Abstract
Immune cell populations determine the balance between ongoing damage and repair following tissue injury. Cells responding to a tissue-damaged environment have significant bioenergetic and biosynthetic needs. In addition to supporting these needs, metabolic pathways govern the function of pro-repair immune cells, including regulatory T cells and tissue macrophages. In this Review, we explore how specific features of the tissue-damaged environment such as hypoxia, oxidative stress, and nutrient depletion serve as metabolic cues to promote or impair the reparative functions of immune cell populations. Hypoxia, mitochondrial DNA stress, and altered redox balance each contribute to mechanisms regulating the response to tissue damage. For example, hypoxia induces changes in regulatory T cell and macrophage metabolic profiles, including generation of 2-hydroxyglutarate, which inhibits demethylase reactions to modulate cell fate and function. Reactive oxygen species abundant in oxidative environments cause damage to mitochondrial DNA, initiating signaling pathways that likewise control pro-repair cell function. Nutrient depletion following tissue damage also affects pro-repair cell function through metabolic signaling pathways, specifically those sensitive to the redox state of the cell. The study of immunometabolism as an immediate sensor and regulator of the tissue-damaged environment provides opportunities to consider mechanisms that facilitate healthy repair of tissue injury.
Introduction
The emerging field of immunometabolism studies the functional interactions between metabolic signals and the myriad cells and molecular networks that constitute the immune system (1). Tissue damage triggers a complex immune response that determines the balance between ongoing injury and a return to homeostasis. Pro-repair cell populations have substantial bioenergetic and biosynthetic requirements; however, our primary goal in this Review is to explore how metabolism governs pro-repair cell function in the tissue-damaged environment. Indeed, biochemical features of the tissue-damaged environment serve as metabolic inputs that signal myeloid and lymphoid cell populations to either sustain damaging inflammation or direct pro-resolution and pro-repair functional programs. Although robust causal links between tissue injury, metabolism, and immune cell function are lacking, several features of damaged tissues, such as hypoxia, oxidative stress, and nutrient depletion, represent powerful modulators of cellular metabolism and thus immune cell function.
While nearly every immune cell type plays a role in resolution of inflammation and repair of tissue damage, in this Review we focus on two key cell types that have emerging roles in coordinating resolution and repair: CD4+Foxp3+ regulatory T cells (Tregs) and tissue-resident macrophages, both of which are under the control of metabolic programming. The specific metabolic features of these cells in their resting and activated states have been reviewed elsewhere (2). Here, our objectives are to examine how Tregs and tissue macrophages respond to injury and how the tissue-damaged environment provides metabolic cues to regulate their fate and function. We discuss metabolic programming, mitochondrial DNA stress, and redox balance as prototypical metabolic inputs regulating pro-repair cell function in tissue-damaged environments. Where possible, we speculate on these mechanisms as clinical biomarkers or targets for therapeutic intervention and discuss possibilities for future investigation.
Metabolism determines cell fate and function
Historically, the utilization of different carbon fuel sources to generate ATP (i.e., catabolism) and generation of macromolecules to support growth (i.e., anabolism) have been considered the major functions of metabolism. In contrast, the central premise of our Review is that major metabolic pathways generate molecules that control key immune cell fates and functions (3). An exciting development in the past decade is that metabolism, beyond catabolism and anabolism, can determine immune cell fate and function through a variety of mechanisms, including the release of tricarboxylic acid (TCA) cycle intermediates, reactive oxygen species (ROS), and DNA from mitochondria into the cytoplasm, extracellular milieu, and circulation (4, 5). In this section, we introduce important pathways involved in immunometabolism and highlight how they influence protein and cell function using examples detailed in the other sections of our Review (Figure 1).
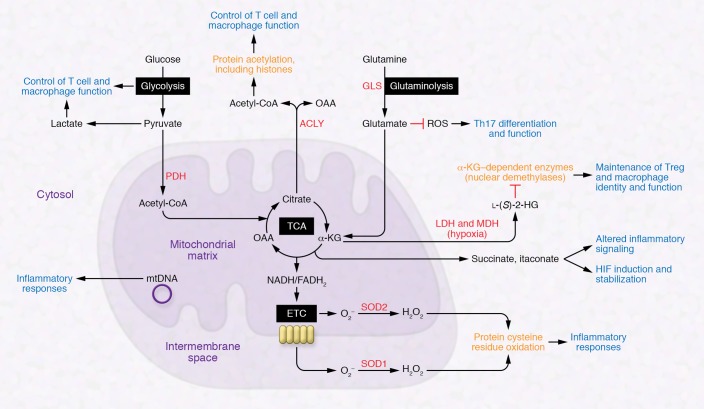
Figure 1. Overview of pathways involved with immunometabolism and their links to protein and cell function.
Both cytosolic and mitochondrial reactions generate important molecules that can modulate protein structure and function, regulate enzymatic reactions, and control cell fate and function. ACLY, ATP-citrate lyase; ETC, electron transport chain; GLS, glutaminase; α-KG, α-ketoglutarate; LDH, lactate dehydrogenase; l-(S)-2-HG, l-(S)-2-hydroxyglutarate; MDH, malate dehydrogenase; mtDNA, mitochondrial DNA; OAA, oxaloacetate; PDH, pyruvate dehydrogenase.
Glucose, through a series of metabolic reactions known as glycolysis, generates pyruvate. Subsequently, pyruvate can either be converted into lactate or enter the mitochondrial matrix, where it is oxidized to acetyl-CoA by pyruvate dehydrogenase to enter the TCA (also known as citric acid or Krebs) cycle. Aerobic glycolysis, a hallmark of many immune cells, including effector T cells and macrophages, can modulate the function of immune cell populations in addition to generating ATP and intermediates that feed macromolecular synthesis and the TCA cycle. For example, the glycolytic intermediate phosphoenolpyruvate promotes calcium signaling essential for murine T cell activation (6). IFN-γ production by effector T cells can be controlled by the glycolytic enzyme GAPDH, which binds to the 3′-UTR of IFN-γ mRNA (7). Consequently, gain or loss of function of the key glucose transporter GLUT1 in T cells can respectively promote or dampen their effector function (8–10). Toll-like receptor (TLR) signaling in murine macrophages also promotes aerobic glycolysis, which generates inflammation through mechanisms that are not fully understood (11, 12). In contrast, the cytokine IL-10 exerts its antiinflammatory effects by inhibiting LPS-induced glucose uptake and promoting oxidative phosphorylation (13). T cells also use glutaminase-mediated glutaminolysis to promote differentiation and function of Th17 cells over Th1 cells via modulation of ROS and TCA cycle intermediates (14).
Although many immune cells display robust glycolysis, they also use the TCA cycle to control immune responses. Mitochondrial acetyl-CoA enters the TCA cycle through reaction with oxaloacetate (OAA) to produce citrate. Citrate can also be exported into the cytoplasm to regenerate acetyl-CoA and OAA by ATP-citrate lyase. This process controls cytoplasmic and nuclear acetyl-CoA levels for protein acetylation, which modulates the structure and function of many proteins, including nuclear histones. For example, acetyl-coA promotes histone acetylation that facilitates T cell–dependent IFN-γ production in mice and humans (15, 16). Other TCA cycle intermediates also undergo cytoplasmic reactions, producing molecules that inhibit key enzymes involved with controlling cell fate and function. As discussed below, cytoplasmic lactate dehydrogenase and malate dehydrogenase can promiscuously use the TCA cycle metabolite α-ketoglutarate (α-KG) to produce l-(S)-2-hydroxyglutarate (2-HG) under hypoxic conditions (17, 18). Histone demethylases and the ten-eleven translocation (TET) family of enzymes that participate in DNA demethylation reactions utilize α-KG as a substrate (19, 20) and are thus susceptible to inhibition by 2-HG as well as the TCA cycle intermediate succinate (17, 21, 22). Recent studies indicate that 2-HG can suppress Treg function and enhance CD8+ T cell function and generation of memory cells (23–25).
