Abstract
Motivation
The application of constraint-based modeling to functionally analyze metagenomic data has been limited so far, partially due to the absence of suitable toolboxes.
Results
To address this gap, we created a comprehensive toolbox to model (i) microbe–microbe and host–microbe metabolic interactions, and (ii) microbial communities using microbial genome-scale metabolic reconstructions and metagenomic data. The Microbiome Modeling Toolbox extends the functionality of the constraint-based reconstruction and analysis toolbox.
Availability and implementation
The Microbiome Modeling Toolbox and the tutorials at https://git.io/microbiomeModelingToolbox.
1 Introduction
Microbial community sequencing data are increasingly available for numerous environmental niches (Mitchell et al., 2018). The analysis of this data often relies on investigating which microbes are present in a given sample. However, to further our understanding of the functional contribution of individual microbes in a community as well as the overall functional differences between communities, advanced analysis approaches, such as computational modeling, are required.
One possible approach is the constraint-based reconstruction and analysis (COBRA) approach, which builds genome-scale reconstructions of an organism and enables the prediction of, e.g. phenotypic properties (Palsson, 2006). Through the application of condition-specific constraints, an organism’s metabolic reconstruction can be converted into many condition-specific models, which can be analyzed using available toolboxes, such as the Matlab (Mathworks, Inc.)-based COBRA Toolbox (Heirendt et al., 2017a). Metabolic reconstructions have been assembled for many organisms, including hundreds of gut microbes (Magnúsdóttir et al., 2017) and human (Brunk et al., 2018). Although the COBRA Toolbox encapsulates many tools developed by the community for biotechnological and biomedical applications, it is currently focused on modeling single organisms or cells. Here, we present the Microbiome Modeling Toolbox, which enables the generation, simulation and interpretation of (i) pairwise microbe-microbe and host-microbe interactions, and (ii) sample-specific microbial community models. By integrating sample-specific metagenomic data, the Microbiome Modeling Toolbox facilitates its analysis in the context of microbial reconstructions.
2 Features
2.1 Pairwise interactions
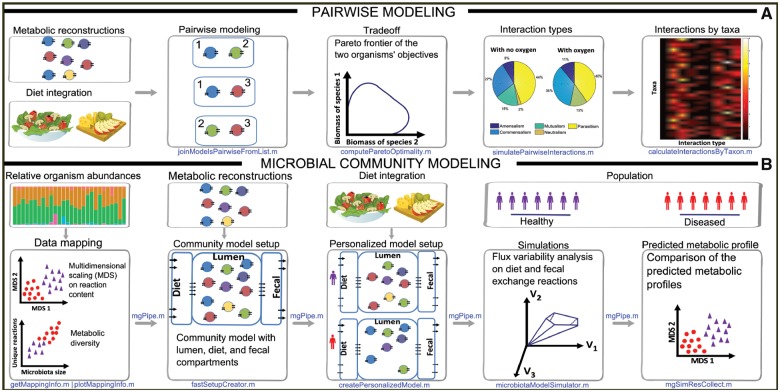
The pairwise interaction analysis determines metabolic exchange between two metabolic reconstructions. A joint matrix of two individual genome-scale reconstructions is generated, which enables them to freely exchange metabolites (Fig. 1A). Defined nutrient input, e.g. a particular medium formulation, can be applied via the shared compartment using the corresponding exchange reactions. The pairwise microbial models can be investigated for six possible interaction types (i.e. competition, parasitism, amensalism, neutralism, commensalism and mutualism) and Pareto optimality frontiers can be calculated. The tutorials MicrobeMicrobeInteractions and HostMicrobeInteractions illustrate the implemented functionalities.
Fig. 1.
