Recent work has shown that sRNAs play crucial roles in S. pneumoniae pathogenesis, as inactivation of nearly one-third of the putative sRNA genes identified in one study led to reduced fitness or virulence in a murine model. Yet our understanding of sRNA-mediated gene regulation in S. pneumoniae has been hindered by limited knowledge about these regulatory RNAs, including which sRNAs are synthesized by different S. pneumoniae strains. We sought to address this problem by developing a sensitive sRNA detection technique to identify sRNAs in S. pneumoniae D39. A comparison of our data set reported here to those of other RNA-seq studies for S. pneumoniae strain D39 and TIGR4 has provided new insights into the S. pneumoniae sRNA transcriptome.
KEYWORDS: RNA-seq, dRNA-seq, pneumococcus, sRNA transcriptome, serotype 2 D39, serotype 4 TIGR4, small RNA identification
ABSTRACT
Streptococcus pneumoniae (pneumococcus) is a major human respiratory pathogen and a leading cause of bacterial pneumonia worldwide. Small regulatory RNAs (sRNAs), which often act by posttranscriptionally regulating gene expression, have been shown to be crucial for the virulence of S. pneumoniae and other bacterial pathogens. Over 170 putative sRNAs have been identified in the S. pneumoniae TIGR4 strain (serotype 4) through transcriptomic studies, and a subset of these sRNAs has been further implicated in regulating pneumococcal pathogenesis. However, there is little overlap in the sRNAs identified among these studies, which indicates that the approaches used for sRNA identification were not sufficiently sensitive and robust and that there are likely many more undiscovered sRNAs encoded in the S. pneumoniae genome. Here, we sought to comprehensively identify sRNAs in Avery’s virulent S. pneumoniae strain D39 using two independent RNA sequencing (RNA-seq)-based approaches. We developed an unbiased method for identifying novel sRNAs from bacterial RNA-seq data and have further tested the specificity of our analysis program toward identifying sRNAs encoded by both strains D39 and TIGR4. Interestingly, the genes for 15% of the putative sRNAs identified in strain TIGR4, including ones previously implicated in virulence, are not present in the strain D39 genome, suggesting that the differences in sRNA repertoires between these two serotypes may contribute to their strain-specific virulence properties. Finally, this study has identified 66 new sRNA candidates in strain D39, 30 of which have been further validated, raising the total number of sRNAs that have been identified in strain D39 to 112.
IMPORTANCE Recent work has shown that sRNAs play crucial roles in S. pneumoniae pathogenesis, as inactivation of nearly one-third of the putative sRNA genes identified in one study led to reduced fitness or virulence in a murine model. Yet our understanding of sRNA-mediated gene regulation in S. pneumoniae has been hindered by limited knowledge about these regulatory RNAs, including which sRNAs are synthesized by different S. pneumoniae strains. We sought to address this problem by developing a sensitive sRNA detection technique to identify sRNAs in S. pneumoniae D39. A comparison of our data set reported here to those of other RNA-seq studies for S. pneumoniae strain D39 and TIGR4 has provided new insights into the S. pneumoniae sRNA transcriptome.
INTRODUCTION
Small regulatory RNAs (sRNAs) are an emerging class of bacterial posttranscriptional regulators that have been implicated in controlling a wide variety of physiological responses in bacteria, ranging from stress responses to virulence (1–4). Many sRNAs regulate gene expression by base pairing with their respective mRNA targets, altering their transcription (5), translation (6), or stability (7). For example, the FasX sRNA of Streptococcus pyogenes increases the expression of the secreted virulence factor streptokinase by increasing the stability of the encoding transcript as a consequence of sRNA-mRNA base pairing (8). Alternatively, sRNAs can act to titrate a nucleic acid binding protein, blocking it from binding and acting on a DNA or RNA substrate. For example, the EutX/Rli55 sRNA found in Enterococcus faecalis and Listeria monocytogenes binds the phosphorylated form of the transcriptional antiterminator regulator EutV, precluding it from binding the eut transcript encoding proteins involved in ethanolamine utilization; binding of EutV to the eut transcript would otherwise stabilize an antiterminator in its 5′ untranslated region (UTR), preventing premature transcription termination (9, 10). In Escherichia coli and Bacillus subtilis, the 6S RNA binds σ70- and σA-bound forms of the RNA polymerase holoenzyme, respectively, blocking them from binding their cognate promoters (11).
The earliest global searches for sRNAs in bacteria utilized bioinformatic approaches in which intergenic regions were examined for potential promoters and rho-independent terminators (12–14); many of those candidate sRNAs were subsequently validated by Northern blot analysis. As a consequence of the success of these studies in identifying novel sRNAs that were either intergenic or antisense (i.e., on the opposite strand of an open reading frame [ORF]), it appeared that the majority of sRNAs were produced from their own promoter as independent transcripts. Moreover, a majority of sRNAs that have been widely characterized to date are expressed from intergenic regions. However, the adoption of tiling microarrays and high-throughput RNA sequencing (RNA-seq)-based approaches along with global transcription start site (TSS) mapping has revolutionized our understanding of bacterial transcriptomes, including sRNA transcriptomes. In particular, these studies have revealed an abundance of small transcripts consisting of 5′ or 3′ UTRs of mRNAs or internal fragments of mRNAs, rRNAs, or tRNAs (15–18). Some of these sRNAs located in the 5′ UTRs are produced as a consequence of premature transcription termination (19, 20) and in some cases contain a riboswitch (20). Other sRNAs were shown to be stable fragments generated by the cleavage of an mRNA by an endoribonuclease (21). Functional characterization of these riboswitch-, tRNA-, and mRNA-derived fragments has revealed a second life for this “junk RNA” as sRNAs (20, 22–24). Given this complexity, that sRNAs can be produced from 3′ UTRs, 5′ UTRs, or internal RNA fragments of transcripts or as independent transcripts, that genes can be expressed by multiple promoters producing transcripts with different lengths of leaders, and that the 3′ UTRs of mRNAs can extend into downstream genes, delineating sRNAs from vast amounts of RNA sequencing data can be challenging.
Streptococcus pneumoniae (pneumococcus) is a Gram-positive human commensal bacterium and a respiratory pathogen that is a major cause of pneumonia and other respiratory tract infections. It is one of the most important bacteria clinically causing more morbidity and mortality worldwide than any other infection (25, 26). We have previously identified the presence of 15 sRNAs in the S. pneumoniae serotype 2 strain D39 from the Winkler laboratory (D39W) (27). Very recently, Slager et al. predicted the presence of 34 putative sRNAs in D39V, the strain used by the Veening laboratory, which exhibited several differences compared to D39W (28). Thirty-two out of 34 predicted sRNAs were identified via their deep-sequencing analysis in D39V (28). Among the 32 sRNAs that were detected by Slager et al., 3 sRNAs were housekeeping sRNAs (ssrA, rnpB, and small cytoplasmic RNA [scRNA]) known to be conserved across bacterial species, 4 sRNAs (Spd_sr14, Spd_sr17, Spd_sr37, and CcnA) were previously validated by us in D39W (27), and another 4 sRNAs (CcnB, CcnC, CcnD, and CcnE) are conserved across all pneumococcal serotypes (29). Genome-wide high-throughput sequencing studies of S. pneumoniae strain TIGR4, a more recently isolated serotype 4 strain, independently identified the presence of more than 170 sRNAs in total (3, 30, 31). In the TIGR4 strain, 50 sRNAs were identified in one study using tiling microarrays (31), 88 sRNAs were discovered by another group using a pyrosequencing-based approach (30), and 89 sRNAs were delineated using Illumina-based RNA sequencing (3). However, only a small fraction of these putative sRNAs were identified by all three studies. This dissimilarity in the sRNA transcriptomes among these studies could be due to differences in RNA isolation, library preparation, detection platforms, and the analysis programs and tools utilized, which use different parameters and thresholds.
Here, we have redefined the sRNA transcriptome of S. pneumoniae strain D39W using data from two independent RNA sequencing experiments. By combining data sets from two parallel sequencing studies (small RNA-seq [sRNA-seq] and differential RNA-seq [dRNA-seq]), we have significantly increased the sensitivity of sRNA detection in strain D39. We present an unbiased approach for identifying sRNAs genome-wide using a combination of new algorithms, which can be applied to a wide variety of prokaryotic RNA-seq data to identify new sRNAs. This study has raised the total number of sRNA candidates in strain D39W to 112, of which 66 sRNAs are novel and were not detected by any other previous studies in D39. We validated a total of 62 sRNAs in D39W by Northern blotting, which reflects the robustness of our analysis method. We also report a thorough reevaluation of the sRNA transcriptome in S. pneumoniae in regard to how the sRNA expression profiles compare between the two serotypes, using D39 (serotype 2) and TIGR4 (serotype 4), the genomes of which differ by approximately 10% at the nucleotide level.
RESULTS
Mapping of the S. pneumoniae D39W transcriptome.
To profile the transcriptome of D39W, we first extracted total RNA from three independent replicates of the wild-type strain grown to exponential phase (optical density at 620 nm [OD620] of ∼0.15) in brain heart infusion (BHI) medium at 37°C in an atmosphere of 5% CO2. Extracted RNA was subjected to mRNA-seq library preparation, and the reads obtained from strand-specific sequencing were then mapped to the D39W reference genome (see Materials and Methods). Total reads with numbers averaging between 3.2 million and 7.5 million were obtained per sample, with nearly 95% of the reads mapping on the D39W genome. Most reads mapped to open reading frames; however, many reads mapped to intergenic noncoding regions on the genome or locations antisense to ORFs, suggesting that these represent transcriptionally active regions that possibly synthesize regulatory RNAs, like sRNAs. Applying the regression model analysis previously developed by Wagner et al. (32), we first determined the transcript expression noise threshold to be 8 reads of coverage per bp; below this read threshold value, the probability that the reads reflect active transcription is low. Based on this expression threshold, we found that ∼82% of the S. pneumoniae genome was expressed under our experimental conditions. Approximately 72% of the genome was expressed as transcripts from known ORFs or annotated gene regions. Additionally, a significant number of reads mapped to intergenic regions lacking annotated genes, and ∼8% of the genome was expressed from such regions. Additionally, the D39W genome showed significant antisense expression, comprising nearly 2.3% of the genome at the nucleotide level. Genes with expression below the threshold value were considered unexpressed and corresponded to ∼13% of the genome. The expression of these genes may be growth phase dependent or may be induced under specific stress conditions. The remaining ∼4.5% of the genome consisted of intergenic regions that were transcriptionally silent during exponential growth of D39W in BHI broth. The transcriptome map of the D39 genome is summarized in Fig. S1 in the supplemental material.