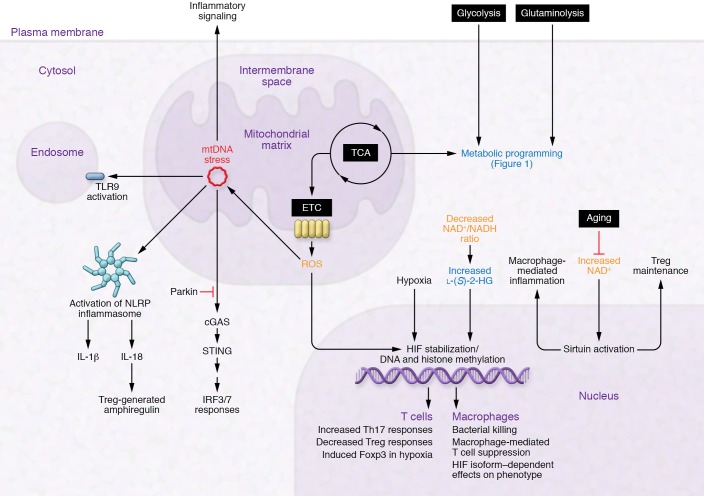
An interesting part of mitochondrial biology is that mitochondria maintain a double-stranded circular DNA genome reminiscent of their primordial origins as α-proteobacteria. Mitochondrial DNA (mtDNA) contains 37 genes, including 13 that encode for critical subunits of the mitochondrial electron transport chain (ETC); the majority of ETC subunits are encoded by the nuclear genome. Because of its unusual biochemistry, the mitochondrial genome, particularly when damaged, can serve as a signaling molecule to alter cell function in the setting of oxidative stress. Indeed, mtDNA is viewed as foreign — as opposed to self — DNA, likely as a result of its similar features to bacterial DNA (26). Mitochondrial DNA released into the cytosol, extracellular milieu, or circulation activates the NLRP3 inflammasome, TLR9, and cytosolic cyclic GMP–AMP synthase (cGAS) stimulator of interferon genes (STING) pathway to induce proinflammatory and type I interferon responses as well as inflammasome activation even in the absence of an infection by a pathogen (i.e., sterile inflammation) (Figure 2) (27–29).
Figure 2. Immunometabolic mechanisms at play in the tissue-damaged environment.
Hypoxia, oxidative stress, and nutrient depletion combine to cause significant changes in metabolism. Metabolic programming (blue text), mtDNA stress (red text), and redox state (orange text) are highlighted in their effects on pro-repair cells.
The TCA cycle also generates the reducing equivalents NADH and FADH2, which provide electrons to the ETC. Mitochondrial ROS are produced by ETC complexes I, II, and III through the reduction of molecular oxygen (O2) to superoxide (O2–). Superoxide is then converted to H2O2 in the mitochondrial matrix or intermembrane space by the superoxide dismutases SOD2 and SOD1, respectively, and released into the cytosol, where it oxidizes cysteine residues within proteins to alter their function. Mitochondrial ROS promote T cell receptor–dependent activation of IL‑2, NLRP3-dependent inflammasome activation, and TLR- and TNF receptor–dependent induction of inflammatory cytokines and bactericidal activity (30–38). Altogether, these studies reinforce our core theme that metabolism controls immune cell fate and function beyond ATP generation and biosynthesis. Below, we explore how metabolic features of damaged tissues arise and subsequently provide biochemical cues that modulate the function of pro-repair cells, particularly Tregs and macrophages.
Metabolic features of damaged tissues
Tissue hypoxia and oxidative stress result from hypoxemia or vascular ischemia or from inflammation and tissue damage themselves (39, 40). Acute lung injury and its clinical manifestation, the acute respiratory distress syndrome (ARDS), cause severe pulmonary tissue hypoxia as well as systemic arterial hypoxemia that leads to extrapulmonary tissue hypoxia in the absence of mechanical ventilation or extracorporeal life support (41–43). Acute myocardial infarction and ischemic stroke represent acute vascular ischemic states that induce local tissue hypoxia and result in inflammatory injury (39). In addition, the normally hypoxic intestinal epithelium becomes even more hypoxic in the setting of inflammatory bowel disease (44). These common clinical settings suggest self-amplifying links between hypoxia, tissue injury, and inflammation. Although specific causal mechanisms underlying these links remain largely undefined, we hypothesize that metabolic responses to hypoxia, oxidative stress, and nutrient depletion modulate the pro-repair functions of immune cell populations responding to (and residing in) tissue-damaged environments.
Prolyl hydroxylases sense intracellular oxygen tension and hydroxylate prolyl residues within the α subunit of the hypoxia-inducible transcription factor (HIF), which coordinates the physiologic response to hypoxia (45). Under normoxia, oxygen-dependent hydroxylation of prolyl residues within HIF-1α or HIF-2α generates a binding site for the von Hippel–Lindau (VHL) protein, which is a component of the E3 ubiquitin ligase complex that ubiquitinates HIF’s α subunit and targets it for proteosomal degradation. Hypoxia prevents prolyl hydroxylation, leading to stabilization of HIF. In addition to hypoxia, HIF is transcriptionally activated or stabilized under normoxic conditions such as LPS exposure (46) or via inhibition of prolyl hydroxylases by ROS or reduced intracellular iron (47, 48).
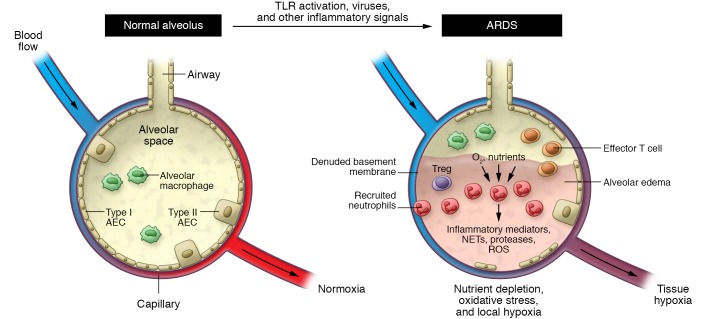
In addition to decreased oxygen delivery, immune cells responding to tissue damage consume substantial oxygen and other nutrients, thus promoting tissue hypoxia and an oxidative, nutrient-depleted environment following infection or sterile injury (ref. 49 and Figure 3). Myeloid cells of the innate immune system, including macrophages and mast cells, serve as tissue sentinels, initiating and regulating neutrophil recruitment to damaged tissues (50, 51). It remains unclear whether tissue-resident cells themselves contribute to tissue hypoxia in a cell-intrinsic fashion. Regardless, numerous investigations have shown that pathogen- and damage-associated molecular patterns released from invading pathogens and damaged and dying cells signal via pattern recognition receptors such as TLRs on these myeloid sentinels (52), attracting circulating neutrophils and promoting their vascular adhesion and tissue transmigration (53, 54). Activated neutrophils dominate the acute inflammatory response by phagocytosing pathogens, releasing inflammatory mediators (55, 56), DNA- and histone-laden neutrophil extracellular traps (57), and tissue-remodeling proteases (58) as well as generating ROS via NADPH oxidases such as NOX4 (59–61). Neutrophil extravasation and activation in response to infectious or sterile injury represent an early event in the immune response to tissue damage (62), and neutrophils also contribute to chronic inflammation and tissue injury via secretion of serine and matrix metalloproteases and other tissue-remodeling enzymes (63). Collectively, the cellular and molecular events associated with tissue injury provide a metabolic environment characterized by hypoxia, oxidative stress, and nutrient depletion, which can serve as signals to modulate the function of both tissue-resident and recruited cells. The balance between tissue-resident and recruited cells in generating these environmental features remains undefined. Future work involving bone marrow chimeras, adoptive transfer experiments, and selective loss and gain of specific tissue-resident and recruited cell populations will help clarify the relative contributions of these populations in creating the tissue-damaged environment.
Figure 3. Metabolic features of the tissue-damaged environment.
Acute lung injury and its clinical correlate the acute respiratory distress syndrome (ARDS) represent a prototypical tissue-damaged environment. Inflammatory signals, including TLR activation, lead to neutrophil recruitment. Neutrophils and other cells consume oxygen and nutrients and cause tissue damage by release of various substances, importantly ROS. Combined with the physiologic effects of tissue injury, the result is an environment characterized by hypoxia, oxidative stress, and nutrient depletion. AEC, alveolar epithelial cell; NET, neutrophil extracellular trap.
Tregs and macrophages in tissue repair
Tregs in tissue repair.
