 A modular chemistry toolbox was developed for cereblon-directed PROTACs.
A modular chemistry toolbox was developed for cereblon-directed PROTACs.
Abstract
A modular chemistry toolbox was developed for cereblon-directed PROTACs. A variety of linkers was attached to a CRBN ligand via the 4-amino position of pomalidomide. We used linkers of different constitution to modulate physicochemical properties. We equipped one terminus of the linker with a set of functional groups, e.g. protected amines, protected carboxylic acids, alkynes, chloroalkanes, and protected alcohols, all of which are considered to be attractive for PROTAC design. We also highlight different opportunities for the expansion of the medicinal chemists' PROTAC toolbox towards heterobifunctional molecules, e.g. with biotin, fluorescent, hydrophobic and peptide tags.
A major pathway for protein degradation in eukaryotes is ubiquitin-dependent and relies on the sequential action of ubiquitin-activating (E1), -conjugating (E2), and -ligating (E3) enzymes. Out of the variety of human E3 ligases, cereblon (CRBN) has received particular attention as the primary target of immunomodulatory imide drugs (IMiDs), i.e. thalidomide, lenalidomide, and pomalidomide.1,2 IMiDs enhance the interaction between CRBN and the lymphoid transcription factors Ikaros (IKZF1) and Aiolos (IKZF3) what results in the ubiquitination and the subsequent proteasomal degradation of these neo-substrates.3–5
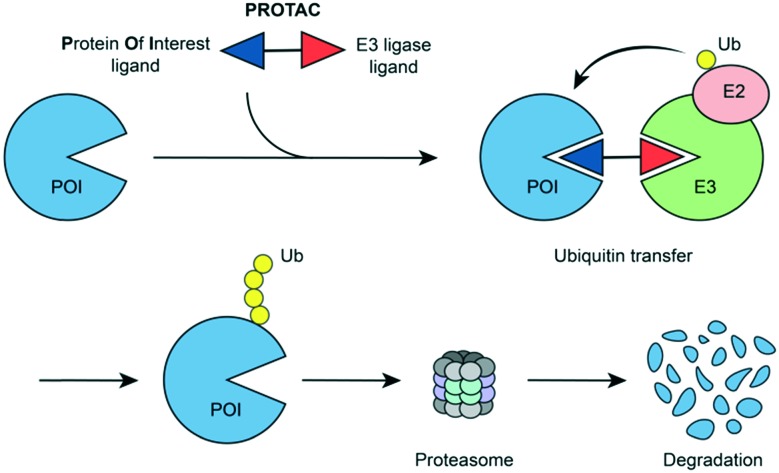
Tailored heterobifunctional molecules, which are referred to as proteolysis targeting chimeras (PROTACs), act through a chemical-induced degradation of disease-causing proteins. PROTACs recruit an E3 ligase to a protein of interest and induce the ubiquitination and proteasomal degradation (Fig. 1). The pioneering PROTAC technology has received exceptional attention in the field of drug discovery.6–10
Fig. 1. PROTAC-induced degradation of target proteins.
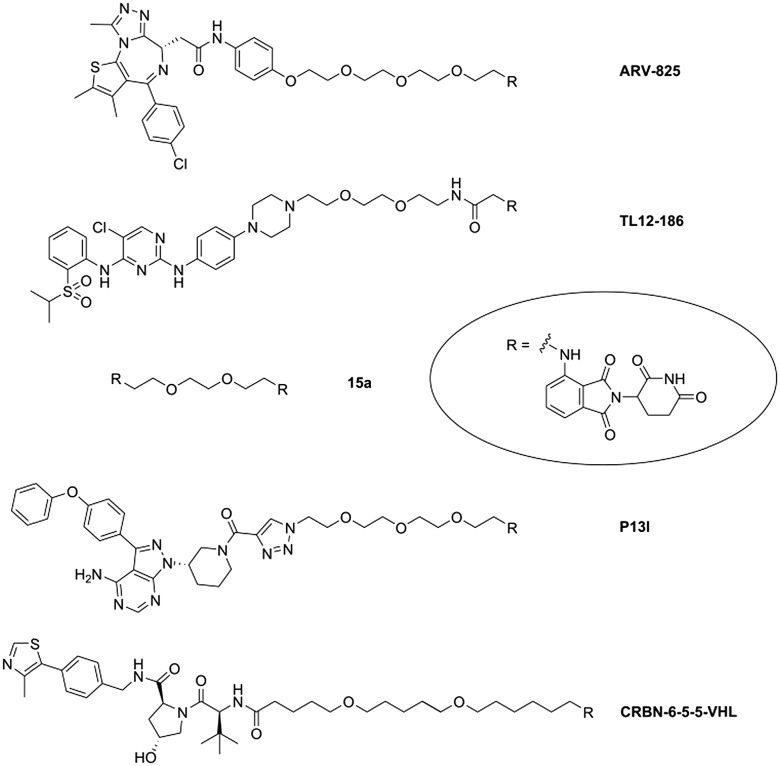
The molecular architecture of PROTACs comprises a target ligand and E3 ligase recruiting component, connected via a variable linker. Current PROTAC developments have been focused on two E3 ligases, i.e. Von-Hippel–Lindau (VHL) and CRBN. Strong affinity for the IMiD binding site of CRBN was achieved with derivatives of lenalidomide,9 4-hydroxy-thalidomide,11 and 4-aminothalidomide (pomalidomide).12,13 In numerous cases, the aromatic amino group of the latter bears the terminal aliphatic linker carbon, while different functional groups have been employed to connect the other linker terminus with the target ligand. Examples for such PROTACs with an N-alkylated pomalidomide unit (Fig. 2) include degraders of BRD4 (ARV-825),14 multiple kinases (TL12-186),15 or Bruton's tyrosine kinase (P13I).16,17 CRBN-6-5-5-VHL containing ligands for two ligases, VHL and CRBN, caused an unrequited ubiquitin transfer to CRBN and its degradation.18
Fig. 2. Examples of bifunctional CRBN-directed PROTACs.
Amino- and carboxy-functionalized building blocks can be utilized for amide coupling reactions to assemble heterobifunctional compounds, as it was also demonstrated for several PROTACs, e.g.ref. 14 and 18–21. Chloroalkyl moieties are easily deployable, through in situ generation of iodo analogues under Finkelstein conditions, for reactions with various nucleophilic partners. There are several corresponding examples in the PROTAC field, e.g.ref. 22.
When reacting haloalkyl groups with sodium azide, the resulting azido compounds can be subjected to Click reactions,23–25 an exquisitely convenient opportunity to produce heterobifunctional compounds, e.g.ref. 16, 26 and 27. As an alternative, Click chemistry would be possible if the linker structure is terminated with an alkyne moiety.28 A primary alcoholic group is convertible to esters and ethers, the latter transformation through Mitsunobu reaction is particularly attractive. Moreover, conversion of terminal alcohols to sulfonates confers electrophilic properties to the linker, and sulfonates can be used to generate haloalkyl derivatives, e.g.ref. 29. The Appel reaction can also be performed to obtain haloalkyl termini.30
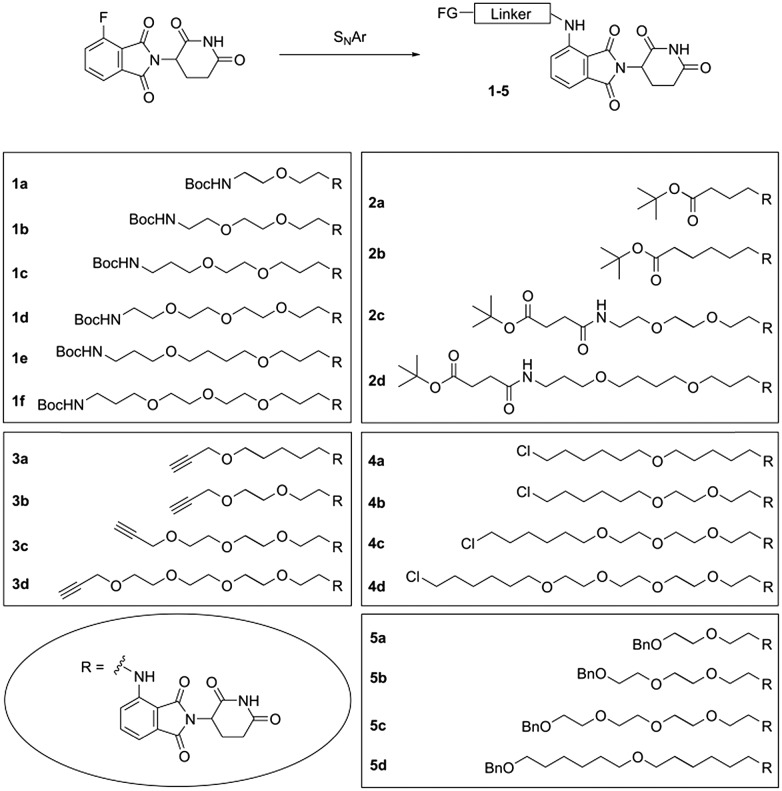
We have considered the aforementioned functionalities and designed a series of toolbox compounds. The common feature of all these toolbox members is the connection of the linker at the 4-amino group of pomalidomide, as it has already been successfully realized in a variety of PROTACs (Fig. 2). Our toolbox compounds with different polyalkylene ether linkers and several terminal functional groups are outlined in Fig. 3. We employed distinct synthetic routes to generate linker building blocks of various chemical nature.
