Summary
mRNAs form ribonucleoprotein complexes (mRNPs) by association with proteins that are crucial for mRNA metabolism. While the mRNP proteome has been well characterized, little is known about mRNP organization. Using a single molecule approach, we show that mRNA conformation changes depending on its cellular localization and translational state. Compared to nuclear mRNPs and lncRNPs, association with ribosomes decompacts individual mRNAs, while pharmacologically dissociating ribosomes or sequestering them into stress-granules leads to increased compaction. Moreover, translating mRNAs rarely show co-localized 5’ and 3’ ends, indicating that mRNAs are either not translated in a closed-loop configuration, or that mRNA circularization is transient, suggesting that a stable closed-loop conformation is not a universal state for all translating mRNAs.
Graphical Abstract
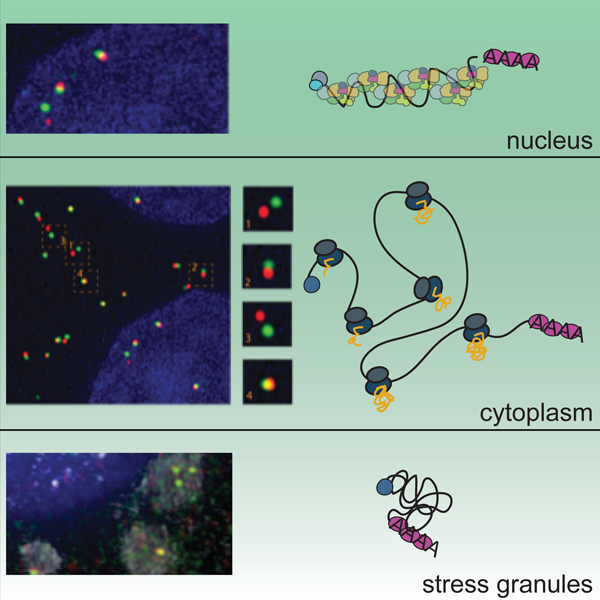
eToc Blurb
Adivarahan et al. show that mRNA compaction varies depending on subcellular localization and mRNA translation state. Whereas translational inhibition and sequestration to stress granules leads to mRNA compaction, translation induces the separation of 5’ and 3’ ends, suggesting that mRNA translation does not occur in a stable circularized conformation.
Introduction
RNAs are single-stranded nucleic acid polymers. Intramolecular base pairing and binding of RNA binding proteins (RBPs), many of which contain homo and hetero-dimerization domains, assemble mRNAs into RNPs (Singh et al., 2015). Assembly of mRNPs is initiated co-transcriptionally, and mRNP composition is thought to constantly change during the different processing and maturation steps, as well as upon translocation to the cytoplasm when mRNAs meet with ribosomes for translation. Proteomic approaches have identified many RBPs assembling to mRNA at these different stages, and recent crosslinking approaches have identified binding sites for many of these proteins, leading to comprehensive maps of mRNP composition (Hentze et al., 2018; Marchese et al., 2016). Similarly, recent transcriptome-wide chemical mapping approaches have identified single and double stranded regions within mRNAs revealing extensive internal secondary structures (Strobel et al., 2018). More broadly, mRNA organization is crucial for many aspects of mRNA metabolism, especially steps where different regions within (pre-) mRNAs are thought to communicate, such as splicing, translation regulation or miRNA-mediated regulation (Fabian and Sonenberg, 2012; Imataka et al., 1998; TarunSachs, AB, 1996) Despite the importance of mRNA organization, little is known on how mRNPs are organized as three-dimensional assemblies.
Much of our understanding on mRNP organization comes from in vivo and in vitro electron microscopy studies. Electron tomography studies of the 35kB-long Balbiani Ring (BR) mRNA in C. tentans salivary glands revealed a dense nuclear particle with a diameter of about 50nm where 5’ and 3’ are in close proximity (Skoglund et al., 1986). A different architecture was observed for nuclear mRNAs purified from yeast and analyzed by electron microscopy (EM) that revealed particles with a homogenous width but variable length (5 nm wide, 20–30 nm long), suggesting a linear assembly with the ends separated (Batisse et al., 2009). Organization of cytoplasmic mRNAs, on the other hand, has been primarily studied by visualizing ribosomes as a proxy for visualizing mRNA. Polysomes containing various numbers of ribosomes and in different conformations have been observed in vivo, as well as in vitro. Polysomes are found either in spiral arrangements, forming double rows of ribosomes, arranged in circles as well as in less defined, open conformations, however, how the mRNA is organized within these polysomes is not visible in these experiments (Afonina et al., 2015; Brandt et al., 2010; Lu et al., 2016; Ramani et al., 2015; Rech, 1995). Considering a ribosome footprint of about 30nt and an average ribosome density of about one ribosome per 200–900nt, large regions of the mRNA must be exposed between individual ribosomes as well as in their 5’ and 3’ UTRs (Hendrickson et al., 2009; Steitz, 1969; C. Wang et al., 2016; Yan et al., 2016). However, the organization of this higher order structure is not known.
The best studied example for a role of intramolecular mRNA organization in gene regulation is the communication between 5’ and 3’ ends during translation (Gallie, 1991). The current model of initiation is that mRNAs are organized in a circular conformation, mediated by a series of interactions between the 5’ cap, the cap binding protein eIF4E, the adaptor protein eIF4G, poly(A) binding protein (PABPC1), and the poly(A) tail. These interactions have been proposed to bring together the ends of the transcript to stimulate translation (Jackson et al., 2010). This closed-loop model is supported by many studies showing physical interactions between eIF4E, eIF4G and PABPC1, in vitro experiments reconstituting mRNA circularization using purified components, as well as the observation of circular polysomes in cells (Christensen et al., 1987; Imataka et al., 1998; TarunSachs, AB, 1996; Wells et al., 1998). However, many polysomes in cells observed by EM show configurations that do not suggest a closed-loop, and so it is unclear whether closed-loops represent transient states, polysomes with mRNAs with connected ends but where ribosomes are positioned distant from the 5’ and 3’ ends, or different classes of transcripts for which translation of only some might occur in a closed-loop configuration. Furthermore, although there are examples of factors that repress gene expression by connecting the 5’ and 3’ ends, how the ends are physically brought together to establish these complexes is not known (Jonas and Izaurralde, 2015).
Thus, the fundamental issue of how mRNAs are organized as mRNPs in vivo remains unresolved. In this study, we investigate mRNA organization within cells by combining Structured Illumination Microscopy (SIM) with single molecule resolution fluorescent in situ hybridization (smFISH) to investigate the spatial relationship of various regions within mRNAs in different cellular compartments. We observe that mRNAs exist in different levels of compaction depending on their cellular localization and translation state and show that translation, at least for a subset of mRNAs studies here, results in the separation of 5’ and 3’ ends, suggesting that these RNAs are not translated in a stable closed-loop.
Results
Visualizing/resolving different regions within mRNAs using smFISH and SIM
To determine whether combining smFISH and SIM allows to spatially resolve different regions within single mRNAs, we first measured co-localization precision by hybridizing a mix of 44 20nt-long DNA probes, alternatingly labeled with cy3 and cy5, and spanning a 1.2 kb region within the 18,413 nt-long MDN1 mRNA in paraformaldehyde-fixed HEK293 cells (Figure 1A). Probes spreading the 1.2kB region were used to ensure enough single smFISH probes, each with similar annealing temperature, bind the mRNA and together result in sufficient signal for robust detection and localization of individual mRNAs. Images were acquired spanning the entire cell volume, and 3D datasets reduced to 2D by maximum intensity projection. We then determined the center of each signal by 2D Gaussian fitting and measured the distance between signals from both channels. 2D Gaussian fitting calculates the centroid of the signal emitted from individual fluorescent particles spread over multiple pixels on the detector and allows sub-diffraction localization precision (R. Thompson et al., 2002; Zenklusen et al., 2008). Measuring the distances between co-localizing spots showed a co-localization precision of 21 nm, indicating that we can resolve discrete regions within mRNAs when separated by more than 20 nm (Figure 1C).
Figure 1. See also Figure S2: Visualizing single mRNA reveals open conformations of cytoplasmic mRNAs.
