Abstract
Mesial temporal lobe epilepsy (mTLE) is a chronic neurological disease characterized by recurrent seizures. The antiepileptic drugs currently available to treat mTLE are ineffective in one-third of patients and lack disease-modifying effects. miRNAs, a class of small noncoding RNAs which control gene expression at the post-transcriptional level, play a key role in the pathogenesis of mTLE and other epilepsies. Although manipulation of miRNAs at acute stages has been reported to reduce subsequent spontaneous seizures, it is uncertain whether targeting miRNAs at chronic stages of mTLE can also reduce seizures. Furthermore, the functional role and downstream targets of most epilepsy-associated miRNAs remain poorly understood. Here, we show that miR-135a is selectively upregulated within neurons in epileptic brain and report that targeting miR-135a in vivo using antagomirs after onset of spontaneous recurrent seizures can reduce seizure activity at the chronic stage of experimental mTLE in male mice. Further, by using an unbiased approach combining immunoprecipitation and RNA sequencing, we identify several novel neuronal targets of miR-135a, including Mef2a. Mef2 proteins are key regulators of excitatory synapse density. Mef2a and miR-135a show reciprocal expression regulation in human (of both sexes) and experimental TLE, and miR-135a regulates dendritic spine number and type through Mef2. Together, our data show that miR-135a is target for reducing seizure activity in chronic epilepsy, and that deregulation of miR-135a in epilepsy may alter Mef2a expression and thereby affect synaptic function and plasticity.
SIGNIFICANCE STATEMENT miRNAs are post-transcriptional regulators of gene expression with roles in the pathogenesis of epilepsy. However, the precise mechanism of action and therapeutic potential of most epilepsy-associated miRNAs remain poorly understood. Our study reveals dramatic upregulation of the key neuronal miRNA miR-135a in both experimental and human mesial temporal lobe epilepsy. Silencing miR-135a in experimental temporal lobe epilepsy reduces seizure activity at the spontaneous recurrent seizure stage. These data support the exciting possibility that miRNAs can be targeted to combat seizures after spontaneous seizure activity has been established. Further, by using unbiased approaches novel neuronal targets of miR-135a, including members of the Mef2 protein family, are identified that begin to explain how deregulation of miR-135a may contribute to epilepsy.
Keywords: antagomirs, epilepsy, Mef2, mesial temporal lobe epilepsy, miRNA, RNA sequencing
Introduction
Epilepsy is a chronic neurological disease that is characterized by recurrent unprovoked seizures and that affects 65 million people worldwide (Chang and Lowenstein, 2003; Moshé et al., 2015). Temporal lobe epilepsy (TLE) is a subclass of epilepsy and accounts for approximately one-third of all epilepsies (Engel, 2001). It consists of several subgroups, of which mesial TLE with hippocampal sclerosis (mTLE-HS) is most severe and is resistant to pharmacological treatment (Wieser, 2004). For many mTLE-HS patients, surgical removal of the hippocampus is the only effective treatment for achieving seizure control (Semah et al., 1998; Blümcke et al., 2013). While anticonvulsant and antiepileptic drugs are used to treat mTLE patients, these drugs reduce the occurrence of seizures but do not treat the underlying pathophysiology. Hence, there is an urgent need to develop novel treatment strategies for treating TLE and other epilepsies (Löscher et al., 2013).
The pathological mechanisms underlying mTLE are still incompletely understood, but animal models of epilepsy and human tissue studies suggest that epileptogenesis involves a cascade of molecular and cellular network alterations (Becker et al., 2003; Staley, 2004; Wetherington et al., 2008; Rakhade and Jensen, 2009). During the past several years, miRNAs have emerged as important post-transcriptional regulators of gene expression, providing a completely new level of control over gene expression. miRNAs are small, noncoding RNAs (18–25 nt long) that are generated from longer RNA precursors transcribed from the genome. miRNAs recognize complementary target sequences in cognate mRNAs and inhibit protein expression by either destabilizing their mRNA targets or by inhibiting protein translation (Kosik, 2006; Bartel, 2018). A single miRNA can have many different targets, and it can regulate several genes in a pathway or single genes in multiple pathways (Ebert and Sharp, 2012; Bartel, 2018). Thus, miRNAs may be used to robustly disrupt single pathways or to simultaneously interfere with multiple pathways (Ebert and Sharp, 2012; Henshall et al., 2016).
Deregulation of miRNAs has been linked to several of the pathological mechanisms underlying TLE (Aronica et al., 2010; Kan et al., 2012; Jimenez-Mateos and Henshall, 2013; Gorter et al., 2014; Cattani et al., 2016). Manipulation of 14 of the 16 miRNAs that have been functionally validated in vivo has been found to elicit beneficial effects at the histopathology level and on seizure activity (Gross et al., 2016; Henshall et al., 2016; Iori et al., 2017). Despite this progress, the mechanisms through which most miRNAs affect seizures and/or epileptogenesis remain unknown. Furthermore, whether manipulation of miRNAs at later, chronic stages of epilepsy has therapeutic effects is an important but largely unresolved question. Previously, we have shown that a significant number of miRNAs are upregulated or downregulated in hippocampal tissue of human mTLE patients (Kan et al., 2012). Of those miRNAs, miR-135a is of particular interest as it is known to control neuronal morphology and synaptic function. For example, miR-135a modulates glutamatergic neurotransmission by regulating Complexin1/2 in the amygdala (Mannironi et al., 2018). Furthermore, miR-135a promotes developmental axon growth and branching, cortical neuronal migration, and regeneration of RGC axons following optic nerve injury in adult mice (van Battum et al., 2018). Because of these biological effects of miR-135a and the strong increase in miR-135a in mTLE patients, we further investigated the potential role of miR-135a in mTLE pathogenesis. Our data show that miR-135a expression is specifically increased during the chronic stage of experimental TLE and that inhibiting miR-135a at this stage reduces spontaneous seizure activity. These data show one of the first examples that inhibiting an miRNA at chronic stages of experimental TLE has therapeutic effects on spontaneous seizure activity. As a first step toward understanding how miR-135a influences seizure activity, we identify the activity-dependent transcription Mef2a as a direct neuronal miR-135a target. Further, our results confirm reciprocal regulation of miR-135a and Mef2a expression in epilepsy and reveal that miR-135a can regulate dendritic spine morphology and number through Mef2.
Materials and Methods
Animals.
All animal experiments were performed according to the institutional guidelines and approved by (1) the Research Ethics Committee of the Royal College of Surgeons in Ireland; ethics: REC 842; and HPRA (Health Products Regulatory Authority) AE19127/P001, or (2) the local ethical animal experimentation committee (Dierexperimenten Ethische Commissie) of the University Medical Center Utrecht (protocol numbers DEC 2014.I.01.005, 527-16-532-03-07). C57bl6J mice (male and female) were obtained from Charles Rivers Laboratories.
Intra-amygdala kainate (IAK) mouse model.
Animals were handled according to institutional guidelines, and experiments were reviewed and approved by Royal College of Surgeons in Ireland (REC 842), under a license from the Department of Health (Health Products Regulatory Authority, AE19127/001), Dublin, Ireland and reviewed and approved by the ethical animal experimentation committee (Dierexperimenten Ethische Commissie) of University Medical Center Utrecht under the project license AVD115002016532 (protocol 527-16-532-03-07). Status epilepticus (SE) induction, EEG recording, and analysis were performed as previously described (Mouri et al., 2008; Jimenez-Mateos et al., 2012). Briefly, for the long-term monitoring (24/7 video/EEG), male mice were implanted with telemetric EEG transmitters (Data Systems International) for bilateral recording on both brain hemispheres with four measuring electrodes. EEG data were acquired using Ponemah acquisition software (version 5.20, Data Systems International), and F20-EET EEG transmitters (Data Systems International) were used. For PBS/scrambled control experiments, EEG data were acquired only unilaterally using the Dataquest A.R.T Gold acquisition software (version 4.33, Data Systems International) and TA11ETA-F10 EEG transmitters (Data Systems International). Two measuring electrodes were affixed over the dorsal hippocampi/temporal cortex (coordinates 2.0 mm posterior to bregma and 1.5 mm from the midline) and over the cerebellum midline. In both experiments, 2 d after mice underwent surgery, SE was induced by the intra-amygdala administration of kainic acid (KA; 0.3 μg in 0.2 μl in PBS). Control animals received the same volume of PBS. Forty minutes after microinjection, mice received an intraperitoneal injection of lorazepam (8 mg/kg) to reduce morbidity and mortality. Mice were video- and EEG-monitored for 24 h to confirm they were presented with similar SE.
Intracerebroventricular injections.
For antagomirs, intracerebroventricular injections were performed as described previously (Jimenez-Mateos et al., 2012; Reschke et al., 2017). From day 7 after SE induction, an epileptic baseline EEG was recorded. At day 14 (D14), mice received an infusion of 1.0 nmol/2 μl of antagomir-135a (ant-135a) LNA modified and 3′-cholesterol-modified oligonucleotides (Exiqon) in PBS. Controls received the same volume of PBS. Similarly, for control experiments, 1.0 nmol/1 μl of scrambled (Scr) LNA modified and 3′-cholesterol-modified oligonucleotides (Exiqon) in PBS were injected, and compared with controls that received the same volume of PBS. During this period, mice were continuously EEG- and video-monitored for another week. EEG data analysis was performed using Neuroscore (version 2.1.0, Data Systems International) and LabChart 8 software (ADInstruments).
RNA isolation and qPCR.
Hippocampal tissue samples from pharmaco-resistant mTLE patients were obtained as described previously (Kan et al., 2012), at the University Medical Center Utrecht. Informed consent was obtained from all patients for procedures approved by the Institutional ethics board. Postmortem human tissue material was obtained from the Netherlands Brain Bank. Samples from 7 patients (male and female) with mTLE-HS and 8 postmortem control samples (male and female) were used (Table 1). Patient tissue representing all hippocampal regions was selected using Nissl staining. Approximately 20 mg of tissue was collected by slicing 25-μm-thick sections on a cryostat. For IAK mice, hippocampus was dissected, frozen, and stored at −80°C. Total RNA was isolated using the miRNeasy kit (QIAGEN), according to the manufacturer's instructions. RNA quantity was determined using Nanodrop (Thermo Fisher Scientific). For miRNA qPCR, first-strand cDNA synthesis was performed using a universal cDNA synthesis kit (Exiqon) according to the manufacturer's recommendation. qPCRs were run in a Quantstudio 6 flex Real-Time PCR system (Applied Biosystems) using microRNA LNA PCR primer sets (miR-135a, miR-124) and SYBR Green master mix (Exiqon). For pre-miRNA qPCR, primer sequences (pre-miR-135a1 and a2) were designed using Primer3 software. Primer sequences for each target are provided in Table 2; 100 ng of RNA was reverse-transcribed using Superscript III first-strand synthesis kit (Thermo Fisher Scientific). Similarly, for validation of bio-immunoprecipitation (bio-IP) targets, 100 ng of inputs RNA and equal amount of IP RNA were reverse-transcribed as above. qPCRs were run on Quantstudio 6 flex Real-Time PCR system (Applied Biosystems) using Fast start universal SYBR Green master mix (Roche Diagnostics). All samples were run in duplicates. Ct values were determined using Quant studio real-time pcr software, version 1.1. For miRNA, expression levels were estimated by normalizing to 5 s rRNA. Pre-miRs were normalized to GAPDH (human) and β-actin (mouse). For determining the fold enrichment of bio-IP target genes, the IP sample was estimated after normalizing to input ΔCt. ΔCt and fold changes were calculated, and the statistical significance was analyzed by Mann–Whitney U test and Student's t test. p < 0.05 was considered as significant.
Table 1.
Details of control and mTLE patients used in this studya
| Number | Sample | Age (yr) | Sex | PMD | Age of onset | Years of epilepsy | AEDs |
|---|---|---|---|---|---|---|---|
| 1 | Control | 58 | M | 7 h | NA | NA | NA |
| 2 | Control | 73 | F | 6,5 h | NA | NA | NA |
| 3 | Control | 71 | M | 9 h | NA | NA | NA |
| 4 | Control | 62 | M | 7 h | NA | NA | NA |
| 5 | Control | 64 | F | 4.5 h | NA | NA | NA |
| 6 | Control | 74 | M | 8 h | NA | NA | NA |
| 7 | Control | 94 | F | 4 h | NA | NA | NA |
| 8 | Control | 70 | M | 20.5 h | NA | NA | NA |
| 9 | Control | 82 | M | 4 h | NA | NA | NA |
| 10 | Control | 94 | M | 5 h | NA | NA | NA |
| 11 | Control | 78 | F | 7 h | NA | NA | NA |
| 12 | Control | 93 | M | 7.5 h | NA | NA | NA |
| 13 | Control | 72 | F | 7 h | NA | NA | NA |
| 14 | Control | 75 | F | 9 h | NA | NA | NA |
| 1 | TLE-HS | 41 | M | NA | 1 | 40 | CBZ |
| 2 | TLE-HS | 36 | F | NA | 14 | 22 | OXC, LZP |
| 3 | TLE-HS | 42 | M | NA | v0.45 | 41 | LEV, LTG |
| 4 | TLE-HS | 52 | F | NA | 20 | 32 | CBZ, CLO, DZP |
| 5 | TLE-HS | 50 | M | NA | 2.5 | 47 | LTG, CBZ, CLO |
| 6 | TLE-HS | 41 | M | NA | 10 | 31 | PHT, CLO, CBZ, LTG |
| 7 | TLE-HS | 49 | F | NA | 12 | 37 | OXC, CLO, SER |
| 8 | TLE-HS | 58 | F | NA | 36 | 22 | LEV, LTG |
| 9 | TLE-HS | 23 | F | NA | 14 | 9 | LTG |
| 10 | TLE-HS | 60 | F | NA | 15 | 45 | LTG, CBZ, LEV |
| 11 | TLE-HS | 41 | M | NA | 16 | 25 | PGB, RES, CBZ |
aAED, anti-epileptic drug; LTG, lamotrigine; PHT, phenytoin; CBZ, carbamazepine; LEV, levetiracetam; OXC, oxcarbazepine; CLO, clobazam; DZP, diazepam; LZP, lorazepam; SER, Seroquel; PGB, pregabaline; RES, Restoril.