Fig. 3. Toolbox compounds 1–5 with different linkers and various functional groups (FG).
It is well known that the biological activity of PROTACs is strongly affected by the physicochemical properties of linkers, in particular their length and hydrophobicity. Accordingly, to address the latter feature, different C/O ratios within the linker chains have been realized.
Toolbox compounds 1–5 were synthesized through a final nucleophilic aromatic substitution (SNAr) of 4-fluoro-thalidomide with the corresponding primary amines under conditions compatible with the present functional groups. To obtain the linker building blocks for compounds 1, di-tert-butyl dicarbonate was reacted with excess α,ω-diamine and such a hemiprotected intermediate (Scheme S1, ESI†) was subjected to the SNAr. Compounds 1 can easily be deprotected under acidic conditions, employed for amide bond formations to attach a target ligand and thus be incorporated into a PROTAC molecule. With respect to deprotection, Boc-protected tool compounds 1a–f were superior to the Cbz analogues 1g–k, which were also synthesized in the course of our study (see ESI†).
Compounds 2a–d (Scheme S2, ESI†) were newly prepared in addition to a previously reported series of tert-butyl protected precursor compounds.18 Prior to SNAr, a free amino group was liberated by hydrogenolysis of a Cbz group. As exemplified for 2c and 2d, succinic acid semi tert-butyl ester was prepared, connected with the respective mono-Cbz-protected diamine (Scheme S2, ESI†), Cbz-deprotected and finally subjected to the SNAr reaction.
For alkyne compounds 3, alkanolamines were used as starting materials (Scheme S3, ESI†). Tailored alkanolamines can be prepared by applying a route from diols through semi-mesylation, azidolysis, and Staudinger reaction.31 Next, the Boc-protected intermediates were converted to mesylates, which in turn were reacted with propargyl alcohol to obtain the desired alkyne linkers. The acidic conditions necessary for Boc removal were compatible with the C–C triple bond.
N-Boc-protected alkanolamines were again employed for the synthesis of chloro compounds 4 (Scheme S4, ESI†). A Williamson ether synthesis, when carried out with α-iodo-ω-chloroalkanes afforded chloroalkyl building blocks. Through deprotection and SNAr, compounds 4 were produced which allow for a variety of further chemical modifications.
O-Monobenzylated diols were subjected to the mesylation, azidolysis, Staudinger sequence (Scheme S5, ESI†) and applied to produce the protected hydroxyl derivatives 5a–c. In the case of 5d, 6-benzyloxy-1-hexanol was alkylated with 1-bromo-6-chlorohexane, followed by conversion to the corresponding azido and amino compound. The final SNAr reaction provided compounds 5.
The toolbox components 1 and 2, as well as 5, were approved for deprotection under appropriate acidic or hydrogenolytic conditions, which are tolerated by the phthalimide portion.18,32 The entire set of compounds 1–5 represents a valuable toolbox and is expected to support the synthetic entry to new heterobifunctional probes and degraders of therapeutically relevant proteins. The SMILES strings and calculated TPSA values for all compounds are given in Table S1 (ESI†).
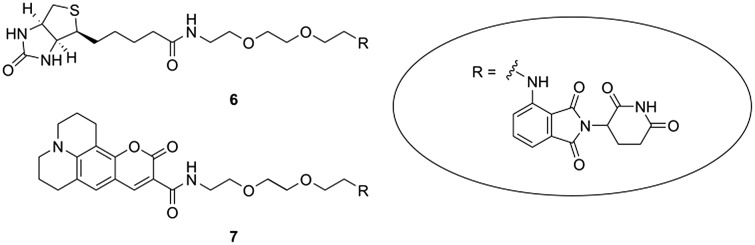
In the following section, we describe a few possible applications of these chemical tools for the generation of biologically applicable molecules. Biotinylation, the functional appendage of a biotin moiety to a bioactive compound, represents an attractive opportunity to elucidate and characterize intracellular binding partners. Biotin labelling takes advantage of the exceedingly strong interaction between biotin and either avidin or streptavidin.33,34 Only one biotinylated thalidomide analogue was accomplished so far by introducing a linker-connected biotin at the glutarimide nitrogen. This compound was developed for efforts towards the identification of thalidomide's targets.35 However, for CRBN-related research, the molecular architecture of the probe is not suitable since the CRBN binding moiety is rendered inoperative with a substituent at this position.18,32,36 Instead, we tethered biotin to one of our chemical tools, i.e.1b, and received the biotin-labelled, cell-permeable 6 (Fig. 4, Scheme S6, ESI†) with a preserved affinity for CRBN (Fig. S1, ESI†).
