Summary
RfaH is required for virulence in several Gram-negative pathogens including Escherichia coli and Klebsiella pneumoniae. Through direct interactions with RNA polymerase (RNAP) and ribosome, RfaH activates the expression of capsule, cell wall and pilus biosynthesis operons by reducing transcription termination and activating translation. While E. coli RfaH has been extensively studied using structural and biochemical approaches, limited data are available for other RfaH homologs. Here we set out to identify small molecule inhibitors of E. coli and K. pneumoniae RfaHs. Results of biochemical and functional assays show that these proteins act similarly, with a notable difference between their interactions with the RNAP β subunit gate loop. We focused on high-affinity RfaH interactions with the RNAP β’ subunit clamp helices as a shared target for inhibition. Among the top 10 leads identified by in silico docking using ZINC database, 3 ligands were a o inhibit E. coli RfaH recruitment in vitro. The most potent lead was active against both E. coli and K. pneumoniae RfaHs in vitro. Our results demonstrate the feasibility of identifying RfaH inhibitors using in silico docking and pave the way for rational design of antivirulence therapeutics against antibiotic-resistant pathogens.
Introduction
Klebsiella pneumoniae, a leading cause of pneumonia in hospitalized patients, has been identified as an urgent public health threat by the World Health Organization and the US Centers for Disease Control and Prevention (Chen et al., 2014; Viaggi et al., 2015). Rapidly spreading resistance to carbapenems, antibiotics of last resort, necessitates the development of novel therapeutics effective against K. pneumoniae and other multidrug-resistant Gram-negative pathogens. In an attempt to identify novel K. pneumoniae targets for intervention, one study utilized transposon insertion mutagenesis to identify genes required for K. pneumoniae strain KPPR1 fitness in a murine model of pneumonia (Bachman et al., 2015). Among these genes, a null mutant of rfaH displayed a greater than 10,000-fold fitness defect in the lung, an effect surpassed only by the disruption of wzi, a gene from the capsular biosynthesis operon which is likely activated by RfaH (Rahn et al., 1999). Bachman et al. demonstrated that RfaH was required for capsule production and resistance to complement-mediated serum killing in KPPR1 (Bachman et al., 2015). The contribution of RfaH to pathogen virulence is widespread, as it is known to be required in Escherichia coli (Nagy et al., 2002), Salmonella enterica serovar Typhimurium (Nagy et al., 2006) and possibly Vibrio vulnificus (Garrett et al., 2016). Additionally, RfaH paralogs encoded on conjugative plasmids could directly activate the spread of antibiotic-resistance genes encoded on these plasmids (NandyMazumdar and Artsimovitch, 2015). Mounting evidence identifies RfaH as a promising, wide-ranging target for drug discovery.
In addition to its essential role in virulence, RfaH utilizes a unique mechanism to activate both transcription and translation of its target genes. RfaH belongs to a ubiquitous NusG family of transcription factors but, in contrast to NusG which plays housekeeping roles, RfaH-like proteins are highly specialized (NandyMazumdar and Artsimovitch, 2015). RfaH homologs are required for expression of long operons that encode biosynthesis of capsules, LPS core, antibiotics, toxins and pili in diverse bacterial species ranging from E. coli to Bacillus amyloliquefaciens (Goodson et al., 2017). While limited functional information exists for most RfaHs, including that from K. pneumoniae, E. coli (Eco) RfaH is one of the best-characterized transcription factors. RfaH is recruited to the transcribing RNA polymerase (RNAP) through specific interactions with the single-stranded ops element in the non-template DNA strand within the transcription bubble (Artsimovitch and Landick, 2002). Following recruitment, RfaH interacts with RNAP and the ribosome to activate the expression of horizontally acquired target genes, which are inefficiently translated and thus silenced by the transcription termination factor Rho. RfaH abrogates Rho-dependent termination by three mechanisms. First, RfaH inhibits RNAP pausing, which is a prerequisite to termination (Artsimovitch and Landick, 2002; Sevostyanova et al., 2011). Second, RfaH excludes NusG, which is required for the efficient RNA release at suboptimal Rho sites (Peters et al., 2012), via competition for a shared binding site (Kang et al., 2018). Third, RfaH activates translation by recruiting the 30S ribosome subunit to mRNA via protein-protein contacts with S10 (Burmann et al., 2012). This alternative recruitment mechanism is essential for the expression of Eco RfaH-controlled operons which lack Shine-Dalgarno (SD) mRNA elements (Burmann et al., 2012) and thus cannot recruit 30S via basepairing with a complementary sequence in the 16S rRNA (Gualerzi and Pon, 2015).
The available high-resolution X-ray, NMR and cryo-EM structures of Eco RfaH alone or bound to the ops DNA, the transcription elongation complex (TEC) or the ribosomal protein S10 (Belogurov et al., 2007, Burmann et al., 2012; Kang et al., 2018; Zuber et al., 2018) could be used to guide the design of RfaH inhibitors. Many mutants of key amino acid residues of RfaH (Belogurov et al., 2010) and ops bases (Zuber et al., 2018) as well as RNAP residues that make contacts to RfaH (Sevostyanova et al., 2011) are also available. Reporter assays and in vitro transcription assays of RfaH mechanism have been extensively validated (Artsimovitch and Landick, 2002; Belogurov et al., 2007; Burmann et al., 2012). In this work, we wanted to assess whether K. pneumoniae (Kpn) and Eco RfaH work similarly enough to justify the use of Eco RfaH as a template for the development of Kpn RfaH inhibitors. In particular, we wanted to find out whether all functional interactions characterized for Eco RfaH are essential in K. pneumoniae, thus representing shared targets for potential inhibitors.
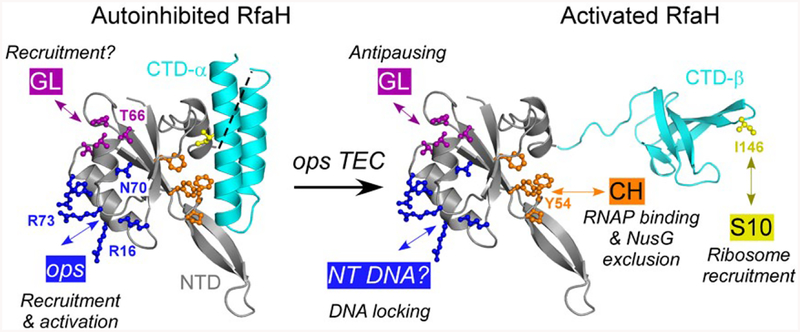
Eco RfaH consists of two domains connected by a flexible linker. The N-terminal domain (NTD) interacts with RNAP in all characterized RfaH homologs. The C-terminal domain (CTD) has an unprecedented ability to reversibly switch folds between the α-helical hairpin that masks the RNAP-binding site on the NTD in the absence of the ops signal (Belogurov et al., 2007) and the β-barrel that interacts with S10 (Burmann et al., 2012). The available data demonstrate that Eco RfaH activation of gene expression relies on four sets of interactions (Fig. 1). Binding of a cluster of charged NTD residues (that includes Arg73) to the ops DNA is necessary to relieve autoinhibition by triggering the dissociation of the CTD to expose the RNAP- and the ribosome-binding sites (Belogurov et al., 2007). Binding of a hydrophobic surface (that includes Tyr54) to the β’ clamp helices (β’ CH) is thought to make high-affinity interactions that persist throughout transcription (Belogurov et al., 2009). Contacts between the HTTT motif of RfaH (residues His65 through Thr68; Fig. S1) and the β subunit gate loop (GL) motif are required for antipausing effects (Sevostyanova et al., 2011). Finally, contacts between the Leu145 and Ile146 CTD residues and the ribosomal protein S10 enable ribosome recruitment in the absence of a SD element (Burmann et al., 2012). While one could assume that these interactions are preserved in all RfaH orthologs, these proteins are quite divergent (Carter et al., 2004) and studies of the highly conserved housekeeping paralog NusG revealed significant differences among bacterial species (Mooney et al., 2009; Sevostyanova and Artsimovitch, 2010; Czyz et al., 2014; Yakhnin et al., 2016).
Fig. 1.
Key residues in Eco RfaH. In the autoinhibited state, the NTD (gray) and CTD (α-helices; cyan) interact to bury the NTD residues that bind to the β’CH domain (orange). In the active state, the domains are connected by a (modeled) flexible linker; the NTD and the β-barrel CTD bind to RNAP and S10 respectively. In both states, the NTD can interact with the nontemplate (NT) ops DNA element (blue) and the βGL (dark magenta).