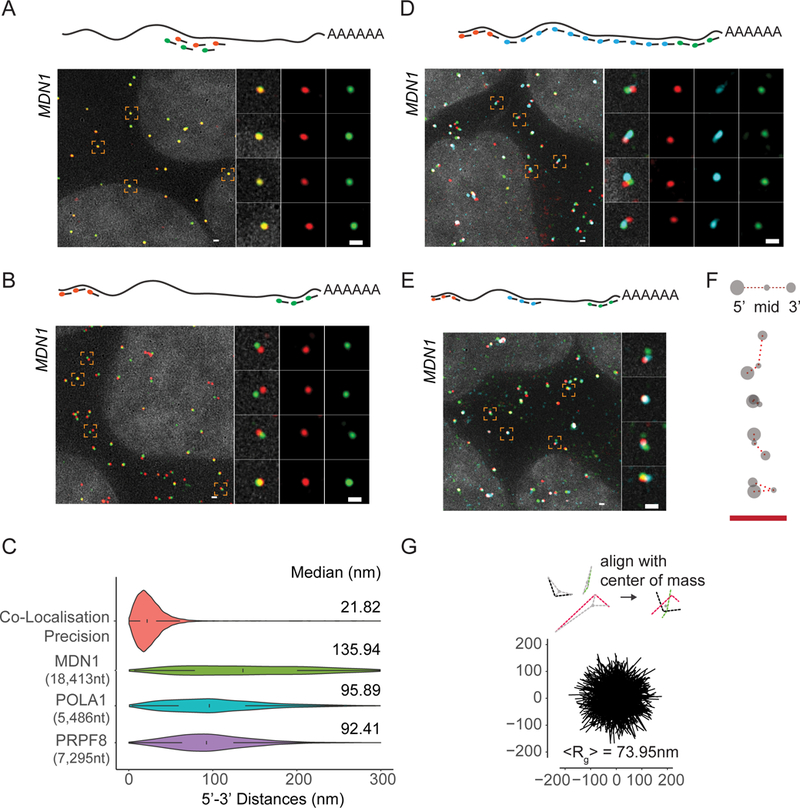
(A) smFISH images using alternating probes labeled in cy3 (red) and cy5 (green) to middle region of MDN1 mRNA (Probe Set#1, Table S3) in paraformaldehyde fixed HEK 293 cells. Nuclei are visualized by DAPI staining (grey). Magnified images of individual RNAs marked by dashed squares are shown on the right. Schematic position of probes shown on top. (B) smFISH using probes targeting the 5’ (red) and 3’ (green) ends of MDN1 mRNAs (Probe Set#2, Table S3). (C) Violin plots showing distance distribution of co-localization precision of co-localizing spots from A, and 5’−3’ distances for MDN1, POLA1, PRPF8 mRNAs determined by Gaussian fitting. White box plot inside the violin plot shows first quartile, median and third quartile. Median distances are shown on the right. (D, E) smFISH using 5’ (red), 3’ (green), and tiling or middle probes (cyan) respectively (Probe Sets#3,4, Table S3). (F) Cartoon depicting different mRNA conformations from E. (G) Projections of superimposed conformations with their centers of mass in registry, n=563. Mean Radius of gyration (<Rg>). Scale bars, 500 nm
We then positioned labeled probes to the 5’ and 3’ ends of the MDN1 mRNA to determine RNA extension in cells (Figure S1 and Table S2, S3), which, in a hypothetical scenario with 0.59nm spacing between nucleotides for a rigid ssRNA, would measure about 10.8µm in length when fully extended (Liphardt et al., 2001). However, as a flexible polymer, mRNA unlikely exists in such a conformation, which will depend on different parameters, including the stiffness of the polymer chain, as well as by thermodynamics and intra-molecular interactions, which will reduce the overall extension of the mRNA (Borodavka et al., 2016; Chen et al., 2012; Gopal et al., 2012; Liphardt et al., 2001). Analyzing cytoplasmic MDN1 mRNAs, we observed few overlapping 5’ and 3’ signals; instead, a majority of 5’ signals had a 3’ signal within close proximity (Figure 1B), with distances up to 300 nm between the two signals (Figure 1C). A similar distribution was observed when measured in 3D, and distances were indistinguishable when the EtOH step in the hybridization protocol was omitted (Figure S2A, B). To determine if 5’ and 3’ signals were part of the same mRNA molecule, we used a third set of FISH probes tiling the entire length of the mRNA between the 5’ and 3’ regions in 500 nt intervals. The tiling signal overlapped with either one of the two regions and connected the 5’ and 3’ within the 300 nm radius, confirming that 5’ and 3’ end signals belonged to the same molecule and, moreover, pointing towards an elongated conformation of cytoplasmic MDN1 mRNAs (Figure 1D). To better understand the spatial relationship between different regions within these mRNAs, we replaced the tiling probes with a probe set hybridizing to the middle region of the MDN1 mRNA (Figure 1E, S1). Using these probes, we observed cytoplasmic mRNAs where the three different regions could be spatially resolved (Figure 1E and F). To measure the average volume of these cytoplasmic mRNAs, we aligned individual mRNAs using their center of mass and calculated the mean radius of gyration (<Rg>) as measure of the global size of the mRNP and found an <Rg> of 73.95 nm (Figure 1G). These dimensions are comparable to the size of polysomes imaged by electron microscopy and super-resolution microscopy, in which polysomes containing 6–10 ribosomes, as suggested for the ribosome occupancy for MDN1 mRNAs, typically have a diameter ranging from around 90–150nm (Brandt et al., 2010; Christensen et al., 1987; Floor et al., 2016; Viero et al., 2015).
To determine whether such open conformations are particular to the long MDN1 mRNA or a more common feature of cytoplasmic mRNAs, we measured compaction of two shorter mRNAs encoding for the splicing factor PRPF8 (7,295 nt) and the DNA polymerase alpha catalytic subunit POLA1 (5,486 nt) and found similar open conformations (Figure S2C). End-to-end distances showed narrower distributions compared to MDN1 mRNAs, indicating that maximum expansion in cells scales with mRNA length (Figure 1C). Together, these data show that cytoplasmic mRNAs predominantly exist in an open conformation where 5’ and 3’ are rarely found in close proximity.
Open mRNP conformation correspond to translating mRNA
Translating mRNPs are thought to exist in a closed-loop conformation where 5’ and 3’ ends of the mRNA are brought together through interactions between the cap binding eIF4F complex and the polyA binding protein PABPC1 (Imataka et al., 1998; TarunSachs, AB, 1996; Wells et al., 1998). Surprisingly, we rarely observed 5’−3’ conformations consistent with such a closed-loop configuration. One possibility could be that most mRNAs with separated 5’ and 3’ ends are not in the process of being translated and that only the fraction with co-localizing ends represents the pool of translating mRNAs. If that were the case, interfering with translation should further reduce the fraction of mRNAs with co-localizing 5’ and 3’ ends. To test this hypothesis, we treated cells, prior to fixation, with drugs that affect translation via different mechanisms: cycloheximide, which inhibits elongation by binding to the E-site of the 60S ribosomal unit and stabilizes polysomes, and puromycin, which causes premature chain termination and disassembles polysomes (Bhat et al., 2015). Treatment with cycloheximide only modestly affected the distribution of 5’−3’ MDN1, PRPF8 and POLA1 mRNA end distances when compared to untreated cells, with slightly lower end to end distances, suggesting that elongating ribosomes contribute to the openness of the mRNA (Figure 2C). In contrast, the disassembly of polysomes following a short treatment with puromycin (10 min) resulted in an unexpected phenotype where the 5’−3’ ends of most transcripts were co-localizing (Figure 2A). For MDN1 mRNA distance measurements showed a narrow distribution with a median of 36 nm. A one-hour treatment with the translation inhibitor homoharringtonine, which stalls ribosomes at the initiation site, yielded similar results (Figure S3A). Similarly, POLA1 and PRPF8 mRNA ends also showed a high degree of co-localization with similar median 5’−3’ end distances (Figure 2C and S3B).
Figure 2. See also Figure S3: Open mRNP conformation correspond to translating mRNA.
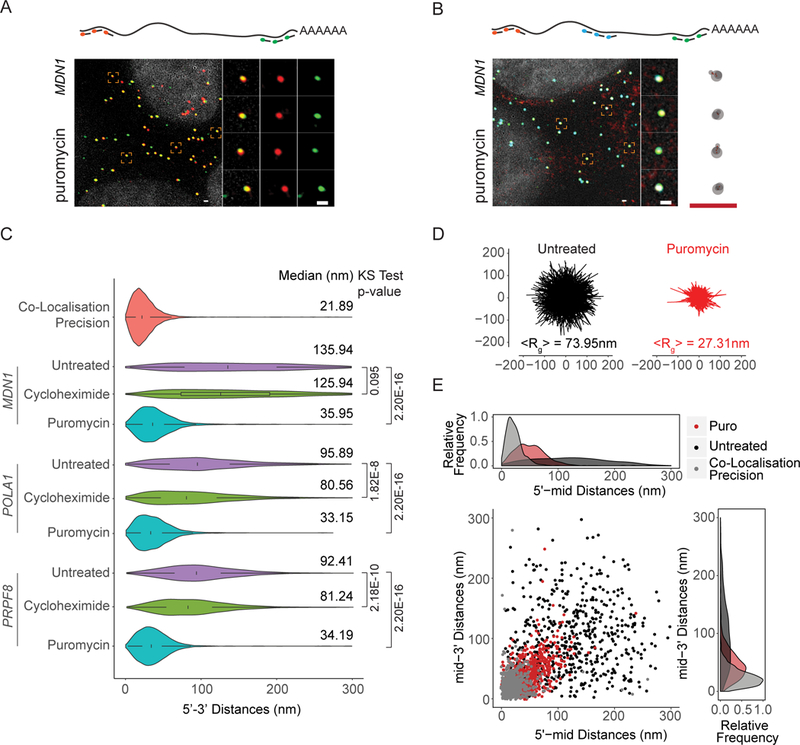
(A, B) 5’ and 3’ (Probe Set#2, Table S3) or three color MDN1 mRNA smFISH (Probe Set#4, Table S3) in HEK 293 cells treated with puromycin (10min, 100µg/ml). (C) Violin plots showing 5’−3’ distances for MDN1, POLA1, PRPF8 mRNAs in untreated and cells treated with cycloheximide and puromycin. White box plot inside the violin plot shows first quartile, median and third quartile. Median distances and p-values calculated using Kolmogorov-Smirnov test are shown on the right. (D) Projections of superimposed conformations from three color MDN1 mRNA smFISH (Probe Set#4, Table S3) in untreated and puromycin treated cells with their centers of mass in registry, n=563. Mean Radius of gyration (<Rg>). (E) Scatter plot showing 5’mid and mid-3’ distances for individual RNAs. Frequency distribution are shown on top and on the right. Scale bars, 500 nm.
These observations could be due to either a change in mRNP conformation resulting in increased levels of 5’−3’ interaction or the result of a general compaction of the mRNP because of the loss of bound ribosomes. To distinguish between these possibilities, we repeated the experiment, this time using probes that hybridize to the middle region of MDN1 mRNA or tile along its entire length and found that puromycin treatment resulted in a general compaction of the mRNPs (Figure 2B, S3C). Overlaying mRNA conformations revealed a less extended form of these mRNPs compared to untreated cells (Figure 2D and E). These observations suggest that most of these cytoplasmic mRNAs are translating, that mRNAs within translating mRNPs are not arranged in a closed-loop conformation, and that disassembly of polysomes results in highly compact mRNAs.
