Abstract
Activating Transcription Factor 4 (ATF4) has been postulated as a key regulator of learning and memory. We previously reported that specific hippocampal ATF4 downregulation causes deficits in synaptic plasticity and memory and reduction of glutamatergic functionality. Here we extend our studies to address ATF4's role in neuronal excitability. We find that long-term ATF4 knockdown in cultured rat hippocampal neurons significantly increases the frequency of spontaneous action potentials. This effect is associated with decreased functionality of metabotropic GABAB receptors (GABABRs). Knocking down ATF4 results in significant reduction of GABABR-induced GIRK currents and increased mIPSC frequency. Furthermore, reducing ATF4 significantly decreases expression of membrane-exposed, but not total, GABABR 1a and 1b subunits, indicating that ATF4 regulates GABABR trafficking. In contrast, ATF4 knockdown has no effect on surface expression of GABABR2s, several GABABR-coupled ion channels or β2 and γ2 GABAARs. Pharmacologic manipulations confirmed the relationship between GABABR functionality and action potential frequency in our cultures. Specifically, the effects of ATF4 downregulation cited above are fully rescued by transcriptionally active, but not by transcriptionally inactive, shRNA-resistant, ATF4. We previously reported that ATF4 promotes stabilization of the actin-regulatory protein Cdc42 by a transcription-dependent mechanism. To test the hypothesis that this action underlies the mechanism by which ATF4 loss affects neuronal firing rates and GABABR trafficking, we downregulated Cdc42 and found that this phenocopies the effects of ATF4 knockdown on these properties. In conclusion, our data favor a model in which ATF4, by regulating Cdc42 expression, affects trafficking of GABABRs, which in turn modulates the excitability properties of neurons.
SIGNIFICANCE STATEMENT GABAB receptors (GABABRs), the metabotropic receptors for the inhibitory neurotransmitter GABA, have crucial roles in controlling the firing rate of neurons. Deficits in trafficking/functionality of GABABRs have been linked to a variety of neurological and psychiatric conditions, including epilepsy, anxiety, depression, schizophrenia, addiction, and pain. Here we show that GABABRs trafficking is influenced by Activating Transcription Factor 4 (ATF4), a protein that has a pivotal role in hippocampal memory processes. We found that ATF4 downregulation in hippocampal neurons reduces membrane-bound GABABR levels and thereby increases intrinsic excitability. These effects are mediated by loss of the small GTPase Cdc42 following ATF4 downregulation. These findings reveal a critical role for ATF4 in regulating the modulation of neuronal excitability by GABABRs.
Keywords: ATF4, Cdc42, GABAB receptors, GIRK channels, neuronal excitability
Introduction
Normal cognitive functions rely on the balance of neuronal excitability properties throughout the brain as well as on synaptic plasticity (Beck and Yaari, 2008). Among the many proteins reported to influence cognition, mounting evidence suggests a pivotal role for Activating Transcription Factor 4 (ATF4), a ubiquitously expressed member of the ATF/CREB transcription factor family of basic leucine zipper proteins. In addition to its well-known functions as a stress-induced protein (Ameri and Harris, 2008), a number of studies have implicated ATF4 in synaptic plasticity and in learning and memory. Depending on cellular context, ATF4 has been characterized as either an inhibitor or promoter of synaptic plasticity (Pasini et al., 2015). Similarly divergent suggestions about ATF4's functions in learning and memory have been advanced, but these are largely based on indirect and nonselective manipulation of ATF4 activity or expression (Chen et al., 2003; Costa-Mattioli et al., 2007). To probe directly ATF4's role in normal brain function, we have monitored the consequences of its knockdown or knock-out in neuronal culture and in animals. This has led to observations that ATF4 plays a role in regulation of mushroom dendritic spine density as well as in synaptic glutamatergic function (Liu et al., 2014). These effects appeared to be due to ATF4's direct transcriptional regulation of RhoGDIα (product of the Arhgdia gene), which in turn affects stability of the Rho family member Cdc42 that is involved in regulation of the actin cytoskeleton (Pasini et al., 2016). At the physiological level, loss of ATF4 manifested in reduced frequency and amplitude of mEPSCs, followed by defective LTP and LTD as well as in memory deficits (Pasini et al., 2015). Of relevance, similar deficiencies in plasticity and memory have been observed after conditional Cdc42 knock-out in brain (Kim et al., 2014). In the context of ATF4's role in neuronal functionality, one area of interest is in its relation to GABAB receptors (GABABRs), the G-protein-linked metabotropic receptors for the inhibitory neurotransmitter GABA. Several studies have shown direct association of GABABRs with ATF4 (Nehring et al., 2000; White et al., 2000; Vernon et al., 2001; Ritter et al., 2004), whereas another study reported that ATF4 differentially regulates activity of promoters for the GABABRs subunits 1a and 1b (Steiger et al., 2004). GABABRs are widely expressed in brain and regulate neuronal excitability by modulating activity of G-protein-gated inwardly rectifying K+ channels (GIRKs), voltage-gated Ca2+ channels, and adenylyl cyclase (Gassmann and Bettler, 2012). Activation of GABABRs has been reported to hyperpolarize and decrease the threshold, whereas deactivation of the receptors increases the threshold required to generate an action potential (Ladera et al., 2008). Thus, alterations of GABABR trafficking/functionality have the potential to significantly alter intrinsic neuronal excitability and brain function. In this work, we have investigated the role of ATF4 in neuronal excitability. We find that ATF4 knockdown in cultured hippocampal neurons significantly increases their firing rate and that this appears to be due to reduced trafficking of GABABR to the cell surface. These effects in turn appear to be a consequence of ATF4's regulation of Cdc42 stability.
Materials and Methods
Cell cultures.
Primary hippocampal cultures were prepared as previously described (Liu et al., 2014). Briefly, hippocampi were dissected from E18 rat embryos of either sex; after dissociation, neurons were plated on poly-D-lysine-coated 12-well plates at the density of 3 × 105 cells/well. Neurons were maintained in Neurobasal medium (Invitrogen) supplemented with 2% B-27 (Invitrogen) and 0.5 mm glutamine (Invitrogen). ELISA measurements of both cell media and cell lysates revealed the presence of both GABA and glutamate in the culture (glutamate = 32.2 μg, GABA = 13.9 μg).
DNA constructs, lentivirus preparation, and infection.
All shRNAs were cloned in the pLVTHM vector (Addgene), which contains an EF-1α promoter for target gene expression, using the following oligo DNA pairs as previously described (Liu et al., 2014).
Lenti-shRNA control: 5′-CGCGTCACAGCCCTTCCACCTCCATTCAAGAGATGGAGGTGGAAGGGCTGTGTTTTTTA-3′ and 5′-CGCGTAAAAAACACAGCCCTTCCACCTCCATCTCTTGAATGGAGGTGGAAGGGCTGTGA-3′. Lenti-shATF4: 5′-CGCGTGCCTGACTCTGCTGCTTATTTCAAGAGAATAAGCAGCAGAGTCAGGCTTTTTTA-3′ and 5′-CGCGTAAAAAAGCCTGACTCTGCTGCTTATTCTCTTGAAATAAGCAGCAGAGTCAGGCA-3′.
Lenti-shATF4 addback was generated using the QuikChange Site-directed Mutagenesis kit (Stratagene). Point mutations were introduced into the Lenti-ATF4 at the recognition site for shATF4 (CCTGACTCTGCTGCTTAT to CCAGAGTCAGCTGCTTAC).
Lenti-shATF4 mut/addback was generated from shATF4addback by introducing point mutations at the DNA binding site (292RYRQKKR298 to 292GYLEAAA298).
Lenti-shCdc42 was generated according to a published siRNA sequence 5′-GAUAACUCACCACUGUCCATT-3′ (Deroanne et al., 2005). A scrambled shRNA (lenti-shCdc42scr) was generated by using the following oligo DNA pair: 5′-CGCGTGTCCAACGTCCATATACCATTCAAGAGATGGTATATGGACGTTGGACTTTTTTA-3′ and 5′-CGCGTAAAAAAGTCCAACGTCCATATACCATCTCTTGAATGGTATATGGACGTTGGACA-3′. Lentiviral particles were produced using the second-generation packaging system. Briefly, HEK293T cells were transfected with the respective lentiviral constructs for shRNA together with the packaging vectors psPAX2 and pMD2.G (Addgene) using calcium phosphate. Two and three days after transfection, cell supernatants were collected and lentiviral particles were concentrated 20–30× by centrifugation in Amicon Ultracentrifugal filters (100 kDa) (Millipore). Viral titer ranged between 1 and 5 × 106 virions/μl. Primary neuronal cultures were infected with viral particles on 7 DIV, and RNA and protein extraction was performed at the indicated time points.