Table 2.
List of primers and their sequences used in the study
| Species | Target | Primer sequence |
|
|---|---|---|---|
| Forward | Reverse | ||
| Mouse | Nr3c1 (GR) | GGGGAAGCGTGATGGACTTG | CAGCAGCCACTGAGGGTGAA |
| Mouse | Klf6 | GAGTTCCTCCGTCATTTCCA | GTCGCCATTACCCTTGTCAC |
| Mouse | Mef2a | AGCAGCACCATCTAGGACAA | CTGCTGTTGGAAGCCTGATG |
| Mouse | Mtss1 | ACAGCACCCAGACCACCACC | TGCCTCCTGGTCGCCACTTA |
| Mouse | PlxnA4 | TCTCAGTACAACGTGCTG | TAGCACTGGATCTGATTGC |
| Mouse | Slit2 | CAGTCATTCATGGCTCCCTC | TTCCCTCGGCAGTCTACAAT |
| Mouse | Tsc1 | CAGGAGTTACAGACAAAGCTGG | AGCTTCTGAGAGACCTGGCT |
| Mouse | Gapdh | CCCCAATGTGTCCGTCGTG | GCCTGCTTCACCACCTTCT |
| Human | Pre-miR-135a1 | TCGCTGTTCTCTATGGCTTTT | CGGCTCCAATCCCTATATGA |
| Human | Pre-miR-135a2 | TGCTTTATGGCTTTTTATTCCT | TGGCTTCCATCCCTACATGA |
| Human | GAPDH | TGGAAGGACTCATGACCACA | GGGATGATGTTCTGGAGAGC |
| Mouse | Pre-miR-135a1 | GCCTCACTGTTCTCTATGGCTTT | CCACGGCTCCAATCCCTATATGA |
| Mouse | Pre-miR-135a2 | TGCTTTATGGCTTTTTATTC | CATCCCTACATGAGACTTTATT |
| Mouse | β-actin | AGCCATGTACGTAGCCATCC | CTCTCAGCTGTGGTGGTGAA |
Nonradioactive ISH.
Nonradioactive ISH was performed as described previously (Kan et al., 2012). Three patients from each group (control and mTLE) were used for ISH. Similarly, for IAK mice sections, 3 mice per group were used. Briefly, 16-μm-thick sections from fresh frozen hippocampal tissue were collected on glass slides and stored at −80°C until use. Sections were fixed (4% PFA for 10 min at room temperature [RT]), acetylated (10 min at RT), and treated with proteinase K (5 μg/ml for 5 min at RT). Prehybridization was performed for 1 h at RT. Hybridization was performed with 10 nm of double-DIG (3′ and 5′)-labeled locked nucleic acid (LNA) probe for human-miR-135a-5p (Exiqon) or LNA-DIG scrambled-miR probe overnight at 50°C. Slides were washed at 55°C in 0.2× SSC for 1 h, followed by blocking with 10% FCS in B1 buffer (0.1 m Tris, pH 7.5, 0.15 m NaCl) for 1 h at RT. For ISH on antagomir-injected mice, a custom-made double DIG labeled miR-135a inhibitor (miR-135a.inh) probe (Exiqon), which specifically recognizes ant-135a, was used. Hybridization was performed with 20 nm of miR-135a.inh probe overnight at 55°C followed by stringency washes at 60°C in B1 buffer. Sections were incubated with anti-digoxigenin-AP Fab fragments (1;2500, Roche Diagnostics) in 10% FCS in B1 buffer overnight at 4°C. Slides were treated with BCIP and NBT substrates (NBT/BCIP stock solution, Roche Diagnostics) in B3 (0.1 m Tris, pH 9.5, 0.1 m NaCl, 50 mm MgCl2) for 5–20 h at RT. Staining was stopped by washes in PBS, and slides were mounted using Vectashield (Vector Labs). No staining was observed in sections hybridized with scramble probe. Images were acquired with brightfield microscope and processed using ImageJ.
A similar protocol was used for FISH, except that hybridization was performed at 55°C and washes at 60°C. After blocking, slides were coincubated with anti-Digoxigenin-POD (1;500, Roche Diagnostics) and mouse anti-NeuN (1;400, Millipore, RRID:AB_2298772) or rabbit anti-GFAP (1;1000, Dako Cytomation, RRID:AB_10013482) antibodies overnight at 4°C. Signal was amplified using the TSA Cyanine 3 System (1:50 in amplification diluent, PerkinElmer) for 10 min at RT. After washes with PBS, slides were incubated with secondary antibodies (AlexaFluor-488, AlexaFluor-647; Invitrogen) against the primary antibody for 1.5 h at RT. Nuclei were stained with DAPI for 10 min at RT, and slides were mounted using ProLong Gold (Invitrogen). Images were acquired using a confocal laser scanning microscope (LSM880, Carl Zeiss) and processed using ImageJ.
RNA coimmunoprecipitation with biotinylated mimics.
N2A cells were cultured in DMEM low glucose supplemented with l-glutamine, penicillin/streptomycin (100 U/ml and 100 mg/ml, respectively), and 10% FCS (Invitrogen) at 37°C with 5% CO2. For each condition (miR-135a, negative control [NC-1] and no transfection), three 10 cm dishes with 2 × 10−6 cells/dish were plated and transfected with 37.5 nm of 3′ biotinylated miRNA mimics (miR-135a and NC-1, Dharmacon) using HighPerfect Transfection reagent (QIAGEN). RNA coimmunoprecipitation was performed as described previously (Wani and Cloonan, 2014) with some modifications. Briefly, 24 h after transfection cells were collected and lysed in lysis buffer (10 mm Tris-Cl, pH 7.5, 10 mm KCl, 1.5 mm MgCl2, 5 mm DTT, 0.5% NP-40, 60 U/ml SUPERase-in RNase inhibitor (Invitrogen), protease inhibitor tablet (Roche Diagnostics) in MilliQ, and the cleared cell lysates were incubated with Dynabeads M-280 Streptavidin beads (Invitrogen) for 30 min at RT. Beads were washed three times in wash buffer (lysis buffer containing 1 m NaCl) and stored in Qiazol at −80°C. Total RNA was extracted using miRNeasy kit (QIAGEN). One part of the beads was incubated with 4× Nu-PAGE sample buffer (with 10% β-mercaptoethanol in MilliQ) for 10 min at 70°C to extract bound proteins. Proteins were then separated in a 8% SDS-PAGE gel, and the subsequent transferred blot was incubated with rabbit anti-Ago2 antibody (1;1000, Cell Signaling Technology, RRID:AB_2096291) and mouse anti-β-actin (1;2000, Sigma-Aldrich, RRID:AB_476743) in blocking solution (5% milk in 1× TBS-T) overnight at 4°C, finally signal was detected as above.
Library preparation and total RNA sequencing.
For input samples, libraries for total RNA sequencing were prepared using the TruSeq Stranded Total RNA (with RiboZero Gold) sample prep kit (Illumina). The starting material (100 ng) of total RNA was depleted of rRNAs using Ribo-Zero Gold (removes both cytoplasmic and mitochondrial rRNA) magnetic bead-based capture-probe system (Illumina). The remaining RNA, including mRNAs, lincRNAs, and other RNA species, was subsequently purified (RNAcleanXP) and enzymatically fragmentated. For IP samples, libraries were prepared using the TruSeq stranded mRNA sample prep kit (Illumina) according to the manufacturer's instructions with some modifications: the starting material (37.5–50.0 ng) of total RNA was not mRNA-enriched nor fragmented before library synthesis. First-strand synthesis and second-strand synthesis were performed and double-stranded cDNA was purified (Agencourt AMPure XP, Beckman Coulter). The cDNA was end repaired, 3′-adenylated, and Illumina sequencing adaptors were ligated onto the fragments ends, and the library was purified (Agencourt AMPure XP). The polyA+ RNA stranded libraries were preamplified with PCR and purified (Agencourt AMPure XP). Library size distribution was validated and quality inspected using the 2100 Bioanalyzer (high sensitivity DNA chip, Agilent Technologies). High-quality libraries were quantified using the Qubit Fluorometer (Invitrogen). Single-end sequencing was performed using the NextSeq500 instrument according to the manufacturer's instructions (Illumina).
Read mapping and differential expression analysis.
Following trimming of low-quality bases and adapter sequences with FASTQ-MCF (version 0.0.13), processed reads were mapped to the GRCm38.p6 reference mouse genome (Ensembl) with TopHat2 (version 2.0.13). 'fr-secondstrand' option was chosen for the alignments of the total RNA sequencing data. Mapped counts were summarized for each gene using the python script htseq-count (Anders et al., 2015). For differential expression analysis, count data for genes and transcripts were analyzed for differential expression in R using the Bioconductor package EdgeR version 3.12.1 (Robinson et al., 2010) with the trimmed mean of M values (TMM) normalization method (Robinson and Oshlack, 2010). Gene expression levels were corrected for batch effects by including the series of sequencing rounds. Adjusted p values for multiple testing were calculated using the Benjamini-Hochberg false discovery rate (FDR), and only genes with an FDR < 0.05 were considered significantly differentially expressed. Data visualization was performed in R using the ggplot2 library (version 2.1.0). Gene expression heatmaps with hierarchical clustering of expression profiles were created in R with the Bioconductor heatmap package. Enrichment analysis was performed using the R package goseq (Young et al., 2010) to correct for bias due to transcript length. All the raw, and processed RNAseq data are deposited at NCBI Gene Expression Omnibus (GEO) with reference number GSE123000 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE123000).
In silico prediction of miRNA binding sites.
miRanda software version 3.3a was used to predict microRNA signatures. The following parameters were used in this study: match with a minimum threshold score of 150; target mRNA duplex with minimum folding free energy threshold −7 kcal/mol; gap opening penalty −8; gap extension penalty −2; scaling parameter 4 for complementary nucleotide match score.
Target validation: luciferase assay and Western blotting.
HEK293 (RRID:CVCL_0045) and N2A (RRID:CVCL_0470) cells were cultured according to the guidelines provided by ATCC. Luciferase assays were performed in HEK293 cells, and target validations by Western blot were performed in N2A cells.
For luciferase assays, miRNA recognition elements (MREs) for miR-135a present in the 3′-untranslated region (UTR) of Mef2a were identified in RNAseq data and also predicted by Targetscan. Oligonucleotides with these sites were cloned into the psi-Check2 vector (Promega). Oligonucleotides with WT (MEF2A-135a-forward: TCG AGA GCA GAA CCT TGG AAA AAA AAA GCC ATG GC, reverse: GGC CGC CAT GGC TTT TTT TTT CCA AGG TTC TGC TC) and MUT (MEF2A-135aM-forward: TCG AGA GCA GAA CCT TGG AAA AAA AAA GGC TTG GC; reverse: GGC CGC CAA GCC TTT TTT TTT CCA AGG TTC TGC TC) miR-135a binding sites were phosphorylated, annealed, and ligated into the NotI and XhoI sites of the multiple cloning site. Cells (8 × 10−4) were transfected using Lipofectamine 2000 (Invitrogen) with 250 ng of reporter construct together with 25 pmol of miRIDIAN miRNA mimic or Negative control (NC-1, Dharmacon), a nontargeting negative control mimic from Caenorhabditis elegans. Cells were harvested after 24 h, and luciferase assays were performed using the dual-luciferase assay system (E1960, Promega) on a Luminometer. Normalization against Renilla luciferase activity was used to determine relative luciferase activity.
