Abstract
RNA alternative polyadenylation contributes to the complexity of information transfer from genome to phenome, thus amplifying gene function. Here, we report the first X. tropicalis resource with 127,914 alternative polyadenylation (APA) sites derived from embryos and adults. Overall, APA networks play central roles in coordinating the maternal–zygotic transition (MZT) in embryos, sexual dimorphism in adults and longitudinal growth from embryos to adults. APA sites coordinate reprogramming in embryos before the MZT, but developmental events after the MZT due to zygotic genome activation. The APA transcriptomes of young adults are more variable than growing adults and male frog APA transcriptomes are more divergent than females. The APA profiles of young females were similar to embryos before the MZT. Enriched pathways in developing embryos were distinct across the MZT and noticeably segregated from adults. Briefly, our results suggest that the minimal functional units in genomes are alternative transcripts as opposed to genes.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03036-1) contains supplementary material, which is available to authorized users.
Keywords: Whole transcriptome termini site sequencing (WTTS-seq), Gene biotypes, APA site types, Genomic neighborhoods, RNA origin
Introduction
Endonucleolytic cleavage and polyadenylation of pre-mRNAs are two essential steps required for nascent RNAs to become mature and subsequently functional [1]. In fact, a gene often executes multiple cleavage and polyadenylation events using alternative polyadenylation (APA) sites, thus producing more than one transcript isoform per gene [2]. For instance, approximately 70% of annotated human genes use more than one APA site to form variable 3′ untranslated regions (UTRs) [3]. Because the 3′UTRs are often rich in regulatory elements, APA usage can dramatically influence RNA processing, stability, localization, translation and degradation [4]. When the same protein is produced after APA, quantitative effects on the protein occur due to gain/loss of regulatory elements in 3′ UTRs [2]. When APA changes the open reading frame, qualitative differences can be encoded in the protein [5]. In cases where a protein-coding gene is converted into a non-coding gene due to APA, epigenetic effects may ensue [6]. In brief, APA promotes RNA structural complexity and functional diversity, thus complicating genetic information transfer from genome to phenome.
Due to its diploid genome, relatively short life cycle and high biological similarity with X. laevis, X. tropicalis has many experimental advantages as a biomedical model for understanding the cellular, molecular and developmental biology underlying complex phenotypes [7]. More than 30 years ago, two APA sites 46 nucleotides apart were observed in the Xenopus beta 1 globin mRNA with the proximal site preferentially expressed over the distal site (approximately 99% vs. 1% of mRNA molecules) [8]. Other APA cases include Xenopus alpha-tubulin, integrin alpha 5, poly(A) polymerase, transcription elongation factor TFIIS (S-II) and alpha-tropomyosin [9–13]. In addition, Xenopus has been used to investigate effects of downstream sequences, upstream sequences and developmental stages on polyA sites, usage efficiency and pathways in polyadenylation [14–19]. Despite this history, genome-wide characterization of APA usage and patterns has not been conducted in X. tropicalis.
Here, we aim to develop the first APA resource in the species using our newly developed method called whole transcriptome termini site sequencing (WTTS-seq) [20]. X. tropicalis embryos collected at five stages, and adult males and females harvested at two age groups were used as experimental materials in the present study. We also carried out a large-scale genome-wide validation of X. tropicalis APA sites using polyA reads retrieved from expressed sequence tag sequencing (EST-seq), and generated from head and tail tag sequencing (HATT-seq), RNA sequencing (RNA-seq) and isoform sequencing (Iso-seq). Furthermore, we developed a whole transcriptome start site sequencing (WTSS-seq) method to validate pathways enriched with differentially expressed (DE)-APA sites between growing males and females. In brief, we observed striking differences in use of APA sites between maternal and zygotic sources of RNAs during the maternal–zygotic transition (MZT) period, between males and females and between embryos and adults. These resources and methods will improve gene annotation, accelerate epigenome mapping and facilitate the ENCODE project in X. tropicalis.
Materials and methods
Animals, RNA extraction, WTTS-seq libraries and read mapping
All X. tropicalis embryos and adult animals were handled in accordance with protocols approved by the Animal Care and Use Committee of Washington State University. Formation of pooled embryo RNA at stages 6 [before midblastula transition (MBT)], 8 (during MBT), 11 (gastrula), 15 (neurula) and 28 (tailbud) from two families were previously described [20]. Eight adult males and eight adult females, approximately 6 months of age (young adults), were purchased from Nasco (Fort Atkinson, WI, USA). Immediately upon arrival, two young males and two young females were euthanized, while the remaining 12 animals were raised in our vivarium facility until they were approximately 18 months of age (growing adults). Animal whole-body collection, tissue disruption, RNA extraction, removal of contaminating DNA and assessment of total RNA quantity and quality were described previously [20]. Our WTTS-seq protocol [20] was used to construct libraries for all 16 adult samples, but with a minor modification. Total RNA fragments between 250 and 500 bp were selected before polyA+ was enriched. All libraries were sequenced using an Ion PGM™ Sequencer at Washington State University.
Raw reads with a minimum of 50% bases covered at 99% identity (Q20) were filtered with the FASTX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) to produce high-quality reads. WTTS-seq is designed to have strand-specific sequencing complementary to polyA ends, so that reads have polyTs at their 5′-ends. In-house Perl scripts [20] were applied to remove the T-rich stretches and reads with at least 16 nt were preserved for mapping to the NCBI X. tropicalis genome (v9.1) using TMAP (version 3.4.1, https://github.com/iontorrent/TMAP). The high-quality aligned reads were filtered using a MAPping Quality (MAPQ) value equal to or greater than 2. Furthermore, the aligned reads were then used to identify poly(A) clustered sites in the X. tropicalis genome using a 24-nt window. At least, 25 mapped reads were required as evidence to define a clustered APA site. The genome coordinates for every site were then determined based on the positions of most of the mapped reads (Table S1).
Genome-wide validation of X. tropicalis APA sites using five different methods
First, we cross-referenced approximately 1.3 million ESTs (expressed sequence tags) from NCBI databases. Next, we retrieved all potential polyA reads (at least 8 consecutive Ts or As at 5′-ends or 3′-ends, respectively) from three sets of RNA-seq data, including (1) libraries created from pooled embryo RNA (collected from stages 6, 8 and 11) and sequenced on an Illumina HiSeq™ 2000 with single 50 bp reads; (2) a library prepared from a pooled RNA sample from three young male and three young female frogs and sequenced on an Ion PGM™ Sequencer; and (3) libraries created from RNA of two egg and 34 embryo pools and sequenced on Illumina platforms. The first two sets of data were generated by our own research [20], while the third was reported previously by Owens et al. [21].
Third, we used an RNA pool from three young female frogs to construct a HATT-seq library to simultaneously capture the 5′-end (head) and the 3′-end (tail) of a transcript. The assay includes synthesis of circularized full-length cDNA molecules, random shearing, dA tailing, ligation, PCR amplification, size selection and sequencing on an Illumina HiSeq™ 2500 with 100 bp reads. Our HATT-seq procedure is illustrated in Figure S1 and the protocol is detailed in the figure legend. This library produced a total of 16 million mapped reads (Table S1) with Ts at their 5′-ends (complementary to polyA tails), which were used in the present study.