RNA extraction and qRT-PCR.
Total RNA was extracted from rat primary hippocampal cultures 4, 8, and 12 d after lentiviral infection according to the RNeasy Mini Protocol (QIAGEN). RNA concentration and purity were determined using a NanoDrop 8000 (Thermo Fisher Scientific). mRNA was then reverse-transcribed into cDNA using the first-strand cDNA synthesis system for qRT-PCR (Origene) following the manufacturer's instructions. Reaction mixtures were diluted fivefold and subjected to qRT-PCR amplification (Eppendorf) using FastStart SYBR Green Master mix (Roche). The following primers were used: ATF4: forward 5′-ATGCCAGATGAGCTCTTGACCAC-3′ and reverse 5′-GTCATTGTCAGAGGGAGTGTCTTC-3′; α-tubulin: forward 5′-TACACCATTGGCAAGGAGAT-3′ and reverse 5′-GGCTGGGTAAAT GGAGAACT-3; GABABR 1a: forward 5′-CACACCCAGTCGCTGTG-3′ and reverse 5′-GAGGTCCCCACCCGTCA-3′; and GABABR 1b 5′-GGGACCCTGTACCCCGGTG-3′ and reverse 5′-GGAGTGAGAGGCCCACACC-3′. Relative product quantities for each transcript were performed in triplicate, normalized to α-tubulin mRNA as an endogenous control, and determined using the comparative CT method.
Electrophysiology.
Primary hippocampal neurons (19–21 DIV, 2 weeks after lentiviral infection) were used for tight-seal conventional whole-cell patch clamp. All the currents were recorded from pyramidal-like neurons, based on the their large (~15 μm) triangular-shaped somas. Coverslips were placed in a recording chamber with bath solution containing the following (in mm): 119 NaCl, 5 KCl, 20 HEPES, 30 glucose, 2 CaCl2, 2 MgCl2. The pH and osmolarity of the bath solution were adjusted to 7.3 and 330 mOsm/L, respectively. For spontaneous action potential recordings, glass pipettes were filled with intracellular electrode solution, pH 7.3 (285 mOsm/L) containing the following (in mm): 130 K-gluconate, 10 KCl, 10 HEPES, 1 MgCl2, 0.06 CaCl2, 0.1 EGTA, 3 MgATP, 0.3 Na2GTP, and typically registered 4–8 mΩ pipette resistances. Following acquisition of electrical access, cells were held in current-clamp mode at I = 0. For mIPSC experiments, glass pipettes were filled with intracellular electrode solution, pH 7.3 (285 mOsm/L), containing the following (in mm): 130 KCl, 10 HEPES, 0.5 CaCl2, 1 EGTA, 3 MgATP, 0.3 Na2GTP. Furthermore, 1 μm TTX, 10 μm CNQX, and 50 μm d-APV were continuously perfused during the experiment. All the cells were recorded at −70 mV for 10 min, and a 5 mV hyperpolarizing test pulse was applied periodically during recordings to ensure that the access resistance did not change significantly and was <25 mΩ. If not, the recordings were discarded. Signals were filtered at 2 kHz, digitized at 10 kHz, stored and analyzed offline using MiniAnalysis Software (Synaptosoft, version 6.0.7). The threshold for event detection was set at 5 pA. Recordings were performed at room temperature under constant perfusion (2 ml/min) and acquired using Clampex software and a microamplifier (MultiClamp 700B, Molecular Devices). For baclofen-induced GIRK currents, hippocampal neurons were bathed initially with a solution containing the following (in mm): 119 NaCl, 5 KCl, 20 HEPES, 30 glucose, 2 CaCl2, 2 MgCl2, pH 7.3 (330 mOsm/L), and then switched to a high potassium solution (hK) containing the following (in mm): 85 NaCl, 60 KCl, 2 MgCl2, 2 CaCl2, 10 HEPES, 10 glucose, pH 7.3, to determine the amplitude of the basal potassium current. When the basal current reached equilibrium, baclofen diluted in hK was applied. The hK-induced current was subtracted from the total current to obtain the baclofen-induced GIRK current. Membrane potential was held at −70 mV throughout the duration of the experiment.
Surface and total protein isolation.
Membrane-bound and total protein isolation was conducted using the EZ-Link NHS-PEO4-Biotinylation Kit (Pierce), following the manufacturer's instructions. Briefly, cells were gently washed three times with ice-cold PBS containing 1 mm MgCl2 and 0.1 mm CaCl2 (PBS/CM) and then incubated with 500 μg/ml of EZ-link NHS-PEO4-biotin dissolved in ice-cold PBS at 4°C for 1 h. Cells were then washed once with ice-cold PBS, and the reaction quenched by adding 500 μl of 100 mm glycine for 10 min, followed by 3 washes in ice-cold PBS. Cells were then harvested in RIPA buffer supplemented with protease/phosphatase inhibitor and centrifuged at 14,000 rpm for 15 min at 4°C; 30 μl of the resulting supernatant was collected for total protein input and the rest incubated with 50 μg of streptavidin beads, rotating overnight at 4°C. Beads were washed 5 times with RIPA buffer and bound proteins eluted with 1× sample buffer by boiling for 5 min.
Immunoblotting.
After adding NuPAGE LDS Sample Buffer (Invitrogen) and 5% β-mercaptoethanol, samples were boiled for 15 min and proteins were separated by electrophoresis on 4%–12% BisTris SDS-acrylamide gels (Invitrogen). After transfer, the membranes were blocked for 1 h at room temperature with 5% milk and then incubated overnight with primary antibody. The following primary antibodies were used: rabbit anti-GABAB R1 (1:1000, Abcam, #55051) (Zapata et al., 2017), GABAB R2 (1:1000, Cell Signaling Technology, #3839), rabbit anti-GABAAR β2 (1:1000, Synaptic Systems, #224803), rabbit anti-GABAAR γ2 (1:1000, Synaptic Systems, #224003), rabbit anti-ATF4 (1:1000, Cell Signaling Technology, #11815), rabbit anti-Cdc42 (1:1000, Cell Signaling Technology, #2462S), rabbit anti-GIRK1 (1:500, Abcam, #129182), rabbit anti-GIRK2 (1:200, Sigma-Aldrich, #P8122), rabbit anti-GIRK3 (1:400, Sigma-Aldrich, #P8247), mouse anti-GAPDH (1:10,000, Invitrogen, #MA1-16757), rabbit anti-Kv1.1 (1:400, Sigma-Aldrich, #P8247), mouse anti-Kv2.1 (1:500, Abcam, #ab192761), rabbit anti-Kv4.2 (1:200, Sigma; Millipore, #07-491), and rabbit anti-Cav2.1 (1:500, Abcam, #ab32642). Densitometric quantification of the bands was performed using ImageJ software (National Institutes of Health). Total level of proteins (input) was normalized to GAPDH, whereas membrane-bound samples were normalized with the ratio input/GAPDH. GAPDH was undetectable in the membrane-bound fraction, therefore excluding the possibility that the membrane was leaky or compromised.
Statistical analysis.
Data are shown as mean ± SEM. Comparison between two groups was performed with a two-tailed unpaired Student's t test. Comparison between multiple groups was performed using two-way ANOVA, followed by a Bonferroni post hoc test when applicable. Statistical significance was set at p < 0.05.