Inhibiting eIF4G1-PABPC1 interaction does not alter open mRNP conformations
Despite the fact that most of the mRNAs exist in an open configuration, we noted that a small fraction of MDN1 mRNAs in untreated cells had ends in close proximity. If we consider 50nm as an upper limit for a closed-loop configuration, 12.5% of MDN1 mRNAs show ends that are closer than 50nm as judged by 2D analysis. Because 2D analysis projection analysis overestimates proximity due to the projection of the z dimension, we re-analyzed the data in 3D to refine our estimate of mRNAs potentially in the closed-loop confirmation and found that only 4.4% are found closer than 50nm, further suggesting that the close proximity of the ends is a rare event (Figure S2A). To determine whether this small fraction indeed represents closed-loop conformations mediated by PABPC1–eIF4G1 interactions, we used CRISPR/Cas9 gene editing to construct two different cell lines mutating key residues in PABPC1 or eIF4G1 needed for the interaction as well as matched wild-type controls (Figure 3A). Although other paralogues of PABPC1 and eIF4G1 are present in the human gene expressed at lower levels in HEK293 cells, these proteins are not sufficient to compensate for a knock-out of eIF4G1 and are expressed at lower levels (Hart et al., 2015). Both mutant cell lines showed reduced interactions, but these mutations had minimal impact on cell survival and overall translation, although there was a slight increase in the monosome:polysome ratio in the eIF4G1 mutant cell line (Figure 3B-D). When we analyzed the conformation of cytoplasmic MDN1 mRNAs in these mutant cell lines, we observed a 5’−3’ distance distribution similar to those in WT cells (Figure 3E), although we observe a modest increase in end-to end distances for the mutant cell lines. Importantly, the fraction of MDN1 mRNAs with 5’–3’ distances below 50 nm remained unchanged, indicating that the small 5’−3’ colocalizing fraction are not dependent on the PABPC1–eIF4G1 interaction. Treatment with puromycin resulted in increased proximity of 5’ and 3’ ends indicating that the open conformations that we observe represent translating mRNAs (Figure S4 A, B). Although we cannot formally exclude compensation by other paralogues, our data strongly suggest that the colocalization fraction may instead be non-translating mRNAs or mRNPs where the ends are close to each other independent of the PABPC1–eIF4G1 interaction, possibly due to the flexibility of the RNA polymer.
Figure 3. See also Figure S4: Inhibiting eIF4G1-PABC1 interactions does not alter 5’−3’ distances.
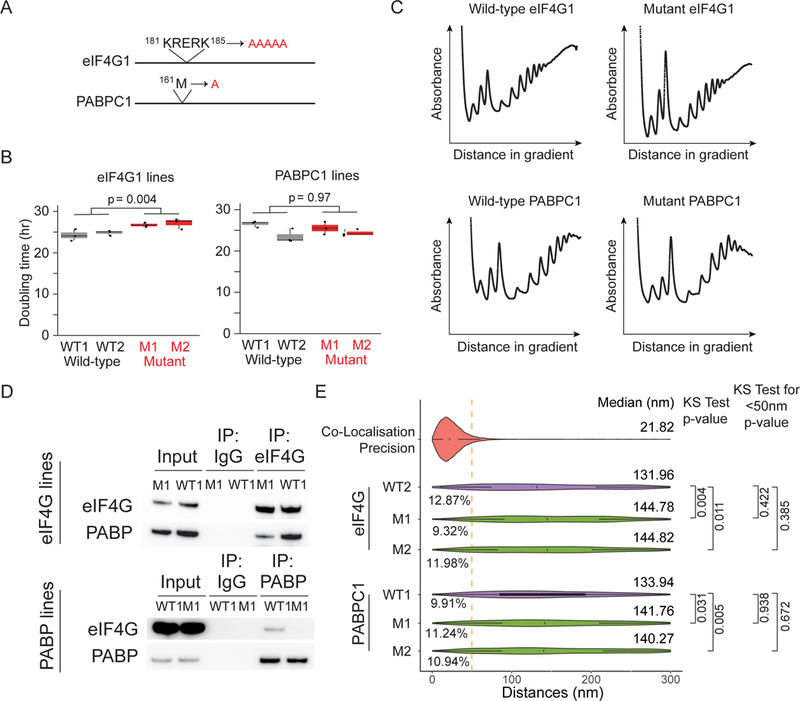
(A) Sites of amino acid substitutions in eIF4G1 and PABPC1 cell lines. (B) Doubling time for eIF4G1 and PABPC1 CRISPR-edited lines. Shown are the doubling times calculated for three independent biological replicates for two independent wild-type and mutant eIF4G1 and PABPC1 lines. (C) Polysome profiles for wild-type eIF4G1, wild-type PABPC1, mutant eIF4G1, and mutant PABPC1 lines. (D) Immuno-precipitation of eIF4G1 and PABPC1 from wild-type and mutant cell lines using anti-eIF4G1 and PABPC1 anti-bodies. (E) Violin plots showing distance distribution of co-localization precision from 5’−3’ distances for MDN1 mRNAs in wild type and mutant cell lines (Probe Set#2, Table S3). White box plot inside the violin plot shows first quartile, median and third quartile. Median distances and p-values calculated using Kolmogorov-Smirnov test are shown on the right. WT1/WT2/M1/M2 represent different clonal cell lines.
Ribosome occupancy determines mRNP compaction
To further investigate the role of ribosome occupancy on mRNP compaction, we performed a ribosome run-off experiment using the translation inhibitor homoharringtonine. Treatment with homoharringtonine inhibits new initiation but allows elongating ribosomes to continue translating until reaching the stop codon, allowing us to determine local compaction within the MDN1 mRNA upon a short drug treatment. Translation elongation in human cells is thought to occur at about 5aa per second, therefore for the 16,791 nt MDN1 mRNA open-reading frame, after a 10 min treatment the first half will be devoid of ribosomes, whereas the second half will still contain ribosomes (Bin Wu et al., 2016). Consistent with the requirement of ribosome occupancy for RNA decompaction, the 5’-to-mid region of MDN1 mRNA became compacted after the 10-min homoharringtonine treatment, whereas the mid-to-3’ region remained in an open conformation (Figure 4A, B, S5).
Figure 4. See also Figure S5: Ribosome occupancy determines mRNP compaction.
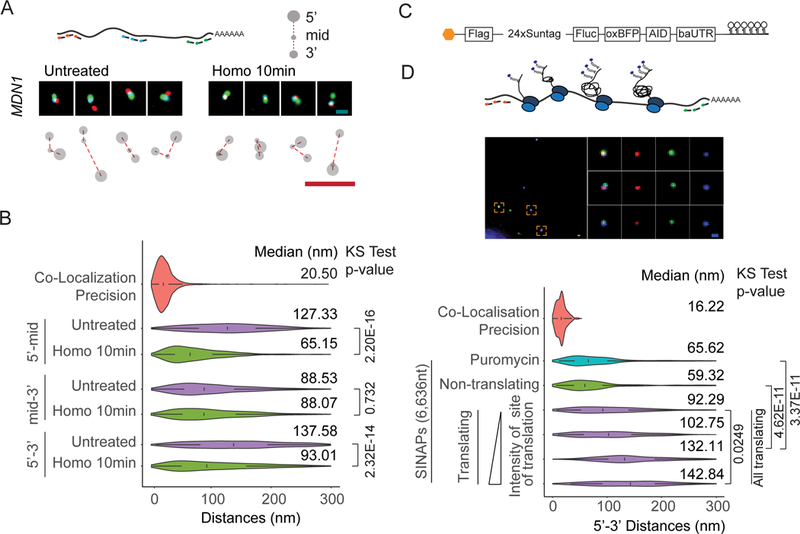
(A) smFISH using 5’ (red), 3’ (green), and middle probes (cyan) respectively (Probe Set#4, Table S3) for untreated and homoharringtonine (100µg/ml, 10min) treated cells and cartoon depicting different mRNA conformations. (B) Violin plots showing 5’-mid, mid-3’ and 5’−3’distance distribution of cytoplasmic MDN1 mRNAs in untreated and homoharringtonine treated cells. (C) Cartoon depicting the SINAPs construct (D) Images showing 5’ and 3’ smFISH and anti-GFP immunofluorescence (Probe Set#15, Table S3) (top), and violin plots depicting 5’−3’ distances for puromycin treated, non-translating and translating mRNAs. Translating mRNAs were clustered in 4 groups (k-means) according to intensity of anti-GFP signal (bottom). White box plot inside the violin plot shows first quartile, median and third quartile. Median distances and pvalues calculated using Kolmogorov-Smirnov test are shown on the right. Scale bars, 500 nm.
To further investigate the relationship between translation and 5’–3’ proximity, we employed a reporter system developed for Single Molecule Imaging of Nascent Peptides (SINAPs), where nascent proteins are rapidly bound at the ribosome exit channel by a fluorescently labeled single-chain antibody (scFv-sfGFP) and fluorescence intensity, therefore, is proportional to the number of ribosomes on a specific mRNA (Figure 4C) (Bin Wu et al., 2016; Pichon et al., 2016; C. Wang et al., 2016; Yan et al., 2016). The SINAPs reporter was transfected into U2OS cells stably expressing the scFv-sfGFP fusion, cells were fixed after 24 hours and ribosome occupancy and mRNA conformation were simultaneously measured by smFISH and immunofluorescence targeting scFv-sfGFP using an anti-GFP antibody (Figure 4D). Consistent with our previous analysis, translating mRNAs had more open conformations relative to non-translating mRNAs, as judged by both nascent peptide signal and puromycin treatment. Importantly, the RNA 5’ −3’ distance increased with the relative intensity of nascent peptides. Taken together, our data indicate that ribosome occupancy decompacts mRNA and separates the ends.
