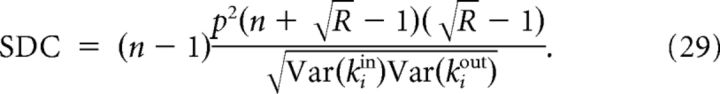Abstract
The structure in cortical microcircuits deviates from what would be expected in a purely random network, which has been seen as evidence of clustering. To address this issue, we sought to reproduce the nonrandom features of cortical circuits by considering several distinct classes of network topology, including clustered networks, networks with distance-dependent connectivity, and those with broad degree distributions. To our surprise, we found that all of these qualitatively distinct topologies could account equally well for all reported nonrandom features despite being easily distinguishable from one another at the network level. This apparent paradox was a consequence of estimating network properties given only small sample sizes. In other words, networks that differ markedly in their global structure can look quite similar locally. This makes inferring network structure from small sample sizes, a necessity given the technical difficulty inherent in simultaneous intracellular recordings, problematic. We found that a network statistic called the sample degree correlation (SDC) overcomes this difficulty. The SDC depends only on parameters that can be estimated reliably given small sample sizes and is an accurate fingerprint of every topological family. We applied the SDC criterion to data from rat visual and somatosensory cortex and discovered that the connectivity was not consistent with any of these main topological classes. However, we were able to fit the experimental data with a more general network class, of which all previous topologies were special cases. The resulting network topology could be interpreted as a combination of physical spatial dependence and nonspatial, hierarchical clustering.
SIGNIFICANCE STATEMENT The connectivity of cortical microcircuits exhibits features that are inconsistent with a simple random network. Here, we show that several classes of network models can account for this nonrandom structure despite qualitative differences in their global properties. This apparent paradox is a consequence of the small numbers of simultaneously recorded neurons in experiment: when inferred via small sample sizes, many networks may be indistinguishable despite being globally distinct. We develop a connectivity measure that successfully classifies networks even when estimated locally with a few neurons at a time. We show that data from rat cortex is consistent with a network in which the likelihood of a connection between neurons depends on spatial distance and on nonspatial, asymmetric clustering.
Keywords: clustering, cortical connectivity, microcircuits, multiple patch-clamp, networks, statistics
Introduction
The organization of cortical microcircuits varies across brain areas and species and undergoes continual plastic modifications (Trachtenberg et al., 2002; Zuo et al., 2005; Le Bé and Markram, 2006; Hofer et al., 2009). However, these circuits also exhibit certain regularities, the canonical example of which is a well defined vertical organization into layers. The existence of conserved connectivity principles suggests the notion of a neocortex composed of a juxtaposition of similarly structured building blocks (Szentágothai, 1978; Mountcastle, 1997; Silberberg et al., 2002), which are dynamically adjusted to respond to the precise demands of every subsystem.
Intracellular recording techniques can detect synaptic connections between pairs of neurons in cortical slices directly (Mason et al., 1991; Markram et al., 1997; Holmgren et al., 2003; Song et al., 2005; Perin et al., 2011). A limitation of these techniques is that they currently allow for the study of only small groups of neurons simultaneously. Therefore, circuit reconstructions require an inference process from partial data. Despite this, experimental studies have brought to light some fundamental common principles; for example, connections tend to be sparse, with connection rates between pyramidal neurons in the range of 5–15% (Mason et al., 1991; Markram et al., 1997; Holmgren et al., 2003; Le Bé and Markram, 2006; Wang et al., 2006; Ko et al., 2011). Interestingly, there is increasing evidence that the connectivity between pyramidal neurons is far from the Erdös–Rényi (ER) model, in which connections appear independently with a fixed probability p. These so-called “nonrandom” features include an excess of reciprocal connections, which can be quantified by the ratio between the number of bidirectional connections and the expected number of such connections in ER networks with equivalent connection rates (R). R has been reported to be ∼2–4 in visual cortex (Mason et al., 1991; Song et al., 2005; Wang et al., 2006), 3–4 in somatosensory cortex (Markram et al., 1997; Le Bé and Markram, 2006), and 4 in mPFC (Song et al., 2005; Wang et al., 2006). In addition, there is an overrepresentation of highly connected motifs (Song et al., 2005; Perin et al., 2011) and the connection probability between neuron pairs increases with the number of shared neighbors (Perin et al., 2011). Some initiatives are seeking to leverage these data to construct realistic microcircuit models for numerical simulation (Hill et al., 2012; Markram et al., 2015; Ramaswamy et al., 2015; Reimann et al., 2015). Conversely, a recent theoretical study has shown that some of these features arise naturally in network models that maximize the number of stored memories (Brunel, 2016).
Here, we studied several broad classes of network structure that could potentially explain the observed nonrandomness. These include clustered networks (Litwin-Kumar and Doiron, 2014), spatially structured networks (Holmgren et al., 2003; Perin et al., 2011; Jiang et al., 2015), and networks defined by strong heterogeneity in the number of incoming and outgoing connections of neurons (Roxin, 2011; Timme et al., 2016).
Surprisingly, all of these network classes were compatible with the reported nonrandomness. In fact, we found that networks with qualitatively distinct global structure could yield similar statistical features when all of the available information came from the study of small groups of neurons, as in electrophysiological experiments in slice. However, we found that a particular combination of motifs, known as the sample degree correlation (SDC), provides a unique fingerprint for each network class based only on the analysis of small samples of neurons. Using the SDC, we showed that microcircuit data from rat somatosensory cortex (Perin et al., 2011) and from rat visual cortex (Song et al., 2005) were incompatible with any of these network classes. Rather, the data led us to develop a more general network class that reduces to the previous models under certain constraints. Our results suggest that the nonrandom features of cortical microcircuits reflect a combination of spatially decaying connectivity and additional nonspatial structure, which, however, is not simple clustering.
Materials and Methods
Network models
All of the networks are treated as directed graphs with N neurons. We assume that the network's size N is large and that the network is sparse, meaning that its connection density p is “small.” We use the following notations: i → j: a connection exists from neuron i to neuron j; i ↛ j: a connection exists from i to j but not from j to i; and i ↔ j: there is a bidirectional connection between i and j.
ER.
Connections are generated independently with probability p.
ER bidirectional (ER-Bi).
Connections between a pair of neurons (i, j) are generated independently according to the following:
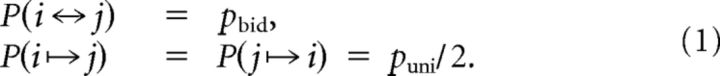 |
The sparseness and the number of bidirectional connections relative to random are as follows:
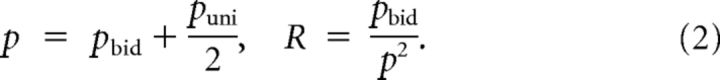 |
Clusters (Cl).
Each neuron belongs to one or more clusters and cluster membership is homogeneous across the network. This means that, for any neuron i, the number of other neurons that share a cluster with i is almost constant. More precisely, if ni denotes the number of neurons that are at least in one of the clusters of i, then the following is true:
 |
as N → ∞. The typical example is a network with a fixed number of clusters C ≪ N where each neuron belongs to one cluster that is chosen uniformly at random. In this case, ni ∼ Binomial(N − 1, 1/C), so
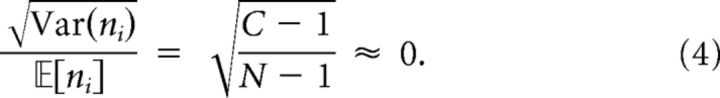 |
Connections are generated independently with probability p+ when neurons are in the same cluster and p− otherwise, p− < p+. Defining f+ and f− = 1 − f+ as the expected fraction of pairs in the same and in different clusters, respectively, p and R are as follows:
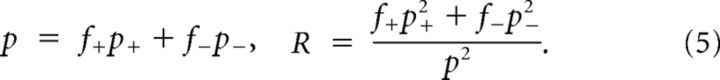 |
In our simulations, each neuron belongs to one cluster which is chosen uniformly at random, so the expected cluster size is N/C and:
 |
Clusters with heterogeneous membership (Cl-Het).
Each neuron belongs to zero, one, or more clusters, but now cluster membership is heterogeneous across neurons, which means that Equation 3 does not necessarily hold. Connections are defined as in the previous model. In our simulations, we have considered networks with C ≪ N clusters where each neuron has a probability pc = 1/C of belonging to any given cluster. Therefore, neurons can be simultaneously in different clusters and clusters may have nonempty overlap. p and R are given by Equation 5, as before, but now the expected fraction of pairs in the same cluster is as follows:
Defining again ni as the number of neurons that are at least in one of the clusters of i, then:
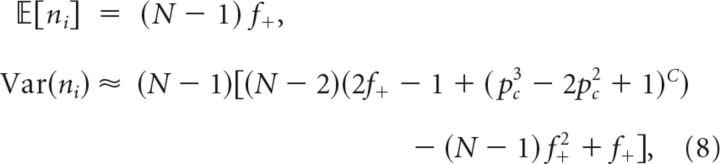 |
so, if C is fixed and N is large, then:
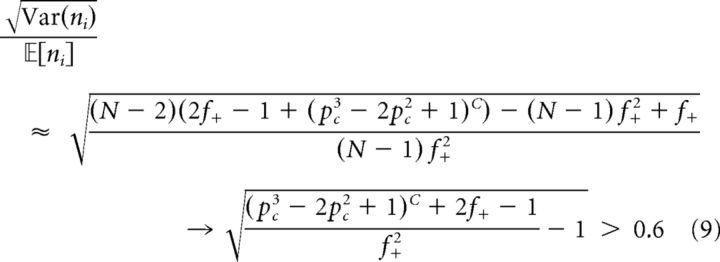 |
for C ≥ 2. This means that there is a non-negligible variability across neurons in terms of cluster membership, which has important consequences for the statistics that we will consider later.
