Abstract
For more than 40 years, protein-polymer conjugates have been widely used for many applications, industrially and biomedically. These bioconjugates have been shown to modulate the activity and stability of various proteins while introducing reusability and new activities that can be used for drug delivery, improve pharmacokinetic ability, and stimuli-responsiveness. Techniques such as RDRP, ROMP and “click” have routinely been utilized for development of well-defined bioconjugate and polymeric materials. Synthesis of bioconjugate materials often take advantage of natural amino acids present within protein and peptide structures for a host of coupling chemistries. Polymer modification may elicit increased or decreased activity, activity retention under harsh conditions, prolonged activity in vivo and in vitro, and introduce stimuli responsiveness. Bioconjugation has resulted to modulated thermal stability, chemical stability, storage stability, half-life and reusability. In this review we aim to provide a brief state of the field, highlight a wide range of behaviors caused by polymer conjugation, and provide areas of future work.
GRAPHICAL ABSTRACT:
This review article discusses the impact of polymer modification on bioconjugate performance, including both activity and stability, with a focus on how the polymer structure and functionality impact these parameters.
Introduction
Biological macromolecules have been studied as potential targets for the treatment of human diseases1, 2, biofuel synthesis3, commercial detergents4, production of food5, and gene delivery.6 Synthetically modulating the inherent activity and stability of these biomolecules has received substantial attention. In biochemistry, post-translational modifications are known to mediate proper protein folding, improved stability, facilitate specific interactions, and increase function.7 The conjugation of polymers, controllable chains of repeating monomer units, to biomolecules is assumed to be an effective synthetic mimic of post-translational modification, which occurs naturally in eukaryotic cells.
The synthesis of the first industrial polymer is credited to John Wesley Hyatt for his simplified synthesis of celluloids in 1863. 90 years later, Hermann Staudinger received a Nobel Prize in Chemistry for the macromolecules he characterized as polymers.8 Since this time, synthetic polymers have been imperative to many medical and infrastructural advances.9, 10 Hybrid polymers, biomolecule-polymer conjugates, are produced upon the conjugation of biological polymers to synthetic polymers, resulting in new functionalities. Davis and Abuchowski’s ground-breaking work in 1977, showed that amino acid side chains are available for polymer and small molecule conjugation.11 The group covalently conjugated polyethylene glycol (PEG) to bovine serum albumin (BSA), a process that has been since referred to as pegylation; which showed increased protein activity, proteolytic resistance, thermal stability, and pH stability.12 This work laid the groundwork for the work of the pioneering work of Ruth Duncan13–18 and Helmut Ringsdorf18–21. In addition to laying the foundation to a field that has revolutionized the use of polymers, most polymer conjugation methods are effective and facile while producing highly desirable results.
Protein-polymer bioconjugates exhibit a unique array of properties and can be tuned to produce desired effects for specific biomaterials (Fig 1). Klok and Gauthier highlight the opportunities offered by new trends in polymer conjugation, including polymer functionality and coupling strategy.22 The pair also discuss the influence of polymer conjugation on biological activity. These hybrid polymers are synthesized using grafting from and grafting to approaches which can be site-specific or randomized. Previous research has shown these conjugations have effects similar to post-translational modification and has been shown to influence protein localization and activity. Since the production of BSA-PEG by Davis and Abuchowski, scientists have studied the effects of polymer conjugates on a plethora of biomolecules for specific applications targeted toward disease treatments, bioimaging, drug delivery, bioactive surfaces, and tissue engineering.
Fig 1.
Polymer conjugation effects many properties of the native protein and is capable of increasing applicability
A significant feature of bioconjugate chemistry is its interface with precision polymer chemistry and synthesis. A target for polymer bioconjugate chemistry is to introduce the chemical and structural diversity and precision of modern synthetic polymer chemistry to biological materials. Indeed, for much of the history of polymer bioconjugation, the functionality was relatively limited to PEG like materials, or polymers with poorly controlled underlying polymer structure.23 However, with the advent of controlled and living polymerization methods that are compatible with a wide range of monomers under bio-friendly conditions, it is possible to precisely engineer the structure and functionality of the polymer attached to the biomolecule of interest. This ability to precisely define and grow the polymer is important for both industrial and biomedical applications of polymer bioconjugates. PEG is typically synthesized by a ring opening reaction of ethylene oxide, which typically is done under strictly anhydrous conditions, making the synthesis of PEG and other related polymers challenging under biologically relevant conditions.24 Indeed, the development of reversible deactivation radical polymerization (RDRP) methods in the 1990s, and as well as ring opening metathesis polymerization (ROMP) enabled well defined polymers to be synthesized under mild conditions and for biocompatible monomers.25, 26 Of the RDRP techniques, atom transfer radical polymerization (ATRP), and reversible addition fragmentation chain transfer polymerization (RAFT) have been extensively explored in bioconjugation processes. 27, 28
Concurrent with advances in polymer chemistry, there have been advances in organic chemistry relevant to bioconjugates. “Click” chemistry, or reactions which are mild, high yielding and proceed with specificity and minimal to no required purification are especially well suited to bioconjugation processes.29 The possibility of post-polymerization modification enabled by “click” chemistry and the ability to efficiently ligate polymers to biomolecules through these “click” reactions has greatly expanded the types of bioconjugation reactions and biohybrid materials possible.
RDRP, ROMP and “click” techniques have been shown to be well suited to the development of well-defined bioconjugate and polymeric materials with access to a diverse range of monomers and polymer architectures. As part of this review, we will highlight how the polymer chosen, often as enabled through these modern techniques, enables protein function to be modulated through bioconjugation. This review will explore the effect of polymer conjugation on protein activity and stability, identifying key examples where polymer modification has impacted the biomolecule. However, other types of bioconjugates including nucleotide-polymer conjugates will be discussed as well to highlight the versatility of polymer chemistry in bioconjugation applications. A key feature of this review is a focus on how polymer chemistry, and often precision in polymer chemistry, can enable powerful bioconjugate materials with distinct advantages in activity or stability compared to the native protein or other biopolymer. This review article highlights key examples of polymer modifications of enzymatic proteins, which make substantial impacts on the biomolecules performance, be it activity, stability or both. This serves as an article that highlights the potential of protein modification by synthetic polymers on the performance of the biomolecule.
Synthesis of Bioconjugates
Polymerization
Since the discovery of PEGylation, PEG has been widely used to enhance various proteins. The polymer chains used in these experiments have traditionally been synthesized using ring-opening polymerization (ROP) of ethylene oxide.30 ROP is a polymerization mechanism in which cyclic monomers are converted to polymeric chain.31 Pegylation is the covalent or non-covalent attachment of PEG to amino acids residues present on the protein of interest. The methods used for PEGylation is highly dependent on available amino acids within protein of interest and functional end-groups available on polymer chains purchased or synthesized. In addition, more complex PEG structures can be grown using Ring-Opening Metathesis Polymerization (ROMP), as highlighted by Pokorski and Isarov.32 The ROMP method allows polymer chains to be grown from the surface of the protein under aqueous conditions, which would be challenging for ionic ROP of ethylene oxide.
In addition to PEGylation, non-PEG polymers have been used in polymer conjugations to achieve more specific results, increased solubility, and stimuli responsive behaviors. RDRP methods have become very popular techniques for bioconjugations due to their relatively simple reactions, mild conditions, compatibility with aqueous media and ability to control the structure of synthesized polymers. These techniques include nitroxide-mediated radical polymerization (NMP), reversible addition-fragmentation chain transfer polymerization (RAFT), and atom transfer radical polymerization (ATRP).33–35 NMP, discovered in 1982, was the first technique for controlling radical polymerization.33 These techniques takes advantage of the radical trapping nature of nitroxides and the radical stabilizing potential of alkoxyamines.36 ATRP is a commonly used controlled polymerization technique, first discovered in 1995.35 The technique utilized a transition metal catalyst that determines the equilibrium between dormant and active species.27 RAFT was first reported in 1998 and has become a popular technique can be used with a wide variety of monomers and reaction conditions while maintaining controlled molecular weight polymers.34 This polymerization technique utilizes chain transfer agents to control the rate of polymerization.28
Biomolecule-polymer conjugates can be synthesized via several approaches: grafting to, grafting from and grafting through. These methods most commonly utilize naturally occurring amino acid residues within the protein of interest, although there is important work on the incorporation of non-natural amino acid residues, and the attachment of various types of polymers.
Choice of Polymer and Linker
The types of polymers that can be used in bioconjugation are limited only by the polymerization method and solubility under application conditions. Therefore, water-soluble polymers are typically used due to the vast majority of bioconjugate applications occurring in aqueous media. However, it is important to note that polymers with hydrophobic character can be important for applications that involve hydrophobic substrates or oil rich environments.37 The excellent compatibility of RDRP and ROMP methods with water-soluble polymers makes these RDRP reactions attractive for various bioconjugate applications. In addition to the choice of polymer, the type of linker must be taken into consideration. Chen and coworkers attached two types of chain transfer agents to pyrophosphatase, CTA with a maleimide end-group and CTA with a pyridine end-group.38 After grafting poly(NIPAm) from the linkers, conjugation of the CTA with a pyridine end-group showed full retention of activity while conjugation of CTA with a maleimide end-group showed inactivation of the enzyme.
Grafting to Method
Grafting to refers to the conjugation approach in which the polymer is synthesized first and subsequently attached to the protein or peptide through the utilization of efficient organic chemistry reactions (Fig. 2A).39 These reactions can include well established organic reactions such as amidation or Michael additions, as well as more recently explored “click” reactions. Traditional reactions take advantage of naturally occurring residues, which will be discussed subsequently, while “click” approaches often require incorporation of bioorthogonal reactive handles such as azide or alkyne groups.40 This technique can be engineered for site-specific or random conjugation and, when compared to other conjugation methods, provides simple and thorough characterization of polymer before conjugation.41 Also, when using this method, the biomolecule remains unaffected by polymerization methods. However, high molecular weight polymers may inhibit effective conjugation, and purification after conjugation can be challenging.
Fig. 2.
(A) Grafting to and (B) Grafting from approaches for protein-polymer conjugation. mages made in PyMOL using BSA from PDB 3V03.43
Grafting from Method
Grafting from refers to the conjugation approach in which a small molecule initiator or chain transfer agent is attached to the biomolecule at specific or random locations and subsequently grows a polymer directly from the biomaterial in the presence of monomer in an appropriate solution (Fig 2B).39 Similarly, to the earlier discussed techniques, grafting from can be designed for site-specific or random attachment.41 The initiator or transfer agent can be attached to either naturally occurring or non-natural functional groups. Appropriately designed traditional amidation and Michael addition, as well as “click” reactions can be used to efficiently attach the initiating site to the biomolecule. When compared to other conjugation methods, grafting-from provides simple purification after conjugation and higher grafting density. However, this method may lead to protein instability based on polymerization method used and polymer chains that are not always well-controlled if the polymerization conditions needed for biomolecule stability leads to suboptimal polymerization conditions.
Grafting through Method
In the case of grafting through a polymerizable group, such as a double bond, is attached to the biomolecule. The grafting through method, is similar to the grafting from method, in that polymerization happens during bioconjugation. However, in grafting through, the protein containing the polymerizable unit is added to the growing polymer chain as a pendant group.42 However, this method is not as common as either grafting to or grafting from.
Common Amino acids used in bioconjugations
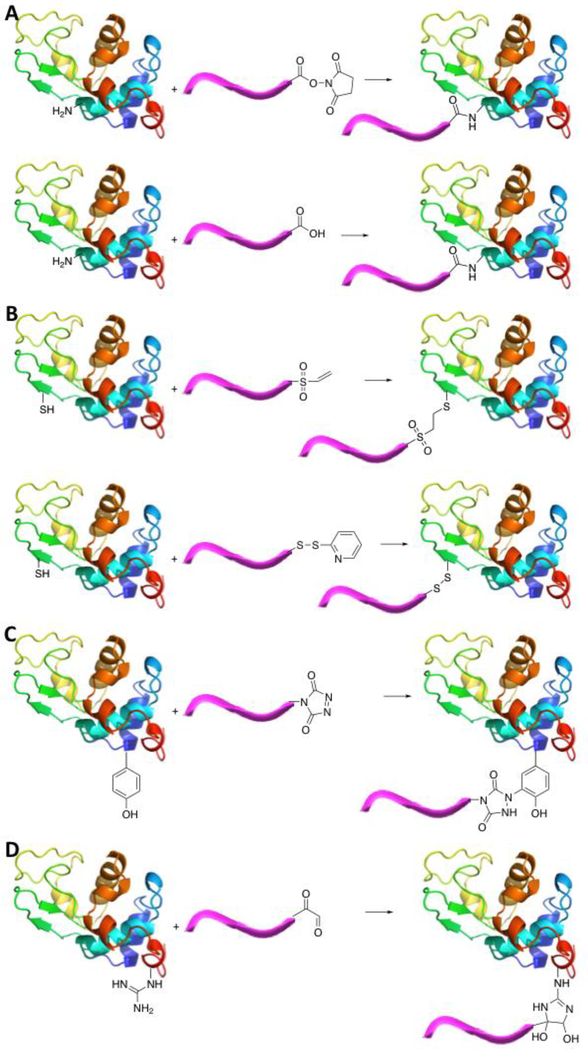
Davis and coworkers first attached activated PEG chains to solvent exposed primary amines within BSA. Lysine residues and the amino-terminus contain primary amines which, when solvent exposed, allow for polymer conjugation. These primary amines can be used in grafting-to, grafting-from and grafting-through polymer conjugation methods. Amidation, which is the formation of an amide bond between the polymer and protein, usually takes place using the following methods: N-succinimidyl ester functionalized polymers coupled to primary amines and EDC/NHS carbodiimide crosslinking chemistry (Fig. 3A).44, 45 Russell et al. elegantly developed a strategy for predicting the sequence of modification of amine residues using tertiary structure information, and it is possible this method could be used in the future for other functional groups.46
Fig. 3.
