Abstract
The growing threat of antibiotic resistance necessitates the discovery of antibiotics that are active against resistant pathogens. Calcium-dependent antibiotics are a small family of structurally diverse acidic lipopeptides assembled by non-ribosomal peptide synthetases (NRPSs) that are known to display various modes of action against antibiotic-resistant pathogens. Here we use NRPS adenylation (AD) domain sequencing to guide the identification, recovery and cloning of the cde biosynthetic gene cluster from a soil metagenome. Heterologous expression of the cde biosynthetic gene cluster led to the production of cadasides A (1) and B (2), a sub-family of acidic lipopeptides that is distinct from previously characterized calcium-dependent antibiotics in terms of both overall structure and acidic residue rich peptide core. The cadasides inhibit the growth of multidrug-resistant Gram-positive pathogens by disrupting cell wall biosynthesis in the presence of high concentrations of calcium. Interestingly, sequencing of AD domains from diverse soils found that sequences predicted to arise from cadaside-like gene clusters are predominantly found in soils containing high levels of calcium carbonate.
Keywords: calcium-dependent antibiotic, metagenomics, NRPS, eDNA, antibacterial resistance
Graphical Abstract
INTRODUCTION
Antibiotic resistance is an increasingly serious global health concern. To address this impending threat to our healthcare system, new antimicrobials with activity against antibioticresistant pathogens must be identified. Natural products have historically been a rewarding source of new antibiotics.1 One intriguing class of natural product antibiotics is the calciumdependent antibiotics, which are non-ribosomal peptide synthetase (NRPS) encoded acidic lipopeptides that exhibit potent antimicrobial activity in the presence of calcium. They are a promising source of potentially novel antibiotics with activity against antibiotic-resistant pathogens as characterized family members present considerable structural variation2 and disparate antimicrobial mechanisms.3–5 One member, daptomycin, is clinically employed to treat Gram-positive infections caused by methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococci (VRE),6 and another member, friulimicin B, has proceeded to phase I clinical trials.7
The vast majority of bacteria present in the environment are not cultured under standard laboratory conditions and therefore remain inaccessible to traditional culture-based natural product discovery programs. To explore the biosynthetic potential of the soil microbiome in a culture-independent manner, it is possible to extract DNA directly from environmental samples (eDNA, environmental DNA). The extracted eDNA can then be cloned into model cultured bacteria where it can be tested for the ability to encode the production of natural products.8,9 Bioinformatic analysis of NRPS adenylation (AD) domains amplified and sequenced from diverse soils indicates that soil metagenomes likely contain a large number of as yet uncharacterized calcium-dependent antibiotic biosynthetic gene clusters.3 Here we report on the use of large-scale sequencing of AD domains from soil metagenomes to guide the tracking, cloning and heterologous expression of a biosynthetic gene cluster that encodes the cadaside antibiotics. The cadasides are calcium-dependent cyclic acidic lipopeptide antibiotics that disrupt cell wall biosynthesis and show activity against multiple drug-resistant Gram-positive pathogens. They are structurally distinct from known members of this family and contain an unusual arrangement of acidic residues. Interestingly, the presence of cadaside-like biosynthetic gene clusters in the environment correlates with locations predicted to contain high concentrations of calcium, suggesting a unique ecological role for these antibiotics. The discovery of the cadasides adds to the growing body of evidence that culture independent discovery approaches can provide access to novel antibiotics. The systematic application of culture-independent discovery methods to the global soil microbiome provides a means of shedding light on this untapped biosynthetic potential and potentially provides the means of combatting the growing problem of resistance to our current arsenal of antibiotics.
RESULTS AND DISCUSSION
Cloning of the cadaside (cde) gene cluster from the soil metagenome
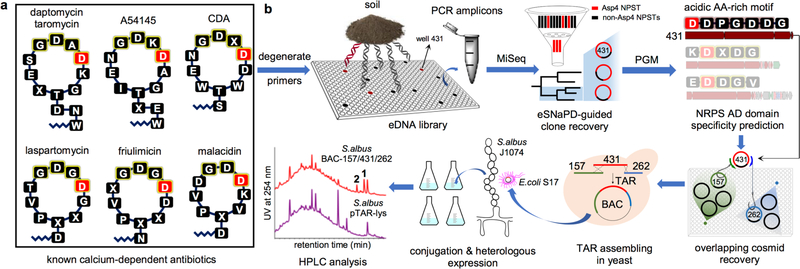
As part of our ongoing culture-independent natural product discovery efforts, we have generated a collection of soil eDNA libraries to use as starting material for the identification of novel biosynthetic gene clusters.3,10–14 Each library contains in excess of 20,000,000 unique cosmid clones which we have empirically shown is sufficient to saturate the biosynthetic diversity present in a soil sample.15 To facilitate the recovery of clones containing biosynthetic gene clusters of interest, each library was partially arrayed in pools of ~25,000 unique cosmid clones, and each library sub-pool was screened with degenerate PCR primers targeting key biosynthetic domains. The resulting PCR amplicons were sequenced to generate Natural Product Sequence Tags (NPSTs) and used to predict the biosynthetic gene cluster content of each library sub-pool. Our pursuit of new calcium-dependent antibiotics started with a search of AD domain NPSTs associated with eight archived soil eDNA libraries. Our analysis of known calcium-dependent antibiotic gene clusters found that AD domains responsible for incorporating the aspartic acid at the fourth position (Asp4) in the ring is the most conserved AD domain sequence among characterized gene clusters. As it is the most conserved AD domain among characterized gene clusters we predicted that it would serve as the most reliable domain to use in an NPST-based search of environmental samples for calcium-dependent antibiotic gene clusters.3
Using the eSNaPD (environmental Surveyor of Natural Product Diversity) software tool,16 which automates the comparison of raw NPST data to domains from functionally characterized biosynthetic gene clusters, AD domain NPSTs generated from each library sub-pool were screened against AD domains from known calcium-dependent antibiotic gene clusters (including daptomycin, friulimicin, CDA, laspartomycin, A54145, taromycin, and malacidin) to identify NPSTs associated with potential calcium-dependent antibiotic gene clusters. NPSTs annotated as being closely related (e-value < 10−40) to a calcium-dependent antibiotic Asp4 domain were used to generate a phylogenetic tree that served as a guide to identify calcium-dependent antibiotic gene clusters (Figure S1). Cosmids associated with representative NPSTs from clades that diverged from those containing reference Asp4 sequences were recovered from the appropriate library sub-pools, sequenced and annotated. As expected, sequencing of the recovered cosmids showed that they each encoded NRPS genes, and specifically contained an AD domain predicted to incorporate an aspartic acid.
The NRPS genes captured on one cosmid of particular interest to us (cosmid DFD0097–431) were bioinformatically predicted to encode the octapeptide Val-Asp-Asp-Pro-Gly-Asp-Asp-Gly, a peptide sequence that is not found in any previously described calcium-dependent antibiotics (Figure 1a). To complete this potentially novel calcium-dependent antibiotic biosynthetic gene cluster, we returned to the AD domain sequences generated for the library from which clone DFD0097–431 was recovered, and looked for wells that contained NPSTs that would indicate the presence of the clones that overlap with DFD0097–431. Two cosmid clones, DFD0097–157 and DFD0097–262, predicted to overlap each end of DFD0097–431 were recovered from the library and sequenced (Figure 1b). These three overlapping cosmid clones were computationally assembled into a single 108.4 kb contig. Annotation of this contig revealed a large NRPS biosynthetic gene cluster that we have called the cadaside or cde gene cluster. BLAST analysis showed that the closest relatives of the cde NRPS genes (cdeI – K) were found in known calcium-dependent antibiotic gene clusters (Figure S2). Based on AD domain amino acid specificity predictions, the thirteen AD domains found in the cde gene cluster are predicted to biosynthesize a peptide with the following sequence: X1-Gly2-Asp3-Tyr4-Thr5-Val6-Asp7-Asp8-Pro9-Gly10-Asp11-Asp12-Gly13. While this peptide is highly divergent from the characterized calcium-dependent antibiotic structures, the fact that it contains multiple acidic residues, which are known to be important for calcium dependence, and that the closest sequenced relatives of the cde NRPS genes encode calcium-dependent antibiotics led us to believe that the metabolite(s) encoded by the cde gene cluster might in fact be a calcium-dependent antibiotic.