Fourth, mRNA molecules were purified from an adult pool (3 young males and 3 young females) and an embryo pool (assembled from the 10 embryo pools mentioned above) and libraries were prepared for Iso-seq (Isoform sequencing) analysis. The purified mRNA was used to make full-length cDNA with the SMARTer cDNA kit (Invitrogen). The resulting cDNA from the young adult pool was then amplified 16× prior to duplex-specific nuclease (DSN) normalization (Evrogen). Normalized cDNA was amplified 18× and size fractionated on a BluePippen (Sage Science, Beverly MA). Three cDNA fractions of 1–2 kb, 2–3 kb, 3+ kb were collected and used for PacBio library construction and sequencing. However, the cDNA for the embryo pool was not DSN normalized and only two cDNA fractions of 1–2 kb and 2–3 kb were constructed and sequenced. Overall, 24 SMRT (Single Molecule, Real-Time) cells were used to sequence the adult pool and 15 SMRT cells were used to sequence the embryo pool.
Lastly, we developed a whole transcriptome start site sequencing (WTSS-seq) method to capture alternative transcriptional start (ATS) sites at the 5′-ends of transcripts. Total RNA samples derived from six growing male and six growing female frogs (18 months old) were used to construct individual WTSS-seq libraries. Briefly, WTSS-seq library construction started with depletion of rRNA from total RNA, followed by purification of rRNA-depleted RNA, which was then mixed with 5′ and 3′ adaptors for first-strand cDNA synthesis with Superscript III reverse transcriptase (Invitrogen, ThermoFisher Scientific, Inc., Waltham, MA, USA). After synthesis, cDNA was treated with RNases I and H, so only single-stranded cDNA remained. Solid-phase reversible immobilization (SPRI) beads (A63880, Beckman Coulter) were used to select cDNA fragments that were approximately 200–500 bp in size. The size-selected cDNA was finally amplified by PCR, purified with SPRI beads and sequenced on an Ion PGM™ Sequencer. Our WTSS-seq procedure is illustrated in Figure S2 and the protocol is detailed in the Figure S2 legend. These libraries were used to test our hypothesis that WTTS-seq and WTSS-seq would yield extremely similar pathways that show differences between growing males and growing females.
Characterization of APA sites in X. tropicalis
The APA sites were characterized by gene biotypes, site types (location within gene), genomic neighborhoods (adenine composition in the sequences adjacent to poly(A) site), poly(A) signal (PAS) motifs and RNA sources of origin in early development.
Gene biotypes were downloaded from the NCBI X. tropicalis genome (v9.1) assembly, which were then aligned to APA sites using the Cuffcompare (v2.2.1) program [22]. In the present study, we focused on protein-coding, long non-coding RNAs (lncRNAs), pseudogenes, microRNAs (miRNAs), and transfer RNAs (tRNAs) gene biotypes. When APA sites had no associated gene biotypes, they were classified as unknown.
The Cuffcompare (v2.2.1) program [22] was used to determine locations of APA site types within genes. We targeted six major regions provided by the program, including exonic (confined in exonic regions), extended exonic (at least 10 bp extended from exonic regions to intronic regions), intronic (confined in intronic regions), distal (located in exonic regions with extension), extended distal (located within 2 kb downstream of reference transcript) and antisense (located in exonic regions, but with opposite direction) APA sites, respectively (Figure S3).
The genomic neighborhoods included two types: an A-rich stretch (ARS) and non-A-rich stretches (NARS). We applied a 10-nt sliding window in the 30 bases downstream of poly(A) sites and counted the number of adenines (As) in the sense orientation or thymines (Ts) in the antisense orientation. An ARS-APA site contained either 7 consecutive As/Ts or ≥ 8 As/Ts out of 10 nt. Otherwise, the site was considered a NARS-APA. Next, we collected sequences from regions between 50 bases upstream and downstream of poly(A) sites and characterized the nucleotide distribution of both ARS- and NARS-APA sites.
We used 28 PAS motifs collected from Homo sapiens, Mus musculus, Danio rerio, Caenorhabditis elegans, and Drosophila melanogaster [3, 23–26] as references to screen PAS motifs located within 100 bases upstream of each APA site.
Raw counts of each APA site were then normalized using DEseq in the R platform [27] and used to classify the RNA sources of origin across the five developmental stages as maternal, maternal–zygotic, zygotic or mixed type. Maternal APA sites were expressed at early stage(s), but were undetectable from either stage 8, 11, 15 or 28, so that they had four sub-types. Zygotic APA sites were not detected at early stage(s), but were expressed from either stage 8, 11, 15 or 28, forming four sub-types. Maternal–zygotic APA sites were those in which maternal sites were expressed at early stage(s) and then totally degraded until zygotic sites were reactivated from either stage 11, 15 or 28. Mixed APA sites were expressed at all of these five stages so that they had five subtypes according to their peak stage. The details on origin and sub-type classification can be seen in Table S2.
Finally, statistical, visual and functional tools were used to characterize APA and ATS sites. The DEseq package in R [27] was used to determine DE-APA and DE-ATS sites with DE genes assigned. Pearson’s Chi squared test was used to examine differences between or among categories or groups. The Metascape program [28] was employed to pursue “GO Biological Processes” pathway enrichment between or among different sets of DE-APA or DE-ATS sites. The chisq.test function in R was applied to evaluate variable independence. A Spearman’s correlation matrix between expression values over different developmental stages was generated with the corrplot package in R (https://github.com/taiyun/corrplot). Custom Venn diagrams were drawn using an online tool provided by the Bioinformatics and Evolutionary Genomics Laboratory at Ghent University (http://bioinformatics.psb.ugent.be/webtools/Venn/). Volcano plots were generated with ggplot2 in R and used to visualize WTTS-seq profiles between a pair of samples. To visualize the total effect of experimental covariates and batch effects, a principal component comparison of samples was generated by plotPCA in R. All tests with p < 0.05 were considered significant.
Data availability
The raw reads for the constructed libraries (44 total) described above are available under accession numbers GSE74919, GSE74919 and GSE74919.
Results
The first X. tropicalis APA resource and effects of gene biotypes on their usage
We generated approximately 160 million WTTS-seq reads and identified 127,914 APA sites with ≥ 25 mapped reads per cluster. Among them, 97,411 APA sites had evidence from other sources, such as 19,116, 80,759, 28,144 and 82,699 APA sites supported by EST-seq, HATT-seq, Iso-seq and RNA-seq reads, respectively (Table S1). Of the 30,503 APA sites without additional support, we found that 5169 were DE-APA sites (adjusted p < 0.05) based on at least one of 36 pairs of DEseq tests. As such, we kept all APA sites to form the first APA resource in X. tropicalis, including 101,183 sites assigned to 17,975 currently annotated genes in this species (Table S1).