For protein analysis, Western blotting was performed. N2A cells were transfected with miRIDIAN mimics for miR-135a or a negative control using Lipofectamine 2000. After 48 h, cells were harvested and lysed in RIPA buffer (50 mm Tris, pH.7.5, 150 mm NaCl, 0.5% NP-40, 0.5% NaDoc, 1% Triton, Protease inhibitor, Roche Diagnostics, in MilliQ). Equal amounts of protein samples were separated in SDS-PAGE gels (8%) and transferred onto nitrocellulose blotting membranes (GE Healthcare), following which blots were blocked for 1 h at RT in 5% milk powder in 1×TBS-Tween. Blots were incubated overnight at 4°C with rabbit-anti-NR3C1 (glucocorticoid receptor [GR]) (1;1000, Santa Cruz Biotechnology, RRID:AB_2155786), rabbit-anti-PlxnA4 (1;250, Abcam, RRID:AB_944890), and mouse-anti-β actin (1;2000, Sigma-Aldrich, RRID:AB_476743). Blots were stained with peroxidase-conjugated secondary antibodies for 1 h at RT, and signal was detected by incubating blots with Pierce ECL substrate (Thermo Fisher Scientific). Images were acquired using a FluorChem M imaging system (Protein Simple). Using ImageJ, individual band intensities for each sample were measured and normalized to corresponding β-actin levels. Statistical significance of the relative expression between conditions of each protein was estimated by t test (GraphPad Prism version 6 software, RRID:SCR_002798). Except for Mef2a experiments, blots were blocked in Supermix blocking solution (Tris 50 mm, NaCl 154 mm, 0.25% gelatin, 0.5% Triton X-100 in MilliQ, pH 7.4) for 10 min at RT and incubated overnight at 4°C with rabbit anti-Mef2a (1;50,000, Abcam, RRID:AB_10862656) and mouse anti-β actin (1;2000, Sigma-Aldrich, RRID:AB_476743). Blots were washed in 1× TBS-Tween and incubated with secondary antibodies coupled with IR dyes (anti-rabbit-IRdye 800 1;5000 and anti-mouse-IRdye700 1;2000 in 1× TBS-Tween) for 1 h at RT. Finally, blots were washed in 1× TBS-Tween and scanned on Odyssey Clx imaging system (Li-COR Biosciences) using Li-COR Image studio version 3.1 software (RRID:SCR_015795), and band intensities were measured and statistical significance of the relative expression between conditions was estimated by t test (GraphPad Prism version 6 software, RRID:SCR_002798).
Immunohistochemistry and Western blotting for Mef2a.
Mef2a immunostainings were performed on resected human hippocampal mTLE sections and 2 week IAK mouse tissue and compared with corresponding controls; 16 μm sections were blocked in 3% NGS, 0.2% Triton in 1× PBS, pH 7.4, for 1 h at RT followed by incubation in anti-Mef2a antibody (1;150, Abcam) and anti-NeuN antibody (1;400, Millipore) in blocking solution overnight at 4°C. Sections were washed and incubated with corresponding AlexaFluor-conjugated (Thermo Fisher Scientific) secondary antibodies for 1.5 h at RT, followed by washes in 1× PBS, and stained for nuclei with DAPI and mounted using ProLong gold (Thermo Fisher Scientific). High-resolution images were acquired using a confocal microscope (LSM880, Carl Zeiss) and processed using ImageJ. For measuring Mef2a fluorescence intensities in ant-135a and control mice, confocal images from the CA3 region were acquired using similar settings for both groups. Internal densities (IntDen) were estimated by using the particle analysis plugin on ImageJ.
For analyzing Mef2a protein levels in human mTLE and IAK mice hippocampal tissue, protein lysates were prepared in RIPA buffer, and equal amounts of proteins were separated in SDS-PAGE gels (8% gel for mouse samples and 10% gel for human samples), and transferred onto nitrocellulose membranes, blocked, and incubated overnight at 4°C with rabbit anti-Mef2a (for human 1;20,000, for mice 1;50,000, Abcam, RRID:AB_10862656) and mouse anti-β-actin (1;2000, Sigma-Aldrich, RRID:AB_476743). Blots were stained, developed, and quantified as described above.
Culturing and transfection of primary mouse hippocampal neurons.
Dissociated hippocampal neurons were cultured as described previously (Van Battum et al., 2014). Briefly, C57bl6J (P0–1) mouse (male or female) pups were decapitated, and brains were quickly isolated in ice-cold dissection medium (Leibovitz's L-15 supplemented with 7 mm HEPES, Thermo Fisher Scientific). Hippocampus was isolated, trypsinized in 0.25% trypsin in L15-HEPES medium for 20 min at 37°C, followed by trituration using fire-polished Pasteur pipettes in growth medium (Neurobasal medium supplemented with B27, penicillin/streptomycin, l-glutamine, and β-mercaptoethanol). Dissociated cells were plated onto glass coverslips coated with PDL (20 μg/ml) and laminin (40 μg/ml) in growth medium and incubated at 37°C with 5% CO2. Half of the growth medium was refreshed twice a week. On DIV14, neurons were transfected with 0.5 μg of pre-miR-135a1 (cloned into the pJEBB vector with CMV promoter, contains GFP reporter) or pJEBB vector only. For rescue experiments, pJEBB-pre-miR-135a1 and the constitutively active mutant Mef2-vp16 (Fiore et al., 2009) were cotransfected. Transfected neurons were fixed on DIV16 with 4% PFA and 4% sucrose in PBS for 20 min. Immunocytochemistry was performed by blocking neurons in blocking buffer (4% NGS, 0.1% BSA, 0.1% Triton X-100 in 1× PBS, pH 7.4) for 1 h at RT followed by incubation with primary antibody chicken anti-GFP (1;1000, Abcam, RRID:AB_300798) diluted in blocking buffer. The next day, washes in 1× PBS were performed followed by incubation with appropriate secondary antibodies in blocking buffer for 1 h at RT. Sections were mounted using ProLong Gold (Thermo Fisher Scientific). High-resolution images were acquired using an oil-immersion 63× objective of a confocal laser scanning microscope (LSM880, Carl Zeiss). Six or seven z-stack images of each apical dendrites close to the soma were captured. Using ImageJ software (RRID:SCR_003070) with cell counter plugin, different types of spines categorized as immature to mature: filopodium, thin, stubby, mushroom, and cup-shaped on secondary dendrite were identified and counted. Spine density was determined by dividing the number of spines on a branch with the length of the branch.
Experimental design and statistical analysis.
C57bl6J mice were used in this study. Statistical analysis was performed using Prism version 7.05 (GraphPad, RRID:SCR_002798), and a p value <0.05 was considered as significant for all statistical tests. Seizure frequencies before (baseline) and after ant-135a were analyzed using paired t test, the number of seizures per day using F statistics mixed-design, repeated-measures GLM. Seizure duration and total time spent in seizures were analyzed using t test. Differences between two groups were tested using either two-tailed t test or Wilcoxon Mann–Whitney test. For comparing more than two groups, one-way ANOVA was used. Exact p values, t values, and degrees of freedom are provided in the text and the n in the figure legends.
In our previous study, miR-135a was found to be upregulated in mTLE-HS condition using microarray (Kan et al., 2012). We started with validating the expression of miR-135a in a different set of hippocampal human patient samples and tested for both mature and pre-miR-135a levels (see Figs. 1A, 2E). n = 8 controls and 7 patient RNA samples, analyzed statistically using unpaired two-tailed t test. Further, LNA ISH and FISH were performed to establish cell type specificity. At least three patient tissue samples were used for ISH (see Fig. 1B).
Figure 1.
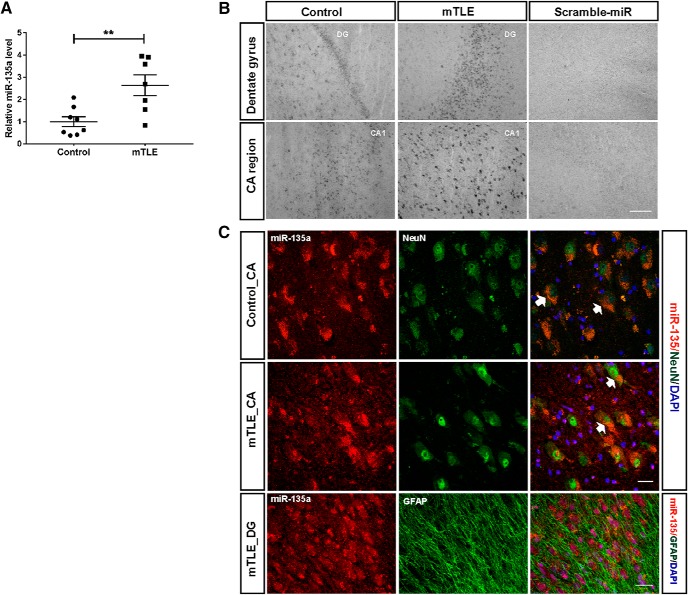
Increased miR-135a expression in human TLE. A, Expression levels of miR-135a in human TLE patients determined by qPCR. Controls, n = 8; mTLE, n = 7. Normalized to 5 s rRNA. Data are mean ± SEM. **p < 0.01 (t test). B, LNA ISH showing localization of miR-135a in control and mTLE groups. Scale bar, 200 μm. C, Cell type-specific localization of miR-135a. Colocalization with neuronal marker, NeuN. Specific localization of miR-135a was observed in neuronal soma in the CA regions. Arrows indicate colabeled cells. No colocalization with the astrocytic marker GFAP was observed. Scale bar, 25 μm.
Figure 2.
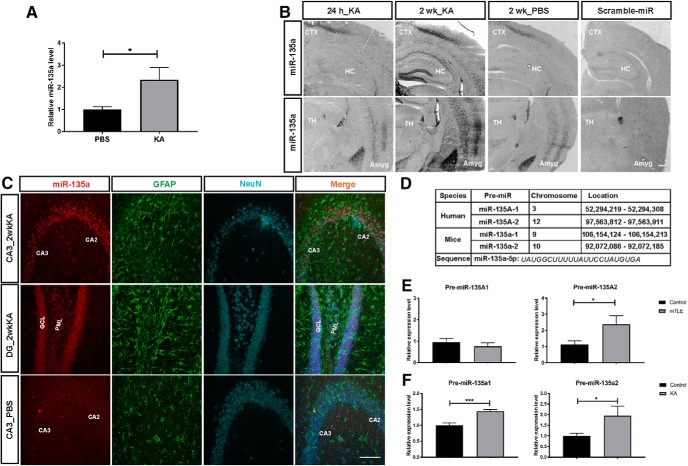
Increased miR-135a in a mouse model of TLE. A, Increased miR-135a levels in the hippocampus of IAK mice 2 weeks after SE. n = 4 PBS and n = 3 IAK mice. Normalized to 5 s rRNA. Data are mean ± SEM. *p < 0.05 (t test). B, Representative images of ISH showing strong miR-135a expression in hippocampus and amygdala regions at 2 weeks after SE induction compared with 24 h and PBS injections. Scramble-stained images were devoid of signal. CTX, Cortex; HC, hippocampus; TH, thalamus; Amyg, amygdala. Scale bar, 300 μm. C, FISH for miR-135a in combination with immunohistochemistry for the astrocytic marker GFAP and neuronal marker NeuN. miR-135a showed no specific colocalization with GFAP. Dentate gyrus PML, Polymorph layer; ML, molecular layer; GCL, granule cell layer; CA, cornu ammonis region. Scale bar, 100 μm. D, Genomic location and sequence of miR-135a in human and mice. Mature miR-135a–5p is spliced from two presequences in mice and human. E, Increased levels of miR-135A-2 in mTLE condition but no change in miR-135A-1. n = 8 controls and n = 7 mTLE samples. Data are mean ± SEM. *p < 0.05 (t test). F, Both miR-135a-1 and miR-135a-2 levels are increased in IAK mice compared with PBS-injected controls. n = 4 PBS and n = 3 KA mice. Data are mean ± SEM. ***p < 0.001 (t test). *p < 0.05 (t test).
Next, we checked whether the expression of miR-135a is also regulated in experimental TLE in an IAK model. Only male mice were used for inducing SE, as previously described (Mouri et al., 2008; Jimenez-Mateos et al., 2012). Hippocampi from 4 PBS and 3 KA mice were used to test the expression levels of mature miR-135a and pre-miR-135a by qPCR at 2 weeks after SE (see Fig. 2A,F); unpaired t test was performed for statistics. Changes in expression were also confirmed by ISH at 24 h and 2 weeks after SE (see Fig. 2B), and cell type specificity was determined by FISH (see Fig. 2C) in at least 3 IAK and control mice. DAPI-positive cell number in the DG suprapyramidal and infrapyramidal blades was estimated using the ImageJ cell counter plugin in a similar ROI in KA and PBS control mice. Statistical analysis was performed using Student's t test, and n = 3 mice per group were used.
Next, to understand whether in vivo targeting of miR-135a in epileptic mice can alter seizure occurrence, SE-induced mice were treated with antagomirs. First, we confirmed whether ant-135a is able to reduce endogenous levels of miR-135a at a given dose without any off-target effects (see Fig. 3A), and tested localization of ant-135a in the hippocampus using an LNA ISH probe designed to recognize ant-135a (see Fig. 3B). One-way ANOVA was used for statistics to compare differences between groups; n = 3 mice per group were used for qPCR and ISH.
Figure 3.