In this work, we found that the ops element and contacts to the β’CH and S10 are required for RfaH function in K. pneumoniae in vivo, whereas contacts to GL appear to be partially dispensable. This conclusion is supported by footprinting and in vitro transcription analyses. Based on these findings, we carried out an in silico search for small molecules that could interfere with RfaH interactions with RNAP. We successfully identify a lead molecule predicted to bind at the NTD-β’CH interface and demonstrate inhibition of Eco and Kpn RfaH recruitment to RNAP in vitro.
Results
Eco RfaH and Kpn RfaH substitute for each other in vivo
In contrast to Eco RfaH, which has been extensively studied, little mechanistic information is available for its orthologs, even in closely related species. In our early work, we demonstrated that Eco and Kpn RfaHs had similar stimulatory effects on the expression of the plasmid-borne hemolysin (hly) operon, the best characterized Eco RfaH target at the time, in E. coli (Carter et al., 2004). In this operon, RfaH appears to reduce termination at an unusual weak hairpin-dependent terminator between the hlyA and hlyB genes (Koronakis et al., 1988), an effect that is distinct from other characterized RfaH-dependent operons in which RfaH counteracts Rho-mediated polarity (NandyMazumdar and Artsimovitch, 2015). We wanted to test whether Eco and Kpn RfaH proteins have similar effects on the expression of chromosomal operons activated by RfaH. In E. coli, deletion of rfaH confers dramatic sensitivity to SDS, an effect that is phenocopied by an early polar mutation in the RfaH-activated waa LPS biosyn-thesis operon (Moller et al., 2003) and suppressed by mutations in rho (Hu and Artsimovitch, 2017). In K. pneumoniae, the deletion of rfaH leads to decreases in capsule production (Bachman et al., 2015); similar effects, attributed to a significant similarity between the capsule biosynthesis clusters, were observed in E. coli (Navasa et al., 2014). Eco RfaH has been shown to inhibit Rho-dependent termination within capsule operons (Stevens et al., 1997).
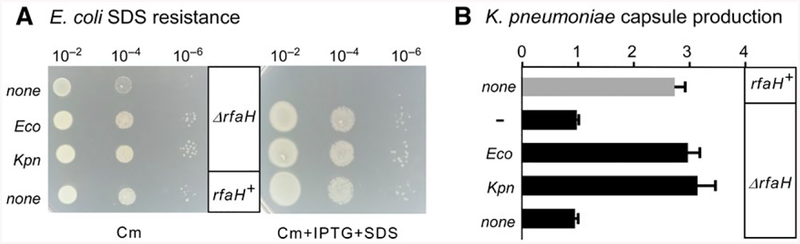
We expressed Eco and Kpn RfaH from an IPTG-inducible Ptrc promoter on a plasmid and tested whether they complemented the SDS sensitivity and abrogated capsule production phenotypes in ΔrfaH E. coli and K. pneumoniae strains, respectively. We observed that both proteins behaved indistinguishably in these assays (Fig. 2). The E. coli MG1655 strain lacking rfaH was unable to grow at 0.5% SDS, whereas the induction of either Eco or Kpn RfaH restored growth to the levels observed with the wild-type MG1655 (Fig. 2A); no growth was observed with an empty vector or in the absence of IPTG (not shown). Similarly, expression of either Eco or Kpn RfaH complemented the loss of the chromosomal gene, restoring capsule production in K. pneumoniae TOP52 (P < 0.0001 for both Eco and Kpn RfaH relative to vector control, Fig. 2B). We conclude that Eco and Kpn RfaH proteins act similarly in both species.
Fig. 2.
Plasmid-encoded Eco and Kpn RfaH complement rfaH deletions in E. coli and K. pneumoniae. A. Dilutions of exponentially growing cultures of MG1655ΔrfaH strain transformed with plasmids expressing Eco RfaH, Kpn RfaH, or a control vector were plated on LB-chloramphenicol (left) or LB-Cm supplemented with 0.5% SDS and 0.2 mM IPTG (right) and incubated at 37°C overnight. A representative set from three independent experiments is shown B. Relative capsule production in K. pneumoniae TOP52 or TOP52ΔrfaH strains transformed with plasmids containing Eco RfaH, Kpn RfaH, or no insert. Data are combined from three independent experiments, normalized to TOP52ΔrfaH without an RfaH plasmid, and error bars represent standard deviation.
Contributions of key RfaH regions to in vivo activity in K. pneumoniae
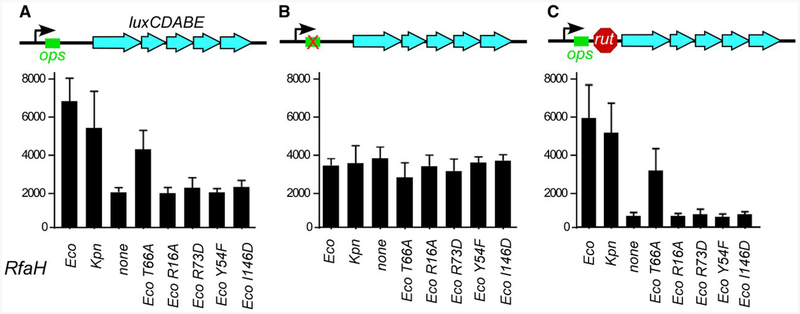
We next wanted to determine whether all Eco RfaH regions identified previously as critical for gene activation in E. coli are also necessary for its activity in K. pneumoniae. We first tested the ability of plasmid-borne wild-type Eco and Kpn RfaHs to activate the expression of a lux reporter in a ΔrfaH K. pneumoniae TOP52 strain. In this reporter (Fig. 3A), the Photorhabdus luminescens lux operon is positioned downstream from an ops element, which is known to recruit both Eco and Kpn RfaH (Rahn et al., 1999; Carter et al., 2004). We have used a nearly identical reporter to identify the key functional residues of Eco RfaH (Belogurov et al., 2010); in this work, we switched the antibiotic-resistance determinants to enable experiments in ampicillin-resistant K. pneumoniae. We observed that, similarly to their effects on activation of LPS and capsule biosynthesis operons (Fig. 2), the wild-type Eco or Kpn RfaH led to similar increases in lux expression (P < 0.0001 for both Eco and Kpn RfaH relative to vector control; Fig. 3A).
Fig. 3.
Reporter assays in K. pneumoniae. Plasmids encoding wild-type RfaH proteins or Eco RfaH variants with single-residue substitutions under the control of Ptrc promoter were co-transformed into TOP52ΔrfaH strain with reporter vectors containing the Photorhabdus luminescens luxCDABE operon under the control of PBAD promoter, with ops and rut elements in the leader region as indicated in the schematics. The results are expressed as luminescence corrected for the cell densities of individual cultures. Data are combined from three independent experiments and error bars represent standard deviation.
We have shown that substitutions of RfaH residues that mediate contacts with β’CH (Tyr54), ops DNA (Arg73), βGL (Thr66) and S10 (Ile146) (Fig. 1) abolish RfaH-dependent activation in E. coli (Belogurov et al., 2010; Burmann et al., 2012). Here we tested whether these RfaH regions contribute similarly to its activity in K. pneumoniae. As could be expected, disruptions of contacts with β’CH, S10 and ops DNA reduced lux activity to background levels observed in the absence of RfaH (Fig. 3A). In contrast, the loss of contacts with βGL led to only a small but significant decrease in the lux expression relative to Eco RfaH (P < 0.0001; Fig. 3A).
RfaH orthologs from a variety of bacteria, including those from V. cholerae and E. coli which are only 43% identical, bind to the ops element in vitro (Carter et al., 2004). Together with conservation of ops sequences in diverse bacteria (Bailey et al., 1997), this suggests that even phylogenetically diverse RfaHs make functionally important contacts to ops. To confirm this conclusion, we tested whether a G8C substitution in the ops element, which eliminates Eco RfaH recruitment to the TEC in vitro (Shi et al., 2017), interferes with RfaH function in K. pneumoniae in vivo. We found that the lux expression from a reporter in which the ops element contained a G8C substitution was reduced with all RfaH variants to the levels observed with an empty vector (Fig. 3B).