Coupling chemistries used for protein-polymer conjugation utilizing (A) lysine residues and amino-terminus, (B) cysteine residues, (C) tyrosine residues, and (D) arginine residues. Image made in PyMOL using Hen Egg White Lysozyme from PDB 1AKI.58
Organic chemistry reactions to attach polymers chains to a variety of amino acids such as cysteine, tyrosine, arginine, histidine, and non-natural amino acids have proven quite successful. Cysteine residues are composed of free thiols which allow for facile polymer conjugation through click chemistry and disulfide formation. The thiol functional groups are available for disulfide bond formation with 2-methacryloyloxyethyl phosphorylchlorine (MPC) functionalized polymers or initiators and thiol-ene click chemistry with maleimide- or divinyl sulfone-functionalized polymers or initiators (Fig. 3B).47–49
Tyrosine residues possess side chains with a phenol group, which can undergo ligation with modified polymer chains. The phenol group is available for electrophilic aromatic substitution reactions with triazolinedione functionalized polymers (Fig. 3C).50
The arginine side chain harbors a guanidine group, and while less reactive than lysine, it can be used for selective modification. Arginine residues are inherently less reactive than the amino terminus and lysine residues. Conjugation using these residues allows for site-specific conjugation through modification with α-oxo-aldehyde-functionalized polymer chains (Fig. 3D).51
Non-natural amino acids have been genetically incorporated into proteins as a method of specific polymer conjugation. These amino acids are often comprised of polymer initiators which are used as stable linkages on the protein into promote polymer chain growth.52 Alternatively, non-natural amino acids containing azide or alkyne functional groups which are compatible with Cu catalyzed “click” cycloaddition chemistry, as well as other functional groups compatible with the family of “click” reactions.53 There are a number of polymer attachment techniques, in addition to the methods discussed above, discussed in reviews by Maynard et al.23, Weck et al.54, Klok et al.55, Perrier et al.56, and Wu et al.57
Confirmation of Bioconjugate Synthesis
Protein-polymer conjugation has typically been confirmed by evaluating the size and mass of the resulting biohybrid material. Polyacrylamide Gel Electrophoresis (PAGE) is a biochemical technique traditionally used to approximate protein molecular weight and purity. Polymer conjugation tends to significantly increase molecular weight which can easily be seen using PAGE. Interactions between PEG and sodium dodecyl sulfate (SDS) presents challenges with this technique. The smeared or broaden bands associated with modified protein result in the inability to give clear separation of mixtures containing proteins modified at various sites, free polymer, and unmodified protein.59 Su and coworker found that native PAGE greatly reduced these interactions and sharpens bands associated with modified proteins, this results in overall greater resolution.59
Matrix assisted laser desorption/ionization-time of flight (MALDI-ToF) is an analytical technique used to ionize samples into charged molecules which allows the mass-to-charge (m/z) ratio to be measured to determine the mass of the ionized protein. Polymer conjugation tends to cause an increased and broadened m/z of ionized protein.60 As mentioned before, one of the major challenges associated with characterization of modified proteins is heterogeneity of conjugation site and degree of polymer modification. MALDI-ToF can be used to calculate degree of polymer modification based on separation of mass of peaks.
Polymer conjugation has also been confirmed using liquid chromatography-mass spectrometry (LC/MS),61 high performance liquid chromatography (HPLC),62 and reversed-phase chromatography (RPC).63 Roffler and co-workers examine a host of analytical techniques to characterize pegylated molecules in their 2011 review.61 LC/MS is an analytical technique used to physically separate compounds based on size while determining mass. As seen with PAGE and MALDI-TOF, polymer conjugation increases in molecular weight, which is shown as an increase in m/z.64
In addition to conjugation confirmation, the purity of the bioconjugate should be considered. Researchers have used techniques such as filtration65, dialysis66, 67, and chromatography23, 68 to remove unreacted polymers.
Effect of Polymer Conjugation on Activity
The work of Davis and Abuchowski showed PEGylation was capable of increasing esterase activity of BSA which in time has led researchers to use PEG and non-PEG synthetic polymers in the hopes of increasing the activity of various proteins with important biological or industrial applications. These studies have shown that the conjugation of PEG and non-PEG polymers to biomolecules can result in modulated and tunable activity. This section will investigate the effect of polymer modification on the activity of various proteins with targeted industrial and biomedical applications.
Model Proteins
Proteins that have well-understood and characterized functions and structures have served as models for polymer conjugations, some of the most popular proteins used are hen egg white lysozyme (HEWL), bovine serum albumin (BSA), and α-chymotrypsin. The proteins are some of the most studied protein-polymer conjugates due to lots of literature being published about their individual substrate preferences, optimum assay conditions, crystal structures, and amino acid sequence. These proteins also tend to be stable, and purified proteins can usually be purchased in crystallized or lyophilized forms and are stable over a wide range of conditions.
Chymotrypsin (CT)
α-Chymotrypsin (α-CT) is a well-studied and characterized protein that catalyzes site-specific hydrolysis of peptide bonds.69 This hydrolysis occurs at the carboxyl ends of large hydrophobic or aromatic amino acids, which including tyrosine, phenylalanine, methionine, and tryptophan. However, while α-CT cleaves the carboxyl side of substrate that has been introduced, it will also begin to undergo autolysis simultaneously.70 This ultimately degrades the enzyme and reduces its long-term activity. Griebenow and coworkers attempt to address this issue through the pegylation of α-CT with PEG chains of varying lengths.71 This work found that while pegyalation caused an initial decrease in activity, long-term activity was significantly increased. The native enzyme lost most of its activity within the first 30 minutes of incubation at a higher temperature while the conjugates retained activity after 2.5 hours of incubation.
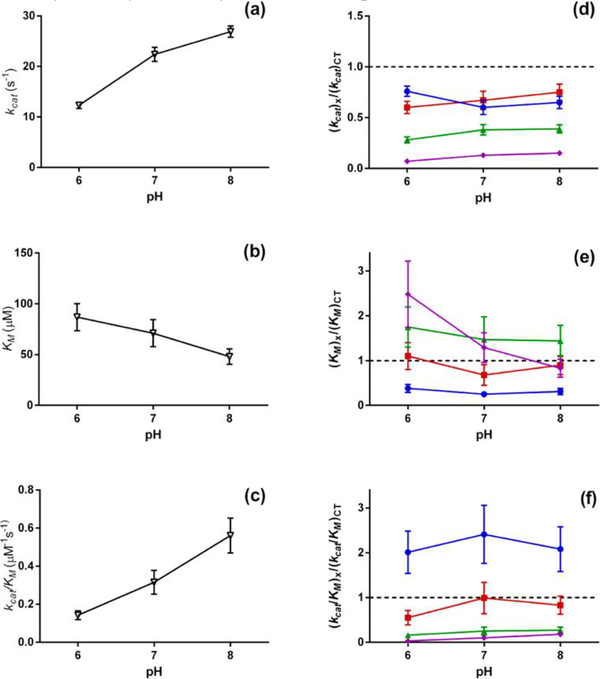
Polymer chains composed of monomers other than ethylene oxide or ethylene glycol can provide charge, hydrophobicity, or tunable responses to stimuli that PEG cannot boast. Russell and coworkers utilize the inherent chemistries of cationic, anionic, and zwitterionic polymers to influence protein activity, using α-CT as a model protein.72 The group constructed four different chymotrypsin-polymer conjugates utilizing an ATRP based grafting-from approach. Polymers conjugated to α-CT include an uncharged poly(oligoethylene glycol methacrylate) (pOEGMA), a cationic poly(quaternary ammonium methacrylate) (pQA), an anionic poly(sulfonate methacrylate) (pSMA), and a zwitterionic poly(carboxybetaine acrylamide) (pCBAA) chain. The activity of α-CT and the bioconjugates were determined using a short peptide substrate, the rate of hydrolysis of the peptide was used to determine enzymatic activity and the effect of polymer conjugation. These activity assays showed a decreased kinetic rate for all conjugates, which has often been found in enzymatic polymer bioconjugates. However, the substrate affinity of these bioconjugates varies compared to the substrate affinity of native α-CT; the cationic polymer conjugate showed an increased affinity, the uncharged and anionic polymer conjugates showed decreased affinity, and the zwitterionic polymer conjugate exhibited no significant change (Fig 4). The results of Russell and coworkers indicate that α-CT substrate affinity is likely caused by electrostatic repulsion and attraction.
Fig 4.
pH-Dependence of kinetic constants (a)kcat, (b)KM, and (c)kcat/KM for native chymotrypsin. Relative kinetic constants for pSMA (purple diamond), pOEGMA (green triangle), pQA (blue circle), and pCBAm (red square) conjugated chymotrypsin. Reprinted with permission.72 Copyright 2017, American Chemical Society.
Bovine Serum Albumin (BSA)
Bovine serum albumin (BSA) is a well-known protein routinely used in laboratories for protein concentration assays,73 a nutrient in cell culture,74 and to stabilize some restriction enzymes during DNA digestion.75 BSA has been shown to have esterase activity, allowing it to serve as a model protein for polymer conjugation. Lavignac and Garcia attempt to increase the activity of BSA through mono- and di-conjugation of poly(amidoamine).76 Mono-conjugated BSA showed a 4% increase in activity when compared to the native protein while di-conjugated BSA reduced activity to 35%. When exposed to denaturing conditions, including incubation at 50 °C and incubation in urea, activity of these conjugates are significantly reduced. After incubation at 50 °C, the activity of mono- and di-conjugated BSA are reduced to 71 and 20%, respectively. In the presence of urea, activity is reduced to approximately 20% for both conjugates. This work shows significant differences in activity based on molecular weight and number of polymers attached.
Sumerlin and coworkers attempted to modulate BSA activity through polymer conjugation.48 The group modified BSA with a maleimide-functionalized chain transfer agent (CTA) followed by RAFT polymerization of N-isopropylacrylamide (NIPAM) to synthesize NIPAM conjugated BSA. Circular Dichroism (CD) showed no significant change in molar ellipticity following conjugation, suggesting retention of secondary structure. CTA modification and polymer conjugation showed activity retention of 97% and 95%, respectively. After 5 cycles of heating and cooling, there was no significant reduction in residual activity. This suggests polymer conjugation via a grafting from approach does not cause significant change in protein activity.
Lysozyme
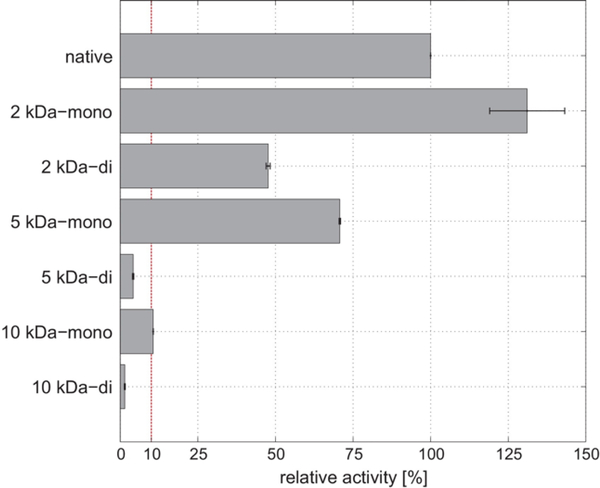
Lysozyme degrades the polysaccharide cell wall of gram positive bacteria, exposing the lipid bilayer, by hydrolyzing peptidoglycan.77 Hubbuch and coworkers attempted to show pegylation is capable of increasing stability and activity while investigating the effect of polymer molecular weight once conjugated to Lysozyme through mono- and di-pegylation.78 The group conjugated 2kDa, 5kDa and 10kDa PEG chains to lysozyme. The activity of these bioconjugates toward Micrococcus lysodeikticus when conjugated with one 2 kDa PEG chain increased nearly 30% when compared to the native enzyme, decreased approximately 30% when conjugated with one 5 kDa PEG chain, and decreased 90% when conjugated with one 10 kDa PEG chain (Fig 5).
Fig 5.
Relative activity of 2 kDa, 5 kDa, and 10 kDa mono- and di-pegylated lysozyme. Reproduced with permission.78 Copyright 2017, Elsevier.
The data suggest molecular weight of PEG chains attached to Lysozyme can significantly increase or greatly inhibit activity. This suggestion is further supported through the activity of di-conjugated Lysozyme conjugates, the activity of these conjugates when compared to their mono-conjugated counterparts showed a significant decrease. For example, when lysozyme is conjugated with one 2kDa PEG chain it increases activity to approximately 130%; however, when the enzyme is conjugated to with two 2kDa PEG chains activity is reduced to approximately 50%.
Berberich and coworkers attempted to improve the antibacterial properties of HEWL through conjugations with polymers of varying lengths and functionalities.79 The group conjugated hetero-block and homo-block copolymers of varying lengths composed of acrylamide, dimethyl acrylamide, dimethylaminoethoxy methacrylate, and oligo(ethylene oxide)methyl ether acrylate to the enzyme through amidation. The activity of this enzyme was tested for all lysozyme-polymer conjugates against M. lysodeikticus which showed modulated activity for all conjugates and reduced activity with increasing polymer molecular weight. The effect of polymer charge on activity was also investigated, showing that modification with anionic polymers led to significantly reduced enzymatic-activity while modification with cationic polymers increased activity compared to conjugates of similar molecular weight. This suggests that electrostatic attractive between functional groups inherent within the polymer and the substrate are important for improving activity of conjugate. Activity was also tested against small molecule analogues of the native substrate to determine the impact of substrate size. The data suggested minimal variation in activity of almost all conjugates against the small molecule analogue.
Proteins with Biomedical Applications
Proteins have been used as effective medical treatments for various diseases for many years. These proteins can be introduced when diseases result from the deficiency of or reduced activity of specific proteins or as a method of inhibition of specific biological processes.80 The activity, half-life, and circulation of these proteins can be diminished in vivo due to proteolytic degradation, an issue which can be addressed through polymer modification.
Recombinant Human Interferon-alpha (IFN-α)
Interferons are a family of signaling proteins that are secreted as a response to pathogens such as viruses and parasites.81 Recombinant human interferon-alpha (IFN-α) is a protein known to effectively inhibit viral replication and tumor cell growth. IFN-α has been used clinically for treatment of cancers and viral diseases such as hepatitis B, hepatitis C, and HIV.82, 83
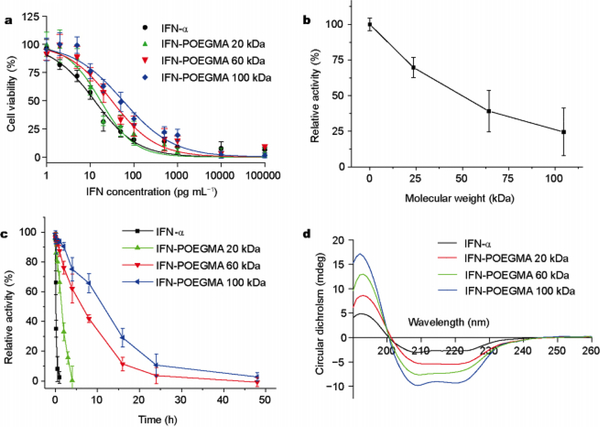
Bordens and coworkers attempted to directly study the effects of size and site of pegylation through the attachment of 12 and 40 kDa linear PEG chains to specific primary amine sites along the exterior of the protein.84 IFN-α conjugated with 12 kDa and 40 kDa PEG showed a signification reduction in activity to 25 and 1%, respectively. The effect of the location of pegylation was determined by measuring ED50 and residual activity which showed highest after conjugation to the histidine residue at location 34. This work suggested that location and size of PEG chain significantly influence activity. Though IFN-α has very effective antitumor and antiviral functions it has poor movement within the body, known as pharmacokinetics. Gao and coworkers showed addressed this issue through the conjugation of 20, 60, and 100 kDa POEGMA which showed a reduction of antiproliferative activity to 73, 40, and 24%, respectively.84 The pharmacokinetic activity of the conjugates were tested by in vivo in mouse models, native IFN-α showed a terminal half-life of 1.5 hours. However, the 20, 60, and 100 kDa POEGMA conjugations showed a terminal half-life of 30.4, 48.1, and 62.8 hours, respectively (Fig 6). This work suggests pegylation of IFN-α decreases antiproliferative activity while significantly increases half-life and pharmacokinetics. The work also showed site specific conjugations at positions distant from the active site increased activity.
