell death or senescence to an extent that precluded further experimentation (Supplementary Fig. S6B). A comparable, albeit less dramatic effect was observed with cells from an MLL-AF9 driven AML model (Fig. 5C). In this model, knock-down of Socs2 also decelerated leukemia onset in mice, promoted LC differentiation, and reduced the abundance, quiescence, and activity of LSCs (Fig. 5D–H), the LC subpopulation that acts as driver of the disease and is responsible for therapy resistance and relapse1. Together with the observation that high SOCS2 expression is associated with a poor prognosis (Supplementary Fig. S1), these data provide strong support for an oncogenic role of this gene in AML. This may reflect a role of SOCS2 not only as a negative regulator of the JAK-STAT pathway, but also as a downstream target of it. Oncogenic roles of SOCS2 were also reported in other tumor entities: SOCS2 levels were elevated in colon and prostate cancer, and high SOCS2 expression was associated with a poor prognosis in the latter23,24. The transcription of SOCS2 was repressed by wild type p53, and it promoted proliferation, anchorage independent growth, resistance against basal and drug induced apoptosis, and tumor growth in xenograft assays in prostate and colon cancer cell lines23–25. Because our data regarding Socs2 expression and growth regulatory effects of Socs2 derived from TP53 wt mice/cells and TP53 mutant human myeloid cells, we suggest that these effects are independent of p53 and TP53 mutations in AML. Other examples of a negative signalling regulator associated with poor prognosis include the dual-specificity phosphatase 6 (DUSP6) which regulates the basal levels of phosphorylated ERK in the cytosol41,42. High DUSP6 expression was found to be associated with poor survival in ALL and breast cancer patients43,44.
As for the other genes in the 4-GES, the interleukin 2 receptor subunit alpha (IL2RA, also named CD25) gene was specifically expressed on LSCs in a subset of patients with AML45, and was previously reported as an independent prognostic parameter able to further improve prognostication based on cytogenetic and mutational data46. An ongoing phase I study of patients with relapsed/refractory CD25-positive AML/ALL treated with the human monoclonal anti-CD25 antibody ADCT-301 (NCT02588092) showed an acceptable safety profile. Data about treatment response are not available yet47. However, an other trial (NCT02432235) tested efficacy of ADCT-301 in relapsed/refractory Hodgkin/Non-Hodgkin lymphoma patients and reported an overall response rate of 56%48.
The neural proliferation, differentiation and control 1 (NPDC1) gene was cloned as a gene associated with contact inhibition of neuronal precursor cells49. It is one of the less intensely studied genes, but, interestingly, was significantly up-regulated at relapse of AML38.
Phosphoglycerate dehydrogenase (PHGDH) catalyzes the rate limiting step of serine synthesis from the glycolytic intermediate 3-phosphoglycerate; serine metabolism plays roles in both nucleotide biosynthesis and antioxidant defense. PHGDH is overexpressed in triple negative breast cancer, lung adenocarcinoma, and other tumors, and is associated with a poor prognosis. It contributes to tumor formation, stemness, metastasis, and chemotherapy resistance50,51, and is actively pursued as a therapeutic target52,53. Its role is far less well studied in leukemia, but serine deprivation and PHGDH inhibition were reported to cooperate to inhibit growth of the malignant human myeloid cell line HL6054.
In summary, we present here a gene expression signature (4-GES) that is a highly significant independent prognostic parameter throughout several independent AML cohorts. A major advantage over previously published signatures is that it is composed of only four genes, which makes its application in routine diagnostics feasible. Moreover, the 4-GES has the potential to refine the ELN classification, which is the current state of the art risk classification of AML. This refinement was associated with re-assignment of substantial numbers of patients into the prognostically poor subgroup. However, as outlined above, the genes constituting the 4-GES are likely to contribute to the pathological features of AML, raising the possibility that the 4-GES may not only be a prognosticator of poor outcome under standard therapy, but even represent a target for rationally designed therapies maybe improving outcome of the subgroup of AML that currently has the poorest prognosis.
