Abstract
Background
WNT and TGFβ signaling pathways play critical regulatory roles in cardiomyocyte fate determination and differentiation. MiRNAs are also known to regulate different biological processes and signaling pathways. Here, we intended to find candidate miRNAs that are involved in cardiac differentiation through regulation of WNT and TGFβ signaling pathways.
Methods
Bioinformatics analysis suggested hsa-miR-335-3p and hsa-miR-335-5p as regulators of cardiac differentiation. Then, RT-qPCR, dual luciferase, TOP/FOP flash, and western blot analyses were done to confirm the hypothesis.
Results
Human embryonic stem cells (hESCs) were differentiated into beating cardiomyocytes, and these miRNAs showed significant expression during the differentiation process. Gain and loss of function of miR-335-3p and miR-335-5p resulted in BRACHYURY, GATA4, and NKX2-5 (cardiac differentiation markers) expression alteration during the course of hESC cardiac differentiation. The overexpression of miR-335-3p and miR-335-5p also led to upregulation of CNX43 and TNNT2 expression, respectively. Our results suggest that this might be mediated through enhancement of WNT and TGFβ signaling pathways.
Conclusion
Overall, we show that miR-335-3p/5p upregulates cardiac mesoderm (BRACHYURY) and cardiac progenitor cell (GATA4 and NKX2-5) markers, which are potentially mediated through activation of WNT and TGFβ signaling pathways. Our findings suggest miR-335-3p/5p to be considered as a regulator of the cardiac differentiation process.
Electronic supplementary material
The online version of this article (10.1186/s13287-019-1249-2) contains supplementary material, which is available to authorized users.
Keywords: miRNA, Cardiomyocyte differentiation, hESC, WNT, TGFβ
Introduction
Human embryonic stem cells (hESCs) are pluripotent cells derived from the inner cell mass of blastocysts. These cells have self-renewal capacity and are able to differentiate into all derivatives of three germ layers [1]. The first lineage decision is chosen following gastrulation in which cells differentiate into the mesoderm, endoderm, and ectoderm [2]. Differentiation of hESCs is an ideal model for cardiogenesis research as these cells mimic the behavior of cells at early stages of embryonic development [3, 4]. Proper differentiation occurs by precise regulation of different transcription networks, signaling pathways, and epigenetic modifications [5, 6]. WNT/β catenin and TGFβ signaling pathways are known as two main and critical signaling pathways that regulate different stages of cardiac differentiation including mesendoderm cell commitment, cardiac progenitor differentiation, and final maturation [7–10]. Numerous factors tightly orchestrate these signaling pathways during differentiation [11]. MicroRNAs (miRNAs) are small (22 to 25 nucleotide in length) non-coding RNAs [12] that regulate gene expression and play important roles in proliferation, differentiation, cell fate decision, and various physiological processes [13–15]. Accumulating evidence has shown that miRNAs regulate signaling pathways related to cardiac differentiation at posttranscriptional and posttranslational levels [16, 17]. For instance, miR-15, miR-16 [18], and miR-430 [19] are identified as regulators of cell fate acquisition through targeting the TGFβ signaling pathway. miR-1 and miR-133 promote mesoderm formation [20] and miR-499 promotes CPC differentiation into cardiomyocytes [21]. Here, we screen miRNAs that might be involved in cardiac differentiation through regulation of WNT/β catenin and TGFβ signaling pathways. Bioinformatics analyses indicate that miR-335-3p and miR-335-5p might regulate these two signaling pathways through targeting core members of the pathways. Gain- and loss-of-function studies were performed to verify the exact role of these two miRNAs in cardiac differentiation. Our findings demonstrate that these two miRNAs might regulate cardiac differentiation by activating WNT and TGFβ signaling pathways. This activation led to enhanced mesoderm cell commitment and promoted cardiac progenitor cell differentiation.
Materials and methods
Cell culture and differentiation
HEK293 and SW480 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco), supplemented with 10% heat-inactivated fetal bovine serum and 1% antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin) (Gibco). Cells were grown at 37 °C in a humidified atmosphere with 5% CO2. The hESC line RH5 [22] was expanded under feeder-free conditions on Matrigel-coated plates. Cardiomyocyte differentiation occurred in a chemically defined medium, as previously described [23, 24] with minor modifications. Cells were stimulated with 20 ng/mL fibroblast growth factor 2 (FGF2), 20 ng/mL activin A, and 10 ng/mL BMP4 in the first 36 h for mesoderm induction; then, cells were treated with 20 ng/mL FGF2, 50 ng/mL BMP4, 0.5 mM retinoic acid, and 5 mM WNT inhibitor (IWP2) from day 1.5 to day 5. Finally, cells were treated with 5 ng/mL FGF2 and 10 ng/mL BMP4 which resulted in cardiomyocyte differentiation. Samples were collected at different time points (0, 0.5, 1, 1.5, 2, 5, and 12 days) of differentiation for expression analysis.
Transfection of hESCs
Gain- and loss-of-function studies were done in (day 0) D0 of differentiation. The miRCURY LNA™ microRNA mimic (Exiqon, Denmark) for miR-335-3p (MIMAT0004703), miR-335-5p (MIMAT0000765), and mimic control as well as miRIDIAN microRNA miR-335-3p and miR-335-5p hairpin inhibitors and miRIDIAN microRNA hairpin inhibitor control (Dharmacon) were used for gain- and loss-of-function studies in which 8 × 105 cells were plated in each 3.5-cm tissue culture dish, 24 h before transfection. When cells reached 80% confluence, they were transfected by 50 nM siRNA or 5 nM mimic structures using Lipofectamine® 3000 reagent, based on the manufacturer’s instructions. The efficiency of siRNAs and microRNA mimics transfection was evaluated using BLOCK-iT Alexa Fluor Red fluorescent oligo (Invitrogen).
RNA extraction and quantitative RT-PCR
Total RNA of harvested cells was extracted using TRIzol™ reagent (Invitrogen, USA) according to the manufacturer’s protocol. The total RNA was used for cDNA synthesis after being treated with RNase-free DNase (Takara, Japan) in order to remove any DNA contamination. cDNAs were synthesized using RevertAid™ Reverse Transcriptase (Fermentase, Lithuania) according to the manufacturer’s instructions. For miRNA detection, polyA tail was added to 3′ end of RNAs before cDNA synthesis. RT-qPCR was performed using specific primers (Table S1) by StepOne Real-Time PCR system (Applied Biosystems). GAPDH and small nucleolar RNA, C/D box 48 (SNORD48) were used as internal controls for normalization of mRNAs and miRNA expression.
Immunocytochemistry (ICC)
The seeded cells were washed once with phosphate buffered saline (PBS) and fixed with 4% (w/v) paraformaldehyde (PFA) for 15 min at room temperature. The cells were permeabilized using PBS containing 0.2% Triton X-100 for 10 min followed by blocking in a solution containing PBS and 10% Donkey serum for 1 h at room temperature. Next, cells were treated with primary antibody overnight at 4 °C. After three washes in PBS, cells were incubated with secondary antibody for 1 h at room temperature in the dark. Finally, cells were incubated with DAPI for nucleic acid staining and imaged with a fluorescent microscope (IX71, Olympus, Japan). Image overlays and contrast enhancement were performed using ImageJ software.