CD4+Foxp3+ Tregs are required to maintain immune homeostasis and suppress overexuberant immune system activation (64–66). Mice bearing a mutation in their Foxp3 gene (scurfy mice) spontaneously develop fatal autoimmunity (67, 68), and humans with FOXP3 mutations develop the immunodysregulation polyendocrinopathy enteropathy X-linked (IPEX) syndrome (69). Emerging evidence demonstrates that Tregs also serve active, pleotropic roles in response to acute inflammation and tissue injury. Following experimental acute lung injury in mice, Tregs appear and proliferate in the alveolar space, suppress proinflammatory cytokine production, increase neutrophil apoptosis and efferocytosis in a TGF-β–dependent manner, and limit fibroproliferation by decreasing fibrocyte recruitment to the lung (70–74). Mechanistically, the inflammatory cytokine IL-18 and the alarmin IL-33 promote pro-repair Treg functions during experimental influenza A virus infection (75). These molecules signal via T cell receptor–independent pathways to cause release of the EGFR ligand amphiregulin from Tregs, representing a critical determinant of lung tissue protection during the acute phase of lung injury. Tregs also promote alveolar epithelial proliferation and regenerative alveologenesis via secretion of keratinocyte growth factor (76, 77). In skeletal muscle, Tregs induce repair after both acute and chronic injury in an IL-33–dependent manner (78, 79). Thus, Tregs, in addition to their canonical role in maintenance of immune homeostasis, exert critical functions in both resolution of inflammation and repair of tissue damage. Clinical protocols exist to expand and administer Tregs to manage chronic inflammatory disorders (66); these protocols could be modified to exploit Treg pro-repair function in the setting of acute and chronic tissue injury.
Macrophages in tissue repair.
Tissue-resident macrophages serve as sensors, effectors, and resolvers of inflammation and tissue damage. Historically defined as M1-like (classical/inflammatory) and M2-like (alternative/pro-repair) (80), macrophage polarization and phenotype are nuanced, tissue-specific phenomena that regulate tissue repair following injury (81). As noted above, tissue-resident macrophages coordinate the innate immune response to initial injury but can also worsen tissue damage through generation of ROS and other toxic mechanisms (82). Nevertheless, polarized pro-repair macrophages also produce antiinflammatory cytokines such as IL-10 (83–85) and growth factors that can stimulate epithelial and endothelial repair, including TGF-β (86, 87), VEGF (88, 89), and Wnt proteins (90). The complex dynamics between tissue-resident and monocyte-derived macrophages in tissue repair represent an area of active inquiry and have been reviewed elsewhere (81, 91).
Metabolic programming during tissue damage and repair
Oxygen tension, oxidative stress, and nutrient depletion serve as key modulators of immune system function, and T cells and macrophages both display differential responses under these conditions. HIF-1α activity drives a crucial glycolytic program in proinflammatory Th17 cell generation, and blocking glycolysis with 2-deoxyglucose promotes generation of Tregs over that of Th17 cells in mice (92). Induction of HIF-1α in this setting requires signaling via the mammalian target of rapamycin (mTOR), a key nutrient sensor and regulator of cellular metabolism. In Tregs, TLR signaling and Foxp3 oppose mTORC1 signaling, diminishing glycolysis and promoting their proliferation and suppressive function (93). Under normoxic conditions, HIF-1α displays a reciprocal relationship in Th17 and Tregs, with HIF-1α directly binding the master Treg transcription factor Foxp3 and targeting it for proteosomal degradation (94). HIF-1α also induces transcription of the canonical Th17 transcription factor RORγt and complexes with RORγt and the lysine acetyltransferase p300 at the Il17 promoter, establishing expression of the key proinflammatory Th17 cytokine, IL-17. IL-17 has an important role in coordinating neutrophilic inflammation that can further tissue injury (95). In contrast, under hypoxic conditions in mice, HIF-1α can induce Foxp3 expression, forming a negative-feedback loop for proinflammatory T cell populations in hypoxic inflammatory microenvironments (96).
In human tissue macrophages, hypoxia drives polarization toward a phenotype characterized by production of proinflammatory and proangiogenic cytokines and chemokines (97). Macrophages in inflammatory microenvironments require HIF-1α for glycolytic ATP production, which is necessary for multiple functions, including bacterial killing (98). Macrophage-mediated T cell suppression in hypoxic cancer microenvironments also requires HIF-1α (99), demonstrating an important feature of myeloid-lymphoid crosstalk. The HIF isoforms HIF-1α and HIF-2α also have antagonistic functions in control of macrophage metabolism, with HIF-1α driving induction of inducible nitric oxide synthase and HIF-2α driving induction of arginase-1 and antiinflammatory functional programs (100, 101). Taken together, the metabolic regulation of macrophages by hypoxia appears to be context-dependent but could be manipulated to promote a pro-repair polarized state. Ideal oxygenation goals for patients with acute hypoxemic respiratory failure and ARDS remain uncertain (102), and consideration of the effect of tissue hyper- or hypo-oxygenation on immune cell function could help define optimal targets.
Oxygen tension also regulates important changes in the output of metabolic pathways independent of HIF stabilization. Under hypoxic conditions, lactate dehydrogenase and malate dehydrogenase promiscuously use α-KG as a substrate to generate l-(S)-2-HG (17, 18). Similar to the oncometabolite d-(R)-2-HG generated by mutant isocitrate dehydrogenase 1 or 2 (IDH1/2) enzymes in human cancers, l-(S)-2-HG can inhibit α-KG–dependent enzymes, including the Jumonji family of histone lysine demethylases (JMJDs) and TET DNA demethylases (17, 21, 22). These enzymes serve as erasers of epigenetic marks that exert powerful effects on gene transcription programs in immune cell populations. Establishment and maintenance of the Treg lineage depends on epigenetic mechanisms, particularly DNA methylation (19, 103–105). Stable expression of the Foxp3 locus requires maintenance of DNA hypomethylation at critical noncoding sequences (106), and disruption of this epigenetic state leads to loss of Foxp3 expression and Treg suppressive function (107). Epigenetic state also determines Treg identity and function after injury (108, 109). Pharmacologic inhibition of DNA methyltransferase activity induces and stabilizes Foxp3 expression (110) and promotes resolution of experimental acute lung injury in a Treg-dependent manner (111). Accumulation of 2-HG in CD4+ T cells causes DNA hypermethylation of the Foxp3 locus, decreased Foxp3 transcription, and skewing toward a proinflammatory Th17 cell phenotype (23). We recently demonstrated that Treg-specific deletion of mitochondrial ETC complex III in mice causes generation of 2-HG, DNA hypermethylation, and ablation of Treg suppressive function without altering their proliferation or survival (25), illustrating the important effects of metabolic substrates in controlling T cell identity and function.
In human primary macrophages, pharmacologic inhibition of the histone 3 lysine 27 trimethyl–specific (H3K27me3-specific) demethylase subfamily (KDM6 subfamily members JMJD3 and UTX) reduces LPS-induced proinflammatory cytokine production (112). Hypoxia, upregulation of HIF via the α-KG analog and prolyl hydroxylase inhibitor dimethyloxalylglycine (DMOG), and ROS generation by 2,3-dimethoxy-1,4-naphthoquinone (DMNQ) have been associated with a reduction in murine macrophage JMJD activity and proinflammatory cytokine expression (113). Monocyte-to-macrophage differentiation depends on derepression of a phagocytic gene network that is repressed by DNA methylation in monocytes, and 2-HG can inhibit the DNA demethylation events that occur during primary human monocyte-to-macrophage differentiation (114). Thus, similarly to T cells, tissue macrophages display sensitivity to metabolic disruption via epigenetic mechanisms. We speculate that therapeutic approaches to increase intracellular α-KG levels could mitigate the detrimental effects of 2-HG generation in pro-repair cells. Investigators will need to establish methods to metabolically target specific cell types in vivo in order to translate this type of metabolic therapy to the bedside.
Lactate, a frequently measured metabolite in the clinical setting, accumulates in damaged and hypoxic tissues. Efferocytosis of apoptotic cells activates an aerobic glycolysis program that leads to release of lactate into the extracellular environment (115). Extracellular lactate inhibits motility of murine and human CD4+ and CD8+ T cells (116). Furthermore, lactate induces a switch toward Th17-polarized cells and causes loss of cytolytic function in CD8+ T cells. Lactate also shifts tumor macrophages toward an M2-like macrophage phenotype (117), which could promote fibrosis in nonmalignant clinical settings. Interestingly, fluorodeoxyglucose-PET scan activity, which measures cellular glucose uptake, can predict progression-free survival and provide risk stratification for patients with idiopathic pulmonary fibrosis (118, 119). One hypothesis is that an increased glycolytic state in fibrosis could lead to local elevations in tissue lactate, M2-like macrophage polarization, and further fibrogenesis. Lactate also inhibits histone deacetylases (HDACs) and can promote gene expression changes in an HDAC-dependent manner in vitro and in vivo (120, 121). While the specific transcriptional effects of HDAC inhibition depend on a number of factors, including the preexisting chromatin landscape, HDAC inhibition by short-chain molecules in CD4+ T cells causes skewing toward a Treg profile (122). Lactate also shifts tumor macrophages toward an antiinflammatory phenotype (123), signaling via activation of the ERK/STAT3 pathway in vitro (124). Elevated lactate levels are associated with worse outcomes among patients with sepsis (125), stimulating interest in lactate clearance as a resuscitation marker. Guidelines for the care of septic patients recommend normalization of lactate levels during sepsis treatment (126), although the benefit of lactate-guided resuscitation strategies remains unclear (127, 128).