Figure 1.
Metagenome-directed antibiotic discovery pipeline. a) Structures of known calcium-dependent antibiotics. b) For this study, DNA was initially extracted directly from soil samples and cloned into E. coli. The resulting metagenomic libraries were arrayed and DNA from each well in the arrayed libraries was used as template for PCR reactions with adenylation (AD) domain specific degenerate primers. Sequenced amplicons were bioinformatically filtered using eSNaPD and Asp4 AD domains from known biosynthetic gene clusters. Cosmids predicted to contain calcium-dependent antibiotic biosynthetic gene clusters were recovered and sequenced from Asp4 positive library wells. One cosmid identified in this analysis, cosmid DFD0097–431 was bioinformatically predicted to encode an acidic amino acid rich NRPS derived peptide that we believed might represent a novel calcium-dependent antibiotic. In a second round of library screening, the complete biosynthetic gene cluster associated with cosmid DFD0097–431 was recovered on two additional overlapping cosmid clones (DFD0097–157 and DFD0097–262). Assembly of these three clones using TAR in yeast and the shuttling of the resulting BAC clone into S. albus led to the heterologous expression of clone specific metabolites 1 and 2.
Heterologous expression and structure elucidation of the cadasides
As a first step toward accessing the metabolite(s) encoded by the cde gene cluster (65.8 kb, Figure 2a and Table S1), the three overlapping eDNA cosmids (DFD0097–431, DFD0097–157, DFD0097–262) containing the cde gene cluster were assembled into a single large fragment of DNA using transformation-associated recombination (TAR) in yeast.17,18 This was done using an Escherichia coli: Streptomyces: yeast bacterial artificial chromosome (BAC) shuttle capture vector (pTAR-lys) containing homology arms flanking the cde gene cluster (Figure 1b). The correct assembly of the cde gene cluster into the BAC (BAC-DFD0097–157/43½62) generated by TAR was confirmed by restriction digest and full sequencing of the assembled product. For heterologous expression, BAC-DFD0097–157/43½62 and the empty pTAR-lys vector were separately conjugated into Streptomyces albus J1074. The resultant strains were fermented in parallel in R5A broth for 10 days and the metabolites were collected from each culture broth using HP-20 resin. The adsorbed metabolites were eluted with methanol and analyzed by LCMS. Two cde gene cluster specific metabolites were identified from the BAC-DFD0097–157/43½62 extract (m/z 780 and 773 [M + 2H]2+) (Figure 1b). These two metabolites, which we called cadasides A (1) and B (2), were purified from 25 liters of S. albus-BAC-DFD0097–157/43½62 culture broth (0.34 and 0.20 mg/L, respectively).
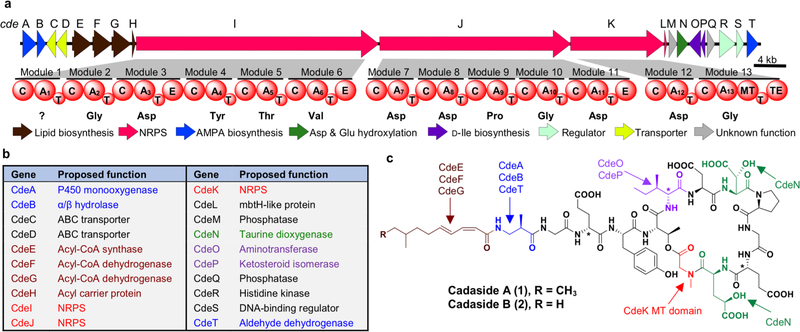
Figure 2.
a) Cadaside (cde) biosynthetic gene cluster. b) Predicted functions for proteins encoded by the cde biosynthetic gene cluster. c) Chemical structures of cadasides A (1) and B (2).
The structures of compounds 1 and 2 were unambiguously determined using the combination of bioinformatic prediction, collision-induced dissociation tandem mass spectrometry (CID-MS/MS), and 1D and 2D high-resolution NMR (see Supplementary Data). The cadasides are cyclic lipopeptides consisting of 13 amino acids, nine of which make the macrocycle. The four exocyclic amino acids are coupled to a medium-chain fatty acid through the N-terminus of the peptide. Based on HRESI(+)MS data (Figure S3) the predicted molecular formulas for 1 and 2 (C69H99N13O28 and C68H97N13O28, respectively) support the final structures shown in Figure 2c. The amino acid sequences of cadasides A (1) and B (2) are identical. They each contain four non-proteinogenic amino acids: 3-amino-2-methylpropionic acid (AMPA), D-isoleucine (D-Ile), β-hydroxy-aspartic acid (β-hyAsp), and γ-hydroxy-glutamic acid (γ-hyGlu). Interestingly, both γ-hyGlu and AMPA have rarely been seen in nonribosomal peptides characterized using culture-based methods.19–24 Advanced Marfey’s analysis of the cadaside A (1) acid hydrolysate revealed that Glu3, Ile6 and Glu11 are D-amino acids while the remaining alpha amino acids are in L-form (Table S3 and Figure S8). The absolute configuration of AMPA was assigned to be R based on the comparison of 2,3,4,6-tetra-O-acetyl-β-D-glucopyranosyl isothiocyanate (GITC) derivatized cadaside A hydrolysate with GITC derivatives of AMPA standards (Table S4 and Figure S9). Finally, the lipid chain was elucidated as 8-methyldeca-2,4-dienoic acid (MDDA) in 1 and 8-methylnona-2,4-dienoic acid (MNDA) in 2 with two conjugated double bonds at cis-Δ2 and trans-Δ4 on the basis of diagnostic coupling constants (J2,3 = 11.4 Hz and J4,5 = 15.3 Hz) and NOE correlations (Figure S4).
The cde gene cluster and predicted biosynthesis of the cadasides
In a BLASTn search, most genes in the cde gene cluster (e.g., NRPS, regulator, transporter, fatty acid biosynthesis, etc.) returned top hits that corresponded to homologs in biosynthetic gene clusters known to encode calcium-dependent antibiotics. Genes exclusive to the cde cluster were primarily predicted to encode for the biosynthesis of the non-proteinogenic amino acid AMPA and D-Ile (Figures S2 and S10), which are not seen in other calcium-dependent antibiotics. A detailed analysis of the predicted functions of the cde genes (Figure 2b and Table S1) supports the analytically defined cadaside structures. Based on an NRPS domain analysis, the cadasides are predicted to follow an orthodox colinear extension model of modular NRPS systems. In our proposed biosynthetic scheme (Figure S10), cadaside biosynthesis is initiated by loading of a fatty acid onto the N-terminal AMPA by CdeH, followed by sequential incorporation of 12 amino acids by the 12 modules present in CdeI, J and K. N-methylation of Gly13 is predicted by the presence of a methyltransferase (MT) domain in the second module of CdeK. Cyclization between the hydroxyl on Thr5 and the terminal carboxylate is predicted to occur via the thioesterase (TE) domain on CdeK. Overall, the structure of the cadaside peptide correlates well with the predicted AD specificities of CdeI – K (X1-Gly2-Asp3-Tyr4-Thr5-Val6-Asp7-Asp8-Pro9-Gly10-Asp11-Asp12-Gly13). Discrepancies between the spectroscopically determined structures and this bioinformatic prediction occur at amino acids 3, 6, 11 and 12. Glutamic acid or glutamic acid derivatives were found at positions 3, 11, and 12 instead of aspartic acid, and D-isoleucine was found at the sixth position instead of the predicted valine. These minor discrepancies are not unexpected based on the accuracy of existing AD domain amino acid specificity prediction algorithms.25–29 The absence of an empirical prediction for AMPA1 was not surprising as it has only been seen in a small number of previously characterized NRPs.