Materials and Methods
Publicly available gene expression data sets
Four independent microarray data sets, 2 of which consist of 2 cohorts each, were obtained from the Gene Expression Omnibus (GEO) database, and The Cancer Genome Atlas (TCGA) RNA-seq data were downloaded from the Cancer Browser database (Table 1). For GSE37642, risk classification according to the ELN 2010 score was kindly provided by T. Herold, University of Munich, Department of Internal Medicine III, Munich, Germany. All samples with French American British (FAB) type M3, which mandates a substantially different treatment protocol than used for all other AML patients, and samples with unknown FAB type or myelodysplastic syndrome (MDS) were excluded. To optimize comparability among the microarray data sets, raw Affymetrix data were processed, normalised and log2 transformed using the frozen robust multiarray (fRMA) algorithm and R 3.4.2 software55. Pre-processed Illumina BeadArray data (GSE71014) and RNA-seq data (TCGA_LAML) were used as provided by the databases. Microarray data of DMSO/sorafenib treated Ba/F3-ITD cells and of human hematopoietic cells were obtained from ArrayExpress database (E-MTAB-4487, E-MEXP-1242) and GEO database (GSE42519, GSE17054, GSE19599, GSE11864). Heatmaps were generated on scaled log2 values using ClustVis56.
Prognostic model selection and survival analyses
A prognostic gene expression signature was calculated by applying rbsurv, a robust likelihood-based survival modeling approach57, to cohort 1 of GSE12417. The samples in this data set were randomly distributed between a training and a validation set. Each gene was fitted to the training set and the parameter estimate for the association between expression of this gene and overall survival (OS) was calculated. With the parameter estimate, log likelihood to predict OS was evaluated in the validation set. This procedure was repeated 100 times, yielding 100 log-likelihoods per gene. The gene with the largest mean log likelihood (SOCS2) in the survival model was selected as the first top gene. By evaluating all possible two-gene models, the gene that together with SOCS2 yielded the largest mean log likelihood was selected as the second top gene. This forward gene selection process was continued and resulted in a set of candidate genes for the prognostic model. Finally, an optimal prognostic model was selected based on the minimal Akaike information criterion (AIC) which takes the goodness of the model fit and the complexity (number of genes) into account.
For each patient in a data set, a score was calculated as the sum of the products of log2 transformed expression values of the model genes and their β-coefficients from a multivariable Cox regression, which was performed on cohort 1 of data set GSE12417 and included all genes. Optimal cut-offs for classification of patients into high-risk or low-risk groups were calculated for each data set using maximally selected rank statistics (R package maxstat). The Kaplan Meier method was used to estimate survival distributions and the log-rank test was used to evaluate statistical significance in OS between risk groups. P-values from these analyses were adjusted for multiple testing as described previously58. Uni- and multivariable Cox regressions were calculated using the coxph function in R. Univariate analyses were performed for all parameters provided along with the respective gene expression data (Supplementary Table S1). Only parameters which were significant in univariate analyses were included in multivariable analyses. A p-value of <0.05 was considered statistically significant. The R packages survival and survminer were used for these calculations and for data visualization. All statistical tests were performed using R 3.4.2.
Ethics statement
Animal experiments were approved by the Animal Ethics Committee of the Medical University of Vienna and the Austrian Federal Ministry of Education, Science, and Research (GZ66.009/0309-WF/V/3b/2015). Federation of European Laboratory Animal Science Associations guidelines to minimize animal distress and suffering were followed.
Isolation, culture, transduction, and transplantation of primary murine cells
The packaging cell lines Platinum-E (PlatE) and Phoenix-GP were maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and 1% Penicillin/Streptomycin (Sigma-Aldrich). Primary murine hematopoietic or leukemic cells (LCs) were cultured in IMDM medium (Thermo Fisher Scientific) containing 10% FBS, 1% L-Glutamine (Thermo Fisher Scientific), 50 ng/ml mSCF, 10 ng/ml mIL-3, 10 ng/ml mTPO, 10 ng/ml mFlt3L (all from Peprotech), and 10 ng/ml mIL-6 (Biolegend). To generate mice with MLL-AF9 driven AML, a retroviral transduction/transplantation approach was used. Lin− Sca-1+ c-Kit+ (LSK) cells were isolated from bone marrow (BM) of 6–8 week old C57BL/6 mice (Department of Laboratory Animal Science & Genetics, Himberg, Austria). Retroviral particles were produced by calcium phosphate mediated co-transfection of PlatE cells with pMSCV_MLL-AF9_IRES_Venus34 (kindly provided by Dr. Johannes Zuber, Research Institute of Molecular Pathology, Vienna, Austria) and the ecotropic packaging plasmid psi2 (gag, pol, env). LSK cells (100,000 cells in 500 µl culture medium) were spinoculated with 1500 µl retroviral supernatant in the presence of 4 µg/ml polybrene (Sigma-Aldrich) and cytokines (as in the culture medium) for 1 h at 1300 rpm and 32 °C in a 12-well plate precoated with Retronectin (Takara). The infection was repeated with new retroviral supernatant after 5, 10, and 24 h, followed by a 48 h incubation in culture medium.