Results
ATF4 knockdown increases neuronal excitability
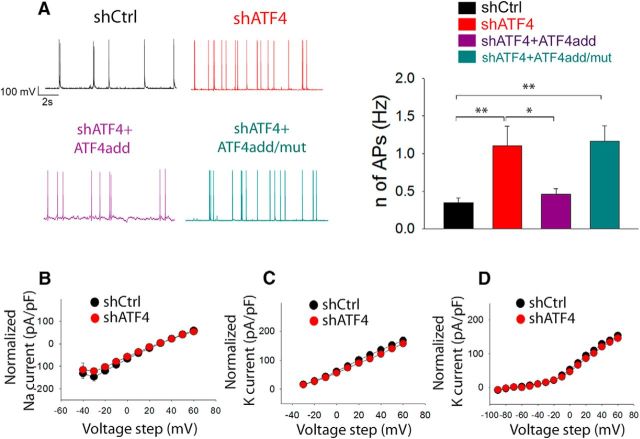
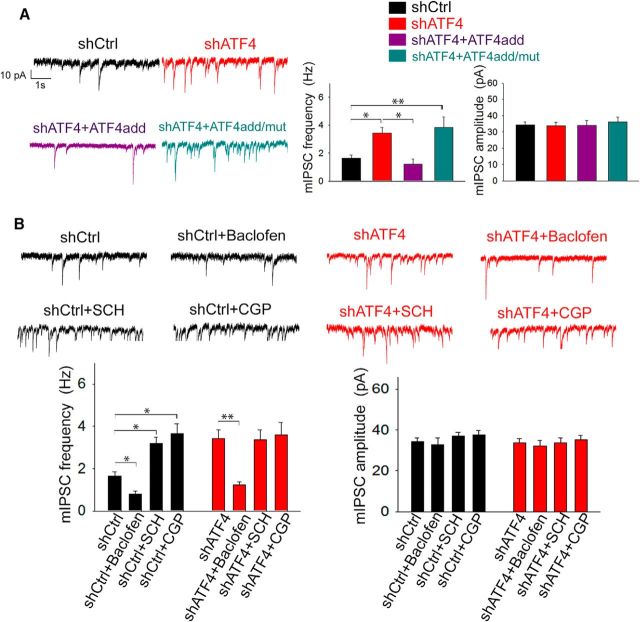
We previously described a key role for ATF4 in modulating glutamatergic neurotransmission both in vitro and in vivo and in regulating dendritic spines (Liu et al., 2014; Pasini et al., 2015). Given the pivotal roles of these two aspects in controlling the excitability properties of neurons, we next set out to directly investigate the role of ATF4 in intrinsic neuronal excitability. For this purpose, we used lentivirally delivered shRNAs to specifically downregulate ATF4 expression in 7 DIV cultured rat hippocampal neurons and performed whole-cell patch clamp 2 weeks after the infection to record the frequency of spontaneously occurring action potentials (sAPs). At this time in culture (3 weeks total), the neurons have formed extensive synaptic connections (Liu et al., 2014). As shown in Figure 1A, B, knockdown of ATF4 resulted in an ~3-fold increase in the frequency of sAPs compared with neurons infected with a control shRNA (shCtrl = 0.35 ± 0.07 Hz, shATF4 = 1.11 ± 0.26 Hz; post hoc Bonferroni, shCtrl vs shATF4, p < 0.01) To confirm that this result was not due to off-target effects, we performed a rescue experiment in which the neurons were coinfected with lentiviruses expressing shATF4 and an ATF4 construct (ATF4add) conservatively mutated to make it unresponsive to shATF4. This resulted in knockdown of endogenous and overexpression of exogenous ATF4, respectively (Liu et al., 2014). Our results indicate that ATF4add restored the firing rate to the control level (Fig. 1A; shATF4+ATF4add = 0.46 ± 0.07 Hz; post hoc Bonferroni, shCtrl vs shATF4+ATF4add, p > 0.05). However, while ATF4 over-expression rescued the firing rate, it did not reduce it below that seen in control cultures. Next, to test whether the effects of ATF4 on sAP frequency requires its transcriptional activity, we co-infected cultured hippocampal neurons with shATF4 and a mutant ATF4 construct, ATF4add/mut, that is not recognized by shATF4 and that encodes a mutant ATF4 that does not bind DNA and thus is transcriptionally inactive. This results in knockdown of endogenous ATF4 and overexpression of inactive exogenous ATF4 (Liu et al., 2014). In contrast to ATF4add, ATF4add/mut failed to rescue the effect of ATF4 knockdown (Fig. 1A; shATF4+ATF4add/mut = 1.17 ± 0.21 Hz; post hoc Bonferroni, shCtrl vs shATF4+ATF4add/mut, p < 0.01), suggesting that ATF4 must retain its transcriptional capability to regulate the frequency of neuronal firing. Because APs are generated by voltage-gated sodium (NaV) and potassium (KV) channels (Bean, 2007), we investigated whether ATF4 down-regulation could affect the neuronal firing rate by influencing these major AP constituents. However, our results show no differences in either NaV or KV I–V curves obtained from shCtrl- or shATF4-infected hippocampal cultures (Fig. 1B–D), suggesting that ATF4 regulates neuronal excitability by a mechanism independent of effects on Nav or Kv.
Figure 1.
ATF4 knockdown increases neuronal excitability. A, The frequency of spontaneously-occurring action potentials (sAPs) is increased upon ATF4 down-regulation; this effect appears to be mediated by ATF4's transcriptional capability. Left panel, representative traces of sAPs recorded from cultured hippocampal neurons infected at DIV7 for 2 weeks with lentivirus expressing either shCtrl (black trace), shATF4 (red trace), shATF4+ATF4add (purple trace), or shATF4+ATF4add/mut (cyan trace). Right panel, summary bar graph showing the mean frequencies (± SEM) of sAPs of hippocampal neurons infected with shCtrl (black bars, n = 24), shATF4 (red bars, n = 22), shATF4+ATF4add (purple bars, n = 18), shATF4+ATF4add/mut (cyan bars, n = 18) constructs. B–D, ATF4 down-regulation does not affect Na2+ and K+ voltage-gated channels in shATF4-infected neurons (red dots) when compared to shCtrl neurons (black dots). B, Voltage-gated Na2+ currents were evoked by 1s step depolarizations from −40 to +60 mV with 10-mV increments (shCtrl n = 15; shATF4 n = 16). C, A-type K+ currents were evoked by 1s step depolarizations from −30 to +60 mV, with 10-mV increments (shCtrl n = 29; shATF4 n = 33), and successively isolated. D, Delayed-rectifying K+ currents were evoked by 1s step depolarizations from −90 to +60 mV, with 10 mV increments (shCtrl n = 30; shATF4 n = 34), and successively isolated. Values are expressed as mean ± SEM. *p < 0.05; **p < 0.01.
ATF4 regulates trafficking of GABABRs to the membrane
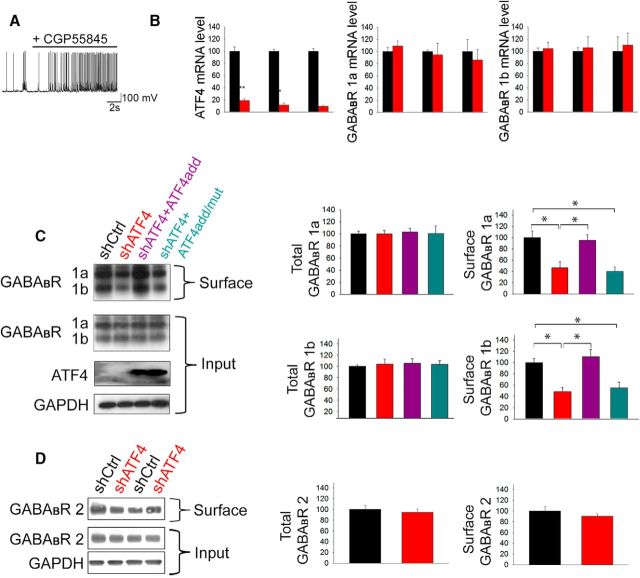
Among the many proteins reported to modulate the excitability of neurons, we focused on GABABRs, the metabotropic (G-protein coupled) receptors for GABA. Postsynaptic GABABRs induce a slow IPSC by gating Kir-type K+ channels. This in turn hyperpolarizes the membrane and shunts excitatory currents, thereby inhibiting generation of action potentials (Leung and Peloquin, 2006). In this light, we confirmed that blocking GABABR activity in our cultures increases the neuronal firing rate. As shown in Figure 2A, application of the specific GABABR antagonist CGP55845 (10 μm) produced a rapid eightfold rise (from ~0.4 to 3.2 Hz) of sAP frequency. Of potential relevance, ATF4 has been reported to directly bind GABABRs as well as to differentially regulate promoter activity of the subunits 1a and 1b of GABABRs (Steiger et al., 2004). We therefore first investigated whether ATF4 downregulation affects expression of GABABRs at the transcriptional level. To achieve this, we infected cultured hippocampal neurons with either shATF4 or shCtrl for 4–15 d and then performed qRT-PCR. As shown in Figure 2B, knockdown of ATF4 did not significantly affect the transcript levels of either GABABR 1a or 1b subunits (ATF4, 4 d: shCtrl = 100 ± 7.1%, shATF4 = 19.1 ± 2.4%, t test, p < 0.01; ATF4, 8 d: shCtrl = 100 ± 3%, shATF4 = 11.6 ± 2.8%, t test, p < 0.01; ATF4, 12 d: shCtrl = 100 ± 4.19%, shATF4 = 9.47 ± 1.63%, t test, p < 0.01).
Figure 2.