Distance (Dis).
Connections are made independently with a probability that decays with the distance rij between the neurons i and j as follows:
And:
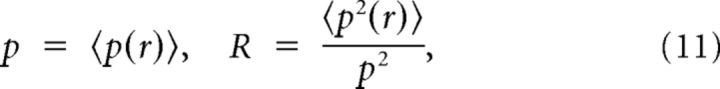 |
where 〈〉 denotes an average over the distribution of distances in the network. We assume that distances are homogeneously distributed in the network; that is, that the proportion of neurons that are a given distance away from a neuron does not vary substantially from i to i. This condition is analogous to Equation 3 for clustered networks. When it does not hold, the model belongs to the Cl-Het class in terms of the properties studied here.
Degree (Deg).
We consider networks defined by a given joint in-/out-degree distribution f(in, out)(k, k′). One realization of the model is obtained by generating a degree sequence {(Kiin, Kiout)}i=1N from N independent instantiations of f(in, out) and uniformly selecting one network among the family of directed graphs that have {(Kiin,Kiout)}i=1N as their degree sequence.
Because the number of edges in any directed network equals the sum of the in-degrees and the sum of the out-degrees, the expectation of the in-degree and the out-degree have to be equal:
The sparseness is as follows:
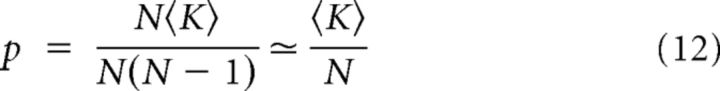 |
in the large N limit.
In this model, the connection probability once the network degrees are known can be approximated by the following:
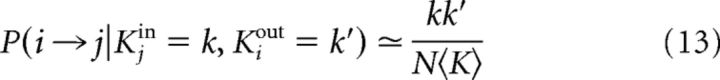 |
and because, once conditioned to the degrees of neurons i and j, i → j, and j → i can be considered independent events,
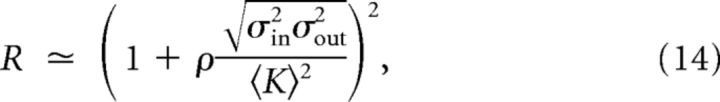 |
where σin2, σout2 and ρ stand for the in-/out-degree variances and the Pearson correlation coefficient of individual in-/out-degrees, respectively.
Modulator (Mod) model.
It is possible to consider a very general class of network models in which each neuron i has an associated parameter xi and the connections are made independently with probability:
where {xi}i=1N are independent and identically distributed random variables. All the previous models except the ER-Bi can be interpreted, at least locally, as particular cases of this model.
In clustered networks (Cl and Cl-Het), xi denotes the cluster membership of neuron i, whereas in the Dis model, xi represents the “position” of neuron i. In both of these cases, the connection probability depends on a notion of distance between pairs, so the function g is symmetric: g(x, y) = g(y, x). Moreover, in a random sample of the Cl and Dis models, coexistence in a cluster or distance can be assumed to be independent from pair to pair, as long as the sample size is small compared with the network size. In the Cl-Het model, this is not the case by virtue of the neuron-to-neuron heterogeneity in cluster membership: the likelihood of a connection from a neuron i is highly dependent on the number of other neurons in the network that share a cluster with i (the quantity ni defined before). Because this quantity varies significantly from neuron to neuron, connections from neuron i cannot be assumed to appear independently. In the particular case in which the clusters of neuron i are chosen independently with a fixed probability, this heterogeneity is captured by the number of clusters to which each neuron belongs, which can be considered the effective modulatory variable.
In the Deg model, the connection probability from neuron i to neuron j once the degrees are known can be approximated by Equation 13. Additional connections from neuron i can be assumed to be made independently as long as k ≫ 1. This independence assumption can be extended up to a group of n neurons as long as the degrees are large compared with n and n ≪ N. Then, the Deg model becomes a special case of the Mod model in which xi = (xiin, xiout) is the 2D vector of the degrees of i and g(x, y) = g1(x)g2(y), where g1(a, b) = , g2(a, b) = .
Generation of distance-dependent networks
In the simulations of Figures 1, 2, 3, 4, 5, and 6A, we considered neurons arranged in periodic rings where r ∈ {0, 1, …, [N/2]} and:
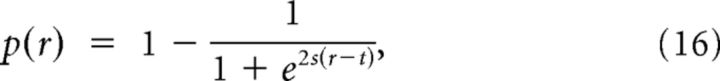 |
which, for s < 0, defines a decreasing sigmoid function with an absolute slope that is maximal at r = t and its value is −s. In the simulations of Figure 6, C and D, we also included 2D periodic lattices where r ∈ {0, 1, …, [/2]} and p(r) was given by Equation 16.
Figure 1.
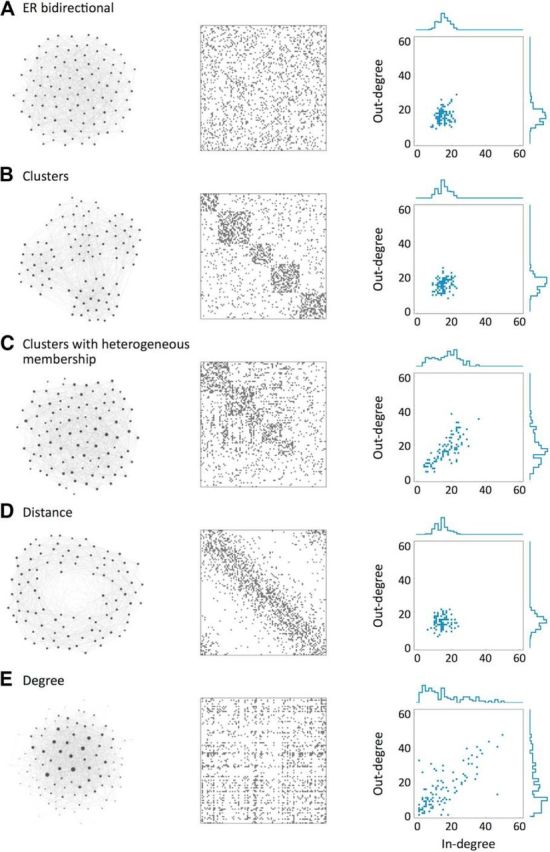
Schematic representation of the models: connectivity (left), adjacency matrix (middle) and in-/out-degree distribution (right). The nodes in the left column are arranged according to the ForceAtlas algorithm using Gephi software (Bastian et al., 2009). The size of each node is proportional to the sum of its degrees and the direction of the connections has been omitted for simplicity. In all the networks, N = 100, p = 0.15, r = 2. A, An ER network in which the fraction of bidirectional connections can be varied independent of the sparseness. B, A network with clusters. C, A network with clusters in which the cluster membership is heterogeneous. D, A network with distance-dependent connectivity. E, A network with prescribed joint degree distribution.
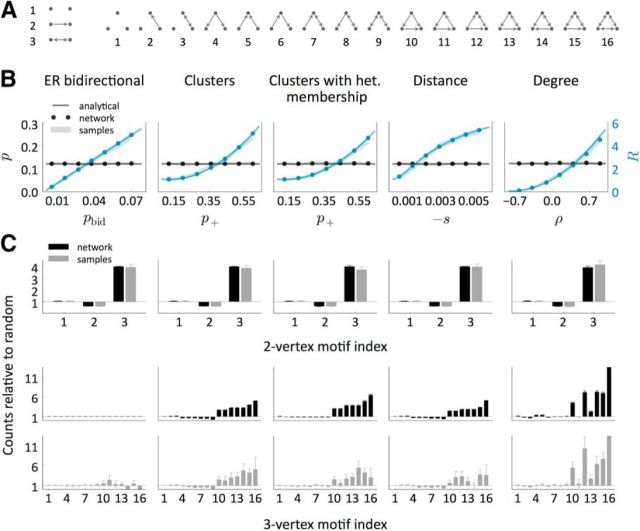
Figure 2.
Counts of two- and three-neuron motifs relative to random models. A, Representation of all the possible two- and three-neuron motifs. B, Sparseness (p) and expected number of reciprocal connections relative to random (R) as a function of a model parameter. In all of the models except the Deg, an additional parameter was varied (puni, p−, p−, t, respectively) to keep p constant. In the Dis model, neurons are arranged in a ring and the connection probability as a function of distance r is defined by the sigmoid function p(r) = 1 − , so t is the point where the absolute slope is maximal and −s is this absolute slope. C, Counts of all the two- and three-neuron motifs relative to random models (ER and ER-Bi, respectively) in networks with p = 0.12, R = 4. We used five different networks of size N = 2000 per condition. The computations were performed both on the whole network and on 163 samples of size 4 per network. Shaded regions and error bars indicate mean ± SEM.
Figure 3.
