Abstract
Introduction
Immunotherapy is regarded as one of the major breakthroughs in cancer treatment. Despite its success, only a subset of patients responds—urging the quest for predictive biomarkers. We hypothesize that artificial intelligence (AI) algorithms can automatically quantify radiographic characteristics that are related to and may therefore act as noninvasive radiomic biomarkers for immunotherapy response.
Patients and methods
In this study, we analyzed 1055 primary and metastatic lesions from 203 patients with advanced melanoma and non-small-cell lung cancer (NSCLC) undergoing anti-PD1 therapy. We carried out an AI-based characterization of each lesion on the pretreatment contrast-enhanced CT imaging data to develop and validate a noninvasive machine learning biomarker capable of distinguishing between immunotherapy responding and nonresponding. To define the biological basis of the radiographic biomarker, we carried out gene set enrichment analysis in an independent dataset of 262 NSCLC patients.
Results
The biomarker reached significant performance on NSCLC lesions (up to 0.83 AUC, P < 0.001) and borderline significant for melanoma lymph nodes (0.64 AUC, P = 0.05). Combining these lesion-wide predictions on a patient level, immunotherapy response could be predicted with an AUC of up to 0.76 for both cancer types (P < 0.001), resulting in a 1-year survival difference of 24% (P = 0.02). We found highly significant associations with pathways involved in mitosis, indicating a relationship between increased proliferative potential and preferential response to immunotherapy.
Conclusions
These results indicate that radiographic characteristics of lesions on standard-of-care imaging may function as noninvasive biomarkers for response to immunotherapy, and may show utility for improved patient stratification in both neoadjuvant and palliative settings.
Keywords: immunotherapy, medical imaging, response prediction, artificial intelligence, machine learning, radiomics
Key Message
This study investigated the performance of machine learning algorithms to quantify radiographic characteristics of cancer lesions predictive of response to immunotherapy. The resulting noninvasive radiomic biomarker showed predictive performance and association with proliferative potential.
Introduction
Cancer immunotherapy has made promising strides as a result of improved understanding of biological interactions between tumor cells and the immune system. Both the EMA and the FDA have approved anti-PD1 antibodies to treat melanoma or non-small-cell lung cancer (NSCLC) patients with unresectable or metastatic disease, which progressed under platinum-based chemotherapy or display high expression of PD-L1 [1–4]—with overall response rates of 44% and 32% in first and second line in melanoma [5, 6] and 19% in second line in lung cancer [7–9]. Unlike traditional cancer treatment, anti-PD1 antibodies potentiate the antitumor immune response.
Despite their remarkable success, clinical benefit remains limited to only a subset of patients [10]. As immunotherapy is expensive and could bring toxicity, there is a need to stratify patients according to likely benefit before therapy. Different biomarkers have been investigated with variable success, such as levels of PD-L1 [11–13], presence of tumor-infiltrating lymphocytes [14, 15], genetic mutations [16–18] and inflammatory cytokines [19].
Recent emergence of quantitative imaging biomarkers provide promising opportunities. Unlike traditional biopsy-based assays that represent only a sample of the tumor, images reflect the entire tumor burden, providing information on each cancer lesion with a single noninvasive examination. This is of particular importance in immunotherapy, where different lesions can have different microenvironments potentially leading to heterogeneous response patterns [20]. Previously, radiolabeled anti-PD1 antibodies were used to visualize specific immunological expressions [21].
Computational imaging approaches originating from artificial intelligence (AI) have achieved impressive successes in automatically quantifying radiographic characteristics of tumors [22]. AI-based characterization on radiology is referred to as ‘radiomics’ and can provide more detailed characterization than possible by eye [22–24]. Radiomics-based biomarkers have shown success in different tumor types [25–31]; but to the best of our knowledge, there is no evidence yet in immunotherapy. Tumor morphology, visualized on imaging, is likely influenced by several aspects of tumor biology. We hypothesize that a set of morphological characteristics, quantified by radiomics, are related to and may therefore act as predictive markers.
In this study, we analyzed all visible cancer lesions to evaluate the potential predictive value of CT-derived radiomic biomarkers in metastatic NSCLC and melanoma patients receiving immunotherapy. A biologic evaluation was carried out in an independent validation set of surgical NSCLC patients with imaging and gene-expression data.