Dual luciferase assay
Dual luciferase assay was utilized to validate the direct interaction between miR-335-3p and miR-335-5p and their target genes 3′-UTRs. To this aim, HEK293 cells were co-transfected with a psiCHECK-2 vectors including 3′-UTR of APC, AXIN-I, and SMAD7 and miR-335-3p, miR-335-5p mimics, and siRNA structures in 48-well plates. As negative controls, mimic and siRNA scramble were used in co-transfection. Luciferase activity was measured 48 h after transfection using the Dual Luciferase Reporter Assay System (Promega, USA) according to the manufacturer’s instructions.
TOP/FOP reporter assays
TOP/FOP reporter assays were carried out using the Dual-Glo luciferase assay kit (Promega), based on the manufacturer’s instructions. SW480 cells were transfected with 1μg of constitutively active vector encoding Renilla luciferase, responsive firefly luciferase reporter plasmid Top Flash, each mimic and siRNA corresponding to the miR-335-3p, miR-335-5p and their related scrambled. Cells were harvested after 48 h, and both firefly and Renilla luciferase activity were measured in three biological replicates according to the manufacturer’s instructions. The firefly luciferase activity was normalized against Renilla luciferase activity.
Western blotting
The protein was extracted from the samples. Samples containing 40 μg purified protein were separated by 12% SDS/PAGE, transferred to PVDF membranes (Santa Cruz), and run at 100 V for 1.5 h at room temperature. The PVDF membrane was subsequently blocked by 5% BSA in phosphate-buffered saline (PBS) containing 0.1% Tween for 1 h at room temperature, followed by overnight incubation at 4 °C with phospho BRACHYURY primary antibodies (1:500, Cell Signaling). The blot membrane was washed and incubated with anti-rabbit secondary antibody (1:1000, Santa Cruz) for 1 h at room temperature. The protein bands were visualized by an enhanced chemiluminescence (ECL) detection system (Amersham, Piscataway, NJ). The membranes were stripped and re-probed with β-actin for verification of protein loading. The bands were quantified using an image analyzer program (ImageJ).
Results
Bioinformatics analyses introduced miR-335 as a potential regulator of cardiac differentiation
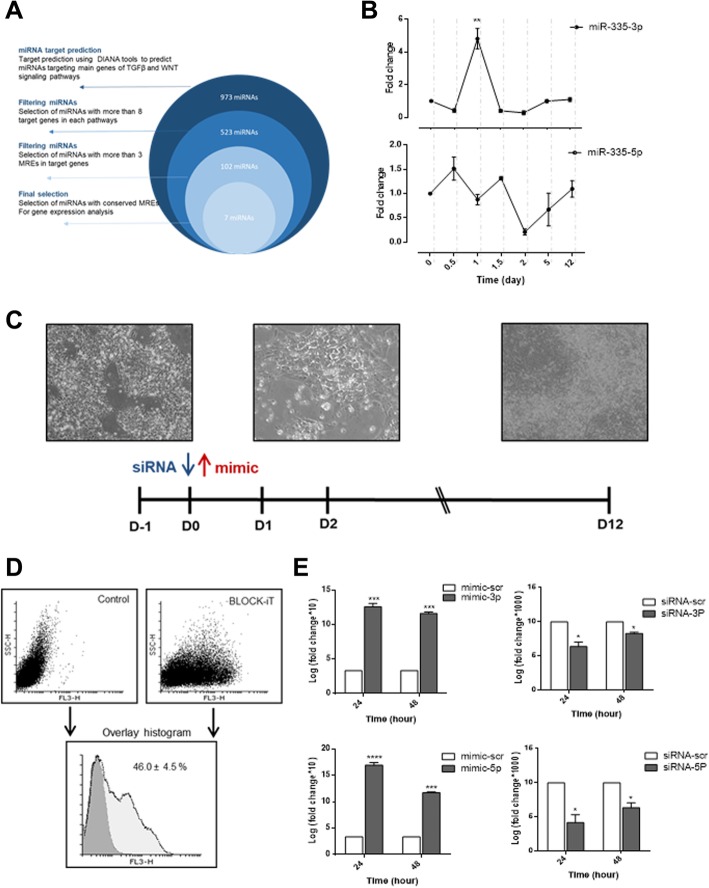
WNT and TGFβ signaling pathways are known as main players in cardiac differentiation. In this study, we aimed to find out miRNAs that are crucially important for driving cardiac differentiation through regulation of WNT and TGFβ signaling pathways. To this aim, a four-step filtering approach was performed to nominate some miRNAs (Fig. 2a). At the first step, 973 miRNAs were predicted to target WNT and TGFβ signaling pathways using miRWalk, a target prediction resource that uses several miRNA target prediction tools. Then, considering the number of genes which are targeted by a miRNA in each pathway, the numbers of MREs (miRNA recognition elements) in 3′UTR sequences of each target gene, and the annealing and conservation status of each MRE, seven candidate miRNAs were chosen for further analyses (Additional file 1: Table S2). Among candidate miRNAs, miR-335-3p/5p hosted by mesoderm-specific transcript (MEST) gene were chosen for further investigations concerning its role in cardiogenesis.
Fig. 2.
MiR-335 expression alteration during cardiac differentiation. a Schematic presentation of different stages of miRNA screening. b miR-335-3p (top) and miR-335-5p (bottom) expression pattern during hESC cardiac differentiation. c Gain- and loss-of-function strategy in which miR-335-specific mimics and siRNAs were transfected on day 0 of differentiation process. d Flow cytometry results showed that hESCs were efficiently transfected by miR-335 specific mimics or siRNAs along with 50 nM BLOCK-iT Alexa Fluor Red Fluorescent oligonucleotide. e The RT-qPCR confirmed significant (P value < 0.05) overexpression of miR-335-3p (top-left) and miR-335-5p (bottom-left) but significant downregulation of miR-335-3p (top- right) and miR-335-5p (bottom-right), 24 and 48 h after transfection. RT-qPCR data are presented as mean ± SEM normalized against day 0 data. GAPDH was used as a housekeeping gene
Differentiation of hESCs into cardiomyocytes
In vitro differentiation of hESC, RH5, to cardiomyocyte-like cells was successfully performed (Fig. 1a). Different time points representing different cell fates during cardiac differentiation were considered (i.e., pluripotent stem cells (day 0), mesendoderm (day 1.5), cardiac progenitor (day 5), and cardiomyocyte cells (day 12)). NANOG stem cell marker expression was substantially decreased while the BRACHYURY (mesendodermal marker) expression level reached its highest level in mesendoderm stage (day 1.5) (Fig. 1b). In addition, NKX2-5 and ISL1 (early cardiac differentiation markers) expression levels elevated from day 2 and substantially increased during the process. As expected, cardiac-specific markers (TNNT and MYH6) were significantly increased at the final stage (day 12) (Fig. 1b), followed by spontaneous beating of the cardiac cells (Additional file 2: Movie S1). The flow cytometry analysis indicated that ~ 51% of the cells were BRACHYURY positive on day 1.5, ~ 87% of the cells were NKX2-5 positive on day 5 (cardiac progenitor cells) and ~ 96% of the cells were MYH6 positive on day 12 (the final step of cardiomyocyte differentiation) (Fig. 1c). Furthermore, immunostaining indicated that hESCs were positive for OCT4, whereas hESC-CMs stained positive for cardiac-specific markers MYH6 (Fig. 1d), approving that these cells were successfully differentiated into cardiomyocytes.