The biosynthesis of the four non-proteinogenic amino acids found in the cadasides can be rationalized based on the predicted functions of genes found in the cde gene cluster. Based on a comparison to other gene clusters that encode AMPA,20,30 as well as the characterized AMPA biosynthesis pathways in eukaryotes31–37 and bacteria,38,39 AMPA is predicted to arise from thymine through the action of a dehydrogenase encoded by cdeT, a monooxygenase encoded by cdeA, and a hydrolase encoded by cdeB (Figure S10).32,34,39 To test this hypothesis, we fed 13CH3-labeled thymine to cultures of S. albus transformed with BAC-DFD0097–157/43½62. We compared the cadaside produced by these cultures to that produced by control cultures supplemented with unlabeled thymine. MS analysis of the cadaside from these cultures revealed ~30% enrichment of the peak corresponding to a single 13C incorporation (m/z 780.34) in 13CH3-thymine fed cultures. Comparison of the MS/MS fragmentation pattern of the m/z 780 peak from both culture conditions confirmed 13C incorporation at the AMPA residue of cadaside (Figure S11). The putative 2-ketoglutaratedependent dioxygenase, CdeN, is predicted to be responsible for hydroxylation of Asp and Glu to give β-hyAsp and γ-hyGlu, respectively.40 D-Ile is predicted to arise from L-Ile through action of an aminotransferase CdeO and an isomerase CdeP followed by epimerization by the epimerase (E) domain in Module 6.41 Finally, the acyl-CoA synthase CdeE in conjunction with two putative acyl-CoA dehydrogenases CdeF and CdeG are predicted to carry out the biosynthesis of the unsaturated fatty acid that acylates the N-terminus of the cadasides (Table S1 and Figure S10).3,42,43
Antimicrobial activity and mode of action of the cadasides
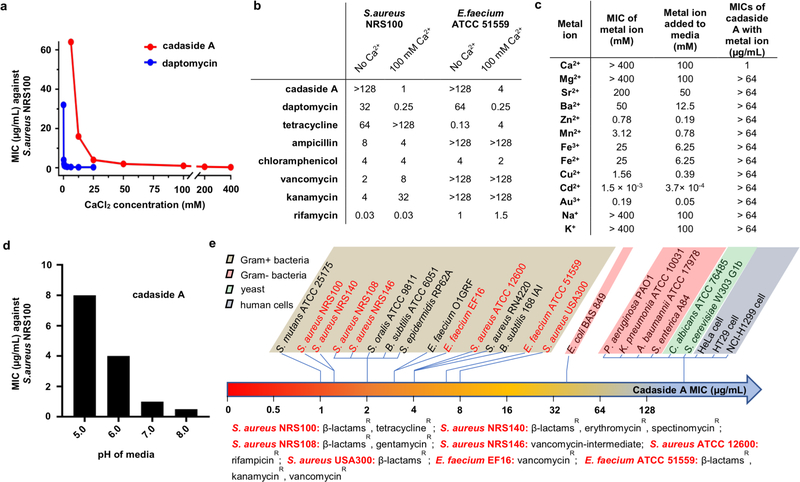
Although structurally distinct from known natural products and devoid of the DXDG motif classically associated with calcium-dependent antibiotics, the cadasides, like other antibiotics in this family, are rich in acidic residues (Figure 2c). We therefore tested cadaside A for antibiosis activity against multidrug-resistant Staphylococcus aureus NRS100 across a wide range of calcium concentrations (0 – 400 mM). As with other calcium-dependent antibiotics, cadaside A only exhibited antibiosis activity in the presence of calcium (Figure 3a). Compared to daptomycin, cadaside A requires more calcium to achieve its maximum antibacterial potency. No other antibiotics we tested showed significantly enhanced antibiosis in the presence of comparable high levels of calcium (Figure 3b). At the highest concentrations, we tested (0.25× the MIC of each cation), no other cations induced cadaside A antibiosis (Figure 3c). As might be expected for chelation of calcium by acidic residues in the peptide, the potency of cadaside A increased under basic conditions (Figure 3d). Cadasides A and B are active against all Gram-positive bacteria we tested with minimum inhibitory concentrations (MICs) in the low micromolar range (Figure 3e). Interestingly, cadaside A was active against Gram-positive bacteria that are resistant to mechanistically diverse, clinically relevant antibiotics, including tetracycline, β-lactams, erythromycin, spectinomycin, rifampicin and vancomycin (Figure 3e). At the highest concentration tested cadaside A was not toxic to human cells (Figure 3e).
Figure 3.
a) Antibacterial activity of cadaside is dependent on calcium. b) Effect of 100 mM calcium on the potency of diverse antibiotics. Concentrations are reported in μg/mL, whereby 1 μg/mL is equal to 0.64 μM for cadasides A and B. c) Ability of different cations to induce cadaside antibiosis. d) pH dependence of cadaside antibiosis. e) Cadaside A spectrum of activity.
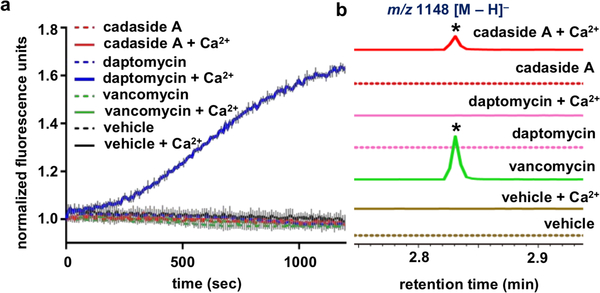
Known calcium-dependent antibiotics induce antibiosis by either disrupting membrane integrity5 or inhibiting cell wall biosynthesis through the sequestration of peptidoglycan intermediates.3,4 In neither the presence nor the absence of calcium did we observe cadaside A induced membrane leakage (Figure 4a) or depolarization (Figure S12), suggesting antibiosis is not the result of disrupting membrane integrity. Antibiotics that cause antibiosis by sequestration of peptidoglycan intermediates result in the accumulation of the early cell wall biosynthetic intermediate undecaprenyl-N-acetylmuramic acid-pentapeptide (UDP-MurNAc-pentapeptide), which can be easily detected by LCMS analysis of antibiotic treated cultures. As seen with other antibiotics that interfere with peptidoglycan biosynthesis, LCMS analysis of S. aureus cultures exposed to cadaside A showed the accumulation of UDP-MurNAc-pentapeptide (Figure 4b), indicating that cadaside A inhibits cell wall (peptidoglycan) biosynthesis. The exact mechanism by which this inhibition occurs necessitates further investigation. While the high level of calcium required for cadaside antibiosis would likely prevent their use systemically, they could prove useful as topical antibiotics. Inhibition of cell wall biosynthesis has proved to be a therapeutically robust antibacterial mode of action.44 Two of the most clinically significant antibiotic classes to arise from culture-based screening programs, the glycopeptides and the β-lactams, inhibit cell wall biosynthesis. The discovery of the cadasides suggests that the full diversity of natural cell wall biosynthesis inhibitors has not yet been discovered and that this therefore remains a promising area to explore for the identification of novel biomedically relevant natural product antibiotics.