Methods–patients
Immunotherapy dataset
Patients with metastatic melanoma or NSCLC receiving 3 mg/kg/2 weeks of anti-PD1 at the Netherlands Cancer Institute (NKI) between 2014 and 2016 were retrospectively analyzed (for more information see supplementary material S1, available at Annals of Oncology online). Contrast-enhanced computed tomography (CE-CT) scans were acquired before (baseline) and around 12 weeks after start of treatment (follow-up) (see supplementary material S2, available at Annals of Oncology online). The study protocol was approved by the Medical Ethics Committee and Board of Directors of the NKI and informed consent was waived.
Genomics dataset
To provide biological validation, we evaluated an independent, dataset of surgical NSCLC patients between 2006 and 2009 treated at the H. Lee Moffitt Cancer Center. Pre-surgical CE-CT (within 60 days of diagnosis) and gene expression data were available for 262 patients. The University of South Florida IRB approved and waived informed consent (IRB#16069); in accordance with HIPAA (more information in supplementary material S4, available at Annals of Oncology online [32]).
Chemotherapy dataset
To study the specificity of the radiomic biomarker for immunotherapeutic response prediction, we retrospectively collected a cohort of 39 patients with stage IV NSCLC treated with cytotoxic chemotherapy at NKI between 2015 and 2016 (IRBd18079, more information in supplementary material S4, available at Annals of Oncology online).
Imaging data and lesion segmentations
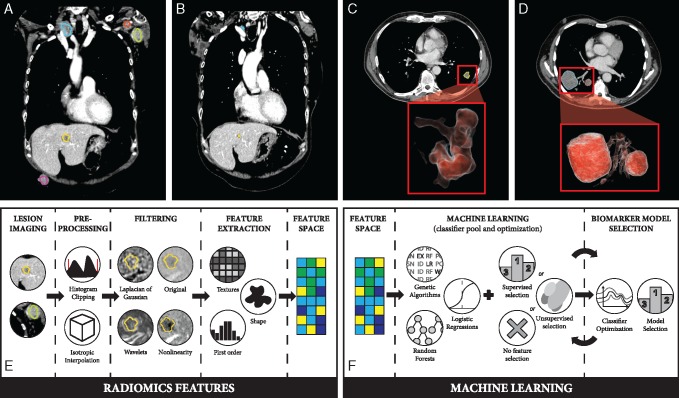
Experienced readers manually delineated lesions on baseline and follow-up scans. Target lesions were defined as any tumor that was well-demarcated on both baseline and follow-up with diameter ≥5 mm (see supplementary material S5, available at Annals of Oncology online). Examples are shown in Figure 1A and B.
Figure 1.
(A) Baseline contrast-enhanced CT scan of melanoma patient presenting with metastases in the liver and lymph nodes in the axilla and subclavicular area. (B) Follow-up scan of the same patient showing complete response in the axillary region and partial response of the lesions in the liver and neck. (C) Baseline CT scan of an NSCLC patient presenting lesion in the left lung, that showed progression at a later FU CT (data not shown). (D) Baseline CT scan of a melanoma patient presenting lesions in the right lung that showed response at a later FU CT (data not shown). (E) Schematic representation of the radiomics feature extraction process. (F) Schematic of the machine learning process.
Response kinetics
To assess the effects of mixed response, we carried out a lesion-per-lesion assessment of relative change in diameter between baseline and follow-up, using RECIST criteria. Furthermore, in patients with >1 lesion, we classified response patterns on a patient basis as mixed for patients presenting both responding and progressive lesions and uniform for patients presenting only responding or progressive lesions (irrespective of stable lesions). This setup allows for the characterization of overall tumor burden.
Radiographic differences between responding and progressive lesions
To generate radiomic sequences for each lesion at baseline, a set of radiomic features was defined [33] (see Figure 1E and supplementary material S6, available at Annals of Oncology online). Radiomic features of responding and progressive lesions were directly compared with identify differences. To reduce redundancy, 10 complementary features were selected using unsupervised feature selection [34]. Statistical significance was assessed using generalized mixed-effect models—controlling for patient, tumor type and organ. False discovery rate was at 10% to correct for multiple comparisons.
Radiomic biomarkers to predict immunotherapy response of cancer lesions
To assess the performance of the radiomic biomarker, we developed a machine learning model [35–37]. We trained the model on all lesions (i.e. progressive, stable and responding) to discern progressive disease.