Measurement of other circulating metabolites, including the TCA cycle metabolites succinate and itaconate, could also guide clinical care if validated in a prospective fashion. Indeed, succinate and itaconate have garnered substantial attention due to their release from inflammatory macrophages and ability to serve as an inflammatory signal (129). Murine macrophages stimulated by TLR4 signaling upregulate glycolytic metabolism, resulting in increased intracellular succinate levels (130). These elevated succinate levels cause an increase in mitochondrial ETC complex I–dependent ROS production, ultimately resulting in IL-1β production (12). Succinate released into the extracellular milieu can signal via the GPR91 (SUCNR1) receptor on macrophages in both an autocrine and a paracrine fashion (131). GPR91 receptor signaling also promotes generation of IL-1β, which further upregulates GPR91 in a feed-forward loop that amplifies inflammation. In a counterregulatory mechanism, TLR4 signaling induces immune-responsive gene 1–dependent (IRG-1–dependent) accumulation of the metabolite itaconate to limit overproduction of IL-1β (132–134). Itaconate acts as an electrophile to dampen inflammation (135). Interestingly, derivatives of itaconate can be used therapeutically in mice to limit proinflammatory cytokine production in disease models, including endotoxemia and psoriasis (136). Collectively, the tissue-damaged environment — through hypoxia, ROS accumulation, and nutrient depletion — provides important metabolic cues that program the function of responding myeloid and lymphoid cells. These mechanisms hold promise as biomarkers or potential therapeutic targets in the clinical setting.
Mitochondrial DNA stress in tissue damage and repair
Hypoxia and other features of the tissue-damaged environment, including neutrophil recruitment and activation, cause increased generation of ROS, and mitochondria are the major intracellular site of ROS generation (137–140). ROS exert pleotropic signaling functions and can damage mitochondrial DNA (mtDNA), causing oxidative mtDNA stress (141). ROS-mediated mtDNA damage tends to be repaired more slowly than in the nucleus, positioning mtDNA as a key sensor and mediator of ROS species. In murine macrophages, danger signals such as extracellular ATP lead to mitochondrial dysfunction and mtDNA stress and oxidation (142). Under mtDNA stress, aberrant mtDNA packaging leads to escape of mtDNA into the cytosol, a process that can be inhibited by the autophagy proteins LC3B and beclin-1 (35). The consequences of releasing mtDNA into the cytoplasm and circulation discussed below underscore our theme of mitochondrial biomolecules as signals that determine cell function.
After release into the cytoplasm, mtDNA interacts with double-stranded DNA sensors including cGAS (29). Upon binding to DNA, cGAS generates the second messenger cGAMP and AMP to activate STING, which is present on the endoplasmic reticulum. Downstream signaling from STING via TBK1 engages inflammatory cellular functions via activation of IRF3/7 responses, promoting expression of antiviral transcripts (type I interferons) that suppress viral replication and promote resistance to viral injury (31). The apoptotic caspases 3 and 7 block induction of the type I interferon response in mice (34, 143), placing mitochondria in the center of a system that determines whether a damaged cell dies in a proinflammatory or a noninflammatory (apoptotic) manner. Oxidized mtDNA derived from damaged, ROS-producing mitochondria also activates the NLRP3 inflammasome (38), leading to secretion of IL-1β from murine macrophages (142). In addition, the NLRP3 inflammasome triggers IL-18 release (144), which serves as a major signal promoting the pro-repair function of Tregs in mice (75). These observations provide mechanistic links between tissue damage, oxidative stress, ongoing injurious inflammation, and repair.
Mitochondrial DNA is also released into the circulation following trauma and induces a systemic inflammatory response (28). Mitochondrial DNA activates TLR9 as a result of similarities with bacterial DNA (26) and, in contrast with nuclear DNA, stimulates inflammatory reactions when introduced into mouse joints or the circulation (27, 28). TLR9 activation of gut dendritic cells suppresses Treg induction from CD4+Foxp3– cells, leading to inflammation and disruption of intestinal homeostasis (145). In the gut, commensal bacteria represent the main source of TLR9 ligands, although mtDNA could serve a similar function in the gut and other tissues. Human Tregs themselves also express high levels of TLR9, particularly the subset producing the antiinflammatory cytokine IL-10 (146), suggesting that Tregs are poised to sense tissue damage in part via mtDNA release. Because of its unique sequence, mtDNA could serve as a circulating biomarker of tissue damage and systemic inflammation (147).
Parkin (an E3 ubiquitin ligase) and PINK1 (a ubiquitin kinase), enzymes linked to familial early-onset parkinsonism, mitigate inflammation generated via the cGAS/STING pathway and mitochondrial antigen-driven, MHC class I–dependent adaptive immune responses (148). PINK1 is required for normal Treg suppressive function even in the setting of normal Foxp3 levels (149). In CD4+ T cells, PINK1 deficiency impairs phosphorylation of AKT, which is a critical step in expression of the canonical Treg surface molecule CD25 (the IL-2 receptor α subunit). Parkin and PINK1 function to remove damaged mitochondria from cells in a modified form of autophagy known as mitophagy, which serves to maintain cellular metabolic homeostasis (150). Parkin and PINK1 also assist in repairing damaged mitochondria by facilitating localized translation of nuclear-encoded respiratory chain complexes at the mitochondrial outer membrane, promoting oxidative phosphorylation in failing mitochondria (151). In the lung, TGF-β1 induces mitochondrial ROS in epithelial cells, which promotes PINK1 stabilization (152, 153); there is evidence that PINK1 deficiency impairs mitochondrial homeostasis and contributes to pulmonary fibrosis (154). Additionally, mitochondrial ROS induced by TGF-β1 augment Smad-mediated transcription of TGF-β target genes, including NOX4 (155). NOX4 is itself a source of intracellular ROS, creating a positive-feedback loop whereby ROS maintain and amplify expression of profibrotic genes. The antifibrotic medication pirfenidone attenuates TGF-β signaling (156, 157), and may therefore limit mtDNA damage in fibrotic disorders such as idiopathic pulmonary fibrosis. Taken together, mtDNA serves as a critical hub that integrates hypoxic and oxidative signals and regulates the function of pro-repair cell types.
Redox balance in tissue injury and repair
Release of ROS and oxidant mediators as well as nutrient depletion in the tissue-damaged environment leads to disruptions in redox balance, including alterations in NAD+ levels and the NAD+/NADH ratio. In this section we highlight examples of oxidant mediators and NAD+ controlling pro-repair cell function, consistent with our overall theme of metabolites as causal regulators of cell function. Aging, a state of decreased resilience to stress and impaired capacity for repair, is associated with a decline in NAD+ levels (158). Mitochondrial dysfunction accompanies advancing age, although many of the precise mechanisms driving age-related mitochondrial dysfunction remain unknown. As noted above, the nuclear genome encodes most of the oxidative phosphorylation system, necessitating nuclear-mitochondrial communication to maintain mitochondrial homeostasis. Aging is associated with specific loss of mitochondrial genome-encoded subunits of the oxidative phosphorylation system (159). With this decline in oxidative phosphorylation, a decrease in nuclear NAD+ causes reduced activity of the lysine deacetylase Sirt1 in the nucleus, which leads to decreased levels of VHL protein and accumulation of HIF-1α. The result is a state of cellular pseudohypoxia and Warburg-like metabolism: a shift away from oxidative phosphorylation toward glycolysis even in the presence of normoxia (160). A decreased NAD+/NADH ratio also increases production of 2-HG (161, 162), which can modulate the function of Tregs and tissue macrophages via mechanisms discussed above.