Fig. 1.
Characterization of hESC-derived cardiomyocyte. a Schematic description of cardiomyocyte differentiation protocol. Activin A and BMP4 compounds were used to induce differentiation of hESCs into mesoderm. Then, WNT inhibitor reagent (IWP2) was used to induce cardiac progenitor cell development followed by cardiomyocyte formation. b Time-dependent expression of NANOG (pluripotency marker), BRACHYURY (mesoderm marker), NKX2-5, ISL1 (cardiac progenitor cell markers), TNNT2, and MYH6 (cardiomyocyte markers) during the cardiac differentiation process. RT-qPCR data are presented as mean ± SEM normalized against day 0 data, for n = 3 independent experiments. GAPDH was used as a housekeeping gene. c Flow cytometry results confirmed the expression of BRACHYURY (day 1.5), NKX2-5 (day 5), and MYH6 (day 12) during the differentiation process. d Immunocytochemistry analysis of OCT4 (pluripotent hESCs) and MYH6 (cardiomyocytes) revealed the stemness potency of hESCs and successful differentiation of cardiomyocyte. Differentiated cardiomyocytes are shown in two magnitudes
MiR-335 expression pattern during the human cardiac differentiation process
MiR-335-3p and miR-335-5p expression status was measured at seven time points of human cardiac differentiation process, using RT-qPCR. Both miR-335-3p and miR-335-5p had relatively high levels of expression during this process (data not shown); however, their expression pattern was different. While miR-335-3p was transiently upregulated at day 1 and substantially downregulated to the end of differentiation process, miR-335-5p showed no significant alteration during the differentiation (Fig. 2b).
The effect of MiR-335 expression alteration on cardiac differentiation
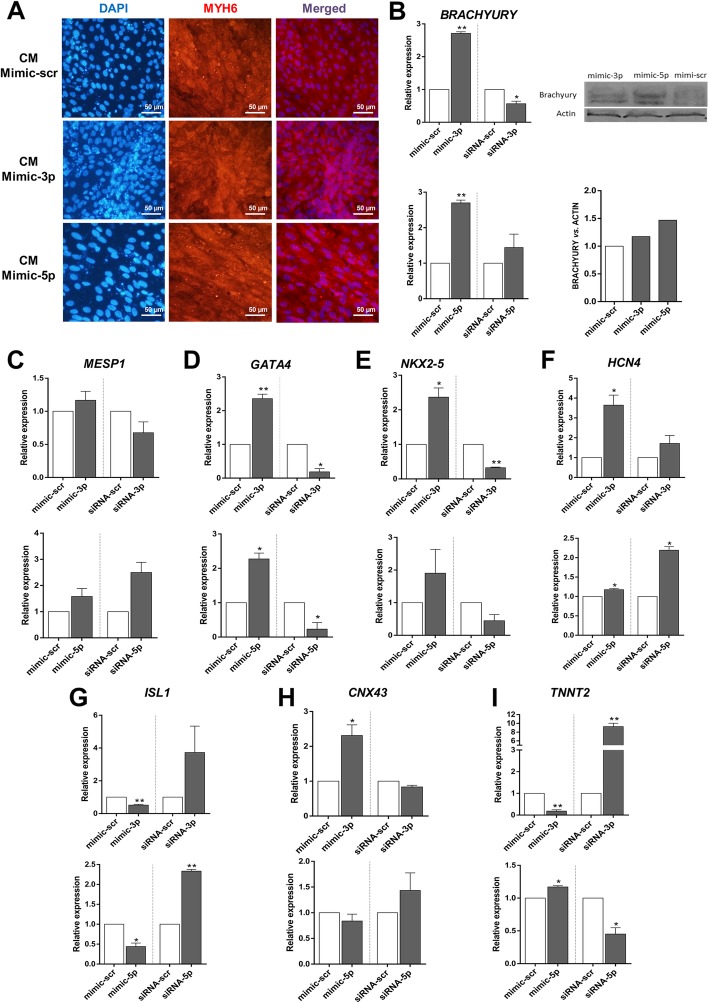
In order to examine the effect of miR-335-3p/5p expression alteration on cardiac differentiation, gain- and loss-of-function study was performed according to miR-335-3p and miR-335-5p expression profiles (Fig. 2c). In this part, miR-335-3p and miR-335-5p mimics or their corresponding siRNAs were transfected into differentiating hESCs on D0 to induce specific overexpression and downregulation, respectively. Red fluorescent oligo transfection indicated ~ 45% transfection efficiency (Fig. 2d), and specific up- and downregulation of each miRNA was further confirmed using RT-qPCR, 24 and 48 h post transfection (Fig. 2e). Immunostaining of the cells transfected by mimic-scr, mimic-3p, or mimic-5p indicated successful cardiac differentiation (Fig. 3a). To determine the differentiation stage which has been most affected by miR-335 expression alteration, the expression level of stage-specific markers was measured by RT-qPCR. Here, the expressions of BRACHUYRY and MESP1 as mesodermal markers, NKX2-5 and GATA4 as cardiac progenitor markers, HCN4 and ISL1 as progenitor markers of the first and second heart field, and TNNT2 and CNX43 as cardiomyocyte markers were analyzed. RT-qPCR data indicated that 48 h post-transfection of mimic-3p or mimic-5p, the expression level of BRACHYURY was significantly increased (Fig. 3b). Consistently, downregulation of miR-335-3p was followed by significant reduction of BRACHUYRY expression (Fig. 3b). The results of western blotting also confirmed the increased protein level of BRACHYURY following mimic-3p and mimic-5p treatments compared to the mimic-scr control (Fig. 3b, right). Increased or decreased level of miR-335 did not show a significant effect on MESP1 expression level (Fig. 3c). Interestingly, GATA4 and NKX2-5 expression levels were significantly increased following miR-335-3p overexpression while they decreased after using their related siRNA (Fig. 3d, e). Similar significant results were obtained for miR-335-5p against GATA4, but results were non-significant against NKX2-5 expression (Fig. 3d, e).
Fig. 3.