ATF4 regulates GABABR surface expression. A, GABABR blockade increases the frequency of sAPs in control neurons. Representative trace of sAPs recorded from cultured hippocampal neurons infected with shCtrl-carrying lentivirus treated with the specific GABABR antagonist CGP55845 (10 μm). B, Time course experiment showing no changes in the mRNA levels for GABABR subunits 1a and 1b upon ATF4 downregulation. Bar graphs represent qPCR analysis of ATF4 (left), GABABR 1a (middle), and GABABR 1b (right) mRNA levels extracted 4, 8, and 12 d after infection with lentivirus carrying either shCtrl (black bars) or shATF4 (red bars) constructs. Data are mean ± SEM from three independent experiments. C, ATF4 downregulation decreases membrane levels of GABABR. The 7 DIV hippocampal cultured neurons were infected with indicated lentiviral constructs (black represents shCtrl; red represents shATF4; purple represents shATF4+ATF4add; cyan represents shATF4+ATF4add/mut) for 14 d before undergoing extraction of total and membrane proteins. Left, Representative Western immunoblot. Right, Bar graphs represent densitometric analysis of total (left) or membrane (right) GABABR 1a and 1b protein normalized to GAPDH. D, ATF4 downregulation does not affect total or surface levels of GABABR 2. Left, Representative Western immunoblot. Right, Bar graphs represent densitometric analysis of total (left) or membrane (right) GABABR 2 protein normalized to GAPDH. Data are mean ± SEM from three independent experiments. *p < 0.05; **p < 0.01.
Next, we determined whether knocking down ATF4 would alter total or membrane-bound protein expression of GABABRs. To achieve this, we performed biotinylation of plasma membrane proteins on cultured hippocampal neurons with or without ATF4 knockdown (infected at 7 DIV for 2 weeks), isolated the biotinylated proteins on streptavidin-bound beads, and performed Western immunoblotting analysis for GABABR subunits 1a and 1b on both the input (total cell lysate) and membrane fractions. Densitometric quantification from multiple experiments showed that ATF4 downregulation did not affect total GABABR 1a and 1b protein levels (Fig. 2C) but significantly decreased the levels of GABABR subunits 1a and 1b in the biotinylated membrane fraction (Fig. 2C; GABABR 1a: shCtrl = 100 ± 11.8%, shATF4 = 46.7 ± 10.7%; post hoc Bonferroni test: shCtrl vs shATF4, p < 0.01; GABABR 1b: shCtrl = 100 ± 7.2%, shATF4 = 48.2 ± 7.9%; post hoc Bonferroni test: shCtrl vs shATF4, p < 0.01), thus indicating a role for ATF4 in regulating GABABR trafficking, but not overall expression.
To address the question of whether the effect of ATF4 on membrane trafficking of GABABRs involves ATF4's transcriptional activity, we performed a rescue experiment as above and found that ATF4add/mut, in contrast to ATF4add, failed to reverse the effects of ATF4 knockdown (Fig. 2C; GABABR 1a, shATF4+ATF4add = 95.1 ± 10.3%, shATF4+ATF4add/mut = 39.8 ± 8.2%; post hoc Bonferroni test: shCtrl vs shATF4+ATF4add, p > 0.05; shATF4 vs shATF4+ATF4add, p < 0.05; shCtrl vs shATF4+ATF4add/mut, p < 0.01; GABABR 1b, shATF4+ATF4add = 110.5 ± 12.7%, shATF4+ATF4add/mut = 55.2 ± 10%; post hoc Bonferroni test: shCtrl vs shATF4+ATF4add, p > 0.05; shATF4 vs shATF4+ATF4add, p < 0.001; shCtrl vs shATF4+ATF4add/mut, p < 0.05). This indicates, as with neuronal excitability, that ATF4 has a transcriptional role in regulating trafficking of GABABRs. The data also show, as with excitability, that ATF4 overexpression is not sufficient to drive surface expression of GABABRs beyond that seen with basal endogenous expression.
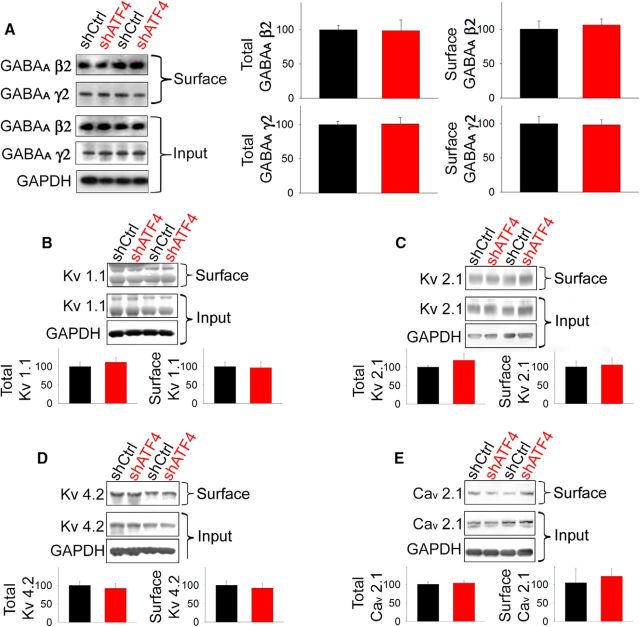
We next asked whether knocking down ATF4 might produce a more general or nonspecific effect on membrane proteins. Interestingly, both the total and membrane-bound levels of GABABR 2 (biotin labeled as above) were unaffected by ATF4 downregulation (Fig. 2D). We also examined the effect of shATF4 on membrane-exposed β2 and γ2 subunits of GABAA receptors and found no effects on either the total or membrane-exposed protein levels (Fig. 3A). In addition to GABABRs, a wide variety of voltage-sensitive K and Ca channels has been described to regulate excitability properties of neurons (Chen et al., 2006; Hsiao et al., 2009; Rossignol et al., 2013; Speca et al., 2014). We therefore queried whether the effect of ATF4 on neuronal excitability was in part due to its capability to regulate the expression or localization of Kv1.1, Kv2.1, Kv4.2, and Cav2.1. As shown in Figure 3B–E, neither the total nor the membrane-bound levels of these proteins were affected by ATF4 downregulation. These findings thus indicate that ATF4 has a selective role in regulation of membrane-bound proteins involved in neuronal excitability and that this includes GABABRs.
Figure 3.
ATF4 downregulation does not affect total and membrane levels of GABAARs and voltage-sensitive K or Ca channels. DIV 7 cultured hippocampal neurons were infected with indicated lentiviral constructs (black represents shCtrl; red represents shATF4) for 14 d before undergoing extraction of total and membrane proteins. A, Left, Representative Western immunoblot. Bar graphs represent densitometric analysis of total (left) or membrane (right) GABAAR β2 and γ2 protein normalized to GAPDH. B, Top, Representative Western immunoblot. Bar graphs represent densitometric analysis of total (left) or membrane (right) Kv1.1 protein normalized to GAPDH. C, Top, Representative Western immunoblot. Bar graphs represent densitometric analysis of total (left) or membrane (right) Kv2.1 protein normalized to GAPDH. D, Top, Representative Western immunoblot. Bar graphs represent densitometric analysis of total (left) or membrane (right) Kv4.2 protein normalized to GAPDH. E, Top, Representative Western immunoblot. Bar graphs represent densitometric analysis of total (left) or membrane (right) Cav2.1 protein normalized to GAPDH. Data are mean ± SEM from three independent experiments, each run in duplicate. The differences between shCtrl and shATF4 were not significant (p > 0.05) in all cases.