Identification of sRNAs in S. pneumoniae D39 using sRNA-seq.
The genomes of two isolates of S. pneumoniae strain D39 have been sequenced, D39W from our laboratory (33) and D39V from the Veening laboratory (28). D39W and D39V are derived from the same ancestral strain, NCTC 7466, but genome assembly comparisons revealed the presence of several differences between the two strains at the sequence level, including 14 single nucleotide polymorphisms (SNPs), 3 insertions, and 2 deletions (28). Streptococcus pneumoniae D39W expresses at least 15 sRNAs, which we previously discovered by bioinformatics analyses and validated by Northern blotting (27). Subsequent deep-genome annotation of S. pneumoniae D39V identified a total of 34 sRNAs, including 3 housekeeping RNAs, scRNA, ssrA, and rnpB (28). We previously identified 9 of these sRNA candidates in D39W (27). To more comprehensively identify sRNAs in the S. pneumoniae D39W genome, we developed our own sRNA-seq-based approach. We performed sRNA-seq as described in Materials and Methods and Fig. S2 in the supplemental material and then applied our sRNA-seq analysis pipeline, nicknamed DROOM, to identify sRNA candidates based on an unbiased approach where the expression over a 100-bp window (test) was compared to the expression of 400-bp flanking regions (background) on either side beyond a 50-bp buffer. This analysis was performed genome-wide with a 50-bp sliding window, and a Z-score was determined for 100-bp test regions. Z-score values represent the probability of expression of a test region relative to its background. This analysis was based on the assumption that background regions reflect normally distributed noise and that the Z-score for the test window is inversely proportional to the probability that a given test region was observed by chance due to noisy expression. Using this methodology, 200 sRNA candidates were identified, which were ranked by their Z-score values and then manually curated. Many of the sRNAs were larger than the 100-bp test window and were initially recognized by the algorithm as multiple adjacent sRNAs. We combined those adjacent sRNAs that were from the same transcriptionally active region, thus condensing 200 predictions down to 119 putative sRNAs. Sixty-eight out of 119 candidate sRNAs mapped within known ORFs and were filtered out of our list of predicted sRNAs in D39W, as we could not differentiate based on our sequencing analysis whether or not these were merely stable intermediates in mRNA decay. Thus, our sRNA-seq analysis identified 51 sRNA candidates in D39W (Table 1). Manual curation of the sequencing data revealed that this automated method failed to detect some sRNAs when two or more sRNAs were transcribed within the 1,000-bp analysis window due to the lack of generation of significant Z-score values corresponding to those individual sRNAs relative to the background. Thus, a manual revision of our sRNA-seq data was performed to find any sRNAs that might have been missed due to the caveats of this automated approach, and this led to the identification of 6 additional candidate sRNAs in D39W, Spd_sr36, Spd_sr64, Spd_sr78, Spd_sr81, Spd_sr109, and Spd_sr111 (Table 1).
TABLE 1.
sRNAs identified in S. pneumoniae D39W and their characteristicsa
| sRNA | Coordinates | Flanking genes | Genetic context(s) | TSS position (classification) | Detection method(s)d | Overlapping sRNA in D39V | Overlapping sRNA(s) in TIGR4 |
|---|---|---|---|---|---|---|---|
| ▶Spd_sr1 | 15002–15144 (+) | spd_0015, rrsA | 5′/intergenic | 15002 (pTSS) | Both | ||
| ▶Spd_sr2 | 16653–16698 (−) | spd_0017, rrsA | Intergenic | 16698 (pTSS) | dRNA-seq | ||
| ▶Spd_sr3 | 19398–19577 (−) | rrfA, spd_0017 | Antisense | 19577 (sTSS) | dRNA-seq | ||
| CcnC | 23967–24065 (+) | spd_0024, spd_0025 | Intergenic | 23967 (pTSS) | dRNA-seq | CcnC | trn0012 |
| ▶Spd_sr4 | 34536–34631 (−) | spd_0040, spd_0039 | 3′/intergenic | 34631 (sTSS) | dRNA-seq | ||
| Spd_sr5 | 39980–40082 (+) | spd_0047, spd_0048 | Intergenic | 39980 (sTSS) | Both | srf-02 | |
| Spd_sr6 | 41494–41559 (+) | spd_0048, comA | Antisense | 41494 (pTSS) | Both | srf-03 | |
| ▶Spd_sr8 | 78855–79001 (+) | spd_0077, spd_0078 | 5′/intergenic | 78855 (sTSS) | dRNA-seq | ||
| ▶Spd_sr9 | 85628–85745 (+) | spd_0082, rpsD | 5′/intergenic and 3′/intergenic | 85628 (pTSS) | Both | ||
| ▶Spd_sr11 | 89748–89828 (−) | spd_0090, spd_0089 | Intergenic | 89828 (sTSS) | dRNA-seq | ||
| ▶Spd_sr13b | 101161–101403 (−) | spd_0101, capD | Antisense | 101403 (sTSS) | dRNA-seq | ||
| ▶Spd_sr15 | 112142–112229 (+) | argH, spd_0112 | Antisense | 112142 (sTSS) | dRNA-seq | ||
| ▶Spd_sr16b | 127264–127488 (+) | spd_0124, spd_0125 | 5′/intergenic | 127264 (ND) | sRNA-seq | ||
| Spd_sr37c | 131773–131842 (+) | tmrU, spd_0128 | 5′/intergenic and 3′/intergenic | 131773 (pTSS) | dRNA-seq | srf-04 | |
| Spd_sr18 | 134413–134576 (−) | spd_0130, gidA | 3′/intergenic | 134576 (sTSS) | dRNA-seq | trn0057 | |
| ▶Spd_sr19b | 136586–136654 (−) | spd_0132, spd_0131 | 5′/intergenic | 136654 (sTSS) | dRNA-seq | ||
| ▶Spd_sr20b | 141329–141397 (+) | spd_0137, spd_0138 | Antisense | 141329 (pTSS) | Both | ||
| Spd_sr14c | 149223–149341 (+) | spd_0143, spd_0144 | Intergenic | 149223 (ND) | sRNA-seq | srf-05 | |
| ▶Spd_sr21 | 164972–165040 (+) | spd_0160, spd_0161 | 3′/intergenic | 164972 (sTSS) | dRNA-seq | ||
| Spd_sr22b | 173841–174014 (−) | ruvA, ribD | 5′/intergenic | 174014 (pTSS) | Both | RNA-switch-1 | R1, srn029 |
| CcnEb | 212278–212425 (+) | spd_0221, spd_0222 | Intergenic | 212278 (pTSS) | Both | CcnE | F7, srn061 |
| CcnAc (Spd_sr56) | 231143–231235 (+) | spd_0240, ruvB | Intergenic | 231143 (pTSS) | dRNA-seq | CcnA | F8 |
| CcnBb | 231331–231427 (+) | spd_0240, ruvB | Intergenic | 231331 (pTSS) | dRNA-seq | CcnB | |
| Spd_sr23b | 231823–232091 (+) | spd_0240, ruvB | Intergenic | 231823 (pTSS) | Both | srf-07 | F9 |
| ▶Spd_sr24b | 231853–232035 (−) | ruvB, spd_0240 | Intergenic | 232035 (pTSS) | Both | ||
| CcnDb | 233715–233809 (+) | spd_0242, uppS | Intergenic | 233715 (pTSS) | dRNA-seq | CcnD | |
| ▶Spd_sr25 | 264619–264738 (−) | spd_0266, spd_0265 | 3′/intergenic | 264738 (sTSS) | dRNA-seq | ||
| ▶Spd_sr26 | 288689–288831 (−) | spd_0288, spd_0287 | Antisense | 288831 (ND) | sRNA-seq | ||
| Spd_sr28 | 376371–376530 (+) | spd_0371, spd_0372 | 5′/intergenic and 3′/intergenic | 376371 (pTSS) | Both | RNA-switch-2 | |