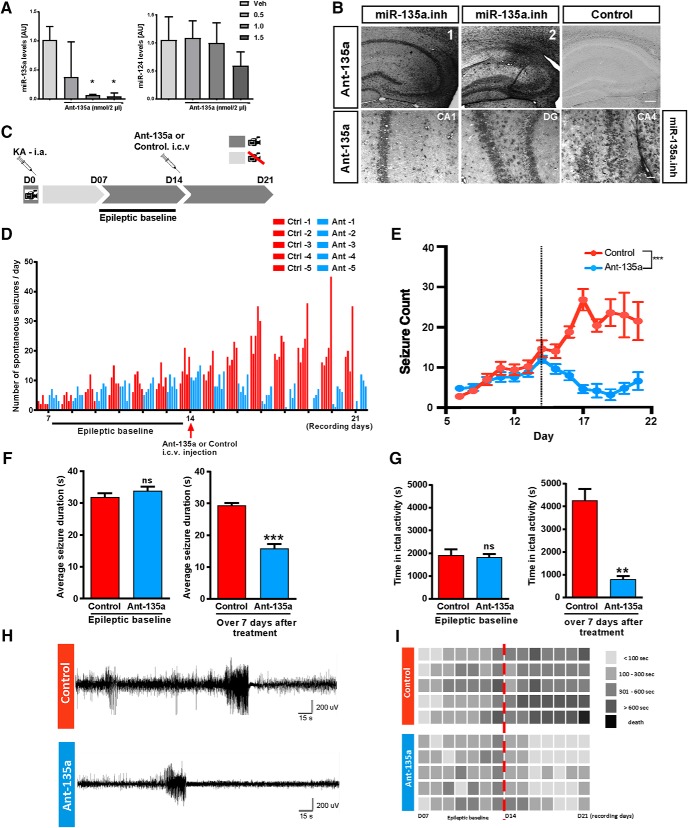
Ant-135a reduces seizures in the mouse IAK model of epilepsy. A, miR-135a expression levels 24 h after administration of ant-135a. Significant reduction of miR-135a levels at 1.0, 1.5 nmol compared with vehicle injection. No off-target effect observed at 1.0 nmol, but reduction in miR-124 levels was observed at 1.5 nmol. n = 3 mice per group. Normalized to RNU6B. Data are mean ± SEM. *p < 0.05 (one-way ANOVA with Sidak post hoc test). B, LNA ISH for the miR-135a inhibitor probe in ant-135a-injected mice. Strong signal for ant-135a is observed in ipsilateral (injected) hippocampus (B1, B2), whereas the control is devoid of specific signal. Scale bar, 200 μm. Ant-135a is taken up by neurons in hippocampal CA1, CA4, and DG regions. Scale bar, 50 μm. C, Male C57BL6 adult mice (∼25 g) were implanted with telemetry devices (Data Systems International) connected to cortical electrodes (both brain hemispheres) for EEG recordings. After appropriate surgical recovery, mice were connected to the EEG equipment and underwent intra-amygdala KA-induced SE on D0. Telemetry devices were turned off and reactivated on D07 to record a 7 d “epileptic baseline.” On D14, mice were intracerebroventricularly injected with ant-135a or its scramble control, and continuously monitored for 7 d (D14 to D21; “after antagomir treatment period”). D, Graph represents the total number of SRSs per day per mouse. Epileptic baseline: No significant difference was detected between treated and control animals in seizure frequency during the 7 d of epileptic baseline (p = 0.743). After antagomir treatment: Following treatment (on D14), a strong decrease in the number of seizures was detected in the antagomir-treated group starting from D15. E, Seizure count is represented as mean ± SD over the EEG recording period. Application of ant-135a at day 14 (dotted line) resulted in a significant decrease in seizure count with respect to time. n = 5 for control and ant-135a. ***p < 0.001 (mixed-design, repeated-measures GLM; day/treatment interaction). F, Average seizure duration: Epileptic baseline: No significant difference between treated and control animals in seizure duration during the 7 d of epileptic baseline (p = 0.4721). ns, not significant. After antagomir treatment: Following treatment (on D14), ant-135a-treated mice presented significantly shorter seizures than the control group; n = 5 mice per group. ***p < 0.001 (t test). G, Time spent in ictal activity: Epileptic baseline: No significant difference between treated and control animals in total time spent in seizures during the 7 d of epileptic baseline (p = 0.7546). ns, not significant. After antagomir treatment: Following treatment (on D14), ant-135a-treated mice spent significantly less time in seizures than control mice; n = 5 mice per group. **p < 0.01 (t test). H, Representative EEG traces of spontaneous seizures 3 d after treatment with ant-135a (bottom) or control (top). I, Total time spent in seizures. Diagram represents seizure burden per day (seconds) per mouse before and after the antagomir treatment (dotted line). A control mouse with high number of seizures died on day 7 after antagomir treatment period.
For in vivo ant-135a experiments, 10 mice were implanted with guide cannulas, and EEG transmitters and SE were induced as described previously (Jimenez-Mateos et al., 2012). Similar SE induction was confirmed by performing continuous EEG recordings until 24 h after SE was induced. Baseline EEG and video recordings were acquired from day 7. On day 14, 5 mice received an intracerebroventricular injection of ant-135a and 5 controls with equal amounts of PBS. Mice were then followed for 1 week with 24/7 EEG and video recordings (see Fig. 3C). Statistical difference in seizure frequency for baseline recordings comparing ant-135a and control group was estimated using Wilcoxon Mann–Whitney statistic; and for overall difference in seizure count (see Fig. 3E), F statistics with mixed-design, repeated-measures GLM test were performed. For average seizure duration and for time spent in ictal activity, paired t test was performed (see Fig. 3F,G). In an independent set of animals, the effect of PBS and Scr injections on seizure activity was assessed. As described above, mice implanted with guide cannulas were injected with KA. On day 14, n = 4 mice per group (injected with either Scr in PBS or an equal volume of PBS and recorded for 1 week). F statistics with mixed-design, repeated-measures GLM test were performed to analyze the results.
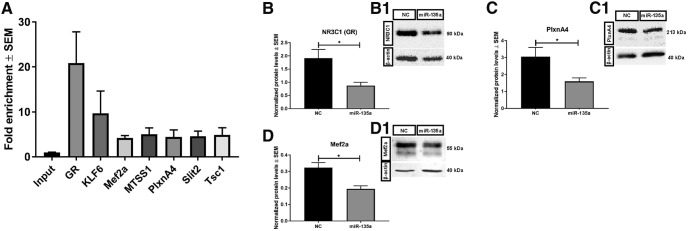
To find new binding partners for miR-135a, immunoprecipitation using biotin-tagged miRNA mimics was performed. Immunoprecipitated RNA was collected from three independent experiments and sent for sequencing; n = 3 samples from each group (sequenced for total RNA; see Fig. 4). Gene expression levels were corrected for batch effects by including a series of sequencing rounds. Adjusted p values for multiple testing were calculated using the Benjamini-Hochberg FDR, and only genes with an FDR < 0.05 were considered significantly differentially expressed and, based on these various plots, were generated. Further, the selected genes were validated for fold enrichment by qPCR (see Fig. 6A), and endogenous protein levels of few selected targets were assessed in N2a cells. Four independent transfections were performed and were repeated twice; normalized means of band intensities were checked statistically using t test.
Figure 4.
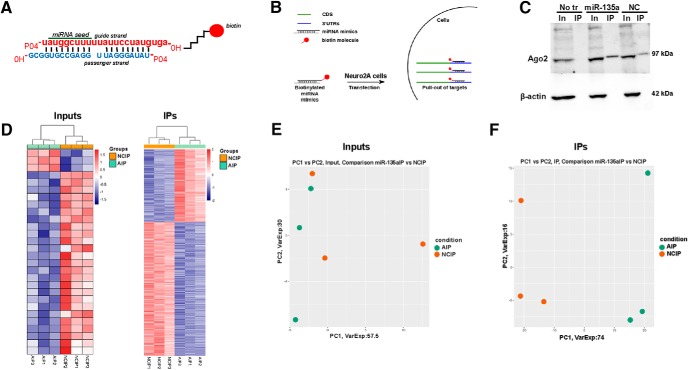
Target identification for miR-135a using biotinylated probes. A, Schematic of miRNA duplex design. The mature strand is labeled with a biotin molecule at the 3′ hydroxyl group via a C6 linker. B, Schematic showing the IP procedure. Neuro2A cells were transfected with biotin-tagged probes. IP was performed using streptavidin beads. Total RNA was extracted and subjected to deep RNA sequencing. n = 3 biological replicates/group. C, Representative Western blot for Ago2 in miR-135a and negative control IP samples. β-actin was used as a loading control and only present in input samples. No tr, No transfection control; NC, negative control; In, inputs; IP, immunoprecipitates. D, Heatmaps of RNA from input and IP samples showing differential gene expression. E, F, Principal component analysis plots of inputs and IPs showing the clustering of samples basing on their differential gene expression. NCIP, Negative control IP; AIP, miR-135a IP.
Figure 6.
Validation of bio-IP targets. A, Several of the selected targets were tested by qPCR and were significantly enriched in the IP samples compared with inputs. n = 3 samples per group. Bar graphs and representative blot images showing GR (B,B1), PlexinA4 (C,C1), and Mef2a (D,D1) protein levels normalized to β-actin after miR-135a overexpression in N2A cells. All of the validated targets were significantly downregulated after miR-135a overexpression compared with negative control (NC) condition. n = 4 independent transfections and were repeated twice. Data are mean ± SEM. *p < 0.05 (t test).
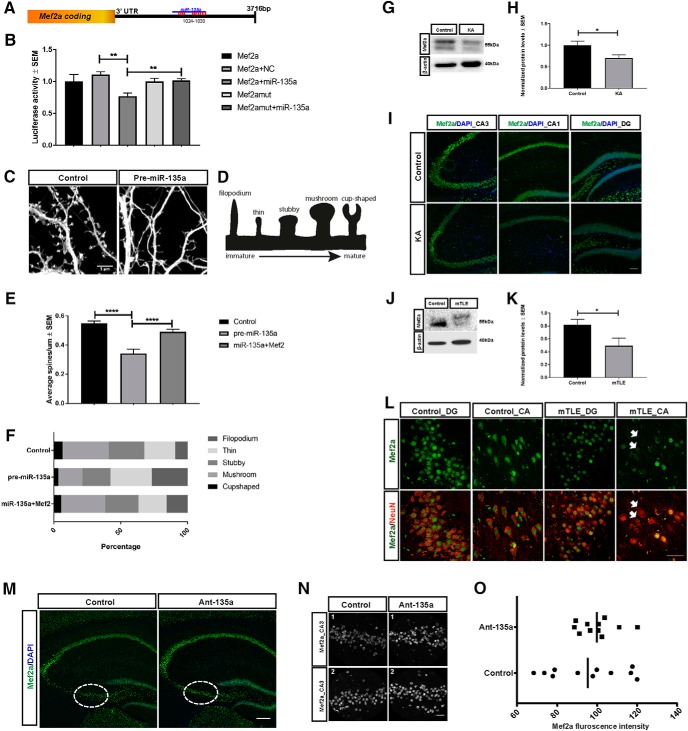
From the immunoprecipitations and RNA-seq data, Mef2a was selected based on its function as a transcription factor regulating several downstream targets that in turn can regulate activity-dependent synaptic density. Direct interaction of miR-135a with Mef2a 3′-UTR was estimated in HEK293 cells by performing luciferase assay in two independent experiments with four transfections each time (see Fig. 7A,B). To assess the role of miR-135a in regulating neuronal spine density, we examined changes in the number of different types of spines (see Fig. 7D) after miR-135a overexpression, and after cotransfecting an Mef2 vector that lacks the 3′-UTR combined with miR-135a. Mouse hippocampal neurons were used and three independent transfections were performed with triplicate of wells each time for each condition. A total of 54 neurons among three conditions were analyzed for different types of spines. One-way ANOVA with Sidak post hoc test was performed (see Fig. 7D,E). Next, we checked whether Mef2a protein levels were altered in vivo in a mice model and in human mTLE patient tissue where miR-135a expression was increased. At least 4 mice per group (see Fig. 7G,H) and 4 patients per group (see Fig. 7J,K) were used. Statistical difference was estimated by Mann–Whitney U test. Next, Mef2a immunostainings were performed in an IAK mouse model (see Fig. 7I) and in human mTLE hippocampus (see Fig. 7L) using at least three tissue samples per group. Finally, to check whether ant-135a-treated KA mice show rescue in Mef2a protein, immunostainings for Mef2a were performed (see Fig. 7M) in 3 mice per group. Fluorescence intensity of Mef2a was analyzed in the CA3 region of the hippocampus (see Fig. 7M, ROI), due to variable cell loss in other regions of the hippocampus between conditions (control and ant-135a mice). Mef2a fluorescence intensity was expressed as ratio of IntDen in a given area. Statistical analysis was performed by Mann–Whitney U test.
Figure 7.
miR-135a regulates dendritic spines through Mef2a, and Mef2a is deregulated in TLE. A, Schematic representation of the 3′-UTR of Mef2a with the miR-135a target site. B, miR-135a target site was ligated into the psiCheck2 vector and used in the Renilla-luciferase assay. Luciferase assay in Hek293 cells transfected with the constructs carrying miR-135a WT and mutant binding sites, cotransfected with and without miR-135a mimic. n = 2 independent experiments, performed with 4 wells/condition each time. Data are mean ± SEM. **p < 0.01 (t test). NC, Negative control. C, Representative image showing secondary apical dendrites quantified for spine density. Dissociated neurons were transfected with miR-135a (with or without Mef2) or control vectors at DIV13 and fixed and analyzed at DIV17. D, Schematic showing the different types of spines quantified. E, Histogram showing the quantification of spine number. Reduced spine density is observed after miR-135a overexpression, and this effect is rescued by cotransfection with Mef2. n = 12–22 neurons, analyzed from three independent transfections. Data are mean ± SEM. ****p < 0.0001 (one-way ANOVA with Sidak post hoc test). F, Graph showing the percentage of different spine types. An increase in immature spines is observed after miR-135a overexpression, which is rescued after Mef2 coexpression. G, Representative Western blot of Mef2a of hippocampal tissue of control and IAK mice at 2 weeks after SE. Mef2a is strongly reduced in experimental mice. H, Quantification of total protein levels normalized to β-actin. n = 5 controls, n = 4 KA mouse hippocampi. Data are mean ± SEM. *p < 0.05 (Mann–Whitney U test). I, Representative image of Mef2a immunostaining in hippocampal subregions of IAK mice. Scale bar, 100 μm. J, Representative Western blot of Mef2a in hippocampi of control and mTLE patients. K, Quantification of total protein levels normalized to β-actin. n = 6 controls, n = 4 mTLE. Data are mean ± SEM. *p < 0.05 (Mann–Whitney U test). L, Representative image of Mef2a immunostaining in controls and mTLE dentate gyrus (DG) and CA regions. Arrows indicate CA neurons that are devoid of Mef2a expression. Scale bar, 50 μm. M, Mef2a immunostaining in PBS control and ant-135a-injected mice. Dotted insets, Region used for quantification. Scale bar, 200 μm. N, Representative ROI from CA3 images used for quantifying Mef2a intensity. N1, mouse 1. N2, mouse 2. Scale bar, 25 μm. O, Mef2a fluorescence intensity in Ant-135a and control mice. n = 3 controls and n = 3 Ant-135a mice. Mean ratio between conditions is shown.