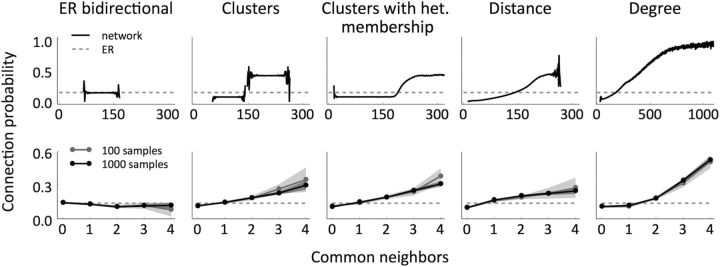
Connection probability as a function of the number of common neighbors for the different models, in the whole network (top) and in samples of size 12 (bottom). In all the cases, N = 2000, p = 0.14, R = 2. The analyses were performed on five networks and the shaded regions indicate the resulting mean ± SEM. In the sample analyses, we took 20 samples per network (100 in total, gray) and 200 samples per network (1000 in total, black). The dotted lines show the expected probability if it were independent of the number of common neighbors, as in the ER and ER-Bi models.
Figure 4.
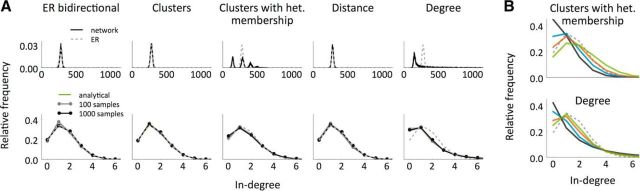
In-degree distribution of the different network models. A, In-degree distribution in the whole network (top) versus in-degree distribution in samples of size 12 (bottom) and comparison with the distributions exhibited by the ER model (dotted lines). The networks and samples used are the same as in Figure 3. The shaded regions indicate mean ± SEM. B, In-degree distributions in samples of size 12 for different networks generated according to the Cl-Het (top) and the Deg (bottom) models, all of them with N = 2000, p = 0.14, R = 2.
Figure 5.
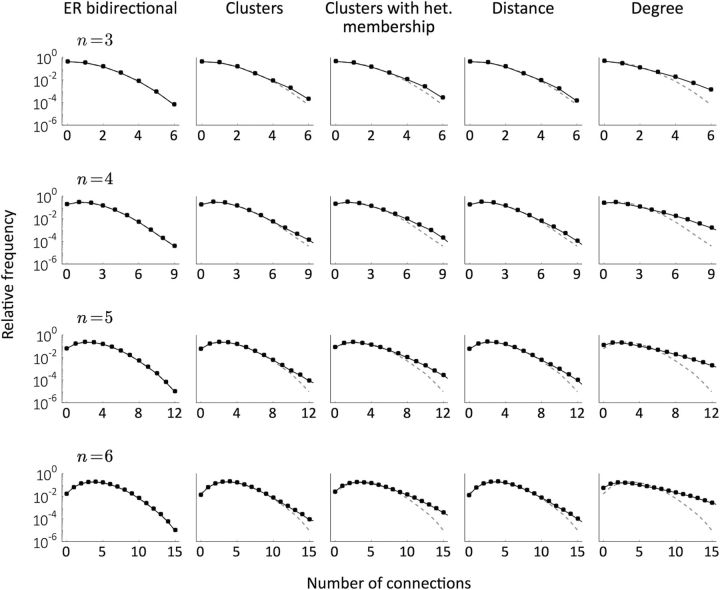
Distribution of the total number of connections in samples of sizes n ∈ {3, … ,6} for the different models (black) compared with the distribution obtained in ER bidirectional networks with the same p and R (dashed gray). The parameters are the same as in Figures 3 and 4. The analyses were performed on five networks per condition and the computations come from 105 random samples for each network.
Figure 6.
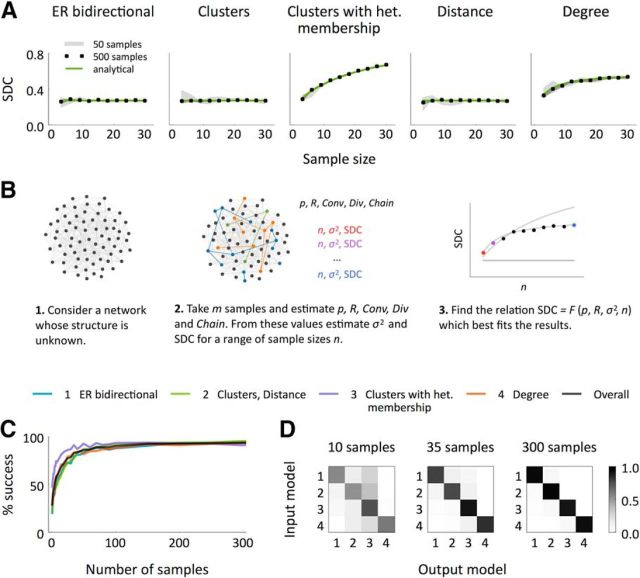
Sample in-/out-degree correlation (SDC) as a measure to distinguish between classes of networks. A, SDC in samples of 3 to 30 neurons in the different models. In all the networks, N = 2000, p = 0.12, R = 3. We computed the empirical correlations using 50 and 500 samples per network for each sample size. Every analysis was performed independently in five different networks and the shaded region indicates the resulting mean ± SEM. B, Schematic representation of the algorithm proposed to distinguish between the model classes: (1) ER-Bi, (2) Cl/Dis, (3) Cl-Het, and (4) Deg. C, Success rate of the algorithm performed on randomly generated networks with N = 2000, p ∈ [0.05, 0.23], R ∈ [1.5, 4.1] as a function of the number of samples considered m. All the samples had size n′ = 12. Each success rate was computed over 2000 experiments. D Frequencies of all the possible input–output combinations in the experiments shown in C for three choices of the number of analyzed samples. Each frequency is normalized by the frequency of the input model so that the sum of every row is 1.
Generation of networks from a prescribed in-/out-degree distribution
To generate networks according to the Deg model, we have used the following method: given a joint distribution defined by f̃(in, out), we independently assign to each node i a pair (K̃iin, K̃iout). Then, we create each connection i → j independently with probability . The final degrees in the network satisfy 〈Kiin|K̃iin〉 = K̃iin and 〈Kiout|K̃iout〉 = K̃iout. Despite the resulting degree distribution in the network is no longer given by f̃(in, out), the statistics 〈K〉 and Cov(Kin, Kout) are preserved (assuming that N is large and K̃in/out ≪ N). The degree variances become larger, in particular σin/out2 = σ̃in/out2 + 〈K̃〉, and this results in the correlation coefficient being smaller, ρ < ρ̃.
In all our simulations, the variables K̃in, K̃out followed Gamma distributions with a shift of magnitude D > 0. In almost all our simulations, they had to be positively correlated and we defined them in the following way: if X ∼ Gamma(κ1, θ) and Y, Z ∼ Gamma(κ2, θ) (κ, θ > 0) are independent random variables, we set the following:
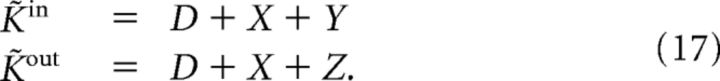 |
K̃in and K̃out follow D-shifted Gamma(κ = κ1 + κ2, θ) distributions and their correlation coefficient is ρ̃ = κ1/κ. In Figure 2B, we also constructed networks with negative degree correlation. In this case we first generated K̃in and K̃out independently and then we inversely ordered the two sequences {K̃iin}i=1N and {K̃iout}i=1N. By reordering a fraction of values in one of the two sequences we could adjust the correlation coefficient.
Parameter values for Mod networks shown in Figure 8
Figure 8.
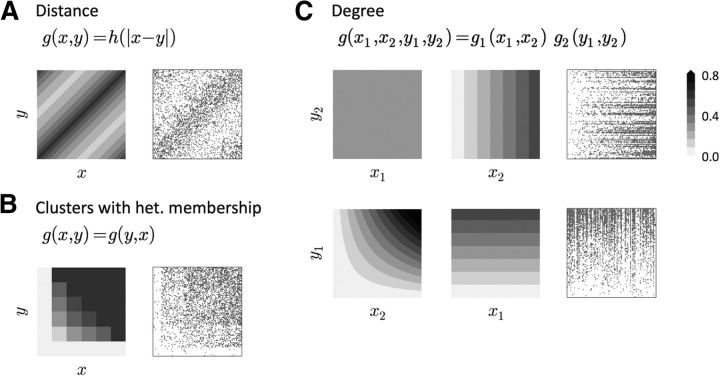
Examples of modulatory functions g and adjacency matrices of the three main classes of networks described here. Notice that the row ordering in the adjacency matrices has been inverted to be coherent with the g plot. A, In the Dis model, the modulators x and y represent the spatial position of presynaptic and postsynaptic neurons, respectively. B, In the Cl-Het model, x and y represent the number of clusters to which presynaptic and postsynaptic neurons belong and g is a symmetric function. C, In the Deg model, (x1, x2) represents the the in-degree and out-degree of the presynaptic neuron and (y1, y2) are the degrees of the postsynaptic neuron. The adjacency matrices result from ordering neurons according to their out-degree (top) and their in-degree (bottom). See Materials and Methods for details and parameter values of the Mod networks.