The dataset was divided into training, tuning and testing sets based on patient identifiers. The training set was used to model data distributions. The tuning set was used during training to control for overfitting. The test set was used for independent evaluation (see Figure 1F and supplementary materials S7 and S8, available at Annals of Oncology online). Mann–Whitney U test was used for statistical testing of AUC curves, one-sided McNeils test was used to test if the radiomic biomarker was outperforming volume and maximum diameter, and log-rank test was used for statistical testing of survival performance.
To test for radiomic association with molecular pathways, Spearman’s rank correlation coefficient was used. Pathways were then ranked by −log10(p), where p is the correlation P-value, and put into a preranked gene set enrichment analysis (GSEA) algorithm [38] version 2.0.14 on the C2 collection version Molecular Signature Database (MSigDB) [39].
Results
Immunotherapy response kinetics
To assess immunotherapy response kinetics, 203 (123 NSCLC, 80 melanoma) patients were analyzed with a total of 1055 target lesions (see supplementary material S1, available at Annals of Oncology online). Lesions were similarly distributed between NSCLC (n = 572, 54%) and melanoma (n = 483, 46%). The most common lesion sites were lung (n = 359, 34%), lymph nodes (n = 312, 30%) and liver (n = 212, 20%). Most lesions (n = 746 versus 309, χ2 test P < 0.01) showed either stable (n = 395) or partial response (n = 351).
Melanoma lesions showed better overall response than NSCLC (40% versus 27% responding, P < 0.01; 23% versus 34% progression, P < 0.01). This trend was more evident for lung lesions, where we observed progression in NSCLC (39% versus 14%, P < 0.01) and response in melanoma (48% versus 26%, P < 0.01). Hepatic melanoma lesions showed response in comparison with NSCLC (22% versus 36%, P = 0.04). Examples are shown in Figure 1C and D.
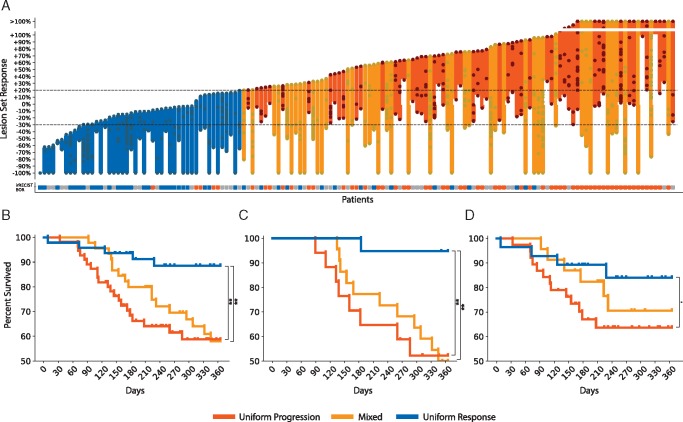
Comparing per-patient response patterns in both cancer types, we observed that 23% (n = 47) showed uniform response, 27% (n = 55) uniform progression and 22% (n = 45) mixed response. The remaining 28% (n = 56) of the patients did not have multiple target lesions or presented only stable lesions. Significantly higher survival rates were seen in uniform response (log-rank test, P < 0.01). This was evident in melanoma (log-rank-test, P < 0.01), while in NSCLC, despite similar trends, did not reach significance (P = 0.08). Per-patient response kinetics are shown in Figure 2A. Kaplan–Meier curves are shown in Figure 2B and D.
Figure 2.
(A) Response kinetics curve depicting individual lesion responses (as dots) on a patient-to-patient basis. (B) One-year survival plot for all analyzed patients (C) for melanoma patients only, (D) for NSCLC patients only.
Radiographic differences between responding and progressive lesions
To investigate radiographic differences between responding and progressing lesions, we compared their radiomic features (see supplementary Figure S9.1A and material S2, available at Annals of Oncology online). Among the most common locations (lung, lymph nodes, liver and adrenal gland), responding lesions presented higher levels of irregular patterns (Wavelet. HLH_GLSZM_ZoneEntropy, Kenward–Roger test P = 0.007) with more compact, spherical profiles (SurfaceVolumeRatio, P = 0.01). Subanalysis on location revealed increased values of morphological heterogeneity in hepatic, nodal and splenic lesions associated with response (P < 0.02).