Fig 6.
(A) In vitro cytotoxicity of native and pOEGMA conjugated IFN-α. (B) Relative activity of pOEGMA conjugated IFN-α as a function of molecular weight. (C) Residual activity of native and pOEGMA conjugated IFN-α after incubation at 50 °C. (D) CD spectra of native and pOEGMA conjugated IFN-α after incubation at 50 °C for 24hr. Reproduced with permission. 84 Copyright 2017, Springer Nature.
Osteoprotegerin (OPG)
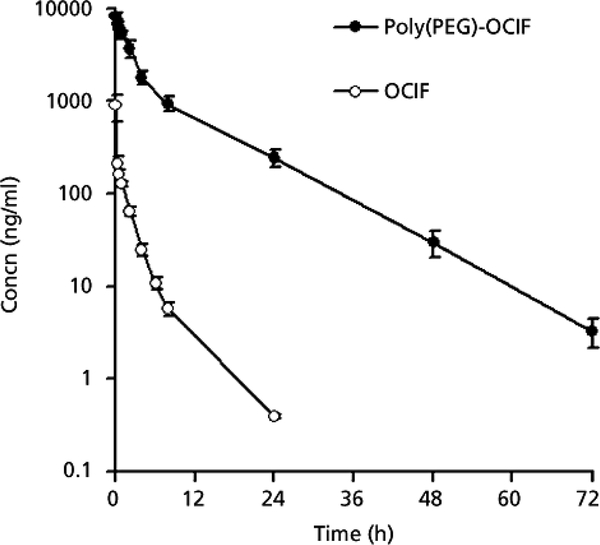
Osteoprotegerin (OPG) is a protein known to inhibit bone resorption, or weakening of the bone, making it a potential therapeutic agent for treatment in bone disorders such as osteoporosis and rheumatoid arthritis.85, 86 The liver is responsible for cleansing toxins and waste from the blood. However, this typically includes efficient uptake of useful proteins such as OPG. To achieve the desired therapeutic benefit high doses of the protein must be introduced to the host in order to account for the portion of protein uptaken by the liver.87 This issue of efficient uptake of OPG, also known as osteoclastogenesis inhibitory factor (OCIF), by the liver was addressed by Okazaki and coworkers through pegylation of the protein.87 The group investigated the effect of pegylation on the uptake of OPG/OCIF in the human liver and various organs in rats. In the rat model, the group studies the uptake of native OPG/OCIF and the bioconjugate in the liver, kidney, and spleen. The most significant uptake of these samples was shown in the liver with nearly no uptake in the kidney or spleen. Pegylation of OPG/OCIF showed significantly higher activity than native OCIF and nearly negligible uptake in the human and rat liver. Through pegylation, the group was also able to significantly increase the half-life of OPG/OCIF in rat models from 3.9 to 7.6 hrs. In addition to this, the group studied the serum circulation of native and pegylated OCIF (Fig 7). This work showed native OCIF having a circulation time of approximately 24 hrs, while pegylated OCIF shows a circulation time over 72 hours.
Fig 7.
Protein concentration in serum after administration of native and pegylated OCIF. Reproduced with permission.87 Copyright 2010, Wiley.
Sumerlin and coworkers attempted to improve in vivo function of OPG via polymer conjugation.88 The group selectively conjugated polymers comprised of poly (ethylene glycol) methyl ether methacrylate (PEGMA) and N-(2-hydroxypropyl) methacrylamide (HPMA) to the amine terminus of OPG. This resulted in non-toxic bioconjugates that retained the activity shown in native OPG. These in vitro studies – outside of living organism – showed polymer structure has no significant effect on function. The group also performed the bone density of rats that had undergone OPG treatment a week prior. The in vivo studies suggested a slight increase in bone mineral density after the loosely branched OPG bioconjugate was administered.
Granulocyte colony-stimulating factor (G-CSF)
Colony-stimulating factors (CSFs) are glycoproteins that are secreted by bone marrow that stimulate the growth and differentiation of stem cells into colonies of specific blood cells, ultimately protecting the host against bacterial, viral, and fungal infections.89 G-CSF has been shown to enhance the antimicrobial functions of mature neutrophilic white blood cells.90 When introduced to humans, recombinant G-CSF has been shown to have low toxicity, induce the production of anti-inflammatory factors, and protect against organ injury induced by endotoxin and sepsis.91–95 G-CSF is available clinically in its recombinant form globally and is FDA approved for uses the include severe chronic neutropenia, peripheral blood progenitor cell transplantation, chemotherapy-induced neutropenia, and bone marrow transplantation.96–102
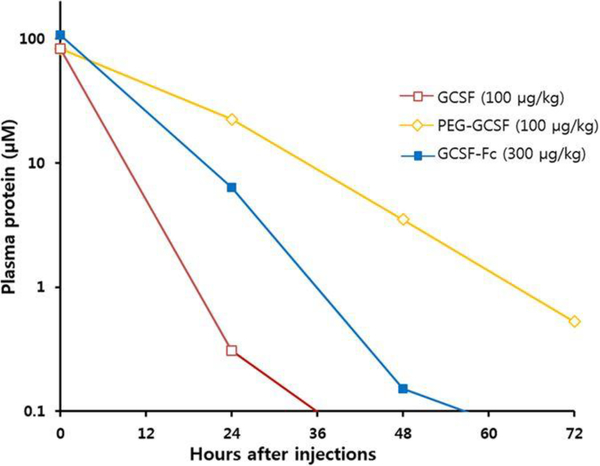
Though G-CSF has been proven as an effective treatment for neutropenia, the protein has short circulation, 24 hour, half-life in the body.103 Choe and coworkers group attempted to address this issue through conjugation 20kDa PEG to the amine terminus of G-CSF.104 The in vitro activity and the half maximal effective concentration (EC50) of the conjugate and native protein samples were investigated through the incubation of mouse myelogenous leukemia cells which ultimately showed similar activity suggesting pegylation does not negatively impact biological activity of G-CSF. The in vivo activity was determined via the injection of the native and conjugated protein into neutropenic rats (Fig 8). This work showed pegylated G-CSF having significantly higher and faster recovery of neutrophils as well as a loner plasma circulation when compared to the native protein. As mentioned before, G-CSF has a circulation time of approximately 24 h while the pegylated protein showed a circulation of more than 72 h. The group also studied the effects of the fused of the Fc domain of IgG1, also known as the crystallizable fragment of the immunoglobulin class G, to G-CSF which had previously been shown to prolong the half-life of the protein.105–107 When compared to pegylated G-CSF, the Fc fused protein has similar in vitro activity but showed slowed recovery of neutrophils in vivo and significantly shortened plasma circulation.
Fig 8.
Protein concentration in plasma after administration of native G-CSF, pegylated G-CSF, and G-CSF fused with Fc domain. Reproduced from Do et al. 2017 licensed under CC BY 4.0104
Zhou and coworkers also attempted to increase plasma half-life of G-CSF.108 However, the group constructed an expression vector consisting of an artificial gelatin-like-protein polymer fused to G-CSF. Similar to the pegylated G-CSF, the purified conjugate showed similar in vitro activity to the native protein, similar in vitro EC50 to the native protein, and in vivo increased plasma circulation.
Human Growth Hormone (hGH)
The pituitary gland attached to the hypothalamus of the brain and responsible for regulation of metabolism, growth, reproduction, and response to the stress through the secretion of various hormones.109 One of the polypeptide hormones secreted is growth hormone (GH) which is released by somatrophs in the pituitary glands.110 Previous research has shown GH regulates somatic growth, energy homeostasis, and carbohydrate and lipid metabolism.111 The FDA approved recombinant human Growth Hormone (rhGH) has been used as therapy for severe growth hormone deficiency, chronic renal insufficiency, turner syndrome, Prader-Willi syndrome, and Noonan syndrome.112–116 HGH was also routinely used by athletes in an attempt to increase muscle growth until forbidden in the early 1990’s.
Though hGH has been proven to serve as an effective treatment for growth hormone deficiency, like most therapeutic proteins it exhibits a short in vivo half-life. The Pasut group attempted to address this issue through the attachment of 20kDa PEG chains to hGH at varying locations.117 The group conjugated the protein at the amine terminus and glutamine at residue 141. In vivo activity was tested by comparing somatic growth produced by the conjugated and native hGH in hypophysectomized rats. These studies showed animals given a single weekly dose of pegylated hGH had similar weights of animals given daily doses of native hGH, this suggests that conjugation prolongs in vivo half-life and has similar activity. In addition to measuring the weight gain in all of the animals in the study, the length of the femur was determined to better understand the effect of the conjugated and native protein on somatic growth. This study showed that animals dosed with pegylated hGH had significantly increased bone length and thicker tibial diaphysis.
Ribonuclease (RNase)
Ribonuclease A (RNase A) is an enzyme that is released by the pancreas, it is responsible for the hydrolysis or degradation of ribonucleic acid (RNA).118 RNase A has been studied for tumoricidal properties due to the link of small noncoding RNAs to the production and formation of malignant tumors.119–123 Hu and coworkers attempted to improve the therapeutic potential of RNase A through mono-pegylation using varying conjugation methods.124 Previous research has shown cell proliferation to be increased in tumors, in this work pegylated RNase A showed increased anti-proliferative activity when compared to the native enzyme.125 However, enzymatic activity showed a slight decrease. Souček and coworkers also attempted to improve the therapeutic potential of bovine pancreatic RNase A through conjugation of classic and star-like hydrophilic poly(N-(2-hydroxtpropyl)methacrylamide) (pHPMA).123 Antitumor activity of the conjugated and native RNase was tested in vivo against human ovarian tumors, human neuroblastoma, and melanoma tumors. When injected in mice bearing tumors, classic and star-like pHPMA conjugated to RNase A showed a significant reduction of tumor volume with reduced toxicity.
Insulin
Insulin is a well-studied hormone that is secreted by the pancreas and responsible for metabolic control - maintaining normal blood glucose levels.126 It is routinely administered as a therapy for insulin-dependent diabetes mellitus, commonly known as type I diabetes.127 Though Insulin is an effective therapy for type I diabetes, it shows poor pharmacokinetic behaviour, requiring multiple injections each day.128 Kim and coworkers attempt to address this issue through the conjugation of 750 and 2000 Da molecular weight methyoxypoly ethylene glycol (mPEG) chains to PheB1 and LysB29.129 The average half-life of insulin once administrated was approximately 12 hours while 750 Da mPEG conjugated at PheB1 and LysB29 showed half-lives of 18.4 and 4.3 days, respectively. 2 kDa mPEG conjugated at PheB1 and LysB29 showed half-lives of 20.7 and 8.6 days. Relative in vivo bioactivity following administration showed a 4 and 12% increase for 750 Da mPEG conjugated at PheB1 and LysB29, respectively. However, a 17 and 15% decrease in bioactivity was observed for 2 kDa mPEG conjugated at PheB1 and LysB29. This suggests mPEG conjugation significantly increases pharmacokinetic behavior when compared to native human insulin, these effects are dependent on both conjugation site and molecular weight of polymers conjugated.
Proteins with Industrial Applications
Proteins have been used many in industrial processes including food processing, clean energy, polymer synthesis, cosmetics, and waste treatment. Though these proteins have catalytic reactions that are very useful in these processes, the activity and half-life of these proteins are often jeopardized under industrial conditions which is addressed in this section through polymer modification.
Laccase
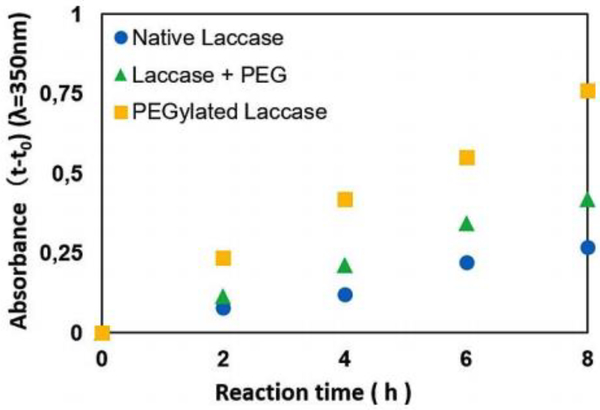
Biologically, laccase is involved in the pigmentation of conidial spores, lignification of cell walls, and delignification during white rot.130–132 Laccase has a wide variety of uses, which include production of ethanol, delignification of biomass, as a sensor for morphine and codeine, and in food and beverage production.133–136 In addition to its industrial properties, laccase has been shown to possess proliferative activity against tumor cells and catalyze the oxidation of various substrates, which has increased interest in enhancing the properties of the enzyme.137, 138 Cavaco-Paulo and coworkers attempt to enhance the polymerase activity of laccase through pegylation.139 Previous studies showed that laccase was able to insufficiently produce polymers, which was caused by reaction products leading to inactivation of the enzyme. The group studied the role of pegylated laccase in the polymerization of catechol when compared to native laccase in the presence or absence of free PEG (Fig 9). In the presence of free PEG, polymerization of catechol was increased to 150%. However, conjugation of PEG to laccase showed an increase of 300%. This work shows free polymer in the presence of laccase significantly increases activity while conjugation to the enzyme has a much greater effect on activity.
Fig 9.
UV absorption at 350 nm during polymerization of catechol in the presence of pegylated laccase, native laccase, and laccease in the presence of free PEG. Reproduced with permission.139 Copyright 2017, Wiley.
Cellulase
Cellulases are used in biofuel production to effectively degrade cellulose to its glucose monomers. However, the chemicals and temperatures used in these industrial processes typically degrade the enzymes or greatly inhibit their function. To address this Zhang and coworkers pegylated commercially available cellulase.140
The activity of pegylated and native cellulase was determined using carboxymethylcellulose and microcrystalline cellulose in the presence of common pretreatment ionic liquid, 1-butyl-3-methylimidazolium chloride [Bmim][Cl]. This ionic liquid is used to dissolve cellulose but has been shown to quickly deactivate enzymes.141, 142 In the presence of [Bmim][Cl], pegylated cellulase showed significantly higher activity at 50 °C and 80 °C. Native cellulase showed a total loss of activity in the presence of 25% [Bmim][Cl] while the activity of pegylated cellulase was reduced to 55% and 45% at 50 °C and 80 °C, respectively, in the presence of 25% [Bmim][Cl].