For all three networks, N = 100, p = 0.3. In the Dis model, the modulatory variable represents spatial position and g is a function of the distance |x − y|. The ordering of neurons in the adjacency matrix corresponds to their position in a ring. In the Cl-Het model, x and y represent the number of clusters to which presynaptic and postsynaptic neurons belong and g is a symmetric function. In particular, g(x, y) = p+(1 − f−(x, y)) + p−f−(x, y), where f− (x, y) is the probability that the two neurons do not coincide in a cluster given x, y. Explicitly, f−(x, y) = if x + y ≤ C and 0 otherwise (where C is the total number of clusters in the network). Neurons in the adjacency matrix have also been ordered according to the number of clusters to which they belong. In this example, C = 5 and each neuron was assigned to each cluster with a fixed probability, so the fraction of neurons that belong to k ∈ {0, 1, …, C} clusters is not uniform. This is why the width of the different domains of the adjacency matrix and the g plot do not coincide. In the Deg model, (x1, x2) represents the in-degree and out-degree of the presynaptic neuron and (y1, y2) are the degrees of the post-synaptic neuron. g(x1, x2, y1, y2) = c x2 y1, so g is separable with respect to the presynaptic and postsynaptic variables. We show different projections of g: g(x1, −, −, y2) (top left), g(−, x2, −, y2) (top right), g(−, x2, y1, −) (bottom left) and g(x1, −, y1, −) (bottom right). The adjacency matrices result from ordering neurons according to their out-degree (top) and their in-degree (bottom).
Definition of the model that fits the data
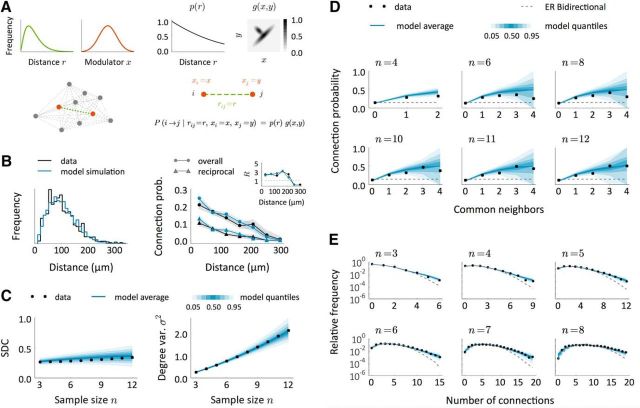
In the proposed model to fit the data of Perin et al. (2011), connections are created independently with probability P(i→j|rij = r, xi = x, xj = y) = p(r) g(x, y). The distance dependency has the following form:
where r is the normalized distance r = that is computed from the real distance d in μm and minimal and maximal distances derived from the data, dmin = 10 μm, dmax = 350 μm. We took a = 1, b = −1.04, c = 0.21. The modulatory part is as follows:
where f1 and f2 have the following form:
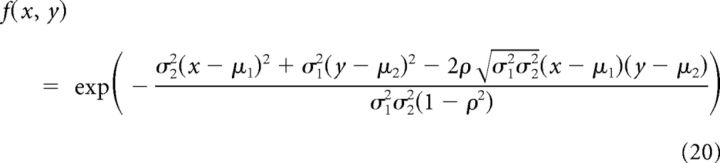 |
and their parameters are shown in Table 1. The modulators {xi}i are independent from neuron to neuron and are drawn from a Gaussian distribution with mean = 0 and SD = 0.5.
Table 1.
Parameters of the modulatory function
| μ1 | μ1 | σ12 | σ22 | ρ | |
|---|---|---|---|---|---|
| f1 | 0.0 | 0.0 | 0.3 | 0.3 | 0.92 |
| f2 | −0.5 | 0.5 | 0.07 | 0.07 | −0.62 |
To obtain a distribution of distances in the simulated data close to the sampled distances in the experiment, we generated samples directly as in the real experiment. In each sample, the first neuron was located in the origin of coordinates and the others were sequentially located on the same plane at a position obtained by drawing a random angle α ∈ [0, 2π) and a radius r from a Gamma(κ, θ) distribution, κ = 3.26, θ = 0.08. The radius was then rescaled as d = d0 + (d1 − d0) * r, d0 = 16 μm, d1 = 250 μm. We avoided having neurons too close in space by checking, at every step, whether the last neuron was closer than a limit distance dlim = 14 μm to the already created neurons in the sample. In this case, we chose a new position.
In-/out-degree correlation in small samples
Given a random sample of a network, we define the SDC as the Pearson correlation coefficient between in-degrees and out-degrees of individual neurons in the sample:
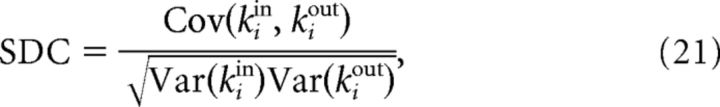 |
where i represents a random neuron and kiin, kiout are the in-degrees and out-degrees of i in the sample.
To compute the SDC in our models, we first need to introduce the following statistics. Given any network and random nodes i, j, k, we define the following:
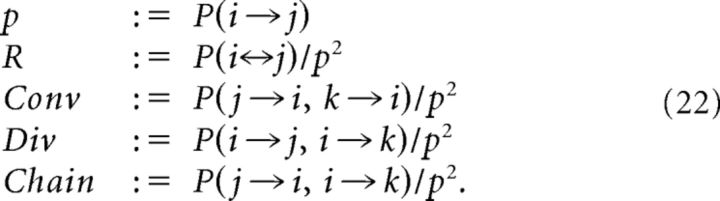 |
Note that these quantities do not trivially coincide with the motifs first defined in Song et al. (2005) and reproduced here in Fig. 2A. For example, the occurrence of the convergent motif number 5 above chance in Fig. 2A can be written , where puni = 2p(1 − pR), pbid = p2R and the factor 3 accounts for the different permutations of i, j, k that produce the same topological configuration. The motifs needed to compute the SDC are not conditioned on the presence or absence of any additional structure in the neuron triplet, merely the existence of, for example, a convergent motif. Therefore, our Conv motif is actually a weighted sum of all motifs in Fig. 2A containing at least one convergent node; that is: 5, 7, 9–10, and 12–16.
The in-degrees and out-degrees of a node i in a sample of size n can be expressed as follows:
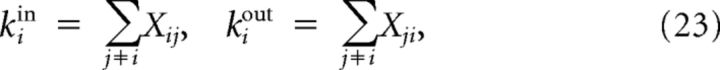 |
where Xij = 1 whenever j → i and Xij = 0 otherwise (the sums in (23) are over the n indices of the neurons in the sample). Explicitly computing the sample degree variances and the covariance between in-degrees and out-degrees of neuron i from expression (23) we find:
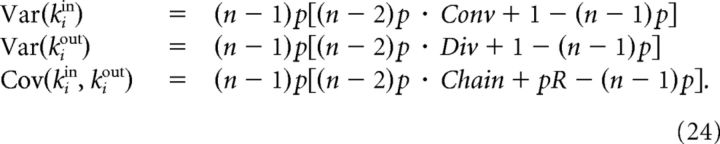 |
In the ER-Bi model, the pair to pair independence implies that Conv = Div = Chain = 1 and:
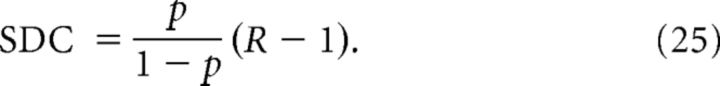 |
In the Mod model, the quantities p, R, Conv, Div, and Chain can be rewritten in terms of moments of g:
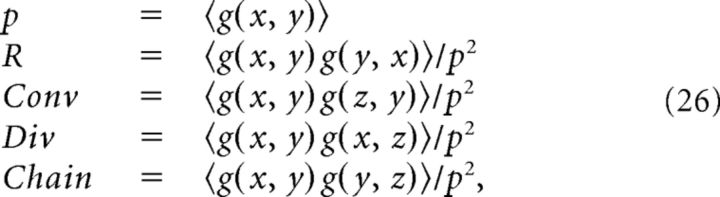 |
where 〈〉 indicates an average over the distribution of x, y, z, which are independent and identically distributed random variables. We have the following particular cases:
- If g(x, y) is independent of g(x, z), g(z, x), g(z, y), and g(y, z), then Conv = Div = Chain = 1 and:
In the Cl and Dis models, the property of being in the same cluster (Cl) and the distance between a pair (Dis) can be assumed to be independent from one pair to another when N is large, so (27) is a good approximation of the sample degree correlation as long as n ≪ N.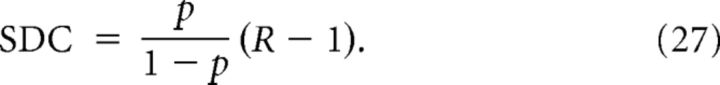
- If g is symmetric, that is, g(x, y) = g(y, x), then Conv = Div = Chain and:
This is the case of the Cl-Het model. Note that, in the Cl/Dis models, g is also symmetric, so this expression for SDC is a generalization of Equation 27, which is recovered whenever .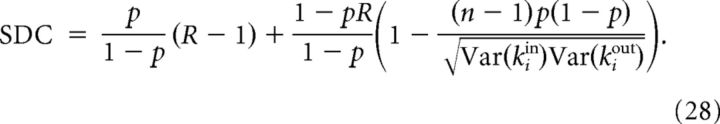
Notice that, because the SDC can be explicitly calculated from p, R, Conv, Div, and Chain, network models that have the same p, R, Conv, Div, and Chain but differ in higher-order statistics that cannot be distinguished by means of the SDC.
Experimental design and statistical analysis
Implementation of the SDC criterion on a random network generator.
In Figure 6, C and D, we applied the SDC criterion on networks generated randomly according to the models ER-Bi, Cl/Dis, Cl-Het, and Deg. We chose a network class and values for p ∈ [0.05, 0.23] and R ∈ [1.5, 4.1] uniformly at random. In the ER-Bi model, these parameters determine puni and pbid. If the chosen class was Cl/Dis, we chose one of these two models with equal probability. In the Cl case, we selected the number of clusters randomly and then computed p+ and p− to get the desired p and R. In the Dis case, we chose a dimension (1 or 2) randomly and then placed neurons in periodic lattices of the given dimension. Then, we determined the parameters s and t of Equation 16 to fit p and R. If the selected model was Cl-Het, we did exactly the same as in the Cl case. Finally, in the Deg model, we chose D and ρ > 0 randomly and then found θ, κ1, and κ2 to fit p and R.