Of the most common NSCLC lesions, similar trends for morphological heterogeneity were seen at the organ level in pulmonary and hepatic lesions, as well as lymph nodes also characterized by the presence of hypodense regions (P = 0.007). No significance was observed in primary NSCLC tumors. Among most common melanoma lesions greater morphological heterogeneity showed association with response (GLCM_DifferenceEntropy, P = 0.006). Similar trends for morphological heterogeneity were seen but lower sample numbers did not allow to pass the patient correction.
Radiomic biomarker to predict immunotherapy responding and stable lesions
To assess the performance of radiomics to rule out progression, we used machine learning to develop a single radiomic biomarker with 133 patients in the discovery set and 70 patients in test (see supplementary material S3, available at Annals of Oncology online). A random forest with wrapper feature selection was used to develop radiomic biomarkers based on the performance in the discovery set (see supplementary material S7, available at Annals of Oncology online) and was validated on the independent test set.
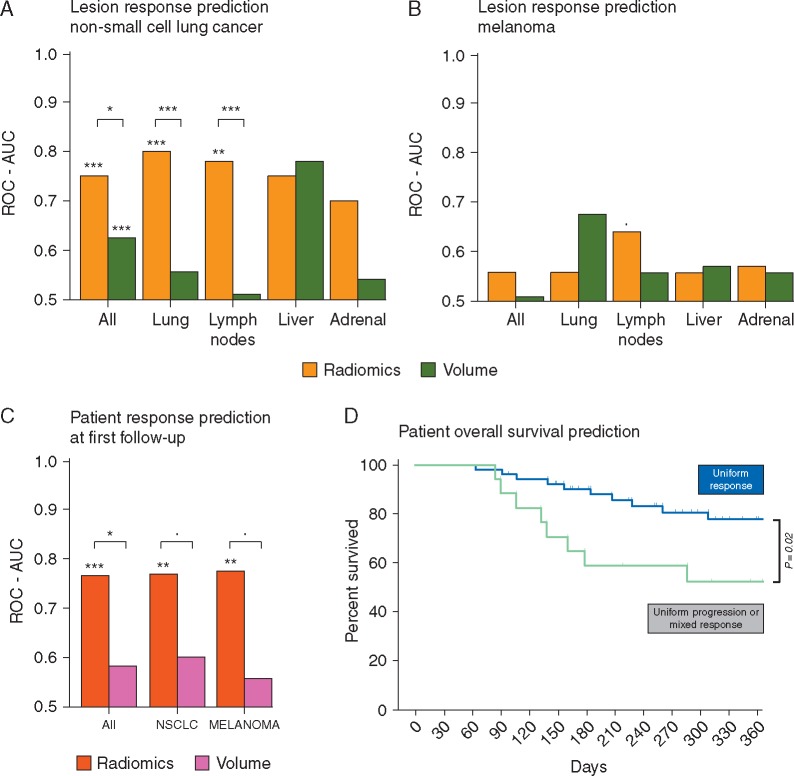
In NSCLC, radiomic biomarker from pulmonary (0.83 AUC, Mann–Whitney U test P < 0.001) and nodal metastases (0.78 AUC, P < 0.001) showed significant performance. Satisfactory performance was observed in NSCLC primary tumors (0.79 AUC, P = 0.05), hepatic (0.75 AUC, P = 0.13) and adrenal lesions (0.70 AUC, P = 0.18) but did not reach significance mostly due to the low number of samples. The model carried out poorly on both pulmonary and hepatic melanoma lesions (0.55 AUC). Despite these results, a trend toward significance is shown in nodal metastases (0.64 AUC, P = 0.05) (see Figure 3A). Evaluation of the radiomic biomarker on all 303 lesions within the test dataset resulted in significant predictive performance (0.66 AUC, P < 0.01; see supplementary material S3, available at Annals of Oncology online).
Figure 3.
Performance of the selected classifier on the independent test set for NSCLC lesions (A) and melanoma lesions (B). (C) Patient level response at first follow-up and (D) prognostic performance of the imaging biomarker on a patient level.
By combining predictions made on individual lesions, it is possible to do a pretreatment patient-wise prediction of immunotherapy response (see Methods section). Significant performances were observed to predict OS for both tumor types (0.76 AUC for all patients, P < 0.01; 0.76 AUC for NSCLC patients, P < 0.01; 0.77 AUC for melanoma patients, P < 0.01; see Figure 3B), with a significant survival difference at 1-year of 25% (77% versus 52%, log-rank-test, P = 0.02; see Figure 3C). Interestingly, in melanoma patients, we observed significant performance to predict OS and response, despite the lower performance on a lesion level.