Eco RfaH has been shown to reduce Rho-dependent termination in vitro and in vivo (Artsimovitch and Landick, 2002; Sevostyanova et al., 2011). To test whether RfaH could overcome the effects of Rho in K. pneumoniae, we used a reporter in which (TC)15 a synthetic Rho-utilization (rut) signal was placed between the ops site and the lux operon (Fig. 3C). Consistent with (TC)15-induced Rho-dependent termination, lux expression from this reporter was reduced ~5-fold in the absence of RfaH. Wild-type Eco or Kpn RfaH restored lux expression to a level observed in the absence of (TC)15 (P<0.0001 for both Eco and Kpn RfaH relative to vector control). Substitutions of Eco RfaH residues interacting with ops, β’CH and S10 failed to activate expression, whereas the T66A RfaH variant restored lux activity to ~50% of the levels obtained with the wild-type Eco RfaH (Fig. 3C). We conclude that Eco and Kpn RfaHs inhibit Rho-dependent termination in K. pneumoniae, with three out of four key functional regions of Eco RfaH being essential for antitermination.
These results suggest that while Eco and Kpn RfaH proteins require the ops element for recruitment and utilize similar mechanisms to activate gene expression, they display one significant mechanistic difference: the disruption of RfaH-βGL contacts is less detrimental for RfaH-mediated activation of gene expression in K. pneumoniae, as compared to E. coli (Belogurov et al., 2010).
Kpn RfaH fails to lock the non-template DNA in the TEC
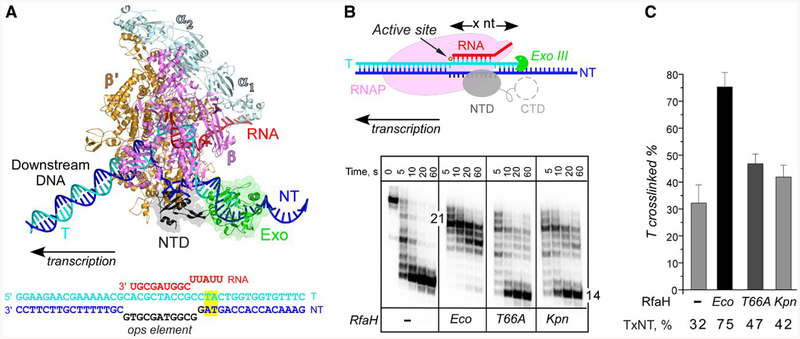
In the ops-paused TEC, the single-stranded non-template DNA in the transcription bubble simultaneously interacts with Eco RfaH and the βGL; these contacts are further stabilized by interactions between the βGL and RfaH HTTT motif (Kang et al., 2018). Disruption of Eco RfaH-βGL interactions do not abolish RfaH recruitment to RNAP but eliminate antipausing activity (Sevostyanova et al., 2011). We proposed that RfaH and βGL act together to constrain the non-template DNA strand to prevent it from assuming nonproductive conformations during elongation, thereby reducing pausing and facilitating transcription (NandyMazumdar et al., 2016). By locking the non-template DNA, RfaH and βGL restrict the mobility of the upstream duplex DNA, inhibiting digestion by Exo III, a double-strand DNA-specific exonuclease that digests DNA in a 3′→5′ direction. When bound to the TEC, Eco RfaH confers protection of 12 bp of DNA upstream of the transcription bubble (Fig. 4A), as compared to 5 bp in a free TEC (Nedialkov et al., 2018). Disruption of RfaH-βGL contacts by the deletion of βGL or substitutions of the HTTT residues weakens the RfaH-dependent Exo III protection (Nedialkov et al., 2018). If Kpn RfaH makes less stable contacts with the βGL, it would be unable to hinder Exo III digestion of the upstream DNA duplex.
Fig. 4.
Probing RfaH-DNA interactions. A. A model of the RfaH-bound TEC. RNAP α (pale cyan), β (magenta) and β’ (orange) subunits, RfaH-NTD (gray), nucleic acids and Exo (green) are shown as cartoons. To provide an unobstructed view of the RfaH-NTD and the exposed nontemplate DNA, the EC is shown inan orientation that is opposite to the conventional left-to-right direction of transcription. The ops TEC scaffold used in these experiments is shown below, with nucleic acid chains colored and oriented as in the model; the ops element is in black. The upstream TA cross-linking motif is highlighted in yellow.
B. Footprinting of the upstream RNAP boundary. The template strand DNA was 5’-end labeled with [γ32P]-ATP. After the addition of Exo III, aliquots were quenched at the indicated times (0 represents an untreated DNA control) and analyzed on a 12% denaturing gel; a representative of three independent experiments is shown. Numbers indicate the distance from the RNAP active site (yellow circle).
C. Probing the upstream fork junction by cross-linking with 8-MP. TECs were supplemented with 100 nM Eco or 250 nM Kpn RfaH (where indicated) and illuminated with the 365 nm UV light. Fractions of the cross-linked DNA were determined after analysis on denaturing gels. Error bars indicate the SDs of triplicate measurements. See also Fig. S2.
To test this idea, we assembled ops TECs on a nucleic-acid scaffold containing the ops element (Fig. 4A). The template strand and the nascent RNA were end-labeled with γ32P-ATP. We incubated the assembled TECs with RfaH variants (or storage buffer) and then added Exo III. Samples were quenched following incubation for indicated times and analyzed on denaturing urea-acrylamide gels. As expected, in the absence of RfaH, RNAP protected 14 nts of the template DNA strand upstream from the RNAP active site from Exo III digestion (Fig. 4B). When added, Eco RfaH strongly protected the upstream DNA from digestion; the footprint boundary was extended by 7 nt, whereas Eco RfaH with the T66A substitution failed to confer protection, as observed previously (Nedialkov et al., 2018). Kpn RfaH displayed an intermediate phenotype even though it was recruited to RNAP as well as the Eco RfaH at the same concentration (Fig. 6).
Fig. 6.
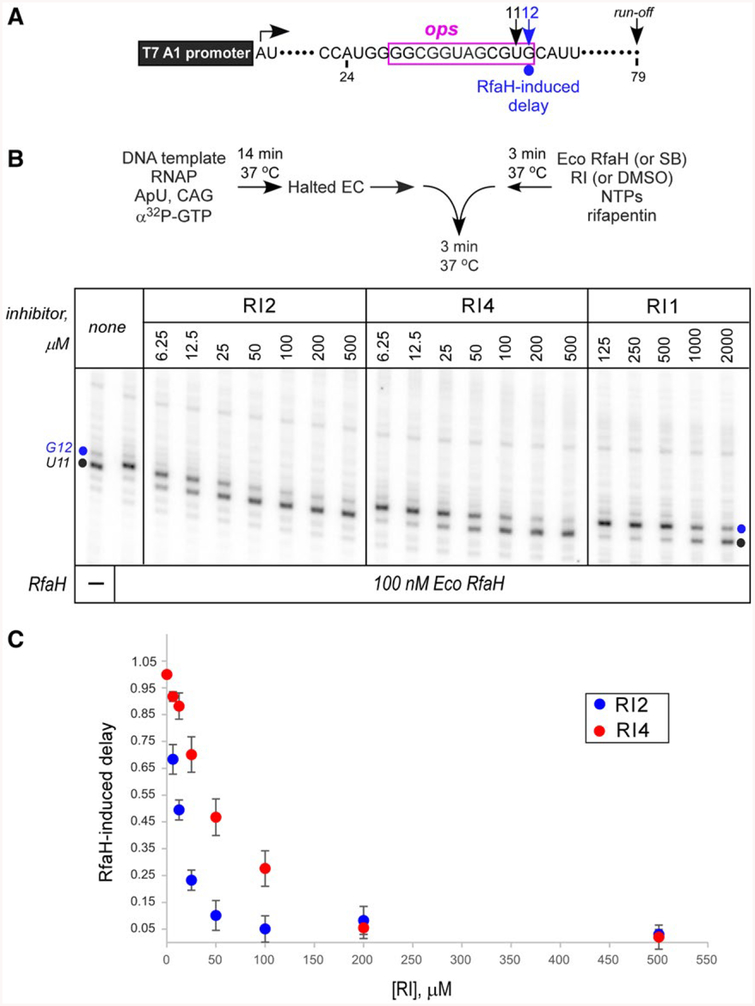
Inhibition of Eco RfaH recruitment by RIs. A. Transcript generated from the T7A1 promoter on a linear DNA template; transcription start site (a bent arrow), ops element (magenta box), pause sites and transcript end are indicated on top.
B. Halted A24 TECs were formed as described in Materials and Methods. Elongation was restarted upon addition of NTPs and rifapentin in the presence of Eco RfaH (100 nM) preincubated with increasing concentrations of RI 1, 2 or 4. Aliquots were withdrawn at selected times and analyzed on a 10% denaturing gel. Positions of the paused and run-off transcripts are indicated; the position of the RfaH-induced RNAP pause at G12 is indicated with a circle.