MiR-335 regulates earlier stages of hESC cardiac differentiation. a Immunocytochemistry analysis showed that overexpression of hsa-miR335-3p or hsa-miR335-5p on day 0 did not affect the final outcome of cardiac differentiation on day 12 (cardiomyocytes), as detected by MYH6 expression in all groups. b Overexpression of both hsa-miR335-3p (top) and hsa-miR335-5p (bottom) resulted in significant elevation of BRACHYURY (mesoderm marker) expression. Consistently, hsa-miR335-3p downregulation resulted in reduced expression of BRACHYURY. Western blot results indicate an increased level of BRACHYURY following the mimic-3p and mimic-5p treatment, compared to mimic-scr control (right). c No significant expression alteration was detected for MESP1 following alterations in hsa-miR335 expression. d, e Expressions of GATA4 and NKX2-5 (main cardiac progenitor markers) were both increased following miR-335-3p (top) and miR-335-5p (bottom) overexpression. Downregulation of miR-335 had reverse effect on both of these cardiac progenitor markers. f Expression of HCN4 (the first heart field marker) was significantly increased following miR-335-3p (top) and miR-335-5p (bottom) overexpression. g Alterations in the expression of miR-335-3p (top) and miR-335-5p (bottom) had a significant reverse effect on ISL1 (the second heart field marker) expression. h, i Has-miR-335-3p (top) overexpression increased CNX43 expression but decreased TNNT2 expression. However, has-miR-335-5p decreased CNX43 but enhanced TNNT2 expressions. RT-qPCR data are presented as mean ± SEM normalized against mimic-scr and siRNA-scr. GAPDH was used as a housekeeping gene
Following transfection of mimic-3p and mimic-5p, HCN4 (the first heart field marker) was significantly upregulated as well; however, miR-335-3p and miR-335-5p downregulation results were not consistent with it (Fig. 3f). MiR-335 expression alteration also has effect on ISL1 (the second heart field marker) expression. Transfection of both mimic-3p and mimic-5p resulted in downregulation of ISL1 expression while, miR-335 downregulation resulted in increased ISL1 expression (Fig. 3g). Overexpression of miR-335-3p significantly upregulated CNX43 expression but suppressed the expression of TNNT2 at the final stage of the differentiation process (Fig. 3h, i). Interestingly, miR-335-5p overexpression could significantly upregulate TNNT2 with no significant effects on CNX43 expression.
MiR-335 as an activator of WNT signaling pathway
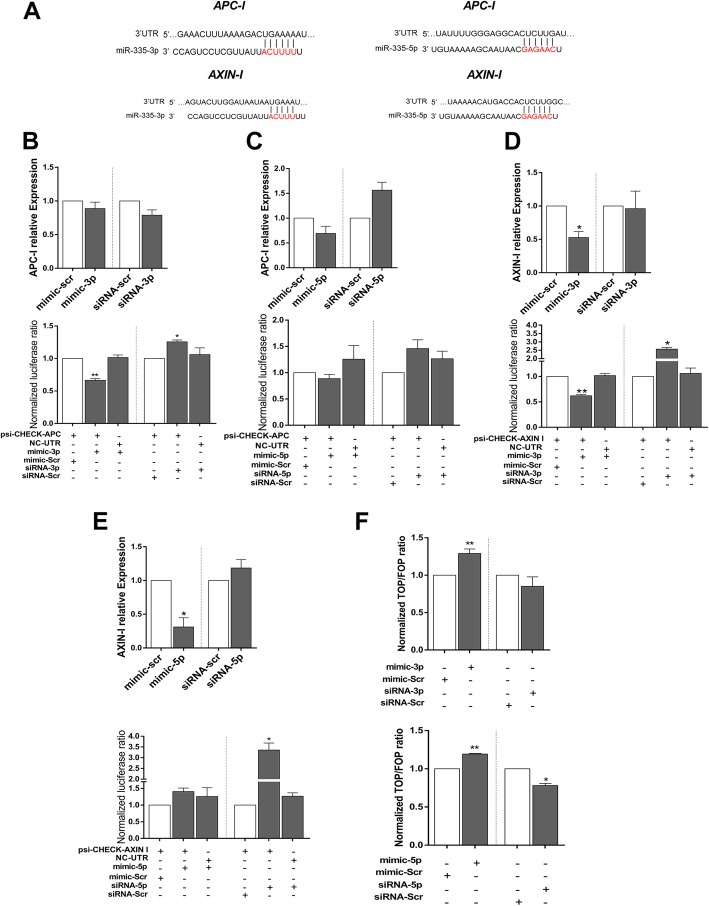
Target prediction tools predicted 5 and 1 potential target sites for miR-335-3p within the 3′UTR sequences of APC and AXIN-I genes, respectively. For miR-335-5p, a single recognition site was predicted within 3′UTR sequences of both APC and AXIN-I genes (Fig. 4a). Alterations in the expression of APC and AXIN-I target genes were investigated 48-h post-transfection of miR-335 mimics or siRNAs on D0 of hESC differentiation. Also, direct interaction of each miRNA mimic and 3′UTR sequences of APC and AXIN-I predicted target genes was investigated in HEK293 cells, using dual luciferase reporter assay.
Fig. 4.
MiR-335 activates WNT signaling through targeting APC and AXIN-I. a Pairing status of miR-335-3p and miR-335-5p with MREs within 3′UTR sequences of APC and AXIN-I. b Alterations in the expression of miR-335-3p did not significantly change the transcription level of APC gene (top); however, dual luciferase assay supported a direct interaction between them (bottom). c Neither RT-qPCR nor dual luciferase assay showed an interaction between miR-335-5p and APC. d Alterations in the expression of miR-335-3p significantly changed the AXIN-I transcript level (top), and dual luciferase assay indicated a direct interaction between them (bottom). e Although RT-qPCR suggested downregulation of AXIN-I following miR-335-5p overexpression (top), dual luciferase assay did not show an interaction between them (bottom). f Overall inductive effect of both miR-335-3p (top) and miR-335-5p (bottom) on WNT signaling as detected by TOP/FOP flash assay. All experiments were done in three biological replicates and results are presented as mean ± SEM
RT-qPCR indicated that up- or downregulation of both miR-335-3p (Fig. 4b, top) and miR-335-5p (Fig. 4c, top) did not significantly affect APC gene expression in differentiating cells. Co-transfection of reporter construct containing APC-3′UTR and miR-335-3p mimic in HEK293 cells resulted in a significant reduction of luciferase activity, and consistently, luciferase activity was increased after the application of siRNA against miR-335-3p, compared to scrambled controls (Fig. 4b, bottom). This effect was abrogated using off target UTR (the same length of UTR with no available target site for miRNA), thereby approving the direct interaction between miR-335-3p and APC 3′UTR. Similar dual luciferase assay results indicated that miR-335-5p was not interacting with APC 3′UTR sequence (Fig. 4c, bottom).
Transfection of differentiating hESCs with miR-335-3p mimic resulted in significant downregulation of AXIN-I expression, compared to scrambled control transfection (Fig. 4d, top). Furthermore, dual luciferase assay confirmed the direct interaction between miR-335-3p mimic and 3′UTR sequence of AXIN-I (Fig. 4d, bottom). Transfection with miR-335-5p mimic also caused a significant reduction in AXIN-I transcripts as examined by RT-qPCR (Fig. 4e, top). However, dual luciferase assay did not show a direct interaction between miR-335-3p and 3′UTR sequence of AXIN-I gene (Fig. 4e, bottom).