This radiomic immunotherapy response biomarker could not significant predict overall survival in patients treated with cytotoxic chemotherapy (P = 0.07), nor in terms of overall patient response (AUC = 0.63; P = 0.09). In terms of lesion response, the biomarker was inversely correlated to response of lung lesions in nonimmunotherapy patients (n = 61, AUC = 0.70, P = 0.04), but did not show any significant predictive value in the remaining nodal (n = 61, AUC = 0.59, P = 0.24) and liver lesions (n = 12, AUC = 0.65, P = 0.29). See supplementary Figure S9.D, available at Annals of Oncology online.
Biological validation of the radiomic biomarker
To evaluate the biological basis of the radiomic biomarker, we evaluated it in an independent dataset of 262 NSCLC patients with matched array-based gene expression data [32]. Through ranked GSEA, we found that the top gene sets showing significant association with the radiomic biomarker were involved in cell cycle progression and mitosis (supplementary Figure S9.1B, available at Annals of Oncology online). This indicates that a link between high tumor proliferation and improved response to immunotherapy may exist, and provides rationale for early-application immunotherapy as a therapeutic option for aggressive rapidly expanding cancers.
Discussion
Our aim was to evaluate radiomics-based models and their potential to predict treatment response in metastatic melanoma and NSCLC patients receiving anti-PD-1 antibodies.
We found that lesions with more heterogeneous morphological profiles with nonuniform density patterns and compact borders are more likely to respond to immunotherapy—irrespective of organ and/or cancer type. Higher levels of SurfaceVolumeRatio in nonresponding lesions in both cancers suggest that more compact and spherical profiles are associated with better response.
Based on these results, it would be prudent to point out that morphological heterogeneity does not necessarily correspond to genetic heterogeneity: infiltration, inflammation, neovascularization and necrosis could also be associated with morphological irregularities. Assuming that a well-vascularized monoclonal tumor growing in the absence of an immune system would expand uniformly in all directions, any deviation could suggest a fault of one of aforementioned characteristics. If we were to relax one of these conditions, e.g. by adding an immune system, we would observe infiltration and inflammatory microenvironment [16] affecting the tumor morphology—now comprising more than solely tumor cells. Irregular vascularization might cause nonhomogeneous growth patterns while hampering T-cell infiltration [40]. The role of the other compartments need to be taken into account in order to explain the overall tumor growth.
Overall results of machine learning model show good predictive performance for NSCLC metastases. In melanoma, the same model carried out poorly. Besides the smaller melanoma cohort, the heterogeneous therapeutic backgrounds likely played a role in the morphological characterization. While NSCLC patients received chemotherapy as first-line, melanoma patients received a variety of different treatments before immunotherapy. This could potentially have led to standardization of defined genetic profiles and tumor microenvironments in NSCLC [23, 32, 41, 42]. In melanoma patients the diversity of therapeutic backgrounds might have induced different genetic profiles and microenvironments. Despite the lower performance on individual melanoma lesions, we still see a correlation with response and overall survival at a patient level, suggesting a relationship between individual lesion response and overall tumor burden.
GSEA on an external cohort revealed associations of the radiomic biomarker to proliferative potential in NSCLC, suggesting that highly proliferative tumors may show preferential response to immunotherapy. While standard of care for patients with aggressive cancer showing rapid expansion is platinum-doublet chemotherapy, these results provides the biological rationale for previous work demonstrating why combination therapy is a viable option in first-line metastatic settings [43].
To the best of our knowledge, this study is the first of its kind to investigate radiomics as a noninvasive biomarker for response to cancer immunotherapy.
We designed the study using a lesion-based approach, reflecting the metastatic condition characterizing patients receiving immunotherapy. This enabled us to investigate lesions individually while avoiding the issue of mixed response. Whenever possible, we limited selection biases and tried to avoid overfitting. Further validation in larger cohorts is warranted.
As imaging can provide information of the total tumor burden which allows the analysis of each lesion individually, its value lies complementary to currently known biomarkers (limited to single lesion samples). Despite the correlations found to overall patient survival and molecular pathways, further studies are needed to investigate the interaction between single (or clusters of) lesions, tumor biology and clinical status. Only a multidisciplinary approach aimed to integrate data from different disciplines can create a fully integrated solution that can be implemented into the clinical workflow.