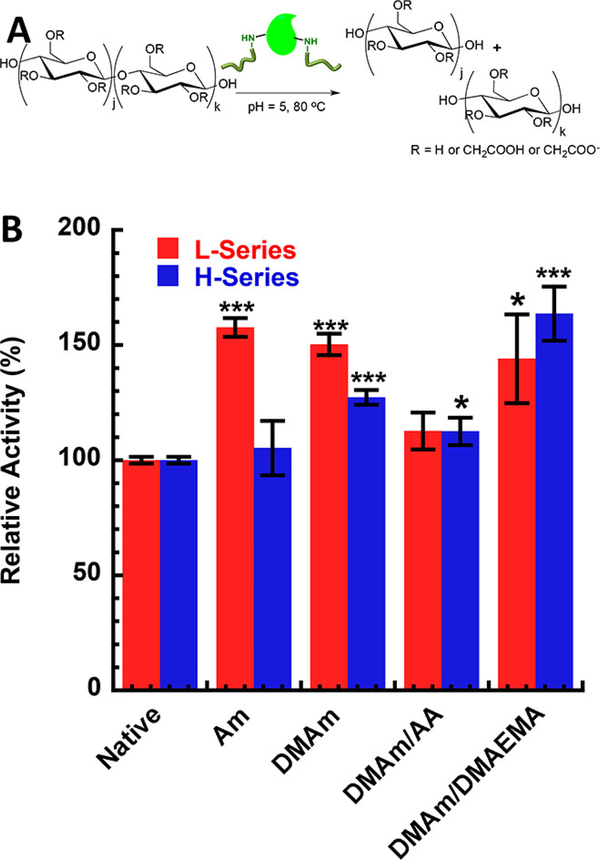
Page and coworkers also attempted to improve the activity of cellulase through the covalent attachment of acrylamide- and dimethyl acrylamide-based polymers to the FnCel5a enzyme.65 FnCel5a is a thermophilic cellulase that efficiently degrades cellulose at an optimum temperature of 80 °C and pH 5.143 The group tested activity of FnCel5a modified with nonionic poly(acrylamide) (Am) and poly(N,N-dimethyl acrylamide) (DMAm) chains and ionic poly(N,N-dimethyl acrylamide – acrylic acid) (DMAm/AA) and poly(dimethyl acrylamide – 2-(N,N-dimethylamino) ethyl methacrylate) (DMAm/DMAEMA) using carboxymethylcelluose (CMC). Conjugations were performed at low and high graft density series with protein:polymer ratios being 1:14 and 1:20 respectively. These series resulted in different numbers of polymer conjugations which ultimately resulted in different effects on activity (Fig 10). Am and DMAm conjugated FnCel5a showed 50% increase while DMAm/AA and DMAm/DMAEMA conjugated FnCel5a showed 12% and 60% increases, respectively, when compared to the native enzyme. This work suggests the use of polymers with functional groups that are complementary to the substrate, can be effective in increasing activity of cellulase.
Fig 10.
(A) Schematic of hydrolysis of CMC by cellulase. (B) Relative activity of Am, DMAm, DMAm/AA, and DMAm/DMAEMA conjugated cellulase compared to native. Reprinted with permission.65 Copyright 2017 American Chemical Society.
Lipase
Lipases, naturally occurring in the stomach and pancreas, are responsible for the hydrolysis of long chain acyl glycerides.144 The enzyme has been historically used in food processing, detergents, wastewater treatment, polymer synthesis, cosmetics, and biodiesel.4, 145–148 Liu and coworkers attempted to enhance the activity of lipase by conjugation of hyperbranched aromatic polyamide (HBPA).149 The activity of the HBPA conjugated and native lipase were measured by the hydrolysis of p-nitrophenylpalmitate (p-NPP) and p-nitrophenylbutyrate (p-NPB). Results from this work showed conjugation increased hydrolysis 20% and 10% of p-NPP and p-NPB, respectively. The activity of conjugated and native lipase was determined after incubation in varying concentrations of DMSO. In the presence of 10, 20, 30 and 40% DMSO the activity of native lipase reduced to 49, 44, 38, and 27%, respectively while conjugated lipase retained 99, 89, 73, and 50%, respectively of its initial activity. This work showed conjugated lipase has significantly higher residual activity in organic solvent compared to the native compound exposed to the same conditions.
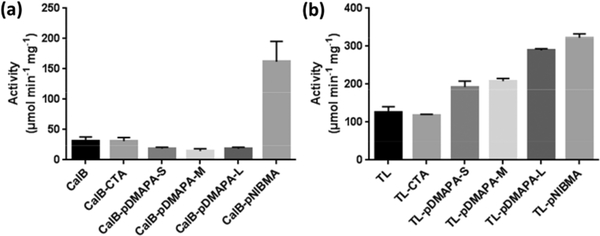
Averick and coworkers attempted to enhance activity of Candida antartica lipase B (CalB) and Thermomyces lanuginose lipase (TL) through grafting-from conjugation.150 These lipases were modified with RAFT CTA followed by photoinduced electron transfer RAFT (PET-RAFT) of N-[3-(Dimethylamino)propyl] acrylamide (DMAPA) and N-(iso-butoxymethyl) acrylamide (NIBMA). This polymerization was done at specific time intervals to make a small, medium and larger chains lengths of pDMAPA. When conjugated to CalB, these polymers showed significant decrease in lipolytic activity (Fig 11). However, when conjugated to TL, these polymers showed a significant increase in activity that correlated with increasing molecular weight. pNIBMA conjugated CalB and TL lipase showed approximately 200% increase in activity. This work shows the effect of polymer conjugation is not general between proteins.
Fig 11.
Activity of native and polymer-conjugated lipase. (A) Candida antartica lipase B (CalB). (B) Thermomyces lanuginose lipase (TL). Reproduced with permission.150 Copyright 2018, Elsevier.
Glucose Oxidase (GOx)
Glucose oxidase (GOx) is a well-characterized enzyme that catalyzes the O2 fueled oxidation of D-glucose to D-gluconolactone, producing hydrogen peroxide (H2O2) which can behave as an antibacterial and antifungal agent.151–154 GOx is industrially used in the food industry and for polymer synthesis.155, 156 McShane and coworkers hypothesized that the pegylation of GOx would increase activity.157 Activity retention of the pegylated and native GOx was tested over a 29 day time period, followed by 24 hour exposure to glucose. Initially, activity of the pegylated enzyme is slightly increased but on the 29th day the activity retention of pegylated and native GOx was 44 and 38%, respectively. The results from this experiment showed statistically equivalent activity between the conjugate and native GOx. Li and coworkers also attempted to modulate the activity of GOx through conjugation of poly[PEG acrylate] (pPEG-A) with increasing mole ratio of polymer conjugated to the enzyme.157 Activity was measured using Horseradish peroxidase through the production of H2O2, which showed a reduction in activity of the conjugate GOx when compared to the native enzyme. With increasing mole ratio of polymer conjugated to the enzyme (5:1, 10:1, and 20:1), overall activity reduced to 72, 64, and 57%, respectively. This work suggests increasing molecular weight results in decreased activity and polymer conjugation is can modulate the activity of GOx.
Russell and coworkers propose efficient electron transfer through the growth of poly(N-3-dimthyl(ferrocenyl)methylammonium bromide)propyl acrylamide (pFcAc) chains from GOx.158 The group synthesized these conjugates in the presence and absence of chitosan for enzyme-based biosensors to improve electron transfer efficiency in enzyme-modified electrodes. Amperometry was used to determine the glucose biosensing behavior of these conjugates at varying concentrations of glucose with constant cell voltage. This work showed an apparat KM of 45.7 and 22.3 mM glucose for GOx conjugate in the absence and presence of chitosan, respectively. This shows the chitosan containing network has a higher affinity for glucose binding.
Xylanase
Xylanases are a class of enzymes responsible for the catalysis of the degradation of xylan, a widely available natural polysaccharide, to produce alcohol, xylose, xylitol and xylooligosaccharides.159, 160 Industrially, xylanase is widely used in animal feed, lignocellulosic biomass processing, textile processing, and baking.161–164 Bordbar and coworkers attempted to increase the catalytic activity of xylanase by immobilizing the enzyme on superparamagnetic graphene oxide nanosheets functionalized with poly(ethylene glycol) bis amine (PEGA).165 Catalytic activity of xylanase was determined by measuring the production of xylose when in the presence of xylan at various temperature and pH. These experiments showed optimum activity for the native enzyme at pH 6.5 at 60 °C while optimum activity for immobilized xylanase was pH 7.5. When compared to the activity of the native enzyme at pH 7.5 and pH 8.5, activity was significantly higher. Activity of the immobilized and native enzyme were similar at 60 °C, while immobilized xylanase showed significantly increased activity at 70 °C and 80 °C. This suggests that PEG functionalized nanosheet immobilization tunes the activity of biocatalysts.
Catalase
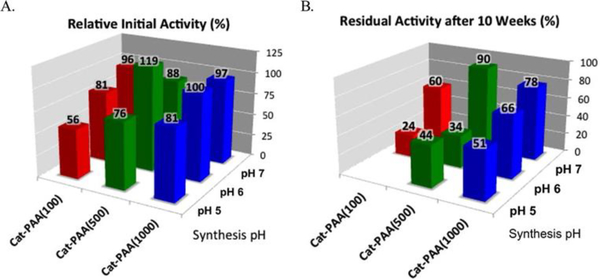
Catalase is a well characterized enzyme responsible for the conversion of hydrogen peroxide to molecular oxygen.166 Industrially, catalase is widely used in shelf-life improvement of food, milk preservation, and hydrogen peroxide removal.167–169 Kumar and coworkers attempted to enhance the enzymatic activity of catalase through the conjugation of poly (acrylic acid) (pAA).170 Various lengths of polymer (100kDa, 500kDa, and 1000kDa) were conjugated to the enzyme with the goal of encapsulating the protein, this synthesis was conducted at various pH values (pH 5, 6, and 7) in order to control the protonation of the pAA chain. The catalytic activity of the conjugated enzyme was measured by studying the rate of decomposition of H2O2 and compared to the native enzyme (Fig 12). These experiments showed increasing activity (55–80% activity retention) with increasing polymer molecular weight for conjugates synthesized at pH 5; however, the activity of these conjugates was decreased when compared to the native enzyme. These experiments also showed an increase in activity with increasing synthesis pH, conjugates synthesized at pH 7 showed 90–100% activity retention when compared to the native enzyme. A nearly 20% increase of activity was observed for the 500kDa pAA conjugated catalase synthesized at pH 6. This work suggests conjugation conditions associated with protein-polymer conjugates significantly effect immediate and long-term activity.
Fig 12.
Relative activities of pAA-catalase conjugates at pH 7. (B) Residual activities of pAA-catalase conjugates after storing at 8 °C for 10 weeks at respective conjugation pH 5, 6, and 7. Reprinted with permission.149 Copyright 2014 American Chemical Society.
General Trends in the Effect of Polymer Conjugation on Activity
Polymer modification of enzymes may elicit the modification of enzyme activity, including increased or decreased activity, activity retention in organic solvents, activity retention at high temperatures, and prolonged activity in vivo and in vitro. A recurring theme among these conjugates is the initial reduction of activity in pegylated proteins. Grienbenow and coworkers attempted to understand structural causes of protein conjugation by studying structural dynamics of chymotrypsin-polymer conjugates using Fourier-Transformed Infrared (FTIR) Hydrogen/ Deuterium exchange experiments, Circular Dichroism (CD) spectroscopy, and Differential Scanning Calorimetry (DSC).171 This work showed a reduction of protein dynamics which had previously been linked to an increase in protein thermodynamic stability.172 Bioconjugate researchers have used these findings as a hypothesis to explain the reduction of kinetic rate observed for enzymatic protein-polymer conjugates. In addition to activity, polymer conjugation has been shown to affect stability. In the case of polymers with more complex functionality than PEG, judicious choice of the polymer can lead to an increase in activity. This is often achieved when there are complementary interactions, such as electrostatic attractions or hydrophobic interactions, between the attached polymer and the substrate of interest. This begins to form guiding principles for the next generation of bioconjugates.
Effect of Polymer Conjugation on Stability
Unlike activity, there are many definitions of stability when referring to proteins which include, but are not limited to: stability against proteases, circulation time in a living system, thermal stability, structural stability, thermodynamic stability, and chemical stability. The work of Davis and Abuchowski showed pegylation can increase proteolytic resistance, thermal stability, and pH stability of BSA – this work led to examining the influence of synthetic linear and branched PEG and non-PEG polymers on the stability and confirmation of various proteins. These studies have shown conjugation of polymers can result in changes in conformational structure, which lead to increased stability and longer half-life.
Model Proteins
Lysozyme
Since its discovery in 1922, lysozyme has served as a model protein for amyloid research, metalation, protein crystallography, and protein-polymer conjugation.173–175 As mentioned before, Hubbuch and coworkers attempted to increase stability and activity of Lysozyme through mono- and di-pegylation.78 Though this work showed significant decrease in activity when conjugated to higher molecular weight PEG and to more than one PEG chain, conjugation proved promising in terms of the enzyme’s stability. The thermal and chemical stability of conjugated and native lysozyme were determined using intrinsic protein fluorescence measurements. These experiments showed similar melting temperatures(Tm), approximately 60 °C, for the modified and native enzyme. NaCl was added to these experiments to test chemical stability of these proteins, the salt has been shown to destabilize conformational and colloidal stability of proteins.176 Upon the addition of this salt, all samples showed a reduction in melting temperature. However, pegylated lysozyme showed significantly higher stability in high concentrations of the salt when compared to the native enzyme that suffered spontaneous precipitation at salt concentrations above 1.59M. At the NaCl concentration of 2.5, the di-pegylated protein samples showed a Tm above 45 °C while the mono-pegylated protein showed a Tm at approximately 40 °C. This suggests that pegylation is capable of increasing stability in the presence of efficient denaturants and precipitants.
Berberich and coworkers attempted to improve the thermal and chemical stability of Lysozyme through the conjugation of various heteropolymers and homopolymers.79 As mentioned before, modification of the enzyme with charged polymers modulates activity based on electrostatic interactions with the substrate, also the molecular weight of the polymer attached may inhibit or stimulate activity. The group determined thermal and chemical stability using differential scanning fluorimetry (DSF) and tryptophan fluorescence. These experiments showed a decrease in Tm for all conjugates, with the most dramatic reduction in Tm occurring for the highest polymer molecular weight bioconjugates. Guanidine Hydrochloride (Gdn-HCl), a common denaturant, was used to test the chemical stability of the conjugated and native lysozyme. These experiments showed increased stability against increasing concentrations of Gdn-HCl for many of the conjugates. This work suggests polymer conjugation is capable of increasing chemical stability and the molecular weight of polymer conjugation is very significant.
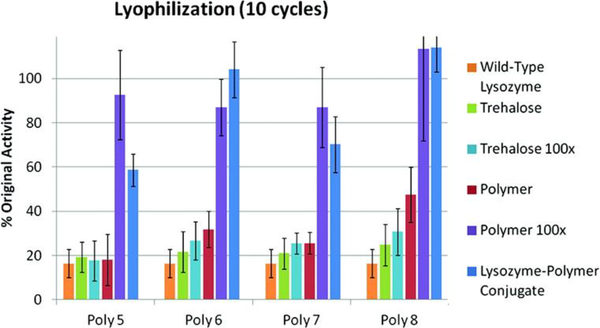
In order to improve activity following thermal stress and overall stability of lysozyme, Maynard and coworkers conjugated 8, 15, 25, and 50 kDa trehalose-based glycopolymers to the enzyme.177 Following 10 cycles of lyophilization, activity for the wild type lysozyme was reduced to 16% while in the presence of 100-fold excess free polymer, the enzyme exhibited full activity retention, regardless of molecular weight (Fig 13). In these conditions, 59, 100, 70, and 100% activity was retained when lysozyme was conjugated with 8, 15, 25, and 50 kDa trehalose-based glycopolymers, respectively. When incubated at 90 °C for 1 hour, the activity of wild type lysozyme reduced to approximately 19% while in the presence of free polymer and when conjugated, the enzyme retained 55–80% of initial activity. This works suggests polymer conjugation is an effective stabilizer for lysozyme for heat and lyophilization stress.