C. The fraction of G12 RNA was quantified as a function of RI concentration and corrected for levels observed in the absence of RfaH; the G12 RNA in the absence of RI (DMSO control) was defined as 1. The results of triplicate measurements for RI2 and RI4 are shown; errors are ± SD. Assays with RI1 were also performed in triplicates, but the observed inhibition was too weak to accurately determine the apparent IC50.
The results of Exo III footprinting suggest that the DNA-lock mechanism is partially disabled in Kpn RfaH. By constraining the upstream DNA duplex, RfaH-βGL contacts could also stabilize the upstream edge of the transcription bubble. To test whether Kpn RfaH is less efficient in stabilizing the upstream fork junction, we used cross-linking with 8-methoxypsoralen (8-MP). 8-MP specifically intercalates into double-stranded 5′-TA-3′ motifs and introduces a T-T inter-strand cross-link upon exposure to UV light. Cross-linking is strongly increased by Eco RfaH (Nedialkov et al., 2018). We assembled ops TEC on a scaffold with a TA motif positioned 12 nucleotides upstream of the RNA 3′ end (Figs. 4A and S2), with 5’-labeled template DNA and RNA (the latter is used as a loading control). We induced cross-linking upon addition of 8-MP and exposure to 365 nM UV light (Fig. 4C) and monitored the inter-strand cross-linking efficiency by gel electrophoresis in denaturing urea-acrylamide gels. Upon addition of Eco RfaH, cross-linking increased more than two fold; 100 nM RfaH (Fig. 4C) gave the same effect as 25 or 50 nM (Nedialkov et al., 2018). By contrast, T66A RfaH and Kpn RfaH (at 250 nM) increased cross-linking only modestly (Fig. 4C); the efficiency did not increase even at 1000 nM Kpn RfaH (Fig. S2). The observed small effects are similar to that of Eco NusG, which acts independently of βGL (Turtola and Belogurov, 2016).
Together, these results are consistent with less stable interactions between the βGL and Kpn RfaH. Observations that Kpn RfaH is recruited to Eco RNAP at 100 nM (Fig. 7) excludes the binding defect as an explanation for weak Exo III protection and upstream duplex stabilization. At present, we do not know the basis for the observed differences. The βGLs are identical between Eco and Kpn RNAPs, but these elements are flexible and their positions, rather than sequence, may determine how they interact with RfaH. Interestingly, Ser84, which interacts with the βGL in Eco RfaH-bound TEC (Kang et al., 2018), is replaced by Leu in Kpn RfaH (Fig. S1). In addition, Asn70, which interacts with the ops DNA (Kang et al., 2018; Zuber et al., 2018), is substituted by Ser in Kpn RfaH. These changes may destabilize the tripartite network of RfaH/non-template DNA/βGL contacts. In contrast, 14 out of 15 residues that make contacts to the β′ subunit are identical between the two proteins (Fig. S1).
Fig. 7.
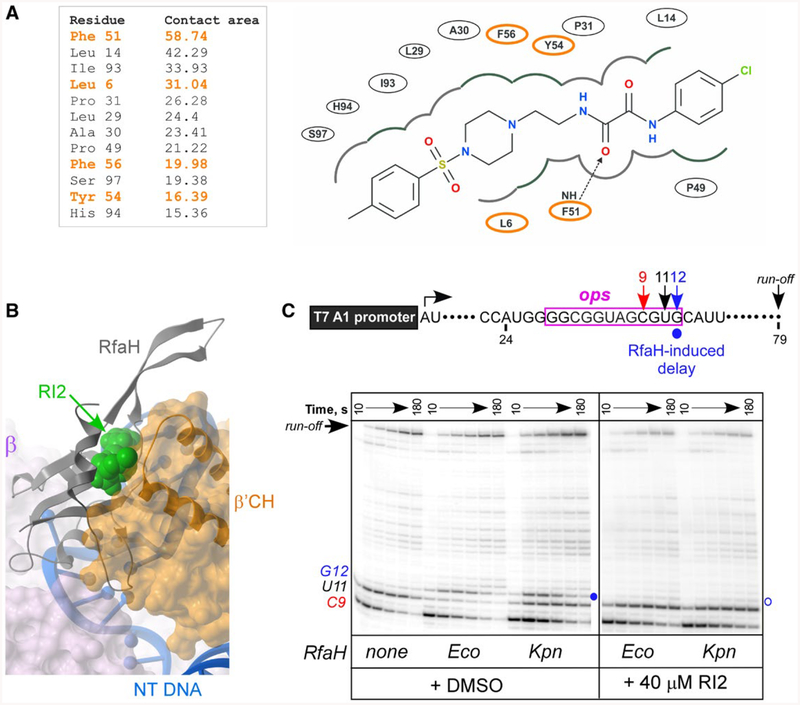
RI2 is predicted to block RfaH interactions with the β’CH domain. A. RfaH residues interacting with RI2 in the predicted binding pose. Residues indicated in orange interact with β’CH of RNAP (Kang et al., 2018). Left, contact areas (Å2) of the RfaH-NTD residues interacting with RI2. Right, a 2D interaction diagram of RfaH-NTD and RI2 in the predicted model. The dashed line with an arrow represents a hydrogen bond between residues and RI2.
B. Superposition of the modeled RfaH-NTD/RI2 complex and the cryo-electron microscopy structure (PDB ID: 6C6T) of the RfaH/TEC complex (Kang et al., 2018) using the RfaH-NTD backbone. Binding of RI2 (green) is incompatible with the β’CH (orange).
C. Effects of RI2 on RfaH recruitment at the ops site. Halted A24 TECs were formed as described in Materials and Methods. Elongation was restarted upon addition of NTPs and rifapentin in the presence of Eco or Kpn RfaH (100 nM) and RI2 (or DMSO). Aliquots were withdrawn at selected times and analyzed on a 10% denaturing gel. Positions of the paused and run-off transcripts are indicated; the position of the RfaH-induced RNAP pause at G12 is indicated with a circle.
In silico design of inhibitors targeting RfaH NTD/CH interactions
We sought to identify druggable pockets on RfaH and use structure-based screening to find small molecule modulators that bind directly to RfaH. This task is extremely challenging since the two domains of RfaH are small and flexible and RfaH biological function does not include binding to a small molecule cofactor or substrate; not surprisingly, small molecules that bind to RfaH have not been identified. Our results show that RfaH contacts with the ops DNA, the β’ subunit of RNAP and the ribo-some appear to play important roles in both E. coli and K. pneumoniae, at least during the activation of the lux reporter operon (Fig. 3). We reasoned that small molecule ligands of the RfaH-NTD that bind at the interface with DNA and β’CH would interfere with RfaH function and that molecules designed to bind to Eco RfaH, for which the structural data are available, could be similarly effective with Kpn RfaH. While ligands that bind to the conserved CTD could abolish its interactions with ribosome and thus compromise function (similarly to I146D substitution; Fig. 3), we currently do not have a suitable in vitro assay for RfaH-dependent activation of translation.