ATF4 knockdown reduces GIRK currents
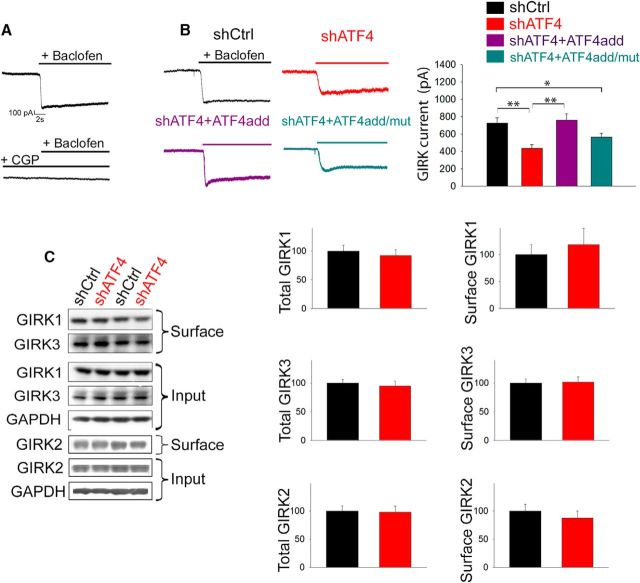
We next queried whether the reduction we observed in the number of membrane-inserted GABABRs after ATF4 downregulation reflected a change in the functionality of the receptors themselves. Postsynaptic GABABRs are associated with, and mediate part of their functions through, GIRK channels (Gassmann and Bettler, 2012). We therefore studied the function of GABABRs by whole-cell patch-clamp recording of GABABR-induced K+ GIRK currents in 7 DIV hippocampal cultured neurons infected for 2 weeks with lentiviruses carrying either shCtrl or shATF4. As shown in Figure 4A, we first calibrated our recordings by applying the GABABR agonist baclofen (100 μm), which elicited sustained K+ currents in control neurons that were prevented by pretreating the cells with a specific GABABR antagonist (SCH50911, 100 μm). Consistent with our finding that ATF4 downregulation reduces cell surface GABABR levels, when we recorded baclofen-induced GIRK currents in ATF4 knockdown neurons, we found them to be significantly reduced compared with those in ShCtrl-infected neurons (Fig. 4B; shCrl 725.9 ± 58.4pA, shATF4 435.1 ± 39.2 pA; post hoc Bonferroni, p < 0.01). This effect did not appear to be mediated by effects on GIRK channels in that shATF4 did not affect total or membrane-bound GIRK1, GIRK2, or GIRK3 protein levels (Fig. 4C). Finally, ATF4add, but not ATF4add/mut, completely restored the currents to the control level (Fig. 4B; shATF4+ATF4add = 786.9 ± 69.2 pA, shATF4+ATF4add/mut = 545.9 ± 43.8 pA; post hoc Bonferroni: shATF4 vs shATF4+ATF4add, p < 0.001; shATF4add vs shATF4add/mut, p < 0.01), further confirming the idea that ATF4 needs to retain its transcriptional capability to regulate the trafficking of GABABRs. As with GABABR membrane trafficking, ATF4 overexpression did not raise GIRK current amplitude beyond that in control cultures.
Figure 4.
ATF4 knockdown reduces GABABR-induced GIRK currents. A, Specific GABABR agonist baclofen (100 μm) elicited a sustained GIRK current in cultured hippocampal neurons infected with shCtrl lentivirus. Top, Representative trace. This effect is abolished by pretreating the culture with the specific GABABR antagonist CGP55845 (10 μm; bottom). B, shATF4 infection reduces baclofen-induced GIRK currents; this effect is dependent on ATF4's transcriptional capability. Left, Representative traces of baclofen-elicited currents recorded from 7 DIV cultured hippocampal neurons infected for 2 weeks with lentivirus expressing shCtrl (black), shATF4 (red), shATF4+ATF4add (purple), or shATF4+ATF4add/mut (cyan). Right, Bar graphs represent measurements of the currents (shCtrl, n = 37; shATF4, n = 39; shATF4+ATF4add, n = 36; shATF4+ATF4add/mut, n = 51). Data are mean ± SEM. *p < 0.05; **p < 0.005. C, ATF4 downregulation does not affect total and membrane levels of GIRK proteins; 7 DIV cultured hippocampal neurons were infected with indicated lentiviral constructs (black represents shCtrl; red represents shATF4) for 14 d before undergoing extraction of total and membrane proteins. Left, Representative Western immunoblot. Bar graphs represent densitometric analysis of total (left) or membrane (right) GIRK proteins normalized to GAPDH. Data are mean ± SEM from three independent experiments. *p < 0.05; **p < 0.01.
ATF4 knockdown increases the frequency, but not amplitude, of mIPSCs
Given that GABABR manipulations have been reported to affect the frequency, but not the amplitude of spontaneous mIPSCs (Ulrich and Huguenard, 1996; Kubota et al., 2003; Kirmse and Kirischuk, 2006), we assessed mIPSCs (confirmed by picrotoxin blockade) in cultured hippocampal neurons infected with either shCtrl or shATF4 as a further readout of GABABR functionality. As shown in Figure 5A, we found that shAT4 significantly increases (by ~2-fold) the frequency, but not the amplitude of mIPSCs (mIPSC frequency: shCtrl = 1.65 ± 0.21 Hz, shATF4 = 3.43 ± 0.41 Hz; post hoc Bonferroni: p < 0.05). In addition, adding back ATF4 completely restored the frequency of mIPSCs to control values (Fig. 5A; shATF4+ATF4add = 1.19 ± 0.37 Hz; post hoc Bonferroni: shATF4 vs shATF4+ATF4add, p < 0.05). As with other properties described above, ATF4add overexpression did not increase mIPSC frequency beyond the level observed in control cultures. Furthermore, our whole-cell patch-clamp recordings showed that, unlike ATF4add, ATF4add/mut was unable to reverse the effect of ATF4 downregulation on mIPSC frequency (Fig. 5A; shATF4+ATF4add/mut = 3.83 ± 0.76 Hz; post hoc Bonferroni: shCtrl vs shATF4+ATF4add/mut, p < 0.01), confirming that the transcriptional activity of ATF4 is required for this action. To further confirm the idea that membrane-bound GABABRs are reduced by shATF4, we treated both shCtrl and shATF4-infected neurons with the specific GABABR agonist baclofen (20 μm) or GABABR antagonists SCH50911 and CGP55845 (used at 100 and 10 μm, respectively). As shown in Figure 5B, baclofen application significantly reduced the frequency of mIPSCs both in shCtrl- and shATF4-infected hippocampal neurons (shCtrl+baclofen = 0.79 ± 0.14 Hz, shATF4+baclofen = 1.23 ± 0.15 Hz; post hoc Bonferroni: shCtrl vs shCtrl+baclofen, p < 0.05; shATF4 vs shATF4+baclofen, p < 0.01), thus confirming that the membrane-bound GABABRs of shATF4-infected hippocampal neurons are properly responding to stimulation. Interestingly, the application of GABABR antagonists CGP55845 and SCH50911 significantly elevated mIPSC frequency in shCtrl but not in shATF4 neurons (Fig. 5B; shCtrl+SCH = 3.19 ± 0.30 Hz, shCrl+CGP = 3.67 ± 0.46 Hz, shATF4+SCH = 3.38 ± 0.44 Hz, shATF4+CGP = 3.59 ± 0.15 Hz; post hoc Bonferroni: shCtrl vs shCtrl+SCH, p < 0.05; shCtrl vs shCtrl+CGP, p < 0.05; shATF4 vs shATF4+SCH, shATF4 vs shATF4+CGP, p > 0.05), which is consistent with the observation that shATF4 reduces membrane-bound levels of GABABRs. As expected, none of the treatments significantly affected the amplitude of mIPSCs (Fig. 5B).
Figure 5.
ATF4 knockdown increases the frequency, but not amplitude, of mIPSCs. A, Left, Representative mIPSC traces recorded from 7 DIV hippocampal neurons infected for 2 weeks with lentivirus expressing shCtrl (black), shATF4 (red), shATF4+ATF4add (purple), or shATF4+ATF4add/mut (cyan). Right, Bar graphs represent the frequency (left) and the amplitude (right) of mIPSCs shown in A (shCtrl, n = 46; shATF4, n = 29; shATF4+ATF4add, n = 16; shATF4+ATF4add/mut, n = 25). Data are mean ± SEM. B, Effect of pharmacological manipulation of GABABRs on mIPSCs recorded from 7 DIV hippocampal neurons infected for 2 weeks with lentivirus expressing either shCtrl,or shATF4. Top, Representative traces of mIPSCs recorded from shCtrl (black traces) or shATF4 (red traces) infected neurons alone (shCtrl, n = 49; shATF4, n = 29) or in the presence of 20 μm baclofen (GABABR agonist; shCtrl, n = 16; shATF4, n = 15), 10 μm SCH50911 (GABABR antagonist; shCtrl, n = 36; shATF4, n = 18), or 100 μm CGP55845 (GABABR antagonist; shCtrl, n = 21; shATF4, n = 17). Bottom, Bar graphs represent the frequency (left) and the amplitude (right) of the mIPSCs. Data are mean ± SEM. *p < 0.05; **p < 0.01.
