Abstract
Motivation
Circular RNAs (circRNAs) originate through back-splicing events from linear primary transcripts, are resistant to exonucleases, are not polyadenylated and have been shown to be highly specific for cell type and developmental stage. CircRNA detection starts from high-throughput sequencing data and is a multi-stage bioinformatics process yielding sets of potential circRNA candidates that require further analyses. While a number of tools for the prediction process already exist, publicly available analysis tools for further characterization are rare. Our work provides researchers with a harmonized workflow that covers different stages of in silico circRNA analyses, from prediction to first functional insights.
Results
Here, we present circtools, a modular, Python-based framework for computational circRNA analyses. The software includes modules for circRNA detection, internal sequence reconstruction, quality checking, statistical testing, screening for enrichment of RBP binding sites, differential exon RNase R resistance and circRNA-specific primer design. circtools supports researchers with visualization options and data export into commonly used formats.
Availability and implementation
circtools is available via https://github.com/dieterich-lab/circtools and http://circ.tools under GPLv3.0.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Circular RNAs (circRNAs) were initially discovered in the 1990s, but the novel class of RNAs was first described as ‘scrambled exons’ (Nigro et al., 1991). Two decades later, new studies employing next generation sequencing discovered a large repertoire of circRNAs in different cell types and provided first hints of potential regulatory functions (Hansen et al., 2013). CircRNAs originate through back-splicing events from linear primary transcripts, are resistant to exonucleases, typically not polyadenylated and are highly specific for cell type and developmental stage. Detection of circRNAs is usually based on the existence of chimeric reads that cover the back splice junction (BSJ) of the circRNA, i.e. the position where the 3’ tail of the linear RNA molecule is fused with the 5’ head to form a circle. To increase the sensitivity for circRNAs in RNA-seq experiments, the CircleSeq protocol was developed based on the circRNAs resistance to exonuclease (RNase R) treatment (Jeck et al., 2013). While circRNA detection tools are available (Gao and Zhao, 2018), workflows for automated analyses including functionality, such as structural analyses or first functional predictions, are still rare.
2 Materials and methods
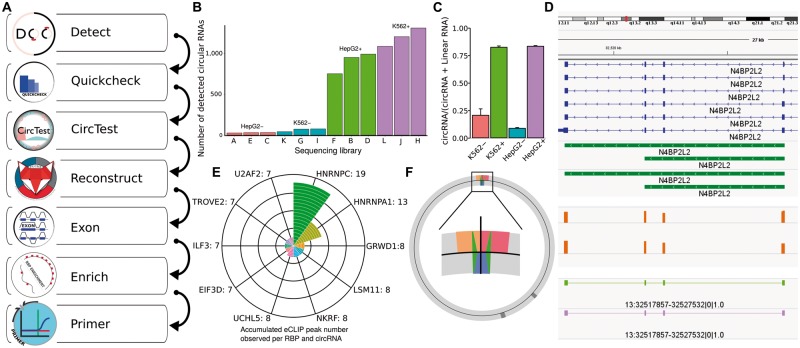
circtools currently offers seven modules shown in Figure 1A. Detection of circRNAs from RNA-seq reads mapped by STAR (Dobin et al., 2013) is based on the DCC software [detect, Cheng et al. (2016)] and generally the first step in the analysis workflow since the TSV-formatted data files generated here are required for subsequent analyses. Briefly, the detection step produces raw counts for circRNAs by exploiting reads that cover the BSJ and additionally generates count tables for the linear host genes. In a second step, these raw counts can be combined with log files of the STAR aligner within the quickcheck module. The diagnostic diagrams generated by the module also show the dramatic effect of the CircleSeq protocol on the number of detected circRNAs in HepG2 and K562 cells (Fig. 1B). Depending on the experimental setup and cell type, the initial detection step usually yields several hundreds to thousands of potential circRNA candidates. This set of candidates should be filtered for subsequent analysis steps. The circtest module of circtools employs a beta-binomial model to model changes in circRNA expression relative to that of the host gene (Cheng et al., 2016). This statistical framework may also be used to test the set of circRNAs for candidates that show a clear enrichment (P < 0.05) in the RNase R-treated samples compared to the untreated samples (Fig. 1C). One crucial point for functional predictions of circRNAs is knowledge of the actual internal RNA sequence, i.e. which exons or introns are part of the processed circRNA. The reconstruct module, based on the FUCHS software (Metge et al., 2017), is able to deliver this information by employing longer RNA-seq reads (e.g. 2 × 150 bp paired-end reads). Results of the module include a global analysis of circRNAs as well as per circRNA level information (exon structure shown in Fig. 1D, bottom, light green and purple for HepG2 and K562). Complementary to the