Fig 13.
Activity of native lysozyme, native lysozyme in the presence of trehalose (1 or 100 equiv), native lysozyme in the presence of polymer (1 or 100 equiv), and 8, 15, 25, and 50 kDa polymer-conjugated lysozyme following 10 cycles of lyophilization. Reprinted with permission.156 Copyright 2012 American Chemical Society.
Bovine Serum Albumin (BSA)
Albumin, human and bovine, is a plasma protein secreted by the liver and exhibits esterase activity, the hydrolysis of esters into an acid and alcohol.74, 178, 179 Nielsen and coworkers attempted to investigate the effect of protein pegylation on the structure, function, and stability of Bovine Serum Albumin (BSA).180 The group conjugated 5, 10, 20, 30, 40 and 60kDa PEG chains to the enzyme and determined the effect of these conjugations on stability utilizing circular dichroism (CD) spectroscopy and differential scanning calorimetry (DSC). CD is an effective measure of secondary structure, these experiments show no significant change in the secondary structure of conjugates when compared to the native enzyme. DSC is an effective measure of thermal stability and can be used to accurately measure Tm and enthalpy, ΔH. These experiments showed native BSA to be slightly more thermal stable than the pegylated BSA. This work suggests that pegylation has little to no effect on the secondary structure of BSA while affecting thermal stability.
Chymotrypsin
Chymotrypsin is well known for undergoing autolysis, which contributes to its overall instability. Russell and coworkers attempt to dramatically enhance pH and thermal stability of chymotrypsin through the conjugation of poly(sulfobetaine methyacrylamide)-block-poly(N-isopropylacrylamide) (pSBAm-block-pNIPAM).181 The group modified chymotrypsin with an ATRP initiator followed by ATRP polymerization of SBAm, followed by ATRP polymerization of NIPAm to form diblock polymer conjugates. This approach resulted in conjugates with 232, 354, and 553 kDa molecular weights. Incubation at 37 °C for 8 hours showed no significant change for the conjugates while native chymotrypsin lost 50% of its initial activity. Incubation in 167 mM HCL of 3 hours showed residual activity of 60% for all conjugates while the native enzyme lost 50% of its initial activity in 30 minutes and all activity after 2 hours. This suggests polymer conjugation is capable of decreasing autolysis and increasing overall stability of chymotrypsin.
Proteins with Biomedical Application
Human Growth Hormone (hGH)
Like many therapeutic proteins, hGH exhibits low circulation half-life, requiring frequent injection for effective treatment of diseases caused by hGH-deficiency. The Pasut group attempted to address this issue while increasing activity and stability through the attachment of PEG chains to hGH at varying locations.117 Conjugation resulted in significantly increased circulation half-life and significant bone growth in rat models. Thermal stability was measured using CD. These experiments showed native hGH having a Tm of 82 °C, which increased to 86 °C upon conjugation at the amine terminus, which suggests a significant increase of thermal stability. The reversibility of thermal unfolding was measured after the samples were heated to 95 °C then cooled to 20 °C. The conjugated hGH able to more easily recover its secondary structure after thermal denaturation, when compared to the native protein. This work suggests pegylation is able to significantly increase thermal stability and recovery following thermal denaturation.
Cocaine Esterase
Cocaine Esterase (CocE) is an enzyme responsible for the hydrolysis of cocaine; however, the enzyme is proven to be unstable at 37 °C limiting its therapeutic potential.182 Sunahara and coworkers attempted to increase the stability of the enzyme through the conjugation of 40kDa PEG.183 Stability was determined by measuring melting temperature and testing in vivo residence over a prolonged period of time. These experiments showed a significant increase in thermal stability of CocE, the native enzyme has a Tm of 34.8 °C while the pegylated enzyme has a Tm of 43.9 °C. Residence time of the pegylated CocE was 72 hours and showed an increased survival rate in rat models. This suggests pegylated CocE increases stability of the enzyme and may be used therapeutically.
Hemoglobin
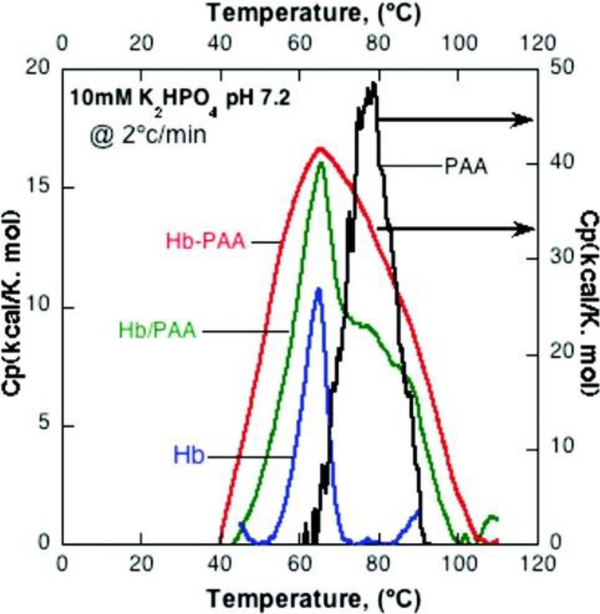
Hemoglobin (Hb) is a well-studied enzyme found in red blood cells transporting oxygen from the lungs to the tissues within the body.184 In the presence of H2O2, the protein also behaves as a peroxidase.185 Kumar and coworkers attempted of improved the stability of Hb through the attachment of 450 kDa poly(acrylic acid) (pAA).67 Structural and thermal stability of the enzyme were measured using circular dichroism (CD) spectroscopy and differential scanning calorimetry (DSC). These experiments showed similar secondary structure of the conjugated and native Hb, suggesting heme coordination is preserved. DSC of the conjugated and native enzyme show no change in Tm (Fig 14). However, the thermographs of these enzymes show significant changes in stability, the native enzyme denatures over the range of 50–70 °C while the conjugate denatures over the range of 40–105 °C. This information was used to calculate the denaturation enthalpy, which were 90 kcal/mol and 1559 kcal/mol for the native and conjugated Hb, respectively. These studies also showed increased room-temperature stability upon the conjugation of pAA.
Fig 14.
DSC Thermogram of Hemoglobin, Hemoglobin in the presence of free pAA, Hemoglobin conjugated with pAA, and pAA. Reprinted with permission.67 Copyright 2011 American Chemical Society.
Proteins with Industrial Application
Papain
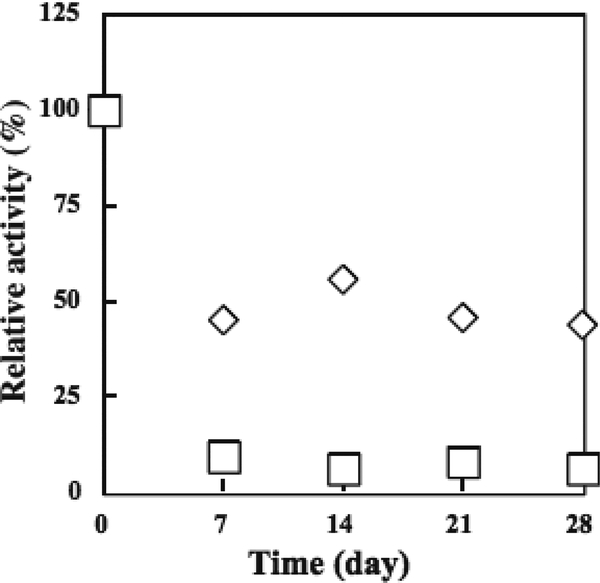
Papain, also known as Papaya proteinase I, is a cysteine endopeptidase. It is widely used in food processing for beer stabilization, meat tenderization, and dairy processing.186–188 Ishihara and coworkers attempted to increase the stability through the conjugation of varying molecular weights of water-soluble phospholipid and ethylene oxide (also known as ethylene glycol) polymers.189 Structural and thermal stability was determined by circular dichroism (CD) spectroscopy and measuring the preservation of enzymatic activity after 28-day incubation at 40 °C, respectively (Fig 15). CD showed no significant change in helical content when conjugated to PEG. A gradual decrease with increasing molecular weight (5, 10, and 20kDa) of phospholipid polymers; however, the 40kDa phospholipid conjugate showed only slightly decreases helical content when compared to the native enzyme.
Fig 15.
Residual activity of native (◻) and pegylated (◇) papain after storage at 40 °C. Reproduced with permission.189 Copyright 2004, Elsevier.
Native enzymatic activity was nearly completely lost after 7 days while pegylated papain activity reduced to approximately 50% and remained constant for the remaining 21 days. When conjugated with the 5kDa phospholipid polymers, enzymatic activity remained nearly unchanged over the 28 day period while the 40kDa conjugate showed a 25% increase in activity over the 28-day period. Like most proteases, Papain is capable of self-digestion, or auto-lysis. This work suggests pegylation or phospholipid polymer conjugation is capable of reducing this phenomenon over a long-period of time, presumably due to steric hinderance, ultimately increasing thermal stability.
Cellulase
Cellulase is responsible for the degradation of cellulose and commercially used in ethanol production. As mentioned before, Page and coworkers found that functional groups inherent to polymer chains can be effective in increasing activity if they are complementary to the target substrate.65 The group also studied the effect of polymer conjugation on thermal, chemical, and thermodynamic stability using differential scanning fluorimetry (DSF) and studying activity following denaturation by DMF. DSF was used to determine thermal and thermodynamic stability of the conjugates when compared to the native enzyme, showing similar Tm values for all samples. The DSF data was also used to calculate the Standard Gibbs free energy of unfolding ΔuG°, entropy of unfolding and enthalpy of unfolding using a method was previously introduced by the group.190 These experiments showed similar Tm and ΔuG° values for the conjugated and native enzyme. Following incubation in 76% DMF, residual activity of the native and conjugates were shown to be approximately 30%. This work suggests polymer conjugation has no adverse effect on the stability of FnCel5a.
Laccase
Laccases is used industrially for a plethora of applications including baking and beverage processing. Hernández-Arana and coworkers attempted to increase stability of the enzyme through pegylation.191 Structural and thermal stability was determined using circular dichroism (CD) spectroscopy and differential scanning calorimetry (DSC). These experiments showed similar CD structure and 2 °C increase in Tm when measured by DSC. The group also measured chemical stability through the rate of inactivation in aqueous-organic solvents such as methanol, ethanol, propanol, and acetonitrile, which showed an overall decreased rate of inactivation. This work suggests pegylation is capable of modestly increasing thermal and chemical stability of laccase.
Xylanase
Xylanase is responsible for the degradation of xylan and commercially used in animal feed and textile processing. As mentioned before, Bordbar and coworkers attempted to increase activity and stability of the enzyme though immobilization on PEG modified superparamagnetic graphene oxide nanocomposite.165 This work showed PEG functionalized nanosheets are capable of tuning activity. Reusability and storage stability of the enzyme was determined by measuring activity following 8 cycles of reuse at varying temperatures and measuring activity after 90 days of storage, respectively. These experiments showed a sharp reduction of activity following the second cycle and the immobilized xylanase retaining the most activity (38%) after 8 cycles at 60 °C. After storage for 90 days, the immobilized xylanase retained approximately 35% of its initial activity while the native enzyme only retained 20%. This work suggests immobilization of catalytic enzymes on PRG functionalized nanosheets may have a cost benefit due to storage stability and reusability.
General Trends in the Effect of Polymer Conjugation of Stability
The work discussed in the section shows polymer modification of proteins is capable of increasing thermal stability, chemical stability, storage stability and reusability. These studies suggest that stability and activity can be improved, reduced, or unchanged independent of conformational, or structural changes. These studies show there are no general trends associated with polymer conjugation. For example, increasing molecular weight of polymers attached in result in increasing stability, decreasing stability, or no change. This is also shown in type of polymer conjugated with no trend in charge, connectivity, or polymerization method. Pokorski and coworkers attempted to better understand the effect of conformation associated with protein-polymer modification through the conjugation of PEG, poly(oligo(ethylene glycol)methyl ether acrylate) and poly (norbornene-(oligo(ethylene glycol)ester)) to Virus-like Particle, Qβ.192 The group utilized small-angle neutron scattering (SANS) to elucidate structures of the conjugates and cryo-electron microscopy (cryo-EM) to enable direct visualization of the conjugate. These experiments showed unique surface polymer conformations for each conjugate. Bioconjugate researchers have used surface polymer configuration to explain changes in activity or stability in the absence of protein conformational change. The impact of polymer structure on stability needs to be expanded in future work in order to provide guidance on how to design bioconjugate for optimal performance.
Stimuli Responsive Bioconjugates
Stimuli-responsive, or smart, polymers are materials that undergo some change based on the environment, which includes but is not limited to temperature, pH, wavelength and humidity.193 The covalent attachment of inert polymer chains has been shown to increase activity and stability; however, the modification of proteins with smart polymers introduces new functionalities and possibilities due to the altered polymer structure with and without stimuli.
pH responsive
Chymotrypsin
α-Chymotrypsin (α-CT) is a digestive enzyme that performs proteolysis, the non-selective degradation of proteins and polypeptide. However, CT has been shown to degrade itself. Russell and coworkers address the stability through the conjugation of poly(2-(dimethylamino)ethyl methacrylate) (pDMAEMA), which has been shown to undergo conformational change with alterations in temperature and pH.194 The work showed the diameter of conjugates increased with lower pH values and decreased at values above 8, due to the deprotonation of DMAEMA. Typically, native CT is nearly inactive at pH values below 8; however, upon modification with pDMAEMA, activity at lower pH values show a nearly 10-fold increase. This work suggests that these properties can be tailed by tunable variables such as pH and temperature.