Figure 4.
a) Effect of antibiotics on S. aureus membrane permeability as measured by SYTOX green nucleic acid stain in the presence and absence of 100 mM calcium. b) Effect of antibiotics on the accumulation of UDP-MurNAc-pentapeptide in S. aureus cultures both in the presence and absence of 100 mM calcium.
Comparison of the cadasides with known calcium-dependent antibiotics
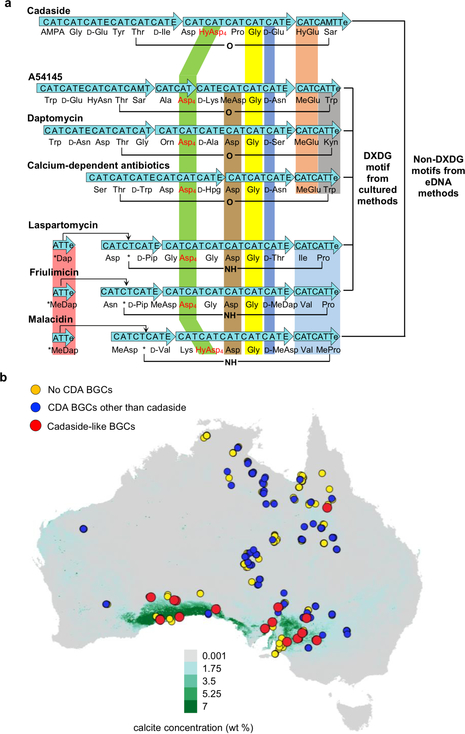
A comparison of known calcium-dependent antibiotics shows that within this family of antibiotics the amino acids at the C-terminus are more conserved than those at the N-terminus (Figure 5a). Interestingly, the previously recognized DXDG motif is not present in either acidic lipopeptide antibiotic we have uncovered using culture-independent methods (i.e., cadaside, malacidin). The only features that are absolutely conserved across all family members are that: 1) the third residue from the C-terminus is a D-amino acid, 2) the fourth residue from the C-terminus is Gly, and 3) the fourth residue in the ring is an aspartic acid derivative. Based on the crystal structure of the laspartomycin C-calcium-geranyl phosphate ternary complex,45 a D-configured amino acid appears to be critical to avoid steric interference of the amino acid side chain with coordination of calcium by the carbonyl of the neighboring Gly.
Figure 5.
a) Comparison of calcium-dependent antibiotic biosynthetic gene clusters and the metabolites they encode. b) DNA was extracted from soils collected at 397 sites across Australia and the presence of calcium-dependent antibiotic biosynthetic gene clusters in soils was predicted based on AD domain sequencing. Each collection site is superimposed on a map of Australia that was color coded according to the predicted environmental calcium carbonate concentration (green for high calcium, grey for low calcium).55 At this scale an individual spot can represent multiple different collection sites. If a calcium-dependent antibiotic AD domain sequence was found at any overlapping site, the spot at that site was colored to represent this observation.
Based on conserved peptide sequence and cyclization pattern, previous calcium-dependent antibiotics can be categorized into two general groups: (Group I) daptomycin/taromycin, calcium-dependent antibiotic and A54145, which cyclize via an ester bond; (Group II) laspartomycin, friulimicin and malacidin, which are cyclized through an amide bond (Figure 5a). Interestingly, these structural distinctions correlate with differences in mode of action. In those cases where mode of action has been determined, Group I antibiotics disrupt the cell membrane,5,46–48 while Group II antibiotics inhibit cell wall biosynthesis.3,4,45,49,50 The cadasides do not cleanly fit into either category. By conserved peptide sequence and cyclization pattern, they more closely resemble Group I antibiotics; however, they inhibit cell wall biosynthesis like Group II antibiotics. Based on these differences and the absence of the classic DXDG sequence, we believe that the cadasides represent a distinct sub-group of calcium-dependent antibiotics.
Iron-binding ability of the cadasides
The cadasides contain five acidic amino acid residues (Glu3, Asp7, β-hyAsp8, Glu11 and γ-hyGlu12), the most among any described natural acidic lipopeptides. Though β-methyl-glutamic acid is commonly seen in calcium-dependent antibiotics, γ-hyGlu is unique to the cadasides. In fact, the cadasides are the only calcium-dependent antibiotics with more than one hydroxy acid moiety. As hydroxy acids are commonly found in siderophores,51,52 the appearance of multiple hydroxy acids in the cadasides led us to explore the possibility that they might also bind iron. Using the chrome azurol S assay,53 a general test used for detecting iron binding by siderophores, we found that cadaside A chelates iron while daptomycin, which does not contain hydroxy acid residues, does not (Figure S13). Although affinity of cadaside A for iron appears to be weaker than that of other siderophores we tested, it clearly has the ability to bind iron. While the biological relevance of the cadasides’ low affinity for iron remains to be determined, this observation suggests that there could be an evolutionary link between calcium dependence by calcium-dependent antibiotics and iron chelation, which is much more commonly observed in bacterial metabolites. In future systematic studies of gene clusters in the global metagenome, it will be interesting to explore whether the cadasides potentially represent a transition state between iron binding and calcium-dependent natural products.
Environmental calcium and cde-like gene clusters
The high levels of calcium required for cadaside antibiosis led us to explore whether the presence of cadaside-like gene clusters in soil metagenomes might correlate with environments containing elevated levels of calcium. In past studies we have mapped natural product biosynthetic gene clusters present in soil metagenomes across many areas of the globe.3,10,12,54 One of the largest uniformly analyzed collections of soil from which we have generated AD sequence tags is a set of 397 samples collected across Australia.54 Using eSNaPD we mapped NPSTs generated from each Australian sample to Asp4 domains found in calcium-dependent antibiotic biosynthetic gene clusters. In this analysis, we used all biosynthetic gene clusters known to encode calcium-dependent antibiotics as well as two eDNA-derived partial gene clusters that were predicted to encode products similar in structure to the cadasides (Figure 2c). Figure 5b shows the location of Australian soil samples containing NPSTs that were associated by eSNaPD with calcium-dependent antibiotic Asp4 domains. We then superimposed on this map modeled calcium carbonate concentrations in sediments across Australia. Cadaside NPST-containing soil samples are almost exclusively located in regions with the highest levels of calcium carbonate. In contrast, the NPSTs of other known calcium-dependent antibiotics are widely distributed across the continent irrespective of the abundance of calcium in the sample. Interestingly, carbonate-rich soil is normally alkaline with pH ranging from 8.0 to 9.5, which correlates to the pH at which the cadasides are most active (Figure 3d).55 Although these observations are correlative in nature, they suggest that the cadasides may have evolved in a narrow niche to play a defense role for their producing organisms.
CONCLUSION
The cadasides represent a new sub-class of calciumdependent antibiotics that differ both in overall structure and acidic residue rich motif from any previously characterized members in this family. While antibiotic resistance is a growing problem, the discovery of the cadasides is further evidence of the existence of a large untapped reservoir of antibiotics in the global soil microbiome. Systematic metagenome sequencing and targeted heterologous expression of biosynthetic gene clusters bioinformatically predicted to encode antibiotics provides a means of accessing these and other classes of antibiotics that have, until now, remained hidden in the global soil microbiome. Cadaside-like gene clusters appear to have been selectively enriched in high calcium environments where we predict they likely play a specific ecological role as defensive metabolites for bacteria that reside in these environments. The observation that cadaside-like gene clusters are found in a narrow ecological range further suggests the global soil microbiome may contain an even larger collection of niche-specific antibiotics whose discovery will require a very systematic survey of ecologically diverse environments using specific niche-relevant assay conditions.