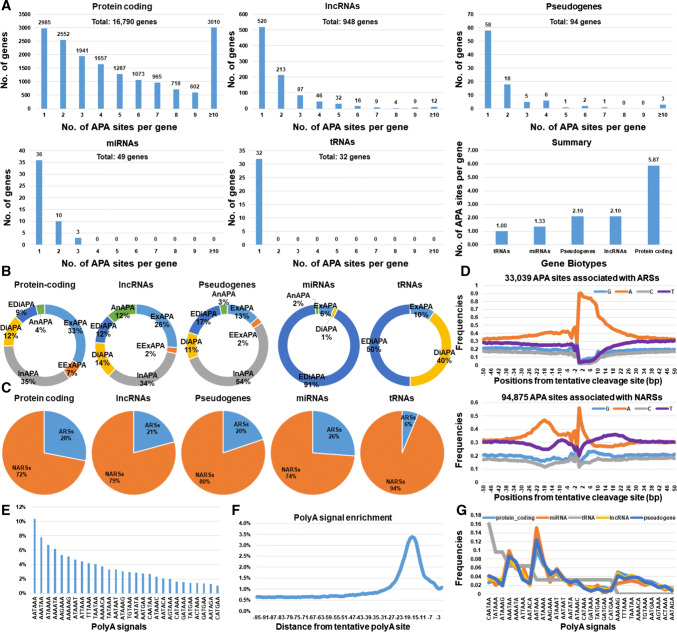
We discovered that numbers of APA sites per gene, site types and genomic neighborhoods were significantly different among gene biotypes (p < 5.632e−11) (Fig. 1a–c). We estimated that at least 82% of protein-coding genes had more than one APA site (Fig. 1a). In comparison, 45% of lncRNAs, 38% of pseudogenes, 27% of miRNAs and 0% of tRNAs had more than one APA site. As such, average APA usage was low in tRNAs and miRNAs with 1.00 and 1.33 site(s) per gene, but moderate in lncRNAs and pseudogenes with 2.10 sites per gene. In contrast, protein-coding genes contained an average of 5.87 APA sites per gene.
Fig. 1.
General characterization of APA sites in X. tropicalis. Gene biotypes had distinct a numbers of APA sites per gene, b site types with ExAPAs (exonic), EExAPAs (extended exonic), InAPAs (intronic), DiAPAs (distal), EDiAPAs (extended distal) and AnAPAs (antisense APA sites) and (C) genome neighborhoods classified as ARSs (“A” rich stretches) and NARS (non “A” rich stretches). d Characterization of nucleotide distributions in the 50 bp sequences flanking each side of a tentative polyA cleavage site associated with ARS-APA and NARS-APA sites. e Usage frequencies of 28 PASs in X. tropicalis. f Locations of PAS motifs within 10–30 bp upstream of poly(A) sites. g Frequencies of PAS motifs among gene biotypes
Intronic APA sites were preponderant in protein-coding genes, lncRNAs and pseudogenes (34–54%), while extended distal APA sites accounted for 91% of APA sites in miRNAs (Fig. 1b). The tRNA genes contained 46% distal, 38% extended distal, 8% exonic and 8% antisense APA sites. Genomic neighborhoods of APA sites accounted for 28, 26, 21, 20 and 6% ARS-APA in protein-coding genes, miRNAs, lncRNAs, pseudogenes, and tRNAs, respectively (Fig. 1c). Globally, we separated these 127,914 APA sites into (1) 33,039 with ARSs and (2) 94,875 with NARSs (Fig. 1d). The frequency of A (adenine) downstream of polyA sites ranged from 0.90 at the + 1 site to 0.75 at the + 10 site in ARS, but dropped dramatically from 0.56 at the + 1 site to 0.27 at the + 10 site in NARS (Fig. 1d). Composition of the 50 nucleotides upstream of polyA sites was also different between ARSs and NARSs (Fig. 1d).
Using 28 PASs as references, we found that approximately 83% of X. tropicalis APA sites possessed at least one of these motifs (Table S1). Overall, the canonical motif AATAAA was most prevalent in X. tropicalis, followed by AAATAA (Fig. 1e). These PAS motifs were mainly found 10–30 bp upstream of poly(A) sites (Fig. 1f). Interestingly, use of PASs by tRNAs was dramatically different from the other gene biotypes (Fig. 1g).
APA transcriptome similarities and their relevance to RNA sources, gender difference and lifespan stages
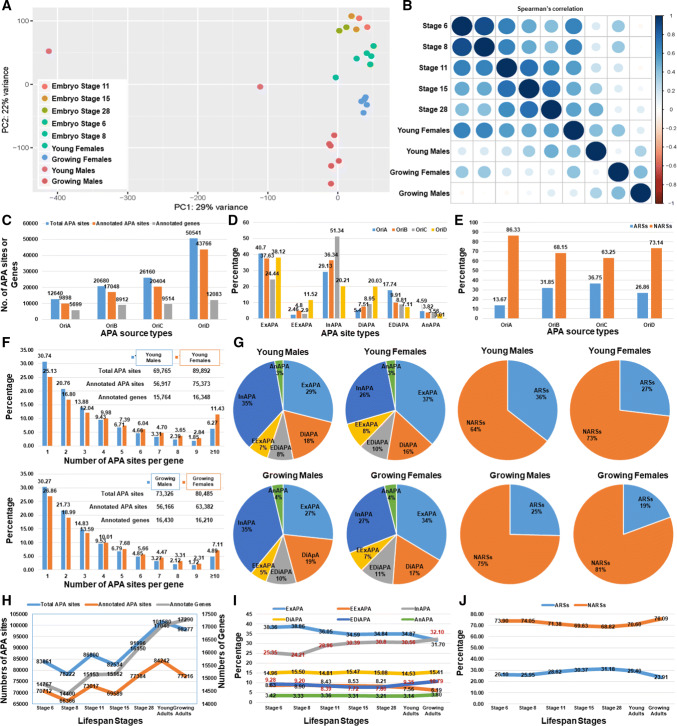
Principle component analysis (PCA) revealed that embryos had two well-defined clusters of APA profiles at stages 6 and 8 before MZT and stages 11, 15 and 28 after MZT (Fig. 2a). Males and females were also distinct from each other at both young and growing ages. However, young frogs had more diverse APA transcriptomes than growing frogs and APA usage was more divergent in males than females (Fig. 2a). Among the four groups of adults, the APA transcriptomes of young females were most similar to before-MZT embryos (Fig. 2a). Spearman’s rank correlation coefficients calculated with normalized expression units of each APA site for all pairs of stages clearly supported the PCA results described above (Fig. 2b).
Fig. 2.
Specific characterization of APA sites in X. tropicalis. a PCA analysis of 26 APA profiles related to five stages of embryos and two stages of adult male and female frogs. b Stage-based Spearman’s correlation estimates. c Classification of embryonic APA sites into maternal (OriA), maternal–zygotic (OriB), zygotic (OriC) and mixed (OriD) RNA sources of origin. Effects of APA origin on d site types and e genome neighborhoods. Effects of gender on f number of APA sites per gene and g site types and ratios between ARSs and NARSs. Effects of longitudinal stages on h APA basics, i site types and j genome neighborhoods
APA usage depends on RNA source of origin. The number of APA sites per gene averaged 1.74, 1.91, 2.15 and 3.62 for maternal, maternal–zygotic, zygotic and mixed transcript types, respectively (Fig. 2c). Of six site types, exonic, extended distal and antisense APA sites were most common in maternal transcripts (40.70, 17.74 and 4.59%, respectively), while intronic APA sites were most common in zygote-activated RNAs (51.34%). Extended exonic (11.52%) and distal (20.03%) APA sites were most common in transcripts derived from mixed type RNA sources (Fig. 2d). Maternal RNAs had the highest percentage (86.33%) of APA sites associated with NARS, while zygotic transcripts had 36.75% adjacent to ARS neighborhoods (Fig. 2e).