| Spd_sr29 | 381067–381205 (−) | spd_0376, serS | 5′/intergenic and 3′/intergenic | 381205 (pTSS) | Both | RNA-switch3 | trn0218 |
| ▶Spd_sr31b | 476084–476234 (+) | spd_0465, spd_0466 | 3′/intergenic | 476084 (sTSS) | Both | ||
| Spd_sr32b | 496899–497104 (+) | spd_0490, spd_0491 | 3′/intergenic | 496899 (pTSS) | dRNA-seq | RNA-switch-5 | F17 |
| Spd_sr33 | 508237–508335 (+) | spd_0500, licT | 5′/intergenic | 508237 (ND) | sRNA-seq | srf-10 | F19 |
| Spd_sr34 | 512889–513087 (+) | bglA-2, pheS | 5′/intergenic | 512889 (pTSS) | Both | RNA-switch-6 | F20, srn157 |
| ▶Spd_sr35b | 518059–518350 (−) | spd_0508, spd_0507 | 5′/intergenic | 518350 (ND) | sRNA-seq | ||
| Spd_sr36b | 522749–522976 (+) | metF, pnp | Intergenic | 522749 (pTSS) | Both | F21 | |
| ▶Spd_sr40 | 557606–557713 (+) | spd_0543, spd_0544 | 5′/intergenic and 3′/intergenic | 557606 (sTSS) | dRNA-seq | ||
| Spd_sr42b | 587439–587538 (+) | spd_0563, spd_0564 | Antisense | 587439 (pTSS) | dRNA-seq | srf-11 | F25, trn0332 |
| Spd_sr43b | 643872–644040 (+) | lctO, spd_0622 | Intergenic | 643872 (pTSS) | dRNA-seq | RNA-switch-8 | srn176 |
| Spd_sr44 | 646754–646926 (+) | thiE1, spd_0625 | Intergenic | 646754 (pTSS) | Both | RNA-switch-9 | F27, trn0358 |
| ▶Spd_sr45b | 653472–653568 (−) | spd_0633, thiD | 5′/intergenic | 653568 (pTSS) | dRNA-seq | ||
| ▶Spd_sr38c | 769926–769992 (+) | spd_0758, spd_0759 | Antisense | 769926 (pTSS) | dRNA-seq | ||
| Spd_sr46 | 781128–781485 (+) | spd_0768, spd_0770 | Intergenic | 781128 (pTSS) | Both | SsrA | F32, srn226 |
| ▶Spd-sr7c | 820183–820245 (+) | spd_0803, spd_0804 | ND | ND | ND* | ||
| ▶Spd_sr47 | 825483–825544 (+) | spd_0807, spd_0808 | Antisense | 825483 (pTSS) | dRNA-seq | ||
| Spd_sr49e | 825802–826129 (+) | spd_0808, cad | Antisense | 825802 (pTSS) | Both | srf-13 | F59, srn235 |
| Spd_sr48c | 862699–862827 (+) | spd_0846, infC | 5′/intergenic | 862699 (pTSS) | Both | F34, srn239 | |
| ▶Spd_sr50 | 882727–882886 (+) | spd_0867, spd_0868 | 5′/intergenic and 3′/intergenic | 882727 (pTSS) | Both | ||
| Spd_sr17c | 912571–912715 (+) | spd_0899, asd | Intergenic | 912571 (pTSS) | Both | srf-17 | F38, srn254 |
| Spd_sr12c | 967941–968177 (+) | ppc, spd_0954 | 5′/intergenic and 3′/intergenic | 967941 (pTSS) | dRNA-seq | RNA-switch-13 | |
| ▶Spd_sr51 | 980052–980117 (+) | spd_0966, murA1 | Antisense | 980052 (pTSS) | dRNA-seq | ||
| ▶Spd_sr53f | 998110–998240 (+) | pta, spd_0986 | 3′/intergenic | 998110 (sTSS) | dRNA-seq | ||
| Spd_sr55b | 999541–999605 (+) | spd_0986, spd_0987 | Antisense | 999541 (pTSS) | dRNA-seq | srn266 | |
| ▶Spd_sr57b | 999977–1000137 (−) | spd_0988, spd_0987 | 5′/intergenic | 1000137 (pTSS) | Both | ||
| Spd_sr58b | 1001180–1001345 (+) | spd_0988, rplU | 5′/intergenic and 3′/intergenic | 1001180 (pTSS) | Both | srn267 | |
| ▶Spd_sr59 | 1073074–1073228 (+) | nrdF, lacR2 | 3′/intergenic | 1073074 (sTSS) | Both | ||
| Spd_sr60 | 1079135–1079199 (−) | lacD, lacT | 5′/intergenic and 3′/intergenic | 1079199 (pTSS) | dRNA-seq | srf-18 | |
| ▶Spd_sr61 | 1110356–1110399 (−) | spd_1080, spd_1079 | 5′/intergenic and 3′/intergenic | 1110399 (pTSS) | dRNA-seq | ||
| Spd_sr62b | 1168377-1168512 (−) | nth, pyrR | 5′/intergenic and 3′/intergenic | 1168512 (pTSS) | dRNA-seq | RNA-switch-15 | R15, srn299 |
| Spd_sr63 | 1170288–1170385 (+) | spd_1136, spd_1137 | 5′/intergenic | 1170288 (pTSS) | Both | srf-19 | F43 |
| Spd_sr64 | 1174647–1174790 (+) | gidB, uraA | 5′/intergenic | 1174647 (sTSS) | Both | RNA-switch-16 | F44 |
| ▶Spd_sr65e | 1189697–1189944 (−) | spd_1161, spd_1160 | Antisense | 1189944 (pTSS) | Both | ||
| ▶Spd_sr66b | 1203549–1203754 (−) | spd_1175, spd_1174 | Intergenic | 1203754 (ND) | sRNA-seq | ||
| ▶Spd_sr67b | 1212229–1212526 (−) | spd_1180, spd_1179 | 5′/intergenic | 1212526 (ND) | sRNA-seq | ||
| Spd_sr54c | 1215844–1215967 (−) | spd_1190, rplJ | Antisense | 1215967 (pTSS) | Both | R3, srn308 | |
| Spd_sr69 | 1217390–1217501 (−) | spd_1191, spd_1190 | 5′/intergenic | 1217501 (sTSS) | dRNA-seq | R17 | |
| Spd_sr70b | 1249557–1249736 (−) | spd_1216, spd_1217 | 5′/intergenic | 1249736 (pTSS) | Both | RNA-switch-17 | trn0634 |
| ▶Spd_sr71b | 1264468–1264569 (−) | spd_1233, spd_1232 | 3′/intergenic | 1264569 (pTSS) | dRNA-seq | ||
| Spd_sr72 | 1300078–1300167 (−) | guaA, spd_1273 | 3′/intergenic | 1300167 (pTSS) | Both | R18 | |
| ▶Spd_sr73b | 1310945–1311101 (−) | spd_1289, spd_1288 | 5′/intergenic and 3′/intergenic | 1311101 (pTSS) | dRNA-seq | ||
| Spd_sr74b | 1326066–1326252 (−) | spd_1308, spd_1307 | 5′/intergenic and 3′/intergenic | 1326252 (pTSS) | Both | RNA-switch-18 | R19, trn0663 |
| Spd_sr76 | 1356924–1356967 (+) | spd_1342, spd_1343 | Intergenic | 1356924 (pTSS) | dRNA-seq | trn0696 | |
| ▶Spd_sr77 | 1383075–1383210 (−) | asnS, rpsF | 5′/intergenic and 3′/intergenic | 1383210 (ND) | sRNA-seq | ||
| Spd_sr78b | 1404038–1404161 (−) | spd_1384, spd_1383 | 5′/intergenic | 1404161 (pTSS) | Both | R21, srn351 | |
| ▶Spd_sr79 | 1444623–1444702 (−) | spd_1426, spd_1425 | 3′/intergenic | 1444702 (sTSS) | dRNA-seq | ||
| Spd_sr80b | 1458804–1458975 (+) | spd_1441, spd_1442 | 5′/intergenic | 1458804 (ND) | sRNA-seq | RNA-switch-20 | F47 |
| ▶Spd_sr81b | 1464370–1464684 (−) | spd_1448, spd_1447 | 3′/intergenic | 1464684 (ND) | sRNA-seq | ||
| ▶Spd_sr82b | 1468939–1469239 (−) | spd_1455, spd_1454 | Intergenic | 1469239 (pTSS) | dRNA-seq | ||
| Spd_sr83b | 1528061–1528186 (−) | recG, spd_1506 | 3′/intergenic | 1528186 (pTSS) | Both | srf-21 | |
| ▶Spd_sr84b | 1595445–1595563 (−) | spd_1578, spd_1577 | 5′/intergenic | 1595563 (pTSS) | Both | ||
| Spd_sr85b | 1597869–1598142 (+) | spd_1580, spd_1582 | Intergenic | 1597869 (pTSS) | Both | SsrS (6S) | srn395 |
| Spd_sr88b | 1619052–1619299 (−) | spd_1605, spd_1604 | Intergenic | 1619299 (pTSS) | Both | RNA-switch-22 | R6, srn400 |
| ▶Spd_sr89b | 1673200–1673322 (−) | spd_1662, murI | 5′/intergenic | 1673322 (pTSS) | dRNA-seq | ||
| ▶Spd_sr90 | 1675536–1675603 (−) | spd_1664, treC | 3′/intergenic and 5′/intergenic | 1675603 (sTSS) | dRNA-seq | ||
| ▶Spd_sr 39c | 1678583–1678648 (−) | spd_1666, spd_1665 | Antisense | 1678648 (pTSS) | dRNA-seq | ||
| ▶Spd_sr 52c | 1687017–1687151 (−) | spd_1672, amiA | ND | ND | ND* | ||
| ▶Spd_sr91 | 1697610–1697676 (−) | spd_1680, spd_1681 | Antisense | 1697676 (pTSS) | Both | ||
| ▶Spd_sr92 | 1703159–1703205 (+) | spd_1701, rrsB | Intergenic | 1703159 (pTSS) | dRNA-seq | ||
| ▶Spd_sr93 | 1704711–1704858 (−) | spd_1703, rrsB | 5′/intergenic | 1704858 (pTSS) | Both | ||