To knock down Socs2, two validated Socs2 shRNAs (shSocs2 #361 and #362) and a non-target control shRNA (SHC012) in pLKO.1_puro_CMV_TagRFP (Mission© library, Sigma-Aldrich) were transfected into Phoenix-GP cells, along with the packaging plasmids pSPAX2 and pMDG.2, using calcium phosphate precipitation. Lentiviral particles were harvested after 48–72 h and used for spinoculation of spleen cells from mice with MLL-AF9 or FLT3-ITD/NPM1c36 driven AML as described above. The infection was repeated with fresh lentiviral supernatant after 24 h. Three days later, fluorescence marker positive cells were sorted and used for in vitro assays and/or transplantation of recipient mice.
For transplantation, 6–8 week old female C57BL/6 recipient mice were sub-lethally irradiated (5 Gy), anaesthesized on the next day, and injected retro-orbitally with MLL-AF9-transduced LSK cells (160,000 cell/mouse; unsorted because of the strong selective advantage associated with MLL-AF9 expression) or with shCtrl or shSocs2-transduced MLL-AF9+ LCs (300,000 Venus+ RFP+ cells/mouse). BM and spleen cells were harvested from terminally ill mice for use in downstream experiments.
Flow cytometric assays to analyse LC differentiation and the abundance and quiescence of LSC enriched cells
To analyze the differentiation status of LCs, spleen cells from terminally ill mice were stained with fluorophor labelled antibodies against Gr-1 and Mac-1 (Supplementary Table S2) for 30 minutes on ice, washed once with staining buffer, and subjected to flow cytometry.
In the MLL-AF9 AML model, LSCs are highly enriched in the Lin− Sca1− c-Kit+ CD34+ CD16/CD32hi population35,59; we therefore defined Venus+ RFP+ cells with this immunophenotype as LSC enriched cells (LSCe). LSCe abundance among Venus+ RFP+ LCs was determined by flow cytometric analysis of leukemic spleen cells stained with the respective antibodies (Supplementary Table S2). To determine the proportion of quiescent LSCe, spleen cells from leukemic mice were stained with antibodies against LSCe surface markers, fixed and permeabilised in Cytofix/Cytoperm (BD Biosciences), stained with Ki-67 antibody (Supplementary Table S2) and DAPI (1 µg/mL; Sigma-Aldrich) in Perm/Wash™ Buffer (BD Biosciences), washed with Perm/Wash™ Buffer, and subjected to flow cytometry. Ki-67− LSCe with a 2n DNA content were considered quiescent.
Flow cytometric analyses were performed on an LSR Fortessa SORP (BD Biosciences), and data were analysed with FlowJoX software (Treestar).
Colony formation assay
BM cells from leukemic mice were sorted for Venus+ RFP+ positivity, and 2,000 cells from each genotype were seeded into methyl cellulose (MethoCult GF M3434; Stem Cell Technologies). Total colony formation was quantified after 7 days, and 2,000 cells per condition were used for replating.
Quantitative RT-PCR
Total RNA was extracted using Trizol (Life Technologies) and reverse transcribed using random hexamer primers (Life Technologies) and M-MLV reverse transcriptase (Life Technologies). Quantitative RT-PCR (qRT-PCR) was performed on a Step One Plus Real Time PCR system (Life Technologies) using GoTaq qPCR Master Mix (Promega) and the following primers: Socs2 (fwd: 5′-CTGCGCGAGCTCAGTCAAA-3′, rev: 5′-CAATCCGCAGGTTAGTCGGT-3′), ß-2-microglobulin (fwd: 5′_CCTTCAGCAAGGACTGGTCT-3′, rev: 5′-TGTCTCGATCCCAGTAGACG-3′). Assays were performed in triplicate, and Socs2 expression was normalised to ß-2-microglobulin expression using the ΔΔCT method60.
Immunoblot analysis
Preparation of protein lysates, SDS-PAGE, transfer to PVDF membranes (Hybond-P; Amersham), and antibody incubations were performed using standard procedures. Blots were developed using SuperSignal West Femto or Pico Chemiluminescent Substrate (both from Thermo Scientific) and scanned using a ChemiDoc Touch Imaging System (Bio Rad). Densitometric analysis was performed with Image-J software (National Institutes of Health, Maryland, USA).