APA usage rather than gene usage depends on gender. Our evidence showed that young males used 20,127 fewer APA sites than young females and had 18,456 less annotated sites with genes. Growing males had 7159 and 7216 fewer APA sites and annotated sites, respectively, than growing females (Fig. 2f). However, gene usage was not dramatically different between the sexes. For example, there were 584 fewer genes in young males than young females, but 220 more genes in growing males compared to growing females. Young females had 11.41% more genes that expressed ≥ 4 APA sites than young males (Fig. 2f). However, this difference decreased to 7.39% between genders of growing frogs. On the other hand, females had 7–8% more exonic, but 8–9% less intronic APA sites than males. In addition, there were 9% and 6% more APA sites adjacent to ARSs in young and growing males, respectively, than females (Fig. 2g).
APA usage depends on lifespan stage. Numbers of APA sites, annotated APAs and associated genes varied among individual stages of a lifespan from embryos to adults (Fig. 2h). Embryos at stage 8 had the lowest values for these three measurements, while adults had tended to have more APA sites and annotated genes. Exonic APA sites decreased 6.66%, while intronic APA sites increased 6.75% from stage 6 to growing adults (Fig. 2i). In comparison, numbers of extended distal and extended exonic APA sites were relatively stable until stage 28, after which extended distal sites increased to 2.93% and extended exonic sites decreased to 2.02% in growing adults. Both distal and antisense APA sites stayed relatively consistent across all developmental/growth stages (Fig. 2i). NARS-associated APA sites changed during a lifespan, decreasing from 73.90% at embryo stage 6 to 68.82% at stage 28, and increasing to 76.09% in growing adults (Fig. 2j).
APA sources of origin, expression abundance and pathway networks during the maternal–zygotic transition in embryos
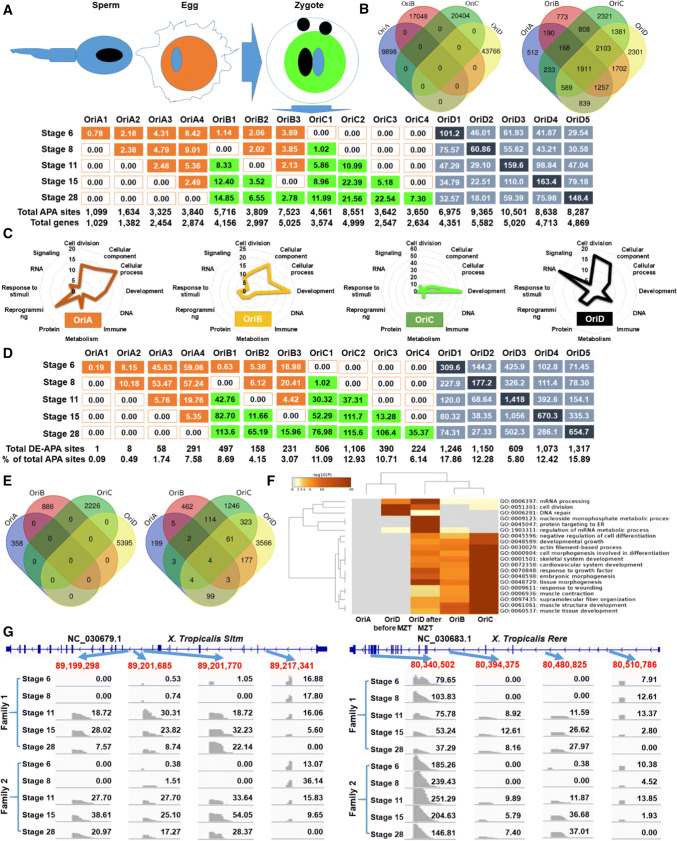
All APA sites expressed in embryos with annotated genes in X. tropicalis (Table S1) were used to determine coordinated networks in early development. The numbers of APA sites, their average expression levels and the numbers of genes are illustrated in Fig. 3a based on 16 RNA origin subtypes. Overall, the average expression levels of APA sites differed dramatically, ranging from 0 to 9.01 normalized units for maternal, from 0 to 14.85 for maternal–zygotic, from 0 to 22.54 for zygotic and from 18.01 to 163.40 for mixed-type RNA origin (Fig. 3a). Clearly, APA sites among these four RNA origins did not overlap, although their associated genes did (Fig. 3b).
Fig. 3.
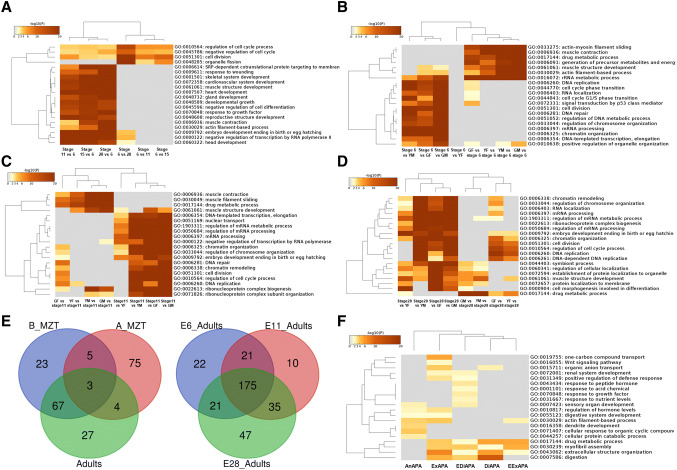
APA roles in MZT. a APA origin sub-types and expression abundance (normalized units in average). Maternal (in orange color), maternal–zygotic, zygotic (in green color) and mixed (in gray color) APA sources included 4 (OriA1–OriA4), 3 (OriB1–OriB3), 4 (OriC1–OriC4) and 5 (OriD1–OriD4) sub-types, respectively. b Sharing of APA sites and genes among four APA sources. c “Summary” pathway clusters enriched for each APA origin based on all APA sites. d Characterization of DE-APA sites among 16 sub-types of origin (see definitions in a). e DE-APA sites and DE genes that were shared among four RNA sources of origin. f “Summary” pathways enriched for each RNA source of origin based on DE-APA sites only. g Examples of genes executing different APA site expression patterns during the MZT period
We observed that APA sites derived from the four RNA origins were involved in different pathway networks. Figure S4 illustrates all 320 “summary” pathways enriched for all 16 subtypes of RNA origin with 20 pathways per subtype. Enriched pathways were subdivided into twelve clusters in a broad sense: cell division, cellular component, cellular process, development, DNA, immune, metabolism, protein, reprogramming, response to stimuli, RNA and signaling (Table S2). The two most significant pathway clusters were cellular processes (19%) and reprogramming (14%) for maternally derived APA sites; development (22%) and cellular components (18%) for APA sites of maternal–zygotic origin; development (53%) and cellular processes (14%) for APA sites from zygotic source; and RNA (19%) and cell division (17%) for APA sites derived from mixed RNA sources (Fig. 3c).
We identified at least 1346 pathways relevant to development in a broad sense (Table S3). Among them, 15, 42, 441 and 84 pathways were exclusively enriched with APA sites derived from maternal, maternal–zygotic, zygotic or mixed origins, respectively. The four most significant developmental pathways exclusively enriched with APA sites of maternal origin were regulation of reproductive process, female gamete generation, oocyte differentiation and positive regulation of myeloid cell differentiation. None of the exclusive developmental pathways coordinated by the maternal–zygotic source of APA sites had more than ten genes per pathway (Table S3).
Fourteen of the 441 developmental pathways exclusively enriched with APA sites of zygotic origin had more than 50 genes per pathway, including myotube differentiation, artery development, regulation of cell activation, ear morphogenesis, regeneration, sarcomere organization, leukocyte migration, regulation of fat cell differentiation, skeletal muscle contraction, musculoskeletal movement, cell maturation, myeloid leukocyte differentiation, regulation of leukocyte activation and epidermis development (Table S3).