The effects of ATF4 on excitability and GABABRs appear to be mediated by changes in Cdc42 expression
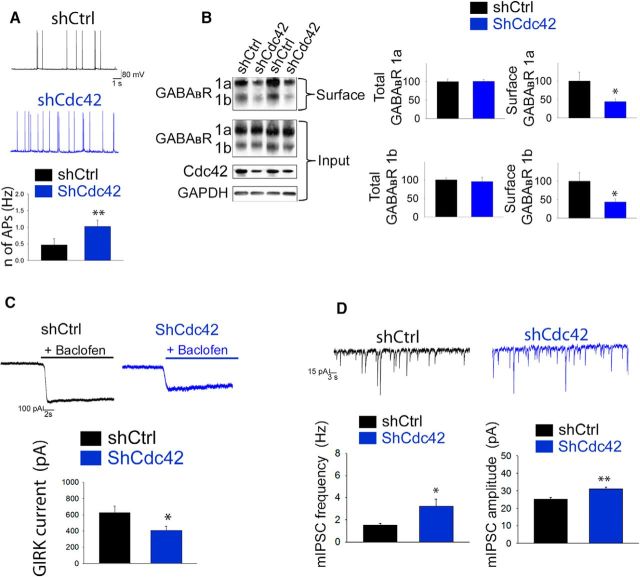
We previously reported that ATF4's modulation of dendritic spine density and glutamatergic functionality is mediated, at least in part, by its capacity to regulate the stability and expression of the total and activated forms of the small Rho GTPase Cdc42 (Liu et al., 2014; Pasini et al., 2015). Of particular relevance here, Cdc42 has been shown to be involved in regulating receptor trafficking (Hussain et al., 2015). We therefore next tested the hypothesis that the effects of ATF4 downregulation on GABABR trafficking and neuronal excitability could be mediated by loss of Cdc42. To achieve this, we used a previously characterized Cdc42 shRNA (Liu et al., 2014) to deplete Cdc42 in cultured hippocampal neurons and determined whether this phenocopies the effects of ATF4 knockdown. We first assessed whether specific Cdc42 downregulation affects neuronal excitability. As in the case of ATF4 knockdown, silencing Cdc42 protein produced a twofold increase of AP frequency (Fig. 6A; shCtrl = 0.43 ± 0.13 Hz, shCdc42 = 1.08 ± 0.15 Hz; t test, p < 0.01). Next, we queried whether Cdc42 downregulation phenocopies the effect of ATF4 knockdown on GABABR trafficking and found that this was sufficient to significantly decrease the levels of membrane-exposed, but not total GABABRs (Fig. 6B; for GABABR 1a shCtrl = 100 ± 23.3%, shCdc42 44.2 ± 7.4%; t test, p < 0.05; GABABR 1b shCtrl = 100 ± 20.3%, shCdc42 38.7 ± 4.7%; t test, p < 0.05). In addition, we found that baclofen-induced GIRK currents were significantly decreased in shCdc42-infected neurons, compared with controls (Fig. 6C; shCtrl = 627.8 ± 77.6 pA, shCdc42 = 406.9 ± 52.9 pA; t test, p < 0.05). Finally, Cdc42 knockdown also increased the frequency mIPSCs (Fig. 6D; shCtrl = 1.55 ± 0.16 Hz, shCdc42 = 3.21 ± 0.67 Hz; t test, p < 0.05) to a degree similar to that observed with ATF4 knockdown. However, with Cdc42 knockdown, there was also a small, but significant, increase in mIPSC amplitude (Fig. 6D; shCtrl = 25.22 ± 1.26 pA, shCdc42 = 31 ± 1.21 pA; t test, p < 0.01). Together, these findings support the idea that regulation of Cdc42 levels mediates the effects of ATF4 on neuronal GABABR trafficking and excitability.
Figure 6.
The effects of ATF4 on excitability and GABABRs are driven by changes in Cdc42 expression. A, Cdc42 downregulation increases intrinsic neuronal excitability. Top, Representative traces of sAPs recorded from cultured hippocampal neurons infected at 7 DIV for 2 weeks with lentivirus expressing either shCtrl (black trace) or shCdc42 (blue trace). Bottom, Summary bar graphs represent the frequency of sAPs of neurons infected with shCtrl (black bars, n = 13) or shCdc42 (blue bars, n = 13) constructs. B, Cdc42 downregulation decreases membrane but not total GABABR levels. The 7 DIV cultured hippocampal neurons were infected with different lentiviral constructs (black represents shCtrl; blue represents shCdc42) for 2 weeks before undergoing extraction of total and membrane proteins. Left, Representative Western immunoblot. Right, Bar graphs represent densitometric analysis of immunoblots for total (left) or membrane (right) GABABR 1a and 1b protein, normalized to GAPDH. C, Cdc42 downregulation decreases baclofen-elicited GIRK currents. Top, Representative traces of baclofen-elicited currents recorded from 7 DIV cultured hippocampal neurons infected for 2 weeks with lentivirus carrying either shCtrl (black) or shCdc42 (blue). Bottom, Bar graph represents the measurements of the currents (shCtrl, n = 22; shCdc42, n = 26). D, Cdc42 downregulation increases mIPSCs. Top, Representative mIPSC traces recorded from 7 DIV hippocampal neurons infected for 2 weeks with lentivirus carrying either shCtrl (black) or shCdc42 (blue) constructs. Bottom, Bar graphs represent the frequency (left) and the amplitude (right) of mIPSCs shown in A (shCtrl, n = 34; shCdc42, n = 29). Data are mean ± SEM. *p < 0.05; **p < 0.01.
Discussion
In the present study, we delineate a novel role for the transcription factor ATF4 in regulating GABABR trafficking and neuronal excitability. Our data show that chronic ATF4 downregulation in hippocampal neurons reduces membrane levels of GABABRs, diminishes their functionality, and consequently leads to a substantial increase of neuronal firing rate. In addition, we found that these effects are mediated by ATF4's transcriptional capability and reflect changes in expression of the small GTPase Cdc42. Studies on ATF4's role in the regulation of synaptic plasticity and memory have led to divergent views. ATF4 has been characterized to be either a negative or positive influence on plasticity. Such interpretations appear to be dependent on cellular context (Bartsch et al., 1995; Hu et al., 2015) or have been based on experimental manipulations that are not specific to ATF4, such as use of a dominant-negative construct (Chen et al., 2003) or regulation of eIF2α phosphorylation (Costa-Mattioli et al., 2007). To more directly assess ATF4's function in unstressed brain, we have used the strategy of studying the effects of long-term ATF4 knockdown or deletion in hippocampal and cortical neurons both in vitro and in vivo. This has revealed required roles for ATF4 in maintaining mushroom spines and glutamatergic functionality as well as in long-term spatial memory and behavioral flexibility (Liu et al., 2014; Pasini et al., 2015). The present findings significantly extend the range of cognition-relevant neuronal properties that are dependent on the presence of ATF4.
In the present work, we found that ATF4 plays a novel role in regulating the proportion of GABABRs that are exposed on the neuronal surface. Two subunit isoforms of GABAB 1 receptors have been described, GABABR 1a and GABABR 1b, which differ by the presence of two sushi domains near the N terminus of GABABR 1a (Hawrot et al., 1998). Our findings indicate that ATF4 knockdown leads to comparable reductions of both GABABR 1a and GABABR 1b on the cell surface. There is evidence that GABABR 1a subunits are mainly located presynaptically, whereas GABABR 1b subunits are predominantly expressed postsynaptically (Vigot et al., 2006). Consistent with this localization and with the reductions of both subunit types on the cell surface, we observed both an increase in the frequency of mIPSCs (which is mainly a presynaptic measurement) and a decrease of GIRK currents (mainly postsynaptic in origin) after ATF4 knockdown. Presynaptic GABABRs act as presynaptic brakes on release of neurotransmitters (Sakaba and Neher, 2003; Laviv et al., 2010) and GABABR modulation consequently affects the frequency, but not the amplitude, of mIPSCs (Ulrich and Huguenard, 1996; Kubota et al., 2003; Kirmse and Kirischuk, 2006). Our observation that ATF4 downregulation increases the frequency, but not amplitude, of mIPSCs is thus consistent with a decrease of presynaptic surface-exposed GABABRs. Further supporting this view, we found that application of the specific GABABR antagonists CGP55845 and SCH50911 significantly elevated mIPSC frequency in shCtrl- but not in shATF4-treated neurons, indicating that presynaptic GABABR activity is compromised in the ATF4 knockdown neurons.
Presynaptic and postsynaptic GABABR function has been widely reported to be associated with GIRKs, which hyperpolarize neurons in response to GABABR activation (Ladera et al., 2008). We therefore measured GABABR-induced K+ currents as a readout of GABABR activity and found a 40% reduction of GIRK current in ATF4 downregulated neurons. These observations are consistent with a decline in trafficking of postsynaptic GABABRs. Thus, our findings suggest effects of ATF4 on both presynaptic and postsynaptic GABABRs. Additional studies will be needed to further characterize these effects and to define their individual influences on synaptic transmission.