Fig. 4. CRBN-addressing probes 6 and 7 with a biotin and fluorescent tag.
Similarly, we prepared the fluorescently labelled compound 7 (Fig. 4, Scheme S6, ESI†) by introducing coumarin 343, a prototypical 7-donor-substituted coumarin, established for activity-based probing and intracellular imaging.37–41 Absorption and emission spectra of 7 were recorded from solutions in CH2Cl2, MeOH and phosphate buffered saline (PBS)/DMSO and Stokes shifts between 36 nm and 51 nm were obtained (Fig. S2, ESI†). Fluorescently labelled IMiDs, e.g. cyanine 5-coupled thalidomide, were shown to be exceptionally helpful in biomedical research, such as for the quantification of protein-compound interactions.2
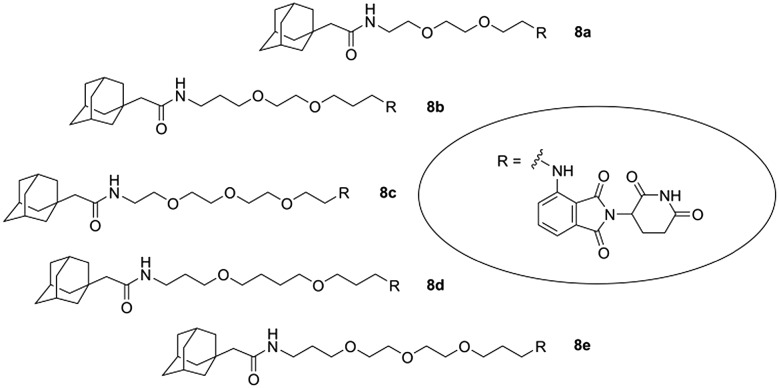
The strategy of hydrophobic tagging relies on the recognition of a hydrophobic moiety, appended to the surface of a protein. The hydrophobic attachment mimics a denatured protein folding state, engages the cellular quality machinery and induces proteasomal degradation. When a small bioactive molecule is fused to a hydrophobic moiety, it can trigger the degradation of a specific target protein.42–46 As a continuation of our PROTAC studies aimed at the degradation of CRBN,18,32 we intended to investigate whether the attachment of a hydrophobic tag via a linker to a CRBN ligand would also induce CRBN degradation. For this purpose, we selected five chemical tool compounds, 1b–f, and coupled them to 1-adamantaneacetic acid (Fig. 5, Scheme S7, ESI†). The adamantane cage constitutes a well-established hydrophobic tag with excellent stability which has repeatedly been employed for the design of cell-permeable degraders.42–46
Fig. 5. Hydrophobically tagged CRBN ligands 8.
An HPLC-based method was employed to determine log P values of hydrophobically tagged compounds 8 and those of the corresponding Boc-protected precursors 1b–f. The same shift to a higher hydrophobicity was observed for all adamantane analogues (Fig. S3, ESI†). We also determined partition coefficients for the new derivatives 2a–d (Fig. S4, ESI†) and eight previously described PROTAC precursors (Fig. S5, ESI†). The latter log P profiles were compared with literature values of the consequent CRBN-VHL PROTACs.18 A similar trend of the linker to influence the log P value was found in both subseries.