Developmental pathways with at least 15 genes singularly enriched with APA sites derived from mixed RNA sources were regulation of hematopoietic progenitor cell differentiation, regulation of hematopoietic stem cell differentiation, hematopoietic stem cell differentiation, spermatid differentiation, spermatid development, male gamete generation, glial cell migration and left/right pattern formation (Table S3). Male and female gamete generation pathways were controlled by different sets of genes (Table S4). The remaining 764 developmental pathways were influenced by APA sites originating from multiple sources of RNA (Table S3). For example, head development contained APA sites stemming from 215 maternal, 540 maternal–zygotic, 728 zygotic and 1198 mixed RNA sources (Table S3).
We examined only the embryonic DE-APA sites (adjusted p < 0.1, Table S1) to determine their functions in early development. This selection dramatically reduced the number of APA sites and their associated genes for pathway analysis, but significantly increased the average levels of APA expression (Fig. 3d). In contrast, the number of genes shared among four sources of APA sites was significantly reduced to 4 (Fig. 3e) compared to the 1911 shown in Fig. 3b. Joint pathway enrichment analysis using these DE genes clearly defined their functions (Fig. 3f). In brief, DE-APA sites from zygotic and maternal–zygotic RNA sources contributed to tissue/organ development, while the DE-APA sites from mixed RNA origin were enriched for cell cycle, cellular components and metabolism.
Detailed examination of DE-APA sites (Table S5) led to identification of essential genes with differential usage of APA sites during early development. For example, two genes: Rere (arginine–glutamic acid dipeptide repeats) and Sltm (SAFB-like transcription modulator) each had four DE-APA sites derived from each of the four RNA sources (Fig. 3g). Such an antagonistic case (at least with one maternal DE-APA “turned off” and another zygotic or maternal–zygotic one “turned on”) also occurred in Atf5.2 (activating transcription factor 5, gene 2), Cnksr2 (connector enhancer of kinase suppressor of Ras 2), Mbtd1 (mbt domain containing 1), Mcph1 (microcephalin 1), Plin3 (perilipin 3), Pou5f3.1 (POU class 5 homeobox 3, gene 1), and Tprn (taperin) (Table S5).
Gender differences in APA usage, differential expression and pathway networks validated with WTSS-seq
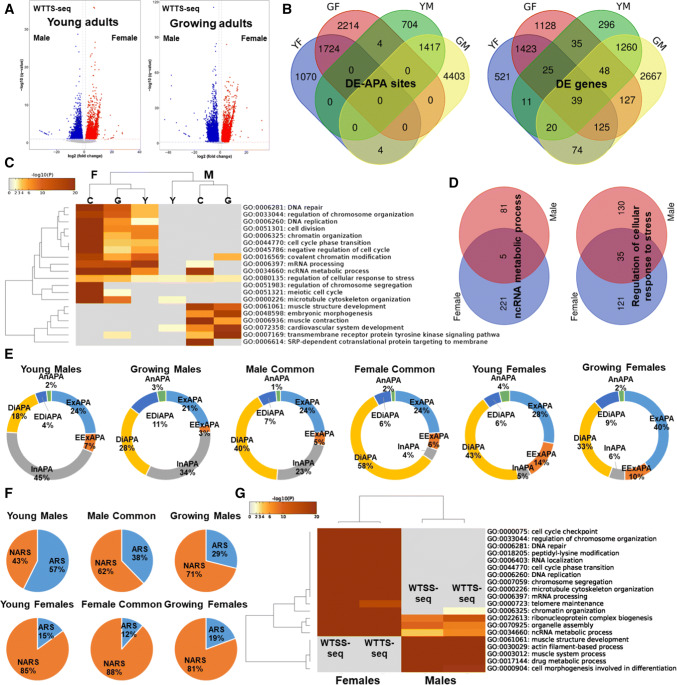
We observed that age had different effects on the overall expression levels of APA sites between both sexes (Fig. 4a). Young females had more up-regulated DE-APA sites than males, but the trend was reversed in the growing groups (Table S5). In summary, 2125 DE-APA sites (adjusted p < 0.05) and 5824 DE-APA sites (adjusted p < 0.005) were up-regulated in young and growing males; while 2798 DE-APA sites (adjusted p < 0.05) and 3942 DE-APA sites (adjusted p < 0.005) were up-regulated in young and growing females; respectively (Fig. 4b and Table S5). There were 1417 up-regulated DE-APA sites in common between young and growing male frogs, and 1724 up-regulated DE-APA sites in common between young and growing females. Males and females had more similar DE genes than DE-APA sites (Fig. 4b).
Fig. 4.
APA differences in sexual dimorphism. a Volcano plots of DE-APA profiles separating males from females at both young and growing ages. b Shared DE-APA sites and DE genes between males and females at different ages (YM, young males; GM, growing males; YF, young females and GF, growing females). c Male- or female-specific and common enriched pathways. d Examples that show the same pathways involved different sets of DE genes between sexes. Gender differences in e APA site types and f genome neighborhoods. g Pathway enrichment using DE-ATS and DE-APA sites on the same set of samples
Pathway enrichment was then performed based on three sets of DE-APA sites for each sex: specific for young, common for both and specific for growing frogs. Among the top 20 “summary” pathways, twelve and six were specifically enriched for females and males, respectively (Fig. 4c). Enriched pathways specific to female frogs were broadly relevant to cell-cycle events, while the enriched pathways exclusive to males were largely related to muscle events. Enriched pathways that were similar between females and males were ncRNA metabolic process and regulation of cellular response to stress (Fig. 4d and Table S4).
There were significant differences in use of DE-APA site types (p < 2.2e−16) and adenine composition of genomic neighborhoods (p < 1.271e−10) within each sex and between sexes. Males tended to use more intronic DE-APA sites than females (23–45% in males vs. 4–6% in females, Fig. 4e). Common DE-APA sites were mostly distal compared to the gender-specific sites in males (common 40% > growing males 28% > young males 18%) and females (common 58% > young females 43% and growing females 33%). There were more DE-APA sites adjacent to ARSs in males (29–57%) than females (12–19%) (Fig. 4f).
For validation, duplicate libraries were prepared from a total RNA sample according to our WTSS-seq procedures and sequenced. The same RNA sample was also sent to DNAFORM Precision Gene Technologies, Yokohama, Japan for sequencing using the traditional CAGE method. Data analysis revealed a total of 69,857 ATS sites in X. tropicalis (Table S6). There were no significant differences in ATS expression levels between WTSS-seq and CAGE (adjusted p > 0.9433, Table S6). DEseq analysis identified 5223 DE-ATS sites (adjusted p < 0.0001) and 3808 DE-ATS sites (adjusted p < 0.0001) that were up-regulated in six males and six females, respectively.
Pathway enrichment analysis using data from WTSS-seq and WTTS-seq revealed the same set of 15 “summary” pathways for females and the same set of 8 “summary” pathways for males (Fig. 4g). However, the WTTS-seq identified one more pathway in males, chromatin organization, than WTSS-seq. Gene sets for each pathway differed between WTSS-seq and WTTS-seq. For example, WTSS-seq identified more genes in females that were involved in telomere maintenance than WTTS-seq (66 genes vs. 52 genes with 38 shared by both methods), but the latter method identified more genes regulating ncRNA metabolic process in males (118 genes vs. 97 genes with 73 commonly revealed by both methods) (Table S4).