Results
Increased expression of miR-135a in human and experimental TLE
Our previous work identified miR-135a as one of the top 20 miRNAs showing increased expression in hippocampal tissue resected from mTLE patients (Kan et al., 2012). Further, we recently showed that miR-135a controls neuronal morphology and migration in vitro and in vivo (van Battum et al., 2018). To begin to characterize a potential role for miR-135a in the pathophysiology of TLE, miR-135a expression was assessed in human TLE hippocampus (mTLE-HS) and controls. qPCR and ISH showed that miR-135a expression levels were increased in mTLE hippocampus compared with controls (t(13) = 3.493, p = 0.0040, unpaired t test; Fig. 1A), as previously shown by microarray analysis (Kan et al., 2012). To verify the spatial distribution of miR-135a in human hippocampal tissue, we performed ISH. In line with the qPCR data, stronger signals for miR-135a were observed in mTLE hippocampus compared with control. Signals were mainly confined to neurons in the CA and DG regions (Fig. 1B). To confirm this cell type-specific localization, FISH was performed in combination with immunohistochemistry for NeuN (neurons) or GFAP (astrocytes). MiR-135a colocalized with NeuN, but not GFAP, indicating that miR-135a expression is predominantly neuronal (Fig. 1C).
Next, we checked whether seizure induction (SE) in an experimental model of TLE (by intra-amygdala microinjection of the glutamate receptor agonist KA) (Mouri et al., 2008) would also lead to increased levels of miR-135a. Indeed, we observed a strong increase in miR-135a expression at day 14 (D14) after SE by qPCR (t(5) = 2.811, p = 0.0375, unpaired t test; Fig. 2A) and ISH (Fig. 2B). ISH revealed a strong signal for miR-135a in the soma of pyramidal neurons in the hippocampus, and also in neurons in the cortex, thalamus, and amygdala at D14 (Fig. 2B). In control mouse, and human, hippocampus, most DG granule cells and CA pyramidal neurons displayed low-to-moderate miR-135a expression. In TLE patients and 2 weeks following KA injections, moderate-to-high expression was found in the majority of DG and CA neurons (Figs. 1B, 2B). Quantification of the number of cells in the DG region of the hippocampus did not reveal significant differences in KA versus PBS control mice (suprapyramidal blade: PBS, 84.44 ± 3.52; KA, 80.33 ± 1.893; t(16) = 1.029, p = 0.319, unpaired t test; infrapyramidal blade: PBS, 72.44 ± 2.352; KA, 67.67 ± 3.997; t(16) = 1.03, p = 0.318, unpaired t test; n = 3 mice and 9 sections/group). This shows that the increase in miR-135a expression is not due to an increase in the number of DG cells (e.g., as a result of increased adult neurogenesis). Similar to our observations in human mTLE hippocampus, miR-135a was predominantly neuronal (i.e., localized in neurons and not in astrocytes; Fig. 2C). The mature form of miR-135a, miR-135a-5p, is spliced from two different pretranscripts in both human and mice that arise from different chromosomal loci (Fig. 2D). To examine whether a specific locus was responsible for the increase in miR-135a expression in TLE, pre-miR levels were studied in human and mouse. Pre-miR-135A2 was significantly increased in human mTLE (t(11) = 2.359, p = 0.0379, unpaired t test; Fig. 2E), whereas in mice both pre-miR-135a1 and pre-miR-135a2 were significantly increased (pre-miR-135a1: t(12) = 4.613, p = 0.0006; pre-miR-135a2: t(12) = 1.828, p = 0.0462, unpaired t test; Fig. 2F). In all, we found increased expression of miR-135a in hippocampal neurons in TLE.
Silencing of miR-135a reduces spontaneous recurrent seizures
Our data show that, at least in a model of experimental TLE, miR-135a levels are high at the time recurrent spontaneous seizures are detected. To link increased miR-135a expression to spontaneous seizures, this miRNA was targeted by antagomirs (LNA 3′-cholesterol-conjugated oligonucleotides; Exiqon). Several studies have shown that antagomirs can effectively reduce the severity of SE when administered before SE induction (Jimenez-Mateos et al., 2012; Gross et al., 2016; Reschke et al., 2017), or the number of spontaneous seizures when administered immediately after SE (Jimenez-Mateos et al., 2012; Reschke et al., 2017). However, whether administering of antagomirs in the spontaneous recurrent seizure (SRS) phase can influence seizure occurrence remains poorly understood. Antagomirs were administered intracerebroventricularly at different concentrations to test for their specific effect on miR-135a. Twenty-four hours after injection, miR-135a levels were significantly reduced at 1.0 nmol of antagomir, whereas expression of another, unrelated miRNA, miR-124, was not affected. Injection of 1.5 nmol of antagomir had a small but nonsignificant effect on miR-124 expression (vehicle vs ant-135a 1.0 nmol, t(8) = 3.545, vehicle vs ant-135a 1.5 nmol, t(8) = 3.645, p = 0.0214, one-way ANOVA with Sidak post hoc test; Fig. 3A). On basis of these data, injections of 1.0 nmol were used in subsequent experiments. However, first ISH was used to detect ant-135a following antagomir or control injection. This analysis showed that ant-135a is taken up by hippocampal neurons in the CA and DG regions (Fig. 3B).
To assess the effect of blocking miR-135a on the occurrence of spontaneous seizures, SE-induced mice were injected with antagomirs for miR-135a or control at D14 and continuously monitored by EEG for 7 d after injection (Fig. 3C). One week prior injections (D7-D14 after SE) baseline EEG recordings were performed, and no significant difference in seizure frequency was observed between treated and control animals (t(9) = 0.34, p = 0.743, Wilcoxon Mann–Whitney test; Fig. 3D,E). We verified in an independent experiment that injecting PBS or a modified Scr version of the antagomir in PBS on D14 yielded similar seizure patterns over 1 week of recording (average number of seizures from D14-D21: PBS = 27 and Scr = 33, mixed-design, repeated-measures GLM; day × treatment interaction; F statistic 0.513; F(7,21) = 2.49 for α = 0.05; p = 0.819; n = 4 mice per group). Following injection of ant-135a at D14, a significant decrease in the number of SRSs per day was detected (Fig. 3D). A significantly different and strong reduction in seizure count was observed in ant-135a-treated compared with control mice (mixed-design, repeated-measures GLM; F statistic 13.858; F(13,39) = 1.98 for α = 0.05; p < 0.001; Fig. 3E). The average seizure duration was not different between the groups, before ant-135a injection (t(4) = 0.7931, p = 0.4721, paired t test), whereas it was significantly lower after injection (t(4) = 10.22, p = 0.0005, paired t test; Fig. 3F) and reduced spontaneous seizures were found when analyzing EEG traces (Fig. 3H). Similarly, the total amount of time spent in seizures was reduced following ant-135a injection (t(4) = 7.715, p = 0.0015, paired t test; Fig. 3G). Control injection at D14 caused an increase in time spent in seizures, which may be due to an additional insult caused by the injection of PBS. Conversely, time spent in seizures was strongly reduced following ant-135a injection mice at D14 (Fig. 3G). On average, ant-135a-injected mice spent less time (<300 s) in seizures per day, compared with control mice (>300 s) (Fig. 3I). Together, these data show that blocking elevated expression of miR-135a during the period of recurrent spontaneous seizures has an acute seizure suppressive effect.
Identification of miR-135a targets
Our previous work shows that miR-135a can affect axon growth and regeneration by controlling KLF4 expression (van Battum et al., 2018). However, the acute nature of the effects of ant-135a injection on seizure activity in vivo hints at interference with cellular processes that regulate neuronal activity, such as intracellular signaling, synaptic transmission, or synaptic morphology. miRNAs function by binding specific sequences known as MREs in the 3′-UTR of target transcripts. Upon binding, miRNAs repress translation or induce target RNA degradation. Prediction tools are available that predict targets based on a few empirical rules derived experimentally (Brennecke et al., 2005; Lewis et al., 2005), but many of these computational prediction tools perform poorly in experimental validation due to high false positive rates (Krek et al., 2005). To identify targets that are physically interacting with miR-135a, we performed miRNA immunoprecipitation in neuronal mouse Neuro2A cells using biotin-tagged mimics. miR-135a and scrambled mimics were tagged with a biotin molecule at their 3′ end (Fig. 4A,B), as 3′ molecule tagging was reported to not interfere with seed recognition, miRNA binding, and function (Ørom and Lund, 2007; Ørom et al., 2008). Although applying previously reported protocols for bio-miR IP (Wani and Cloonan, 2014), we validated the IP procedure by immunoblotting for Ago2 (the main component of RISC complex) following IP of miR-135 and scrambled mimics. Ago2 was detected in both input and IP samples, whereas the cytoskeletal protein β-actin was detected only in input samples (Fig. 4C). The presence of Ago2 confirms that the bio-miRNA mimic has been immunoprecipitated with the RISC complex, and presumably bound RNA targets. The sequence of the negative control is based on a C. elegans miRNA with minimal sequence identity with human, mouse, and rat. The presence of Ago2 in the negative control IP sample can most likely be explained by the fact that Argonaute proteins are very conserved among species (Höck and Meister, 2008).
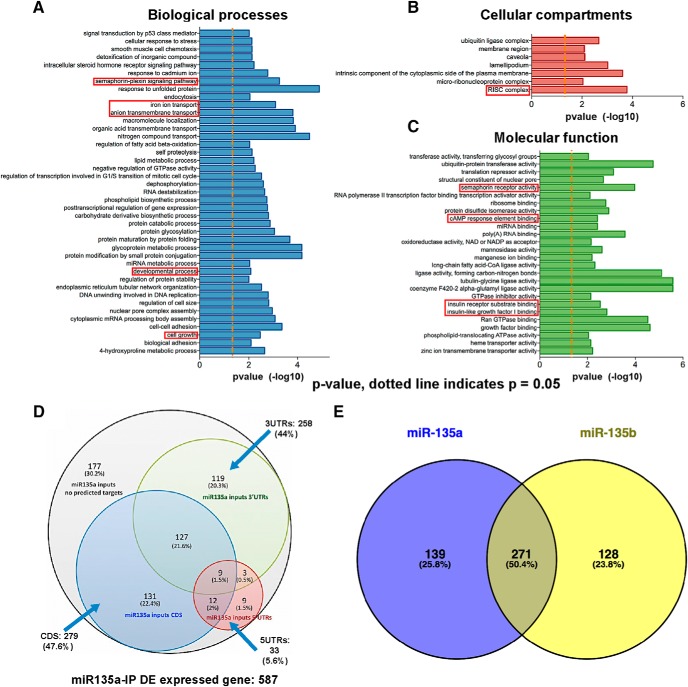
Following IP, total RNA sequencing was performed. For input samples, on average, 58.5 million and for IP samples 48.7 million high-quality reads were obtained. For input samples, 80%–90% of the reads could be aligned with the mouse reference genome, but for IP samples 39.7% of reads could be aligned with the reference genome and the rest with ribosomal RNA. The presence of ribosomal RNA can be explained by the lack of polyA+ enrichment or ribosomal RNA depletion in the sample preparation. Analysis of input samples revealed only few significantly changed transcripts, including validated miR-135a targets, such as Complexins (Cplx1 and Cplx2; data deposited at GEO; GSE123000) (Hu et al., 2014; Mannironi et al., 2018). In IP samples, levels of 587 transcripts were significantly altered (using a cutoff of FDR < 0.05 and p < 0.01) (Fig. 4D; data deposited at GEO GSE123000). These observations were supported by principal component analyses, which showed clear segregation of gene expression profiles for IP samples (NC vs miR-135a IP), but no clear segregation for inputs (Fig. 4E,F). Furthermore, IP samples contained many previously reported miR-135a targets (Table 3).
Table 3.