To classify a network according to the SDC, we took m random samples of size n′ = 12 each. From them, we estimated p, R, Conv, Div, and Chain (Eq. 22) and computed the connection probability as a function of the number of common neighbors. From p, R, Conv, Div, and Chain, we predicted σ2 = and Cov(kiin, kiout) for any sample size n through Equation 24. We compared the resulting SDC (seen as a function of n) with the SDC that would result in each of the model classes given the observed p, R, and σ2 (Eqs. 27–29). We determined which of these relationships between SDC and n better described the results by computing the sum of the squared distances between the actual SDC and the model predictions while varying n. The range of n values used to make this comparison is arbitrary. We chose n ∈ {3, …, 12}, but the results are essentially the same for other choices. Because the formula for the Cl-Het model generalizes the formula for ER-Bi/Cl/Dis, the SDC of a network of the class ER-Bi/Cl/Dis will be fitted equally well by these two formulas. Therefore, whenever the best fit corresponded to the Cl-Het class, we further investigated whether the SDC increased significantly with n by computing the slope of its linear regression and deciding if it was larger than a critical value s*, which had been determined previously by means of simulations. If the slope was smaller than s*, then the network was reclassified as ER-Bi/Cl/Dis. Finally, to distinguish between ER-Bi and Cl/Dis networks, we determined whether the connection probability in the n′ samples increased significantly with the number of common neighbors. Again, this was done by computing a linear regression and comparing the slope with a previously defined threshold.
We further checked that the same algorithm works if σ2 and Cov(kiin, kiout) are calculated directly for each n on n-neuron samples instead of being estimated from p, R, Conv, Div, and Chain. The n-neuron samples in this case are subsamples of the original samples of size n′. The only limitation of this procedure is that the original sample size n′ has to be large enough to make it possible to compute σ2 for n in the desired range, whereas the estimation of p, R, Conv, Div, and Chain only requires three-neuron samples. A study based on sampling from triplets or quadruplets, however, would not allow us to distinguish between the ER-Bi and Cl/Dis classes using the common neighbor rule.
Implementation of the SDC criterion on data.
To apply the SDC criterion on the experimental data from Perin et al. (2011), we considered all of the possible subsamples of the original samples. For each subsample size, we used in-degrees and out-degrees of all the neurons to compute σ2 = and Cov(kiin, kiout). Because the expected SDCs for each model class are functions of p, R, and σ2, which in a real situation are estimated quantities, they are prone to estimation errors, as well as the real SDC. We estimated the data SDC, the predicted SDC for the model classes and their standard errors by means of the bootstrap method with 1000 resamplings, as detailed below. We created 1000 artificial samples with replacement from the set of in-/out-degrees for each sample size. From each of these samples, we computed σ2. The mean of this collection of values gives the estimated σ2 and the SD, a measure of the SE. The same was done to estimate the real SDC and its SE. We estimated the mean and SE for p, R and the different functions of p, R that participate in Equations 27–29 in a similar way (in this case, by resampling over the different neuronal pairs in the network). For formulas that involve both σ2 and p, R (i.e., Eqs. 28 and 29), we computed the upper bounds of the resulting errors from the previous partial errors.
We repeated the same procedure considering the predicted σ2 and Cov(kiin, kiout) from p, R, Conv, Div, and Chain, where these statistics were computed using all of the information in the original samples (i.e., using all the pairs and triplets). The results were almost identical.
It is important to notice that this exhaustive data analysis might introduce biases in the estimation of Conv, Div, Chain, σ2, and SDC because the triplets and the nodes involved in computing in-/out-degrees partially overlap. To cope with this, we used exactly the same procedure in all of the analyses of Figures 6, 7, and 9. The fact that the classification algorithm is fairly accurate even when the number of studied samples is small (see Fig. 6C) indicates that such correlations do not play a very important role. Despite this, we wondered whether the deviation in σ2 from the Cl/Dis model seen in the data could be due to these effects and not to the fact that the real underlying structure deviates from this simple model. To investigate this issue, we simulated networks with the same distance-dependent component exhibited by the data with an additional modulatory component based on clustering. The repetition of many replicas of the real experiment on this model indicated that the observed deviation of σ2 is statistically significant (p < 0.05, data not shown). This suggests that the discrepancy from a symmetric Mod model is not due to sparse sampling or correlations derived from data overlaps.
Figure 7.
Sample in-/out-degree correlation SDC and geometric mean of the sample degree variances σ2 as a function of the sample size n. A, Values calculated directly from the data of Perin et al. (2011). B, Inferred values from the motif counts presented in Song et al. (2005). The black curves correspond to the observed SDC and σ2, whereas colors show the expected SDC (σ2) in networks generated according to the studied models with the same p, R, σ2 (p, R) as in the data. Shaded regions in A indicate mean ± SEM computed with the bootstrap method.
Figure 9.
A modulator network model to reproduce connectivity data from (Perin et al., 2011). A, Schematic of a model to explain the observed data. First, neurons are arranged in space so that distances between neuronal pairs follow a given distribution (green). Each neuron has also an associated modulator whose distribution is shown in red. Given a distance-decaying probability p(r) and a function g = g(x, y), connections are created independently with probability P(i → j|rij = r, xi = x, xj = y) = p(r)g(x, y). B, Intersomatic distance distribution and connection probabilities as a function of distance in the data (black) and in the model (blue). Inset, Number of reciprocal connections relative to random R as a function of distance. The model results come from a single replica of the real experiment and shaded regions indicate mean ± SEM. C, SDC and geometric mean of the sample degree variances σ2 as a function of sample size n in the data (black) and in the model (blue). The blue-shaded regions indicate quantiles computed from a set of 200 replicas of the real experiment, each performed on an independent network. D, E, Comparison between model and data in terms of the common neighbor rule (D) and the distribution of the total number of connections (E) in samples of size n. Dashed lines show the prediction for the ER-Bi networks.
The analysis of the data from Song et al. (2005) was done by computing Conv, Div, and Chain directly from the statistics of three-neuron motifs.
Results
Canonical network models for cortical circuits
We asked ourselves to what extent simple, canonical models of network topology could reproduce the salient statistics from actual cortical circuits in slice experiments. The simplest possible sparsely connected network model is the so-called ER network, for which connections between neurons are made with a fixed probability p. However, data show that cortical circuits are not well described by the ER model and, in particular, the occurrence of certain cortical motifs is above what would be expected from ER. Therefore, we consider other candidate network models that go beyond ER (Fig. 1). First, we consider an ER network with additional bidirectional connections (ER-Bi). This model has just two parameters: the probability of a unidirectional connection puni and that of a bidirectional connection pbid. The second model is a network with C clusters in which cluster membership is homogeneous across neurons (Cl). The probability of connection between neurons within the same cluster is p+ and, between clusters, it is p− < p+. The third network has C clusters and heterogeneous membership (Cl-Het), in which neurons belong to a variable number of clusters. The probability of connection within and between clusters is as for the Cl model. The fourth network has distance-dependent connectivity (Dis), that is, the probability of connection between two cells at a distance r is p(r), which is a decreasing function of r. The fifth network model is defined by the distribution of in-degrees and out-degrees (Deg), with mean degree 〈K〉, variances σin2, σout2, and degree correlation ρ (see Materials and Methods for details).
Representation of two- and three-neuron motifs relative to random
We first investigated whether the deviation in the number of two-neuron motifs relative to random that has been reported previously (Song et al., 2005) could be explained by any of the models presented here. Given the sparseness p of a network model (i.e., the expected number of connections divided by the total number of possible connections), we denote by R the expected number of reciprocal connections relative to that in ER(p), which can be calculated for each model as shown in Table 2 (see Materials and Methods for details). The expected number of unidirectionally connected and unconnected pairs is then determined uniquely once p and R are known.
Table 2.
Sparseness (p) and fraction of bidirectional connections relative to random (R) in the different models
| Model | p | R |
|---|---|---|
| ER bidirectional | ||
| Clusters | f+p+ + f−p− | |
| Distance | 〈p(r)〉 | |
| Degree |
In the models with clusters f− = 1 − f+ and f+ is the fraction of neuronal pairs that are in the same cluster. The brackets 〈 〉 in the Dis model represent averages over the distribution of distances in the network. See the main text for a description of the other parameters.
Once p has been fixed, all models can account for a wide range of values in R, including the specific values reported in Song et al. (2005); Wang et al. (2006); Mason et al. (1991); Markram et al. (1997); and Le Bé and Markram (2006) (Fig. 2B,C; in Fig. 2C, we have used the values of p and R reported in Song et al., 2005). The numbers of three-neuron motifs relative to ER-Bi are also qualitatively similar across models and consistent with experiment, with the exception of ER-Bi, which has no additional structure beyond two-neuron motifs (Fig. 2C, bottom).
An important question to be addressed here is to what extent the experimental results are sensitive to the sampling procedure. Data are collected through simultaneous patch-clamp recordings and thus can only record from a small number of cells at a time. The motif counts are local properties whose averages do not depend on the sample size, but the results can be highly variable if the number of samples studied is not large enough. To mimic the experiment by Song et al. (2005), we computed p and R, not only from the study of the whole network, but also through 163 samples of four neurons per network over five networks. As shown in Figure 2, B and C (gray bars), the estimates of the two-neuron motif counts are quite close to the real counts in networks of N = 2000 neurons, which suggests that the magnitudes p and R are well approximated even when only a small fraction of the total network is known. Although the results of three-neuron motifs were approximately consistent between the full analysis and that from small sample sizes, they were much more variable than the two-neuron motifs.