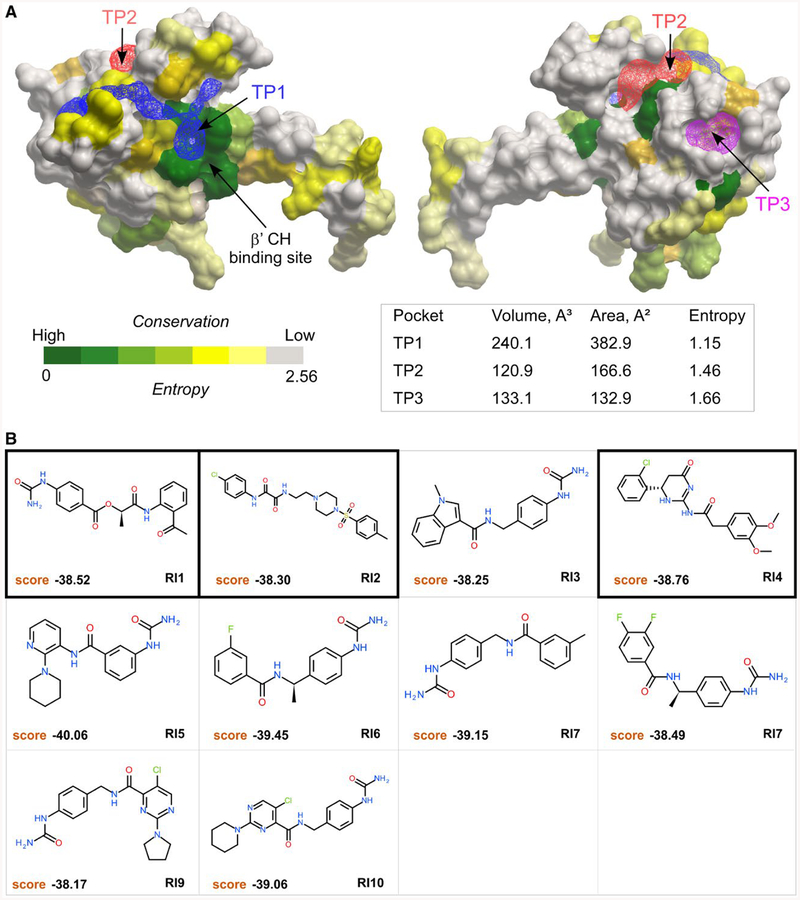
The X-ray structure of Eco RfaH (PDB: 2OUG) was used to identify hypothetical pockets for potential RfaH inhibitors via in silico ligand screening. The RfaH-CTD was removed to unmask the β’CH binding interface of the NTD and a pocket-finding algorithm based on the mathematical transformation of the surface attraction fields called ICMPocketFinder (An et al., 2005; Abagyan and Kufareva, 2009) was applied to the NTD model. Three tentative pockets (TP) on the RfaH-NTD were identified (Fig. 5A). The largest pocket, TP1, located near the interface of the NTD and β’CH, was chosen for further analysis (Fig. 5A).
Fig. 5.
Tentative pockets on the RfaH-NTD and structures of potential inhibitors. A. Three tentative pockets (TP1, blue; TP2, red; and TP3, magenta) identified by ICMPocketFinder tool in ICM-Pro v3.8–6a are shown as transparent meshes; the volume and area data are shown in the table below. The RfaH-NTD is shown as a molecular surface where residues are colored by the alignment conservation Entropies (see Methods and Materials), with highly conserved (low Entropy) residues shown in green. The Entropy of each pocket was calculated as the average Entropy of residues around the pocket.
B. Structures and docking scores of the top 10 hits from virtual ligand screening predicted to bind to TP1. Three molecules that show inhibitory activity against RfaH are indicated by thick borders.
To evaluate how residues around TP1 are conserved among all RfaH sequences, we aligned 751 RfaH sequences from various bacteria and quantified diversity of each position (see Materials and Methods). This analysis reveals that residues around TP1 are quite conserved (Fig. 5A), indicating the structural or functional importance of those residues and the feasibility of modulating diverse RfaHs by targeting TP1.
The RfaH-NTD structure needed to be optimized to achieve better docking results. To take the induced fit effect of protein pocket into account, we applied the SCARE method that generates a set of conformers with systematic omissions of pairs of interacting flexible residues (Bottegoni et al., 2008), an approach that partially takes the induced fit effect into consideration. The ZINC database containing over 20 million (potentially commercially available) small molecules was chosen as the small molecule library for virtual ligand screening (Irwin et al., 2012). After docking and scoring 20 million compounds, 10 putative RfaH inhibitors (RI 1–10) were selected based on docking score and availability for further experimental validation (Fig. 5B).
Three small molecules inhibit Eco RfaH recruitment
In initial experiments, we tested whether RIs inhibited Eco RfaH effects on transcription when present at 1 and 2 mM. We used single-round in vitro transcription assays on a template that contains the 12-nt ops element downstream from a strong T7 A1 promoter (Fig. 6A). On this template, RNAP can be stalled at position A24 in the absence of UTP; the inclusion of an α32P-labeled NTP allows for the formation of radiolabeled halted TEC. The synchronized halted A24 TECs are restarted upon the addition of all NTPs. Rifapentin, which blocks re-initiation, is added to restrict transcription to a single round. In the absence of RfaH, RNAP pauses after the addition of C9 and U11 within the ops element (Figs 6B and 7C), before making the full-length RNA of 79 nt. Addition of Eco RfaH reduces pausing at U11 ~3-fold, a reflection of RfaH antipausing activity, but not at U9 because RfaH is not yet recruited to the TEC (Nedialkov et al., 2018). In contrast, Eco RfaH delays RNAP escape from the G12 position, a well-documented consequence of RfaH recruitment which is presumably due to RfaH NTD-DNA interactions that must be broken to allow RNAP escape (Belogurov et al., 2010). This delay is commonly used as a reporter of RfaH binding to the TEC (Belogurov et al., 2010). Here, we used a one-point assay (Fig. 6) and a six-point time course (Fig. 7C) to assay RfaH-dependent delay at G12.
We found that three compounds (RI1, RI2 and RI4) inhibited Eco RfaH recruitment to the transcribing RNAP; the remaining compounds did not exhibit any effect. RI2 and RI4 exhibited apparent IC50 of ~12 and 50 μM, respectively, whereas RI1 was only marginally active with IC50 of ~1 mM. For subsequent experiments, we focused on RI2, the most potent among the three ligands.
RI2 is predicted to block RfaH binding to the β’ clamp helices
The hypothetical mode of RI2 binding to the RfaH-NTD suggests that it would sterically occlude the β′CH-binding site (Fig. 7A and B). RI2 has two amide groups connected though the two carbonyl groups, which form a large conjugation system with the benzene ring connected. The delocalization of electrons in the π bonds in two carbonyl groups and lone pairs of secondary amines makes the molecule rigid to some degree, potentially aiding its binding to RfaH-NTD. Several RfaH residues may be involved in the interaction with RI2 (Fig. 7A), and a subset of these residues (highlighted in orange) interact with the β′CH (Fig. S1 and (Kang et al., 2018)). Substitutions of RfaH residues that interact with the β′CH, including Tyr54 and Phe56, abolish RfaH activity (Belogurov et al., 2007, 2010). In a model, RI2 is positioned between the NTD and the tip of the β′CH (Fig. 7B), the high-affinity RfaH binding on the TEC (Kang et al., 2018).
All but one residue that interact with the β’ clamp domain are identical between Eco and Kpn RfaH, suggesting that RI2 may also inhibit Kpn RfaH. We found that, similarly to Eco RfaH, Kpn RfaH was recruited to E. coli RNAP, delaying its escape from G12. However, in contrast to Eco RfaH, Kpn RfaH did not reduce pausing at U11 (Fig. 7C). This observation is consistent with the lack of productive interactions with GL inferred from Exo III and cross-linking experiments (Fig. 4) because the loss of GL contacts abolishes antipausing activity of Eco RfaH (Sevostyanova et al., 2011). The addition of RI2 (at 40 μM) completely abolished the recruitment of either RfaH (Fig. 7C).
To confirm that RI2 is a specific inhibitor of RfaH interactions with RNAP, and not a promiscuous inhibitor that could nonspecifically “stick” to nonpolar surfaces of diverse proteins and block their functional interactions, we tested if RI2 inhibits GreB-assisted transcript cleavage. Like RfaH, which binds to the N-terminal β’ subunit coiled-coil domain (CH), GreB is a similarly sized (22 kDa), two-domain protein that uses hydrophobic interactions with the C-terminal β’ subunit coiled-coil domain (a.k.a. rim helices) to bind to RNAP (Vassylyeva et al., 2007). We found that, despite similar modes of binding of GreB and RfaH, RI2 did not inhibit GreB cleavage (Fig. S3A).
We next tested whether RI2 inhibits Rho-dependent termination and NusG-stimulation of Rho (Fig. S3B). While Rho-binding site on RNAP remains to be identified, NusG binds to the same region of β′CH as does RfaH, yet most residues that make contacts are different (Fig. S1). NusG potentiates early Rho-mediated RNA release, shifting the termination window upstream (Belogurov et al., 2009). We found that RI2 had no effect on Rho-dependent termination and marginally inhibited NusG (Fig. S3B). These observations support a model in which RI2 binds to the CH-docking site on RfaH, which is also nonpolar but rather different in NusG (Kang et al., 2018). Neither RI had any effect on RNA synthesis by RNAP (Fig. S3 and data not shown).
These compounds are only modestly active, and their chemical characteristics suggest that they will not accumulate in Gram-negative bacteria (Richter et al., 2017). Nonetheless, we tested RI2 for the ability to inhibit RfaH function in E. coli. RfaH is not essential, but its deletion in MG1655 confers extreme sensitivity to SDS (Fig. 2A). However, RI2 did not sensitize E. coli cells to SDS in a disk-diffusion assay, even when a tolC derivative of MG1655 strain was used (data not shown), suggesting that these compounds are not able to cross the cell wall to gain access to the cytoplasmic location of transcription and translation.