List of several experimentally validated targets of miR-135a that were enriched in the bio-IP samples
| Target | Name | Reference |
|---|---|---|
| Mtss1 | Metastasis suppressor protein 1 | Zhou et al., 2012 |
| Cdk4 | Cyclin-dependent kinase 4 | Dang et al., 2014 |
| Vcan | Proteoglycan versican | Zhao et al., 2017 |
| Zfp217 | Zinc finger protein 217 | Xiang et al., 2017 |
Gene ontology analysis REVIGO (Supek et al., 2011) demonstrated that differentially expressed transcripts found in IP samples are involved in neuron-related functions as semaphorin-plexin signaling, semaphorin receptor activity, ion transport, and cAMP response element binding (Fig. 5A–C). As a first step to identify targets of miR-135a relevant for the observed effect of ant-135a treatment in vivo, we selected transcripts from the IP samples with predicted miR-135a MREs, using miRanda software. MREs were found to be present not only in the 3′-UTR (258), but also in the 5′ UTR (33) and the coding sequence (CDS) (279) of the 587 transcripts, 177 putative targets had no predicted target site (Fig. 5D; data deposited at GEO GSE123000). miR-135a and miR-135b are highly similar and have an identical seed region, so in principle these miRNAs could target a similar set of mRNAs (van Battum et al., 2018). Comparison of targets in IP samples of miR-135a and miR-135b (data not shown) revealed 50% overlap, whereas 25.8% of targets were unique for miR-135a and 23.8% for miR-135b (Fig. 5E).
Figure 5.
Gene ontology analysis of miR-135a targets. A–C, Gene ontology terms for miRNA bio-IPs highlighting various processes that could potentially be regulated by miR-135a. D, Venn diagram showing the overlap of predicted binding sequence location (targeting site for miR-135a) in various segments of a transcript. E, Venn diagram showing the common (50.4%) and unique targets of miR-135a and miR135b. miR-135a and miR-135b contain the same mature sequence with only one mismatch outside the seed region.
Using the approach outlined above, we identified several new targets of miR-135a with reported roles in the regulation of neuronal development and function (Table 4). For further validation, seven targets were selected on basis of their function in neurons and/or implication in epilepsy (e.g., Tuberous sclerosis complex 1). All targets tested were enriched in IP compared with input samples (Fig. 6A). The effect of overexpression of miR-135a mimics in N2A cells on the expression of a few of the selected targets was tested and showed a significant downregulation of NR3C1 (GR), PlxnA4, and Mef2a protein expression (NR3C1, GR: NC vs miR-135a, t(6) = 2.889, p = 0.0277, unpaired t test; PlxnA4: NC vs miR-135a, t(6) = 2.488, p = 0.0473, unpaired t test; Mef2a: NC vs miR-135a, t(6) = 3.611, p = 0.0112, unpaired t test; Fig. 6B–D). This experiment confirms that targets identified by IP can be regulated by miR-135a.
Table 4.
Selection of targets identified by bio-IP with important roles in the regulation of epilepsy-relevant processes and pathwaysa
| Gene | Function | Log FC | p | FDR |
|---|---|---|---|---|
| Nr3c1 (GR) | Glucocorticoid receptor | 3.37865036 | 8.86E-16 | 2.17E-13 |
| Tsc1 | Tuberous sclerosis complex | 1.37190519 | 0.00050763 | 0.00589048 |
| Nrp1 | AG, MFS | 1.3675057 | 7.45E-06 | 0.00017204 |
| Tgfbr1 | TGFβ signaling | 1.35700473 | 1.27E-06 | 3.73E-05 |
| Mtss1 | Spine density | 1.3275843 | 0.00238517 | 0.02018217 |
| PlxnA4 | AG, MFS | 1.20703562 | 0.00012557 | 0.00190651 |
| Cacna1c | Calcium channel | 1.1901732 | 0.00076192 | 0.00819715 |
| Ncam1 | Neurite outgrowth | 1.05245567 | 5.18E-05 | 0.00092656 |
| Slit2 | AG, MFS | 1.04442804 | 0.00033258 | 0.00418342 |
| Mef2a | Spine density | 0.983455 | 0.00203267 | 0.01769962 |
| Creb1 | Transcription factor | 0.90909684 | 0.00363345 | 0.02809798 |
aThese targets were selected based on their function involved in key neuronal functions. AG, Axon guidance; MFS, mossy fiber sprouting.
The miR-135a target Mef2a is regulated in TLE
MEF2 proteins (MEF2A-D) form a family of transcription factors that are spatially and temporally expressed in the brain (Lyons et al., 1995), with most prominent expression for MEF2A, MEF2C, and MEF2D. MEF2s mediate activity-dependent synaptic development and are activated by neurotrophin stimulation and calcium influx resulting from increased neurotransmitter release at synapses (Flavell et al., 2008). Mutations in MEF2C are described in patients with severe mental retardation and epilepsy (Nowakowska et al., 2010; Bienvenu et al., 2013). In addition, MEF2A is deregulated in the temporal cortex of epilepsy patients and following experimental TLE (Huang et al., 2016). Based on our ant-135a experiments (Fig. 3), the reported functions of Mef2a and its deregulation in TLE, and its specific enrichment by miR-135a IP, we focused subsequent experiments on Mef2a. Mef2a 3′-UTR contains one specific conserved binding site for miR-135a (seed sequence from 1024–1030nt) (Fig. 7A). This site is targeted by miR-135a as shown by luciferase assay. Coexpression of miR-135a mimics with the miR-135a binding site in a luciferase reporter vector led to reduced luciferase activity. Mutation of the site abolished the effect of miR-135a (Mef2a+NC vs Mef2a+miR-135a: t(6) = 5.291, p = 0.0018, unpaired t test; Mef2a+miR-135a vs Mef2amut+miR-135a: t(6) = 3.951, p = 0.0075, unpaired t test; Fig. 7B).
Both MEF2 proteins and miR-135a have previously reported effects on synapse development and function. Mef2 proteins mediate activity-dependent synaptic development by regulating genes that control synapse number (Flavell et al., 2006), while miR-135 was shown to promote NMDA-induced spine retraction and long-lasting spine shrinkage (Hu et al., 2014). Furthermore, significant spine loss has been reported in pathological tissue specimens from human epilepsy patients (Multani et al., 1994; Aliashkevich et al., 2003) and in experimental models (Isokawa, 1998; Gibbs et al., 2011). Further, spine loss is directly correlated to the extent of SE induced in animal models, and spine number remains altered for weeks (Guo et al., 2012). To verify whether miR-135a also regulates spine number, miR-135a was overexpressed in mouse primary hippocampal neurons. Spine density was measured at a distance of 100 μm from the first secondary dendritic branch on the apical dendrite (Fig. 7C) and five different spine types (cup-shaped, mushroom, stubby, thin, and filopodium) were counted (Fig. 7D). Overexpression of miR-135a led to a significant reduction in the number of spines (0.34 ± 0.13 spines/μm) compared with the control (0.55 ± 0.06 spines/μm). Overexpression of miR-135a in vitro caused spine defects that resembled pathological neuron changes observed in vivo in TLE. Thus, increased miR-135a expression in epileptic brain could be directly or indirectly contributing to the neuronal spine loss observed. This effect was rescued to control levels when Mef2 vector lacking the 3′-UTR was coexpressed with miR-135a (0.49 ± 0.08 spines/μm) (control vs pre-miR-135a: t(51) = 5.738, p < 0.0001; pre-miR-135a vs pre-miR-135a+Mef2: t(51) = 4.89, p < 0.0001, one-way ANOVA with Sidak post hoc test; Fig. 7E). Interestingly, miR-135a overexpression led to a specific reduction in the number of mature spines (cup-shaped (3.32%), mushroom (18.05%), stubby (20.81%)), but led to increase in immature type of spines (thin (31.00%) and filopodium (26.82%)) compared with control (cup-shaped, 6.60%; mushroom, 34.51%; stubby, 26.53%; thin, 23.16%; filopodium, 9.2%). The reduction in mature spines and increase in immature spine type due to miR-135a overexpression were normalized to control levels when miR-135a was coexpressed with Mef2 (cup-shaped, 5.49%; mushroom, 32.77%; stubby, 25.00%; thin, 21.12%; filopodium, 15.63%) (Fig. 7F). Thus, increased expression of miR-135a leads to an MEF2-dependent change in spine number and type.
Our previous results revealed that miR-135a levels are increased in human and experimental TLE. To examine whether miR-135a and Mef2a could interact in TLE, we tested Mef2a expression in mouse and human TLE hippocampus. In line with our model, Mef2a protein expression was significantly reduced in the hippocampus of D14 IAK mice as detected by Western blotting (Mann–Whitney U = 1, p = 0.0317; Fig. 7G,H) and immunohistochemistry (Fig. 7I). Similarly, in patients with mTLE, MEF2A expression was strongly reduced in mTLE hippocampal samples compared with controls (Mann–Whitney U = 22.5, p = 0.0316, Mann–Whitney U test; Fig. 7J,K), and weaker immunostaining was observed in mTLE condition compared with controls in the dentate gyrus and CA region (Fig. 7L). Finally, blocking miR-135a in vivo using antagomirs resulted in increased Mef2a expression (Fig. 7M). Quantification of Mef2a fluorescence intensity in the CA3 region of the hippocampus revealed an increase in Mef2a expression, which, as a result of large differences in cell death between animals, was not statistically significant (Mann–Whitney U = 51, p = 0.5619, Mann–Whitney U test; Fig. 7N,O). Together, these results suggest that increased miR-135a expression in hippocampal neurons in mTLE may lead to decreased Mef2a levels. Loss of MEF2 in mTLE could lead to abnormal spine formation and thereby contribute to aberrant firing patterns and cell death observed in epilepsy.
Discussion
An ever-increasing number of studies implicates altered miRNA expression in epilepsy and identifies these noncoding RNAs as promising therapeutic targets. However, despite this progress, the mechanism of action and therapeutic potential of most epilepsy-associated miRNAs remain unknown. Here, we dissected the role of the brain-enriched miRNA miR-135a in TLE. Expression of miR-135a was increased in neurons in human and experimental TLE, particularly during the stage of spontaneous recurrent seizures. Remarkably, silencing miR-135a by intracerebroventricular treatment with antagomirs (ant-135a) during the stage of SRS had potent anticonvulsant effects. These data show that silencing a single miRNA at the SRS stage can rescue mice from spontaneous seizures. To begin to understand how miR-135a deregulation may cause epilepsy and seizures, immunoprecipitation in combination with RNAseq was used to identify miR-135a target RNAs. We report that the activity-dependent transcription factor Mef2a is a neuronal miR-135a target in vitro and in vivo, that Mef2 is required for miR-135a-induced dendritic spine changes, and that miR-135a and Mef2a show reciprocal expression regulation in experimental and human TLE.
Targeting spontaneous recurrent seizures with miRNA treatment
miRNAs are emerging as a novel class of therapeutic targets in epilepsy, due to their broad effects on neuronal structure, inflammation, ion channels, and gliosis (Cattani et al., 2016; Henshall et al., 2016). Our previous work showed increased expression of miR-135a in human TLE hippocampal samples, an observation subsequently confirmed by others (Kan et al., 2012; Alsharafi and Xiao, 2015). The molecular mechanisms that cause increased miR-135a expression in TLE remain unknown. miR-135a is generated from two different genomic loci in human and mice. Pre-miR-135A2 levels were increased in TLE patients, and both pre-miR-135a1 and pre-miR135a2 were increased in IAK mice, suggesting species-specific gene regulation (Fig. 2). Wnt signaling is an attractive candidate for partly explaining this miR-135a regulation, as aberrant Wnt signaling is linked to experimental TLE (Qu et al., 2017) and Wnt signaling is known to control pre-miR-135a expression. For example, during mouse development, canonical Wnt signaling induces the expression of pre-miR-135a2 in dorsal forebrain (Caronia-Brown et al., 2016) and pre-miR-135a2 and its host gene, the long noncoding RNA RMST, are induced by Wnt/β-catenin signaling (Anderegg et al., 2013; Caronia-Brown et al., 2016). Several Wnt pathway genes were enriched in the miR-135a bio-IP (e.g., Lrp6, Lgr5, Jade1), suggesting an autoregulatory loop between miR-135a and the Wnt pathway in TLE, as reported previously (Anderegg et al., 2013).
miRNA antagomirs and mimics have been applied successfully to modulate seizures in experimental TLE (Gross et al., 2016; Henshall et al., 2016). For example, targeting miR-134 before or immediately after SE reduces SRSs in multiple animal models of experimental TLE and alters underlying pathological hallmarks (Jimenez-Mateos et al., 2012; Reschke et al., 2017). Further, repeated administration of miR-146a mimics after SE reduces seizures (Iori et al., 2017), whereas silencing miR-324 delays seizure onset and protects from cell death (Gross et al., 2016). Similarly, targeting miR-203 by antagomirs reduced SRS frequency (Lee et al., 2017). Our study shows that targeting miRNAs at a stage at which SRSs have been established, long after SE, also has seizure reducing effects. Antagomirs against miR-135a caused strong reduction of the total number of seizures and average seizure time. Further, treated mice spent less time seizing (Fig. 3). In this study, we did not focus on potential long-term disease-modifying effects of ant-135a treatment. No neuroprotective effect was found in the first 7 d after ant-135a application (data not shown), but this is in line with the observation that in IAK mice cell death is generally completed in the first week after SE (Mouri et al., 2008). Thus, while further work is needed to establish whether ant-135a treatment has disease-modifying effects, our observation that this treatment has anticonvulsant effects has interesting therapeutic implications as it hints at the possibility of using miRNAs as therapeutic targets after SRSs have been established.
miR-135a regulates dendritic spines through the epilepsy-associated transcription factor Mef2
In the brain, several functions have been reported for miR-135a. It is required for stress resiliency, intact serotonergic activity, and has a potential role as an endogenous antidepressant (Issler et al., 2014). Further, miR-135 is required for sustained spine remodeling and induction of synaptic depression by regulating the SNARE complex proteins Complexin1 and 2 (Hu et al., 2014). By targeting Complexin1 and 2, miR-135a regulates synaptic transmission and anxiety-like behavior in the amygdala (Mannironi et al., 2018). Finally, miR-135a induces axon growth and neuronal migration, and axon regeneration after optic nerve injury (van Battum et al., 2018).