TOP/FOP flash assay indicated that overexpression of miR-335-3p or miR-335-5p led to significant upregulation of WNT signaling pathway in SW480 cells, compared to mimic-scr controls (Fig. 4f). Similar results to those of Top/Fop flash assay were observed when miR-335 was downregulated by specific siRNA (Fig. 4f). The RT-qPCR analysis against CYCLIND1 and c-MYC (downstream targets of WNT signaling pathway) also confirmed the upregulation of WNT signaling, following miR-335-5p overexpression (Additional file 1: Figure S1). Alterations in miR-335-3p expression did not significantly affect CYCLIND1 and c-MYC expression (Additional file 1: Figure S1). Altogether, these data suggested that miR-335-3p might enhance the WNT signaling through targeting the inhibitory components of WNT signaling pathway (i.e., APC and AXIN-I) while miR-335-5p could activate this pathway indirectly without any direct effect on these two target genes.
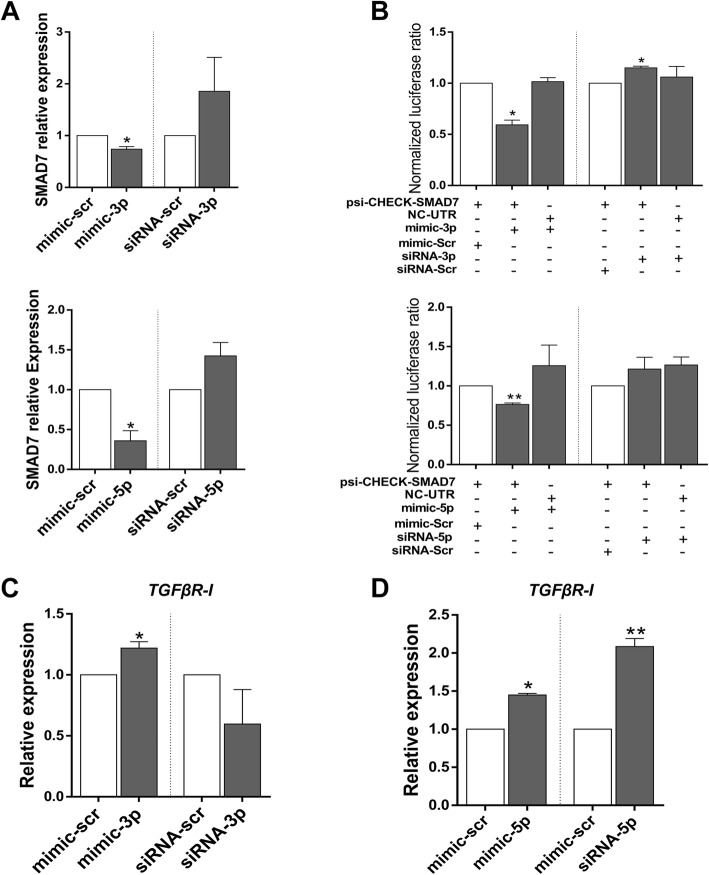
MiR-335 as an inducer of TGFβ signaling pathway
The in silico analyses suggested that miR-335 could regulate TGFβ signaling through targeting SMAD7 transcripts. It was predicted that both miR-335-3p and miR-335-5p target inhibitory SMAD7 gene (Additional file 1: Figure S2A). RT-qPCR indicated that SMAD7 expression was significantly reduced following miR-335-3p and miR-335-5p overexpression, 48 h after transfection in differentiating hESCs. Consistently, downregulation of these miRNAs had reverse effects (though non-significant) on SMAD7 expression (Fig. 5a, top).
Fig. 5.
MiR-335 activates TGFβ signaling pathway through targeting SMAD7. a Alterations in the expression of SMAD7 followed by overexpression and downregulation of miR-335-3p (top) and miR-335-5p (bottom). b Dual luciferase assay study confirmed a direct interaction between miR-335-3p (top) and miR-335-5p (bottom), and SMAD7 3′UTR. Alteration in the expression of TGFβR-I was assessed by RT-qPCR followed by miR-335-3p and miR-335-5p overexpression (c) and downregulation (d) in hESC. RT-qPCR data are presented as mean ± SEM normalized against mimic-scr and siRNA-scr. GAPDH was used as a housekeeping gene
Luciferase activity was decreased following miR-335-3p overexpression, and this suppression was relieved by downregulation of miR-335-3p as detected by dual luciferase assay (Fig. 5b, top). Significant reductions in luciferase activity were also detected after miR-335-5p overexpression (Fig. 5a, bottom), while miR-335-5p downregulation could not rescue luciferase activity, significantly (Fig. 5b, bottom). Overall, these data suggested that both miR-335-3p and miR-335-5p were capable of targeting SMAD7 transcript. The effect of MiR-335 overexpression on other TGFβ signaling pathway components was also investigated using RT-qPCR. Results indicated that both miR-335-3p and miR-335-5p overexpression led to TGFβR-I upregulation (Fig. 5c, d). Alterations in the expression of miR-335 had no significant effect on SMAD2 and SMAD3 expression (Additional file 1: Figure S2B-C). Altogether, these data suggest that both miR-335-3p and miR-335-5p might activate TGFβ signaling pathway through targeting SMAD7 transcript.
Discussion
TGFβ and WNT signaling pathways have some pleiotropic and multifunctional effects and regulate several biological processes including embryonic development, as well as cell differentiation, proliferation, and survival [25–27]. These two pathways play critical roles in orchestration of cardiac development and differentiation [28–30]. It is known that regulation of TGFβ and WNT signaling pathway is essential for mesoderm cell fate commitment, as well as cardiomyocyte progenitor cell formation and cardiomyocyte maturation [31]. These cardiogenesis steps are closely regulated by different mechanisms at posttranscriptional and posttranslational levels. MiRNAs are known to regulate different bioprocesses through targeting related signaling pathways [32–34]. For example, hsa-miR-590-5p was shown to be involved in cardiogenesis through regulating TGFβ signaling [35]. Also, hsa-miR-23b cluster was shown to induce proliferation in hepatocytes through TGFβ pathway inhibition [36] and hsa-miR-126 is known to induce VEGF signaling and promote angiogenesis [37]. Here, we intended to introduce miRNAs which fine-tune TGFβ and WNT signaling pathways, during the cardiogenesis process. Bioinformatics analyses introduced 7 miRNAs which had multiple targets in both WNT and TGFB signaling pathways (Additional file 1: Table S2). Finally, has-miR-335 that is well conserved in mammals and is under the control of mesoderm-specific enhancers in the second intron of MEST [38] (Additional file 1: Figure S3), was chosen for further investigation. Also, miR-335 was reported to be involved in the fate commitment of mesoderm during the mouse embryonic differentiation [38]. MiR-335-3p (miRBase ID: MIMAT0004703) and miR-335-5p (miRBase ID: MIMAT0000765) were both highly expressed in mature heart [38]. It is predicted that these two miRNAs target different genes in WNT and TGFβ signaling pathways (Additional file 1: Tables S3–S4, Figure S4).