Sirt1 and the other members of the sirtuin family of lysine deacetylases, which are NAD+-dependent enzymes and thus activated in nutrient-limiting conditions (163), serve other functions beyond regulation of the VHL/HIF-1α axis. In Tregs, Sirt1 colocalizes with Foxp3 in the nucleus and deacetylates Foxp3 (164). Deacetylated Foxp3 is rapidly degraded by cytoplasmic proteasomes, contributing to Treg cellular dysfunction (165). In an opposing role, Sirt1 regulates Notch signaling in Tregs, deacetylating the Notch1 intracellular domain and retaining it in the cytosol to promote antiapoptotic signaling and Treg survival (166). Indeed, Treg-specific loss of Sirt1 leads to antigen-induced effector T cell proliferation, inflammation, and tissue damage. In murine macrophages, loss of SIRT1 causes activation of the JNK and IKK inflammatory pathways and increased LPS-stimulated secretion of inflammatory cytokines (167), similar to the effect of age-related decreases in de novo NAD+ synthesis (168). Accordingly, there has been interest in dietary supplementation with nicotinamide (NAM), nicotinamide mononucleotide (NMN), and nicotinamide riboside (NR), which boost NAD+ levels and restore sirtuin activity to potentially treat age-related inflammatory disorders (169).
The transcription factor Nrf2 coordinates a cellular defense program in the setting of oxidative damage (170). In the steady state, the Cul3-Keap1 ubiquitin E3 ligase complex ubiquitinates Nrf2, targeting it for rapid proteosomal degradation. Oxidative conditions modify reactive cysteine residues on Keap1, decreasing its E3 ligase activity and allowing Nrf2 to activate transcription of cytoprotective genes, including those involved with glutathione synthesis. Systemic activation of Nrf2 via knockdown of Keap1 mitigates tissue inflammation in scurfy mice (171), which lack Tregs and spontaneously develop fatal inflammation due to mutation of the Foxp3 gene (172). Nrf2 expression within Tregs promotes their tissue-protective function following acute kidney injury (173), and Treg-specific induction of Nrf2 via CRISPR-Cas9–mediated deletion of Keap1 holds therapeutic potential for treatment of autoimmune and inflammatory disorders (174). Macrophages also respond to oxidative environments with increased Nrf2 stabilization, which suppresses their inflammatory response by blocking the transcription of proinflammatory cytokines (175). Thus, the Keap1-Nrf2 system represents an important link between oxidative stress and pro-repair cell functions in damaged tissues.
Glutathione (GSH) is the major molecular defense against oxidation, participating in the reduction of disulfides and other oxidized species and protecting against the deleterious effects of ROS (176). Glutamate cysteine ligase (GCL) serves as the rate-limiting enzymatic step in GSH synthesis, and conditional deletion of the GCL-encoding gene Gclc in T cells prevents their ability to execute inflammatory responses (177). Tregs interact with dendritic cells via the CTLA4-CD80/CD86 synapse, leading to signaling events that inhibit GSH synthesis in the dendritic cell as well as local effector T cells (178). GSH contributes to the induction of nitric oxide synthase in IFN-γ– and LPS-stimulated macrophages, which has implications for control of intra- and extracellular pathogens (179). Antioxidants have received significant interest in promoting clinical pro-resolution and pro-repair immune responses (180), although clinical trials of antioxidant compounds have been almost universally negative, with some antioxidants, such as vitamin E and β-carotene, associated with increased mortality (181). In summary, the redox state of pro-repair cells represents an important governor of their function as they respond to tissue injury.
Conclusion and future directions
The tissue-damaged environment generates numerous immunometabolic inputs that influence the function of pro-repair cells. Hypoxia, oxidative stress, and nutrient depletion signal via complex networks that involve metabolic programming, damage to mtDNA, and redox balance among other pathways. As the initial sensor of environmental cues, metabolic pathways and their constituents can serve as both the sentinels of tissue damage and the effectors of processes that lead to either persistent damaging inflammation or resolution and repair. A detailed mechanistic understanding of these pathways in specific pro-repair cell types could lead to biomarker discovery and pharmacotherapies that leverage immunometabolism to promote tissue repair following injury and mitigate the diminished repair capacity associated with aging.
Acknowledgments
BDS was supported by NIH K08HL128867 and U19AI135964. NSC was supported by NIH R35CA197532, P01AG049665, and P01HL071643.
Version 1. 05/13/2019
Electronic publication
Version 2. 07/01/2019
Print issue publication
Footnotes
Conflict of interest: BDS has a pending US patent entitled “Compositions and Methods to Accelerate Resolution of Acute Lung Inflammation” (patent application no. 15/542,380).
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(7):2597–2607.https://doi.org/10.1172/JCI124613.
Contributor Information
Benjamin D. Singer, Email: benjamin-singer@northwestern.edu.
Navdeep S. Chandel, Email: nav@northwestern.edu.
References
- 1.Phan AT, Goldrath AW, Glass CK. Metabolic and epigenetic coordination of T cell and macrophage immunity. Immunity. 2017;46(5):714–729. doi: 10.1016/j.immuni.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42(3):406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olenchock BA, Rathmell JC, Vander Heiden MG. Biochemical underpinnings of immune cell metabolic phenotypes. Immunity. 2017;46(5):703–713. doi: 10.1016/j.immuni.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West AP, Shadel GS. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat Rev Immunol. 2017;17(6):363–375. doi: 10.1038/nri.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehta MM, Weinberg SE, Chandel NS. Mitochondrial control of immunity: beyond ATP. Nat Rev Immunol. 2017;17(10):608–620. doi: 10.1038/nri.2017.66. [DOI] [PubMed] [Google Scholar]
- 6.Ho PC, et al. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell. 2015;162(6):1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang CH, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153(6):1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michalek RD, et al. Cutting edge: Distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186(6):3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macintyre AN, et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014;20(1):61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freemerman AJ, et al. Metabolic reprogramming of macrophages: glucose transporter 1 (GLUT1)-mediated glucose metabolism drives a proinflammatory phenotype. J Biol Chem. 2014;289(11):7884–7896. doi: 10.1074/jbc.M113.522037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang A, et al. Opposing effects of fasting metabolism on tissue tolerance in bacterial and viral inflammation. Cell. 2016;166(6):1512–1525.e12. doi: 10.1016/j.cell.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tannahill GM, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496(7444):238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ip WKE, Hoshi N, Shouval DS, Snapper S, Medzhitov R. Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science. 2017;356(6337):513–519. doi: 10.1126/science.aal3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson MO, et al. Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell. 2018;175(7):1780–1795.e19. doi: 10.1016/j.cell.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng M, Yin N, Chhangawala S, Xu K, Leslie CS, Li MO. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science. 2016;354(6311):481–484. doi: 10.1126/science.aaf6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bantug GR, et al. Mitochondria-endoplasmic reticulum contact sites function as immunometabolic hubs that orchestrate the rapid recall response of memory CD8(+) T cells. Immunity. 2018;48(3):542–555 e546. doi: 10.1016/j.immuni.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Intlekofer AM, et al. Hypoxia induces production of L-2-hydroxyglutarate. Cell Metab. 2015;22(2):304–311. doi: 10.1016/j.cmet.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oldham WM, Clish CB, Yang Y, Loscalzo J. Hypoxia-mediated increases in L-2-hydroxyglutarate coordinate the metabolic response to reductive stress. Cell Metab. 2015;22(2):291–303. doi: 10.1016/j.cmet.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morales-Nebreda L, McLafferty FS, Singer BD. DNA methylation as a transcriptional regulator of the immune system. Transl Res. 2019;204:1–18. doi: 10.1016/j.trsl.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, et al. TET2 coactivates gene expression through demethylation of enhancers. Sci Adv. 2018;4(11):eaau6986. doi: 10.1126/sciadv.aau6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao M, et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26(12):1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu T, et al. Metabolic control of TH17 and induced Treg cell balance by an epigenetic mechanism. Nature. 2017;548(7666):228–233. doi: 10.1038/nature23475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyrakis PA, et al. S-2-hydroxyglutarate regulates CD8(+) T-lymphocyte fate. Nature. 2016;540(7632):236–241. doi: 10.1038/nature20165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinberg SE, et al. Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature. 2019;565(7740):495–499. doi: 10.1038/s41586-018-0846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.West AP, Shadel GS, Ghosh S. Mitochondria in innate immune responses. Nat Rev Immunol. 2011;11(6):389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins LV, Hajizadeh S, Holme E, Jonsson IM, Tarkowski A. Endogenously oxidized mitochondrial DNA induces in vivo and in vitro inflammatory responses. J Leukoc Biol. 2004;75(6):995–1000. doi: 10.1189/jlb.0703328. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Q, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464(7285):104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong Z, et al. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature. 2018;560(7717):198–203. doi: 10.1038/s41586-018-0372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.West AP, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520(7548):553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.West AP, et al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472(7344):476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sena LA, et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38(2):225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rongvaux A, et al. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159(7):1563–1577. doi: 10.1016/j.cell.2014.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakahira K, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12(3):222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mills EL, et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell. 2016;167(2):457–470.e13. doi: 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandel NS, Schumacker PT, Arch RH. Reactive oxygen species are downstream products of TRAF-mediated signal transduction. J Biol Chem. 2001;276(46):42728–42736. doi: 10.1074/jbc.M103074200. [DOI] [PubMed] [Google Scholar]