To begin to understand miR-135a's role in epilepsy and why reducing miR-135a decreases seizure activity, we performed an unbiased biotin-IP screen that revealed several predicted and novel targets of miR-135a (Fig. 4). Several of these targets have relevant biological functions and had been implicated in epilepsy (e.g., GR, Mef2a, and plexinA4). The GRs NR3C1 and Mef2a both regulate neuronal plasticity (Speksnijder et al., 2012) and are deregulated in epilepsy (Huang et al., 2016; Martínez-Levy et al., 2018). Similarly, changes in axon guidance receptors, such as PlexinA4, can contribute to mossy fiber sprouting in the dentate gyrus, a critical feature of the epileptic hippocampus leading to hyperexcitability and cell death (Van Battum et al., 2015). The effect of ant-135a treatment on seizure activity was not only strong but also fast (i.e., already after 1 d significant differences between treated and control mice were found; Fig. 3). Explanations for such a swift response are that ant-135a treatment impacts on processes, such as synaptic function, perhaps through local regulation of gene expression. For example, activity-dependent local translation regulation of target mRNAs by miRNAs occurs in single synapses (Sambandan et al., 2017). Therefore, loss of miR-135a may lead to derepression of its target mRNAs in distal dendrites, leading to changes in excitatory neurotransmission observed in epileptic networks (McNamara et al., 2006). Interestingly, one of the targets of miR-135a that we identified was Mef2a, an activity-dependent transcription factor that regulates excitatory and inhibitory synaptic strength both locally at synapses and in the nucleus (Flavell et al., 2008). Overexpression of miR-135a in cultured mouse hippocampal neurons reduced spine number and increased the relative number of immature spines, in line with the profound loss of dendritic spines found in epilepsy (Swann et al., 2000). Both defects were rescued by reexpression of miR-135a-insensitive Mef2, implicating an miR-135a-Mef2 pathway in the control of spine maturation and number (Fig. 7). This, together with the observation that miR-135a and Mef2a show reciprocal expression regulation in TLE, invites the speculation that miR-135a may regulate Mef2a to induce synaptic defects in TLE.
Mef2 proteins function as synapse eliminating factors in development and disease (Pfeiffer et al., 2010; Tsai et al., 2012; Wang, 2016) but can also regulate genes known to mediate synapse weakening (e.g., Homer1a, Arc, kcna1) or strengthening (e.g., leucine-rich glioma-inactivated 1 [Lgi1], BDNF, adenyl cyclase 8) (Flavell et al., 2008). Several of these genes have been implicated in epilepsy. For example, loss of Lgi1 in glutamatergic neurons induces epileptic seizures due to increased synaptic glutamate levels, leading to hyperexcitable neuronal networks (Boillot et al., 2014, 2016). In addition, BDNF has a dual role, as a proepileptic and antiepileptogenic factor (Simonato et al., 2006). BDNF expression is increased immediately after SE but reduced during the chronic stage. It selectively localizes to dendritic compartments after chemoconvulsant seizures (Tongiorgi et al., 2004). A recent study shows that continuous release of BDNF into the epileptic hippocampus reduces the frequency of generalized seizures and rescues from histological alterations observed in chronic epilepsy (Falcicchia et al., 2018). Further work is needed to explore whether the morphological and seizure-suppressive effects of ant-135a derive from effects on these or other Mef2 targets, and whether these effects originate from altered functioning of glutamatergic synapses (e.g., Lgi1) or of both glutamatergic and GABAergic synapses (e.g., BDNF) (Simonato et al., 2006; Gu et al., 2017). Mef2c KO mice show a reduction in excitatory synapse number but an increase in inhibitory synapse number. It has therefore been proposed that Mef2c simultaneously regulates spine density on both inhibitory and excitatory neurons to maintain balanced activity in neuronal networks (Harrington et al., 2016). Disruption of this regulation may lead to abnormal synaptic activity leading to seizure activity in TLE.
In conclusion, a deeper understanding of the roles, targets, and mechanisms of action of miRNAs in the pathogenesis of epilepsy may lead to the development of novel diagnostic biomarkers and the identification of therapeutic targets for treatment. Here, we identify miR-135a as a target for reducing SRSs once these seizures have already been established. This is important as the majority of studies so far have focused on miRNA manipulation during or at least starting in the acute stages of the disease. It will be interesting to explore whether manipulation of other miRNAs at the SRS stage also has seizure-suppressive effects or even disease-modifying properties. Further insight into how miR-135a expression is regulated in epilepsy and which miR-135a targets, in addition to Mef2a, are affected by ant-135a administration in experimental epilepsy will provide more insight into the mechanism of action of miR-135a in TLE.
Footnotes
This work was supported by European Union's 7th Framework Programme Grant Agreement 602130, EpimiRNA, and Dutch Epilepsiefonds (WAR 12-08, 15-05), and Netherlands Organization for Scientific Research to R.J.P. We thank Prof. Jorgen Kjems for providing the pJEBB vector backbone; and Prof. Gerhard Schratt for the Mef2-vp16 vector.
R.Q.J.S. is a shareholder in InteRNA Technologies B.V. R.Q.J.S. is a stock option holder in InteRNA Technologies B.V. The remaining authors declare no competing financial interests.
References
- Aliashkevich AF, Yilmazer-Hanke D, Van Roost D, Mundhenk B, Schramm J, Blümcke I (2003) Cellular pathology of amygdala neurons in human temporal lobe epilepsy. Acta Neuropathol 106:99–106. 10.1007/s00401-003-0707-0 [DOI] [PubMed] [Google Scholar]
- Alsharafi W, Xiao B (2015) Dynamic expression of microRNAs (183, 135a, 125b, 128, 30c and 27a) in the rat pilocarpine model and temporal lobe epilepsy patients. CNS Neurol Disord Drug Targets 14:1096–1102. 10.2174/1871527314666150317225945 [DOI] [PubMed] [Google Scholar]
- Anderegg A, Lin HP, Chen JA, Caronia-Brown G, Cherepanova N, Yun B, Joksimovic M, Rock J, Harfe BD, Johnson R, Awatramani R (2013) An Lmx1b-miR135a2 regulatory circuit modulates Wnt1/Wnt signaling and determines the size of the midbrain dopaminergic progenitor pool. PLoS Genet 9:e1003973. 10.1371/journal.pgen.1003973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq-A python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronica E, Fluiter K, Iyer A, Zurolo E, Vreijling J, van Vliet EA, Baayen JC, Gorter JA (2010) Expression pattern of miR-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Eur J Neurosci 31:1100–1107. 10.1111/j.1460-9568.2010.07122.x [DOI] [PubMed] [Google Scholar]
- Bartel DP. (2018) Metazoan microRNAs. Cell 173:20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker AJ, Chen J, Zien A, Sochivko D, Normann S, Schramm J, Elger CE, Wiestler OD, Blümcke I (2003) Correlated stage- and subfield-associated hippocampal gene expression patterns in experimental and human temporal lobe epilepsy. Eur J Neurosci 18:2792–2802. 10.1111/j.1460-9568.2003.02993.x [DOI] [PubMed] [Google Scholar]
- Bienvenu T, Diebold B, Chelly J, Isidor B (2013) Refining the phenotype associated with MEF2C point mutations. Neurogenetics 14:71–75. 10.1007/s10048-012-0344-7 [DOI] [PubMed] [Google Scholar]
- Blümcke I, Thom M, Aronica E, Armstrong DD, Bartolomei F, Bernasconi A, Bernasconi N, Bien CG, Cendes F, Coras R, Cross JH, Jacques TS, Kahane P, Mathern GW, Miyata H, Moshé SL, Oz B, Özkara C, Perucca E, Sisodiya S, et al. (2013) International consensus classification of hippocampal sclerosis in temporal lobe epilepsy: a Task Force report from the ILAE Commission on Diagnostic Methods. Epilepsia 54:1315–1329. 10.1111/epi.12220 [DOI] [PubMed] [Google Scholar]
- Boillot M, Huneau C, Marsan E, Lehongre K, Navarro V, Ishida S, Dufresnois B, Ozkaynak E, Garrigue J, Miles R, Martin B, Leguern E, Anderson MP, Baulac S (2014) Glutamatergic neuron-targeted loss of LGI1 epilepsy gene results in seizures. Brain 137:2984–2996. 10.1093/brain/awu259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boillot M, Lee CY, Allene C, Leguern E, Baulac S, Rouach N (2016) LGI1 acts presynaptically to regulate excitatory synaptic transmission during early postnatal development. Sci Rep 6:1–9. 10.1038/srep21769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM (2005) Principles of microRNA-target recognition. PLoS Biol 3:e85. 10.1371/journal.pbio.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caronia-Brown G, Anderegg A, Awatramani R (2016) Expression and functional analysis of the Wnt/beta-catenin induced mir-135a-2 locus in embryonic forebrain development. Neural Dev 11:1–15. 10.1186/s13064-016-0065-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattani AA, Allene C, Seifert V, Rosenow F, Henshall DC, Freiman TM (2016) Involvement of microRNAs in epileptogenesis. Epilepsia 57:1015–1026. 10.1111/epi.13404 [DOI] [PubMed] [Google Scholar]
- Chang BS, Lowenstein DH (2003) Epilepsy. N Engl J Med 349:1257–1266. 10.1056/NEJMra022308 [DOI] [PubMed] [Google Scholar]
- Dang Z, Xu WH, Lu P, Wu N, Liu J, Ruan B, Zhou L, Song WJ, Dou KF (2014) MicroRNA-135a inhibits cell proliferation by targeting Bmi1 in pancreatic ductal adenocarcinoma. Int J Biol Sci 10:733–745. 10.7150/ijbs.8097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA (2012) Roles for MicroRNAs in conferring robustness to biological processes. Cell 149:505–524. 10.1016/j.cell.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J., Jr (2001) A proposed diagnostic scheme for people with epileptic seizures and with epilepsy: report of the ILAE task force on classification and terminology. Epilepsia 42:796–803. 10.1046/j.1528-1157.2001.10401.x [DOI] [PubMed] [Google Scholar]
- Falcicchia C, Paolone G, Emerich DF, Lovisari F, Bell WJ, Fradet T, Wahlberg LU, Simonato M (2018) Seizure-suppressant and neuroprotective effects of encapsulated BDNF-producing cells in a rat model of temporal lobe epilepsy. Mol Ther Methods Clin Dev 9:211–224. 10.1016/j.omtm.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, Greenberg ME, Schratt G (2009) Mef2-mediated transcription of the miR379–410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J 28:697–710. 10.1038/emboj.2009.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME (2006) Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311:1008–1012. 10.1126/science.1122511 [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME (2008) Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60:1022–1038. 10.1016/j.neuron.2008.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs S, Chattopadhyaya B, Desgent S, Awad PN, Clerk-Lamalice O, Levesque M, Vianna RM, Rébillard RM, Delsemme AA, Hébert D, Tremblay L, Lepage M, Descarries L, Di Cristo G, Carmant L (2011) Long-term consequences of a prolonged febrile seizure in a dual pathology model. Neurobiol Dis 43:312–321. 10.1016/j.nbd.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Gorter JA, Iyer A, White I, Colzi A, van Vliet EA, Sisodiya S, Aronica E (2014) Hippocampal subregion-specific microRNA expression during epileptogenesis in experimental temporal lobe epilepsy. Neurobiol Dis 62:508–520. 10.1016/j.nbd.2013.10.026 [DOI] [PubMed] [Google Scholar]