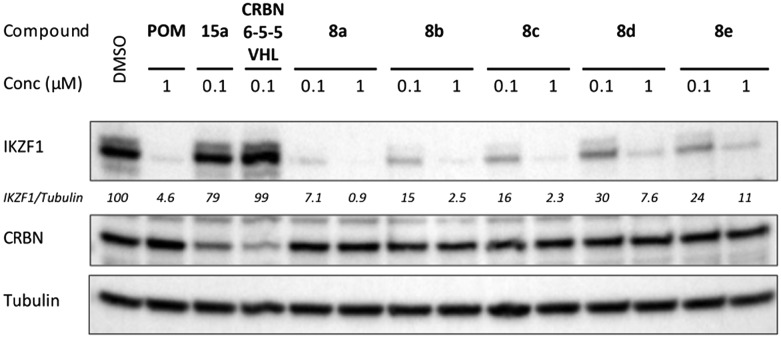
To study the ability of chimeras 8a–e for hydrophobic tagging of CRBN and inducing its degradation, MM1S cells were treated with each compound and western blot analyses were performed for CRBN and the neo-substrate IKZF1. No decrease of the CRBN protein level was observed after a 16 h treatment with any of the compounds at concentrations of 100 nM and 1 μM (Fig. 6). This finding suggests that the hydrophobic tag in compounds 8 did not cause proteasomal CRBN degradation, unlike the correspondingly incorporated second CRBN and VHL ligand in 15a (ref. 32, Fig. 2) and CRBN-6-5-5-VHL,18 respectively. However, IKZF1 levels were decreased in all cases, clearly indicating the cellular uptake of these compounds. Compound 8a decreased IKZF1 levels to a somewhat stronger extent than pomalidomide as it was shown at lower concentrations (Fig. S6, ESI†) and by analysing the time course of IKZF1 degradation (Fig. S7, ESI†).
Fig. 6. Hydrophobically tagged CRBN ligands 8 induce degradation of IKZF1, but not of CRBN. MM1S cells were treated for 16 h at the indicated concentrations (μM). For comparison, pomalidomide (POM), the CRBN homo-PROTAC 15a and the CRBN-VHL hetero-PROTAC CRBN-6-5-5-VHL were used. The quantification was performed with IKZF1 levels normalized to tubulin, then to DMSO control (100%).
In line with these data, the hydrophobically tagged compound 8a exhibited higher toxicity for the IMiD-sensitive multiple myeloma cell line MM1S than for LP1 cells (Fig. S8, ESI†). Hydrophobically tagged compounds of type 8 might be useful tools to study CRBN-dependent degradation of other cellular proteins, for example chaperons which recognize the hydrophobic tag and might be subjected to ubiquitination.
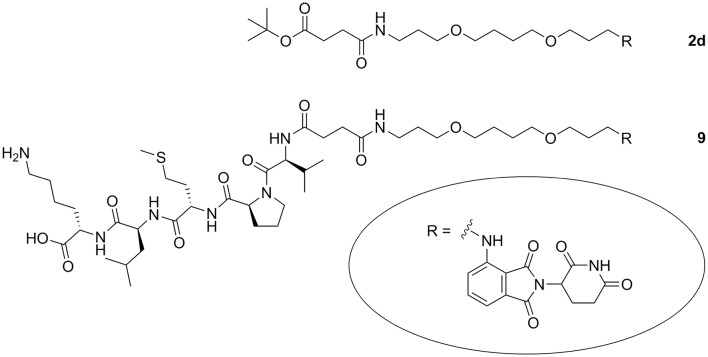
In order to investigate whether a peptidic label can be employed to facilitate cellular uptake of CRBN ligands, we designed a compound with a CRBN ligand as cargo which was linker-connected to a cell-penetrating peptide. The pentapeptide VPMLK based on the N-terminal sequence of the Bax binding domain of the human protein Ku-70 is a known membrane permeable peptide of very low toxicity.47,48 The peptide is synthetically accessible through Fmoc solid-phase chemistry.49 In order to generate the peptide as tert-butyl ester, we accomplished a new convergent solution-phase synthesis. After cleaving the N-Cbz group of the peptide and the tert-butyl ester of the toolbox compound 2d, both building blocks were combined. Final C-terminal deprotection provided the pomalidomide–peptide conjugate 9 (Fig. 7, Scheme S8, ESI†). An effect of 9 on the cellular protein level of the pro-apoptotic protein Bax was not observed. However, we took advantage of the neo-substrate degradation to estimate the cellular uptake of 9 in comparison with pomalidomide and compound 2d. These three CRBN binders are clearly distinguished by their physicochemical properties, but did all induce IKZF1 degradation (Fig. S9, ESI†). Although 9 was not superior to pomalidomide and 2d (Fig. S10, ESI†), we still consider the utilization of cell penetrating tags to be a promising opportunity for PROTAC development.
Fig. 7. CRBN ligand coupled to a linker (2d) and to a cell-penetrating peptide (9).
The comprehensive chemical toolbox introduced herein was developed to take account of the different chemistries of target protein ligands onto which the toolbox compounds have to be coupled. The set of compounds is expected to be useful owing to its broad applicability for PROTAC design. Moreover, toolbox members can be employed to generate heterobifunctional molecules beyond protein degraders, which was demonstrated in this study.