Compaction state of lncRNAs and mRNA sequestered to stress-granules
If translation is a main cause for an open mRNP conformation, we hypothesized that non-translating RNAs, such as cytoplasmic long non-coding RNAs (lncRNAs), might show a level of compaction similar to non-translating mRNAs, and, moreover, their compaction should be unaffected by translation inhibitors. To test this model, we measured end-to-end distances for two lncRNAs, TUG1 (7,469nt) and OIP5-AS1 (8,829nt), previously found to be present in the nucleus and the cytoplasm (Cabili et al., 2015). Both lncRNAs contain short putative ORFs that could lead to their association with ribosomes, however, their translation will be limited to the very 5’-end of the transcript (van Heesch et al., 2014). As shown in Figure 5A, 5’ and 3’ labeled cytoplasmic TUG1 and OIP5-AS1 lncRNAs displayed a more compact conformation compared to the similarly sized PRPF8 mRNA. In addition, 5’−3’ distances of OIP5-AS1 lncRNA was unaffected by puromycin, further suggesting that decompaction of cytoplasmic mRNAs requires the formation of polysomes (Figure 5B). Interestingly, we observe a small, but significant, change of end-to-end distance for TUG1 lncRNAs upon puromycin treatment (Figure 5B). Unlike OIP5-AS1 lncRNAs, TUG1 lncRNAs has been shown to associate with higher polysome fractions despite its very short putative ORFs, which could explain this observation (Floor et al., 2016).
Figure 5. See also Figure S6: lncRNAs in the cytoplasm and mRNAs sequestered to stress granules show compact conformations.
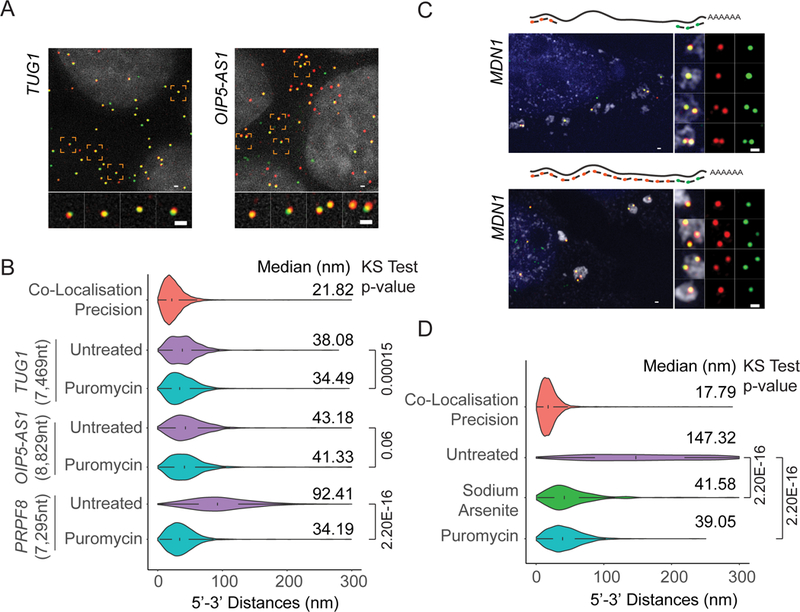
(A) smFISH visualizing 5’ and 3’ ends of TUG1 and OIP5-AS1 lncRNAs (Probe Sets#7,8, Table S3). Nuclei are visualized by DAPI staining (grey). (B) Violin plots showing 5’−3’ distance distribution of cytoplasmic TUG1 and OIP5-AS1 lncRNAs in untreated and puromycin treated cells compared to PRPF8 mRNAs. (C) 5’ - 3’ (Probe Set#9, Table S3) or 3’ and tiling (Probe Set#10, Table S3) MDN1 mRNA smFISH in U2OS cells treated with arsenite (1hour, 2 mM). Stress granules are visualized using an oligo dT probe (grey). Nuclei are visualized by DAPI staining (blue). (D) Violin plots comparing MDN1 mRNA 5’−3’ distance distribution for untreated, arsenite and puromycin treated U2OS cells. For arsenite treated cells, only mRNAs in stress granules were considered. White box plot inside the violin plot shows first quartile, median and third quartile. Median distances and p-values calculated using Kolmogorov-Smirnov test are shown on the right. Scale bars, 500 nm.
We next hypothesized that, if eviction of ribosomes from translating mRNAs by puromycin results in a strong compaction of mRNA, then mRNAs that are translationally repressed in response to external stimuli or environmental triggers should also acquire a compact conformation. Treatment with sodium arsenite inhibits translation through phosphorylation of eIF2α and results in disassembly of polysomes and sequestration of mRNAs in stress granules (Buchan and Parker, 2009; Panas et al., 2016). We induced stress granule assembly in U2OS cells upon treatment with arsenite for 1 hour and found that this treatment relocalized cytoplasmic MDN1 mRNAs to stress granules (Figure 5C). Furthermore, mRNAs show a highly compact conformation, observed by measuring end to end distances using 5’- 3’ probes, as well as using tiling probes spanning the entire transcript up to the 3’ region, which was hybridized using differently labeled probes (Figure 5C). End-to-end measurements for MDN1 mRNAs in stress-granules showed a level of compaction similar to that seen in puromycin-treated cells (Figure 5D), and similar compaction was also observed for POLA1 and PRPF8 mRNAs under the same conditions (Figure S6A). Interestingly, not all POLA1 and PRPF8 mRNAs accumulated in stress granules, but those mRNAs that remained outside showed the same level of compaction as those within stress granules, suggesting that translation inhibition occurs independently of mRNA sequestration to stress granules, as previously suggested (Mollet et al., 2008; Panas et al., 2016; Souquere et al., 2009; Khong et al., 2017). Moreover, a fraction of TUG1 and OIP5-AS1 lncRNAs were also found localized to stress granules, and this localization did not alter their compaction (Figure S6B).
Organization of nuclear mRNAs
We finally asked whether the compacted state of mRNAs found within stress granules, or after puromycin treatment, reflects a default state for non-translating cellular mRNPs. In the nucleus, nascent mRNAs are co-transcriptionally spliced and assembled into mRNPs resulting in the binding of a large set of RBPs, including the exon-junction complex and SR proteins (Le Hir et al., 2000; Müller-McNicoll and Neugebauer, 2013; Singh et al., 2012). During translation in the cytoplasm, many RBPs bound to the open reading frame are evicted by the ribosome. mRNAs that have been translated and then go into a translationally silent state might therefore be bound by fewer proteins than cytoplasmic mRNAs prior to their first round of translation, or nuclear mRNPs before their export to the cytoplasm.
To determine whether a default compaction state exists for non-translating mRNPs, we investigated the organization of nuclear MDN1 mRNAs. Compared to cytoplasmic MDN1 mRNAs upon puromycin treatment, nuclear MDN1 mRNAs were found in an extended conformation, although they were more compacted than translating cytoplasmic mRNAs (Figure 6A–E). Moreover, 5’ to mid and mid to 3’ distances were shorter than the 5’ to 3’ distance and larger than cytoplasmic mRNAs upon puromycin treatment (Figure S7A). Unlike for cytoplasmic mRNAs, open mRNP conformations of nuclear MDN1 mRNAs were still observed upon puromycin (10 min) or homoharringtonine (1 h) treatment, although we measured a small overall reduction in 5’−3’ distances (Figure S7B, C). This might in part be due to the difficulty to accurately segment nuclear-cytoplasmic borders so that our analysis of nuclear mRNAs includes a small fraction of cytoplasmic mRNAs. Together, these observations suggest that assembly of nuclear mRNPs results in more extended mRNP compared to translationally inhibited mRNPs.
Figure 6. See also Figure S7: Organization of nuclear MDN1 mRNAs.
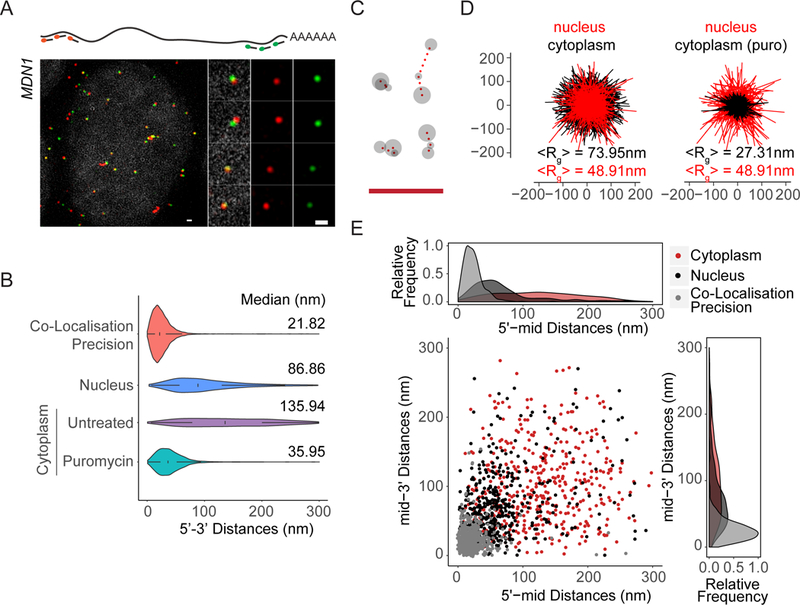
(A) 5’-3’ MDN1 mRNA smFISH (Probe Set#2, Table S3) of nuclear mRNAs. The nucleus was stained with DAPI (gray). (B) Violin plots comparing MDN1 mRNA 5’−3’ distance distribution of nuclear and cytoplasmic mRNAs. White box plot inside the violin plot shows first quartile, median and third quartile. Median distances are shown on the right. (C) Representative conformations of nuclear MDN1 mRNAs observed by 5’, middle and 3’ labeling as in 1E. (D) Projections of superimposed conformations with their centers of mass in registry, compared to untreated or puromycin treated cytoplasmic MDN1 mRNAs, n=452. Mean Radius of gyration (<Rg>). (E) Scatter plot comparing 5’-mid and mid-3’ distances for individual nuclear and cytoplasmic MDN1 mRNAs. Frequency distribution are shown on top and on the right. Scale bars, 500 nm.