H5 human ESCs were successfully differentiated into beating cardiomyocytes after 12 days (Fig. 1a). Then, the expression pattern of mesoderm, cardiac progenitor, and mature cardiomyocyte molecular markers defined the span of each stage (Fig. 1b) which was further confirmed by flow cytometry (Fig. 1c) and ICC results (Fig. 1d). RT-qPCR results indicated a distinct expression pattern for miR-335-3p and miR-335-5p during the cardiac differentiation process. While miR-335-3p expression was significantly altered (up to six folds) during the mesoderm stage (before day 1.5), miR-335-5p was changed much less (1.5-fold) at this stage. miR-335-5p expression was strongly altered at the progenitor stage (after day 1.5) (Fig. 2b). Up- and downregulation of miR-335 since D0 of the differentiation process lasted until the progenitor stage of the process, 2 days post-transfection (Fig. 2e). ICC results indicated that miR-335 expression alterations did not change the fate of differentiating cells towards cardiomyocytes (Fig. 3a). RT-qPCR as well as western blot results indicated that both miR-335-3p and miR-335-5p gain of functions enhanced mesodermal cell commitment, as mirrored by the increased BRACHYURY expression level. Results of loss-of-function study also supported gain-of-function results for miR-335-3p (Fig. 3b). It was consistent with a previous report showing that miR-335 stabilizes the lineage decision in mouse ESCs for mesendoderm formation [38]. Also, Schoeftner et al. consistently reported that miR-335 targets OCT4 and Rb to control mESC proliferation and induced differentiation [39].
Following miR-335-3p or miR-335-5p gain- and loss-of-functions, cardiac progenitor markers (GATA4 and NKX2-5) were also up- and down-regulated, respectively (Fig. 3d, e). Upregulation of GATA4 and NKX2-5 progenitor markers could enhance the first heart field progenitor cell commitment through upregulating HCN4 (Fig. 3f) and downregulating ISL1 expression (Fig. 3g). RT-qPCR results against late cardiac differentiation markers (TNNT2 and CNX43) suggest that miR-335-3p and miR-335-5p might have complementary effects on the process.
While miR-335-3p downregulated TNNT2 expression, miR-335-5p significantly upregulated it within the cells (Fig. 3i). Similar results were also obtained for CNX43 expression after alterations in miR-335-3p and miR-335-5p expression (Fig. 3h). Altogether, these data suggest that miR-335 enhances cardiac differentiation through upregulation of the expression of mesoderm (BRACHYURY) and cardiac progenitor marker (GATA4 and NKX2-5) genes. Nevertheless, molecular mechanism(s) via which these miRNAs regulate the expression of these markers remained undiscovered.
Bioinformatics analyses indicated that miR-335 regulates WNT and TGFβ signaling pathways. Moreover, RT-qPCR (Fig. 4b–e, top) and dual luciferase assay (Fig. 4b–e, bottom) showed that miR-335-3p specifically target 3′UTR sequences of APC and AXIN-I, which are two main members of WNT inhibitor complex. These data suggested that miR-335-3p but not miR-335-5p could regulate APC expression at the post-transcriptional level.
Although miR-335-5p had no direct interaction with 3′UTR of these two target genes, but RT-qPCR results indicated a significant downregulation of AXIN-I transcript (Fig. 4e, top). Thus, it could be concluded that miR-335-5p indirectly downregulated AXIN-I expression without interacting with its transcripts. Consistently, both of these miRNAs were capable of activating WNT signaling pathway as detected by Top/Fop flash assay (Fig. 4f). This is also consistent with a previous report which showed that miR-335-5p activates WNT signaling pathway through DKK1 downregulation [40].
Interaction between both miR-335-3p and miR-335-5p and SMAD7 3′UTR was also confirmed by RT-qPCR (Fig. 5a) as well as dual luciferase assay (Fig. 5b). Results indicated that these two miRNAs could enhance TGFβR-I expression (Fig. 5c, d). There are two SMAD-binding elements within TGFβR-I promoter recognized by SMAD7, which inhibit TGFβR-I expression [41]. Downregulation of SMAD7 by these two miRNAs could lessen the inhibitory effect of SMAD7 and enhance TGFβR-I expression. Altogether, the results proposed that miR-335-3p and miR-335-5p could activate WNT and TGFβ signaling pathways.
LEF-1/β-catenin complex is reported to bind to the TCF binding site at the BRACHYURY promoter sequence and enhance its expression [42]. TGFβ1 also induces the expression of BRACHYURY in human carcinoma cells [43]. In other words, activation of WNT and TGFβ signaling pathways could enhance BRACHYURY expression, as also observed in the current experiment. Furthermore, WNT signaling pathway activation was shown to enhance GATA4 [44] as well as NKX2-5 expression through hindering HDAC1 inhibitory effect [45]. There is also evidence showing that TGFβ activation enhances GATA4 and NKX2-5 expression [46–48].
Conclusion
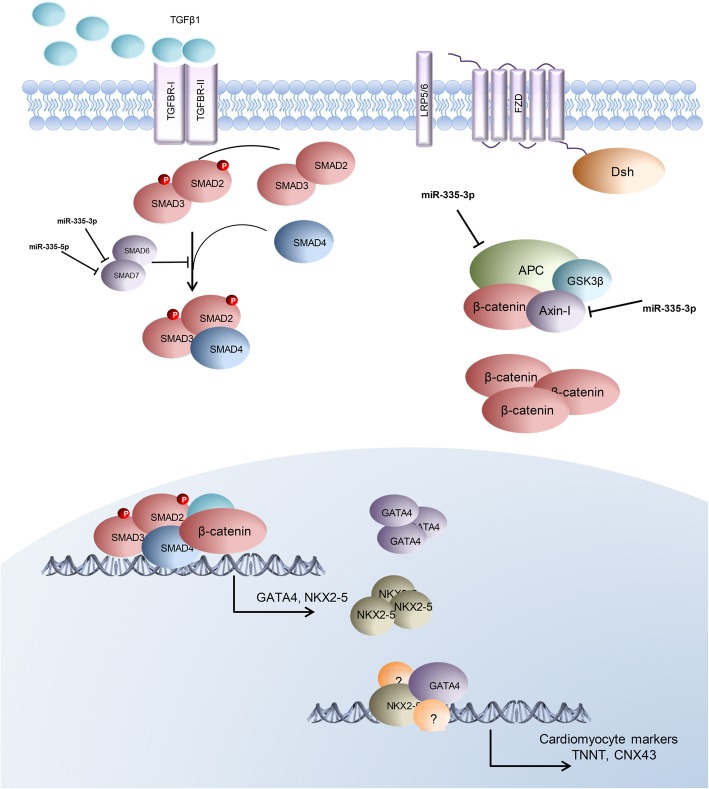
Based on the bioinformatics and experimental validations, we propose a regulatory network for miR-335-3p and miR-335-5p during cardiomyocyte differentiation (Fig. 6). Accordingly, both miR-335-3p and miR-335-5p might activate WNT and TGFβ signaling pathways that, in turn, induce mesoderm (BRACHYURY) and progenitor (GATA4 and NKX2-5) marker expression and cardiac differentiation.
Fig. 6.