Pyrophosphatase
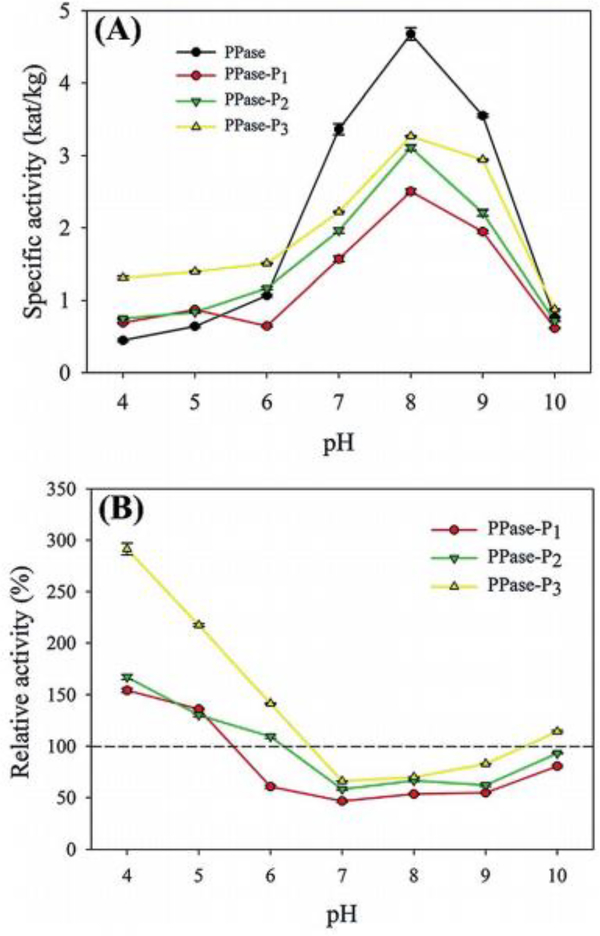
Pyrophosphatase (PPase) is an enzyme responsible for the hydrolysis of inorganic pyrophosphate to phosphate ions.196 Brash and coworkers attempted to improve the activity and stability of PPase through the site-specific conjugation of varying molecular weights of pH-responsive pDMAEMA.195 The effect of pDMAEMA conjugation was investigated by measuring the activity, hydrodynamic diameter, and stability of the conjugates. Activity was measured over a range of pH values (4–10) for PPase conjugated to 8kDa, 14.8kDa, and 21.2kDa pDMAEMA and compared to the native enzyme (Fig 16). These experiments showed significantly increased activity for all conjugates at pH 4 and 5, activity was increased 300% when modified with 21.2kDa pDMAEMA at pH 4. Native PPase is optimum at pH 8; however, the activity modified enzyme was significantly reduced at pH 7, 8, and 9. pDMAEMA has been shown to undergo conformational change with alternations in pH, this is shown in the hydrodynamic size distributions of the conjugates over a range of pH values (4–10).197 The hydrodynamic diameter of PPase reduced from 3531nm to 5.3nm at pH 4 and 10, respectively, while the conjugated enzymes at pH 4 have an average of 10.9nm, at pH 8 have an average hydrodynamic diameter of 722.5nm and is again reduced at pH 10 to approximately 9.1nm. This work suggests conjugation of pH-responsive polymers are capable of introducing pH-responsive activity to the enzyme.
Fig 16.
(A) Activity of native and pDMAEMA conjugated PPase. (B) Relative activity of 8kDa (red), 14.8kDa (green), and 21.2kDa (yellow) pDMAEMA conjugates PPase as a function of pH. Reproduced with permission.195 Copyright 2015, The Royal Society of Chemistry.
Temperature responsive
Lysozyme
The conjugation of stimuli responsive polymers to enzymes has received a lot of attention, Mann and coworkers attempted to use this chemistry to produce stimulus-responsive protein-based micro-compartments, also known as proteinosomes.198 The group conjugated 8.8kDa poly(N-isopropylacrylamide) (pNIPAm) polymers to BSA. The conjugate was used for spontaneous self-assembly of proteinosomes (aqueous media encapsulated in protocelle) in oil. Cooling of these proteinosomes resulted in changes in the conformational and hydrophobic conformations pNIPAm. This ultimately resulted in temperature mediated release of aqueous material encapsulated within the proteinosomes.
Endoglucanase
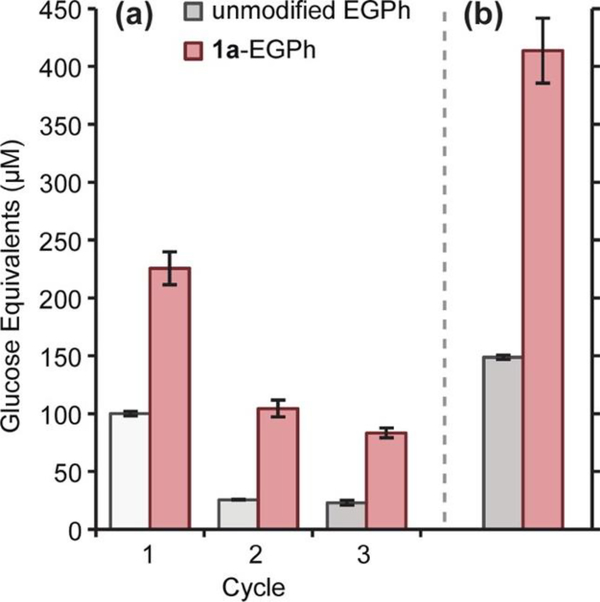
In addition to enzyme instability affected with industrial conditions associated with biofuel productions, enzymatic costs are a substantial portion of overall biofuel costs.199–201 To address this, scientists have attempted to reduce costs through the collection and reusability of these enzymes within multiple cycles of processing (Fig 17). Francis and coworkers address this need through the conjugation of poly(N-isopropylacrylamide) (pNIPAm) to endoglucanase (EGPh).202 Lower critical solution temperature (LCST) and high critical solution temperature (HCST) polymers are a class thermal responsive polymers. LCSTs, such as pNIPAm, are soluble at lower temperatures but precipitate as temperatures increase, this work shows that after conjugation, the responsive activity of the polymer remains. Following two cycles of heating and cooling, NIPAm modified EGPh retained approximately 60% of its initial activity. This work suggests thermos-responsive polymers can be used to increase reusability of enzymes.
Fig. 17.
Hydrolytic activity of native and pNIPAm conjugated EGPh. (A) 12 hour reusability assay for EGPh. soluble reducing sugar was measured at the start and end of each cycle, the difference is shown. (B) Total glucose equivalents produced over all cycles. Sum of values over three cycles, from A, are shown. Reprinted with permission.202 Copyright 2013 American Chemical Society.
Photoresponsive
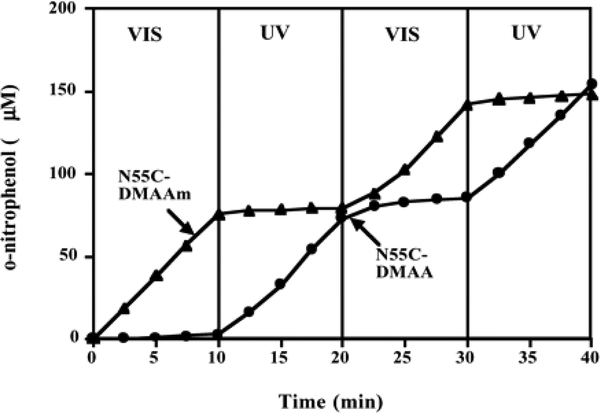
Endoglucanasse 12A
In addition to thermo- and pH- responsive polymers, photo-responsive polymers have received attention for protein conjugation. Stayton and coworkers conjugated photoresponsive Dimetheyl acrylamide-co-4-phenylazophenyl acrylate (DMAA) and Dimethyl acrylamide-co-N-4-phenylazophenyl acrylamide (DMAAm) polymers to Endoglucanase 12A (EG 12A).203 EG 12A is commercially used for its hydrolysis of cellulose. The enzymatic activity of the conjugates were determined in the presence of o-nitrophenyl-β-D-cellobioside (ONPC). These experiments showed sequential photoswitching activity (Fig 18). DMAAm conjugates only showed activity under visible photoirradiation while DMAA conjugates only showed activity under ultraviolet photoirradiation. Surprisingly, conjugation reduced activity to approximately 55% for the conjugates while EG 12A in the presence of these polymers showed no reduction in activity or photoswitching capabilities. This work suggests the possibility of regulating enzyme activity based on photoirradiation.
Fig 18.
Sequential photoswitching of the activity of pDMAA and pDMAAm conjugated EG 12A. Reproduced with permission.203 Copyright 2002, National Academy of Sciences, USA.
Conclusions
Since the first documented protein-polymer conjugate synthesized by Davis and coworkers in 1977, the bioconjugate field has grown substantially and includes non-PEG conjugations that result in a variety of new behaviors.11 Polymers have been conjugated to or grown from a number of biomolecules with a plethora of behaviors and applications. NMP, ATRP, RAFT, and ROMP are common polymerization methods utilized in biomolecule-polymer conjugation. Researchers have demonstrated these methods can be used in traditional chemical or through photoinduction using both grafting-to and grafting-from conjugation approaches. Coupling chemistries to synthesize bioconjugates most commonly utilize amino acids present within the native structure of the protein or introduce polymerization initiators to non-natural amino acids.
The protein-polymer conjugates discussed on this review exhibit a vast array of behaviors, which is independent of polymer length, monomer composition, or conjugation site. This review highlights the ability of pegylation to modestly and significantly increasing stability and reusability while impacting activity in a number of ways. This includes but not limited: initial reduction, decreasing with increasing polymer molecular weight, increasing when conjugated to sites away from active site, significantly increasing, and causing no statistically different change. Conjugation of charged polymers to various proteins showed a range of effects, including: increased or decreased substrate affinity, decreased activity with polymer increasing molecular weight, significantly increased chemical and storage stability, and no observed significant changes. However, these changes in activity and stability are often specific to a given pair of polymer and protein, with prediction of polymer conjugation of biohybrid perfoamce being difficult to assess prior to conjugation. For instance, Averick and coworkers modified two lipases (CalB and TL) with small, medium, and large DMAPA polymers.150 Under the same reaction conditions, both enzymes showed very dissimilar behaviors. The lypolytic activity of CalB was reduced but had not effect based on length while TL showed significantly increased activity correlating with polymer length. This suggests a need for expansion in this field with the goal of understanding the general rules and principles that guide the behaviors caused by polymer conjugation. Future directions for this field are an expansion of the smart or responsive polymers attached to biological materials to regulate biomolecule activity in real time. Additionally, conjugation to increase stability and solubility of proteins in organic solvents for more efficient industrial applicability and increase the utility of protein-polymer conjugations is an ongoing area of research. Within the next ten years, we expect to have a better understanding of the effect of polymer attachment to protein activity and stability. This would entail understanding the effect of the polymer length, composition, and attachment site, as well as if the type of linker, conjugation chemistry, and polymerization technique (ATRP, RAFT, photo-induced polymerization, or PEG) plays any role in changes to protein activity or stability. Ultimately, this would result in more predictive power as to the effects of polymer conjugation and better utilization of resources, so that polymers can be designed from first principles to provide a protein with a given functionality.
Acknowledgements
The authors would like to acknowledge Jamie M. Stewart and Kevin M. Burridge for productive discussions
Funding Sources
D.K. acknowledges support from the National Science Foundation under Grant No. DMR-1749730. R.C.P. acknowledges support from the National Institutes of Health under Grant No. R35 GM128595, and institutional support from Miami University through the Robert H. and Nancy J. Blayney professorship.
Footnotes
Conflicts of interest
Richard C. Page has recently served as an expert in a case associated with Granulocyte colony-stimulating factor.
Footnotes relating to the title and/or authors should appear here.
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here].