Nonetheless, at least in the example networks shown in Fig. 2C, it seems that the particular distribution of triplet motifs might provide a means of classifying the different models. In subsequent sections, we will show that there is a particular combination of dual and triplet motifs from which we can extract information about the network class independently of the choice of other parameters.
Connection probability as a function of the number of common neighbors
A common neighbor to neurons i and j is a third neuron that is connected to both i and j. Perin et al. (2011) have shown that the probability of connection between pairs of cortical neurons increases with the number of common neighbors that they have (the so-called “neighbor rule”). Figure 3 (top) shows the connection probability as a function of common neighbors for examples from each model class from the analysis of a network of N = 2000 neurons where p and R are close to the values reported in Perin et al. (2011). In the ER-Bi model, as in the classical ER model, all the pairs are connected independently and according to the same rule, so the number of common neighbors does not provide any information about the “laws” controlling a given connection. All of the other models, however, exhibit the common neighbor rule for a general choice of the network parameters. Interestingly, the precise shape of this dependence is quite distinct for different models, indicating that it might provide a signature for inferring the full network structure from this one measure. However, these qualitative differences between models largely vanish when realistic sample sizes are analyzed (Fig. 3, bottom). It is important to keep in mind that the curves shown in Figure 3 are for a particular choice of network from each model class; the exact shape of the curves will depend on that choice. In general, we can say that, given small sample sizes, one will observe a monotonically increasing dependence of the connection probability on the number of common neighbors for all models but ER-Bi. Specifically, for clustered (distance dependent) models, neuron pairs with more common neighbors are more likely to belong to the same cluster (be closer together), which increases the probability of connection. In the Deg model neuron pairs with more common neighbors are more likely to have large degrees, which again increases the probability of connection.
Degree distributions and higher-order connectivity
Figure 4A (top) shows the in-degree distributions exhibited by example networks from the different models for physiological values of p and R. For both the Cl-Het and Deg models, the distribution differs dramatically from that of the equivalent ER network. Nonetheless, and as was the case with the common neighbor rule, when the distributions are constructed from realistic sample sizes (here 12), all models are qualitatively similar (Fig. 4A, bottom). In fact, due to additional degrees of freedom that both the Cl-Het and the Deg models have, it is possible to define networks with a fixed p and R with distributions that are nevertheless very different (Fig. 4B). In some situations, the distribution is quite close to ER/ER-Bi cases.
Finally, real data also exhibit a significant overrepresentation of densely connected groups (Perin et al., 2011). We therefore also studied the distribution of the number of connections in small groups of neurons and found that all models, with the exception of ER-Bi, could account for these findings (Fig. 5).
Method for distinguishing between network models using measures from small sample sizes
We sought a measure, based on small sample sizes, that would allow us to distinguish between the classes of topological models defined here. In other words, we looked for a way to infer general topological properties of the network when only local information is available. We found such a measure in the sample in-/out-degree correlation:
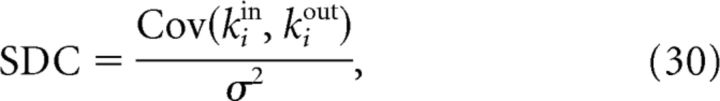 |
where σ2 = and i represents a random neuron in the sample. The SDC therefore depends on the variances and covariances of the sample degrees. The in- (resp out-) variance in turn depends on the occurrence of convergent (resp. divergent) motifs, whereas the covariance depends on the occurrence of chain and reciprocal motifs. All of these quantities can be calculated analytically for the network classes that we have considered here and the SDC is finally expressed as a function of p, R, σ2, and the sample size n.
In particular, we can group the five network types into three classes based on the functional form of the SDC: (1) The ER-Bi, Cl, and Dis models; (2) the Cl-Het model; and (3) the Deg model (see Materials and Methods for details). We can additionally use the common neighbor rule to distinguish between the ER-Bi (which shows no dependence) and the Cl and Dis models (which do). Note that all of these classes of networks have SDC  0 whenever R = 1, which means that networks that do not show an overrepresentation of bidirectional connections cannot be distinguished in terms of the SDC. Therefore, as long as R > 1, in principle, we can distinguish between all models except for the Cl and Dis models. This is not surprising given that the Cl is nothing but a particular case of the Dis in which the distance is binary.
0 whenever R = 1, which means that networks that do not show an overrepresentation of bidirectional connections cannot be distinguished in terms of the SDC. Therefore, as long as R > 1, in principle, we can distinguish between all models except for the Cl and Dis models. This is not surprising given that the Cl is nothing but a particular case of the Dis in which the distance is binary.
We applied this “SDC criterion” to networks of size N = 2000 generated randomly according to the four classes of models presented here (grouping Cl and Dis), with p and R chosen uniformly in the ranges [0.05, 0.23] and [1.5, 4.1], respectively. We then used the SDC to distinguish between the different model classes by taking samples of size n′ = 12. We estimated σ2 and the SDC over a range of sample sizes n by computing p, R, and the occurrence of convergent, divergent, and chain motifs (through the quantities Conv, Div, and Chain defined in Eq. 22; see Fig. 6B and Materials and Methods for details). An alternative approach is to generate n-neuron subsamples from the original samples of size n′ and compute σ2 and the SDC directly for each n ≤ n′. We also checked that the performance is almost the same for the second method when n′ = 12 (data not shown). The advantage of estimating σ2 and the SDC instead of calculating them directly is that it allows one to implement the criterion even when the original samples are small (e.g., n′ = 3, 4). To further distinguish between the ER-Bi and Cl/Dis classes, we investigated whether the connection probability increases with the number of common neighbors in the n′ samples.
The efficacy of this classification criterion increases with the number of samples considered, m. Figure 6, C and D, shows the performance as a function of m. The rate of success is above the chance level (chance here is 25%) for all models already for m = 2 samples and reaches 94% for m = 300. As long as the network size is large compared with the sample size, the classification accuracy is independent of system size (data not shown). This simply means that it can be applied to real data without the need to worry about the true size of functional cortical circuits.
Analysis of the SDC in data from rat somatosensory cortex
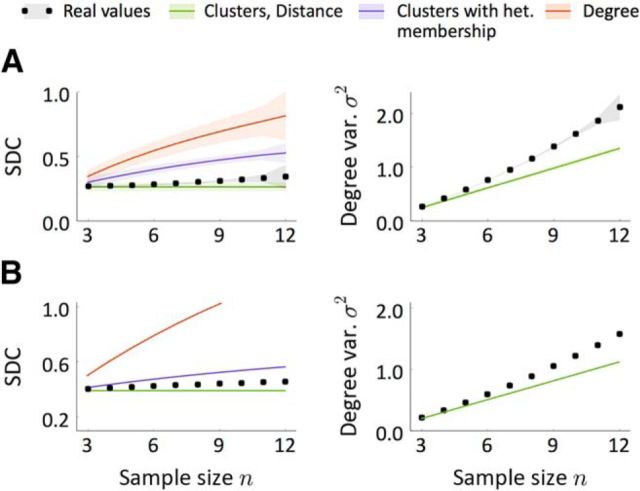
We implemented our SDC criterion in the data obtained by Perin et al. (2011) from pyramidal neurons of the rat somatosensory cortex. The data come from 6, 9, 5, 10, and 10 groups of 8, 9, 10, 11, and 12 neurons, respectively. As reported previously in Perin et al. (2011), these data show a clear dependency of connection probability on intersomatic distance. The estimated connection density and number of reciprocal connections relative to random were p = 0.144, R = 2.575. The analysis of the SDC revealed a relationship that deviates from any of the previously defined models (Fig. 7A). Although the form of the SDC appears close to that of the Dis model (Fig. 7A, left), the degree variance from the data σ2, which should be that of a binomial distribution, differs strongly from the theoretical value (Fig. 7A, right). Note that the degree variance for the other two classes of network is a free parameter and thus here is estimated directly from the data.
Because the SDC can be extrapolated when the counts of two- and three-neuron motifs are known, we calculated the expected SDC in putative samples of three to 12 neurons from the motif distribution described in Song et al. (2005) (Fig. 7B), which corresponds to layer 5 pyramidal neurons in rat visual cortex. The connection density and the number of reciprocal connections relative to random in this case are p = 0.116 and R = 4. The results are qualitatively similar to the ones computed directly from the data of Perin et al. (2011). This suggests the underlying network structure itself may be similar.
General class of network model
We discovered that all of the models, with the exception of the ER-Bi model, which could be rejected already by its failure to capture triplet motifs and the neighbor rule, belong to a more general class of model. Specifically, in what we dub Modulator (Mod) networks, the probability of a connection from neuron i to neuron j is P(i→j|xi = x, xj = y) = g(x, y), where xi and xj, the modulators, are properties associated with neurons i and j. These properties (or sets of properties) might represent spatial position, axonal/dendritic length, neuronal type defined by the expression of some proteins, presence of neuromodulators in the medium, amount of input received from other brain areas, stimulus selectivity, or even information related to the past history of neurons, to cite just some possibilities. The Mod model, therefore, represents any general scenario in which connections appear with higher or lower probability depending on features of the two neurons involved. The models we have considered so far are special cases of this more general modulator framework. This is illustrated in Figure 8, which shows three sample networks from the Dis, Cl-Het, and Deg classes.