Statistical analyses of experimental data
Differences between two independent groups were calculated using Student’s t-test, and differences between multiple groups were determined by 2-way ANOVA followed by Bonferroni’s post-hoc test. The log-rank test was used to evaluate survival differences between groups of mice. p-values < 0.05 were considered statistically significant. Analyses were performed using GraphPad Prism 6 software.
Ethics approval
Animal experiments were approved by the Animal Ethics Committee of the Medical University of Vienna and the Austrian Federal Ministry of Education, Science, and Research (GZ66.009/0309-WF/V/3b/2015). Federation of European Laboratory Animal Science Associations guidelines to minimize animal distress and suffering were followed.
Supplementary information
Acknowledgements
We thank the Acute Myeloid Leukemia Cooperative Group (AMLCG) Munich for providing clinical data to data set GSE37642. Dr. Johannes Zuber, Research Institute of Molecular Pathology, Vienna, Austria, is gratefully acknowledged for kindly providing pMSCV_MLL-AF9_IRES_Venus. This work was funded by the Austrian Science Fund (FWF) through projects P28013 and P28256 to R.W., by a Research Grant of the “Initiative Krebsforschung” of the Medical University of Vienna to G.H., and by a Doc stipend of the Austrian Academy of Sciences to A.G.
Author Contributions
C.H.N., T.G., A.S., K.B. and A.M.G. performed wet lab experiments; G.H., H.H., O.D., J.L.C., G.S.V. and D.S. performed data analyses; R.W., G.H. and C.H.N. wrote the manuscript; R.W., G.H. and S.Z.M. supervised the project team; all authors carefully proof-read the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rotraud Wieser, Email: rotraud.wieser@meduniwien.ac.at.
Gerwin Heller, Email: gerwin.heller@meduniwien.ac.at.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-45579-0.
References
- 1.Wiseman DH, Greystoke BF, Somervaille TC. The variety of leukemic stem cells in myeloid malignancy. Oncogene. 2014;33:3091–3098. doi: 10.1038/onc.2013.269. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research, N. et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liersch R, Muller-Tidow C, Berdel WE, Krug U. Prognostic factors for acute myeloid leukaemia in adults–biological significance and clinical use. Br J Haematol. 2014;165:17–38. doi: 10.1111/bjh.12750. [DOI] [PubMed] [Google Scholar]
- 4.Grimwade D, Ivey A, Huntly BJ. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood. 2016;127:29–41. doi: 10.1182/blood-2015-07-604496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papaemmanuil E, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimwade D, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–365. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 7.Valk PJ, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 8.Laverdiere I, et al. Leukemic stem cell signatures identify novel therapeutics targeting acute myeloid leukemia. Blood Cancer J. 2018;8:52. doi: 10.1038/s41408-018-0087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe-Okochi N, et al. The shortest isoform of C/EBPbeta, liver inhibitory protein (LIP), collaborates with Evi1 to induce AML in a mouse BMT model. Blood. 2013;121:4142–4155. doi: 10.1182/blood-2011-07-368654. [DOI] [PubMed] [Google Scholar]
- 10.Jin G, et al. Trib1 and Evi1 cooperate with Hoxa and Meis1 in myeloid leukemogenesis. Blood. 2007;109:3998–4005. doi: 10.1182/blood-2006-08-041202. [DOI] [PubMed] [Google Scholar]
- 11.Wang M, et al. Development and Validation of a Novel RNA Sequencing-Based Prognostic Score for Acute Myeloid Leukemia. J Natl Cancer Inst. 2018;110:1094–1101. doi: 10.1093/jnci/djy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heuser M, et al. Gene-expression profiles and their association with drug resistance in adult acute myeloid leukemia. Haematologica. 2005;90:1484–1492. [PubMed] [Google Scholar]
- 13.Metzeler KH, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008;112:4193–4201. doi: 10.1182/blood-2008-02-134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentles AJ, Plevritis SK, Majeti R, Alizadeh AA. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA. 2010;304:2706–2715. doi: 10.1001/jama.2010.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eppert K, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 16.Li Z, et al. Identification of a 24-gene prognostic signature that improves the European LeukemiaNet risk classification of acute myeloid leukemia: an international collaborative study. J Clin Oncol. 2013;31:1172–1181. doi: 10.1200/JCO.2012.44.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng SW, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540:433–437. doi: 10.1038/nature20598. [DOI] [PubMed] [Google Scholar]