This work was supported by the DFG (Emmy-Noether Program Kr-3886/2-1 and SFB-1074 to JK and FOR2372 to MG). C.S. received a scholarship from the Bonn International Graduate School of Drug Sciences. I.S. acknowledges a Short-Term Fellowship from FEBS and support from the Slovenian Research Agency (ARRS programme P1-0208). We thank M. Schneider, S. Terhart-Krabbe, A. Reiner and Dr. E. Gilberg for support.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Footnotes
†Electronic supplementary information (ESI) available: Fig. S1–S10 and Table S1; biological and chemical methods; synthetic Schemes S1–S8 and procedures; log P, 1H NMR, 13C NMR and MS data. See DOI: 10.1039/c9md00185a
References
- Ito T., Ando H., Suzuki T., Ogura T., Hotta K., Imamura Y., Yamaguchi Y., Handa H. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- Fischer E. S., Böhm K., Lydeard J. R., Yang H., Stadler M. B., Cavadini S., Nagel J., Serluca F., Acker V., Lingaraju G. M., Tichkule R. B., Schebesta M., Forrester W. C., Schirle M., Hassiepen U., Ottl J., Hild M., Beckwith R. E., Harper J. W., Jenkins J. L., Thomä N. H. Nature. 2014;512:49–53. doi: 10.1038/nature13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krönke J., Udeshi N. D., Narla A., Grauman P., Hurst S. N., McConkey M., Svinkina T., Heckl D., Comer E., Li X., Ciarlo C., Hartman E., Munshi N., Schenone M., Schreiber S. L., Carr S. A., Ebert B. L. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Middleton R. E., Sun H., Naniong M., Ott C., Mitsiades C. S., Wong K. K., Bradner J. E., Kaelin, Jr. W. G. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krönke J., Fink E. C., Hollenbach P. W., MacBeth K. J., Hurst S. N., Udeshi N. D., Chamberlain P. P., Mani D. R., Man H. W., Gandhi A. K., Svinkina T., Schneider R. K., McConkey M., Järås M., Griffiths E., Wetzler M., Bullinger L., Cathers B. E., Carr S. A., Chopra R., Ebert B. L. Nature. 2015;523:183–188. doi: 10.1038/nature14610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins I., Wang H., Caldwell J. J., Chopra R. Biochem. J. 2017;474:1127–1147. doi: 10.1042/BCJ20160762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churcher I. J. Med. Chem. 2018;61:444–452. doi: 10.1021/acs.jmedchem.7b01272. [DOI] [PubMed] [Google Scholar]
- Zoppi V., Hughes S. J., Maniaci C., Testa A., Gmaschitz T., Wieshofer C., Koegl M., Riching K., Daniels D. L., Spallarossa A., Ciulli A. J. Med. Chem. 2019;62:699–726. doi: 10.1021/acs.jmedchem.8b01413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Ding N., Song Y., Yang Z., Liu W., Zhu J., Rao Y., Leukemia, 2019. 10.1038/s41375-019-0440-x , , Epub ahead of print . [DOI] [PubMed] [Google Scholar]
- Dobrovolsky D., Wang E. S., Morrow S., Leahy C., Faust T., Nowak R. P., Donovan K. A., Yang G., Li Z., Fischer E. S., Treon S. P., Weinstock D. M., Gray N. S. Blood. 2019;133:952–961. doi: 10.1182/blood-2018-07-862953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohbeck J., Miller A. K. Bioorg. Med. Chem. Lett. 2016;26:5260–5262. doi: 10.1016/j.bmcl.2016.09.048. [DOI] [PubMed] [Google Scholar]
- Boichenko I., Deiss S., Bär K., Hartmann M. D., Hernandez Alvarez B. J. Med. Chem. 2016;59:770–774. doi: 10.1021/acs.jmedchem.5b01735. [DOI] [PubMed] [Google Scholar]