| ▶Spd_sr94 | 1708422–1708593 (−) | spd_1708, spd_1707 | 5′/intergenic | 1708593 (sTSS) | Both | ||
| ▶Spd_sr95 | 1730706–1730807 (−) | dinF, lytA | Intergenic | 1730807 (pTSS) | dRNA-seq | ||
| ▶Spd_sr10c | 1750985–1751149 (+) | spd_1756, ndk | ND | ND | ND* | ||
| ▶Spd_sr96b | 1759319–1759411 (−) | spd_1760, rpoB | Intergenic | 1759411 (pTSS) | Both | ||
| ▶Spd_sr97 | 1759807–1760010(−) | spd_1761, spd_1760 | 3′/intergenic | 1760010 (sTSS) | dRNA-seq | ||
| ▶Spd_sr98 | 1773206–1773384 (−) | cbf-1, purR | 5′/intergenic and 3′/intergenic | 1773384 (pTSS) | Both | ||
| ▶Spd_sr99b | 1802263–1802409 (−) | spd_1817, rrsC | 5′/intergenic | 1802409 (pTSS) | Both | ||
| ▶Spd_sr100b | 1804114–1804220 (−) | pbp2A, secE | 5′/intergenic and 3′/intergenic | 180422 (pTSS) | Both | ||
| ▶Spd_sr101 | 1819112–1819298 (−) | spd_1834, spd_1833 | 3′/intergenic | 1819298 (sTSS) | dRNA-seq | ||
| ▶Spd_sr103 | 1862635–1862681 (+) | spd_1892, rrsD | Intergenic | 1862635 (pTSS) | dRNA-seq | ||
| ▶Spd_sr104 | 1864190–1864334 (−) | spd_1894, rrsD | 5′/intergenic | 1864334 (pTSS) | Both | ||
| Spd_sr105 | 1873276–1873324 (−) | mutS, spd_1902 | 5′/intergenic and 3′/intergenic | 1873324 (pTSS) | dRNA-seq | srf-24 | R10, srn477, trn0978 |
| Spd_sr106b | 1892400–1892550 (−) | spd_1924, spd_1923 | 5′/intergenic and 3′/intergenic | 1892550 (pTSS) | Both | srf-25 | srn491 |
| ▶Spd_sr107 | 1903127–1903247 (−) | malP, spd_1931 | 3′/intergenic | 1903247 (sTSS) | dRNA-seq | ||
| ▶Spd_sr108b | 1913211–1913442 (−) | spd_1939, malR | Antisense | 1913442 (sTSS) | dRNA-seq | ||
| Spd_sr109b | 1972859–1973060 (+) | spd_1996, adcA | Intergenic | 1972859 (ND) | sRNA-seq | F66, srn502 | |
| Spd_sr110 | 1973000–1973113 (−) | adcA, spd_1996 | Intergenic | 1973113 (pTSS) | dRNA-seq | srf-27 | |
| Spd_sr111b | 1973154–1973403 (+) | spd_1996, adcA | Intergenic | 1973154 (ND) | sRNA-seq | F67, srn503 | |
| Spd_sr112 | 1973342–1973456 (−) | adcA, spd_1996 | Intergenic | 1973456 (pTSS) | dRNA-seq | srf-28 | |
| ▶Spd_sr113 | 2000298–2000399 (−) | spd_2022, spd_2021 | 3′/intergenic and 5′/intergenic | 2000399 (sTSS) | dRNA-seq | ||
| Spd_sr114b | 2006263–2006428 (−) | cbpD, spd_2027 | 5′/intergenic and 3′/intergenic | 2006428 (pTSS) | Both | RNA-switch-24 | |
| ▶Spd_sr115 | 2016672–2016759 (−) | spd_2038, cysK | 3′/intergenic | 2016759 (sTSS) | dRNA-seq | ||
| Spd_sr116b | 2020112–2020228 (−) | spd_2043, rpsB | 5′/intergenic | 2020228 (pTSS) | dRNA-seq | srf-30 |
▶, new sRNA species identified in D39W but not in D39V or TIGR4. Columns from left to right represent sRNA identifications in S. pneumoniae D39W; the coordinates to which the sRNAs mapped on the D39 genome, including strand information for the sRNAs, where (+) and (−) indicate that the sRNA is transcribed from the plus strand and the minus strand, respectively; their flanking genes; the genetic context of the sRNAs in D39W; TSS information; and whether there are overlaps between sRNAs identified in D39W and D39V (28) or those previously identified in TIGR4 by Mann et al. (3) and Acebo et al. (30). sRNAs in boldface type were validated for expression. ND, not determined.
sRNAs validated in this study (Fig. 2 and 3; see also Fig. S3 and S4 in the supplemental material).
sRNA validated by Tsui et al. (27).
ND*, not detected by either sRNA-seq or dRNA-seq but previously validated in D39W by Tsui et al. (27).
Spd_sr49 and Spd_sr65 could also be transcribed as 5′/intergenic sRNAs.
Spd_sr53 could also be transcribed as an antisense sRNA.
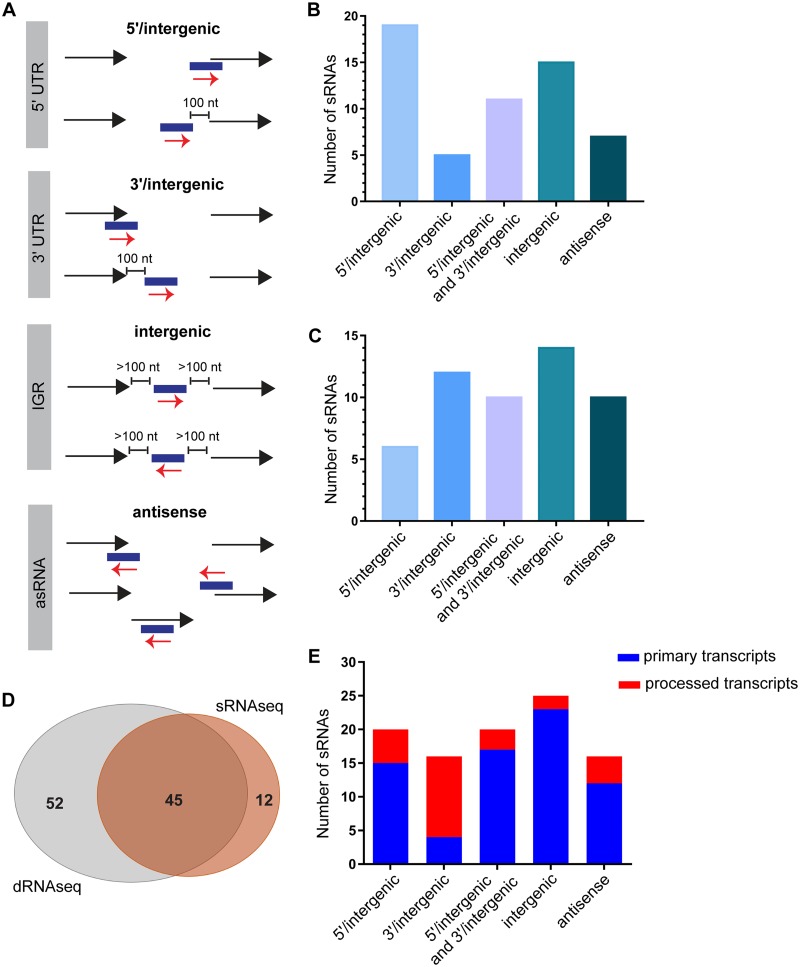
We next determined the genetic context of these 57 sRNAs that were identified in D39W. These sRNAs were classified based on their location relative to previously annotated genes in the D39W genome into four different categories: (i) 5′/intergenic, (ii) 3′/intergenic, (iii) intergenic, and (iv) antisense (Fig. 1A). Out of 57 sRNAs, 19 sRNAs were 5′/intergenic, 5 were 3′/intergenic, 15 were intergenic, and 7 were antisense sRNAs (asRNAs). The remaining 11 sRNAs were transcribed from a genomic locus within 100 nucleotides (nt) from the 5′ end of one ORF and the 3′ end of another and accordingly were classified as 5′/intergenic and 3′/intergenic sRNAs (Fig. 1B and Table 1). Out of these 57 sRNAs identified by our new computational analysis method, 3 sRNAs (Spd_sr17, Spd_sr48, and Spd_sr54) were previously identified in strain D39W (27). Additionally, only one (CcnE) of the five Ccn sRNAs (CcnA, CcnB, CcnC, CcnD, and CcnE), which are conserved among different serotypes of S. pneumoniae, including D39 and TIGR4 (29), was detected by our sRNA-seq analysis. CcnA, CcnB, CcnC, and CcnD were also not identified by the two previously reported high-throughput RNA sequencing studies in the TIGR4 strain (3, 30). sRNA-seq analysis thus led to the identification of 53 novel candidate sRNAs in D39W, of which 31 were validated for expression by Northern blotting (Fig. S3).
FIG 1.
Overview of the sRNAs identified in D39W using sRNA-seq and dRNA-seq. (A) Genomic context of the sRNAs identified in D39W. sRNAs identified in this study were classified into four major classes based on their location in the D39W genome relative to flanking genes. sRNAs partially overlapping the 5′ end of an annotated gene or located within a 100-nt distance of the start codon of the downstream ORF were classified as 5′/intergenic. sRNAs partially overlapping the 3′ end of an annotated gene or within a 100-nt distance from the stop codon of the upstream ORF were classified as 3′/intergenic. sRNAs were classified as intergenic if they were located at least 100 nt away from the flanking genes. sRNAs encoded on the opposite strand of an ORF and overlapping by at least 1 nt were classified as antisense sRNAs. IGR, intergenic region. (B) Genomic contexts of sRNAs identified by sRNA-seq analysis as defined above for panel A. (C) Genomic contexts of sRNAs identified by dRNA-seq analysis, but not sRNA-seq analysis, as defined above for panel A. (D) Venn diagram showing the overlap between the sRNAs identified by sRNA-seq and those identified by dRNA-seq analyses. (E) Transcription start site (TSS) characteristics of the sRNAs in each category defined above for panel A that were identified by dRNA-seq in D39W. sRNAs determined by dRNA-seq analysis to have unprocessed 5′ ends or to contain processed 5′ ends are indicated in blue and red, respectively.
Identification of the primary sRNA transcriptome in S. pneumoniae D39W using dRNA-seq.