Notes and references
- 1.Dimitrov DS, Methods Mol Biol, 2012, 899, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaczmarek JC, Kowalski PS and Anderson DG, Genome Med, 2017, 9, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hess M, Trends Microbiol, 2008, 16, 414–419. [DOI] [PubMed] [Google Scholar]
- 4.Jurado E, García-Román M, Luzón G, Altmajer-Vaz D and Jiménez-Pérez JL, Ind Eng Chem Res, 2011, 50, 11502–11510. [Google Scholar]
- 5.Visuri K and Klibanov AM, Biotechnol Bioeng, 1987, 30, 917–920. [DOI] [PubMed] [Google Scholar]
- 6.Uherek C and Wels W, Adv. Drug Deliv. Rev, 2000, 44, 153–166. [DOI] [PubMed] [Google Scholar]
- 7.Muller MM, Biochemistry, 2018, 57, 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mülhaupt R, Angewandte Chemie International Edition, 2004, 43, 1054–1063. [DOI] [PubMed] [Google Scholar]
- 9.Maitz MF, Biosurface and Biotribology, 2015, 1, 161–176. [Google Scholar]
- 10.Pendhari SS, Kant T and Desai YM, Composite Structures, 2008, 84, 114–124. [Google Scholar]
- 11.Abuchowski A, van Es T, Palczuk NC and Davis FF, J Biol Chem, 1977, 252, 3578–3581. [PubMed] [Google Scholar]
- 12.Abuchowski A, McCoy JR, Palczuk NC, van Es T and Davis FF, J Biol Chem, 1977, 252, 3582–3586. [PubMed] [Google Scholar]
- 13.Duncan R, Nat. Rev. Drug Discov, 2003, 2, 347. [DOI] [PubMed] [Google Scholar]
- 14.Duncan R, Dimitrijevic S and Evagorou EG, S. T. P. Pharma Science 1996, 6, 237–263. [Google Scholar]
- 15.Satchi R, Connors TA and Duncan R, Brit. J. Cancer, 2001, 85, 1070–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan R, Advanced Drug Delivery Reviews, 2009, 61, 1131–1148. [DOI] [PubMed] [Google Scholar]
- 17.Hardwicke J, Ferguson EL, Moseley R, Stephens P, Thomas DW and Duncan R, Journal of Controlled Release, 2008, 130, 275–283. [DOI] [PubMed] [Google Scholar]
- 18.Duncan R, Ringsdorf H and Satchi-Fainaro R, in Polymer Therapeutics I, eds. Satchi-Fainaro Rand Duncan R, Springer Berlin Heidelberg, Berlin, Heidelberg, 2006, DOI: 10.1007/12_037, pp. 1–8. [DOI] [Google Scholar]
- 19.Bader H, Dorn K, Hupfer B and Ringsdorf H, Berlin, Heidelberg, 1985. [Google Scholar]
- 20.Gros L, Ringsdorf H and Schupp H, Ang Chem Int Ed Eng, 1981, 20, 305–325. [Google Scholar]
- 21.Ringsdorf H, J. Polym. Sci, 1975, 51, 135–153. [Google Scholar]
- 22.Gauthier MA and Klok H-A, Polym. Chem, 2010, 1, 1352–1373. [Google Scholar]
- 23.Pelegri-O’Day EM, Lin E-W and Maynard HD, J Am Chem Soc, 2014, 136, 14323–14332. [DOI] [PubMed] [Google Scholar]
- 24.Nuyken O and Pask S, 2013, 5, 361. [Google Scholar]
- 25.Grubbs RB and Grubbs RH, Macromolecules, 2017, 50, 6979–6997. [Google Scholar]
- 26.Ogawa KA, Goetz AE and Boydston AJ, J Am Chem Soc, 2015, 137, 1400–1403. [DOI] [PubMed] [Google Scholar]
- 27.Matyjaszewski K, Macromolecules, 2012, 45, 4015–4039. [Google Scholar]
- 28.Perrier S, Macromolecules, 2017, 50, 7433–7447. [Google Scholar]
- 29.Kolb HC, Finn MG and Sharpless KB, Angew Chem Int Edit, 2001, 40, 2004–2021. [DOI] [PubMed] [Google Scholar]
- 30.Herzberger J, Niederer K, Pohlit H, Seiwert J, Worm M, Wurm FR and Frey H, Chem Rev, 2016, 116, 2170–2243. [DOI] [PubMed] [Google Scholar]
- 31.Wagener KB, Boncella JM and Nel JG, Macromolecules, 1991, 24, 2649–2657. [Google Scholar]
- 32.Isarov SA and Pokorski JK, ACS Macro Letters, 2015, 4, 969–973. [DOI] [PubMed] [Google Scholar]
- 33.Moad G, Rizzardo E and Solomon DH, Polym Bull, 1982, 6, 589–593. [Google Scholar]
- 34.Chiefari J, Chong YK, Ercole F, Krstina J, Jeffery J, Le TPT, Mayadunne RTA, Meijs GF, Moad CL, Moad G, Rizzardo E and Thang SH, Macromolecules, 1998, 31, 5559–5562. [Google Scholar]
- 35.Wang J-S and Matyjaszewski K, J Am Chem Soc, 1995, 117, 5614–5615. [Google Scholar]
- 36.Grubbs RB, Polym Rev, 2011, 51, 104–137. [Google Scholar]
- 37.Kovaliov M, Cheng C, Cheng B and Averick S, Polym Chem, 2018, DOI: 10.1039/C8PY01026A. [DOI] [Google Scholar]
- 38.Yang W, Zhu L, Cui Y, Wang H, Wang Y, Yuan L and Chen H, ACS Appl. Mater. Interfaces, 2016, 8, 15967–15974. [DOI] [PubMed] [Google Scholar]
- 39.Averick S, Mehl RA, Das SR and Matyjaszewski K, J Control Release, 2015, 205, 45–57. [DOI] [PubMed] [Google Scholar]
- 40.Liang L and Astruc D, Coordin Chem Rev, 2011, 255, 2933–2945. [Google Scholar]
- 41.Lutz J-F and Börner HG, 2008, 33, 1–39. [Google Scholar]
- 42.Borchmann DE, Carberry TP and Weck M, Macromol Rapid Comm, 2013, 35, 27–43. [DOI] [PubMed] [Google Scholar]
- 43.Majorek KA, Porebski PJ, Dayal A, Zimmerman MD, Jablonska K, Stewart AJ, Chruszcz M and Minor W, Mol Immunol, 2012, 52, 174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lecolley F, Tao L, Mantovani G, Durkin I, Lautru S and Haddleton DM, Chem Commun 2004, DOI: 10.1039/b407712a, 2026–2027. [DOI] [PubMed] [Google Scholar]
- 45.Keleştemur S, Altunbek M and Culha M, Appl Surf Sci, 2017, 403, 455–463. [Google Scholar]
- 46.Carmali S, Murata H, Amemiya E, Matyjaszewski K and Russell AJ, ACS Biomater Sci Eng, 2017, 3, 2086–2097. [DOI] [PubMed] [Google Scholar]
- 47.Seo JH, Matsuno R, Lee Y, Konno T, Takai M and Ishihara K, Acta Biomater, 2011, 7, 1477–1484. [DOI] [PubMed] [Google Scholar]
- 48.De P, Li M, Gondi SR and Sumerlin BS, J Am Chem Soc, 2008, 130, 11288–11289. [DOI] [PubMed] [Google Scholar]
- 49.Grover GN, Alconcel SN, Matsumoto NM and Maynard HD, Macromolecules, 2009, 42, 7657–7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vandewalle S, De Coen R, De Geest BG and Du Prez FE, ACS Macro Lett, 2017, 6, 1368–1372. [DOI] [PubMed] [Google Scholar]
- 51.Gauthier MA and Klok HA, Biomacromol, 2011, 12, 482–493. [DOI] [PubMed] [Google Scholar]
- 52.Peeler JC, Woodman BF, Averick S, Miyake-Stoner SJ, Stokes AL, Hess KR, Matyjaszewski K and Mehl RA, J Am Chem Soc, 2010, 132, 13575–13577. [DOI] [PubMed] [Google Scholar]
- 53.Lee TC, Kang M, Kim CH, Schultz PG, Chapman E and Deniz AA, ChemBioChem, 2016, 17, 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borchmann DE, Carberry TP and Weck M, Macromol. Rapid Commun, 2014, 35, 112–112. [DOI] [PubMed] [Google Scholar]
- 55.Gauthier MA and Klok H-A, Chem. Commun, 2008, DOI: 10.1039/B719689J, 2591–2611. [DOI] [PubMed] [Google Scholar]
- 56.Cobo I, Li M, Sumerlin BS and Perrier S, Nat. Mater, 2014, 14, 143. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y and Wu C, Biomacromolecules, 2018, 19, 1804–1825. [DOI] [PubMed] [Google Scholar]
- 58.Artymiuk PJ, Blake CCF, Rice DW and Wilson KS, Acta Crystallogr. B, 1982, 38, 778–783. [Google Scholar]
- 59.Zheng C, Ma G and Su Z, Electrophoresis, 2007, 28, 2801–2807. [DOI] [PubMed] [Google Scholar]
- 60.Seyfried BK, Siekmann J, Belgacem O, Wenzel RJ, Turecek PL and Allmaier G, J Mass Spectrom, 2010, 45, 612–617. [DOI] [PubMed] [Google Scholar]
- 61.Cheng T-L, Chuang K-H, Chen B-M and Roffler SR, Bioconjugate Chem, 2012, 23, 881–899. [DOI] [PubMed] [Google Scholar]
- 62.Li N, Ziegemeier D, Bass L and Wang W, Journal of Pharmaceutical and Biomedical Analysis, 2008, 48, 1332–1338. [DOI] [PubMed] [Google Scholar]
- 63.Park EJ and Na DH, Analytical Chemistry, 2016, 88, 10848–10853. [DOI] [PubMed] [Google Scholar]
- 64.Huang L, Gough PC and Defelippis MR, Anal Chem, 2009, 81, 567–577. [DOI] [PubMed] [Google Scholar]
- 65.Wright TA, Lucius Dougherty M, Schmitz B, Burridge KM, Makaroff K, Stewart JM, Fischesser HD, Shepherd JT, Berberich JA, Konkolewicz D and Page RC, Bioconjugate Chemistry, 2017, 28, 2638–2645. [DOI] [PubMed] [Google Scholar]
- 66.Broyer RM, Grover GN and Maynard HD, Chem. Commun. (Camb). 2011, 47, 2212–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thilakarathne V, Briand VA, Zhou Y, Kasi RM and Kumar CV, Langmuir, 2011, 27, 7663–7671. [DOI] [PubMed] [Google Scholar]
- 68.Bouvier ESP and Koza SM, TrAC Trends Anal. Chem, 2014, 63, 85–94. [Google Scholar]
- 69.Fastrez J and Fersht AR, Biochem, 1973, 12, 2025–2034. [DOI] [PubMed] [Google Scholar]
- 70.Hofstee BHJ, Archives of Biochemistry and Biophysics, 1965, 112, 224–232. [Google Scholar]
- 71.Rodriguez-Martinez JA, Rivera-Rivera I, Sola RJ and Griebenow K, Biotechnol Lett, 2009, 31, 883–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cummings CS, Campbell AS, Baker SL, Carmali S, Murata H and Russell AJ, Biomacromol, 2017, 18, 576–586. [DOI] [PubMed] [Google Scholar]
- 73.Han XX, Xie Y, Zhao B and Ozaki Y, Anal Chem, 2010, 82, 4325–4328. [DOI] [PubMed] [Google Scholar]
- 74.Francis GL, Cytotechnology, 2010, 62, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang XF, Yang GY, Zhang Y, Xie Y, Withers SG and Feng Y, Sci Rep, 2016, 6, 33797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garcia DR and Lavignac N, Polym. Chem, 2016, 7, 7223–7229. [Google Scholar]
- 77.Imoto T, in eLS, 2017, DOI: doi:10.1002/9780470015902.a0000869.pub3. [Google Scholar]
- 78.Morgenstern J, Baumann P, Brunner C and Hubbuch J, Int J Pharm, 2017, 519, 408–417. [DOI] [PubMed] [Google Scholar]
- 79.Lucius M, Falatach R, McGlone C, Makaroff K, Danielson A, Williams C, Nix JC, Konkolewicz D, Page RC and Berberich JA, Biomacromolecules, 2016, 17, 1123–1134. [DOI] [PubMed] [Google Scholar]
- 80.Russell CS and Clarke LA, Clinical Genetics, 2001, 55, 389–394. [DOI] [PubMed] [Google Scholar]
- 81.Fensterl V and Sen GC, BioFactors, 2009, 35, 14–20. [DOI] [PubMed] [Google Scholar]
- 82.Tarhini AA, Gogas H and Kirkwood JM, J Immunol, 2012, 189, 3789–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Finter NB, Chapman S, Dowd P, Johnston JM, Manna V, Sarantis N, Sheron N, Scott G, Phua S and Tatum PB, Drugs, 1991, 42, 749–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grace MJ, Lee S, Bradshaw S, Chapman J, Spond J, Cox S, Delorenzo M, Brassard D, Wylie D, Cannon-Carlson S, Cullen C, Indelicato S, Voloch M and Bordens R, J Biol Chem, 2005, 280, 6327–6336. [DOI] [PubMed] [Google Scholar]
- 85.Li M, Yang S and Xu D, J Biol Chem, 2016, 291, 24160–24171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schett G, Redlich K and Smolen JS, Arthritis Res Ther, 2003, 5, 239–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saito-Yabe M, Kasuya Y, Yoshigae Y, Yamamura N, Suzuki Y, Fukuda N, Honma M, Yano K, Mochizuki S, Okada F, Okada A, Nagayama Y, Tsuda E, Fischer T, Hopner U, Zaja S, Mueller J, Okada J, Kurihara A, Ikeda T and Okazaki O, J Pharm Pharmacol, 2010, 62, 985–994. [DOI] [PubMed] [Google Scholar]
- 88.Tucker BS, Stewart JD, Aguirre JI, Holliday LS, Figg CA, Messer JG and Sumerlin BS, Biomacromolecules, 2015, 16, 2374–2381. [DOI] [PubMed] [Google Scholar]
- 89.Metcalf D, Cancer Immunol Res, 2013, 1, 351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Root RK and Dale DC, J Infect Dis, 1999, 179 Suppl 2, S342–352. [DOI] [PubMed] [Google Scholar]
- 91.Martinez C, Urbano-Ispizua A, Marin P, Merino A, Rovira M, Carreras E and Montserrat E, Bone Marrow Transplant, 1999, 24, 1273–1278. [DOI] [PubMed] [Google Scholar]
- 92.Martins A, Han J and Kim SO, IUBMB Life, 2010, 62, 611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gardner JC, Noel JG, Nikolaidis NM, Karns R, Aronow BJ, Ogle CK and McCormack FX, J Immunol, 2014, 192, 2405–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hustinx K, Heezius Burgers, Lammers and Hoepelman, Clinical & Experimental Immunology, 2001, 112, 334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weiss M, Moldawer LL and Schneider EM, Blood, 1999, 93, 425. [PubMed] [Google Scholar]
- 96.Mehta HM, Malandra M and Corey SJ, The Journal of Immunology, 2015, 195, 1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Skokowa J, Dale DC, Touw IP, Zeidler C and Welte K, Nature Reviews Disease Primers, 2017, 3, 17032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hill GR, Morris ES, Fuery M, Hutchins C, Butler J, Grigg A, Roberts A, Bradstock K, Szer J, Kennedy G, Morton J and Durrant S, Biology of Blood and Marrow Transplantation, 2006, 12, 603–607. [DOI] [PubMed] [Google Scholar]
- 99.Pelus LM, Curr Opin Hematol, 2008, 15, 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lalami Y and Klastersky J, Crit Rev Oncol Hematol, 2017, 120, 163–179. [DOI] [PubMed] [Google Scholar]
- 101.Himmelmann B, Himmelmann A, Furrer K, Halter J and Schanz U, Bone Marrow Transplant, 2002, 30, 491–496. [DOI] [PubMed] [Google Scholar]
- 102.Deotare U, Al-Dawsari G, Couban S and Lipton JH, Bone Marrow Transpl, 2015, 50, 1150–1156. [DOI] [PubMed] [Google Scholar]
- 103.Almenar D, Mayans J, Juan O, Bueno JM, Lopez JI, Frau A, Guinot M, Cerezuela P, Buscalla EG, Gasquet JA and Sanchez J, Eur J Cancer Care 2009, 18, 280–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Do BH, Kang HJ, Song JA, Nguyen MT, Park S, Yoo J, Nguyen AN, Kwon GG, Jang J, Jang M, Lee S, So S, Sim S, Lee KJ, Osborn MJ and Choe H, Sci Rep, 2017, 7, 6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lobner E, Traxlmayr MW, Obinger C and Hasenhindl C, Immunol Rev, 2016, 270, 113–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cox GN, Smith DJ, Carlson SJ, Bendele AM, Chlipala EA and Doherty DH, Exp Hematol, 2004, 32, 441–449. [DOI] [PubMed] [Google Scholar]
- 107.Cox GN, Chlipala EA, Smith DJ, Carlson SJ, Bell SJ and Doherty DH, PLoS One, 2014, 9, e91990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang YS, Wen XF, Wu YL, Wang YF, Fan M, Yang ZY, Liu W and Zhou LF, Eur J Pharm Biopharm, 2010, 74, 435–441. [DOI] [PubMed] [Google Scholar]
- 109.Davis SW, Ellsworth BS, Peréz Millan MI, Gergics P, Schade V, Foyouzi N, Brinkmeier ML, Mortensen AH and Camper SA, in Current Topics in Developmental Biology, ed. Thomas P, Academic Press, 2013, vol. 106, pp. 1–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vijayakumar A, Yakar S and Leroith D, Front Endocrinol (Lausanne), 2011, 2, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bartke A, NPJ Aging Mech Dis, 2017, 3, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ayyar VS, Indian J Endocr Metab, 2011, 15 Suppl 3, S162–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nissel R, Lindberg A, Mehls O, Haffner D and Pfizer B International Growth Database International, J Clin Endocrinol Metab, 2008, 93, 1359–1365. [DOI] [PubMed] [Google Scholar]
- 114.Bolar K, Hoffman AR, Maneatis T and Lippe B, J Clin Endocrinol Metab, 2008, 93, 344–351. [DOI] [PubMed] [Google Scholar]
- 115.Carrel AL, Myers SE, Whitman BY and Allen DB, J Clin Endocrinol Metab, 2002, 87, 1581–1585. [DOI] [PubMed] [Google Scholar]
- 116.Ferreira LV, Souza SA, Arnhold IJ, Mendonca BB and Jorge AA, J Clin Endocrinol Metab, 2005, 90, 5156–5160. [DOI] [PubMed] [Google Scholar]
- 117.Grigoletto A, Mero A, Zanusso I, Schiavon O and Pasut G, Macromol Biosci, 2016, 16, 50–56. [DOI] [PubMed] [Google Scholar]
- 118.Messmore JM, Fuchs DN and Raines RT, J Am Chem Soc, 1995, 117, 8057–8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M and Croce CM, Cancer Res, 2005, 65, 7065–7070. [DOI] [PubMed] [Google Scholar]
- 120.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F and Croce CM, Proc Natl Acad Sci U S A, 2002, 99, 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mironova N, Patutina O, Brenner E, Kurilshikov A, Vlassov V and Zenkova M, PLoS One, 2013, 8, e83482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mironova NL, Petrushanko IY, Patutina OA, Sen’kova AV, Simonenko OV, Mitkevich VA, Markov OV, Zenkova MA and Makarov AA, Cell Cycle, 2013, 12, 2120–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pouckova P, Zadinova M, Hlouskova D, Strohalm J, Plocova D, Spunda M, Olejar T, Zitko M, Matousek J, Ulbrich K and Soucek J, J Control Release, 2004, 95, 83–92. [DOI] [PubMed] [Google Scholar]
- 124.Liu S, Sun L, Wang J, Ma G, Su Z and Hu T, Mono-PEGylation of ribonuclease A: High PEGylation efficiency by thiolation with small molecular weight reagent, 2012.