- Boichenko I., Bär K., Deiss S., Heim C., Albrecht R., Lupas A. N., Hernandez Alvarez B., Hartmann M. D. ACS Omega. 2018;3:11163–11171. doi: 10.1021/acsomega.8b00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Qian Y., Altieri M., Dong H., Wang J., Raina K., Hines J., Winkler J. D., Crew A. P., Coleman K., Crews C. M. Chem. Biol. 2015;2:755–763. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. T., Dobrovolsky D., Paulk J., Yang G., Weisberg E. L., Doctor Z. M., Buckley D. L., Cho J. H., Ko E., Jang J., Shi K., Choi H. G., Griffin J. D., Li Y., Treon S. P., Fischer E. S., Bradner J. E., Tan L., Gray N. S. Cell Chem. Biol. 2018;25:88–99. doi: 10.1016/j.chembiol.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Zhao X., Ding N., Gao H., Wu Y., Yang Y., Zhao M., Hwang J., Song Y., Liu W., Rao Y. Cell Res. 2018;28:779–781. doi: 10.1038/s41422-018-0055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Wang J., Yao X., Zheng W., Mao Y., Lan T., Wang L., Sun Y., Zhang X., Zhao Q., Zhao J., Xiao R., Zhang X., Ji G., Rao Y. Cell Discovery. 2019;5:10. doi: 10.1038/s41421-018-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinebach C., Kehm H., Lindner S., Vu L. P., Köpff D., López Mármol Á, Weiler C., Wagner K. G., Reichenzeller M., Krönke J., Gütschow M. Chem. Commun. 2019;55:1821–1824. doi: 10.1039/c8cc09541h. [DOI] [PubMed] [Google Scholar]
- Ishoey M., Chorn S., Singh N., Jaeger M. G., Brand M., Paulk J., Bauer S., Erb M. A., Parapatics K., Müller A. C., Bennett K. L., Ecker G. F., Bradner J. E., Winter G. E. ACS Chem. Biol. 2018;13:553–560. doi: 10.1021/acschembio.7b00969. [DOI] [PubMed] [Google Scholar]
- Olson C. M., Jiang B., Erb M. A., Liang Y., Doctor Z. M., Zhang Z., Zhang T., Kwiatkowski N., Boukhali M., Green J. L., Haas W., Nomanbhoy T., Fischer E. S., Young R. A., Bradner J. E., Winter G. E., Gray N. S. Nat. Chem. Biol. 2018;14:163–170. doi: 10.1038/nchembio.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L., Zhou B., Yang C. Y., Ji J., McEachern D., Przybranowski S., Jiang H., Hu J., Xu F., Zhao Y., Liu L., Fernandez-Salas E., Xu J., Dou Y., Wen B., Sun D., Meagher J., Stuckey J., Hayes D. F., Li S., Ellis M. J., Wang S. Cancer Res. 2017;77:2476–2487. doi: 10.1158/0008-5472.CAN-16-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A. C., Toure M., Hellerschmied D., Salami J., Jaime-Figueroa S., Ko E., Hines J., Crews C. M. Angew. Chem., Int. Ed. 2016;55:807–810. doi: 10.1002/anie.201507634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkel S. S., Dirks A. T., Debets M. F., van Delft F. L., Cornelissen J. J., Nolte R. J., Rutjes F. P. ChemBioChem. 2007;8:1504–1508. doi: 10.1002/cbic.200700278. [DOI] [PubMed] [Google Scholar]
- Meldal M., Tornøe C. W. Chem. Rev. 2008;108:2952–3015. doi: 10.1021/cr0783479. [DOI] [PubMed] [Google Scholar]
- Debets M. F., van Hest J. C., Rutjes F. P. Org. Biomol. Chem. 2013;11:6439–6455. doi: 10.1039/c3ob41329b. [DOI] [PubMed] [Google Scholar]
- Schiedel M., Herp D., Hammelmann S., Swyter S., Lehotzky A., Robaa D., Oláh J., Ovádi J., Sippl W., Jung M. J. Med. Chem. 2018;61:482–491. doi: 10.1021/acs.jmedchem.6b01872. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Lan T., Su S., Rao Y. Chem. Commun. 2019;55:369–372. doi: 10.1039/c8cc07813k. [DOI] [PubMed] [Google Scholar]
- Wurz R. P., Dellamaggiore K., Dou H., Javier N., Lo M. C., McCarter J. D., Mohl D., Sastri C., Lipford J. R., Cee V. J. J. Med. Chem. 2018;61:453–461. doi: 10.1021/acs.jmedchem.6b01781. [DOI] [PubMed] [Google Scholar]
- Smith B. E., Wang S. L., Jaime-Figueroa S., Harbin A., Wang J., Hamman B. D., Crews C. M. Nat. Commun. 2019;10:131. doi: 10.1038/s41467-018-08027-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kalkeren H. A., van Delft F. L., Rutjes F. P. Pure Appl. Chem. 2013;85:817–828. [Google Scholar]