Conclusions
Our findings suggest associations between radiomics characteristics and immunotherapy response showing consistent trends across cancer types and anatomical location. Lesions that are more likely to respond to immunotherapy typically present with more heterogeneous morphological profiles with nonuniform density patterns and compact borders. Moreover, we provide a predictive machine learning model that could be used within the context of lesion response to treatment, patient treatment response, and response pattern characterization. Furthermore, we evaluated the biological basis of the proposed biomarker and found to be correlated with cell proliferative potential. Motivated by the results and the wide availability of routine clinical CT scans for cancer immunotherapy patients, we aim to expand this research further to the design of clinically applicable automatic computer models that could support the oncological decision-making process.
Funding
This work was supported by the Dutch national e-infrastructure with the support of the SURF Cooperative. The authors acknowledge financial support from the Informatics Technology for Cancer Research (ITCR) program (NIH-USA U24CA194354) and the Quantitative Imaging Network (QIN) program (NIH-USA U01CA190234) of the NIH.
Disclosure
CS reports grant support from Cancer Research UK, UCLH Biomedical Research Council, and Rosetrees Trust, AstraZeneca and personal fees from Boehringer Ingelheim, Novartis, Eli Lilly, Roche, GlaxoSmithKline, Pfizer, Servier, MSD, BMS, AstraZeneca, Illumina, Sarah Canon Research Institute and Celgene. CS also reports stock options in GRAIL, APOGEN Biotechnologies, and EPIC Bioscience and has stock options and is co-founder of Achilles Therapeutics. HJWLA reports shares from Genospace and Sphera, outside of the submitted work. All remaining authors have declared no conflicts of interest.
Supplementary Material
References
- 1.U.S. Food and Drug Administration. Drugs (KEYTRUDA Label). https://www.accessdata.fda.gov/drugsatfda_docs/label/2017/125514s014l bl.pdf (1 April 2019, date last accessed).
- 2.U.S. Food and Drug Administration. Drugs (OPDIVO Label). https://www.accessdata.fda.gov/drugsatfda_docs/label/2017/125554s055lbl.pdf (1 April 2019, date last accessed).
- 3.Opdivo: EPAR—Product Information. http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Product_Information/human/00398 5/WC500189765.pdf (1 April 2019, date last accessed).
- 4.Keytruda: EPAR—Product Information. http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Product_Information/human/003820/WC500190990.pdf (1 April 2019, date last accessed).
- 5. Wolchok JD, Chiarion-Sileni V, Gonzalez R. et al. Efficacy and safety results from a phase III trial of nivolumab (NIVO) alone or combined with ipilimumab (IPI) versus IPI alone in treatment-naive patients (pts) with advanced melanoma (MEL) (CheckMate 067). JCO 2015; 33(Suppl 18): LBA1. [Google Scholar]
- 6. Weber JS, D'Angelo SP, Minor D. et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol 2015; 16(4): 375–384. [DOI] [PubMed] [Google Scholar]
- 7. Borghaei H, Paz-Ares L, Horn L. et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med 2015; 373(17): 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brahmer J, Reckamp KL, Baas P. et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med 2015; 373(2): 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Herbst RS, Baas P, Kim D-W. et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016; 387(10027): 1540–1550. [DOI] [PubMed] [Google Scholar]
- 10. Hodi FS, O’Day SJ, McDermott DF. et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010; 363(8): 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ma W, Gilligan BM, Yuan J, Li T.. Current status and perspectives in translational biomarker research for PD-1/PD-L1 immune checkpoint blockade therapy. J Hematol Oncol 2016; 9(1): 47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meng X, Huang Z, Teng F. et al. Predictive biomarkers in PD-1/PD-L1 checkpoint blockade immunotherapy. Cancer Treat Rev 2015; 41(10): 868–876. [DOI] [PubMed] [Google Scholar]
- 13. Kerr KM, Tsao M-S, Nicholson AG. et al. Programmed death-ligand 1 immunohistochemistry in lung cancer: in what state is this art? J Thorac Oncol 2015; 10(7): 985–989. [DOI] [PubMed] [Google Scholar]
- 14. Zito Marino F, Ascierto PA, Rossi G. et al. Are tumor-infiltrating lymphocytes protagonists or background actors in patient selection for cancer immunotherapy? Expert Opin Biol Ther 2017; 17(6): 735–746. [DOI] [PubMed] [Google Scholar]