Schematic presentation of miR-335 involvement in cardiomyocyte differentiation procedure (details are discussed in the text)
Additional files
Table S1. Primer sequences used in this research. Table S2. Final list of candidate miRNAs. Table S3. List of potential miR-335-3p target genes related to WNT and TGFβ signaling pathways. Table S4. List of potential miR-335-5p target genes related to WNT and TGFβ signaling pathways. Figure S1. RT-qPCR results of C-MYC and CCND1 after mimics and siRNA treatment for miR-335-3p (A) and miR-335-5p (B). All experiments were done in three biological replicates and presented as mean ± SEM. Figure S2. RT-qPCR results of SMAD2 and SMAD3 expression. A) Pairing status of miR-335-3p (left) and miR-335-5p (right), with 3′UTR of SMAD7 gene. B) SMAD2 expression was not significantly changed followed by miR-335-3p (top) and miR-335-5p (bottom) overexpression. C) RT-qPCR data also showed no significant alterations in SMAD3 expression following miR-335-3p (top) and miR-335-5p (bottom) overexpression. All data are presented as mean ± SEM normalized against mimic-scr and siRNA-scr. GAPDH was used as a housekeeping gene. Figure S3. Genomic location of mir-335 presented in UCSC genome browser. miR-335 is located within the second intron of MEST gene, containing two conserved mature miRNAs (highlighted in red) including miR-335-3p and miR-335-5p. Figure S4. The potential targets of miR-335 in TGFβ (A) and WNT (B) signaling pathways according to the KEGG pathway. The target genes are marked with red stars. (DOCX 474 kb)
Movie S1. (AVI 2586 kb)
Acknowledgements
Authors would like to thank all members of both departments for their kind comments.
Funding
This work was supported by a grant from Royan Institute and the Iran National Science Foundation (INSF, grant no. 96001316) to H.B., and NIMAD and TMU to B.M.S.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Authors’ contributions
MK contributed to do experiment design and carried out the experiment, and also contributed in the manuscript preparation. HA and FHA did the experiment. BMS and HB did the experiment design, result interpretation, and manuscript preparation. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethical/Scientific Committee of Royan Institute.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Maryam Kay, Email: Maryam_kay2001@yahoo.com.
Bahram Mohammad Soltani, Phone: +98-21-82884703, Email: soltanib@modares.ac.ir.
Fahimeh Hosseini Aghdaei, Email: hoseiny6965@gmail.com.
Hassan Ansari, Email: hassanansaritat@yahoo.com.
Hossein Baharvand, Phone: +98 21 22306485, Email: Baharvand@RoyanInstitute.org.
References
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. nature. 1981;292(5819):154. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Burdon T, Smith A, Savatier P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 2002;12(9):432–438. doi: 10.1016/s0962-8924(02)02352-8. [DOI] [PubMed] [Google Scholar]
- 3.Keller GM. In vitro differentiation of embryonic stem cells. Curr Opin Cell Biol. 1995;7(6):862–869. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
- 4.Vidarsson H, Hyllner J, Sartipy P. Differentiation of human embryonic stem cells to cardiomyocytes for in vitro and in vivo applications. Stem Cell Rev Rep. 2010;6(1):108–120. doi: 10.1007/s12015-010-9113-x. [DOI] [PubMed] [Google Scholar]
- 5.Verma V, Purnamawati K, Shim W. Steering signal transduction pathway towards cardiac lineage from human pluripotent stem cells: a review. Cell Signal. 2013;25(5):1096–1107. doi: 10.1016/j.cellsig.2013.01.027. [DOI] [PubMed] [Google Scholar]
- 6.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313(5795):1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol. 2009;10(2):91. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- 8.Gadue P, Huber TL, Paddison PJ, Keller GM. Wnt and TGF-β signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci. 2006;103(45):16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujimori K, Matsumoto T, Kisa F, Hattori N, Okano H, Akamatsu W. Escape from pluripotency via inhibition of TGF-β/BMP and activation of Wnt signaling accelerates differentiation and aging in hPSC progeny cells. Stem Cell Reports. 2017;9(5):1675–1691. doi: 10.1016/j.stemcr.2017.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakaki-Yumoto M, Katsuno Y, Derynck R. TGF-β family signaling in stem cells. Biochimica et Biophysica Acta (BBA)-General Subjects. 2013;1830(2):2280–2296. doi: 10.1016/j.bbagen.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chew CL, Conos SA, Unal B, Tergaonkar V. Noncoding RNAs: master regulators of inflammatory signaling. Trends Mol Med. 2017;24(1):66–84. doi: 10.1016/j.molmed.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432(7014):231. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 13.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao S. MicroRNA biogenesis and their functions in regulating stem cell potency and differentiation. Biological Procedures Online. 2016;18(1):8. doi: 10.1186/s12575-016-0037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shenoy A, Blelloch RH. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat Rev Mol Cell Biol. 2014;15(9):565. doi: 10.1038/nrm3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espinoza-Lewis RA, Wang D-Z. MicroRNAs in heart development. Curr Top Dev Biol 100: Elsevier; 2012. p. 279–317. [DOI] [PMC free article] [PubMed]
- 17.Gama-Carvalho M, Andrade J, Brás-Rosário L. Regulation of cardiac cell fate by microRNAs: implications for heart regeneration. Cells. 2014;3(4):996–1026. doi: 10.3390/cells3040996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martello G, Zacchigna L, Inui M, Montagner M, Adorno M, Mamidi A, et al. MicroRNA control of nodal signalling. Nature. 2007;449(7159):183. doi: 10.1038/nature06100. [DOI] [PubMed] [Google Scholar]
- 19.Choi W-Y, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of nodal agonist and antagonist by miR-430. Science. 2007;318(5848):271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 20.Ivey KN, Muth A, Arnold J, King FW, Yeh R-F, Fish JE, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2(3):219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sluijter JP, van Mil A, van Vliet P, Metz CH, Liu J, Doevendans PA, et al. MicroRNA-1 and-499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler Thromb Vasc Biol. 2010;30(4):859–868. doi: 10.1161/ATVBAHA.109.197434. [DOI] [PubMed] [Google Scholar]
- 22.Baharvand H, Ashtiani SK, Taee A, Massumi M, Valojerdi MR, Yazdi PE, et al. Generation of new human embryonic stem cell lines with diploid and triploid karyotypes. Develop Growth Differ. 2006;48(2):117–128. doi: 10.1111/j.1440-169X.2006.00851.x. [DOI] [PubMed] [Google Scholar]