Discussion
Although the proteome of mRNPs have been studied extensively, the understanding of how mRNA and proteins organize into mRNPs is still poorly understood. Here, using a single molecule super-resolution microscopy approach to describe features of mRNP organization in cells, our data shows that mRNA in cells are found at different levels of compaction depending on their subcellular localization and translation state, with actively translating mRNAs and mRNAs sequestered to stress granules representing two extremes of open and compacted mRNAs states in vivo. Furthermore, we show that decompaction during translation results in the separation of the 5’ and 3’ ends of mRNAs, indicating that at least for the mRNAs investigated here, translation does not occur in a stable closed-loop conformation.
Nuclear mRNPs show a linear organization
EM studies visualizing the 35kb-long nuclear BR mRNPs show mRNPs assembled as compact particles with a croissant shape where 5’ and 3’ ends are in close proximity (Mehlin et al., 1995). The formation of this particle occurs sequentially and co-transcriptionally, starting with the formation of a rod-like structure with about a 12 nm diameter, that further compacts into stalk and finally results in a croissant-shaped mRNP with a ~50 nm diameter and ~15 nm thickness. Considering a hypothetically fully extended, linear mRNA in which a spacing between nucleotides of 0.59nm, and the 50 nm diameter of the BR mRNPs as the maximal extension of the BR mRNP, BR mRNPs are compacted about ~413-fold. We also observe a high degree of compaction of about 111-fold for nuclear MDN1 mRNAs, considering a diameter of 97 nm (double the radius of gyration), suggesting mRNPs are generally highly compact in the nucleus.
However, in contrast to BR mRNAs, we do not observe a 5’ and 3’ ends in close proximity, but rather 5’−3’ end further apart than the 5’ to the middle region and middle to the 3’, suggesting a more linear conformation of the nuclear mRNP (Figure 6 and S7). This structure could be the result of the sequential assembly of RNPs, such as the EJC to nascent mRNAs and the further compaction through binding to other proteins containing homo- and heterodimerization domains, such as SR proteins, as suggested in (Singh et al., 2012). Indeed, in agreement with such a model, a recently developed RNA ImmunoPrecipitation and Proximity Ligation in Tandem (RIPPLiT) approach investigating the proximity of different regions within an mRNAs identified only local intramolecular contacts, but failed to observe long range intramolecular mRNA interactions (Metkar et al., 2018). These observations suggest that mammalian nuclear mRNPs may be organized as rod-like structures, similar to the nuclear mRNPs previously purified from yeast (Batisse et al., 2009). Interestingly, we also observed a small fraction in nuclear MDN1 mRNAs with a more open conformation. One possibility could be that these mRNAs are not fully spliced, although we view this explanation as unlikely because analysis of nuclear mRNA sequencing datasets from HEK293 cells does not suggest inefficiently spliced introns for MDN1 mRNAs (not shown) (Neve et al., 2016). Alternatively, if mRNPs assemble linearly, mediated by the binding of EJC and other RBPs, inefficient assembly of these complexes might result in more open mRNPs.
Variable levels of RNP compaction in cells
The compaction state of nuclear mRNA represents an intermediate state relative to the compacted and extended states observed for cytoplasmic mRNAs. Only a few examples of large RNP structures have been described that allow a direct comparison of the different levels of compaction found for cytoplasmic mRNAs. For instance, the 80S eukaryotic ribosome is a highly compact RNP with a diameter of about 30nm and containing 7,216nt, resulting in an RNA compaction of ~142-fold. Nonetheless, this is less compact than MDN1 mRNAs upon puromycin treatment, where we observe compaction of ~199-fold. Interestingly, the compaction of MDN1 mRNA upon puromycin treatment is similar to packaged viruses. For instance, the 7.5 kB RNA genome of the Hepatitis A virus is packed into a capsid with an inner diameter of about 22nm, leading to a ~200-fold compaction of its genome (X. Wang et al., 2017), and the Zika genome (11kb, 30nm capsid inner diameter) gets similarly compacted (Sirohi et al., 2016). Interestingly, viral RNAs when transcribed in vitro were shown to acquire a condensed conformation, as measured using cryo-EM or SAXS, but the volume occupied by these RNAs in vitro is larger than when the RNA gets packaged into the viral capsid, consistent with the idea that compaction into the capsid is an active packaging mechanism (Gopal et al., 2012). Finally, a recent study showed that different in vitro transcribed mRNAs and lncRNA get compacted in vitro to a level similar, or sometimes even greater, than rRNA (Borodavka et al., 2016). Together, these results suggests that different levels of RNA compaction in vivo are likely mediated by a combination of RNA sequence as well as associated proteins, and it will be interesting to determine whether the high level of mRNA compaction observed for mRNAs upon ribosome eviction or sequestration into stress granules, is an active process that requires specific proteins, or whether it rather reflects the collapse of the RNA polymer onto itself due to the absence of ribosomes and other RBPs.
Closed-loop translation and regulation of gene expression
Our end-to-end measurements revealed that translating mRNAs rarely show co-localizing 5’ and 3’ ends, and the sun-tag reporter data further suggest that separation of the ends increases as a function of ribosome occupancy. Similar results were recently observed in another study using a similar approach but different mRNAs, suggesting that open conformations of translating mRNAs is a widespread phenomenon (Khong and Parker, 2018). Thus, our results are seemingly at odds with the current view that translating mRNAs exist in a stable closed-loop conformation. One possibility is that the eIF4G–PABP interaction may be transient and only occurs during specific steps of the translation cycle. Recent studies have shown that translation of many transcripts occurs in a bursting pattern, and the variable ribosome occupancy during ‘on’ and ‘off’ times of translation bursts is likely to cause altered mRNA compaction (Bin Wu et al., 2016; Pichon et al., 2016; Yan et al., 2016). Translation bursting could therefore induce structural reorganization of mRNAs that facilitate 5’−3’ proximity during ‘off’ times, allowing transient eIF4G–PABP interactions to occur. Interestingly, in vitro transcribed mRNAs were shown to obtain conformations where the 5’ and 3’ end are close in space, which is also suggested using computational predictions (Lai et al., 2018; Leija-Martínez et al., 2014; Yoffe et al., 2011). It will be interesting to investigate whether this occurs for mRNAs in vivo, maybe as a result of translation bursting, or as a result of translation inhibition in response to an external stimulus, and whether this will facilitate transient, eIF4G–PABP dependent, closed-loop configurations.
Closed-loop interactions could also occur during the pioneer round of translation. However, arguing against such a model is that pre-translation, EJC-containing mRNAs have a rod-like organization where the 5’ and 3’ are not in proximity, making it unlikely that these mRNP would be able to acquire a closed-loop conformation without further reorganization (Metkar et al., 2018). Nevertheless, mRNP reorganization at the cytoplasmic side of the nuclear pore has been shown in yeast, and recent studies suggest two populations of EJC containing mRNAs, with a cytoplasmic EJC-mRNP fraction that contains far fewer proteins and therefore possibly a different architecture (Mabin et al., 2018; Tran et al., 2007).
An alternate possibility to bring ends together could be a long poly A tail. As it is not possible to design probes for the tail that do not hybridize to all polyadenylated RNAs, our probes targeting to the 3’ of the RNA only hybridize up to the start of the poly A tail. However, it is unlikely that the tail is long enough to bridge a gap of up to 300nm, even if fully extended, as recent TAIL-seq studies revealed that polyA tails in mammals are on average only 50–100nt long (Chang et al., 2014; Subtelny et al., 2014).
Alternatively, it may be that only a subset of mRNAs are translated in a closed-loop conformation. EM and cryo-ET have shown polysomes in various conformation and only some of these conformations are compatible with a possible closed-loop conformation of the mRNAs (Brandt et al., 2010; Christensen et al., 1987; Christensen and Bourne, 1999). Interestingly, recent studies demonstrate that not all mRNAs are bound to the same extent by the closed-loop factors, supporting the idea that closed-loop formation might preferentially occur for some mRNA and/or during distinct phases of polysome assembly (Archer et al., 2015; Costello et al., 2015; Rissland et al., 2017; M. K. Thompson et al., 2016). A closed-loop configuration could also be more difficult to achieve for longer mRNAs where the ends could be separated by larger distances. Furthermore, it has been suggested that the formation of a closed loop might be more complex than the interaction of eIF4G and PABPC1, that this interaction might not be a prerequisite for translation at all times, or that it can be mediated by additional factors. Indeed, non-polyadenylated mRNAs can associate with polysomes and produce proteins, S. cerevisiae strains with impaired closed-loop components are viable, and we show here that mammalian cell lines have limited phenotypes upon reduced eIF4G–PABPC1 interactions (Figure 3) (Park et al., 2011; Proweller and Butler, 1997; Wilusz et al., 2012). Together with our observations showing that ribosome occupancy results in a decompaction of the mRNA and separation of the ends, all these observations argue against a model where a stable closed-loop conformation can be considered as a universal state of translating mRNAs.
Finally, some of the strongest functional evidence for 5’–3’ proximity comes from the numerous examples of regulatory elements in the 3’UTR that modulate processes at the 5’ end, such as de-capping or translation repression or initiation (Fabian and Sonenberg, 2012; Rissland, 2016). Signal transmission from the 3’ to the 5’ likely requires the mRNP to be flexible to allow both ends to meet, and it is unclear whether this flexibility is possible when mRNAs are in polysomes, as we show that ribosome occupancy leads to the separation of the ends. In general, we have little understanding of the biophysical properties of mRNPs in vivo. Obtaining a better mechanistic understanding of different aspects of mRNP metabolism involving intramolecular communication will therefore require a detailed understanding of the biophysical properties of RNPs in cells and, with it, new tools that allow us to study mRNP organization in vivo, with single molecule resolution and in real time.
STAR methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Daniel Zenklusen (daniel.r.zenklusen@umontreal.ca)
Method Details
Reagents used, stock concentrations, working concentrations and treatment conditions
Puromycin dihydrochloride (Sigma P8833) – stock at 5 mg/ml in water, Cycloheximide (Sigma C7698–1G) – stock 5 mg/ml in ethanol, Sodium Arsenite (Sigma 35000–1L-R) – stock 50 mM in water, Homoharringtonine (Sigma SML1091–10MG) – stock 10mg/ml in DMSO. The drugs were diluted in warm media to get final working concentrations and cells were treated prior to fixation as follows: Puromycin (100µg/ml for 10 min), Cycloheximide (100µg/ml for 10min), Homoharringtonine – 100µg/ml for 10 mins or 1hr and Sodium Arsenite (2mM for 1 hour).