Although we detected many new sRNA candidates in D39W via sRNA-seq analysis, we could not determine whether these were produced as primary transcripts or as degradation products resulting from RNase-mediated cleavage. To resolve between these possibilities, we performed differential RNA-seq (dRNA-seq) to determine how many of the newly identified sRNAs in D39W are transcribed as independent transcriptional units (primary transcripts) or generated as cleavage products from longer primary transcripts (processed transcripts). Total RNA extracted from cultures of wild-type S. pneumoniae D39W (IU1781) cells grown in BHI broth at 37°C in an atmosphere of 5% CO2 was prepared for dRNA-seq library preparation as described in Materials and Methods and Fig. S2 in the supplemental material. Isolated RNA samples were either treated with 5′-terminator exonuclease (+TEX) or mock treated (−TEX). TEX degrades processed transcripts containing 5′-monophosphates, whereas primary sRNA transcripts containing 5′-triphosphates are protected from degradation. Therefore, the presence of a signal in the TEX-treated samples corresponding to an sRNA in dRNA-seq data indicates that the sRNA is produced as a primary transcript. dRNA-seq generated a total number of ∼1.4 million to 3.3 million paired-end reads per sample after filtering out unpaired reads and reads less than 21 nt long. On average, 57% of the paired reads aligned concordantly to the D39W reference genome.
Next, we determined how many of the sRNAs identified by our sRNA-seq analysis were detected independently by dRNA-seq. Forty-five out of 57 sRNAs that were identified by sRNA-seq were found to be common between the two sequencing experiments (sRNA-seq and dRNA-seq) (Table 1 and Fig. 1D). Interestingly, our dRNA-seq approach identified an additional 52 sRNAs in D39W that were not detected in our sRNA-seq analysis (Fig. 1D). dRNA-seq successfully identified all the Ccn sRNAs, including the four that were not detected in our sRNA-seq analysis. Moreover, dRNA-seq also detected the presence of four out of seven sRNAs (Spd_sr10, Spd_sr37, Spd_sr38, and Spd_sr39) that were previously validated in D39W (27) but not detected by our sRNA-seq analysis. Altogether, dRNA-seq identified 44 novel sRNA candidates in D39W, of which 16 sRNAs have been validated for expression (Fig. S4).
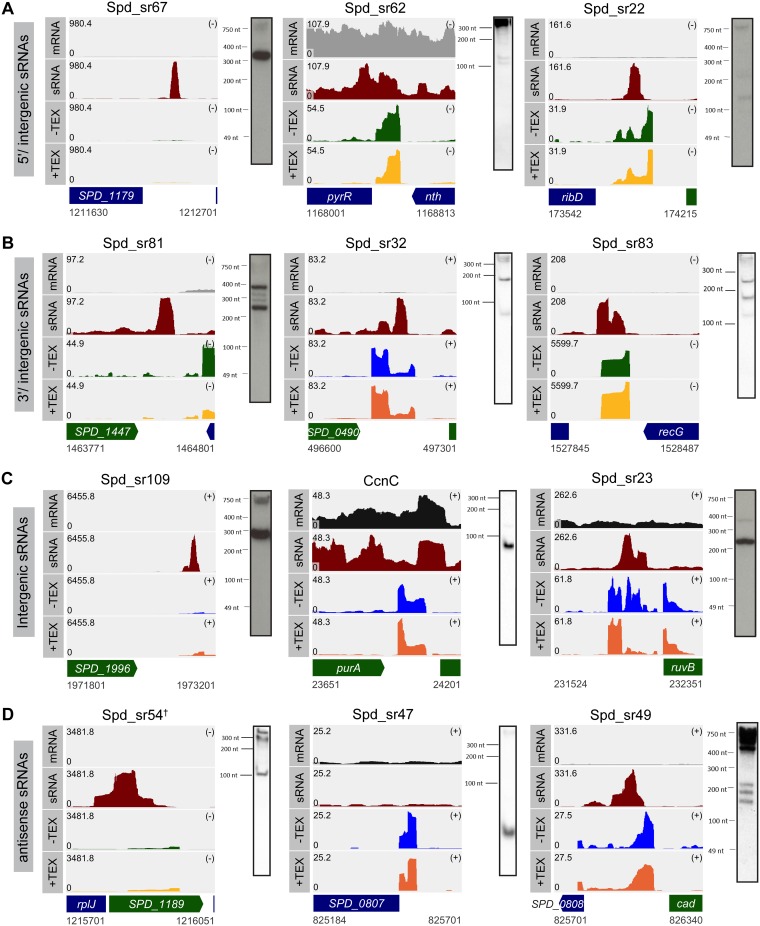
Based on our sequencing data, we can successfully classify the 52 sRNAs identified via our dRNA-seq analysis into different categories as described in the legend of Fig. 1A. Out of 52 sRNAs, 6 sRNAs were 5′/intergenic, 12 were 3′/intergenic, 14 were intergenic, and 10 were antisense sRNAs. The remaining 10 sRNAs were classified under the category called 5′/intergenic and 3′/intergenic sRNAs (Fig. 1C and Table 1), as the reads were mapped within 100 bp of both upstream and downstream genes. Taken together, sRNA-seq and dRNA-seq raised the total number of sRNAs in D39W to 112, which includes 3 sRNAs (Spd_sr7, Spd_sr10, and Spd_sr52) that were not identified by sRNA-seq or dRNA-seq analysis but that were previously validated in D39W (27). Representative examples of sRNAs that are classified as either 5′/intergenic, 3′/intergenic, intergenic, or antisense sRNAs are shown in Fig. 2A to D, respectively, along with corresponding Northern blots that independently validated their existence. Furthermore, Fig. 2 additionally includes examples of sRNAs in each of the above-mentioned categories that were detected only by sRNA-seq (Spd-sr67, Spd-sr81, Spd-sr109, and Spd-sr54) or dRNA-seq (Spd-sr62, Spd-sr32, CcnC, and Spd-sr47) or detected by both sRNA-seq and dRNA-seq (Spd-sr22, Spd-sr83, Spd-sr23, and Spd-sr49).
FIG 2.
Examples of sRNAs transcribed from different genetic contexts in D39W. Shown are read coverage maps of sRNAs identified in each of the following categories: 5′/intergenic (A), 3′/intergenic (B), intergenic (C), and antisense (D). The left panels show examples of sRNAs that were identified only by sRNA-seq. The middle panels show examples of sRNAs that were identified only by dRNA-seq, and the right panels show examples of sRNAs that were identified by both sRNA-seq and dRNA-seq analyses. The corresponding Northern blots for the each sRNA identified by RNA-seq analysis are shown. Tracks labeled mRNA, sRNA, −TEX, and +TEX correspond to the read coverages for the sRNAs and their flanking regions in the wild-type (WT) strain that were obtained from mRNA-seq, sRNA-seq, and mock-treated and TEX-treated dRNA-seq samples, respectively. Coverage represents depth per million reads (mRNA and sRNA) of paired-end fragments (−TEX and +TEX) and was averaged between normalized replicates (see Materials and Methods). In each coverage graph, ORFs encoded on the plus and the minus strands are color-coded in green and blue, respectively. † indicates that the sRNA can also be detected by dRNA-seq analysis, but the read coverage obtained for Spd-sr54 by sRNA-seq analysis is >10-fold higher than that obtained by dRNA-seq analysis. Probes used are listed in Table S5 in the supplemental material.
Using dRNA-seq, we can classify a total of 71 sRNAs as primary transcripts, containing primary transcription start sites (pTSSs), and 26 sRNAs consisting of processed transcripts containing an alternative/secondary transcription start site (sTSS). As shown in Fig. 1E, ∼80% and 92% of the sRNAs that are classified under the 5′/intergenic and intergenic sRNA categories, respectively, possess a pTSS, in contrast to those falling under the category of 3′/intergenic sRNAs, in which 58% are primary transcripts. Taken together, we have detected the presence of a total of 109 sRNAs in this study (Table 1), of which 60 sRNAs have been validated by Northern blot analysis for expression (Fig. 2 and 3, Fig. S3 and S4, and Table 1) and 97 sRNAs have been classified as primary or processed transcripts (Fig. 1E).
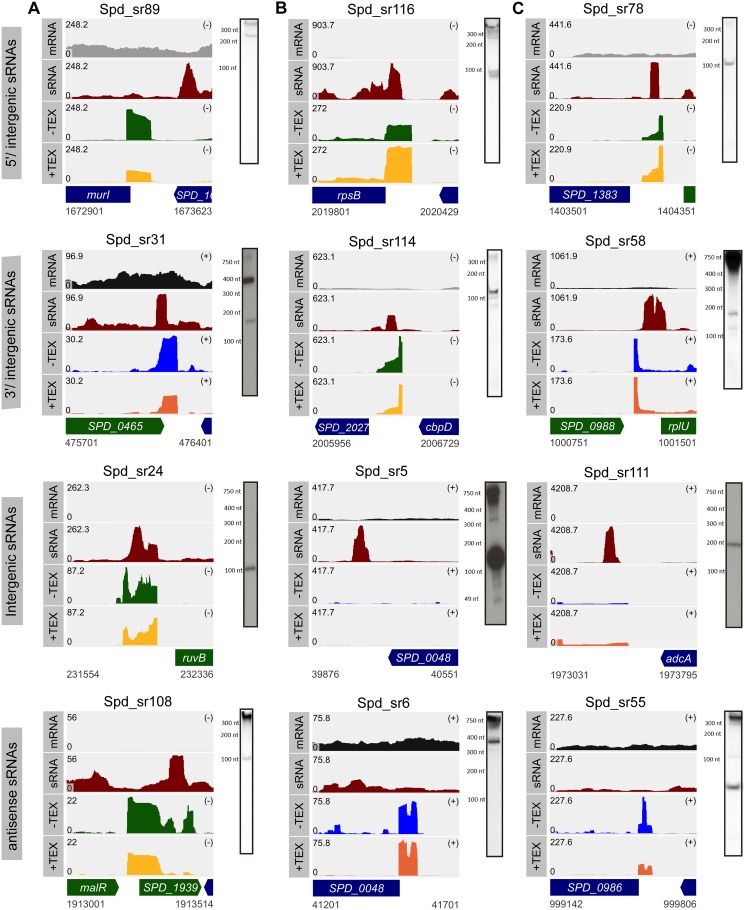
FIG 3.