- 38.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 39.Eltzschig HK, Carmeliet P. Hypoxia and inflammation. N Engl J Med. 2011;364(7):656–665. doi: 10.1056/NEJMra0910283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kruger B, et al. Donor toll-like receptor 4 contributes to ischemia and reperfusion injury following human kidney transplantation. Proc Natl Acad Sci U S A. 2009;106(9):3390–3395. doi: 10.1073/pnas.0810169106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thiel M, et al. Oxygenation inhibits the physiological tissue-protecting mechanism and thereby exacerbates acute inflammatory lung injury. PLoS Biol. 2005;3(6):e174. doi: 10.1371/journal.pbio.0030174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walter JM, Corbridge TC, Singer BD. Invasive mechanical ventilation. South Med J. 2018;111(12):746–753. doi: 10.14423/SMJ.0000000000000905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurihara C, et al. Extracorporeal membrane oxygenation can successfully support patients with severe acute respiratory distress syndrome in lieu of mechanical ventilation. Crit Care Med. 2018;46(11):e1070–e1073. doi: 10.1097/CCM.0000000000003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karhausen J, Furuta GT, Tomaszewski JE, Johnson RS, Colgan SP, Haase VH. Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. J Clin Invest. 2004;114(8):1098–1106. doi: 10.1172/JCI21086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Semenza GL. Life with oxygen. Science. 2007;318(5847):62–64. doi: 10.1126/science.1147949. [DOI] [PubMed] [Google Scholar]
- 46.Blouin CC, Page EL, Soucy GM, Richard DE. Hypoxic gene activation by lipopolysaccharide in macrophages: implication of hypoxia-inducible factor 1α. Blood. 2004;103(3):1124–1130. doi: 10.1182/blood-2003-07-2427. [DOI] [PubMed] [Google Scholar]
- 47.Cash TP, Pan Y, Simon MC. Reactive oxygen species and cellular oxygen sensing. Free Radic Biol Med. 2007;43(9):1219–1225. doi: 10.1016/j.freeradbiomed.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knowles HJ, Mole DR, Ratcliffe PJ, Harris AL. Normoxic stabilization of hypoxia-inducible factor-1α by modulation of the labile iron pool in differentiating U937 macrophages: effect of natural resistance-associated macrophage protein 1. Cancer Res. 2006;66(5):2600–2607. doi: 10.1158/0008-5472.CAN-05-2351. [DOI] [PubMed] [Google Scholar]
- 49.Campbell EL, et al. Transmigrating neutrophils shape the mucosal microenvironment through localized oxygen depletion to influence resolution of inflammation. Immunity. 2014;40(1):66–77. doi: 10.1016/j.immuni.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ajuebor MN, Das AM, Virag L, Flower RJ, Szabo C, Perretti M. Role of resident peritoneal macrophages and mast cells in chemokine production and neutrophil migration in acute inflammation: evidence for an inhibitory loop involving endogenous IL-10. J Immunol. 1999;162(3):1685–1691. [PubMed] [Google Scholar]
- 51.Galli SJ, Borregaard N, Wynn TA. Phenotypic and functional plasticity of cells of innate immunity: macrophages, mast cells and neutrophils. Nat Immunol. 2011;12(11):1035–1044. doi: 10.1038/ni.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34(5):637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 53.Massena S, et al. A chemotactic gradient sequestered on endothelial heparan sulfate induces directional intraluminal crawling of neutrophils. Blood. 2010;116(11):1924–1931. doi: 10.1182/blood-2010-01-266072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stark K, et al. Capillary and arteriolar pericytes attract innate leukocytes exiting through venules and ‘instruct’ them with pattern-recognition and motility programs. Nat Immunol. 2013;14(1):41–51. doi: 10.1038/ni.2477. [DOI] [PubMed] [Google Scholar]
- 55.Scapini P, et al. Proinflammatory mediators elicit secretion of the intracellular B-lymphocyte stimulator pool (BLyS) that is stored in activated neutrophils: Implications for inflammatory diseases. Blood. 2005;105(2):830–837. doi: 10.1182/blood-2004-02-0564. [DOI] [PubMed] [Google Scholar]
- 56.Files DC, et al. Therapeutic exercise attenuates neutrophilic lung injury and skeletal muscle wasting. Sci Transl Med. 2015;7(278):278ra232. doi: 10.1126/scitranslmed.3010283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 58.Pham CT. Neutrophil serine proteases: specific regulators of inflammation. Nat Rev Immunol. 2006;6(7):541–550. doi: 10.1038/nri1841. [DOI] [PubMed] [Google Scholar]
- 59.Sokolovska A, et al. Activation of caspase-1 by the NLRP3 inflammasome regulates the NADPH oxidase NOX2 to control phagosome function. Nat Immunol. 2013;14(6):543–553. doi: 10.1038/ni.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rada BK, Geiszt M, Kaldi K, Timar C, Ligeti E. Dual role of phagocytic NADPH oxidase in bacterial killing. Blood. 2004;104(9):2947–2953. doi: 10.1182/blood-2004-03-1005. [DOI] [PubMed] [Google Scholar]
- 61.Hecker L, et al. NADPH oxidase-4 mediates myofibroblast activation and fibrogenic responses to lung injury. Nat Med. 2009;15(9):1077–1081. doi: 10.1038/nm.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. 2013;13(3):159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 63.Christoffersson G, et al. VEGF-A recruits a proangiogenic MMP-9-delivering neutrophil subset that induces angiogenesis in transplanted hypoxic tissue. Blood. 2012;120(23):4653–4662. doi: 10.1182/blood-2012-04-421040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133(5):775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 65.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30(1):531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singer BD, King LS, D’Alessio FR. Regulatory T cells as immunotherapy. Front Immunol. 2014;5:46. doi: 10.3389/fimmu.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4(4):330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 68.Brunkow ME, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27(1):68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 69.Wildin RS, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27(1):18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- 70.D’Alessio FR, et al. CD4+CD25+Foxp3+ Tregs resolve experimental lung injury in mice and are present in humans with acute lung injury. J Clin Invest. 2009;119(10):2898–2913. doi: 10.1172/JCI36498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garibaldi BT, et al. Regulatory T cells reduce acute lung injury fibroproliferation by decreasing fibrocyte recruitment. Am J Respir Cell Mol Biol. 2013;48(1):35–43. doi: 10.1165/rcmb.2012-0198OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Proto JD, et al. Regulatory T cells promote macrophage efferocytosis during inflammation resolution. Immunity. 2018;49(4):666–677.e6. doi: 10.1016/j.immuni.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.D’Alessio FR, et al. Enhanced resolution of experimental ARDS through IL-4-mediated lung macrophage reprogramming. Am J Physiol Lung Cell Mol Physiol. 2016;310(8):L733–746. doi: 10.1152/ajplung.00419.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aggarwal NR, et al. Immunological priming requires regulatory T cells and IL-10-producing macrophages to accelerate resolution from severe lung inflammation. J Immunol. 2014;192(9):4453–4464. doi: 10.4049/jimmunol.1400146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arpaia N, et al. A distinct function of regulatory T cells in tissue protection. Cell. 2015;162(5):1078–1089. doi: 10.1016/j.cell.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mock JR, et al. Foxp3+ regulatory T cells promote lung epithelial proliferation. Mucosal Immunol. 2014;7(6):1440–1451. doi: 10.1038/mi.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dial CF, Tune MK, Doerschuk CM, Mock JR. Foxp3(+) regulatory T cell expression of keratinocyte growth factor enhances lung epithelial proliferation. Am J Respir Cell Mol Biol. 2017;57(2):162–173. doi: 10.1165/rcmb.2017-0019OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burzyn D, et al. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155(6):1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kuswanto W, et al. Poor repair of skeletal muscle in aging mice reflects a defect in local, interleukin-33-dependent accumulation of regulatory T cells. Immunity. 2016;44(2):355–367. doi: 10.1016/j.immuni.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122(3):787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vannella KM, Wynn TA. Mechanisms of organ injury and repair by macrophages. Annu Rev Physiol. 2017;79(1):593–617. doi: 10.1146/annurev-physiol-022516-034356. [DOI] [PubMed] [Google Scholar]