Cell culture and drug treatment
HEK293 (American Type Culture Collection CRL-1573) and U2OS osteosarcoma (American Type Culture Collection HTB-96) cell lines were maintained at 37°C and 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM) (Wisent, 319–005-CL) supplemented with 10% fetal bovine serum (FBS) (Wisent, 080–150) and passaged every 2–3 days with Trypsin (Wisent 325–043-EL). Cells were plated on poly-L-Lysine (Sigma, P8920) coated coverslips the day before treatment and fixation. On the day of the experiment, media was replaced with fresh warm media containing drug in indicated concentrations and placed back in the incubator. After treatment, the cells were briefly washed with 1xPBS, fixed with 4% paraformaldehyde in 1xPBS (pH 7.4) for 10 minutes at room temperature, washed three times with 1xPBS and stored overnight in 70% ethanol at −20°C for permeabilization. Alternatively, the cells were permeabilized using 0.1% TritonX-100 + 0.5%BSA in 1x PBS for 15mins after which they were washed 2 times with 1x PBS for 5 mins each immediately before using the samples for smFISH (Figure S2B).
Plasmid Preparation:
The phage-ubc-flag-24xSunTag-Fluc-oxBFP-AID-baUTR-24xMS2 plasmid was prepared as described in (Bin Wu et al., 2016).
Generation and screening of eIF4G1 and PABPC1 mutant cell lines
Mutant cell lines were generated using CRISPR-Cas9. To produce sgRNAs targeting either eIF4G1 or PABPC1, annealed DNA oligos (Table S1) were ligated into the BbsI site of plasmid pX330 (Ran 2013). Homology repair constructs containing the intended mutations and upstream and downstream homology arms (~1 kb in total) were ligated into the plasmid Lox-Stop-Lox-TOPO-∆stop (Rakheja 2014), in which homology arms are cloned surrounding a puromycin resistance cassette flanked by loxP sites (Table S1).
HEK293 cells (5 × 105 cells in one well of a 6-well plate) were transfected with 250 ng of the pX330-sgRNA construct and 1 µg of the repair construct using Lipofectamine 2000 according to the manufacturer instructions, and then incubated in EMEM supplemented with 10% FBS in a humidified incubator at 37°C with 5% CO2. Two days following transfection, cells were trypsinized and 10% of the cells were moved into a 15-cm dish. After 24 h, puromycin was added to a final concentration of 3 µg/mL, and the media was changed daily for the next 3 days. The following day, single cells were seeded into each well of a 96-well plate on a MoFlo Astrios cell sorter (Beckman Coulter) at the Flow and Mass Cytometry Facility at SickKids Hospital, Toronto. Following expansion of single colonies, cells were harvested and screened by PCR using primers that anneal to the genome outside of the homology arm region (Table S1). To excise the puromycin resistance cassette from positive clones, the cells were transfected with 1µg of pgk-Cre (Rakheja et al., 2014) and incubated for 3 days before single-cell seeding, expansion, and screening for loss of the puromycin resistance gene by PCR as described above. The PCR products were analyzed by Sanger sequencing to ensure that the intended mutations were present.
Cell viability assays
Cell viability was measured using PrestoBlue Cell Viability Reagent (Invitrogen) according to the manufacturer’s instructions. Briefly, cells were seeded in triplicate in 96-well plates at 1000 cells per well in 90µL of EMEM supplemented with 10% FBS, and then incubated at 37°C with 5% CO2. At 24h, 48h, and 72h after seeding, 10µL of PrestoBlue reagent was added to each well. After a further 6.5-h incubation at 37°C with 5% CO2, the fluorescence of each well was read on a SpectraMax M2 microplate reader (Molecular Devices).
Polysome profiling
To generate polysome profiles, cycloheximide was added to cells in a 10-cm dish to a final concentration of 100 µg/mL, and the cells were incubated for 10 min at 37°C. The cells were then placed on ice and washed twice with ice-cold PBS containing 100 µg/mL cycloheximide. Cells were lysed by shearing four times through a 26-gauge needle in 500 µL of lysis buffer (10 mM Tris-HCl pH 7.4, 5 mM MgCl2, 100 mM KCl, 1% Triton X-100, 2 mM DTT, 500 U/ml Rnasin (Promega), EDTA-free protease inhibitor cocktail (Sigma), 100 µg/mL cycloheximide). Following centrifugation at 1300 × g for 10 min at 4°C, the supernatant was collected, flash frozen in liquid nitrogen, and stored at −80°C until further processing.
Lysates were separated by loading 300 µL onto a 10–50% (w/v) sucrose gradient prepared with a Gradient Master (BioComp Instruments) and centrifuging for 2 h at 36,000 rpm in a SW41Ti rotor (Beckman Coulter) at 4°C. Gradients were fractionated on a Piston Gradient Fractionator (BioComp) coupled to an EM-1 Econo UV detector (Bio-Rad). UV profile data were recorded using Gradient Profiler software v 2.07 (BioComp).
smRNA FISH
Custom DNA probe sets were designed using Stellaris® Probe Designer, synthetized by Biosearch Technologies containing 3’ amine reactive group and labeled with far red dye Cy5 (GEPA25001), red dyes Cy3 (GEPA23001) from Sigma or Dylight 550 (Thermo Scientific 62263) or green dye Dy488 (Thermo Scientific 46403) as described in (Rahman et al., 2017). For the mRNAs and the lncRNAs, the isoforms used to design the probes are mentioned in Figure S1. For the mRNAs, these isoforms were verified as the predominantly expressed transcripts in HEK293 using RNA-seq datasets from human protein atlas. For the lncRNAs, the probes were designed such that they hybridize to the longer isoforms. Probe sequences are shown in Table S2. Probe combination used are shown in Table S3 and the probe combinations used for the experiment is mentioned in the figure legends. smFISH was done as described in (Rahman et al., 2017). Prior to hybridization, cells were rehydrated in 1xPBS, then washed with 10% formamide/2xSSC for 10 minutes at room temperature. The cells were hybridized with 1020 ng of each probe mix plus 40 µg of ssDNA/tRNA resuspended in the hybridization solution (10% dextran sulfate/10% formamide/2xSSC/2 mM VRC/0.1 mg/ml BSA) for 3 hours in the dark at 37°C. Post hybridization washes (2× 30 min) were carried out at 37°C with 10% formamide/2xSSC. Samples were then rinsed with 1xPBS and mounted with ProLong Gold antifade reagent with DAPI (P36935, Invitrogen).
Image Acquisition and pixel shift correction
Images were acquired with a 63x NA 1.46 oil objective on a Zeiss Elyra PS.1 system equipped with an Andor EMCCD iXon3 DU-885 CSO VP461 camera (1004×1002 pixels), the following filter sets: DAPI: BP420–480 + LP750 (Zeiss SR cube 07), Cy2: BP495–590+LP750 (Zeiss SR cube 13), Cy3: LP570 (Zeiss SR cube 14), Cy5: LP655 (Zeiss SR cube 10) and the following lasers: 50 mW 405 nm HR diode, 100 mW 488 nm HR diode, 100 mW 561 nm HR DPSS, 150 mW 642 nm HR diode. Each image was acquired using 3 rotations and a grid size of 42 µm for all channels. The microscope was located in a temperature-controlled room and samples were kept in the room for at least an hour before imaging to minimize thermal fluctuations. To correct for pixel shifts between channels, 0.1 µm TetraSpec beads (Invitrogen T-7279) were imaged in all channels, and the channel shift values and chromatic aberration were calculated and corrected using the built-in channel alignment tool in ZEN 2012 SP5 which uses an affine image alignment algorithm and later applied to the images. This correction was calculated for each day of imaging.
Combined smFISH and Immunofluorescence for simultaneous detection of mRNA conformation and nascent translation
Human U2OS osteoscarcoma cell line (American Type Culture Collection HTB-96) expressing stdMCP-Halotag, phR-scFV-GCN4-sfGFP-GB1-NLS-dWPRE, and pBabe-TIR1–9myc was prepared as described in (Bin Wu et al., 2016). Single-molecule FISH immunofluorescence was performed as described in (Bin Wu et al., 2016). In brief, cells were plated on 18mm diameter, #1 collagen coated coverslips (Fisher) in a 12-well dish. Cells were then transfected with 250 ng of the phage-ubc-flag-24xSunTag-Fluc-oxBFP-AID-baUTR-24xMS2 construct using X-tremeGENE 9 transfection reagent (XTG9-RO ROCHE). Six hours after transfection, IAA (Sigma-Aldrich) was added to a final concentration of 250 µM. 20 hours after transfection, fresh IAA was added to a final concentration of 250 µM. 24 hours after transfection, cells were fixed for 10 minutes in PBS + 5 mM MgCl2 (PBSM), permeabilized for 15 minutes in PBSM + 0.1% Triton-X and 0.5 % BSA, and incubated with 100 nM MS2v5-Cy5 and 50 nM SunTagV4-Qusar 570 smFISH probe sets (Table S2) and a primary antibody against GFP (GFP-1010, Aves labs, Inc.) and incubated for three hours at 37°C. After washing, cells were incubated with Alexa Fluor 488 labeled secondary antibody (ThermoFischer) and mounted in ProLong Diamond antifade reagent with DAPI (Life Technologies). Images were acquired on a custom inverted wide-field Nikon Eclipse Ti-E microscope equipped with three Andor iXon DU897 EMCCD cameras (512×512 pixels), Apochromatic TIRF 100X Oil Immersion Objective Lens/1.49 NA (Nikon MRD01991), encoded Stage with 150 micron Piezo Z (ASI), and LU-n4 four laser unit with solid state 405 nm, 488 nm, 561 nm, and 640 nm lasers (Nikon), a TRF89901-EM ET-405/488/561/640nm Laser Quad Band Filter Set for TIRF applications (Chroma), and Nikon HTIRF system. Images were acquired using in-unit intermediate 1.5x magnification changer for a final magnification of 150x and independent, epi-illumination from the 488, 561, and 640 nm lasers. Image pixel size: XY, 106.7 nm; Z-step, 200 nm. A total of 29 cells without drug treatment (total of individual 396 mRNAs) and 40 cells (97 individual mRNAs) upon puromycin treatment were analyzed.