In the clustered and distance-dependent models that we have considered, g(x, y) = g(y, x) is reflection symmetric. In this case, the modulators are the position or membership in a cluster (or group of clusters, e.g., Fig. 8B). It can be shown that any Mod network with a symmetric g exhibits the same SDC as the Cl-Het model. If, additionally, g(x, y) can be assumed to be independent from one neuronal pair to another (as in our Cl and Dis models when a small sample is considered, where g(x, y) only depends on the distance |x − y|; see Fig. 8A), the formula reduces to the Cl/Dis case. In the Deg model, g is separable, that is, g(x, y) = g1(x)g2(y), and the modulator itself is the pair of in-degrees and out-degrees. The g function is just the product of the presynaptic out-degree and postsynaptic in-degree normalized by the appropriate factor (Fig. 8C). In the Materials and Methods, we show that the SDC of any Mod network with separable g has the form of the SDC of the Deg model.
Therefore, the SDC criterion not only makes it possible to distinguish between the families Cl/Dis, Cl-Het, and Deg, but also allows for a classification into three major types of Mod networks defined by different properties and symmetries. The fact that the data are not fit by any of the models indicates that real cortical circuits have features that violate the reflection symmetry and separability of the function g.
Because the estimated SDC lies in between the predicted SDC for the Dis/Cl and Cl-Het models (Fig. 7A), one would be tempted to think that a hybrid network from these two classes would be compatible with data. Such a model, however, would still belong to the class of Mod networks with symmetric g and would therefore exhibit the same SDC as the Cl-Het class (purple line in Fig. 7A). This suggests that not only is there additional structure in the data beyond the distance dependence of connection probabilities, but that this structure is not simple clustering.
Data are consistent with network with spatial dependence and hierarchical clustering
We were able to obtain an excellent fit to all relevant topological statistics in the data with a Mod network. Specifically, we considered a network in which the probability of connection between pairs was as follows:
where p(r) depends on the physical distance r between pairs and the modulator component g(x, y) is not reflection symmetric. This model is itself a 2D Mod network in which one dimension is physical space and the other represents a property of the neurons not captured by their spatial location (Fig. 9A). We assumed that the distribution of distances in samples obtained from the model is close to the sampled distribution in the data (Fig. 9B, left) and that the {xi}i modulators are independent from neuron to neuron and independent of distances. We assume a Gaussian distribution of the modulator and take g(x, y) to be the weighted sum of the p.d.f. of two bivariate Gaussians, one of which breaks the reflection symmetry (for details, see Fig. 9A and Materials and Methods). This choice is equivalent to other possible distributions of the modulator as long as g is also appropriated transformed. The model successfully captures the observed distance dependency of the connection probabilities (Fig. 9B, right); note in particular that it reproduces the overrepresentation of reciprocal connections as a function of distance (Fig. 9B, right inset). A pure Dis model cannot explain this finding: although the value of R evaluated globally would be >1, for any given distance it would be identically 1. Therefore, the increased R as a function of distance is a clear signature of additional structure, captured here by our modulator function. The Mod model also reproduces the sample degree correlation and variance (Fig. 9C), as well as the common neighbor rule and the density of connections in groups of few neurons (Fig. 9D,E).
What is the interpretation of the modulator in this network? The modulator acts as an identifier for each neuron and neurons with similar modulators will connect in similar ways. Indeed, if the modulator is symmetric, we recover a continuous version of a clustered network with heterogeneous membership (Cl-Het). Therefore, the symmetric part of g(x, y) (see plot in Fig. 9A) can be interpreted as clustering: neurons with similar values of x are more likely to connect to one another than to neurons with different values (although this preference decreases for extreme values of x). However, the presence of asymmetry in g indicates that connections between clusters are actually hierarchical. Specifically, in our example, neurons with low x are likely to connect to similar neurons and also to neurons with large x. Conversely, neurons with large x are likely to connect with similar neurons, but not to neurons with low x. We further checked that this is actually captured by a model in which the distance-independent modulatory component is based on discrete hierarchical clustering. It was sufficient to consider a homogeneous distribution into three clusters where connection probability within cluster 2 is higher than within clusters 1 and 3 and where connection probabilities between different clusters are low except for cluster 1, which has a strong preference to project to cluster 3, as in our continuous model (data not shown). Although these two model versions are essentially the same, the continuous one incorporates a higher variability in the modulatory variables that could resemble real modulatory mechanisms that operate through continuous variables such as concentration of molecules or the amount of input received from other brain areas. In conclusion, the data are consistent with a network in which neurons are connected according to the physical distance between them and their membership in a clustered structure independent of distance, which itself exhibits hierarchical features.
Discussion
We have presented three major classes of network models that are compatible with the “nonrandomness” reported so far in cortical microcircuits (Song et al., 2005; Perin et al., 2011). The first is based on a similarity principle: pairs of neurons have associated a notion of distance that modulates the likelihood of the connections between them in the sense that similar neurons tend to be connected more frequently than different ones. The connections appear independently once the distances between neuronal pairs are known. Distance in this context can represent not only a spatial proximity, but any other measure of similarity, for example, based on input received from other areas or stimulus selectivity. This family also includes networks in which neurons are classified homogeneously into clusters so that connections form preferentially between cells that are in the same cluster. In the second model, neurons are assigned to clusters, but there is heterogeneity both in the cluster size and in the number of clusters to which different neurons belong. Connections form with higher likelihood between neurons that coincide in any of the clusters. The third family corresponds to networks in which in-degrees and out-degrees of single neurons follow a prescribed joint probability distribution.
Our results show that the three classes of networks can exhibit both an excess of reciprocal connections relative to random and the so-called common neighbor rule for a wide range of parameters. In the case of networks with a specified degree distribution, in-degrees and out-degrees must be positively correlated for the bidirectional connections to be overrepresented, meaning that neurons that receive more synapses from the network tend to be the ones that have more outgoing connections; that is, they are hubs. All of the models can also be similar in terms of the marginal degree distribution in small samples and are in qualitative agreement with previously reported results concerning the number of connections in groups of few neurons. The first important conclusion of our study is therefore that these “nonrandom” features, rather than being a footprint of a specific topology, seem to arise naturally from several qualitatively distinct types of models.
One of the major difficulties of inferring structural principles from real data is that functional neuronal networks likely encompass thousands of neurons, whereas simultaneous patch-clamp experiments, which provide ground truth for synaptic connectivity, provide samples of only a few neurons at a time. Although the models presented here are based on very different principles, they can be almost indistinguishable from one another given only small sample sizes. Therefore, even structures that are distinct globally can exhibit similar properties locally. Electron microscopy can also in principle provide ground truth connectivity (Denk and Horstmann, 2004; Bock et al., 2011; Kleinfeld et al., 2011; Kasthuri et al., 2015), although current throughput is too small to allow for the reconstruction of microcircuits. Finally, some studies have sought to infer network connectivity from observations of the neuronal dynamics (Nykamp, 2007; Pajevic and Plenz, 2009; Stetter et al., 2012; Sadovsky and MacLean, 2013; Tomm et al., 2014); the accuracy of such methods generally depends on the how closely the real data might conform to specific model assumptions. In cortical cultures, the functional connectivity inferred from transfer entropy measures of the spiking activity of small assemblies of neurons shows dense connectivity above that expected from ER (Shimono and Beggs, 2015; in agreement with Perin et al., 2011). Nonetheless, the data acquired through slice electrophysiology still currently represent the most accurate picture of cortical microcircuitry available.
A natural question is whether it is possible to define a local measure, that is, a measure that can be estimated from the study of small samples, that could be used to distinguish between models. We have found such a measure in the SDC, the correlation coefficient between sample in-degrees and out-degrees. The SDC is, in fact, a particular nonlinear combination of triplet motifs that allows us to classify network models correctly without recourse to training classifiers numerically. Interestingly, the SDC depends on precisely those second-order network statistics that have been used recently to develop dynamical mean-field models for neuronal networks with structure beyond the ER network (Zhao et al., 2011; Nykamp et al., 2017).
Note that a machine learning approach to this problem would require training a classifier on particular instantiations of networks from a given network class; each class encompasses a vast range of possible networks. Therefore, training sets would not likely be representative of the class as a whole. A major advantage of our approach, in contrast, is that it allows us to classify networks regardless of the details of every model candidate, which can be difficult to estimate in real situations. For example, in the Dis model, the exact shape of the function p(r) is irrelevant for estimating the SDC, which only depends on the overall connection probability and the overrepresentation of reciprocal connections. We have also shown that these three model classes are particular cases of a very general model according to which single neurons have an associated property that modulates the connection probability. We call such a property a “modulator.”
We estimated the SDC for distinct datasets from both rat somatosensory cortex and rat visual cortex and found that the structure in those cortical circuits fell outside of all three classes of model network in a qualitatively similar way (Fig. 7). These observations therefore suggest that, if the underlying network topology can be interpreted in the Mod framework, then the modulatory function g(x, y), which defines the probability of finding a connection from a neuron with modulator x to a neuron with modulator y, can be neither symmetric nor separable. Finally, we obtained an excellent fit to the first dataset by considering a more general Mod network in which the probability of connection between neurons depended both on the physical distance between them, as well as on an additional modulator unrelated to distance. In the second dataset, there is no evidence of distance dependency of connectivity (Song et al., 2005), but the qualitative similarity between datasets in terms of the SDC suggests that a similar nonspatial modulator mechanism might be common to both of them. The structure of this nonspatial modulator could be interpreted as hierarchical clustering, in which connectivity between clusters is asymmetric. However, we cannot rule out that other choices of modulators, that would lead to other interpretations, might provide equally good fits to the data.