MATERIALS AND METHODS
Screening for Asp4 NPSTs of calcium-dependent antibiotics
For this study, we used eight previously archived eDNA cosmid libraries constructed from soils collected in the United States. The construction of these libraries has been described in detail previously.3,10–14 Each library containing > 20,000,000 unique eDNA cosmid clones hosted in E. coli was arrayed as 5,000 to 25,000 sublibrary pools.3,10–14 To identify sub-library pools containing calciumdependent antibiotic gene clusters, all libraries were screened by PCR using barcoded AD domain degenerate primers A3F and A7R (Table S6).10,56 Methods for primer barcoding, PCR conditions, PCR programs, MiSeq sequencing, and sequence data processing have been described in detail previously.57 Using the eSNaPD (environmental Surveyor of Natural Product Diversity) software package,16 the resultant amplicon sequences were screened to identify those related to the Asp4 AD domain from calcium-dependent antibiotics. eDNA AD domains that matched an Asp4 domain present in any known calcium-dependent antibiotic gene clusters (i.e., daptomycin, friulimicin, calcium-dependent antibiotic, laspartomycin, A54145, taromycin, and malacidin) at an e-value < 10−40 were considered hits.
Recovery of clones containing calcium-dependent antibiotic biosynthetic gene clusters
The library well locations for targeted AD domain sequences were identified using well-specific barcodes incorporated into the degenerate primers.16 Specific primers targeting each unique sequence of interest were designed manually. Single cosmid clones of interest were recovered from wells of interest using serial dilution whole-cell PCR.3 Recovered cosmid clones were sequenced using an IonTorrent PGM sequencer (Life Technologies) and reads were assembled into contigs using Newbler 2.6 (Roche).58 The resulting contigs were analyzed bioinformatically (NRPSPredictor2,26 Stachelhaus,27 pHMM-based method,28 and SANDPUMA29) to predict the amino acid specificity of each AD domain. The most common amino acid prediction across algorithms was used in our gene cluster analysis.
Recovery of overlapping clones containing the cde gene cluster
Library wells containing clones overlapping the cosmid DFD0097–431 were identified using PCR primers designed to target the upstream and downstream edges of the cosmid insert. Cosmids DFD0097–157 and DFD0097–262 were recovered from library wells that were positive for the upstream and downstream primer pairs, respectively. The recovered overlapping cosmids were sequenced and assembled in silico with DFD0097–431 into a single continuous fragment using Geneious (version 11.0.3). The assembly of the three cosmids (DFD0097–157/43½62) was again analyzed bioinformatically to predict AD domain specificities.
Transformation-associated recombination (TAR) assembly
The three overlapping cosmids, DFD0097–157, DFD0097–431, and DFD0097–262 that contain the cde gene cluster were assembled into a bacterial artificial chromosome (BAC) using transformation-associated recombination (TAR) in yeast.17,18 Using the primer pairs (cde-157-UPS_Fw/cde-157-UPS_Rv, and cde-262-DWS_Fw2/cde-262-DWS_Rv2) described in Table S6, 1,153 bp upstream (UPS) and 1,116 bp downstream (DWS) homology arms were amplified from the cosmids DFD0097–157 and DFD0097–262, respectively. Gel-purified amplicons (Qiagen) and SphI linearized pTAR-lys shuttle vector were joined in a standard In-Fusion cloning reaction (Clontech) to yield a pathway-specific capture vector to use in TAR.
For TAR assembly, 200 ng of each DraI-cut cosmid (DFD0097–157, DFD0097–431, and DFD0097–262) and 100 ng of the PmeI-digested pathway-specific capture vector were co-transformed into 200 μL of Saccharomyces cerevisiae CRY1−2 spheroplasts prepared according to published methods.18 Transformed spheroplasts were overlaid onto synthetic complete (SC-lys) top agar (1 M sorbitol, 1.92 g/L synthetic complete lysine dropout supplement, 6.7 g/L yeast nitrogen base, 20 g/L glucose, and 2.5% agar) depleted of lysine. Plates were incubated at 30 °C until colonies appeared (~72 h). Ten yeast colonies were picked and grown overnight in 3 mL of SC-lys dropout liquid media without lysine. BAC vectors were miniprepped from each culture using zymolyase assisted cell lysis (Zymo Research). Each BAC construct was screened using sets of PCR primers spaced at 10 kb intervals across the cde gene cluster. A BAC that tested positive for all primer pairs was electroporated into E. coli EPI300 (Epicenter). E. coli EPI300 colonies were again screened with all sets of primers to identify the correct construct. BAC-DFD0097–157/43½62 was isolated from one PCR positive E. coli colony. The sequence of this BAC was confirmed by PGM-sequencing. The resulting data is deposited in GenBank under the accession number MK060022.
Heterologous expression and chemical screening
BAC-DFD0097–157/43½62 and the empty pTAR-lys vector control were separately transformed into E. coli S17–1. The resulting E. coli strains were used to independently integrate each vector into the chromosome of Streptomyces albus J1074 via intergenic conjugation. Exconjugants were selected using 50 μg/mL apramycin. Spore suspensions of the recombinant strains were inoculated into 50 mL of trypticase soy broth (TSB) and shaken (200 rpm) for 48 h at 30 °C. 400 μL of each seed culture was transferred into 150 mL baffled flasks containing 40 mL of R5A broth (100 g/L sucrose, 10 g/L D-glucose, 5 g/L yeast extract, 10.12 g/L MgCl2·6H2O, 0.25 g/L K2SO4, 0.1 g/L casamino acids, 21 g/L MOPS, 2 g/L NaOH, 5.88 mg/L CaCl2, 80 μg/L ZnCl2, 400 μg/L FeCl3·6H2O, 20 μg/L MnCl2, 20 μg/L CuCl2, 20 μg/L Na2B4O7·10H2O, 20 μg/L (NH4)6Mo7O24·4H2O, pH = 6.85). Cultures were shaken (200 rpm) at 30 °C for 10 days. After 10 days, mycelia were removed by centrifugation at 5,000 ×g for 10 min. 2 g of Diaion HP-20 resin was added to the supernatant (~35 mL). After overnight incubation, the resin was collected by filtration. The resin was subsequently washed with 50 mL of H2O, and the captured metabolites were eluted with 10 mL of methanol. After evaporation of the methanol each dried extract was dissolved in 500 μL of 50% aqueous methanol and analyzed by UPLC-MS (Acquity UPLC BEH C18, 2.1 × 50 mm, 1.7 μm, 130 Å, 0.6 mL/min gradient elution from 95% H2O/MeCN to MeCN over 10 min, with a constant 0.1% formic acid; positive and negative ionization modes).
Scale-up cultivation, extraction and isolation of cadasides A (1) and B (2)
625 individual 150 mL baffled flasks containing 40 mL of R5A broth were inoculated with S. albus J1074 containing BAC-DFD0097–157/43½62 as described above. After 10 days of shaking, the cultures (25 L) were combined and the mycelia were removed by centrifugation. 1 kg of Diaion HP-20 resin was added to the supernatant and this slurry was shaken overnight, after which the resin was collected with cheese cloth and dried at 30 °C for 24 h. The dried resin was packed in a column, washed with 2 L of H2O, and eluted with 3 L of methanol.