Immunoprecipitations and western blotting
Cells were washed with 1X PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4, pH 7.4) and then lysed with 1 ml ice-cold lysis buffer A (100 mM KCl, 0.1 mM EDTA, 20 mM Hepes, pH 7.6, 0.4% NP-40, 10% glycerol, with freshly added 1 mM DTT and complete mini EDTA-free protease inhibitors [Roche; one tablet per 25 ml lysis buffer]) per 2.5 million cells. 50 µl was saved as the input sample. Cells were incubated with antibody (diluted according to manufacturer’s instructions) for 2 hours, rotating at 4°C. α-PABPC1 antibody was purchased from Abcam (ab21060), and α-eIF4G1 from MBL International. EZ view protein G Sepharose (Sigma) was washed twice with lysis buffer and added to lysate with 40 µl slurry used per ml of lysate. The beads and lysate were incubated with the lysate for an additional hour rotating at 4°C. The beads were washed 3X with cold lysis buffer. After the first wash, the beads were transferred to a new tube. The beads were then resuspended in protein loading dye (Life Technologies) with freshly added reducing agent, according to manufacturer’s instructions, and boiled for 3 min. 2% lysate and 10% immunoprecipitants were loaded onto an SDS-PAGE gel and probed for PABPC1 and eIF4G1. α-PABPC1 and α-eIF4G1 were used at 1:1000, and α rabbit IgG HRP (at 1:10,000) was used as the secondary antibody.
Quantification and Statistical Analysis
RNA spot detection, spot assignment and distance measurements
For image analysis, 3D datasets were reduced to 2D data using maximum projections in FiJi. Spot detection was done by 2D Gaussian fitting as described in (Thompson et al., 2002; Zenklusen et al., 2008). For 3D analysis, the spots were detected using AIRLOCALIZE as described in (Lionnet et al., 2011). To separate cytoplasmic and nuclear mRNPs, masks were created in FiJi by manual segmentation using DAPI stained nuclei as reference, while ensuring that regions with overlapping spots within the same channel were not included. Assignment of the 5’, 3’ and/or the mid spots to either the cytoplasmic or the nuclear masks was done using MATLAB (MathWorks). To measure distances between different regions of mRNPs, spots from different channels were first grouped to assign neighboring spots corresponding a single RNA. This was achieved by using spots from one channel as a reference and finding spots from the other channels within a defined radius using the coordinates from 2D Gaussian fitting or 3D Gaussian fitting using a custom MATLAB script. 300nm for 2D analysis and 400nm for 3D analysis were chosen as radii to limit assigning signals from neighboring RNAs. These values were chosen as we observed very few RNAs with distances larger than these thresholds. Moreover, a threshold was required to ensure that there was no wrongful assignment of the signals. Groups with more than one spot from each channel, which could correspond to overlapping mRNPs or mRNPs close together in space, were discarded. For 2 color imaging, the 5’ signal was taken as reference and for 3 color imaging, the middle was taken as reference. Switching references yielded comparable results (not shown). 2D or 3D distances between different regions of the mRNPs were then calculated for each signal within a group.
Combined smFISH and Immunofluorescence Data Analysis
All image analysis was performed using existing or custom build packages in MatLab (MathWorks). Gaussian fitting of smFISH and immunofluorescence spot intensities was performed using FISH-quant (Mueller et al., 2013). Briefly, cytoplasmic FISH spots were fit to a 3D Gaussian to determine the mRNA and translation site coordinates in each color. Both 5’-end, 3’-end, and translation site intensities were detected independently by this method. Image registration was performed by imaging 100 nm TetraSpeck Microspheres (ThermoFisher) and calibrating the field correction based on an affine transformation in a custom built MatLab package. The transformation matrix was first verified for reproducibility on other microsphere samples and then applied to mRNA samples (data not shown). Only 2D distances were considered for this analysis. To determine the end-to-end mRNA distance, we first assigned the Quasar 570 channel (SunTag Probes) to FITC channel (Alexa 488 labeled translation site) by setting a colocalization threshold of 300 nm after image correction. We then assigned the Quasar 570 to Cy5 (MS2 Probes), again with a colocalization threshold of 300 nm. We first grouped mRNA with both Cy3 and Cy5 colocalization, and then determined if there was also a colocalized translation site signal. We then binned two-color mRNA based on the presence (translating) or absence (non-translating) of translation site signal. We then determined the end-to-end distance, and, in the case of the translating mRNAs, the associated translation site intensity.
Data Plotting
All measurements were made for at least 2 independent biological replicates and the data plotted are representative from one of the replicates. For each measurement, at least 5 different fields were imaged with each image containing a minimum of 10 cells to make a total of at least 50 cells. For the smFISH plots, a minimum of 593 RNAs were considered for cytoplasmic plots and a minimum of 430 RNAs were considered for the nuclear plots for data from HEK293 cells and a minimum of 308 RNAs were considered for data from U2OS cells, unless mentioned otherwise. For the FISH-IF plots, a total of 323 data points for translating, 97 for puromycin and 73 for non-translating were considered. The translating mRNAs were clustered using k-means algorithm in R according to the intensity of the site of translation. After clustering, the four groups contained 64, 115, 104 and 40 RNAs from lower to higher intensity. The p-values were calculated using Kolmogorov-Smirnov test in R for the data points plotted. The center of mass plots in Figure 1G, 2D, 6D were made using R. The center of mass was calculated as the mean of the coordinates of the three regions. The different conformations were then aligned using their center of masses. For the 3-color scatter plot in Figure 2E, 6E, S5 and S7B, to get a pair of colocalization precision values, two values were chosen randomly from our data. These values were taken as the X and Y coordinates for the scatter plot. The values that served as the X and Y coordinates were used to get density plots in the same figure. The mean Radius of gyration (<Rg->) was calculated using:
where k represents one of the three regions of the mRNP and rk the position of the corresponding position in space as determined by 2D Gaussian fitting.
To calculate cell doubling times, fluorescence readings were taken at 24, 48, and 72 hrs after seeding the cells. The background fluorescence was subtracted, and the values were then normalized to the 24 hr time point. The slope of the line of best fit (after plotting in linear-log space) was determined and used to give the doubling time for each replicate. The doubling times were calculated in three independent replicates for each cell line, and then plotted as box-and-whisker plots in R.
Supplementary Material
Table S2. smFISH probes used in this study, Related to STAR Methods.
Highlights.
mRNA compaction determined by multi-color, single-molecule mRNA FISH
Nuclear mRNAs have extended morphology
Translating mRNAs rarely show interacting 5’ and 3’ ends
mRNAs in stress-granules are more compacted than translating mRNAs
Acknowledgement
We thank members of the Zenklusen and Rissland laboratories, Marlene Oeffinger, Nicole Francis, Steve Michnick, Jeff Kieft and Julie Claycomb for critical discussion and comments on the manuscript, and Melissa Moore, Job Dekker and Roy Parker for sharing data prior to publication. This work has been supported CIHR (Project Grant-366682), FRQ-S (Chercheurboursier Junior 2), CFI (DZ), and the NIH (R35GM128680) to OSR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
Data and Software Availability
The uncompressed imaging files can be found using this link: doi:10.17632/rjwfnvykd5.1
References
- Afonina ZA, Myasnikov AG, Shirokov VA, Klaholz BP, Spirin AS (2015). Conformation transitions of eukaryotic polyribosomes during multi-round translation. Nucleic Acids Res 43, 618–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Shirokikh NE, Hallwirth CV, Beilharz TH, Preiss T (2015). Probing the closed-loop model of mRNA translation in living cells. RNA Biol 12, 248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batisse J, Batisse C, Budd A, Böttcher B, Hurt E (2009). Purification of nuclear poly(A)-binding protein Nab2 reveals association with the yeast transcriptome and a messenger ribonucleoprotein core structure. J. Biol. Chem 284, 34911–34917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I (2015). Targeting the translation machinery in cancer. Nat. Rev. Drug Discov 14, 261–278. [DOI] [PubMed] [Google Scholar]
- Bin Wu, Eliscovich C, Yoon YJ, Singer RH (2016). Translation dynamics of single mRNAs in live cells and neurons. Science 352, 1430–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodavka A, Singaram SW, Stockley PG, Gelbart WM, Ben-Shaul A, Tuma R (2016). Sizes of Long RNA Molecules Are Determined by the Branching Patterns of Their Secondary Structures. Biophys. J 111, 2077–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt F, Carlson L-A, Hartl FU, Baumeister W, Grünewald K (2010). The three-dimensional organization of polyribosomes in intact human cells. Mol. Cell 39, 560–569.. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Parker R (2009). Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, Rinn JL, Raj A (2015). Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol 16, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Lim J, Ha M, Kim VN (2014). TAIL-seq: Genome-wide Determination of Poly(A) Tail Length and 3′ End Modifications. Mol. Cell 53, 1044–1052. [DOI] [PubMed] [Google Scholar]
- Chen H, Meisburger SP, Pabit SA, Sutton JL, Webb WW, Pollack L (2012). Ionic strength-dependent persistence lengths of single-stranded RNA and DNA. Proc. Natl. Acad. Sci. U. S. A 109, 799–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AK, Bourne CM (1999). Shape of large bound polysomes in cultured fibroblasts and thyroid epithelial cells. Anat. Rec 255, 116–129. [DOI] [PubMed] [Google Scholar]
- Christensen AK, Kahn LE, Bourne CM (1987). Circular polysomes predominate on the rough endoplasmic reticulum of somatotropes and mammotropes in the rat anterior pituitary. Am. J. Anat 178, 1–10. [DOI] [PubMed] [Google Scholar]
- Costello J, Castelli LM, Rowe W, Kershaw CJ, Talavera D, Mohammad-Qureshi SS, Sims PFG, Grant CM, Pavitt GD, Hubbard SJ, Ashe MP (2015). Global mRNA selection mechanisms for translation initiation. Genome Biol 16, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N (2012). The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol 19, 586–593. [DOI] [PubMed] [Google Scholar]