In our study, we observed that the total and surface levels of GIRK proteins were unaltered by ATF4 downregulation; this finding suggests the idea that GABABRs and GIRK can be independently trafficked to the cell surface. In addition, our finding that ATF4 downregulation reduces GABABR functionality without affecting GABABR 2 levels is in line with the evidence that the 2 subunits (GABABR 1 and 2) need to heterodimerize to produce a functionally active GABABR (Jones et al., 1998).
A major issue addressed in our study is the mechanism by which ATF4 regulates the surface expression of GABABRs. As a leucine zipper protein, ATF4 has the capacity to undergo direct protein–protein interactions, among which is binding to GABABRs. However, this interaction did not appear to be relevant to the observed regulation of GABABR membrane trafficking because the deficits in this parameter were not rescued by a mutant ATF4 lacking DNA binding capacity but possessing an intact leucine zipper. These observations rather support a transcriptional role for ATF4 in regulating GABABR surface exposure. A prior study reported that ATF4 overexpression elevated expression of a GABABR 1a promoter-reporter construct and decreased expression of a GABABR 1b promoter-reporter construct (Steiger et al., 2004). However, we noted no changes either in GABABR 1a or GABABR 1b mRNA or total protein after ATF4 knockdown. Moreover, ATF4 overexpression also failed to affect surface or total GABABR 1a or GABABR 1b levels in our experiments. Our findings instead indicate that the role of ATF4 in controlling GABABR trafficking stems from its transcription-dependent capacity to regulate Cdc42 expression. In support of this idea, Cdc42 knockdown fully phenocopied the effects of ATF4 knockdown on surface expression of GABABRs. Although the ATF4/Cdc42 pathway appears to be a key player in GABABR trafficking, this does not seem to be a universal mechanism for regulating receptor surface expression in neurons. Thus, ATF4 knockdown did not affect total or membrane expression of GABABR 2, GABAA β2 and γ2 subunits, or Kv1.1, Kv2.1, Kv4.2, and Cav2.1 channels. It remains to be seen whether the ATF4/Cdc42 pathway affects trafficking of other neurotransmitter receptors.
Cdc42-dependent signaling has been implicated in trafficking of GluA1 AMPA receptors (Hussain et al., 2015). One way in which the effect of Cdc42 knockdown differed from ATF4 knockdown was that the former, but not the latter, caused a small, but significant increase in the amplitude of mIPSCs. One possible explanation for this discrepancy may be the different magnitude of Cdc42 protein silencing exerted by the two shRNA constructs. Whereas shATF4 caused an ~40%-50% reduction of Cdc42 protein, shCdc42 depleted 70%–80% of total Cdc42 protein. It may be that the greater loss of Cdc42 with shCdc42 results in postsynaptic modifications that affect GABA sensitivity.
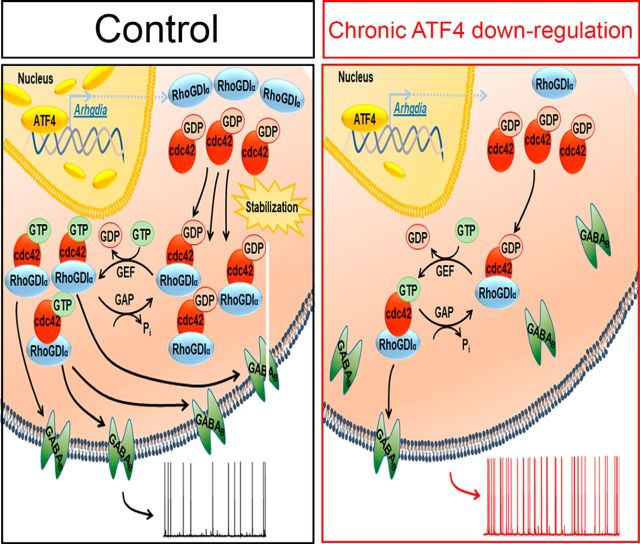
Our findings establish roles for transcriptionally active ATF4 and for Cdc42 in regulation of GABABR trafficking. Our prior work has shown that ATF4 influences Cdc42 levels by promoting its stability and that this in turn reflects the Cdc42-stabilizing activity of RhoGDIα, a direct transcriptional target of ATF4 (Pasini et al., 2016). Figure 7 shows a proposed mechanistic pathway by which ATF4 regulates GABABR trafficking and neuronal excitability via Cdc42 and RhoGDIα. An important feature of this mechanism is that it appears to occur over a prolonged time course. Cdc42 is a relatively stable protein, and knocking down ATF4 in neurons reduces Cdc42's apparent half-life from 31.5 to 18.5 h (Liu et al., 2014). This suggests that sustained changes in ATF4 protein levels/activity over many hours may be required to materially affect Cdc42 expression and thereby to affect synaptic activity. ATF4 itself is a rapidly turning-over protein, and its expression is regulated both by translational and transcriptional mechanisms. Thus, ATF4 may serve as a “sentinel” protein whose ambient expression levels in neurons influence GABABR trafficking as well as other elements of neuronal plasticity.
Figure 7.
Proposed mechanism by which ATF4 regulates neuronal excitability. Left, Control condition where basal levels of ATF4 protein ensure the appropriate amount of RhoGDIα expression to bind and stabilize cytoplasmic Rho GTPase family members, including Cdc42. Appropriate levels of Cdc42 result in basal levels of membrane-bound GABABRs that contribute to control the pace of neuronal firing. Right, Condition of chronic ATF4 downregulation, the consequent decrease of RhoGDIα levels, and augmented Cdc42 turnover. This in turn negatively affects the amount of membrane-bound GABABRs, altering neuronal intrinsic excitability properties, which results in increased sAP frequency.
Footnotes
This work was supported in party by National Institutes of Health National Institute of Neurological Disorders and Stroke Grants R01NS033689 and R01NS072050, Taub Institute for Research in Alzheimer's Disease, and Marilyn and Henry Taub Foundation. We thank Drs. Ottavio Arancio and Asa Abeliovich for providing equipment for electrophysiological experiments.
The authors declare no competing financial interests.