Overview of the Microbiome Modeling Toolbox. (A) Pairwise modeling of microbe–microbe and host–microbe interactions. (B) Microbial community modeling
2.2 Microbial community modeling
Metagenomic data can be analyzed using mgPipe (Fig. 1B), which requires microbe identification and relative abundance data for each sample, obtained with bioinformatic tools, such as QiIME 2 (Caporaso et al., 2010) and MetaPhlAn (Segata et al., 2012). mgPipe is divided into three parts: (i) the analysis of individuals: specific microbes abundances, including metabolic diversity and classical multidimensional scaling of the reactions in the identified microbes. (ii) Construction of a personalized microbial community model using the identified microbes and their relative abundance data. For each personalized (or sample-specific) model, the corresponding microbial reconstructions are joined by adding reactions to each microbial reconstruction transporting metabolites from the extracellular space to the common lumen compartment. Metabolites present in the lumen compartment are connected to a diet and fecal compartment, enabling the uptake and secretion from/to the environment, respectively. Hundreds of reconstructions can be combined and modeled with using static parallelization. In each microbial community model, the community biomass reaction is personalized using the relative abundance data. Finally, coupling constraints (Heinken et al., 2013) are applied to couple the flux through each microbial reaction to its corresponding biomass reaction flux. (iii) Simulation of the personalized microbial community models under different diet regimes, e.g. using flux variability analysis (Heirendt et al., 2017b). The differences between maximal uptake and secretion fluxes provide a metabolic profile for each microbial community sample, which can be analyzed using classical multidimensional scaling analyses. Diet-specific constraints (e.g. obtained from https://vmh.life/#nutrition) can be applied to the corresponding diet exchange reactions.
3 Implementation
The Microbiome Modeling Toolbox is written in MATLAB (Mathworks, Inc.) and accompanied with comprehensive documentation and tutorials. The toolbox allows for the integrative analysis of any number of reconstructions, including the human metabolic reconstruction (Brunk et al., 2018). Metabolic reconstructions can be obtained from, e.g. the VHM (https://vmh.life), BioModels (https://www.ebi.ac.uk/biomodels-main/) and the KBase (https://kbase.us/). A uniform nomenclature of reaction and metabolite abbreviations across the reconstructions is required. The implemented diet constraints require VMH abbreviations. To use higher taxonomical levels create pan-reconstructions (createPanModels). For larger datasets and/or bigger microbial community models, we recommend the use of the MATLAB command line or.m files and of a high-performance computing cluster.
4 Discussion
The Microbiome Modeling Toolbox enables the user to investigate microbial interactions at a large scale (Heinken et al., 2013; Magnúsdóttir et al., 2017). Moreover, metagenomically derived data can be integrated with microbial metabolic reconstructions permitting the prediction of altered functional assessment of different microbial communities, e.g. in health and disease (Heinken et al., 2018; Thiele et al., 2018).
Funding
This study received funding from the Luxembourg National Research Fund (FNR), through the ATTRACT program [FNR/A12/01], and the OPEN grant [FNR/O16/11402054], as well as the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program [grant agreement No 757922].
Conflict of Interest: none declared.
References
- Brunk E. et al. (2018) Recon3d enables a three-dimensional view of gene variation in human metabolism. Nat. Biotechnol., 36, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G. et al. (2010) Qiime allows analysis of high-throughput community sequencing data. Nat. Methods, 7, 335.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinken A. et al. (2013) Systems-level characterization of a host-microbe metabolic symbiosis in the mammalian gut. Gut Microbes, 4, 28–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinken A. et al. (2018) Personalized modeling of the human gut microbiome reveals distinct bile acid deconjugation and biotransformation potential in healthy and IBD individuals. bioRxiv, doi.org/10.1101/229138.
- Heirendt L. et al. (2017a) Creation and analysis of biochemical constraint-based models: the COBRA ToolBox v3. 0. Nature Protocols, in press. [DOI] [PMC free article] [PubMed]
- Heirendt L. et al. (2017b) DistributedFBA. jl: high-level, high-performance flux balance analysis in julia. Bioinformatics, 33, 1421–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnúsdóttir S. et al. (2017) Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol., 35, 81.. [DOI] [PubMed] [Google Scholar]
- Mitchell A.L. et al. (2018) EBI Metagenomics in 2017: enriching the analysis of microbial communities, from sequence reads to assemblies. Nucleic Acid Res., 46, D726–D735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson B. (2006) Systems Biology: Properties of Reconstructed Networks. Cambridge University Press, Cambridge. [Google Scholar]
- Segata N. et al. (2012) Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods, 9, 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele I. et al. (2018) When metabolism meets physiology: Harvey and Harvetta. bioRxiv, doi.org/10.1101/255885.