- Prokorski J. K., Breitenkamp K., Liepold L. O., Qazi S., Finn M. G. J. Am. Chem. Soc. 2011;133:9242–9245. doi: 10.1021/ja203286n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinebach C., Lindner S., Udeshi N. D., Mani D. C., Kehm H., Köpff S., Carr S. A., Gütschow M., Krönke J. ACS Chem. Biol. 2018;13:2771–2782. doi: 10.1021/acschembio.8b00693. [DOI] [PubMed] [Google Scholar]
- Trippier P. C. ChemMedChem. 2013;8:190–203. doi: 10.1002/cmdc.201200498. [DOI] [PubMed] [Google Scholar]
- Embaby A. M., Schoffelen S., Kofoed C., Meldal M., Diness F. Angew. Chem., Int. Ed. 2018;57:8022–8026. doi: 10.1002/anie.201712589. [DOI] [PubMed] [Google Scholar]
- Stewart S. G., Braun C. J., Polomska M. E., Karimi M., Abraham L. J., Stubbs K. A. Org. Biomol. Chem. 2010;8:4059–4062. doi: 10.1039/c0ob00060d. [DOI] [PubMed] [Google Scholar]
- Zhou B., Hu J., Xu X., Chen Z., Bai L., Fernandez-Salas E., Lin M., Liu L., Yang C. Y., Zhao Y., McEachern D., Przybranowski S., Wen B., Sun D., Wang S. J. Med. Chem. 2018;61:462–481. doi: 10.1021/acs.jmedchem.6b01816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnes R. S., Jernigan F., Shell J. R., Sharma V., Lawrence D. S. J. Am. Chem. Soc. 2010;132:6075–6080. doi: 10.1021/ja909652q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens M. D., Schmitz J., Horn M., Furtmann N., Bajorath J., Mareš M., Gütschow M. ChemBioChem. 2014;15:955–959. doi: 10.1002/cbic.201300806. [DOI] [PubMed] [Google Scholar]
- Kohl F., Schmitz J., Furtmann N., Schulz-Fincke A. C., Mertens M. D., Küppers J., Benkhoff M., Tobiasch E., Bartz U., Bajorath J., Stirnberg M., Gütschow M. Org. Biomol. Chem. 2015;13:10310–10323. doi: 10.1039/c5ob01613d. [DOI] [PubMed] [Google Scholar]
- Cottam Jones J. M., Harris P. W., Scanlon D. B., Forbes B. E., Brimble M. A., Abell A. D. Org. Biomol. Chem. 2016;14:2698–2795. doi: 10.1039/c5ob02110c. [DOI] [PubMed] [Google Scholar]
- Schulz-Fincke A. C., Tikhomirov A. S., Braune A., Girbl T., Gilberg E., Bajorath J., Blaut M., Nourshargh S., Gütschow M. Biochemistry. 2018;57:742–752. doi: 10.1021/acs.biochem.7b00906. [DOI] [PubMed] [Google Scholar]
- Neklesa T. K., Tae H. S., Schneekloth A. R., Stulberg M. J., Corson T. W., Sundberg T. B., Raina K., Holley S. A., Crews C. M. Nat. Chem. Biol. 2011;7:538–543. doi: 10.1038/nchembio.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson J. L., Neklesa T. K., Cox C. S., Roth A. G., Buckley D. L., Tae H. S., Sundberg T. B., Stagg D. B., Hines J., McDonnell D. P., Norris J. D., Crews C. M. Angew. Chem., Int. Ed. 2015;54:9659–9662. doi: 10.1002/anie.201503720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromm P. M., Crews C. M. Cell Chem. Biol. 2017;24:1181–1190. doi: 10.1016/j.chembiol.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubner S., Scharow A., Schubert S., Berg T. Angew. Chem., Int. Ed. 2018;57:17043–17047. doi: 10.1002/anie.201809640. [DOI] [PubMed] [Google Scholar]
- Rubner S., Schubert S., Berg T. Org. Biomol. Chem. 2019;17:3113–3117. doi: 10.1039/c9ob00080a. [DOI] [PubMed] [Google Scholar]
- Gomez J. A., Gama V., Yoshida T., Sun W., Hayes P., Leskov K., Boothman D., Matsuyama S. Biochem. Soc. Trans. 2007;35:797–801. doi: 10.1042/BST0350797. [DOI] [PubMed] [Google Scholar]
- Gomez J., Matsuyama S. Methods Mol. Biol. 2011;683:465–471. doi: 10.1007/978-1-60761-919-2_33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Tomioka I., Nagahara T., Holyst T., Sawada M., Hayes P., Gama V., Okuno M., Chen Y., Abe Y., Kanouchi T., Sasada H., Wang D., Yokota T., Sato E., Matsuyama S. Biochem. Biophys. Res. Commun. 2004;321:961–966. doi: 10.1016/j.bbrc.2004.07.054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.