The methanolic elution was concentrated in vacuo at 30 °C and adsorbed onto C18 reversed phase silica gel. The crude extract was initially partitioned by medium-pressure liquid chromatography (100 g Gold HP C18 column, 20 min elution gradient from 95% H2O/MeOH to MeOH with 0.1% formic acid, 60 mL/min). Fractions were analyzed by UPLC-MS (Acquity UPLC BEH C18, 2.1 × 50 mm, 1.7 μm, 130 Å, 0.6 mL/min gradient elution from 95% H2O/MeCN to MeCN over 10 min, with a constant 0.1% formic acid; positive and negative ionization modes) and those containing the cadasides were combined. The combined fractions (226.6 mg) were subjected to Sephadex LH-20 chromatography eluted with methanol. Subsequently, the combined cadaside-containing subfraction (103.3 mg) from the Sephadex LH-20 column was subjected to HPLC chromatography (XBridge Prep C18, 10 × 150 mm, 5 μm, 130 Å, 3.5 mL/min gradient elution from 80% to 50% H2O/MeCN over 45 min, with a constant 0.1% formic acid) to afford cadasides A (1) (tR = 31.5 min) and B (2) (tR = 27.5 min). Finally, cadasides A (1, 8.5 mg) and B (2, 5.0 mg) were obtained as ammonium salts by a second round of HPLC chromatography (XBridge Prep C18, 10 × 150 mm, 5 μm, 130 Å, 3.5 mL/min isocratic elution at 90% H2O/MeCN over 5 min, with a constant 0.02% ammonium hydroxide).
Structural determination by NMR and MS/MS
NMR spectra of cadasides A (1) and B (2) were acquired in CD3OD on a Bruker Avance DMX600 NMR spectrometer with a 5 mm CPTCI 1H-13C/15N/D Z-GRD Z75812/0050 cryoprobe at 25 °C. The HPLC-ESI-HRMS-MS/MS spectra of 1 and 2 were collected on a Thermo Scientific LTQ Orbitrap XL Hybrid Ion Trap-Orbitrap Mass Spectrometer connected to a Dionex UltiMate 3000 HPLC system (Thermo Acclaim 120 RP-C18 column, 2.1 × 150 mm, 5 μm, 0.2 mL/min gradient elution from 99% to 60% H2O/MeCN over 12.4 min, then 60% to 10% H2O/MeCN over 1 min, and finally 10% H2O/MeOH for 3 min, with a constant 0.1% formic acid). The ESI source parameters were set as follows: capillary temperature 300 °C, sheath gas flow 8 units, positive polarity, and source voltage 3.9 kV. For the MS-MS CID spectra, a normalized collision energy of 35% was used. The acquired MS/MS data were analyzed using Thermo Xcalibur 2.0.7 software and mass peaks were annotated manually.
Determination of absolute configuration of cadasides by chiral derivatization
0.9 mg of cadaside A (1) was hydrolyzed in 600 μL of 6 M HCl at 115 °C for 10 h. After being dried in vacuo, the resulting hydrolysate was resuspended in 500 μL of H2O and dried again. This process was repeated three times to remove excess acid. The hydrolysate was divided into three portions (3 × 300 μg) for chemical derivatization with 1-fluoro-2,4-dinitrophenyl-5-L-alanine amide (L-FDAA), 1-fluoro-2,4-dinitrophenyl-5-D-alanine amide (DFDAA),59 and 2,3,4,6-tetra-O-acetyl-β-D-glucopyranosyl isothiocyanate (GITC).60 For FDAA derivatization, two separate aliquots (300 μg) of hydrolysate were treated with 100 μL of 1 M NaHCO3 solution and 100 μL of either L- or D-FDAA (1% solution in acetone) at 40 °C for 1 h. The reaction was then neutralized with addition of 150 μL of 1 M HCl and diluted with 150 μL of 50% MeCN/H2O. The final sample was analyzed by HPLC-ESI-HRMS analysis (Thermo Acclaim 120 RP-C18 column, 2.1 × 150 mm, 5 μm, 0.2 mL/min elution gradient from 80% to 40% H2O/MeCN with 0.1% formic acid over 40 min; positive ion modes). For GITC derivatization, 300 μg of hydrolysate was incubated with 100 μL of GITC (1% solution in acetone) and 100 μL of 6% triethylamine at room temperature for 20 min. The reaction was quenched by adding 100 μL of 5% acetic acid and dried in vacuo. The resulting collection of GITC derivatives was resuspended in 100 μL of MeOH and analyzed by HPLC (XBridge C18, 4.6 × 150 mm, 5 μm, 130 Å, 1.0 mL/min isocratic elution at 80% H2O/MeCN with 0.1% formic acid over 60 min). The L-, D-FDAA and GITC derivatives were detected by either UV or extracted ion chromatograms (EICs). The absolute configurations of amino acid residues in 1 were established by comparing the retention times of derivatized amino acids with those of amino acid standards.
Supplementary Material
ACKNOWLEDGMENT
We thank J. Fernandez and H. Molina (The Proteomics Resource Center, The Rockefeller University) for HPLC-MS and MS/MS acquisition, J. Chu (The Rockefeller University) for cytotoxicity assay, and N. Antonovsky and Z. Wang for valuable discussion. The raw data for calcium carbonate abundance across Australia was kindly provided by Dr. John Wilford (Geoscience, Australia) and Dr. Elisabeth Bui (CSIRO Land and Water, Australia). Australian soil sample were provided by Dr. Ben Sparrow (TERN Ausplots, University of Adelaide, Australia). This work was supported by NIH grants U19AI109713 and R35GM122559. C.W. was supported by the Rubicon grant (019.163LW.018) from The Netherlands Organization for Scientific Research.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Bioassay methods, structure elucidation description, tabulated 1D NMR data, NMR, HRMS and MS/MS spectra, HPLC chromatogram of Marfey’s and GITC analyses, spectrum of activity, isotope feeding experiment, cell membrane depolarization assay, CAS assay, and gene cluster comparison (PDF).
S.F.B. is a founder of Lodo Therapeutics.