- 23.Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, et al. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell. 2011;9(2):144–155. doi: 10.1016/j.stem.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendjan S, Mascetti VL, Ortmann D, Ortiz M, Karjosukarso DW, Ng Y, et al. NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell. 2014;15(3):310–325. doi: 10.1016/j.stem.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Dobaczewski M, Chen W, Frangogiannis NG. Transforming growth factor (TGF)-β signaling in cardiac remodeling. J Mol Cell Cardiol. 2011;51(4):600–606. doi: 10.1016/j.yjmcc.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bujak M, Frangogiannis NG. The role of TGF-β signaling in myocardial infarction and cardiac remodeling. Cardiovasc Res. 2007;74(2):184–195. doi: 10.1016/j.cardiores.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marvin MJ, Di Rocco G, Gardiner A, Bush SM, Lassar AB. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 2001;15(3):316–327. doi: 10.1101/gad.855501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci. 2012;109(27):E1848–E1E57. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozhan G, Weidinger G. Wnt/β-catenin signaling in heart regeneration. Cell Regeneration. 2015;4(1):3. doi: 10.1186/s13619-015-0017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim J-Y, Kim WH, Kim J, Park SI. Involvement of TGF-β1 signaling in cardiomyocyte differentiation from P19CL6 cells. Molecules & Cells (Springer Science & Business Media BV) 2007;24(3):431–6. [PubMed] [Google Scholar]
- 31.Paige SL, Plonowska K, Xu A, Wu SM. Molecular regulation of cardiomyocyte differentiation. Circ Res. 2015;116(2):341–353. doi: 10.1161/CIRCRESAHA.116.302752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11(4):252. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 33.Najafi H, Soltani BM, Dokanehiifard S, Nasiri S, Mowla SJ. Alternative splicing of the OCC-1 gene generates three splice variants and a novel exonic microRNA, which regulate the Wnt signaling pathway. RNA. 2017;23(1):70–85. doi: 10.1261/rna.056317.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dokanehiifard S, Soltani BM. Hsa-miR-11181 regulates Wnt signaling pathway through targeting of APC2 transcripts in SW480 cell line. Gene. 2018;641:297–302. doi: 10.1016/j.gene.2017.10.075. [DOI] [PubMed] [Google Scholar]
- 35.Ekhteraei-Tousi S, Mohammad-Soltani B, Sadeghizadeh M, Mowla SJ, Parsi S, Soleimani M. Inhibitory effect of hsa-miR-590-5p on cardiosphere-derived stem cells differentiation through downregulation of TGFB signaling. J Cell Biochem. 2015;116(1):179–191. doi: 10.1002/jcb.24957. [DOI] [PubMed] [Google Scholar]
- 36.Rogler CE, LeVoci L, Ader T, Massimi A, Tchaikovskaya T, Norel R, et al. MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology. 2009;50(2):575–584. doi: 10.1002/hep.22982. [DOI] [PubMed] [Google Scholar]
- 37.Fish JE, Santoro MM, Morton SU, Yu S, Yeh R-F, Wythe JD, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15(2):272. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang D, Lutter D, Burtscher I, Uetzmann L, Theis FJ, Lickert H. miR-335 promotes mesendodermal lineage segregation and shapes a transcription factor gradient in the endoderm. Development. 2014;141(3):514–525. doi: 10.1242/dev.104232. [DOI] [PubMed] [Google Scholar]
- 39.Schoeftner S, Scarola M, Comisso E, Schneider C, Benetti R. An Oct4-pRb axis, controlled by MiR-335, integrates stem cell self-renewal and cell cycle control. Stem Cells. 2013;31(4):717–728. doi: 10.1002/stem.1315. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, Tu Q, Bonewald LF, He X, Stein G, Lian J, et al. Effects of miR-335-5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J Bone Miner Res. 2011;26(8):1953–1963. doi: 10.1002/jbmr.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao W, Pan Z, Du X, Zhang J, Li Q. miR-181b-induced SMAD7 downregulation controls granulosa cell apoptosis through TGF-β signaling by interacting with the TGFBR1 promoter. J Cell Physiol. 2018;233(9):6807–21. doi: 10.1002/jcp.26431. [DOI] [PubMed] [Google Scholar]
- 42.Arnold SJ, Stappert J, Bauer A, Kispert A, Herrmann BG, Kemler R. Brachyury is a target gene of the Wnt/β-catenin signaling pathway. Mech Dev. 2000;91(1–2):249–258. doi: 10.1016/s0925-4773(99)00309-3. [DOI] [PubMed] [Google Scholar]
- 43.Larocca C, Cohen JR, Fernando RI, Huang B, Hamilton DH, Palena C. An autocrine loop between TGF-β1 and the transcription factor brachyury controls the transition of human carcinoma cells into a mesenchymal phenotype. Mol Cancer Ther. 2013;12(9):1805–1815. doi: 10.1158/1535-7163.MCT-12-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang J, Guo X, Li W, Zhang H. Activation of Wnt/β-catenin signalling via GSK3 inhibitors direct differentiation of human adipose stem cells into functional hepatocytes. Sci Rep. 2017;7:40716. doi: 10.1038/srep40716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Z, Li T, Liu Y, Jia Z, Li Y, Zhang C, et al. WNT signaling promotes Nkx2. 5 expression and early cardiomyogenesis via downregulation of Hdac1. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2009;1793(2):300–311. doi: 10.1016/j.bbamcr.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 46.Jamali M, Karamboulas C, Rogerson PJ, Skerjanc IS. BMP signaling regulates Nkx2-5 activity during cardiomyogenesis. FEBS Lett. 2001;509(1):126–130. doi: 10.1016/s0014-5793(01)03151-9. [DOI] [PubMed] [Google Scholar]
- 47.Haveri H, Ashorn M, Iltanen S, Wilson DB, Andersson LC, Heikinheimo M. Enhanced expression of transcription factor GATA-4 in inflammatory bowel disease and its possible regulation by TGF-β1. J Clin Immunol. 2009;29(4):444–453. doi: 10.1007/s10875-009-9292-x. [DOI] [PubMed] [Google Scholar]
- 48.Li T-S, Komota T, Ohshima M, Qin S-L, Kubo M, Ueda K, et al. TGF-β induces the differentiation of bone marrow stem cells into immature cardiomyocytes. Biochem Biophys Res Commun. 2008;366(4):1074–1080. doi: 10.1016/j.bbrc.2007.12.095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer sequences used in this research. Table S2. Final list of candidate miRNAs. Table S3. List of potential miR-335-3p target genes related to WNT and TGFβ signaling pathways. Table S4. List of potential miR-335-5p target genes related to WNT and TGFβ signaling pathways. Figure S1. RT-qPCR results of C-MYC and CCND1 after mimics and siRNA treatment for miR-335-3p (A) and miR-335-5p (B). All experiments were done in three biological replicates and presented as mean ± SEM. Figure S2. RT-qPCR results of SMAD2 and SMAD3 expression. A) Pairing status of miR-335-3p (left) and miR-335-5p (right), with 3′UTR of SMAD7 gene. B) SMAD2 expression was not significantly changed followed by miR-335-3p (top) and miR-335-5p (bottom) overexpression. C) RT-qPCR data also showed no significant alterations in SMAD3 expression following miR-335-3p (top) and miR-335-5p (bottom) overexpression. All data are presented as mean ± SEM normalized against mimic-scr and siRNA-scr. GAPDH was used as a housekeeping gene. Figure S3. Genomic location of mir-335 presented in UCSC genome browser. miR-335 is located within the second intron of MEST gene, containing two conserved mature miRNAs (highlighted in red) including miR-335-3p and miR-335-5p. Figure S4. The potential targets of miR-335 in TGFβ (A) and WNT (B) signaling pathways according to the KEGG pathway. The target genes are marked with red stars. (DOCX 474 kb)
Movie S1. (AVI 2586 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its supplementary information files].