Combined data from sRNA-seq and dRNA-seq analyses increase the sensitivity of sRNA detection in D39W. Shown are read coverage maps of sRNAs classified as 5′/intergenic, 3′/intergenic, intergenic, or antisense that were detected only in D39W by sRNA-seq and/or dRNA-seq analysis in this study but not previously detected in D39V (28) and TIGR4 (3, 30) (A), identified in both D39W in this study and D39V in another study (28) but not previously detected in TIGR4 (3, 30) (B), and identified in both D39W in this study and previously in TIGR4 (3, 30) but not in D39V by Slager et al. (28) (C). Corresponding Northern blots detecting the sRNAs identified by RNA-seq analysis are presented alongside the read coverage maps. Track labels corresponding to the read coverage maps are described in the legend to Fig. 2. Probes used are listed in Table S5 in the supplemental material.
Identification of antisense RNAs in S. pneumoniae D39.
Our RNA-seq analysis led to the identification of regions exhibiting antisense expression. These different regions can be classified broadly into three different categories: antisense sRNAs, short antisense RNAs (transcripts corresponding to antisense over the entire length of a known ORF), and long antisense RNAs (transcripts corresponding to antisense to an operon consisting of more than one ORF). We have identified the presence of 17 asRNAs (Table 1). Eleven of these asRNAs (Spd-sr6, Spd-sr13, Spd-sr20, Spd-sr38, Spd-sr39, Spd-sr42, Spd-sr47, Spd-sr49, Spd-sr54, Spd-sr55, and Spd-sr108) have been tested for expression and have been validated by Northern blotting (Table 1, Fig. 2 and 3, and Fig. S3 and S4). The antisense sRNAs were identified as regions showing significantly higher expression levels than the background on the opposite strand of an ORF. Next, we determined what type of genes exhibited the presence of short antisense transcripts. Out of 44 ORFs that showed the presence of significant antisense expression, 13 ORFs encoded transposon elements, 3 ORFs were annotated as pseudogenes, 18 ORFs encoded hypothetical proteins, and the remaining 10 ORFs encoded proteins that can be assigned to a variety of functional categories (Table S4). Long antisense transcripts were identified for the following gene clusters, which constitute operon structures: (i) spd_0223 through spd_0225 (spd_0223-spd_0225), (ii) spd_0413-spd_0416, (iii) spd_0470-spd_0472 (blpCBA), (iv) spd_0615-spd_0618, (v) spd_0638-spd_0639, (vi) spd_1213-spd_1214, (vii) spd_1452-spd_1456, and (viii) spd_1628-spd_1629 (xpt-pbuX). Interestingly, spd_0413-spd_0416 constitute pseudogenes, and spd_0638-spd_0639 encode transposase family proteins. The other operons listed above encode putative ABC transporters or conserved hypothetical proteins of unknown function (Table S4). Thus, this study revealed the presence of significant antisense transcription for those regions of the D39W genome that contain transposon elements (functional and truncated versions) and pseudogenes.
Comparative analysis of the sRNAs identified in D39W and D39V.
A recent study by Slager et al. (28) predicted a total of 34 sRNAs in D39V, of which 32 were detected by their RNA-seq analysis in D39V; therefore, we next determined how many of the sRNAs that we identified overlapped those identified previously (28). Our sRNA-seq or dRNA-seq analyses uncovered 25 of the 32 sRNAs previously detected in D39V. These include eight sRNAs highly conserved among S. pneumoniae strains (CcnA, CcnB, CcnC, CcnD, CcnE, scRNA, RnpB, and SsrA) and the highly expressed sRNA Spd_sr85, which we validated by Northern blotting (Fig. S3). We also validated another seven of these sRNAs that overlapped those of the previous study but were never independently verified (Spd_sr5, Spd_sr6, Spd_sr23, Spd_sr42, Spd_sr83, Spd_sr106, and Spd_sr116) (Fig. S3 and S4). In addition to the 32 sRNAs that were identified in D39V, there were two additional sRNAs that were predicted to be present in D39 by Slager et al. (28). Our analysis independently identified one of those two sRNAs in D39W (Spd-sr110 in D39W and Srf-27 in D39V) (Fig. S5). The genomic sequences corresponding to the eight sRNAs (Srf-01, Srf-06, Srf-08, Srf-12, Srf-16, Srf-22, Srf-26, and Srf-29) that were identified via RNA-seq analysis in D39V but were not independently validated by Slager et al. (28) are present in D39W; however, we were not able to detect these sRNAs by our RNA-seq-based approaches. Slager et al. also reported the presence of 24 riboswitches in D39V, of which 22 candidates were successfully identified by RNA-seq analysis in their study (28). We also detected by RNA-seq analysis 15 of the 22 riboswitch elements elucidated by Slager et al. (28), which we have denoted sRNAs (Table S1). In some cases, riboswitches have been found to also function as small regulatory RNAs (9, 10, 20). We have further validated the expression of 10 out of the 15 riboswitch sRNAs in D39W by Northern blotting (Fig. S3 and S4).
Thus, nearly 71% of the putative sRNAs or riboswitches (41 out of 58) uncovered by Slager et al. (28) in D39V overlapped those of our sequencing analysis (Fig. 4A). The 17 remaining sRNAs reported to be present in D39V had not been validated by Northern blot analysis. Our analysis detected another 71 sRNAs in D39W, which were not detected in that previous study (28). We validated the expression of 36 of these new candidate sRNAs in D39W (Table 1 and Fig. S3 and S4). Representative coverage graphs and Northern blot validations for examples of sRNAs that were identified only in D39W or in both D39W and D39V (28) in each of the 4 categories (5′/intergenic, 3′/intergenic, intergenic, and antisense) are shown in Fig. 3A and B, respectively. A majority of the sRNAs that were common between D39W and D39V were classified as intergenic sRNAs, followed by those in the 5′/intergenic category (Fig. 4B).
FIG 4.
Differences in sRNA contents between S. pneumoniae strains D39 and TIGR4. (A) Venn diagram illustrating the overlap between the total numbers of sRNAs identified by RNA-seq in D39W and D39V. (B) Numbers of sRNAs in each category (5′/intergenic, 3′/intergenic, intergenic, or antisense) that were detected in D39W in this study and in D39V by Slager et al. (28). (C) Distribution of sRNAs that were identified in TIGR4 (3, 30), D39V (28), and D39W in this study and their genetic contexts. (D) Overlap of sRNAs identified in TIGR4 and D39W in this study. TIGR4-1 refers to sRNAs identified by Mann et al. (3), and TIGR4-2 refers to sRNAs identified by Acebo et al. (30). Only TIGR4 sRNAs that are also encoded in the genome of D39W are included in this analysis. The number of sRNAs in D39W represents the total number of sRNAs that were identified by sRNA-seq and dRNA-seq in this study. (E and F) Read coverage maps for the TIGR4 sRNAs F14 and F23, which are present in D39W genomes but were not detected as sRNAs by sRNA-seq or dRNA-seq analysis. Northern blots probed for F14 and F23 in D39W (lane 1) and TIGR4 (lane 2) are shown alongside the read coverage maps. (G to J) Read coverage maps for D39W sRNAs Spd-sr71, Spd-sr84, Spd-sr96, and Spd-sr106, which were validated for expression in D39W but were not detected as sRNAs by RNA-seq analysis of TIGR4. Northern blots probed for Spd-sr71, Spd-sr84, Spd-sr96, and Spd-sr106 in D39W (lane 1) and TIGR4 (lane 2) are shown alongside the read coverage maps. Track labels corresponding to coverage graphs are described in the legend of Fig. 2. Probes used are listed in Table S5 in the supplemental material.
Comparison of sRNA expression profiles in D39 and TIGR4.
TIGR4 is the serotype 4 strain of S. pneumoniae that has been subjected to extensive genome-wide analysis for identifying sRNAs, and collectively, 178 putative sRNAs were detected in this strain (3, 30, 31). Next, we determined how well the newly identified sRNAs in our study overlapped those identified in TIGR4. We chose the sRNA data sets from Mann et al. (3) and Acebo et al. (30), since both studies used RNA-seq-based approaches to identify sRNAs. We next mapped the sRNAs that were identified by the above-mentioned two studies onto the D39W genome by using BLAST version 2.2.26 with a postprocessing filter with the requirement that the mapped region had a 90% sequence identity over at least 90% of the length of the TIGR4 region. A total of 148 sRNAs were identified in TIGR4 by Mann et al. (3) and Acebo et al. (30), of which 26 sRNAs were common between the two studies. The conditions under which RNA was isolated as well as the methods used to prepare total RNA for sRNA library preparations differed between the above-mentioned two studies. For example, Mann et al. (3) extracted RNA from late-exponential-phase cultures grown in C+Y medium, which was subsequently processed to enrich for sRNA fragments of <200 nt in length. On the other hand, Acebo et al. (30) enriched for RNAs of 100 to 400 nt in length from total RNA extracted from cells grown in Todd-Hewitt broth supplemented with yeast extract (THY medium) to early exponential phase. Furthermore, RNA sequencing by Mann et al. was performed using an Illumina platform (3), while Acebo et al. (30) used 454 pyrosequencing. These differences, along with differences in the analysis methods to predict sRNAs implemented by Mann et al. (3) and Acebo et al. (30), may have contributed to the identification of unique sRNA data sets in TIGR4. Interestingly, sequences corresponding to 24 sRNAs that were identified in TIGR4 by the previous two sequencing studies were not found in the D39W genome (Table S2). We subsequently determined which of the remaining 124 putative sRNAs overlapped the 109 sRNAs that we detected in D39W by our RNA-seq-based approach; we considered sRNAs to be common between D39W and TIGR4 if their sequences overlapped by at least 40 bp and had an E value cutoff of 0.001. Out of the remaining 124 putative sRNAs in TIGR4, 37 sRNAs were identified in D39W by our sRNA-seq or dRNA-seq approach (Fig. 4D and Table 1). Twelve of these 37 sRNAs were not previously identified by RNA-seq analysis in D39V (28), bringing the total number of sRNAs in common between all TIGR4 and D39 analyses to 25 (Fig. 4C). Of these 12 candidates, we validated 8 in D39W by Northern blotting (Fig. S3 and S4). In Fig. 3C, the mapped reads and representative Northern blots of sRNAs transcribed from four different genetic contexts that were identified in both D39W and TIGR4 but were not previously uncovered in D39V (28) are shown.