Lifespan stage differences in APA usage, differential expression and pathway progression from embryos to adults
DEseq analysis revealed no essential differences in APA profiles between embryos at stages 6 and 8 (Table S5). The DE-APA sites of stage-6 embryos, representing before-MZT events were compared to the after-MZT embryos to identify functional changes that occurred during MZT (adjusted p < 0.05 and fold change ≥ |2.0|). Among the top 20 “summary” pathways (Fig. 5a), three pathways relevant to cell cycles were enriched for both before- and after-MZT embryos. Organelle fission was the only pathway specific to before-MZT embryos. The remaining 14 pathways mainly belonged to the after-MZT embryos and they played roles in development, response to both internal and external stimuli and transcription/translation events.
Fig. 5.
APA roles in longitudinal growth. Pathway progression a from stage 6 to stages 11, 15 and 28 embryos; b from stage 6 embryos to adults (YM, young males; GM, growing males; YF, young females and GF, growing females); c from stage 11 embryos to adults; and d from stage 28 embryos to adults. e Numbers of the uniquely up-regulated DE genes involved in the cell division pathway for before-MZT embryos (B_MZT), after-MZT embryos (A_MZT) and adults; and for embryos at stages 6, 11 and 28 in comparison to adults (E6_Adults, E11_Adults and E28_Adults). f “Summary” pathways enriched with APA sites exclusively expressed in adults. Splitting them into six groups based on their site types made it possible to complete the pathway analysis
Differentially expressed APA sites with adjusted p < 0.001 and fold change ≥ |2.0| of before-MZT embryos were then compared with young and growing males as well as young and growing females. These comparisons revealed that DE-APA sites of before-MZT embryos actively participated in RNA/DNA and cell-cycle events, while the DE-APA sites of adult males and females contributed to muscle and metabolic events (Fig. 5b). Comparisons of DE-APA sites between stage 11 embryos and adults revealed similar enriched pathways (Fig. 5c) as did comparisons between DE-APA sites of stage 28 embryos versus adult (Fig. 5d). However, enriched pathways diverged somewhat between males (muscle and metabolic events) and females (DNA and cell-cycle events).
These results indicate that up-regulated DE-APAs and their associated DE genes play essential roles in RNA/DNA and cell cycle events for the before-MZT embryos; RNA/DNA, cell-cycle and developmental events for the after-MZT embryos; and muscle function, drug metabolism and energy production for adults. Pathway enrichment analysis of the DE-APA sites in growing females indicates that RNA/DNA and cell-cycle functions are reactivated. Examination of cell division as an example indicated that there were APA sites specific to each stage (only three genes shared among before-MZT embryos, after-MZT embryos and adult females) or common to all stages (at least 175 genes shared between before- and after-MZT embryos (Fig. 5e).
Finally, we found that APA sites exclusively expressed in adults are specifically responsible for digestion and digestive systems; response to acid chemical, drug, growth factor, nutrient levels, organic cyclic compounds and peptide hormone; organic anion and compound transport; and maintenance of dendrite, sensory and renal system development (Fig. 5f).
Discussion
During the last two decades, the Xenopus community has invested heavily in development of genetic and genomic resources to enhance this model organism. For example, the first draft genome of X. tropicalis was successfully assembled [29] along with development of ESTs and full-length cDNA sequence resources for gene discovery and genome annotation [30–32]. No doubt, our current study establishes the first genome-wide APA resource in X. tropicalis. This timely, much-needed resource contains a total of 127,914 sites, 97,411 of which were confirmed with additional evidence (Table S1). In addition, 101,183 sites were assigned to 17,975 currently annotated X. tropicalis genes. We firmly believe that this unique APA resource will help the X. tropicalis community to study quantitative, qualitative and epigenetic effects of genes on complex phenotypes and use APA sites to understand genome-wide regulatory blueprints, thus strengthening the organism as a model that can be used to address important biological questions relevant to human health and disease. In addition, the resource also allowed us to understand the genetic information flows associated with MZT, sexual dimorphism and longitudinal growth in X. tropicalis.
The MZT is a unique period during embryonic development where maternal RNA species are progressively degraded and zygotic transcription drives development [33, 34]. This well-orchestrated transition is critical for normal development and when compromised leads to an array of developmental disorders [35]. Xenopus species have been widely used as model organisms to investigate the processes involved in the MZT, because maternal RNA degradation and zygotic genome activation (ZGA) are not tightly coupled and particularly, because ZGA coincides with the so-called mid-blastula transition (MBT) [36, 37]. The present study is the first to classify APA sites into four sources of origin: maternal (10.86%), maternal–zygotic (18.71%), zygotic (22.40%) and mixed type (48.03%) (Fig. 2a). Certainly, this classification allowed us to characterize unique features and roles of APA sites in early development. For example, RNA origins orchestrate APA-site use (Fig. 2d), expression abundance (Fig. 3a) and pathway networks (Fig. 3c), which lead to a smooth and successful MZT.
Our present study revealed that nearly 26% of APA sites were adjacent to ARSs (Fig. 1d). Because ARS regions are rich in As, we must comment about internal priming issues in sequencing, which can lead to erroneous conclusions. When oligo(dT) was employed in construction of libraries in methods such as 3′T-fill, polyA site sequencing (PAS-seq) and polyadenylation sequencing (PolyA-seq) (reviewed in [38]), filters were used to remove the internal priming sites. However, we observed that prevalence of ARS-APA sites largely depended on gene biotype (Fig. 1c), gender (Figs. 2g) and developmental stage (Fig. 2j), clearly indicating that ARS-APA sites do not occur randomly and that WTTS-seq library preparation minimizes internal priming sites. Nunes et al. [39] showed that more than 10,000 poly(A) sites harbor A-rich 3′end processing sites in the human genome and proved core poly(A) sequences were necessary for polyadenylation. Yoon et al. [40] also found low abundance short transcripts harbored A-rich stretches in human B-lymphoblastoid cells. The same phenomenon was observed in C. elegans [25]. Currently, we know that there are two types of RNAs: polyA+ and polyA−. Whether the A-rich stretches give rise to naturally occurring polyA+ RNAs warrants further investigation.
Tian et al. [41] observed that intronic polyadenylation events occurred in ~ 20% of human genes. Using RNA-seq data collected from human and mouse tissues, van Bakel et al. [42] also found that most non-exonic transcribed fragments are located in introns. In the present study, we confirmed that APA usage in introns is relatively frequent. For example, intronic APA sites were dominant in protein-coding genes (35%), lncRNAs (34%) and pseudogenes (54%) (Fig. 1b). Compared with maternal transcripts, studies have shown that a higher percentage of introns are retained in zygotic transcripts among different species [43–45]. As a result, most intronic polyadenylation events occur after the ZGA period, which explains why we found that a large proportion (51.34%) of zygotic APA sites were intronic (Fig. 2d). Because intronic APA sites use proximal polyA sites, zygote-activated transcripts were the shortest.