REFERENCES
- (1).Newman DJ; Cragg GM Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod 2016, 79 (3), 629–661. [DOI] [PubMed] [Google Scholar]
- (2).Strieker M; Marahiel MA The Structural Diversity of Acidic Lipopeptide Antibiotics. ChemBioChem 2009, 10 (4), 607–616. [DOI] [PubMed] [Google Scholar]
- (3).Hover BM; Kim SH; Katz M; Charlop-Powers Z; Owen JG; Ternei MA; Maniko J; Estrela AB; Molina H; Park S; et al. Culture-Independent Discovery of the Malacidins as Calcium-Dependent Antibiotics with Activity against Multidrug-Resistant Gram-Positive Pathogens. Nat. Microbiol 2018, 3 (4), 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Schneider T; Gries K; Josten M; Wiedemann I; Pelzer S; Labischinski H; Sahl HG The Lipopeptide Antibiotic Friulimicin B Inhibits Cell Wall Biosynthesis through Complex Formation with Bactoprenol Phosphate. Antimicrob. Agents Chemother 2009, 53 (4), 1610–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Müller A; Wenzel M; Strahl H; Grein F; Saaki TNV; Kohl B; Siersma T; Bandow JE; Sahl H-G; Schneider T; et al. Daptomycin Inhibits Cell Envelope Synthesis by Interfering with Fluid Membrane Microdomains. Proc. Natl. Acad. Sci 2016, 113 (45), E7077–E7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Steenbergen JN; Alder J; Thorne GM; Tally FP Daptomycin: A Lipopeptide Antibiotic for the Treatment of Serious Gram-Positive Infections. J. Antimicrob. Chemother 2005, 55 (3), 283–288. [DOI] [PubMed] [Google Scholar]
- (7).Butler MS Natural Products to Drugs: Natural Product-Derived Compounds in Clinical Trials. Nat. Prod. Rep 2008, 25 (3), 475–516. [DOI] [PubMed] [Google Scholar]
- (8).Katz M; Hover BM; Brady SF Culture-Independent Discovery of Natural Products from Soil Metagenomes. J. Ind. Microbiol. Biotechnol 2016, 43 (2–3), 129–141. [DOI] [PubMed] [Google Scholar]
- (9).Handelsman J; Rondon MR; Brady SF; Clardy J; Goodman RM Molecular Biological Access to the Chemistry of Unknown Soil Microbes: A New Frontier for Natural Products. Chem. Biol 1998, 5 (10), r245–r249. [DOI] [PubMed] [Google Scholar]
- (10).Owen JG; Charlop-Powers Z; Smith AG; Ternei MA; Calle PY; Reddy BVB; Montiel D; Brady SF Multiplexed Metagenome Mining Using Short DNA Sequence Tags Facilitates Targeted Discovery of Epoxyketone Proteasome Inhibitors. Proc. Natl. Acad. Sci 2015, 112 (14), 4221–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Chang FY; Ternei MA; Calle PY; Brady SF Discovery and Synthetic Refactoring of Tryptophan Dimer Gene Clusters from the Environment. J. Am. Chem. Soc 2013, 135 (47), 17906–17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Chang F-Y; Ternei MA; Calle PY; Brady SF Targeted Metagenomics: Finding Rare Tryptophan Dimer Natural Products in the Environment. J. Am. Chem. Soc 2015, 137 (18), 6044–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Kang HS; Brady SF Arimetamycin A: Improving Clinically Relevant Families of Natural Products through Sequence-Guided Screening of Soil Metagenomes. Angew. Chemie - Int. Ed 2013, 52 (42), 11063–11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Brady SF Construction of Soil Environmental DNA Cosmid Libraries and Screening for Clones That Produce Biologically Active Small Molecules. Nat. Protoc 2007, 2 (5), 1297–1305. [DOI] [PubMed] [Google Scholar]
- (15).Kim JH; Feng Z; Bauer JD; Kallifidas D; Calle PY; Brady SF Cloning Large Natural Product Gene Clusters from the Environment: Piecing Environmental DNA Gene Clusters Back Together with TAR. Biopolymers 2010, 93 (9), 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Reddy BV ija B; Milshteyn A; Charlop-Powers Z; Brady SF ESNaPD: A Versatile, Web-Based Bioinformatics Platform for Surveying and Mining Natural Product Biosynthetic Diversity from Metagenomes. Chem. Biol 2014, 21 (8), 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Kunes S; Botstein D; Fox MS Transformation of Yeast with Linearized Plasmid DNA. Formation of Inverted Dimers and Recombinant Plasmid Products. J. Mol. Biol 1985, 184 (3), 375–387. [DOI] [PubMed] [Google Scholar]
- (18).Kallifidas D; Brady SF Clusters, Reassembly of Functionally Intact Environmental DNA-Derived Biosynthetic Gene. Methods Enzymol. 2012, 517, 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Biggins JB; Kang HS; Ternei MA; Deshazer D; Brady SF The Chemical Arsenal of Burkholderia Pseudomallei Is Essential for Pathogenicity. J. Am. Chem. Soc 2014, 136 (26), 9484–9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Ninomiya A; Katsuyama Y; Kuranaga T; Miyazaki M; Nogi Y; Okada S; Wakimoto T; Ohnishi Y; Matsunaga S; Takada K Biosynthetic Gene Cluster for Surugamide A Encompasses an Unrelated Decapeptide, Surugamide F. ChemBioChem 2017, 18 (17), 1770. [DOI] [PubMed] [Google Scholar]
- (21).Zhang S; Qiu Y; Kakule TB; Lu Z; Xu F; Lamb JG; Reilly CA; Zheng Y; Sham SWS; Wang W; et al. Identification of Cyclic Depsipeptides and Their Dedicated Synthetase from Hapsidospora Irregularis. J. Nat. Prod 2017, 80 (2), 363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Magarvey NA; Beck ZQ; Golakoti T; Ding Y; Huber U; Hemscheidt TK; Abelson D; Moore RE; Sherman DH Biosynthetic Characterization and Chemoenzymatic Assembly of the Cryptophycins. Potent Anticancer Agents from Cyanobionts. ACS Chem. Biol 2006, 1 (12), 766–779. [DOI] [PubMed] [Google Scholar]
- (23).Shiono Y; Tsuchinari M; Shimanuki K; Miyajima T; Murayama T; Koseki T; Laatsch H; Funakoshi T; Takanami K; Suzuki K Fusaristatins A and B, Two New Cyclic Lipopeptides from an Endophytic Fusarium Sp. J. Antibiot. (Tokyo) 2007, 60 (5), 309–316. [DOI] [PubMed] [Google Scholar]
- (24).Bao J; Zhang XY; Xu XY; He F; Nong XH; Qi SH New Cyclic Tetrapeptides and Asteltoxins from Gorgonian-Derived Fungus Aspergillus Sp. SCSGAF 0076. Tetrahedron 2013, 69 (9), 2113–2117. [Google Scholar]
- (25).Challis GL; Ravel J; Townsend CA Predictive, Structure-Based Model of Amino Acid Recognition by Nonribosomal Peptide Synthetase Adenylation Domains. Chem. Biol 2000, 7 (3), 211–224. [DOI] [PubMed] [Google Scholar]
- (26).Röttig M; Medema MH; Blin K; Weber T; Rausch C; Kohlbacher O NRPSpredictor2 - A Web Server for Predicting NRPS Adenylation Domain Specificity. Nucleic Acids Res. 2011, 39 (SUPPL. 2), 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Stachelhaus T; Mootz HD; Marahiel MA The Specificity-Conferring Code of Adenylation Domains in Nonribosomal Peptide Synthetases. Chem. Biol 1999, 6 (8), 493–505. [DOI] [PubMed] [Google Scholar]
- (28).Minowa Y; Araki M; Kanehisa M Comprehensive Analysis of Distinctive Polyketide and Nonribosomal Peptide Structural Motifs Encoded in Microbial Genomes. J. Mol. Biol 2007, 368 (5), 1500–1517. [DOI] [PubMed] [Google Scholar]
- (29).Chevrette MG; Aicheler F; Kohlbacher O; Currie CR; Medema MH SANDPUMA: Ensemble Predictions of Nonribosomal Peptide Chemistry Reveal Biosynthetic Diversity across Actinobacteria. Bioinformatics 2017, 33 (20), 3202–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Sørensen JL; Sondergaard TE; Covarelli L; Fuertes PR; Hansen FT; Frandsen RJN; Saei W; Lukassen MB; Wimmer R; Nielsen KF; et al. Identification of the Biosynthetic Gene Clusters for the Lipopeptides Fusaristatin A and W493 B in Fusarium Graminearum and F. Pseudograminearum. J. Nat. Prod 2014, 77 (12), 2619–2625. [DOI] [PubMed] [Google Scholar]