We also examined by Northern blotting the expression of two sRNAs, F14 and F23, which are encoded in the genomes of strains D39 and TIGR4 but were identified only in TIGR4 (3). We could not detect the presence of these two sRNAs in D39 but instead detected a longer transcript of >1,000 nt in length (Fig. 4E and F). This observation indicates that the sRNAs F14 and F23 are part of a longer transcript, which is why we did not identify these RNAs as sRNAs. In parallel, we tested for the expression of sRNAs F14 and F23 in TIGR4, where the predicted sizes are 120 nt and 143 nt, respectively. Surprisingly, we also could not detect any such RNA fragment in TIGR4 (Fig. 4E and F). Finally, 71 of the 72 sRNAs that we detected in D39W but that were not previously identified in TIGR4 (Fig. 4D) have corresponding DNA sequences in the TIGR4 genome; only Spd-sr65 is not encoded in the TIGR4 genome (Table 1). To determine whether these sRNA genes conserved between D39 and TIGR4 are also transcribed as sRNAs in TIGR4, we isolated RNA from cells from TIGR4 cultures grown under the same conditions as those that we used to grow D39 cultures, i.e., to an OD620 of 0.15 to 0.20 in BHI medium at 37°C under an atmosphere of 5% CO2; we subsequently tested for the expression of four sRNAs (Spd-sr71, Spd-sr84, Spd-sr96, and Spd-sr106) that were highly expressed in D39W under these growth conditions. Northern blot analysis revealed that sRNAs Spd-sr71 and Spd-sr96 are expressed in both D39W and TIGR4 (Fig. 4G and I). Spd-sr106 exhibited a slightly different banding pattern in TIGR4 than in D39, where the RNA fragment corresponding to 200 nt was absent but the ∼150-nt band was present (Fig. 4J). Spd-sr84 was more abundant in D39W than in TIGR4 (Fig. 4H); however, this apparent difference in abundance could be due to a reduced affinity of the probe, which binds a region of the sRNA that is slightly variable between D39 and TIGR4. Based on these results, we predict that most if not all of the 71 sRNA genes that we identified in D39 and that are found in the TIGR4 genome are expressed as sRNAs in TIGR4. Altogether, we have validated by Northern blot analysis 36 out of these 71 sRNA candidates in D39W (Fig. S3 and S4).
DISCUSSION
Advances in RNA sequencing technologies over the last few years have accelerated the discovery of sRNAs in a variety of different bacteria, including bacterial pathogens (34, 35). The identification of novel sRNAs in bacterial pathogens has opened up new avenues of study for virulence gene regulation (36, 37). In this study, we have successfully identified 109 sRNAs in the human pathogen S. pneumoniae D39W using a novel RNA-seq data analysis approach. We have identified 66 new sRNAs in D39 in this study, and we have successfully detected all 60 of the sRNAs in D39 that we tested for by Northern blot analysis, which further validates the robustness of our newly developed computational analysis method. Furthermore, we have successfully determined the TSS characteristics of 87% of the sRNA candidates. Finally, we have performed a comparative analysis between the sRNA data sets in D39 and TIGR4, and our analysis reveals differences in the sRNA transcriptomes between these strains, which may contribute to the underlying differences in virulence properties that are exhibited by the two different strains.
Differences in sRNA contents between S. pneumoniae strains D39 and TIGR4.
This study reveals differences in the sRNA repertoires between the two strains TIGR4 and D39. We can map ∼85% of the sRNAs identified in TIGR4 by RNA-seq analyses onto the D39 genome. Moreover, ∼72% of the sRNAs (Fig. 4C and D) identified in this study were not previously identified in TIGR4, even though the corresponding sequences for 99% of these sRNAs are present in the TIGR4 genome (Table 1 and Fig. 4D). However, we detected both in TIGR4 and D39 the presence of several of the sRNAs that were identified in D39W but not in TIGR4 by high-throughput sequencing studies (Fig. 4G to J). Interestingly, we could not detect F14 and F23 in D39W or TIGR4, although these sRNAs were identified by Mann et al. (3) in TIGR4; instead, we detected a much larger transcript (Fig. 4E and F). Moreover, F14 and F23 are examples of two sRNAs whose DNA sequences are present in both serotypes but were not identified in D39 by our analysis. This apparent discrepancy between studies in which sRNAs are identified via RNA-seq analysis highlights the importance of verification of the existence and size of transcripts identified by these approaches via Northern blotting. While we cannot rule out the possibility that some sRNAs that were detected in TIGR4 but not in our study are an artifact, the observed differences in the sRNA repertoires between D39 and TIGR4 may be attributed to the differences in growth conditions and technical issues such as differences in RNA isolation, library construction, and/or sequencing methodology. Interestingly, recent reports have indicated that the Gram-negative human opportunistic pathogen Pseudomonas aeruginosa as well as the Gram-positive pathogen Staphylococcus aureus exhibit strain-specific differences in their sRNA contents (38, 39).
Functional classification of D39W sRNAs.
In order to gain insight into the potential roles of the D39W sRNAs in gene regulation, we classified them into different functional categories. We defined sRNAs as 5′/intergenic, 3′/intergenic, intergenic, and antisense. We observed that 40% of the total predicted sRNAs in S. pneumoniae constituted leader sequences or can be classified as 5′/intergenic sRNAs, the majority of which consisted of a pTSS (Fig. 1B to D). Spd_sr48, Spd_sr54, and Spd_sr55 were identified as L20, L10, and L21 leader regions, respectively, which constitute a family of autoregulatory structures commonly found in the 5′ UTRs of mRNAs encoding ribosomal proteins (40). In low-GC-content Gram-positive bacteria, L20 leaders constitute ribosomal protein leader autoregulatory structures and are present upstream of the operon infC-rpmL-rplT encoding translation initiation factor 3 and ribosomal proteins L35 and L20, respectively. Ribosomal proteins are autoregulated by their specific leader sequences, and only a specific ribosomal protein can recognize the RNA secondary structure of its leader and inhibit its own translation (41). Thus, Spd-sr48, Spd-sr54, and Spd-sr55 appear to be cis-acting regulatory elements controlling the expression of downstream genes but could have a second life as sRNAs.
Fifteen of the sRNAs that we identified can be classified as riboswitch RNAs and possess riboswitch regulatory elements (Table 1; see also Table S3 in the supplemental material). Riboswitches are another class of cis-regulatory elements, which are present in the 5′ leader sequences of genes and can fold to form secondary structures that change conformation upon binding small molecules. Riboswitches can sense and respond to the availability of various different metabolites and environmental signals, including stalled ribosomes, uncharged tRNAs, elevated temperatures, and small ligands (42). A riboswitch has been previously shown to function as both a riboswitch and an sRNA (20). Spd-sr43, Spd-sr44, and Spd-sr114 were identified as thiamine PPi (TPP) riboswitch RNAs. TPP riboswitches are highly conserved RNA elements in the 5′ UTRs of certain mRNAs which can directly bind to thiamine pyrophosphate or vitamin B1, which is an essential cofactor for several important enzymes and is known to regulate the expression of downstream genes (43). Interestingly, Spd-sr44 is present in the 5′ UTR of the three-gene operon spd_0622-thiM-thiE1, where the last two genes encode enzymes involved in thiamine biosynthesis. Likewise, Spd-sr22 was identified as a reduced flavin mononucleotide (FMN) riboswitch and is present upstream of the ribDEBH operon encoding proteins involved in riboflavin biosynthesis, consistent with other FMN riboswitches, which are typically present in the 5′ UTRs of genes encoding FMN biosynthesis and transport proteins (44, 45). Spd-sr28 was also identified in D39V as an sRNA and was classified as a glycine riboswitch. A total of eight sRNAs (Spd-sr29, Spd-sr32, Spd-sr34, Spd-sr12, spd-sr70, Spd-sr74, Spd-sr80, and Spd-sr88) can be classified as T-box riboswitches, which consist of a cis-regulatory element controlling the expression of amino-acid-related genes by sensing the aminoacylation state of a specific tRNA (46). Accordingly, Spd-sr29, Spd-sr32, and Spd-sr70 are present in the 5′ UTRs of serS (the gene encoding seryl-tRNA synthetase), pheS (gene encoding phenylalanine-tRNA synthetase), and spd_1216 (gene encoding alanyl-tRNA synthetase), respectively, in D39.