- Floor SN, Doudna JA, Green R (2016). Tunable protein synthesis by transcript isoforms in human cells. Elife 5, e10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR (1991). The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev 5, 2108–2116. [DOI] [PubMed] [Google Scholar]
- Gopal A, Zhou ZH, Knobler CM, Gelbart WM (2012). Visualizing large RNA molecules in solution. RNA 18, 284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, Mero P, Dirks P, Sidhu S, Roth FP, Rissland OS, Durocher D, Angers S, Moffat J (2015). High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 163, 1515–1526. [DOI] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO (2009). Concordant Regulation of Translation and mRNA Abundance for Hundreds of Targets of a Human microRNA. PLoS Biol 7, e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Castello A, Schwarzl T, Preiss T (2018). A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol 19, 327–341. [DOI] [PubMed] [Google Scholar]
- Imataka H, Gradi A, Sonenberg N (1998). A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J 17, 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen C, Pestova TV (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol 11, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, Izaurralde E (2015). Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet 16, 421–433. [DOI] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R (2017). The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol. Cell 68, 808–820.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Parker R (2018). mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction bioRxiv 366690. [DOI] [PMC free article] [PubMed]
- Lai W-JC, Kayedkhordeh M, Cornell EV, Farah E, Bellaousov S, Rietmeijer R, Mathews DH, Ermolenko DN (2018). The formation of intramolecular secondary structure brings mRNA ends in close proximity bioRxiv 289496. [DOI] [PMC free article] [PubMed]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ (2000). The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leija-Martínez N, Casas-Flores S, Cadena-Nava RD, Roca JA, Mendez-Cabañas JA, Gomez E, Ruiz-Garcia J (2014). The separation between the 5“−3” ends in long RNA molecules is short and nearly constant. Nucleic Acids Res 42, 13963–13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH (2011). A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat. Methods 8, 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liphardt J, Onoa B, Smith SB, Tinoco I, Bustamante C (2001). Reversible Unfolding of Single RNA Molecules by Mechanical Force. Science 292, 733–737. [DOI] [PubMed] [Google Scholar]
- Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, Mesirov JP, Cech TR, Chang HY (2016). RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 165, 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabin JW, Woodward LA, Patton R, Yi Z, Jia M, Wysocki V, Bundschuh R, Singh G (2018). The exon junction complex undergoes a compositional switch that alters mRNP structure and nonsense-mediated mRNA decay activity bioRxiv, 355495. [DOI] [PMC free article] [PubMed]
- Marchese D, de Groot NS, Gotor NL, Livi CM, Tartaglia GG (2016). Advances in the characterization of RNA binding proteins. Wiley Interdiscip. Rev. RNA 7, 793–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlin H, Daneholt B, Skoglund U (1995). Structural interaction between the nuclear pore complex and a specific translocating RNP particle. J. Cell Biol 129, 1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metkar M, Ozadam H, Lajoie BR, Imakaev M, Mirny LA, Dekker J, Moore MJ (2018). Higher-Order Organization Principles of Pre-translational mRNPs bioRxiv, 278747. [DOI] [PMC free article] [PubMed]
- Mollet S, Cougot N, Wilczynska A, Dautry F, Kress M, Bertrand E, Weil D (2008). Translationally Repressed mRNA Transiently Cycles through Stress Granules during Stress. Mol. Biol. Cell 19, 4469–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-McNicoll M, Neugebauer KM (2013). How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet 14, 275–287. [DOI] [PubMed] [Google Scholar]
- Mueller F, Senecal A, Tantale K, Marie-Nelly H, Ly N, Collin O, Basyuk E, Bertrand E, Darzacq X, Zimmer C (2013). FISH-quant: automatic counting of transcripts in 3D FISH images. Nat. Methods 10, 277–278. [DOI] [PubMed] [Google Scholar]
- Neve J, Burger K, Li W, Hoque M, Patel R, Tian B, Gullerova M, Furger A (2016). Subcellular RNA profiling links splicing and nuclear DICER1 to alternative cleavage and polyadenylation. Genome Res 26, 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MD, Ivanov P, Anderson P (2016). Mechanistic insights into mammalian stress granule dynamics. J. Cell Biol 215, 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EH, Walker SE, Lee JM, Rothenburg S, Lorsch JR, Hinnebusch AG (2011). Multiple elements in the eIF4G1 N-terminus promote assembly of eIF4G1•PABP mRNPs in vivo. EMBO J 30, 302–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon X, Bastide A, Safieddine A, Chouaib R, Samacoits A, Basyuk E, Peter M, Mueller F, Bertrand E (2016). Visualization of single endogenous polysomes reveals the dynamics of translation in live human cells. J. Cell Biol 214, 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proweller A, Butler JS (1997). Ribosome concentration contributes to discrimination against poly(A)- mRNA during translation initiation in Saccharomyces cerevisiae. J. Biol. Chem 272, 6004–6010. [DOI] [PubMed] [Google Scholar]
- Rahman S, Zorca CE, Traboulsi T, Noutahi E, Krause MR, Mader S, Zenklusen D (2017). Single-cell profiling reveals that eRNA accumulation at enhancer-promoter loops is not required to sustain transcription. Nucleic Acids Res 45, 3017–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakheja D, Chen KS, Liu Y, Shukla AA, Schmid V, Chang T-C, Khokhar S, Wickiser JE, Karandikar NJ, Malter JS, Mendell JT, Amatruda JF (2014). Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat. Commun 2, 4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani V, Qiu R, Shendure J (2015). High-throughput determination of RNA structure by proximity ligation. Nat. Biotechnol 33, 980–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech J (1995). An Ultrastructural Characterization of in Vitro-Assembled hnRNP C Protein-RNA Complexes. J. Struct. Biol 114, 84–92. [DOI] [PubMed] [Google Scholar]
- Rissland OS (2016). The organization and regulation of mRNA–protein complexes. Wiley Interdiscip. Rev. RNA 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Subtelny AO, Wang M, Lugowski A, Nicholson B, Laver JD, Sidhu SS, Smibert CA, Lipshitz HD, Bartel DP (2017). The influence of microRNAs and poly(A) tail length on endogenous mRNA–protein complexes. Genome Biol 18, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ (2012). The Cellular EJC Interactome Reveals Higher-Order mRNP Structure and an EJC-SR Protein Nexus. Cell 151, 750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Pratt G, Yeo GW, Moore MJ (2015). The Clothes Make the mRNA: Past and Present Trends in mRNP Fashion. Annu. Rev. Biochem 84, 325–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirohi D, Chen Z, Sun L, Klose T, Pierson TC, Rossmann MG, Kuhn RJ (2016). The 3.8 Å resolution cryo-EM structure of Zika virus. Science 352, 467–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund U, Andersson K, Strandberg B, Daneholt B (1986). Three-dimensional structure of a specific pre-messenger RNP particle established by electron microscope tomography. Nature 319, 560–564. [DOI] [PubMed] [Google Scholar]
- Souquere S, Mollet S, Kress M, Dautry F, Pierron G, Weil D (2009). Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J. Cell Sci 122, 3619–3626. [DOI] [PubMed] [Google Scholar]
- Steitz JA (1969). Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature 224, 957–964. [DOI] [PubMed] [Google Scholar]
- Strobel EJ, Yu AM, Lucks JB (2018). High-throughput determination of RNA structures. Nat. Rev. Genet 19, 615–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP (2014). Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508, 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ, Sachs AB (1996). Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J 15, 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Thompson MK, Rojas-Duran MF, Gangaramani P, Gilbert WV, Hinnebusch AG (2016). The ribosomal protein Asc1/RACK1 is required for efficient translation of short mRNAs. Elife 5, e11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R, Larson D, Webb W (2002). Precise nanometer localization analysis for individual fluorescent probes. Biophys. J 82, 2775–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Zhou Y, Corbett AH, Wente SR (2007). The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell 28, 850–859. [DOI] [PubMed] [Google Scholar]
- van Heesch S, van Iterson M, Jacobi J, Boymans S, Essers PB, de Bruijn E, Hao W, MacInnes AW, Cuppen E, Simonis M (2014). Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol 15, R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viero G, Lunelli L, Passerini A, Bianchini P, Gilbert RJ, Bernabò P, Tebaldi T, Diaspro A, Pederzolli C, Quattrone A (2015). Three distinct ribosome assemblies modulated by translation are the building blocks of polysomes. J. Cell Biol 208, 581–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Han B, Zhou R, Zhuang X (2016). Real-Time Imaging of Translation on Single mRNA Transcripts in Live Cells. Cell 165, 990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhu L, Dang M, Hu Z, Gao Q, Yuan S, Sun Y, Zhang B, Ren J, Kotecha A, Walter TS, Wang J, Fry EE, Stuart DI, Rao Z (2017). Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site. Proc. Natl. Acad. Sci. U. S. A 114, 770–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells S, Hillner P, Vale R, Sachs A (1998). Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2, 135–140. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, JnBaptiste CK, Lu LY, Kuhn C-D, Joshua-Tor L, Sharp PA (2012). A triple helix stabilizes the 3’ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev 26, 2392–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Hoek TA, Vale RD, Tanenbaum ME (2016). Dynamics of Translation of Single mRNA Molecules In Vivo. Cell 165, 976–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoffe AM, Prinsen P, Gelbart WM, Ben-Shaul A (2011). The ends of a large RNA molecule are necessarily close. Nucleic Acids Res 39, 292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH (2008). Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol 15, 1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. smFISH probes used in this study, Related to STAR Methods.