- 18.Herold T, et al. A 29-gene and cytogenetic score for the prediction of resistance to induction treatment in acute myeloid leukemia. Haematologica. 2018;103:456–465. doi: 10.3324/haematol.2017.178442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcucci G, et al. Epigenetics meets genetics in acute myeloid leukemia: clinical impact of a novel seven-gene score. J Clin Oncol. 2014;32:548–556. doi: 10.1200/JCO.2013.50.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilop S, et al. A three-gene expression-based risk score can refine the European LeukemiaNet AML classification. J Hematol Oncol. 2016;9:78. doi: 10.1186/s13045-016-0308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dohner H, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letellier E, Haan S. SOCS2: physiological and pathological functions. Front Biosci (Elite Ed) 2016;8:189–204. doi: 10.2741/E760. [DOI] [PubMed] [Google Scholar]
- 23.Kim JH, et al. Alterations in the p53-SOCS2 axis contribute to tumor growth in colon cancer. Exp Mol Med. 2018;50:3. doi: 10.1038/s12276-017-0001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoefer J, et al. SOCS2 correlates with malignancy and exerts growth-promoting effects in prostate cancer. Endocr Relat Cancer. 2014;21:175–187. doi: 10.1530/ERC-13-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Misawa A, Takayama K, Urano T, Inoue S. Androgen-induced Long Noncoding RNA (lncRNA) SOCS2-AS1 Promotes Cell Growth and Inhibits Apoptosis in Prostate Cancer Cells. J Biol Chem. 2016;291:17861–17880. doi: 10.1074/jbc.M116.718536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen N, et al. SOCS2 is dispensable for BCR/ABL1-induced chronic myeloid leukemia-like disease and for normal hematopoietic stem cell function. Leukemia. 2013;27:130–135. doi: 10.1038/leu.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vitali C, et al. SOCS2 Controls Proliferation and Stemness of Hematopoietic Cells under Stress Conditions and Its Deregulation Marks Unfavorable Acute Leukemias. Cancer Res. 2015;75:2387–2399. doi: 10.1158/0008-5472.CAN-14-3625. [DOI] [PubMed] [Google Scholar]
- 28.Schultheis B, et al. Overexpression of SOCS-2 in advanced stages of chronic myeloid leukemia: possible inadequacy of a negative feedback mechanism. Blood. 2002;99:1766–1775. doi: 10.1182/blood.V99.5.1766. [DOI] [PubMed] [Google Scholar]
- 29.Cook AM, et al. Role of altered growth factor receptor-mediated JAK2 signaling in growth and maintenance of human acute myeloid leukemia stem cells. Blood. 2014;123:2826–2837. doi: 10.1182/blood-2013-05-505735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verhaak RG, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94:131–134. doi: 10.3324/haematol.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chuang MK, et al. An mRNA expression signature for prognostication in de novo acute myeloid leukemia patients with normal karyotype. Oncotarget. 2015;6:39098–39110. doi: 10.18632/oncotarget.5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang M, et al. Validation of risk stratification models in acute myeloid leukemia using sequencing-based molecular profiling. Leukemia. 2017;31:2029–2036. doi: 10.1038/leu.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kogan SC, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100:238–245. doi: 10.1182/blood.V100.1.238. [DOI] [PubMed] [Google Scholar]
- 34.Zuber J, et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011;25:1628–1640. doi: 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krivtsov AV, et al. Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia. 2013;27:852–860. doi: 10.1038/leu.2012.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mupo A, et al. A powerful molecular synergy between mutant Nucleophosmin and Flt3-ITD drives acute myeloid leukemia in mice. Leukemia. 2013;27:1917–1920. doi: 10.1038/leu.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perl AE. The most novel of the novel agents for acute myeloid leukemia. Curr Opin Hematol. 2018;25:81–89. doi: 10.1097/MOH.0000000000000411. [DOI] [PubMed] [Google Scholar]