- 82.Larson-Casey JL, Deshane JS, Ryan AJ, Thannickal VJ, Carter AB. Macrophage Akt1 kinase-mediated mitophagy modulates apoptosis resistance and pulmonary fibrosis. Immunity. 2016;44(3):582–596. doi: 10.1016/j.immuni.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zigmond E, et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity. 2014;40(5):720–733. doi: 10.1016/j.immuni.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 84.Shouval DS, et al. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity. 2014;40(5):706–719. doi: 10.1016/j.immuni.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boehler RM, et al. Lentivirus delivery of IL-10 to promote and sustain macrophage polarization towards an anti-inflammatory phenotype. Biotechnol Bioeng. 2014;111(6):1210–1221. doi: 10.1002/bit.25175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khalil N, Bereznay O, Sporn M, Greenberg AH. Macrophage production of transforming growth factor beta and fibroblast collagen synthesis in chronic pulmonary inflammation. J Exp Med. 1989;170(3):727–737. doi: 10.1084/jem.170.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Harel-Adar T, Ben Mordechai T, Amsalem Y, Feinberg MS, Leor J, Cohen S. Modulation of cardiac macrophages by phosphatidylserine-presenting liposomes improves infarct repair. Proc Natl Acad Sci U S A. 2011;108(5):1827–1832. doi: 10.1073/pnas.1015623108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.D’Alessio S, et al. VEGF-C-dependent stimulation of lymphatic function ameliorates experimental inflammatory bowel disease. J Clin Invest. 2014;124(9):3863–3878. doi: 10.1172/JCI72189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Berse B, Brown LF, Van de Water L, Dvorak HF, Senger DR. Vascular permeability factor (vascular endothelial growth factor) gene is expressed differentially in normal tissues, macrophages, and tumors. Mol Biol Cell. 1992;3(2):211–220. doi: 10.1091/mbc.3.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boulter L, et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med. 2012;18(4):572–579. doi: 10.1038/nm.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wynn TA, Vannella KM. Macrophages in tissue repair, regeneration, and fibrosis. Immunity. 2016;44(3):450–462. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shi LZ, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208(7):1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gerriets VA, et al. Foxp3 and toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat Immunol. 2016;17(12):1459–1466. doi: 10.1038/ni.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dang EV, et al. Control of T(h)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146(5):772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ferretti S, Bonneau O, Dubois GR, Jones CE, Trifilieff A. IL-17, produced by lymphocytes and neutrophils, is necessary for lipopolysaccharide-induced airway neutrophilia: IL-15 as a possible trigger. J Immunol. 2003;170(4):2106–2112. doi: 10.4049/jimmunol.170.4.2106. [DOI] [PubMed] [Google Scholar]
- 96.Clambey ET, et al. Hypoxia-inducible factor-1 alpha-dependent induction of FoxP3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proc Natl Acad Sci U S A. 2012;109(41):E2784–2793. doi: 10.1073/pnas.1202366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Varesio L, et al. Hypoxia reprograms human macrophages towards a proinflammatory direction. J Immunol. 2016;196(1 suppl):201.2 [Google Scholar]
- 98.Cramer T, et al. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112(5):645–657. doi: 10.1016/S0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Doedens AL, et al. Macrophage expression of hypoxia-inducible factor-1 alpha suppresses T-cell function and promotes tumor progression. Cancer Res. 2010;70(19):7465–7475. doi: 10.1158/0008-5472.CAN-10-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takeda N, et al. Differential activation and antagonistic function of HIF-α isoforms in macrophages are essential for NO homeostasis. Genes Dev. 2010;24(5):491–501. doi: 10.1101/gad.1881410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Imtiyaz HZ, et al. Hypoxia-inducible factor 2α regulates macrophage function in mouse models of acute and tumor inflammation. J Clin Invest. 2010;120(8):2699–2714. doi: 10.1172/JCI39506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Aggarwal NR, et al. Oxygen exposure resulting in arterial oxygen tensions above the protocol goal was associated with worse clinical outcomes in acute respiratory distress syndrome. Crit Care Med. 2018;46(4):517–524. doi: 10.1097/CCM.0000000000002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ohkura N, et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37(5):785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 104.Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity. 2013;38(3):414–423. doi: 10.1016/j.immuni.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 105.Delacher M, et al. Genome-wide DNA-methylation landscape defines specialization of regulatory T cells in tissues. Nat Immunol. 2017;18(10):1160–1172. doi: 10.1038/ni.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lal G, et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182(1):259–273. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yang R, et al. Hydrogen sulfide promotes Tet1- and Tet2-mediated Foxp3 demethylation to drive regulatory T cell differentiation and maintain immune homeostasis. Immunity. 2015;43(2):251–263. doi: 10.1016/j.immuni.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McGrath-Morrow SA, et al. DNA methylation regulates the neonatal CD4(+) T-cell response to pneumonia in mice. J Biol Chem. 2018;293(30):11772–11783. doi: 10.1074/jbc.RA118.003589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Walter JM, Helmin KA, Abdala-Valencia H, Wunderink RG, Singer BD. Multidimensional assessment of alveolar T cells in critically ill patients. JCI Insight. 2018;3(17):e123287. doi: 10.1172/jci.insight.123287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Polansky JK, et al. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38(6):1654–1663. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- 111.Singer BD, et al. Regulatory T cell DNA methyltransferase inhibition accelerates resolution of lung inflammation. Am J Respir Cell Mol Biol. 2015;52(5):641–652. doi: 10.1165/rcmb.2014-0327OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kruidenier L, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488(7411):404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tausendschon M, Dehne N, Brune B. Hypoxia causes epigenetic gene regulation in macrophages by attenuating Jumonji histone demethylase activity. Cytokine. 2011;53(2):256–262. doi: 10.1016/j.cyto.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 114.Wallner S, et al. Epigenetic dynamics of monocyte-to-macrophage differentiation. Epigenetics Chromatin. 2016;9:33. doi: 10.1186/s13072-016-0079-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Morioka S, et al. Efferocytosis induces a novel SLC program to promote glucose uptake and lactate release. Nature. 2018;563(7733):714–718. doi: 10.1038/s41586-018-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Haas R, et al. Lactate regulates metabolic and pro-inflammatory circuits in control of T cell migration and effector functions. PLoS Biol. 2015;13(7):e1002202. doi: 10.1371/journal.pbio.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Colegio OR, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513(7519):559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Justet A, et al. [(18)F]FDG PET/CT predicts progression-free survival in patients with idiopathic pulmonary fibrosis. Respir Res. 2017;18(1):74. doi: 10.1186/s12931-017-0556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Win T, et al. Pulmonary (18)F-FDG uptake helps refine current risk stratification in idiopathic pulmonary fibrosis (IPF) Eur J Nucl Med Mol Imaging. 2018;45(5):806–815. doi: 10.1007/s00259-017-3917-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Latham T, et al. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res. 2012;40(11):4794–4803. doi: 10.1093/nar/gks066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brand A, et al. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab. 2016;24(5):657–671. doi: 10.1016/j.cmet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 122.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ohashi T, et al. M2-like macrophage polarization in high lactic acid-producing head and neck cancer. Cancer Sci. 2017;108(6):1128–1134. doi: 10.1111/cas.13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mu X, et al. Tumor-derived lactate induces M2 macrophage polarization via the activation of the ERK/STAT3 signaling pathway in breast cancer. Cell Cycle. 2018;17(4):428–438. doi: 10.1080/15384101.2018.1444305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Casserly B, et al. Lactate measurements in sepsis-induced tissue hypoperfusion: results from the surviving sepsis campaign database. Crit Care Med. 2015;43(3):567–573. doi: 10.1097/CCM.0000000000000742. [DOI] [PubMed] [Google Scholar]
- 126.Rhodes A, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Intensive Care Med. 2017;43(3):304–377. doi: 10.1007/s00134-017-4683-6. [DOI] [PubMed] [Google Scholar]
- 127.Simpson SQ, Gaines M, Hussein Y, Badgett RG. Early goal-directed therapy for severe sepsis and septic shock: a living systematic review. J Crit Care. 2016;36:43–48. doi: 10.1016/j.jcrc.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 128.Gu WJ, Zhang Z, Bakker J. Early lactate clearance-guided therapy in patients with sepsis: a meta-analysis with trial sequential analysis of randomized controlled trials. Intensive Care Med. 2015;41(10):1862–1863. doi: 10.1007/s00134-015-3955-2. [DOI] [PubMed] [Google Scholar]