The classes of networks that we have explored here are simple enough to be treated analytically. Nature is certainly more complex and, clearly, cortical microcircuits are shaped by other principles, including ongoing synaptic plasticity. We have not considered these mechanisms here. Nevertheless, independent of the mechanisms that shape cortical microcircuitry, if the topology of the resultant network can be reduced to a modulatory mechanism, then our results show that this modulation involves both a distance dependence and an additional nonspatial component that is asymmetric.
Footnotes
This work was partially supported by the CERCA program of the Generalitat de Catalunya. M.V. has received financial support through the “la Caixa” Fellowship Grant for Post-Graduate Studies, “la Caixa” Banking Foundation, Barcelona, Spain. A.R. acknowledges Grants BFU2012-33413 and MTM2015-71509-C2-1-R from the Spanish Ministry of Economics and Competitiveness and Grant 2014 SGR 1265 4662 for the Emergent Group “Network Dynamics” from the Generalitat de Catalunya.
The authors declare no competing financial interests.
References
- Bastian M, Heymann S, Jacomy M (2009) Gephi: an open source software for exploring and manipulating networks In Third International AAAI Conference on Weblogs and Social Media. Proceedings of the Third International ICWSM Conference. [Google Scholar]
- Bock DD, Lee WC, Kerlin AM, Andermann ML, Hood G, Wetzel AW, Yurgenson S, Soucy ER, Kim HS, Reid RC (2011) Network anatomy and in vivo physiology of visual cortical neurons. Nature 471:177–182. 10.1038/nature09802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunel N. (2016) Is cortical connectivity optimized for storing information? Nat Neurosci 19:749–755. 10.1038/nn.4286 [DOI] [PubMed] [Google Scholar]
- Denk W, Horstmann H (2004) Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol 2:e329. 10.1371/journal.pbio.0020329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SL, Wang Y, Riachi I, Schürmann F, Markram H (2012) Statistical connectivity provides a sufficient foundation for specific functional connectivity in neocortical neural microcircuits. Proc Natl Acad Sci U S A 109:E2885–E2894. 10.1073/pnas.1202128109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer SB, Mrsic-Flogel TD, Bonhoeffer T, Hübener M (2009) Experience leaves a lasting structural trace in cortical circuits. Nature 457:313–317. 10.1038/nature07487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren C, Harkany T, Svennenfors B, Zilberter Y (2003) Pyramidal cell communication within local networks in layer 2/3 of rat neocortex. J Physiol 551:139–153. 10.1113/jphysiol.2003.044784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Shen S, Cadwell CR, Berens P, Sinz F, Ecker AS, Patel S, Tolias AS (2015) Principles of connectivity among morphologically defined cell types in adult neocortex. Science 350:aac9462. 10.1126/science.aac9462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, Knowles-Barley S, Lee D, Vázquez-Reina A, Kaynig V, Jones TR, Roberts M, Morgan JL, Tapia JC, Seung HS, Roncal WG, Vogelstein JT, Burns R, Sussman DL, Priebe CE, Pfister H, Lichtman JW (2015) Saturated reconstruction of a volume of neocortex. Cell 162:648–661. 10.1016/j.cell.2015.06.054 [DOI] [PubMed] [Google Scholar]
- Kleinfeld D, Bharioke A, Blinder P, Bock DD, Briggman KL, Chklovskii DB, Denk W, Helmstaedter M, Kaufhold JP, Lee WC, Meyer HS, Micheva KD, Oberlaender M, Prohaska S, Reid RC, Smith SJ, Takemura S, Tsai PS, Sakmann B (2011) Large-scale automated histology in the pursuit of connectomes. J Neurosci 31:16125–16138. 10.1523/JNEUROSCI.4077-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko H, Hofer SB, Pichler B, Buchanan KA, Sjöström PJ, Mrsic-Flogel TD (2011) Functional specificity of local synaptic connections in neocortical networks. Nature 473:87–91. 10.1038/nature09880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bé JV, Markram H (2006) Spontaneous and evoked synaptic rewiring in the neonatal neocortex. Proc Natl Acad Sci U S A 103:13214–13219. 10.1073/pnas.0604691103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin-Kumar A, Doiron B (2014) Formation and maintenance of neuronal assemblies through synaptic plasticity. Nat Commun 5:5319. 10.1038/ncomms6319 [DOI] [PubMed] [Google Scholar]
- Markram H, Lübke J, Frotscher M, Roth A, Sakmann B (1997) Physiology and anatomy of synaptic connections between thick tufted pyramidal neurones in the developing rat neocortex. J Physiol 500:409–440. 10.1113/jphysiol.1997.sp022031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H, et al. (2015) Reconstruction and simulation of neocortical microcircuitry. Cell 163:456–492. 10.1016/j.cell.2015.09.029 [DOI] [PubMed] [Google Scholar]
- Mason A, Nicoll A, Stratford K (1991) Synaptic transmission between individual pyramidal neurons of the rat visual cortex in vitro. J Neurosci 11:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountcastle VB. (1997) The columnar organization of the neocortex. Brain 120:701–722. 10.1093/brain/120.4.701 [DOI] [PubMed] [Google Scholar]
- Nykamp DQ. (2007) A mathematical framework for inferring connectivity in probabilistic neuronal networks. Mathematical Biosciences 205:204–251. 10.1016/j.mbs.2006.08.020 [DOI] [PubMed] [Google Scholar]
- Nykamp DQ, Friedman D, Shaker S, Shinn M, Vella M, Compte A, Roxin A (2017) Mean-field equations for neuronal networks with arbitrary degree distributions. Phys Rev E 95:042323. 10.1103/PhysRevE.95.042323 [DOI] [PubMed] [Google Scholar]
- Pajevic S, Plenz D (2009) Efficient network reconstruction from dynamical cascades identifies small-world topology of neuronal avalanches. PLOS Comput Biol 5:e1000271. 10.1371/journal.pcbi.1000271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perin R, Berger TK, Markram H (2011) A synaptic organizing principle for cortical neuronal groups. Proc Natl Acad Sci U S A 108:5419–5424. 10.1073/pnas.1016051108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, et al. (2015) The neocortical microcircuit collaboration portal: a resource for rat somatosensory cortex. Front Neural Circuits 9:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann MW, King JG, Muller EB, Ramaswamy S, Markram H (2015) An algorithm to predict the connectome of neural microcircuits. Front Comput Neurosci 9:120. 10.3389/fncom.2015.00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxin A. (2011) The role of degree distribution in shaping the dynamics in networks of sparsely connected spiking neurons. Front Comput Neurosci 5:8. 10.3389/fncom.2011.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovsky AJ, MacLean JN (2013) Scaling of topologically similar functional modules defines mouse primary auditory and somatosensory microcircuitry. J Neurosci 33:14048–14060, 14060a. 10.1523/JNEUROSCI.1977-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono M, Beggs JM (2015) Functional clusters, hubs, and communities in the cortical microconnectome. Cereb Cortex 25:3743–3757. 10.1093/cercor/bhu252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberberg G, Gupta A, Markram H (2002) Stereotypy in neocortical microcircuits. Trends Neurosci 25:227–230. 10.1016/S0166-2236(02)02151-3 [DOI] [PubMed] [Google Scholar]
- Song S, Sjöström PJ, Reigl M, Nelson S, Chklovskii DB (2005) Highly nonrandom features of synaptic connectivity in local cortical circuits. PLoS Biol 3:e68. 10.1371/journal.pbio.0030068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetter O, Battaglia D, Soriano J, Geisel T (2012) Model-free reconstruction of excitatory neuronal connectivity from calcium imaging signals. PLoS Comput Biol 8:e1002653. 10.1371/journal.pcbi.1002653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentágothai J. (1978) The Ferrier lecture, 1977: the neuron network of the cerebral cortex: a functional interpretation. Proc R Soc Lond B Biol Sci 201:219–248. 10.1098/rspb.1978.0043 [DOI] [PubMed] [Google Scholar]
- Timme NM, Ito S, Myroshnychenko M, Nigam S, Shimono M, Yeh FC, Hottowy P, Litke AM, Beggs JM (2016) High-degree neurons feed cortical computations. PLoS Comput Biol 12:e1004858. 10.1371/journal.pcbi.1004858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomm C, Avermann M, Petersen C, Gerstner W, Vogels TP (2014) Connection-type-specific biases make uniform random network models consistent with cortical recordings. J Neurophysiol 112:1801–1814. 10.1152/jn.00629.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtenberg JT, Chen BE, Knott GW, Feng G, Sanes JR, Welker E, Svoboda K (2002) Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature 420:788–794. 10.1038/nature01273 [DOI] [PubMed] [Google Scholar]
- Wang Y, Markram H, Goodman PH, Berger TK, Ma J, Goldman-Rakic PS (2006) Heterogeneity in the pyramidal network of the medial prefrontal cortex. Nat Neurosci 9:534–542. 10.1038/nn1670 [DOI] [PubMed] [Google Scholar]
- Zhao L, Beverlin B 2nd, Netoff T, Nykamp DQ (2011) Synchronization from second order network connectivity statistics. Front Comput Neurosci 5:28. 10.3389/fncom.2011.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Yang G, Kwon E, Gan WB (2005) Long-term sensory deprivation prevents dendritic spine loss in primary somatosensory cortex. Nature 436:261–265. 10.1038/nature03715 [DOI] [PubMed] [Google Scholar]