To profile the 5′-ends of transcripts, library preparation usually involves first-strand cDNA synthesized using random primers, and cDNAs with cap sites are then trapped [46]. Like the SAGE (serial analysis of gene expression) method [47], a linker that contains a recognition site for MmeI is ligated to the 5′-end of cDNA and then primed for synthesis of the second-strand cDNA. Next, double-stranded cDNA is cleaved with MmeI, generating a 20 bp/21 bp tag for each cDNA. A second linker is added to the 3′-end of the digested tag and the final library is prepared by PCR amplification and purified for sequencing without cloning. NanoCAGE and CAGEscan libraries were first described by Plessy et al. [48], who used reverse transcription in combination with template-switching primers to produce 25 bp tags after EcoP151 digestion. Overall, the library preparation process for these methods has many steps. Like any tag-based sequencing technique, assignment of short tags (20–25 bp) to genes or genome regions is challenging [38]. In the present study, we successfully developed a WTSS-seq method with a few steps: rRNA depletion and first- and second-strand cDNA syntheses (Figure S1). Our results confirmed that both WTTS-seq and WTSS-seq can produce well-matched pathways on the same set of samples (Fig. 4g). As such, our present study also provides the first alternative transcriptional start site resource for X. tropicalis (Table S4).
In summary, the X. tropicalis genome exhibits a high incidence of alternative polyadenylation events. We found that approximately 80% of currently annotated genes have more than one APA site (Table S1). APA diversity, dynamics and timely usage patterns can record the information flow from genome to phenome relevant to the maternal–zygotic transition in embryo development, gender differences responsible for sexual dimorphism and longitudinal growth from embryos to adults. Our results suggest that the minimal functional unit in a genome is the alternative transcript rather than the gene. Profiling of alternative transcriptional start and termination sites using our WTSS-seq and WTTS-seq methods, for example, may efficiently determine multiple functions of genes in response to quantitative, qualitative and epigenetic effects. In addition, genes or alternative transcripts cannot work alone and a complex phenotype requires coordination of many genes/transcripts. Furthermore, alternative transcript profiles can directly link transcriptome flexibility, diversity and dynamics to functional pathways and networks of genes. Together, these features enable a systems-biology approach to discover key elements that are essential to physical, physiological, psychological and pathological events affecting health and disease in human and animals.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 5 (XLSX 28399 kb)
Supplementary material 9 (XLSX 44743 kb)
Supplementary material 10 (XLSX 17414 kb)
Acknowledgements
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under Award Number R21HD076845 and the National Institute of Food and Agriculture, United States Department of Agriculture under Award number 2016-67015-24470 to ZJ. Development of bioinformatics pipelines for the data analysis at Xiamen University, China was supported by the National Science Foundation of China under Award number 61573296 to GJ.
Compliance with ethical standards
Conflict of interest
We have filed a provisional patent for our WTSS-seq method.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiang Zhou and Yangzi Zhang contributed equally to the work.
References
- 1.Wahle E, Keller W. The biochemistry of 3′-end cleavage and polyadenylation of messenger RNA precursors. Annu Rev Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- 2.Tian B, Manley JL. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci. 2013;38(6):312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matoulkova E, Michalova E, Vojtesek B, Hrstka R. The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 2012;9(5):563–576. doi: 10.4161/rna.20231. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y. Alternative polyadenylation: new insights from global analyses. RNA. 2012;18(12):2105–2117. doi: 10.1261/rna.035899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma L, Guo C, Li QQ. Role of alternative polyadenylation in epigenetic silencing and antisilencing. Proc Natl Acad Sci USA. 2014;111(1):9–10. doi: 10.1073/pnas.1321025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harland RM, Grainger RM. Xenopus research: metamorphosed by genetics and genomics. Trends Genet. 2011;27(12):507–515. doi: 10.1016/j.tig.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mason PJ, Jones MB, Elkington JA, Williams JG. Polyadenylation of the Xenopus beta 1 globin mRNA at a downstream minor site in the absence of the major site and utilization of an AAUACA polyadenylation signal. EMBO J. 1985;4(1):205–211. doi: 10.1002/j.1460-2075.1985.tb02337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabbitts KG, Morgan GT. Alternative 3′ processing of Xenopus alpha-tubulin mRNAs; efficient use of a CAUAAA polyadenylation signal. Nucleic Acids Res. 1992;20(12):2947–2953. doi: 10.1093/nar/20.12.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joos TO, Whittaker CA, Meng F, DeSimone DW, Gnau V, Hausen P. Integrin alpha 5 during early development of Xenopus laevis. Mech Dev. 1995;50(2–3):187–199. doi: 10.1016/0925-4773(94)00335-K. [DOI] [PubMed] [Google Scholar]
- 11.Zhao W, Manley JL. Complex alternative RNA processing generates an unexpected diversity of poly(A) polymerase isoforms. Mol Cell Biol. 1996;16(5):2378–2386. doi: 10.1128/MCB.16.5.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plant KE, Hair A, Morgan GT. Genes encoding isoforms of transcription elongation factor TFIIS in Xenopus and the use of multiple unusual RNA processing signals. Nucleic Acids Res. 1996;24(18):3514–3521. doi: 10.1093/nar/24.18.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anquetil V, Le Sommer C, Méreau A, Hamon S, Lerivray H, Hardy S. Polypyrimidine tract binding protein prevents activity of an intronic regulatory element that promotes usage of a composite 3′-terminal exon. J Biol Chem. 2009;284(47):32370–32383. doi: 10.1074/jbc.M109.029314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andres AC, Hosbach HA, Weber R. Comparative analysis of the cDNA sequences derived from the larval and the adult alpha 1-globin mRNAs of Xenopus laevis. Biochim Biophys Acta. 1984;781(3):294–301. doi: 10.1016/0167-4781(84)90096-4. [DOI] [PubMed] [Google Scholar]
- 15.Banville D, Williams JG. Developmental changes in the pattern of larval beta-globin gene expression in Xenopus laevis. Identification of two early larval beta-globin mRNA sequences. J Mol Biol. 1985;184(4):611–620. doi: 10.1016/0022-2836(85)90307-9. [DOI] [PubMed] [Google Scholar]
- 16.Mason PJ, Elkington JA, Lloyd MM, Jones MB, Williams JG. Mutations downstream of the polyadenylation site of a Xenopus beta-globin mRNA affect the position but not the efficiency of 3′ processing. Cell. 1986;46(2):263–270. doi: 10.1016/0092-8674(86)90743-9. [DOI] [PubMed] [Google Scholar]
- 17.Paris J, Richter JD. Maturation-specific polyadenylation and translational control: diversity of cytoplasmic polyadenylation elements, influence of poly(A) tail size, and formation of stable polyadenylation complexes. Mol Cell Biol. 1990;10(11):5634–5645. doi: 10.1128/MCB.10.11.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79(4):617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- 19.Kühl M, Wedlich D. XB/U-cadherin mRNA contains cytoplasmic polyadenylation elements and is polyadenylated during oocyte maturation in Xenopus laevis. Biochim Biophys Acta. 1995;1262(1):95–98. doi: 10.1016/0167-4781(95)00073-P. [DOI] [PubMed] [Google Scholar]