Discussion
RfaH-like regulators are dedicated activators of long operons which depend on antitermination mechanisms for complete synthesis of unusually long, up to 80,000 nts RNAs [reviewed in (NandyMazumdar and Artsimovitch, 2015)]. These operons encode proteins required for the biosynthesis of a plethora of factors including capsule polysaccharides, LPS and toxins. RfaH is critical for virulence in several pathogenic Enterobacteriaceae and its plasmid-encoded homologs are proposed to activate conjugative transfer of antibiotic-resistance genes (NandyMazumdar and Artsimovitch, 2015).
Despite being easily recognizable as orthologs, RfaH proteins are unusually diverse, with identity as low as 43% between Eco and V. cholerae RfaH (Carter et al., 2004). Indeed, our results demonstrate that while mutations in regions that interact with the β’CH, the nontemplate DNA and ribosome compromise Eco RfaH activity in K. pneumoniae, a substitution in the βGL contact site has only a modest effect (Fig. 3). This result indirectly suggests that RfaH-GL contacts may be dispensable in Klebsiella, an idea supported by in vitro transcription, footprinting and cross-linking assays (Figs 4 and 7). Consistently, two residues involved in the RfaH-GL-DNA network of interactions differ between Eco and Kpn RfaHs (Fig. S1). Notably, Eco NusG function is also independent of the βGL (NandyMazumdar et al., 2016; Turtola and Belogurov, 2016), in part due to differences in the NTD region that contacts the βGL (Kang et al., 2018).
In contrast to differences in DNA and β subunit contacts, all NusG homologs utilize the β’CH as a high-affinity site on the TEC (Kang et al., 2018). We reasoned that molecules that bind to the β’CH binding site on the RfaH-NTD could give rise to broad-spectrum RfaH inhibitors. In this work, we identified a druggable pocket at the β’CH interface and used structure-based screening to find small molecule modulators that bind this surface and alter RfaH’s function. This task is compounded by a small size of the interface and the lack of a natural or obvious small molecule binding site on RfaH. Furthermore, identifying the very first small molecule modulator for a conformationally flexible protein presents a significant challenge. These obstacles notwithstanding, two small molecules among the 10 top hits were able to inhibit Eco RfaH in vitro, and the most potent lead, RI2, blocked recruitment of both Eco and Kpn RfaH to RNAP (Fig. 7). The predicted location of the RI2 binding site on the RfaH-NTD is consistent with a competition with the β’CH (Fig. 7B). However, we note that the proposed interactions are tenuous and need extensive validation. Given the uncertainty of the RfaH-RI2 contacts, a systematic analysis using substitutions in RfaH and derivatives of RI2 would be necessary, a goal for future studies.
An obvious question is how RIs gain access to their putative binding site hidden at the RfaH domain interface (Fig. 1). We note that interactions between the NTD and CTD are relatively weak, and single substitutions in either domain have been shown to destabilize the domain interface to expose the β’CH binding site on the NTD, thereby bypassing a need for the ops element for RfaH recruitment to the TEC (Burmann et al., 2012; Shi et al., 2017). We hypothesize that equilibrium between the closed, autoinhibited and an open, activated state of RfaH enables RI recruitment to free RfaH in vitro. Notably, RfaH inhibition by RI is observed only when the inhibitor is preincubated with RfaH in the absence of TEC; the RfaH-bound TEC is insensitive to inhibition, consistent with extensive interactions between the β’CH and the tentative RI docking site. Equilibrium between the two states could explain a lone example of RfaH association with an operon in the absence of the ops site (Belogurov et al., 2009). Rapid and reversible transitions between these states are necessary for RfaH recruitment to the transcribing RNAP and refolding into the autoinhibited state upon dissociation from RNAP at the terminator. We are currently testing this hypothesis using NMR analysis.
Even though the identified leads have modest apparent affinities and do not inhibit Eco RfaH in vivo, these results are encouraging. Several key physicochemical characteristics favor compound accumulation in Gram-negative cells: low globularity, amphiphilic nature, rigidity and the presence of a primary amine (Richter et al., 2017). Extensive modifications would be required to turn our leads, which meet only the first criterion, into promising bioactive molecules. However, recent insights into the structural basis of the RfaH action (Kang et al., 2018; Zuber et al., 2018) and into the rules that govern small molecule accumulation in Gram-negative cells (Richter et al., 2017) could be leveraged to design more potent RfaH inhibitors. It is also worth noting that we have developed a sensitive whole-cell reporter assay that can be used to screen libraries of drug-like molecules for potential RfaH inhibitors. We hope that our efforts to rationally re-design the leads described in this work and find new RfaH ligands by in silico and high-throughput screening will lead to identification of potent RIs. Since RfaH is required for LPS core biogenesis, RIs would be expected to act synergistically with existing antibiotics. Given the urgent concern of mounting Gram-negative antimicrobial resistance, new therapeutics are desperately needed. Inhibitors of Eco, Kpn and related RfaHs could serve as novel antivirulence compounds to inhibit pathogenesis of organisms we are unable to kill with our failing armamentarium of antibiotics.
Experimental procedures
Tentative pocket identification and analysis
Crystal structure of RfaH (PDB code: 2OUG) was used to identify tentative binding pockets for virtual ligand screening. First, the CTD of RfaH (residues 115–156) was removed to unmask the β’CH interface of RfaH-NTD. Tentative pockets were identified using the ICMPocketFinder tool (An et al., 2005; Abagyan and Kufareva, 2009) of ICM-Pro (Abagyan et al., 2017), with default values of input parameter (tolerance = 4.6). Predicted pockets were analyzed by their volumes, position and conservation of surrounding residues, which were used to prioritize pockets for docking. The conservation of each residue was evaluated as residue composition Entropy at each position in an alignment of RfaH sequences as reported earlier (Shi et al., 2017). Briefly, Entropy was calculated according to formula (1), where Pia is calculated from ratios of the observed frequency of amino acid a at position i in the sequence alignment over the expected frequency for the same amino acid, followed by normalizing the probabilities to the total value of one.
| (1) |
Generation of RfaH ligand pocket models using SCARE
From identified tentative pockets, the pocket that has a large volume and conserved surrounding residues was selected for ligand screening. This binding site was used to generate an ensemble of modified conformations with the Scan Alanines and Refine (SCARE) approach in ICM (Bottegoni et al., 2008). SCARE generates 20 conformations where pairs of residues in the binding site are systematically masked out and restored around the bound ligand to mimic the induced fit effect. All conformations were combined to generate potential map ensembles for docking screening without further modifications. The potential map ensembles are calculated on a 0.5 Å 3D grid, containing: (i) van der Waals interaction; (ii) electrostatic interaction; (iii) hydrogen bond; and (iv) hydrophobic potential grids.
In silico screening for RfaH inhibitors
In silico screening was conducted by docking the ZINC library of 20 million small molecules to the pre-defined pockets on RfaH-NTD and ranking them based on their docking scores. The docking and scoring of one molecule was conducted using a stochastic global energy optimization procedure in internal coordinates (Abagyan et al., 1994) implemented in the ICM-Pro v3.8–6a. Ligand docking started with generating multiple starting conformations of the ligand by sampling it in vacuo and placing each sampled conformations to the binding pocket with four principal orientations. Then the ligand was sampled in the pre-calculated potential map ensembles through biased probability Monte Carlo method to optimize the position and internal variables of the ligand. For each ligand, 10 top ranking conformations were re-scored with ICM full atom scoring function (Neves et al., 2012), and conformation with the best docking score was kept for comparison. ZINC molecules were pre-filtered with the molecular weight between 100 and 1000, and ICM ToxScore smaller than 1.5. ICM ToxScore of a compound was calculated based on the number of present bioactive chemical fragments that were identified as structural alerts (Barratt et al., 1994; Gerner et al., 2004). Then the compounds were docked to the selected pockets on RfaH-NTD following the procedures described above with a computing cluster containing 128 cores. After docking, chemicals were ranked by their docking scores and the top 10 hits were tested in in vitro RfaH assay.
Identification of RfaH residues interacting with RI2 in the predicted model
After the docking screen, the model of RI2 binding to RfaH was refined by restrained energy optimization of the full atom model. To identify residues of RfaH interacting with RI2, contact areas of residues with RI2 were calculated as the difference of areas of molecular surface of each residue with and without RI2. Big contact areas implied stronger interactions between residues and RI2. The contact area threshold of 15 Å2 identified 12 residues on RfaH-NTD potentially interacting with RI2.