References
- Ameri K, Harris AL (2008) Activating transcription factor 4. Int J Biochem Cell Biol 40:14–21. 10.1016/j.biocel.2007.01.020 [DOI] [PubMed] [Google Scholar]
- Bartsch D, Ghirardi M, Skehel PA, Karl KA, Herder SP, Chen M, Bailey CH, Kandel ER (1995) Aplysia CREB2 represses long-term facilitation: relief of repression converts transient facilitation into long-term functional and structural change. Cell 83:979–992. 10.1016/0092-8674(95)90213-9 [DOI] [PubMed] [Google Scholar]
- Bean BP. (2007) The action potential in mammalian central neurons. Nat Rev Neurosci 8:451–465. 10.1038/nrn2148 [DOI] [PubMed] [Google Scholar]
- Beck H, Yaari Y (2008) Plasticity of intrinsic neuronal properties in CNS disorders. Nat Rev Neurosci 9:357–369. 10.1038/nrn2371 [DOI] [PubMed] [Google Scholar]
- Chen A, Muzzio IA, Malleret G, Bartsch D, Verbitsky M, Pavlidis P, Yonan AL, Vronskaya S, Grody MB, Cepeda I, Gilliam TC, Kandel ER (2003) Inducible enhancement of memory storage and synaptic plasticity in transgenic mice expressing an inhibitor of ATF4 (CREB-2) and C/EBP proteins. Neuron 39:655–669. 10.1016/S0896-6273(03)00501-4 [DOI] [PubMed] [Google Scholar]
- Chen X, Yuan LL, Zhao C, Birnbaum SG, Frick A, Jung WE, Schwarz TL, Sweatt JD, Johnston D (2006) Deletion of Kv4.2 gene eliminates dendritic A-type K+ current and enhances induction of long-term potentiation in hippocampal CA1 pyramidal neurons. J Neurosci 26:12143–12151. 10.1523/JNEUROSCI.2667-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Gobert D, Stern E, Gamache K, Colina R, Cuello C, Sossin W, Kaufman R, Pelletier J, Rosenblum K, Krnjević K, Lacaille JC, Nader K, Sonenberg N (2007) eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell 129:195–206. 10.1016/j.cell.2007.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deroanne CF, Hamelryckx D, Ho TT, Lambert CA, Catroux P, Lapiére CM, Nusgens BV (2005) Cdc42 downregulates MMP-1 expression by inhibiting the ERK1/2 pathway. J Cell Sci. 118(Pt 6):1173–1183. 10.1242/jcs.01707 [DOI] [PubMed] [Google Scholar]
- Gassmann M, Bettler B (2012) Regulation of neuronal GABA(B) receptor functions by subunit composition. Nat Rev Neurosci 13:380–394. 10.1038/nrn3249 [DOI] [PubMed] [Google Scholar]
- Hawrot E, Xiao Y, Shi QL, Norman D, Kirkitadze M, Barlow PN (1998) Demonstration of a tandem pair of complement protein modules in GABA(B) receptor 1a. FEBS Lett 432:103–108. 10.1016/S0014-5793(98)00794-7 [DOI] [PubMed] [Google Scholar]
- Hsiao CF, Kaur G, Vong A, Bawa H, Chandler SH (2009) Participation of Kv1 channels in control of membrane excitability and burst generation in mesencephalic V neurons. J Neurophysiol 101:1407–1418. 10.1152/jn.91053.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JY, Levine A, Sung YJ, Schacher S (2015) cJun and CREB2 in the postsynaptic neuron contribute to persistent long-term facilitation at a behaviorally relevant synapse. J Neurosci 35:386–395. 10.1523/JNEUROSCI.3284-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain NK, Thomas GM, Luo J, Huganir RL (2015) Regulation of AMPA receptor subunit GluA1 surface expression by PAK3 phosphorylation. Proc Natl Acad Sci U S A 112:E5883–E5890. 10.1073/pnas.1518382112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA, Borowsky B, Tamm JA, Craig DA, Durkin MM, Dai M, Yao WJ, Johnson M, Gunwaldsen C, Huang LY, Tang C, Shen Q, Salon JA, Morse K, Laz T, Smith KE, Nagarathnam D, Noble SA, Branchek TA, Gerald C (1998) GABA(B) receptors function as a heteromeric assembly of the subunits GABA(B)R1 and GABA(B)R2. Nature 396:674–679. 10.1038/25348 [DOI] [PubMed] [Google Scholar]
- Kim IH, Wang H, Soderling SH, Yasuda R (2014) Loss of Cdc42 leads to defects in synaptic plasticity and remote memory recall. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmse K, Kirischuk S (2006) Ambient GABA constrains the strength of GABAergic synapses at Cajal-Retzius cells in the developing visual cortex. J Neurosci 26:4216–4227. 10.1523/JNEUROSCI.0589-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Katsurabayashi S, Moorhouse AJ, Murakami N, Koga H, Akaike N (2003) GABAB receptor transduction mechanisms, and cross talk between protein kinases A and C, in GABAergic terminals synapsing onto neurons of the rat nucleus basalis of Meynert. J Physiol 551:263–276. 10.1113/jphysiol.2003.046524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladera C, del Carmen Godino M, Jose Cabanero M, Torres M, Watanabe M, Lujan R, Sánchez-Prieto J (2008) Pre-synaptic GABA receptors inhibit glutamate release through GIRK channels in rat cerebral cortex. J Neurochem 107:1506–1517. 10.1111/j.1471-4159.2008.05712.x [DOI] [PubMed] [Google Scholar]
- Laviv T, Riven I, Dolev I, Vertkin I, Balana B, Slesinger PA, Slutsky I (2010) Basal GABA regulates GABA(B)R conformation and release probability at single hippocampal synapses. Neuron 67:253–267. 10.1016/j.neuron.2010.06.022 [DOI] [PubMed] [Google Scholar]
- Leung LS, Peloquin P (2006) GABA(B) receptors inhibit backpropagating dendritic spikes in hippocampal CA1 pyramidal cells in vivo. Hippocampus 16:388–407. 10.1002/hipo.20168 [DOI] [PubMed] [Google Scholar]
- Liu J, Pasini S, Shelanski ML, Greene LA (2014) Activating transcription factor 4 (ATF4) modulates post-synaptic development and dendritic spine morphology. Front Cell Neurosci 8:177. 10.3389/fncel.2014.00177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehring RB, Horikawa HP, El Far O, Kneussel M, Brandstätter JH, Stamm S, Wischmeyer E, Betz H, Karschin A (2000) The metabotropic GABAB receptor directly interacts with the activating transcription factor 4. J Biol Chem 275:35185–35191. 10.1074/jbc.M002727200 [DOI] [PubMed] [Google Scholar]
- Pasini S, Corona C, Liu J, Greene LA, Shelanski ML (2015) Specific downregulation of hippocampal ATF4 reveals a necessary role in synaptic plasticity and memory. Cell Rep 11:183–191. 10.1016/j.celrep.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini S, Liu J, Corona C, Peze-Heidsieck E, Shelanski M, Greene LA (2016) Activating transcription factor 4 (ATF4) modulates ρ GTPase levels and function via regulation of RhoGDIalpha. Sci Rep 6:36952. 10.1038/srep36952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter B, Zschüntsch J, Kvachnina E, Zhang W, Ponimaskin EG (2004) The GABA(B) receptor subunits R1 and R2 interact differentially with the activation transcription factor ATF4 in mouse brain during the postnatal development. Brain Res Dev Brain Res 149:73–77. 10.1016/j.devbrainres.2003.12.006 [DOI] [PubMed] [Google Scholar]
- Rossignol E, Kruglikov I, van den Maagdenberg AM, Rudy B, Fishell G (2013) CaV 2.1 ablation in cortical interneurons selectively impairs fast-spiking basket cells and causes generalized seizures. Ann Neurol 74:209–222. 10.1016/j.neuroimage.2013.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaba T, Neher E (2003) Direct modulation of synaptic vesicle priming by GABA(B) receptor activation at a glutamatergic synapse. Nature 424:775–778. 10.1038/nature01859 [DOI] [PubMed] [Google Scholar]
- Speca DJ, Ogata G, Mandikian D, Bishop HI, Wiler SW, Eum K, Wenzel HJ, Doisy ET, Matt L, Campi KL, Golub MS, Nerbonne JM, Hell JW, Trainor BC, Sack JT, Schwartzkroin PA, Trimmer JS (2014) Deletion of the Kv2.1 delayed rectifier potassium channel leads to neuronal and behavioral hyperexcitability. Genes Brain Behav 13:394–408. 10.1111/gbb.12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger JL, Bandyopadhyay S, Farb DH, Russek SJ (2004) cAMP response element-binding protein, activating transcription factor-4, and upstream stimulatory factor differentially control hippocampal GABABR1a and GABABR1b subunit gene expression through alternative promoters. J Neurosci 24:6115–6126. 10.1523/JNEUROSCI.1200-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich D, Huguenard JR (1996) Gamma-aminobutyric acid type B receptor-dependent burst-firing in thalamic neurons: a dynamic clamp study. Proc Natl Acad Sci U S A 93:13245–13249. 10.1073/pnas.93.23.13245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon E, Meyer G, Pickard L, Dev K, Molnar E, Collingridge GL, Henley JM (2001) GABA(B) receptors couple directly to the transcription factor ATF4. Mol Cell Neurosci 17:637–645. 10.1006/mcne.2000.0960 [DOI] [PubMed] [Google Scholar]
- Vigot R, Barbieri S, Bräuner-Osborne H, Turecek R, Shigemoto R, Zhang YP, Luján R, Jacobson LH, Biermann B, Fritschy JM, Vacher CM, Müller M, Sansig G, Guetg N, Cryan JF, Kaupmann K, Gassmann M, Oertner TG, Bettler B (2006) Differential compartmentalization and distinct functions of GABAB receptor variants. Neuron 50:589–601. 10.1016/j.neuron.2006.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JH, McIllhinney RA, Wise A, Ciruela F, Chan WY, Emson PC, Billinton A, Marshall FH (2000) The GABAB receptor interacts directly with the related transcription factors CREB2 and ATFx. Proc Natl Acad Sci U S A 97:13967–13972. 10.1073/pnas.240452197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata J, Moretto E, Hannan S, Murru L, Longatti A, Mazza D, Benedetti L, Fossati M, Heise C, Ponzoni L, Valnegri P, Braida D, Sala M, Francolini M, Hildebrand J, Kalscheuer V, Fanelli F, Sala C, Bettler B, Bassani S, et al. (2017) Epilepsy and intellectual disability linked protein Shrm4 interaction with GABABRs shapes inhibitory neurotransmission. Nat Commun 8:14536. 10.1038/ncomms14536 [DOI] [PMC free article] [PubMed] [Google Scholar]