- 15. He Y, Rozeboom L, Rivard CJ. et al. PD-1, PD-L1 protein expression in non-small cell lung cancer and their relationship with tumor-infiltrating lymphocytes. Med Sci Monit 2017; 23: 1208–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McGranahan N, Furness AJS, Rosenthal R. et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016; 351(6280): 1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rizvi NA, Hellmann MD, Snyder A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015; 348(6230): 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hellmann MD, Ciuleanu T-E, Pluzanski A. et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med 2018; 378(22): 2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ayers M, Lunceford J, Nebozhyn M. et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017; 127(8): 2930–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene 2008; 27(45): 5904–5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu AM. Antibodies and antimatter: the resurgence of immuno-PET. J Nucl Med 2009; 50(1): 2–5. [DOI] [PubMed] [Google Scholar]
- 22. Hosny A, Parmar C, Quackenbush J. et al. Artificial intelligence in radiology. Nat Rev Cancer 2018; 18: 500–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aerts H, Velazquez ER, Leijenaar RTH. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014; 5: 4006.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aerts H, Hugo JW.. The potential of radiomic-based phenotyping in precision medicine. JAMA Oncol 2016; 2(12): 1636.. [DOI] [PubMed] [Google Scholar]
- 25. Coroller TP, Agrawal V, Narayan V. et al. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol 2016; 119(3): 480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kirienko M, Cozzi L, Antunovic L. et al. Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur J Nucl Med Mol Imaging 2017; 45: 207–217. [DOI] [PubMed] [Google Scholar]
- 27. Fave X, Zhang L, Yang J. et al. Using pretreatment radiomics and delta-radiomics features to predict non-small cell lung cancer patient outcomes. Int J Radiat Oncol Biol Phys 2017; 98(1): 249. [Google Scholar]
- 28. Parmar C, Grossmann P, Rietveld D. et al. Radiomic machine-learning classifiers for prognostic biomarkers of head and neck cancer. Front Oncol 2015; 5: 272.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kickingereder P, Burth S, Wick A. et al. Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology 2016; 280(3): 880–889. [DOI] [PubMed] [Google Scholar]
- 30. Prasanna P, Patel J, Partovi S. et al. Radiomic features from the peritumoral brain parenchyma on treatment-naïve multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: preliminary findings. Eur Radiol 2017; 27(10): 4188–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H, Zhu Y, Burnside ES. et al. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Radiology 2016; 281(2): 382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grossmann P, Stringfield O, El-Hachem N. et al. Defining the biological basis of radiomic phenotypes in lung cancer. elife 2017; 6: e23421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van Griethuysen JJM, Fedorov A, Parmar C. et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res 2017; 77: e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Swiniarski RW, Skowron A. Rough set methods in feature selection and recognition. Pattern Recognit Lett 2003; 24(6): 833–849. [Google Scholar]
- 35. Cox DR. The regression analysis of binary sequences. J R Stat Soc Ser B Stat Methodol 1958; 20(2): 215–242. [Google Scholar]
- 36. Breiman L. Machine learning. Random Forests 2001; 45: 5–32. [Google Scholar]
- 37. Olson RS, Bartley N, Urbanowicz RJ, Moore JH.. Evaluation of a tree-based pipeline optimization tool for automating data science In Proceedings of the Genetic and Evolutionary Computation Conference 2016. New York, NY, USA: ACM; 2016; 485–492. [Google Scholar]
- 38. Subramanian A, Tamayo P, Mootha VK. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102(43): 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liberzon A, Subramanian A, Pinchback R. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011; 27(12): 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang Y, Goel S, Duda DG. et al. Vascular normalization as an emerging strategy to enhance cancer immunotherapy. Cancer Res 2013; 73(10): 2943–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Greaves M, Maley CC.. Clonal evolution in cancer. Nature 2012; 481(7381): 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rios Velazquez E, Parmar C, Liu Y. et al. Somatic mutations drive distinct imaging phenotypes in lung cancer. Cancer Res 2017; 77(14): 3922–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gandhi L, Rodríguez-Abreu D, Gadgeel S. et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med 2018; 378(22): 2078–2092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.