- Gross C, Yao X, Engel T, Tiwari D, Xing L, Rowley S, Danielson SW, Thomas KT, Jimenez-Mateos EM, Schroeder LM, Pun RY, Danzer SC, Henshall DC, Bassell GJ (2016) MicroRNA-mediated downregulation of the potassium channel Kv4.2 contributes to seizure onset. Cell Rep 17:37–45. 10.1016/j.celrep.2016.08.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F, Parada I, Shen F, Li J, Bacci A, Graber K, Taghavi RM, Scalise K, Schwartzkroin P, Wenzel J, Prince DA (2017) Structural alterations in fast-spiking GABAergic interneurons in a model of posttraumatic neocortical epileptogenesis. Neurobiol Dis 108:100–114. 10.1016/j.nbd.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Arnspiger S, Rensing NR, Wong M (2012) Brief seizures cause dendritic injury. Neurobiol Dis 45:348–355. 10.1016/j.nbd.2011.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington AJ, Raissi A, Rajkovich K, Berto S, Kumar J, Molinaro G, Raduazzo J, Guo Y, Loerwald K, Konopka G, Huber KM, Cowan CW (2016) MEF2C regulates cortical inhibitory and excitatory synapses and behaviors relevant to neurodevelopmental disorders. Elife 5:1–27. 10.7554/eLife.20059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henshall DC, Hamer HM, Pasterkamp RJ, Goldstein DB, Kjems J, Prehn JH, Schorge S, Lamottke K, Rosenow F (2016) MicroRNAs in epilepsy: pathophysiology and clinical utility. Lancet Neurol 15:1368–1376. 10.1016/S1474-4422(16)30246-0 [DOI] [PubMed] [Google Scholar]
- Höck J, Meister G (2008) The Argonaute protein family. Genome Biol 9:210. 10.1186/gb-2008-9-2-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Wu X, Guo J, Yuan J (2016) Myocyte-specific enhancer binding factor 2A expression is downregulated during temporal lobe epilepsy. Int J Neurosci 126:786–796. 10.3109/00207454.2015.1062003 [DOI] [PubMed] [Google Scholar]
- Hu Z, Yu D, Gu QH, Yang Y, Tu K, Zhu J, Li Z (2014) miR-191 and miR-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nat Commun 5:3263. 10.1038/ncomms4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iori V, Iyer AM, Ravizza T, Beltrame L, Paracchini L, Marchini S, Cerovic M, Hill C, Ferrari M, Zucchetti M, Molteni M, Rossetti C, Brambilla R, Steve White H, D'Incalci M, Aronica E, Vezzani A (2017) Blockade of the IL-1R1/TLR4 pathway mediates disease-modification therapeutic effects in a model of acquired epilepsy. Neurobiol Dis 99:12–23. 10.1016/j.nbd.2016.12.007 [DOI] [PubMed] [Google Scholar]
- Isokawa M. (1998) Remodeling dendritic spines in the rat pilocarpine model of temporal lobe epilepsy. Neurosci Lett 258:73–76. 10.1016/S0304-3940(98)00848-9 [DOI] [PubMed] [Google Scholar]
- Issler O, Haramati S, Paul ED, Maeno H, Navon I, Zwang R, Gil S, Mayberg HS, Dunlop BW, Menke A, Awatramani R, Binder EB, Deneris ES, Lowry CA, Chen A (2014) MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron 83:344–360. 10.1016/j.neuron.2014.05.042 [DOI] [PubMed] [Google Scholar]
- Jimenez-Mateos EM, Henshall DC (2013) Epilepsy and microRNA. Neuroscience 238:218–229. 10.1016/j.neuroscience.2013.02.027 [DOI] [PubMed] [Google Scholar]
- Jimenez-Mateos EM, Engel T, Merino-Serrais P, McKiernan RC, Tanaka K, Mouri G, Sano T, O'Tuathaigh C, Waddington JL, Prenter S, Delanty N, Farrell MA, O'Brien DF, Conroy RM, Stallings RL, DeFelipe J, Henshall DC (2012) Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat Med 18:1087–1094. 10.1038/nm.2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan AA, van Erp S, Derijck AA, de Wit M, Hessel EV, O'Duibhir E, de Jager W, Van Rijen PC, Gosselaar PH, de Graan PN, Pasterkamp RJ (2012) Genome-wide microRNA profiling of human temporal lobe epilepsy identifies modulators of the immune response. Cell Mol Life Sci 69:3127–3145. 10.1007/s00018-012-0992-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS. (2006) The neuronal microRNA system. Nat Rev Neurosci 7:911–920. 10.1038/nrn2037 [DOI] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N (2005) Combinatorial microRNA target predictions. Nat Genet 37:495–500. 10.1038/ng1536 [DOI] [PubMed] [Google Scholar]
- Lee ST, Jeon D, Chu K, Jung KH, Moon J, Sunwoo J, Park DK, Yang H, Park JH, Kim M, Roh JK, Lee SK (2017) Inhibition of miR-203 reduces spontaneous recurrent seizures in mice. Mol Neurobiol 54:3300–3308. 10.1007/s12035-016-9901-7 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20. 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Löscher W, Klitgaard H, Twyman RE, Schmidt D (2013) New avenues for anti-epileptic drug discovery and development. Nat Rev Drug Discov 12:757–776. 10.1038/nrd4126 [DOI] [PubMed] [Google Scholar]
- Lyons GE, Micales BK, Schwarz J, Martin JF, Olson EN (1995) Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J Neurosci 15:5727–5738. 10.1523/JNEUROSCI.15-08-05727.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannironi C, Biundo A, Rajendran S, De Vito F, Saba L, Caioli S, Zona C, Ciotti T, Caristi S, Perlas E, Del Vecchio G, Bozzoni I, Rinaldi A, Mele A, Presutti C (2018) miR-135a regulates synaptic transmission and anxiety-like behavior in amygdala. Mol Neurobiol 55:3301–3315. 10.1007/s12035-017-0564-9 [DOI] [PubMed] [Google Scholar]
- Martínez-Levy GA, Rocha L, Rodríguez-Pineda F, Alonso-Vanegas MA, Nani A, Buentello-García RM, Briones-Velasco M, San-Juan D, Cienfuegos J, Cruz-Fuentes CS (2018) Increased expression of brain-derived neurotrophic factor transcripts I and VI, cAMP response element binding, and glucocorticoid receptor in the cortex of patients with temporal lobe epilepsy. Mol Neurobiol 55:3698–3708. 10.1007/s12035-017-0597-0 [DOI] [PubMed] [Google Scholar]
- McNamara JO, Huang YZ, Leonard AS (2006) Molecular signaling mechanisms underlying epileptogenesis. Sci STKE 2006:re12. 10.1126/stke.3562006re12 [DOI] [PubMed] [Google Scholar]
- Moshé SL, Perucca E, Ryvlin P, Tomson T (2015) Epilepsy: new advances. Lancet 385:884–898. 10.1016/S0140-6736(14)60456-6 [DOI] [PubMed] [Google Scholar]
- Mouri G, Jimenez-Mateos E, Engel T, Dunleavy M, Hatazaki S, Paucard A, Matsushima S, Taki W, Henshall DC (2008) Unilateral hippocampal CA3-predominant damage and short latency epileptogenesis after intra-amygdala microinjection of kainic acid in mice. Brain Res 1213:140–151. 10.1016/j.brainres.2008.03.061 [DOI] [PubMed] [Google Scholar]
- Multani P, Myers RH, Blume HW, Schomer DL, Sotrel A (1994) Neocortical dendritic pathology in human partial epilepsy: a quantitative Golgi study. Epilepsia 35:728–736. 10.1111/j.1528-1157.1994.tb02503.x [DOI] [PubMed] [Google Scholar]
- Nowakowska BA, Obersztyn E, Szymanska K, Bekiesinska-Figatowska M, Xia Z, Ricks CB, Bocian E, Stockton DW, Szczaluba K, Nawara M, Patel A, Scott DA, Cheung SW, Bohan TP, Stankiewicz P (2010) Severe mental retardation, seizures, and hypotonia due to deletions of MEF2C. Am J Med Genet Part B Neuropsychiatr Genet 153:1042–1051. 10.1002/ajmg.b.31071 [DOI] [PubMed] [Google Scholar]
- Ørom UA, Lund AH (2007) Isolation of microRNA targets using biotinylated synthetic microRNAs. Methods 43:162–165. 10.1016/j.ymeth.2007.04.007 [DOI] [PubMed] [Google Scholar]
- Ørom UA, Nielsen FC, Lund AH (2008) MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 30:460–471. 10.1016/j.molcel.2008.05.001 [DOI] [PubMed] [Google Scholar]
- Pfeiffer BE, Zang T, Wilkerson JR, Taniguchi M, Maksimova MA, Smith LN, Cowan CW, Huber KM (2010) Fragile X mental retardation protein is required for synapse elimination by the activity-dependent transcription factor MEF2. Neuron 66:191–197. 10.1016/j.neuron.2010.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z, Su F, Qi X, Sun J, Wang H, Qiao Z, Zhao H, Zhu Y (2017) Wnt/β-catenin signalling pathway mediated aberrant hippocampal neurogenesis in kainic acid-induced epilepsy. Cell Biochem 35:472–476. 10.1002/cbf.3306 [DOI] [PubMed] [Google Scholar]
- Rakhade SN, Jensen FE (2009) Epileptogenesis in the immature brain: Emerging mechanisms. Nat Rev Neurol 5:380–391. 10.1038/nrneurol.2009.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschke CR, Silva LF, Norwood BA, Senthilkumar K, Morris G, Sanz-Rodriguez A, Conroy RM, Costard L, Neubert V, Bauer S, Farrell MA, O'Brien DF, Delanty N, Schorge S, Pasterkamp RJ, Rosenow F, Henshall DC (2017) Potent anti-seizure effects of locked nucleic acid antagomirs targeting miR-134 in multiple mouse and rat models of epilepsy. Mol Ther Nucleic Acids 6:45–56. 10.1016/j.omtn.2016.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11:R25. 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandan S, Akbalik G, Kochen L, Rinne J, Kahlstatt J, Glock C, Tushev G, Alvarez-Castelao B, Heckel A, Schuman EM (2017) Activity-dependent spatially localized miRNA maturation in neuronal dendrites. Science 355:634–637. 10.1126/science.aaf8995 [DOI] [PubMed] [Google Scholar]
- Semah F, Picot MC, Adam C, Broglin D, Arzimanoglou A, Bazin B, Cavalcanti D, Baulac M (1998) Is the underlying cause of epilepsy a major prognostic factor for recurrence? Neurology 51:1256–1262. 10.1212/WNL.51.5.1256 [DOI] [PubMed] [Google Scholar]
- Simonato M, Tongiorgi E, Kokaia M (2006) Angels and demons: neurotrophic factors and epilepsy. Trends Pharmacol Sci 27:631–638. 10.1016/j.tips.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Speksnijder N, Christensen KV, Didriksen M, De Kloet ER, Datson NA (2012) Glucocorticoid receptor and myocyte enhancer factor 2 cooperate to regulate the expression of c-JUN in a neuronal context. J Mol Neurosci 48:209–218. 10.1007/s12031-012-9809-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley K. (2004) Epileptic neurons go wireless. Science 1538:1–30. 10.1126/science.1101133 [DOI] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, Smuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800. 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann JW, Al-Noori S, Jiang M, Lee CL (2000) Spine loss and other dendritic abnormalities in epilepsy. Hippocampus 10:617–625. [DOI] [PubMed] [Google Scholar]
- Tongiorgi E, Armellin M, Giulianini PG, Bregola G, Zucchini S, Paradiso B, Steward O, Cattaneo A, Simonato M (2004) Brain-derived neurotrophic factor mRNA and protein are targeted to discrete dendritic laminas by events that trigger epileptogenesis. J Neurosci 24:6842–6852. 10.1523/JNEUROSCI.5471-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai NP, Wilkerson JR, Guo W, Maksimova MA, DeMartino GN, Cowan CW, Huber KM (2012) Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell 151:1581–1594. 10.1016/j.cell.2012.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Battum EY, Gunput RA, Lemstra S, Groen EJ, Yu KL, Adolfs Y, Zhou Y, Hoogenraad CC, Yoshida Y, Schachner M, Akhmanova A, Pasterkamp RJ (2014) The intracellular redox protein MICAL-1 regulates the development of hippocampal mossy fibre connections. Nat Commun 5:4317. 10.1038/ncomms5317 [DOI] [PubMed] [Google Scholar]
- Van Battum EY, Brignani S, Pasterkamp RJ (2015) Axon guidance proteins in neurological disorders. Lancet Neurol 14:532–546. 10.1016/S1474-4422(14)70257-1 [DOI] [PubMed] [Google Scholar]
- van Battum EY, Verhagen MG, Vangoor VR, Fujita Y, Derijck AA, O'Duibhir E, Giuliani G, de Gunst T, Adolfs Y, Lelieveld D, Egan D, Schaapveld RQ, Yamashita T, Pasterkamp RJ (2018) An image-based miRNA screen identifies miRNA-135s as regulators of CNS axon growth and regeneration by targeting Krüppel-like factor 4. J Neurosci 38:613–630. 10.1523/JNEUROSCI.0662-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. (2016) Myocyte enhancer factor-2 (MEF2) in diseases of central nervous system: a mini review. Explor Res Hypothesis Med 1:4–8. [Google Scholar]
- Wani S, Cloonan N (2014) Profiling direct mRNA-microRNA interactions using synthetic biotinylated microRNA-duplexes. bioRxiv. Advance online publication. Retrieved May 22, 2014. doi: 10.1101/005439. [DOI] [Google Scholar]
- Wetherington J, Serrano G, Dingledine R (2008) Astrocytes in the epileptic brain. Neuron 58:168–178. 10.1016/j.neuron.2008.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser HG. (2004) ILAE Commission Report: mesial temporal lobe epilepsy with hippocampal sclerosis. Epilepsia 45:695–714. 10.1111/j.0013-9580.2004.09004.x [DOI] [PubMed] [Google Scholar]
- Xiang H, Zhong ZX, Peng YD, Jiang SW (2017) The emerging role of Zfp217 in adipogenesis. Int J Mol Sci 18:1–17. 10.3390/ijms18071367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14. 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Sun Z, Li H, Jiang F, Zhou J, Zhang L (2017) MiR-135a–5p modulates biological functions of thyroid carcinoma cells via targeting VCAN 3′-UTR. Cancer Biomark 20:207–216. 10.3233/CBM-170566 [DOI] [PubMed] [Google Scholar]
- Zhou W, Li X, Liu F, Xiao Z, He M, Shen S, Liu S (2012) MiR-135a promotes growth and invasion of colorectal cancer via metastasis suppressor 1 in vitro. Acta Biochim Biophys Sin (Shanghai) 44:838–846. 10.1093/abbs/gms071 [DOI] [PubMed] [Google Scholar]