Reagents and proteins
All general reagents were obtained from Sigma Aldrich (St. Louis, MO) and Fisher (Pittsburgh, PA); NTPs, from GE Healthcare (Piscataway, NJ); [γ32P]-ATP and [α32P]-GTP, from Perkin Elmer (Boston, MA); PCR reagents and modification enzymes, from NEB (Ipswich, MA). Oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA) and Sigma Aldrich. DNA purification kits were from Qiagen (Valencia, CA). E. coli RNAP (Svetlov and Artsimovitch, 2015), RfaH (Belogurov et al., 2007), NusG (Belogurov et al., 2009), Rho (Belogurov et al., 2009) and GreB (Vassylyeva et al., 2007) were purified as described previously. Kpn RfaH was purified as in (Carter et al., 2004). Plasmids and oligonucleotides are listed in Table S1.
Structural probing of the ops TEC
Scaffolds were assembled as described previously (Nedialkov et al., 2018). The template DNA strand was end-labeled with [γ32P]-ATP using T4 polynucleotide kinase (PNK; NEB). The assembled TEC were resuspended in TB40 (20 mM Tris-Cl, 5% Glycerol, 40 mM KCl, 5 mM MgCl2, 10 mM β-mercaptoethanol, pH 7.9). For Exo III probing, it was divided in two aliquots; one was incubated with 100 nM RfaH and the other with storage buffer for 3 min at 37°C. For each time point, 5 μl EC were mixed with 5 μl of Exo III (NEB, 40 U) and incubated at 21°C. At times indicated in figure legends, the reactions were quenched with an equal volume of Stop buffer (8 M Urea, 20 mM EDTA, 1 × TBE, 0.5% Brilliant Blue R, 0.5% Xylene Cyanol FF). For psoralen cross-linking, the TECs were supplemented with 6.3% DMSO and 0.92 mM 8-MP and incubated for 2 min at 37°C, followed by addition of 100 nM Eco RfaH, 250 nM Kpn RfaH, or storage buffer and a 3-min incubation at 37°C. Complexes were then exposed to 365 nm UV light (8W Model UVLMS-38; UVP, LLC) for 20 min on ice. The reactions were quenched as above.
In vitro transcription assays
RfaH recruitment assays were performed as described previously (Zuber et al., 2018). Templates were made by a two-step PCR on pIA1087 plasmid that encodes the wild-type ops signal. Linear DNA template (30 nM), holo RNAP (40 nM), ApU (100 μM) and starting NTP subsets (1 μM GTP, 5 μM ATP and CTP, 10 μCi [α32P]-GTP, 3000 Ci/mmol) were mixed in 100 μl of TGA2 (20 mM Tris-acetate, 20 mM Na-acetate, 2 mM Mg-acetate, 5% glycerol, 1 mM DTT, 0.1 mM EDTA, pH 7.9). Reactions were incubated for 14 min at 37°C; thus, halted TECs were stored on ice. RfaH (100 nM final concentration or an equal volume of storage buffer) was mixed with RIs (at concentrations indicated in figures, or DMSO), chase NTPs (20 μM GTP, 300 μM ATP, CTP and UTP) and rifapentin (50 μg/ml) in TGA2, followed by a 3-min incubation at 37°C. Equal volumes of prewarmed at 37°C halted A24 TEC and RfaH/RI2/NTP mix were combined, followed by incubation at 37°C. Samples were removed at time points indicated in the figures and quenched by the addition of an equal volume of 10 M urea, 60 mM EDTA, 45 mM Tris-borate; pH 8.3. GreB-mediated cleavage (Vassylyeva et al., 2007) and Rho-dependent termination (Belogurov et al., 2009) assays were performed as described previously, with modifications indicated in Fig. S3 legend. Samples were heated for 2 min at 95°C and separated by electrophoresis in denaturing acrylamide (19:1) gels (7 M Urea, 0.5× TBE). The gels were dried and the products were visualized and quantified using a FLA9000 Phosphorimaging System (GE Healthcare), ImageQuant Software, and Microsoft Excel.
Capsule quantification assays
Capsule extraction and uronic acid quantification were performed using a modified protocol (Blumenkrantz and Asboe-Hansen, 1973; Lin et al., 2012). Twenty-milliliter cultures of K. pneumoniae TOP52 or TOP52ΔrfaH (Rosen et al., 2018) transformed with pIA947 (empty vector), pIA957 (Eco RfaH) or pIA1282 (Kpn RfaH) (Table S1) were grown shaking for 16 h in Luria-Bertani (LB) broth. Cultures were titered to determine colony forming units (CFU)/ml for normalization. Five hundred microliters of each culture was mixed with 100 μl of 1% Zwittergent 3–14 in 100 mM citric acid and incubated at 50°C for 20 min. Samples were centrifuged at 10,000 × g for 5 min, and 300 μl of each supernatant was precipitated with 1 ml of cold ethanol for 20 min at 4°C. After centrifugation at 10,000 × g for 5 min, the pellet was dissolved in 200 μl water, and 1.2 ml of 12.5 mM sodium tetraborate in concentrated sulfuric acid was added. Samples were vortexed, boiled at 95°C for 5 min and mixed with 20 μl of 0.15% 3-phenylphenol in 0.5% NaOH. Absorbance was measured at 520 nm and divided by bacterial titer to determine and absorbance/108 CFU; assays were performed in triplicate. Relative capsule production was determined by dividing all absorbance/108 CFU values by that of TOP52ΔrfaH.
In vivo lux assays
Plasmids carrying RfaH variants were co-transformed with a lux reporter vector (pHK2, pIA1297 or pIA1293) into K. pneumoniae strain TOP52ΔrfaH and plated on LB agar containing 50 μg/ml spectinomycin and 20 μg/ml chloramphenicol. Strains containing both a lux reporter and an RfaH variant were grown overnight at 37°C shaking in LB broth. Cultures were subcultured 1:100 into 10 ml of LB broth containing antibiotics and incubated at 37°C with agitation for 6 hours. Neither construct required induction (with IPTG or arabinose) since background expression of rfaH and lux operon was sufficient to generate signal. Two hundred microliters of each culture was added in triplicate to a black polysterene 96-well plate with clear bottoms (Corning 3904). A Biotek Synergy 2 plate reader was used to measure luminescence, with an integration time of 1 s and a vertical offset of 5 mm. Luminescence was corrected for the cell densities of individual cultures.
Statistical analysis
Comparisons between two groups of continuous variables were analyzed using the nonparametric Mann-Whitney U test. All tests were two-tailed, and P values <0.05 were considered significant. Analyses were performed using GraphPad Prism 7.01.
Supplementary Material
Acknowledgements
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health (grant GM067153 to I.A.). Experiments performed by JT and DR used funding from the NIH (award K08-AI127714) and the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital.
Abbreviations:
- CH
clamp helices
- CTD
C-terminal domain
- EC
elongation complex
- Eco
E. coli
- GL
gate loop
- Kpn
K. pneumoniae
- NTD
N-terminal domain
Footnotes
Conflict of interest
The Authors declare no conflicts of interest.