- 38.Hackl H, et al. A gene expression profile associated with relapse of cytogenetically normal acute myeloid leukemia is enriched for leukemia stem cell genes. Leuk Lymphoma. 2015;56:1126–1128. doi: 10.3109/10428194.2014.944523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wingelhofer B, et al. Pharmacologic inhibition of STAT5 in acute myeloid leukemia. Leukemia. 2018;32:1135–1146. doi: 10.1038/s41375-017-0005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kazi JU, Ronnstrand L. Suppressor of cytokine signaling 2 (SOCS2) associates with FLT3 and negatively regulates downstream signaling. Mol Oncol. 2013;7:693–703. doi: 10.1016/j.molonc.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ekerot M, et al. Negative-feedback regulation of FGF signalling by DUSP6/MKP-3 is driven by ERK1/2 and mediated by Ets factor binding to a conserved site within the DUSP6/MKP-3 gene promoter. Biochem J. 2008;412:287–298. doi: 10.1042/BJ20071512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piya S, et al. DUSP6 is a novel transcriptional target of p53 and regulates p53-mediated apoptosis by modulating expression levels of Bcl-2 family proteins. FEBS Lett. 2012;586:4233–4240. doi: 10.1016/j.febslet.2012.10.031. [DOI] [PubMed] [Google Scholar]
- 43.Shojaee S, et al. Erk Negative Feedback Control Enables Pre-B Cell Transformation and Represents a Therapeutic Target in Acute Lymphoblastic Leukemia. Cancer Cell. 2015;28:114–128. doi: 10.1016/j.ccell.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menyhart O, et al. DUSP4 is associated with increased resistance against anti-HER2 therapy in breast cancer. Oncotarget. 2017;8:77207–77218. doi: 10.18632/oncotarget.20430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saito Y, et al. Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells. Sci Transl Med. 2010;2:17ra19. doi: 10.1126/scitranslmed.3000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonen M, et al. CD25 expression status improves prognostic risk classification in AML independent of established biomarkers: ECOG phase 3 trial, E1900. Blood. 2012;120:2297–2306. doi: 10.1182/blood-2012-02-414425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldberg AD, et al. Results from an Ongoing Phase 1 Study Indicate ACDT-301 (Camidanlumab Tesirine) Is Well-Tolerated in Patients with Relapsed or Refractory CD25-Positive Acute Leukemia. Blood. 2017;130:2662. [Google Scholar]
- 48.Horwitz SM, et al. Interim Results from a Phase 1 Study of ADCT-301 (Camidanlumab Tesirine) Show Promising Activity of a Novel Pyrrolobenzodiazepine-Based Antibody Drug Conjugate in Relapsed/Refractory Hodgkin/Non-Hodgkin Lymphoma. Blood. 2017;130:1510. [Google Scholar]
- 49.Galiana E, Vernier P, Dupont E, Evrard C, Rouget P. Identification of a neural-specific cDNA, NPDC-1, able to down-regulate cell proliferation and to suppress transformation. Proc Natl Acad Sci USA. 1995;92:1560–1564. doi: 10.1073/pnas.92.5.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samanta D, Semenza GL. Serine Synthesis Helps Hypoxic Cancer Stem Cells Regulate Redox. Cancer Res. 2016;76:6458–6462. doi: 10.1158/0008-5472.CAN-16-1730. [DOI] [PubMed] [Google Scholar]
- 51.Zhang B, et al. PHGDH Defines a Metabolic Subtype in Lung Adenocarcinomas with Poor Prognosis. Cell Rep. 2017;19:2289–2303. doi: 10.1016/j.celrep.2017.05.067. [DOI] [PubMed] [Google Scholar]
- 52.Mullarky E, et al. Identification of a small molecule inhibitor of 3-phosphoglycerate dehydrogenase to target serine biosynthesis in cancers. Proc Natl Acad Sci USA. 2016;113:1778–1783. doi: 10.1073/pnas.1521548113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pacold ME, et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat Chem Biol. 2016;12:452–458. doi: 10.1038/nchembio.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Polet F, et al. Reducing the serine availability complements the inhibition of the glutamine metabolism to block leukemia cell growth. Oncotarget. 2016;7:1765–1776. doi: 10.18632/oncotarget.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA) Biostatistics. 2010;11:242–253. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43:W566–570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho HJ, Yu A, Kim S, Kang J, Hong SM. Robust Likelihood-Based Survival Modeling with Microarray Data. J Stat Softw. 2009;29:1–16. doi: 10.18637/jss.v029.i01. [DOI] [Google Scholar]
- 58.Altman DG, Lausen B, Sauerbrei W, Schumacher M. Dangers of using “optimal” cutpoints in the evaluation of prognostic factors. J Natl Cancer Inst. 1994;86:829–835. doi: 10.1093/jnci/86.11.829. [DOI] [PubMed] [Google Scholar]
- 59.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 60.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262S1046-2023(01)91262-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.