reconstruction of circRNAs, circtools also offers the exon module to identify individual exons, which are resistant to RNase R digestion (i.e. CircleSeq experiments, Fig. 1D, middle, orange bars). Given the RNA sequence of circRNA candidates, further analyses are possible. The enrich module of circtools can be used to screen a set of circRNAs for significant enrichment of selected sequence features (provided via BED file). One typical use case is an enrichment screen for eCLIP peaks in circRNAs (Fig. 1E, Supplementary Fig. S4), which could be indicative of RNA-protein binding events, which are relevant for circRNA function. Prioritized in silico circRNA candidates should be verified by experiment. A quantitative validation experiment is typically performed using qRT-PCR. Herein, specific primer pairs are required to ensure that only circular target transcripts get amplified. While most primer design tools are not intended for this specific use case, circtools incorporates a primer design tool able to retrieve well-suited, circRNA-specific primers and additionally generates a graphical representation of the circRNA, the designed primers and the expected product (Fig. 1F, Supplementary Fig. S5). circtools is implemented in Python 3 (tested with 3.4, 3.5 and 3.6) and has been tested on major Linux distributions. The software is straightforward to install via pip3 install circtools and only requires a working R installation. Required Python and R packages are automatically installed. While much of circtools core functionality is implemented in Python, most plotting functions have been implemented in R. New modules can be easily added to the software by extending the circtools Python base class. We intend to add more functionality in the future in order to provide a comprehensive bioinformatics toolbox and we also encourage researches to contribute modules to circtools.
Fig. 1.
Overview of the circtools software showing different output visualizations. (A) General workflow of circtools. (B) Initial quality check after read mapping and circRNA detection. (C) Relative enrichment of circN4BP2L2 in four samples. (D) Visualization of BED tracks produced by circtools; circRNA predictions (green), differentially spliced exons for K562/HepG2 cell lines (orange), reconstructed circRNAs for both cell lines (light green, purple). (E) Enrichment of different RBP binding sites within circN4BP2L2. (F) Visualization of a automatically designed primer pair (green) bracketing the back-splice junction (black line) that separates the two fused exons (orange, red)
3 Discussion
circtools provides a well-tested, harmonized workflow for state-of-the-art circRNA research. The software covers different aspects in this endeavor: It performs initial quality checks, detects and reconstructs circRNAs, tests for host gene-independent expression, screens for enriched sequence features (i.e. RBP sites), supports the design of primers for qRT-PCR verification and visualizes and exports all analyses results. A complete experimental workflow and detailed methods are described in the online documentation and the Supplementary Material.
Supplementary Material
Acknowledgements
The authors would like to thank Thiago Britto-Borges, Etienne Boileau and Brandon Malone for their great input and insightful discussions. They thank the Kurian Lab for cell culture experiments and total RNA isolation.
Funding
The work was supported by the Klaus Tschira Stiftung gGmbH and the German Centre for Cardiovascular Research (DZHK) to T.J., A.U. and C.D. were supported by the Klaus Tschira Stiftung gGmbH and the German Centre for Cardiovascular Research (DZHK).
Conflict of Interest: none declared.
References
- Cheng J., et al. (2016) Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics, 32, 1094–1096. [DOI] [PubMed] [Google Scholar]
- Dobin A., et al. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Zhao F. (2018) Computational strategies for exploring circular RNAs. Trends Genet., 34, 389–400. [DOI] [PubMed] [Google Scholar]
- Hansen T.B., et al. (2013) Natural RNA circles function as efficient microRNA sponges. Nature, 495, 384–388. [DOI] [PubMed] [Google Scholar]
- Jeck W.R., et al. (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (New York, N.Y.), 19, 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metge F., et al. (2017) FUCHS-towards full circular RNA characterization using RNAseq. PeerJ., 5, e2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro J.M., et al. (1991) Scrambled exons. Cell, 64, 607–613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.