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher’s web site:
References
- Abagyan R and Kufareva I (2009) The flexible pocketome engine for structural chemogenomics. Methods in Molecular Biology, 575, 249–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abagyan R, Raush E, Totrov M and Orry A (2017) ICM Manual v3.8–6. San Diego, CA: Molsoft LCC. [Google Scholar]
- Abagyan R, Totrov M and Kuznetsov D (1994) ICM – a new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. Journal of Computational Chemistry, 15, 488–506. [Google Scholar]
- An J, Totrov M and Abagyan R (2005) Pocketome via comprehensive identification and classification of ligand binding envelopes. Molecular & Cellular Proteomics, 4, 752–761. [DOI] [PubMed] [Google Scholar]
- Artsimovitch I and Landick R (2002) The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed non-template DNA strand. Cell, 109, 193–203. [DOI] [PubMed] [Google Scholar]
- Bachman MA, Breen P, Deornellas V, Mu Q, Zhao L, Wu W et al. (2015) Genome-wide identification of Klebsiella pneumoniae fitness genes during lung infection. MBio, 6, e00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MJ, Hughes C and Koronakis V (1997) RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Molecular Microbiology, 26, 845–851. [DOI] [PubMed] [Google Scholar]
- Barratt MD, Basketter DA, Chamberlain M, Admans GD and Langowski JJ (1994) An expert system rule-base for identifying contact allergens. Toxicology in Vitro, 8, 1053–1060. [DOI] [PubMed] [Google Scholar]
- Belogurov GA, Mooney RA, Svetlov V, Landick R and Artsimovitch I (2009) Functional specialization of transcription elongation factors. The EMBO Journal, 28, 112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belogurov GA, Sevostyanova A, Svetlov V and Artsimovitch I (2010) Functional regions of the N-terminal domain of the antiterminator RfaH. Molecular Microbiology, 76, 286–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG et al. (2007) Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Molecular Cell, 26, 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenkrantz N and Asboe-Hansen G (1973) New method for quantitative determination of uronic acids. Analytical Biochemistry, 54, 484–489. [DOI] [PubMed] [Google Scholar]
- Bottegoni G, Kufareva I, Totrov M and Abagyan R (2008) A new method for ligand docking to flexible receptors by dual alanine scanning and refinement (SCARE). Journal of Computer-Aided Molecular Design, 22, 311–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmann BM, Knauer SH, Sevostyanova A, Schweimer K, Mooney RA, Landick R et al. (2012) An alpha helix to beta barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell, 150, 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter HD, Svetlov V and Artsimovitch I (2004) Highly divergent RfaH orthologs from pathogenic proteobacteria can substitute for Escherichia coli RfaH both in vivo and in vitro. Journal of Bacteriology, 186, 2829–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA and Kreiswirth BN (2014) Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends in Microbiology, 22, 686–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyz A, Mooney RA, Iaconi A and Landick R (2014) Mycobacterial RNA polymerase requires a U-tract at intrinsic terminators and is aided by NusG at suboptimal terminators. mBio, 5, e00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett SB, Garrison-Schilling KL, Cooke JT and Pettis GS (2016) Capsular polysaccharide production and serum survival of Vibrio vulnificus are dependent on antitermination control by RfaH. FEBS Letters, 590, 4564–4572. [DOI] [PubMed] [Google Scholar]
- Gerner I, Barratt MD, Zinke S, Schlegel K and Schlede E (2004) Development and prevalidation of a list of structure-activity relationship rules to be used in expert systems for prediction of the skin-sensitising properties of chemicals. Alternatives to Laboratory Animals, 32, 487–509. [DOI] [PubMed] [Google Scholar]
- Goodson JR, Klupt S, Zhang C, Straight P and Winkler WC (2017) LoaP is a broadly conserved antiterminator protein that regulates antibiotic gene clusters in Bacillus amyloliquefaciens. Nature Microbiology, 2, 17003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi CO and Pon CL (2015) Initiation of mRNA translation in bacteria: structural and dynamic aspects. Cellular and Molecular Life Sciences, 72, 4341–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K and Artsimovitch I (2017) A screen for rfaH suppressors reveals a key role for a connector region of termination factor Rho. MBio, 8, pii: e00753–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin JJ, Sterling T, Mysinger MM, Bolstad ES and Coleman RG (2012) ZINC: a free tool to discover chemistry for biology. Journal of Chemical Information and Modeling, 52, 1757–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Mooney RA, Nedialkov Y, Saba J, Mishanina TV, Artsimovitch I et al. (2018) Structural basis for transcript elongation control by NusG family universal regulators. Cell, 173(7), 1650–1662.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis V, Cross M and Hughes C (1988) Expression of the E.coli hemolysin secretion gene hlyB involves transcript anti-termination within the hly operon. Nucleic Acids Research, 16, 4789–4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TL, Yang FL, Yang AS, Peng HP, Li TL, Tsai MD et al. (2012) Amino acid substitutions of MagA in Klebsiella pneumoniae affect the biosynthesis of the capsular polysaccharide. PLoS ONE, 7, e46783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller AK, Leatham MP, Conway T, Nuijten PJ, de Haan LA, Krogfelt KA et al. (2003) An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infection and Immunity, 71, 2142–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Schweimer K, Rosch P, Gottesman M and Landick R (2009) Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. Journal of Molecular Biology, 391, 341–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy G, Danino V, Dobrindt U, Pallen M, Chaudhuri R, Emody L et al. (2006) Down-regulation of key virulence factors makes the Salmonella enterica serovar Typhimurium rfaH mutant a promising live-attenuated vaccine candidate. Infection and Immunity, 74, 5914–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy G, Dobrindt U, Schneider G, Khan AS, Hacker J and Emody L (2002) Loss of regulatory protein RfaH attenuates virulence of uropathogenic Escherichia coli. Infection and Immunity, 70, 4406–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NandyMazumdar M and Artsimovitch I (2015) Ubiquitous transcription factors display structural plasticity and diverse functions: NusG proteins – Shifting shapes and paradigms. Bioessays, 37, 324–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NandyMazumdar M, Nedialkov Y, Svetlov D, Sevostyanova A, Belogurov GA and Artsimovitch I (2016) RNA polymerase gate loop guides the nontemplate DNA strand in transcription complexes. Proceedings of the National Academy of Sciences United States of America, 113, 14994–14999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navasa N, Rodriguez-Aparicio LB, Ferrero MA, Monteagudo-Mera A and Martinez-Blanco H (2014) Transcriptional control of RfaH on polysialic and colanic acid synthesis by Escherichia coli K92. FEBS Letters, 588, 922–928. [DOI] [PubMed] [Google Scholar]
- Nedialkov Y, Svetlov D, Belogurov GA and Artsimovitch I (2018) Locking the non-template DNA to control transcription. Molecular Microbiology. doi: 10.1111/mmi.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves MA, Totrov M and Abagyan R (2012) Docking and scoring with ICM: the benchmarking results and strategies for improvement. Journal of Computer-Aided Molecular Design, 26, 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Mooney RA, Grass JA, Jessen ED, Tran F and Landick R (2012) Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes & Development, 26, 2621–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahn A, Drummelsmith J and Whitfield C (1999) Conserved organization in the cps gene clusters for expression of Escherichia coli group 1 K antigens: relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. Journal of Bacteriology, 181, 2307–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter MF, Drown BS, Riley AP, Garcia A, Shirai T, Svec RL et al. (2017) Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature, 545, 299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DA, Twentyman J and Hunstad DA (2018) High levels of cyclic Di-GMP in Klebsiella pneumoniae attenuate virulence in the lung. Infection and Immunity, 86, e00647–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevostyanova A and Artsimovitch I (2010) Functional analysis of Thermus thermophilus transcription factor NusG. Nucleic Acids Research, 38, 7432–7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevostyanova A, Belogurov GA, Mooney RA, Landick R and Artsimovitch I (2011) The beta subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Molecular Cell, 43, 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D, Svetlov D, Abagyan R and Artsimovitch I (2017) Flipping states: a few key residues decide the winning conformation of the only universally conserved transcription factor. Nucleic Acids Research, 45, 8835–8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens MP, Clarke BR and Roberts IS (1997) Regulation of the Escherichia coli K5 capsule gene cluster by transcription antitermination. Molecular Microbiology, 24, 1001–1012. [DOI] [PubMed] [Google Scholar]
- Svetlov V and Artsimovitch I (2015) Purification of bacterial RNA polymerase: tools and protocols. Methods in Molecular Biology, 1276, 13–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turtola M and Belogurov GA (2016) NusG inhibits RNA polymerase backtracking by stabilizing the minimal transcription bubble. Elife, 5, e18096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyeva MN, Svetlov V, Dearborn AD, Klyuyev S, Artsimovitch I and Vassylyev DG (2007) The carboxy-terminal coiled-coil of the RNA polymerase beta’-subunit is the main binding site for Gre factors. EMBO Reports, 8, 1038–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaggi B, Sbrana F, Malacarne P and Tascini C (2015) Ventilator-associated pneumonia caused by colistin-resistant KPC-producing Klebsiella pneumoniae: a case report and literature review. Respiratory Investigation, 53, 124–128. [DOI] [PubMed] [Google Scholar]
- Yakhnin AV, Murakami KS and Babitzke P (2016) NusG Is a sequence-specific RNA polymerase pause factor that binds to the non-template DNA within the paused transcription bubble. Journal of Biological Chemistry, 291, 5299–5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber PK, Artsimovitch I, NandyMazumdar M, Liu Z, Nedialkov Y, Schweimer K et al. (2018) The universally-conserved transcription factor RfaH is recruited to a hairpin structure of the non-template DNA strand. Elife, 7, pii: e36349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.