- 125.Tubiana M, Acta Oncologica, 1989, 28, 113–121. [DOI] [PubMed] [Google Scholar]
- 126.Wilcox G, Clinical Biochemist Reviews, 2005, 26, 19–39. [PMC free article] [PubMed] [Google Scholar]
- 127.Rodger W, CMAJ: Canadian Medical Association Journal, 1991, 145, 1227–1237. [PMC free article] [PubMed] [Google Scholar]
- 128.Morello CM, International Journal of General Medicine, 2011, 4, 827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hinds K, Koh JJ, Joss L, Liu F, Baudyš M and Kim SW, Bioconjugate Chemistry, 2000, 11, 195–201. [DOI] [PubMed] [Google Scholar]
- 130.Hölker U, Dohse J and Höfer M, Folia Microbiologica, 2002, 47, 423–427. [DOI] [PubMed] [Google Scholar]
- 131.Schuetz M, Benske A, Smith RA, Watanabe Y, Tobimatsu Y, Ralph J, Demura T, Ellis B and Samuels AL, Plant Physiol, 2014, 166, 798–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Su Y, Xian H, Shi S, Zhang C, Manik SM, Mao J, Zhang G, Liao W, Wang Q and Liu H, BMC Biotechnol, 2016, 16, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Moreno AD, Ibarra D, Fernandez JL and Ballesteros M, Bioresour Technol, 2012, 106, 101–109. [DOI] [PubMed] [Google Scholar]
- 134.Bagewadi ZK, Mulla SI and Ninnekar HZ, Int J Recycl Org Waste Agricult, 2017, 6, 351–365. [Google Scholar]
- 135.Bauer CG, Kühn A, Gajovic N, Skorobogatko O, Holt P-J, Bruce NC, Makower A, Lowe CR and Scheller FW, Fresenius J. of Anal. Chem, 1999, 364, 179–183. [Google Scholar]
- 136.Osma JF, Toca-Herrera JL and Rodriguez-Couto S, Enzyme Res, 2010, 2010, 918761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang G-Q, Tian T, Liu Y-P, Wang H-X and Chen Q-J, Process Biochem, 2011, 46, 2336–2340. [Google Scholar]
- 138.Xu F, Kulys JJ, Duke K, Li K, Krikstopaitis K, Deussen H-JW, Abbate E, Galinyte V and Schneider P, Applied and Environmental Microbiology, 2000, 66, 2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Su J, Noro J, Loureiro A, Martins M, Azoia NG, Fu J, Wang Q, Silva C and Cavaco-Paulo A, ChemCatChem, 2017, 9, 3888–3894. [Google Scholar]
- 140.Li L, Xie J, Yu S, Su Z, Liu S, Liu F, Xie C, Zhang B and Zhang C, Green Chemistry, 2013, 15, 1624–1630. [Google Scholar]
- 141.De Diego T, Lozano P, Gmouh S, Vaultier M and Iborra JL, Biomacromol, 2005, 6, 1457–1464. [DOI] [PubMed] [Google Scholar]
- 142.Engel P, Mladenov R, Wulfhorst H, Jäger G and Spiess AC, Green Chem, 2010, 12, 1959–1966. [Google Scholar]
- 143.Wang Y, Wang X, Tang R, Yu S, Zheng B and Feng Y, Journal of Molecular Catalysis B: Enzymatic, 2010, 66, 294–301. [Google Scholar]
- 144.Fink CS, Hamosh P and Hamosh M, Pediatr Res, 1984, 18, 248–254. [DOI] [PubMed] [Google Scholar]
- 145.Sarac N and Ugur A, Desalination and Water Treatment, 2016, 57, 19750–19759. [Google Scholar]
- 146.Kobayashi S, Proceedings of the Japan Academy. Series B, Physical and Biological Sciences, 2010, 86, 338–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ansorge-Schumacher MB and Thum O, Chem Soc Rev, 2013, 42, 6475–6490. [DOI] [PubMed] [Google Scholar]
- 148.Zhao X, Qi F, Yuan C, du W and Liu D, Lipase-catalyzed process for biodiesel production: Enzyme immobilization, process simulation and optimization, 2015.
- 149.Ge J, Yan M, Lu D, Zhang M and Liu Z, Hyperbranched polymer conjugated lipase with enhanced activity and stability, 2007.
- 150.Kovaliov M, Allegrezza ML, Richter B, Konkolewicz D and Averick S, Polymer, 2018, 137, 338–345. [Google Scholar]
- 151.Duke FR, Weibel M, Page DS, Bulgrin VG and Luthy J, J Am Chem Soc, 1969, 91, 3904–3909. [Google Scholar]
- 152.Ferri S, Kojima K and Sode K, J. Diabetes Sci. Technol, 2011, 5, 1068–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tiina M and Sandholm M, International Journal of Food Microbiology, 1989, 8, 165–174. [DOI] [PubMed] [Google Scholar]
- 154.Kim KK-A, Fravel DR and Papavizas GC, Canadian Journal of Microbiology, 1990, 36, 760–764. [DOI] [PubMed] [Google Scholar]
- 155.Wong CM, Wong KH and Chen XD, Applied Microbiology and Biotechnology, 2008, 78, 927–938. [DOI] [PubMed] [Google Scholar]
- 156.Berron BJ, Johnson LM, Ba X, McCall JD, Alvey NJ, Anseth KS and Bowman CN, Biotechnol Bioeng, 2011, 108, 1521–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ritter DW, Roberts JR and McShane MJ, Enzyme Microb Technol, 2013, 52, 279–285. [DOI] [PubMed] [Google Scholar]
- 158.Campbell AS, Islam MF and Russell AJ, Electrochimica Acta, 2017, 248, 578–584. [Google Scholar]
- 159.Bibi Z, Ansari A, Zohra RR, Aman A and Ul Qader SA, J. Radiat. Res. Appl. Sci, 2014, 7, 478–485. [Google Scholar]
- 160.Uchida H, Kusakabe I, Kawabata Y, Ono T and Murakami K, Journal of Fermentation and Bioengineering, 1992, 74, 153–158. [Google Scholar]
- 161.Ravindran V, The Journal of Applied Poultry Research, 2013, 22, 628–636. [Google Scholar]
- 162.Shen B, Sun X, Zuo X, Shilling T, Apgar J, Ross M, Bougri O, Samoylov V, Parker M, Hancock E, Lucero H, Gray B, Ekborg NA, Zhang D, Johnson JCS, Lazar G and Raab RM, Nature Biotechnology, 2012, 30, 1131. [DOI] [PubMed] [Google Scholar]
- 163.Battan B, Dhiman SS, Ahlawat S, Mahajan R and Sharma J, Indian Journal of Microbiol, 2012, 52, 222–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ahmad Z, Butt MS, Ahmed A, Riaz M, Sabir SM, Farooq U and Rehman FU, J Food Sci Tech, 2014, 51, 2445–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mehnati-Najafabadi V, Taheri-Kafrani A and Bordbar A-K, International Journal of Biological Macromolecules, 2018, 107, 418–425. [DOI] [PubMed] [Google Scholar]
- 166.Mhamdi A, Queval G, Chaouch S, Vanderauwera S, Van Breusegem F and Noctor G, J Exp Bot, 2010, 61, 4197–4220. [DOI] [PubMed] [Google Scholar]
- 167.Cichello SA, Food Sci J. Technol, 2015, 52, 1889–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pack MY, Vedamuthu ER, Sandine WE and Elliker PR, Journal of Dairy Science, 1968, 51, 511–516. [DOI] [PubMed] [Google Scholar]
- 169.Yoon DS, Won K, Kim YH, Song BK, Kim SJ, Moon SJ and Kim BS, Water Science and Technology, 2007, 55, 27–33. [DOI] [PubMed] [Google Scholar]
- 170.Riccardi CM, Cole KS, Benson KR, Ward JR, Bassett KM, Zhang Y, Zore OV, Stromer B, Kasi RM and Kumar CV, Bioconjug Chem, 2014, 25, 1501–1510. [DOI] [PubMed] [Google Scholar]
- 171.Rodriguez-Martinez JA, Sola RJ, Castillo B, Cintron-Colon HR, Rivera-Rivera I, Barletta G and Griebenow K, Biotechnol Bioeng, 2008, 101, 1142–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sola RJ and Griebenow K, FEBS Lett, 2006, 580, 1685–1690. [DOI] [PubMed] [Google Scholar]
- 173.Kato A, Tanimoto S, Muraki Y, Kobayashi K and Kumagai I, Biosci Biotechnol Biochem, 1992, 56, 1424–1428. [DOI] [PubMed] [Google Scholar]
- 174.Swaminathan R, Ravi VK, Kumar S, Kumar MV and Chandra N, Adv Protein Chem Struct Biol, 2011, 84, 63–111. [DOI] [PubMed] [Google Scholar]
- 175.Strynadka NC and James MN, EXS, 1996, 75, 185–222. [DOI] [PubMed] [Google Scholar]
- 176.Kunz W, Henle J and Ninham BW, Current Opinion in Colloid & Interface Science, 2004, 9, 19–37. [Google Scholar]
- 177.Mancini RJ, Lee J and Maynard HD, Journal of the American Chemical Society, 2012, 134, 8474–8479. [DOI] [PubMed] [Google Scholar]
- 178.Nayar S, Mir A, Ashok A, Guha A and Sharma V, Journal of Bionic Engineering, 2010, 7, 29–34. [Google Scholar]
- 179.Goncharov NV, Belinskaia DA, Shmurak VI, Terpilowski MA, Jenkins RO and Avdonin PV, Molecules, 2017, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Plesner B, Fee CJ, Westh P and Nielsen AD, Eur J Pharm Biopharm, 2011, 79, 399–405. [DOI] [PubMed] [Google Scholar]
- 181.Cummings C, Murata H, Koepsel R and Russell AJ, Biomacromol, 2014, 15, 763–771. [DOI] [PubMed] [Google Scholar]
- 182.Narasimhan D, Woods JH and Sunahara RK, Future Med Chem, 2012, 4, 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Narasimhan D, Collins GT, Nance MR, Nichols J, Edwald E, Chan J, Ko MC, Woods JH, Tesmer JJ and Sunahara RK, Mol Pharmacol, 2011, 80, 1056–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Giardina B, Messana I, Scatena R and Castagnola M, Crit Rev Biochem Mol Biol, 1995, 30, 165–196. [DOI] [PubMed] [Google Scholar]
- 185.Kapralov A, Vlasova II, Feng W, Maeda A, Walson K, Tyurin VA, Huang Z, Aneja RK, Carcillo J, Bayir H and Kagan VE, J Biol Chem, 2009, 284, 30395–30407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hebert JP, Scriban R and Strobbel B, Journal of the American Society of Brewing Chemists, 1978, 36, 31–38. [Google Scholar]
- 187.Bekhit AA, Hopkins DL, Geesink G, Bekhit AA and Franks P, Crit Rev Food Sci Nutr, 2014, 54, 1012–1031. [DOI] [PubMed] [Google Scholar]
- 188.Fernández-Lucas J, Castañeda D and Hormigo D, “New trends for a classical enzyme: Papain, a biotechnological success story in the food industry ”, 2017.
- 189.Miyamoto D, Watanabe J and Ishihara K, Biomaterials, 2004, 25, 71–76. [DOI] [PubMed] [Google Scholar]
- 190.Wright TA, Stewart JM, Page RC and Konkolewicz D, J Phys Chem Lett, 2017, 8, 553–558. [DOI] [PubMed] [Google Scholar]
- 191.Lopez-Cruz JI, Viniegra-Gonzalez G and Hernandez-Arana A, Bioconjug Chem, 2006, 17, 1093–1098. [DOI] [PubMed] [Google Scholar]
- 192.Lee PW, Isarov SA, Wallat JD, Molugu SK, Shukla S, Sun JE, Zhang J, Zheng Y, Lucius Dougherty M, Konkolewicz D, Stewart PL, Steinmetz NF, Hore MJ and Pokorski JK, J Am Chem Soc, 2017, 139, 3312–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wei M, Gao Y, Li X and Serpe MJ, Polymer Chemistry, 2017, 8, 127–143. [Google Scholar]
- 194.Murata H, Cummings CS, Koepsel RR and Russell AJ, Biomacromolecules, 2013, 14, 1919–1926. [DOI] [PubMed] [Google Scholar]
- 195.Wang L, Li X, Yuan L, Wang H, Chen H and Brash JL, Journal of Materials Chemistry B, 2015, 3, 498–504. [DOI] [PubMed] [Google Scholar]
- 196.Kajander T, Kellosalo J and Goldman A, FEBS Lett, 2013, 587, 1863–1869. [DOI] [PubMed] [Google Scholar]
- 197.Scott C, Mitrovic B, Eastwood S and Kinsel G, Polymer (Guildf), 2014, 55, 3551–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Huang X, Li M, Green DC, Williams DS, Patil AJ and Mann S, Nat Commun, 2013, 4, 2239. [DOI] [PubMed] [Google Scholar]
- 199.Sasso F, Natalello A, Castoldi S, Lotti M, Santambrogio C and Grandori R, Biotechnol J, 2016, 11, 954–960. [DOI] [PubMed] [Google Scholar]
- 200.Graham JE, Clark ME, Nadler DC, Huffer S, Chokhawala HA, Rowland SE, Blanch HW, Clark DS and Robb FT, Nat Commun, 2011, 2, 375. [DOI] [PubMed] [Google Scholar]
- 201.Johnson E, Biofuels, Bioproducts and Biorefining, 2016, 10, 164–174. [Google Scholar]
- 202.Mackenzie KJ and Francis MB, J Am Chem Soc, 2013, 135, 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Shimoboji T, Larenas E, Fowler T, Kulkarni S, Hoffman AS and Stayton PS, Proc Natl Acad Sci U S A, 2002, 99, 16592–16596. [DOI] [PMC free article] [PubMed] [Google Scholar]