- (31).Van Gennip, A. H.; Van Bree-blom, E. J.; Nicolaas GGM; Van Erven, A. J.; Vofite PA Beta-Aminoisobutyric Acid as a Marker of Thymine Catabolism in Malignancy. 2002, 165, 1–13. [DOI] [PubMed] [Google Scholar]
- (32).van Kuilenburg ABP; Stroomer AEM; van Lenthe H; Abeling NGGM; van Gennip AH New Insights in Dihydropyrimidine Dehydrogenase Deficiency: A Pivotal Role for Beta-Aminoisobutyric Acid? Biochem. J 2004, 379 (1), 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Gartler SM A Metabolic Investigation of Urinary β-Aminoisobutyric Acid Excretion in Man. Arch. Biochem. Biophys 1959, 80 (2), 400–409. [Google Scholar]
- (34).Fink K; Cline RE; Henderson RB; Fink RM Metabolism of Thymine by Rat Liver in Vitro. J. Biol. Chem 1956, 221, 425–433. [PubMed] [Google Scholar]
- (35).Fink K Excretion of Pyrimidine Reduction Products by the Rat. J. Biol. Chem 1956, 218, 9–14. [PubMed] [Google Scholar]
- (36).Fink RM; McGaughey C; Cline RE; Fink K Metabolism of Intermediate Reduction Products in Vitro. J. Biol. Chem 1956, 218, 1–7. [PubMed] [Google Scholar]
- (37).Fink RM; Fink K; Henderson RB Beta-Amino Acid Formation by Tissue Slices Incubated with Pyrimidines. J. Biol. Chem 1953, 201, 349–355. [PubMed] [Google Scholar]
- (38).Vogels GD; van der Drift C Degradation of Purines and Pyrimidines by Microorganisms. Bacteriol. Rev 1976, 40 (4), 963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Kim S; West TP Pyrimidine Catabolism in Pseudomonas Aeruginosa. FEMS Microbiol Lett 1991, 61 (2–3), 175–179. [DOI] [PubMed] [Google Scholar]
- (40).Kohonen T; Curry B; Davies F; Phillips P; Evans M Mechanistic and Structural Basis of Stereospecific Cβ-Hydroxylation in Calcium-Dependent Antibiotic, a Daptomycin-Type Lipopeptide. ACS Chem. Biol 2007, 2, 187–196. [DOI] [PubMed] [Google Scholar]
- (41).Li Q; Qin X; Liu J; Gui C; Wang B; Li J; Ju J Deciphering the Biosynthetic Origin of L-allo-Isoleucine. J. Am. Chem. Soc 2016, 138 (1), 408–415. [DOI] [PubMed] [Google Scholar]
- (42).Heinzelmann E; Berger S; Müller C; Härtner T; Poralla K; Wohlleben W; Schwartz D An Acyl-CoA Dehydrogenase Is Involved in the Formation of the Δcis3 Double Bond in the Acyl Residue of the Lipopeptide Antibiotic Friulimicin in Actinoplanes Friuliensis. Microbiology 2005, 151 (6), 1963–1974. [DOI] [PubMed] [Google Scholar]
- (43).McCafferty DG; Cudic P; Frankel BA; Barkallah S; Kruger RG; Li W Chemistry and Biology of the Ramoplanin Family of Peptide Antibiotics. Biopolymers 2002, 66 (4), 261–284. [DOI] [PubMed] [Google Scholar]
- (44).Müller A; Klöckner A; Schneider T Targeting a Cell Wall Biosynthesis Hot Spot. Nat. Prod. Rep 2017, 34 (7), 909–932. [DOI] [PubMed] [Google Scholar]
- (45).Kleijn LHJ; Vlieg HC; Wood TM; Sastre Toraño J; Janssen BJC; Martin NI A High-Resolution Crystal Structure That Reveals Molecular Details of Target Recognition by the Calcium-Dependent Lipopeptide Antibiotic Laspartomycin C. Angew. Chemie - Int. Ed 2017, 56 (52), 16546–16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Lake JH; Ptak M Fluorescence Indicates a Calcium-Dependent Interaction between the Lipopeptide Antibiotic LY146032 and Phospholipid Membranes. Biochemistry 1988, 27 (13), 4639–4645. [DOI] [PubMed] [Google Scholar]
- (47).Lakey JH; Lea EJ; Rudd BA; Wright HM; Hopwood DA A New Channel-Forming Antibiotic from Streptomyces Coelicolor A3(2) Which Requires Calcium for Its Activity. J Gen Microbiol 1983, 129 (12), 3565–3573. [DOI] [PubMed] [Google Scholar]
- (48).Zhang TH; Taylor SD; Palmer M; Duhamel J Membrane Binding and Oligomerization of the Lipopeptide A54145 Studied by Pyrene Fluorescence. Biophys. J 2016, 111 (6), 1267–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Kleijn LHJ; Oppedijk SF; T Hart P; Van Harten RM; Martin-Visscher LA; Kemmink J; Breukink E; Martin NI Total Synthesis of Laspartomycin C and Characterization of Its Antibacterial Mechanism of Action. J. Med. Chem 2016, 59 (7), 3569–3574. [DOI] [PubMed] [Google Scholar]
- (50).Rubinchik E; Schneider T; Elliott M; Scott WRP; Pan J; Anklin C; Yang H; Dugourd D; Müller A; Gries K; et al. Mechanism of Action and Limited Cross-Resistance of New Lipopeptide MX-2401. Antimicrob. Agents Chemother 2011, 55 (6), 2743–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Robertson AW; McCarville NG; Macintyre LW; Correa H; Haltli B; Marchbank DH; Kerr RG Isolation of Imaqobactin, an Amphiphilic Siderophore from the Arctic Marine Bacterium Variovorax Species RKJM285. J. Nat. Prod 2018, 81 (4), 858–865. [DOI] [PubMed] [Google Scholar]
- (52).Kreutzer MF; Kage H; Nett M Structure and Biosynthetic Assembly of Cupriachelin, a Photoreactive Siderophore from the Bioplastic Producer Cupriavidus Necator H16. J. Am. Chem. Soc 2012, 134 (11), 5415–5422. [DOI] [PubMed] [Google Scholar]
- (53).Schwyn B; Neilands JB Universal Chemical Assay for the Detection and Determination of Siderophore. Anal. Biochem 1987, 160, 47–56. [DOI] [PubMed] [Google Scholar]
- (54).Lemetre C; Maniko J; Charlop-Powers Z; Sparrow B; Lowe AJ; Brady SF Bacterial Natural Product Biosynthetic Domain Composition in Soil Correlates with Changes in Latitude on a Continent-Wide Scale. Proc. Natl. Acad. Sci 2017, 114 (44), 201710262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Wilford J; de Caritat P; Bui E Modelling the Abundance of Soil Calcium Carbonate across Australia Using Geochemical Survey Data and Environmental Predictors. Geoderma 2015, 259–260, 81–92. [Google Scholar]
- (56).Owen JG; Reddy BVB; Ternei MA; Charlop-Powers Z; Calle PY; Kim JH; Brady SF Mapping Gene Clusters within Arrayed Metagenomic Libraries to Expand the Structural Diversity of Biomedically Relevant Natural Products. Proc. Natl. Acad. Sci 2013, 110 (29), 11797–11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Charlop-Powers Z; Pregitzer CC; Lemetre C; Ternei MA; Maniko J; Hover BM; Calle PY; McGuire KL; Garbarino J; Forgione HM; et al. Urban Park Soil Microbiomes Are a Rich Reservoir of Natural Product Biosynthetic Diversity. Proc. Natl. Acad. Sci 2016, 113 (51), 14811–14816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Zhang T; Luo Y; Chen Y; Li X; Yu J BIGrat: A Repeat Resolver for Pyrosequencing-Based Re-Sequencing with Newbler. BMC Res. Notes 2012, Notes 5, 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Fujii K; Harada KI A Nonempirical Method Using LC/MS for Determination of the Absolute Configuration of Constituent Amino Acids in a Peptide: Combination of Marfey’s Method with Mass Spectrometry and Its Practical Application. Anal. Chem 1997, 69 (24), 5146–5151. [Google Scholar]
- (60).Hess S; Gustafson KR; Milanowski DJ; Alvira E; Lipton MA; Pannell LK Chirality Determination of Unusual Amino Acids Using Precolumn Derivatization and Liquid Chromatography-Electrospray Ionization Mass Spectrometry. J. Chromatogr. A 2004, 1035 (2), 211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.