- 129.Littlewood-Evans A, et al. GPR91 senses extracellular succinate released from inflammatory macrophages and exacerbates rheumatoid arthritis. J Exp Med. 2016;213(9):1655–1662. doi: 10.1084/jem.20160061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jha AK, et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 2015;42(3):419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 131.He W, et al. Citric acid cycle intermediates as ligands for orphan G-protein-coupled receptors. Nature. 2004;429(6988):188–193. doi: 10.1038/nature02488. [DOI] [PubMed] [Google Scholar]
- 132.Cordes T, et al. Immunoresponsive gene 1 and itaconate inhibit succinate dehydrogenase to modulate intracellular succinate levels. J Biol Chem. 2016;291(27):14274–14284. doi: 10.1074/jbc.M115.685792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Michelucci A, et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc Natl Acad Sci U S A. 2013;110(19):7820–7825. doi: 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lampropoulou V, et al. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 2016;24(1):158–166. doi: 10.1016/j.cmet.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bambouskova M, et al. Electrophilic properties of itaconate and derivatives regulate the IκBζ-ATF3 inflammatory axis. Nature. 2018;556(7702):501–504. doi: 10.1038/s41586-018-0052-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mills EL, et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556(7699):113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mansfield KD, Simon MC, Keith B. Hypoxic reduction in cellular glutathione levels requires mitochondrial reactive oxygen species. J Appl Physiol (1985) 2004;97(4):1358–1366. doi: 10.1152/japplphysiol.00449.2004. [DOI] [PubMed] [Google Scholar]
- 138.Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci U S A. 1998;95(20):11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mansfield KD, et al. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-alpha activation. Cell Metab. 2005;1(6):393–399. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Guzy RD, et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1(6):401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 141.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94(2):514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shimada K, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36(3):401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.White MJ, et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159(7):1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ketelut-Carneiro N, et al. IL-18 triggered by the Nlrp3 inflammasome induces host innate resistance in a pulmonary model of fungal infection. J Immunol. 2015;194(9):4507–4517. doi: 10.4049/jimmunol.1402321. [DOI] [PubMed] [Google Scholar]
- 145.Hall JA, et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29(4):637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Urry Z, et al. Ligation of TLR9 induced on human IL-10-secreting Tregs by 1α,25-dihydroxyvitamin D3 abrogates regulatory function. J Clin Invest. 2009;119(2):387–398. doi: 10.1172/JCI32354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kilbaugh TJ, et al. Peripheral blood mitochondrial DNA as a biomarker of cerebral mitochondrial dysfunction following traumatic brain injury in a porcine model. PLoS One. 2015;10(6):e0130927. doi: 10.1371/journal.pone.0130927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sliter DA, et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature. 2018;561(7722):258–262. doi: 10.1038/s41586-018-0448-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 149.Ellis GI, Zhi L, Akundi R, Bueler H, Marti F. Mitochondrial and cytosolic roles of PINK1 shape induced regulatory T-cell development and function. Eur J Immunol. 2013;43(12):3355–3360. doi: 10.1002/eji.201343571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85(2):257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gehrke S, et al. PINK1 and Parkin control localized translation of respiratory chain component mRNAs on mitochondria outer membrane. Cell Metab. 2015;21(1):95–108. doi: 10.1016/j.cmet.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Patel AS, et al. Epithelial cell mitochondrial dysfunction and PINK1 are induced by transforming growth factor-β1 in pulmonary fibrosis. PLoS One. 2015;10(3):e0121246. doi: 10.1371/journal.pone.0121246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Singer BD, et al. Flow-cytometric method for simultaneous analysis of mouse lung epithelial, endothelial, and hematopoietic lineage cells. Am J Physiol Lung Cell Mol Physiol. 2016;310(9):L796–L801. doi: 10.1152/ajplung.00334.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bueno M, et al. PINK1 deficiency impairs mitochondrial homeostasis and promotes lung fibrosis. J Clin Invest. 2015;125(2):521–538. doi: 10.1172/JCI74942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jain M, et al. Mitochondrial reactive oxygen species regulate transforming growth factor-β signaling. J Biol Chem. 2013;288(2):770–777. doi: 10.1074/jbc.M112.431973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Conte E, Gili E, Fagone E, Fruciano M, Iemmolo M, Vancheri C. Effect of pirfenidone on proliferation, TGF-β-induced myofibroblast differentiation and fibrogenic activity of primary human lung fibroblasts. Eur J Pharm Sci. 2014;58:13–19. doi: 10.1016/j.ejps.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 157.King TE, et al. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med. 2014;370(22):2083–2092. doi: 10.1056/NEJMoa1402582. [DOI] [PubMed] [Google Scholar]
- 158.Khan SS, Singer BD, Vaughan DE. Molecular and physiological manifestations and measurement of aging in humans. Aging Cell. 2017;16(4):624–633. doi: 10.1111/acel.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gomes AP, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155(7):1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 161.Mullen AR, et al. Oxidation of α-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep. 2014;7(5):1679–1690. doi: 10.1016/j.celrep.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tateishi K, et al. Extreme vulnerability of IDH1 mutant cancers to NAD+ depletion. Cancer Cell. 2015;28(6):773–784. doi: 10.1016/j.ccell.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306(5704):2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 164.van Loosdregt J, Brunen D, Fleskens V, Pals CE, Lam EW, Coffer PJ. Rapid temporal control of Foxp3 protein degradation by sirtuin-1. PLoS One. 2011;6(4):e19047. doi: 10.1371/journal.pone.0019047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.van Loosdregt J, et al. Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood. 2010;115(5):965–974. doi: 10.1182/blood-2009-02-207118. [DOI] [PubMed] [Google Scholar]
- 166.Marcel N, Perumalsamy LR, Shukla SK, Sarin A. The lysine deacetylase Sirtuin 1 modulates the localization and function of the Notch1 receptor in regulatory T cells. Sci Signal. 2017;10(473):eaah4679. doi: 10.1126/scisignal.aah4679. [DOI] [PubMed] [Google Scholar]
- 167.Yoshizaki T, et al. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am J Physiol Endocrinol Metab. 2010;298(3):E419–E428. doi: 10.1152/ajpendo.00417.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Minhas PS, et al. Macrophage de novo NAD(+) synthesis specifies immune function in aging and inflammation. Nat Immunol. 2019;20(1):50–63. doi: 10.1038/s41590-018-0255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Katsyuba E, Auwerx J. Modulating NAD(+) metabolism, from bench to bedside. EMBO J. 2017;36(18):2670–2683. doi: 10.15252/embj.201797135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16(2):123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 171.Suzuki T, et al. Systemic activation of NRF2 alleviates lethal autoimmune inflammation in scurfy mice. Mol Cell Biol. 2017;37(15):e00063-17. doi: 10.1128/MCB.00063-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299(5609):1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 173.Noel S, et al. T lymphocyte-specific activation of Nrf2 protects from AKI. J Am Soc Nephrol. 2015;26(12):2989–3000. doi: 10.1681/ASN.2014100978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Noel S, Lee SA, Sadasivam M, Hamad ARA, Rabb H. KEAP1 editing using CRISPR/Cas9 for therapeutic NRF2 activation in primary human T lymphocytes. J Immunol. 2018;200(5):1929–1936. doi: 10.4049/jimmunol.1700812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kobayashi EH, et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat Commun. 2016;7:11624. doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Meister A. Selective modification of glutathione metabolism. Science. 1983;220(4596):472–477. doi: 10.1126/science.6836290. [DOI] [PubMed] [Google Scholar]
- 177.Mak TW, et al. Glutathione primes T cell metabolism for inflammation. Immunity. 2017;46(4):675–689. doi: 10.1016/j.immuni.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 178.Yan Z, Garg SK, Banerjee R. Regulatory T cells interfere with glutathione metabolism in dendritic cells and T cells. J Biol Chem. 2010;285(53):41525–41532. doi: 10.1074/jbc.M110.189944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Buchmuller-Rouiller Y, et al. Role of glutathione in macrophage activation: effect of cellular glutathione depletion on nitrite production and leishmanicidal activity. Cell Immunol. 1995;164(1):73–80. doi: 10.1006/cimm.1995.1144. [DOI] [PubMed] [Google Scholar]
- 180.Jain M, Chandel NS. Rethinking antioxidants in the intensive care unit. Am J Respir Crit Care Med. 2013;188(11):1283–1285. doi: 10.1164/rccm.201307-1380CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bjelakovic G, Nikolova D, Gluud LL, Simonetti RG, Gluud C. Antioxidant supplements for prevention of mortality in healthy participants and patients with various diseases. Cochrane Database Syst Rev. 2012;2012(3):CD007176. doi: 10.1002/14651858.CD007176.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]