- 20.Zhou X, Li R, Michal JJ, Wu XL, Liu Z, Zhao H, Xia Y, Du W, Wildung MR, Pouchnik DJ, Harland RM, Jiang Z. Accurate profiling of gene expression and alternative polyadenylation with whole transcriptome termini site sequencing (WTTS-Seq) Genetics. 2016;203(2):683–697. doi: 10.1534/genetics.116.188508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owens ND, Blitz IL, Lane MA, Patrushev I, Overton JD, Gilchrist MJ, Cho KW, Khokha MK. Measuring absolute RNA copy numbers at high temporal resolution reveals transcriptome kinetics in development. Cell Rep. 2016;14:632–647. doi: 10.1016/j.celrep.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Retelska D, Iseli C, Bucher P, Jongeneel CV, Naef F. Similarities and differences of polyadenylation signals in human and fly. BMC Genom. 2006;7:176. doi: 10.1186/1471-2164-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ulitsky I, Shkumatava A, Jan CH, Subtelny AO, Koppstein D, Bell GW, Sive H, Bartel DP. Extensive alternative polyadenylation during zebrafish development. Genome Res. 2012;22:2054–2066. doi: 10.1101/gr.139733.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, Yángüez E, Andenmatten D, Pache L, Manicassamy B, Albrecht RA, Gonzalez MG, Nguyen Q, Brass A, Elledge S, White M, Shapira S, Hacohen N, Karlas A, Meyer TF, Shales M, Gatorano A, Johnson JR, Jang G, Johnson T, Verschueren E, Sanders D, Krogan N, Shaw M, König R, Stertz S, García-Sastre A, Chanda SK. Meta- and orthogonal integration of influenza “OMICs’’ data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18(6):723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, Blitz IL, Blumberg B, Dichmann DS, Dubchak I, Amaya E, Detter JC, Fletcher R, Gerhard DS, Goodstein D, Graves T, Grigoriev IV, Grimwood J, Kawashima T, Lindquist E, Lucas SM, Mead PE, Mitros T, Ogino H, Ohta Y, Poliakov AV, Pollet N, Robert J, Salamov A, Sater AK, Schmutz J, Terry A, Vize PD, Warren WC, Wells D, Wills A, Wilson RK, Zimmerman LB, Zorn AM, Grainger R, Grammer T, Khokha MK, Richardson PM, Rokhsar DS. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328(5978):633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein SL, Strausberg RL, Wagner L, Pontius J, Clifton SW, Richardson P. Genetic and genomic tools for Xenopus research: the NIH Xenopus initiative. Dev Dyn. 2002;225:384–391. doi: 10.1002/dvdy.10174. [DOI] [PubMed] [Google Scholar]
- 31.Morin RD, Chang E, Petrescu A, Liao N, Griffith M, Chow W, Kirkpatrick R, Butterfield YS, Young AC, Stott J, Barber S, Babakaiff R, Dickson MC, Matsuo C, Wong D, Yang GS, Smailus DE, Wetherby KD, Kwong PN, Grimwood J, Brinkley CP, III, Brown-John M, Reddix-Dugue ND, Mayo M, Schmutz J, Beland J, Park M, Gibson S, Olson T, Bouffard GG, Tsai M, Featherstone R, Chand S, Siddiqui AS, Jang W, Lee E, Klein SL, Blakesley RW, Zeeberg BR, Narasimhan S, Weinstein JN, Pennacchio CP, Myers RM, Green ED, Wagner L, Gerhard DS, Marra MA, Jones SJ, Holt RA. Sequencing and analysis of 10,967 full-length cDNA clones from Xenopus laevis and Xenopus tropicalis reveals post-tetraploidization transcriptome remodeling. Genome Res. 2006;16(6):796–803. doi: 10.1101/gr.4871006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fierro AC, Thuret R, Coen L, Perron M, Demeneix BA, Wegnez M, Gyapay G, Weissenbach J, Wincker P, Mazabraud A, Pollet N. Exploring nervous system transcriptomes during embryogenesis and metamorphosis in Xenopus tropicalis using EST analysis. BMC Genom. 2007;8:118. doi: 10.1186/1471-2164-8-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee MT, Bonneau AR, Giraldez AJ. Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev Cell Dev Biol. 2014;30:581–613. doi: 10.1146/annurev-cellbio-100913-013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onichtchouk DV, Voronina AS. Regulation of Zygotic Genome and Cellular Pluripotency. Biochemistry (Mosc) 2015;80:1723–1733. doi: 10.1134/S0006297915130088. [DOI] [PubMed] [Google Scholar]
- 35.Langley AR, Smith JC, Stemple DL, Harvey SA. New insights into the maternal to zygotic transition. Development. 2014;141:3834–3841. doi: 10.1242/dev.102368. [DOI] [PubMed] [Google Scholar]
- 36.Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- 37.Yang J, Aguero T, King ML. The Xenopus maternal-to-zygotic transition from the perspective of the germline. Matern Zygotic Transit. 2015;113:271–303. doi: 10.1016/bs.ctdb.2015.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang Z, Zhou X, Li R, Michal JJ, Zhang S, Dodson MV, Zhang Z, Harland RM. Whole transcriptome analysis with sequencing: methods, challenges and potential solutions. Cell Mol Life Sci. 2015;72:3425–3439. doi: 10.1007/s00018-015-1934-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nunes NM, Li W, Tian B, Furger A. A functional human poly(A) site requires only a potent DSE and an A-rich upstream sequence. EMBO J. 2010;29:1523–1536. doi: 10.1038/emboj.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon OK, Hsu TY, Im JH, Brem RB. Genetics and regulatory impact of alternative polyadenylation in human B-lymphoblastoid cells. PLoS Genet. 2012;8:e1002882. doi: 10.1371/journal.pgen.1002882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian B, Pan ZH, Lee JY. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res. 2007;17:156–165. doi: 10.1101/gr.5532707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biedler JK, Hu WQ, Tae H, Tu ZJ. Identification of early zygotic genes in the yellow fever mosquito Aedes aegypti and discovery of a motif involved in early zygotic genome activation. PLoS One. 2012;7:e33933. doi: 10.1371/journal.pone.0033933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Artieri CG, Fraser HB. Transcript length mediates developmental timing of gene expression across Drosophila. Mol Biol Evol. 2014;31:2879–2889. doi: 10.1093/molbev/msu226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guilgur LG, Prudencio P, Sobral D, Liszekova D, Rosa A, Martinho RG. Requirement for highly efficient pre-mRNA splicing during Drosophila early embryonic development. Elife. 2014;3:e02181. doi: 10.7554/eLife.02181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valen E, Pascarella G, Chalk A, Maeda N, Kojima M, Kawazu C, Murata M, Nishiyori H, Lazarevic D, Motti D, Marstrand TT, Tang MH, Zhao X, Krogh A, Winther O, Arakawa T, Kawai J, Wells C, Daub C, Harbers M, Hayashizaki Y, Gustincich S, Sandelin A, Carninci P. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res. 2009;19:255–265. doi: 10.1101/gr.084541.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270(5235):484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 48.Plessy C, Bertin N, Takahashi H, Simone R, Salimullah M, Lassmann T, Vitezic M, Severin J, Olivarius S, Lazarevic D, Hornig N, Orlando V, Bell I, Gao H, Dumais J, Kapranov P, Wang H, Davis CA, Gingeras TR, Kawai J, Daub CO, Hayashizaki Y, Gustincich S, Carninci P. Linking promoters to functional transcripts in small samples with nanoCAGE and CAGEscan. Nat Methods. 2010;7:528–534. doi: 10.1038/nmeth.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 5 (XLSX 28399 kb)
Supplementary material 9 (XLSX 44743 kb)
Supplementary material 10 (XLSX 17414 kb)
Data Availability Statement
The raw reads for the constructed libraries (44 total) described above are available under accession numbers GSE74919, GSE74919 and GSE74919.