Interestingly, 35% of the predicted sRNAs overlap 3′ UTRs of genes. There are many examples now of sRNAs that overlap the 3′ UTRs of genes that have regulatory functions. For example, CcnA, the best-characterized small regulatory RNA in S. pneumoniae (27, 47), can be produced as part of the 3′ UTR of spd_0240, although our dRNA-seq data suggest that much of it is produced as an independent transcript from its own distinct promoter. A recent deep-sequencing study in Salmonella enterica serovar Typhimurium identified 3′ UTRs as reservoirs of regulatory sRNAs (22). SroC and CpxQ in S. Typhimurium and MicL in E. coli, which are encoded in the 3′ UTRs of mRNAs, have been characterized as sRNAs that regulate gene expression by base pairing with target mRNAs (47–49).
Two of the housekeeping sRNAs, SsrA (Spd_sr46) and 6S RNA (Spd_sr85), were identified in this study. These sRNAs are known to be processed into functional RNA molecules. SsrA is involved in trans-translation control, where it rescues ribosomes from stalled translation complexes (50). 6S sequesters the σA-bound RNA polymerase holoenzyme, blocking it from binding target promoters (11, 50, 51). Spd-sr46, Spd-sr34, Spd-sr49, Spd-sr88, and Spd-sr64 correspond to F32, F20, srn235, srn400, and F44 sRNAs that are present in TIGR4. All of these sRNA mutants in TIGR4 exhibited altered fitness during lung infection in mice (3). sRNA mutants corresponding to the D39W sRNAs Spd-sr34, Spd-sr17, and Spd-sr111 were defective in colonizing the nasopharynx of mice in TIGR4 (3). Additionally, the sRNA mutants corresponding to D39W genes encoding sRNAs Spd-sr44 and Spd-sr42 showed a fitness defect for replicating in the bloodstream of mice (3). Furthermore, deletion mutants of four TIGR4 sRNAs corresponding to the D39W sRNAs Spd-sr34, Spd-sr46, Spd-sr64, and CcnE were shown to be attenuated for virulence in a murine model of invasive pneumonia (3).
Finally, we successfully detected the three type I toxin-antitoxin systems in D39W, containing three distinct toxin modules, Spd-sr23, Spd-sr109, and Spd-sr111, and their corresponding antitoxins, Spd-sr24, Spd-sr110, and Spd-sr112, respectively. The sRNAs identified in this study that can be classified into different functional categories are listed in Table S3.
Altogether, we used RNA-seq and dRNA-seq to redefine the sRNA repertoire in the D39W serotype 2 strain of S. pneumoniae. This analysis led to the identification of numerous new sRNAs (Table 1) and antisense RNAs (Table S4), whose physiological relevance can now be determined in future experiments. In addition, we evaluated the benefits and pitfalls associated with this technique and show that the sRNA contents between strains of at least two different serotypes of S. pneumoniae are distinct.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
IU1781 (wild-type), an rpsL1 derivative of the S. pneumoniae strain D39W (serotype 2), was used in this study. This strain exhibits wild-type phenotypes but possesses a mutation in the rpsL gene and is routinely used as the parental strain to create allelic replacements for constructing mutant strains (52, 53). Liquid cultures of IU1781 were grown statically in BD Difco brain heart infusion (BHI) broth at 37°C in an atmosphere of 5% CO2. For cultures grown overnight, strains were inoculated from freezer stocks into tubes containing 5 ml of BHI broth, and 100-fold serial dilutions were then performed. The serially diluted cultures were grown for 10 to 16 h. Cultures with an optical density at 620 nm (OD620) of 0.1 to 0.4 were subcultured to a starting OD620 of 0.002 in fresh BHI broth and grown to the exponential growth phase. Growth was monitored by measuring the OD620 using a Spectronic 20 spectrophotometer.
RNA isolation.
To isolate RNA for mRNA and sRNA sequencing, strains were grown in 30 ml of BHI broth starting at an OD620 of 0.002 in 50-ml conical tubes. RNA was subsequently extracted from exponential-growth-phase cultures (OD620 of ∼0.15) using the FastRNA Pro Blue kit (MP Biomedicals) according to the manufacturer’s guidelines. Briefly, cells were collected by centrifugation at 14,500 × g for 5 min at 4°C. After culture supernatants were discarded, cell pellets were suspended in 1 ml of RNApro solution (MP Biomedicals), added to lysis matrix B (MP Biomedicals), and processed three times in the Fast Prep instrument (MP Biomedicals) for 40 s at a speed setting of 6.0. Cell debris and the lysing matrix were pelleted by centrifugation at 16,000 × g for 5 min at 4°C, and the supernatant was placed in a new tube. A total of 300 μl of chloroform was added to the supernatant, which was vortexed for 10 s. After incubation at room temperature for 5 min, the aqueous and organic phases of each sample were separated by centrifugation at 16,000 × g for 5 min at 4°C. The aqueous phase was placed in a new tube, 0.5 volumes of 100% ethanol were added, and the samples were then immediately loaded onto an miRNeasy column. On-column DNase I (Qiagen) treatment was performed and RNA was subsequently purified according to the manufacturer’s instructions for the miRNeasy minikit (Qiagen). The amount and purity of all RNA samples isolated were assessed by NanoDrop spectroscopy (Thermo Fisher). The RNA integrity of the samples used for RNA-seq library preparation was further assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies).
To prepare RNA for differential RNA-seq (dRNA-seq), cultures were grown and RNA extraction was performed as mentioned above, with the following modifications. Cell pellets were suspended in 1 ml of RNApro solution and processed five times in the Beadbug homogenizer (Benchmark Scientific) for 40 s at a speed setting of 4,000 rpm. After removal of the lysing matrix and cell debris by centrifugation and separation of the organic and aqueous phases by chloroform addition, vortexing, and centrifugation, RNA was ethanol precipitated from the aqueous phase. After suspension in diethyl pyrocarbonate (DEPC)-treated water, the samples were subjected to DNase treatment (DNase Turbo; Ambion) according to the manufacturer’s protocol. Sample mixtures (total reaction mixture volume of 100 μl) were incubated for 1 h at 37°C, and the reaction was stopped by the addition of 100 μl of DEPC-treated water and 200 μl of neutral phenol-chloroform-isoamyl alcohol (Fisher Scientific). DNase-treated RNA samples were phenol extracted and alcohol precipitated, and the RNA concentration was measured as described above.
Library preparation and sequencing for mRNA-seq.
cDNA libraries were prepared from total RNA by the University of Wisconsin—Madison Biotechnology Center. mRNA was enriched from 2 μg total RNA using a RiboZero rRNA removal (Gram-positive bacteria) kit (Illumina). rRNA-depleted mRNA samples were purified by ethanol precipitation and quantified by fluorometry with the Qubit RNA assay kit (Invitrogen). Double-stranded cDNA synthesis was performed according to the ScriptSeq v2 RNA-seq library preparation guide (Epicentre) in accordance with the manufacturer’s standard protocol. Thirty nanograms of enriched mRNA was fragmented using divalent cations via incubation for 5 min at 85°C. The first strand of cDNA was synthesized by reverse transcription using random-sequence primers containing a tagging sequence at their 5′ ends. Ditagged cDNA was synthesized by random annealing of a terminal-tagging oligonucleotide (TTO) to the 3′ end of the cDNA for extension of the cDNA by DNA polymerase. Ditagged cDNA was purified using Agencourt AMPure XP beads (Beckman Coulter) followed by PCR amplification for 15 cycles using Failsafe PCR enzyme and the ScriptSeq index DNA primer set (Epicentre). This step generated the second strand of cDNA and completed the addition of Illumina adapter sequences incorporating a user-defined barcode. The amplified libraries were purified using Agencourt AMPure XP beads. Quality and quantity were assessed using an Agilent DNA 1000 chip (Agilent Technologies) and a Qubit dsDNA HS assay kit (Invitrogen), respectively. Libraries were standardized to 2 μM. Cluster generation was performed using standard Cluster kits (v3) and an Illumina cluster station. Single-end 100-bp sequencing was performed using standard SBS chemistry (v3) on an Illumina HiSeq2000 sequencer. Images were analyzed using the standard Illumina pipeline, version 1.8.2.
Library preparation and sequencing for sRNA-seq.
Small RNA libraries were created according to Illumina’s TruSeq stranded RNA sample preparation (Rev.C) guide and using the Illumina TruSeq stranded RNA kit (Illumina Inc.), with modifications. cDNA was synthesized as described above. Following synthesis of the double-stranded cDNA, samples were extracted in equal volumes of phenol-chloroform-isoamyl alcohol (25:24:1), and the cDNA was ethanol precipitated. cDNAs were adenylated with a single “A” base, followed by ligation of an adapter. The adapter-ligated material was purified using Agencourt AMPure XP beads (Beckman Coulter). Adapter-ligated cDNA was amplified by linker-mediated PCR (LM-PCR) for 15 cycles and then size selected by gel electrophoresis using 6% Tris-borate-EDTA (TBE) gels (Invitrogen) targeting 145- to 500-bp fragments. Excised gel fragments were macerated using “gel breaker” tubes. cDNA was eluted from the gel debris with DNA storage solution in 1.5-ml tubes with rotation, isolated via an acetate column, and then ethanol precipitated. The quality and quantity of the finished libraries were assessed using an Agilent high-sensitivity DNA chip (Agilent Technologies) and a Qubit dsDNA HS assay kit (Invitrogen), respectively. Libraries were standardized to 2 μM. Cluster generation was performed using standard cluster kits (v3) and the Illumina cluster station. Single-end 50-bp sequencing was performed using standard SBS chemistry (v3) on an Illumina HiSeq2000 sequencer. Images were analyzed using the standard Illumina pipeline, version 1.8.2.
Library preparation and sequencing for dRNA-seq.
Five micrograms of DNase-treated RNA was subjected to rRNA removal (RiboZero rRNA removal for Gram-positive bacteria; Illumina). rRNA-depleted samples were either treated with terminator 5′-phosphate-dependent exonuclease (+TEX) (Epicenter) or mock treated (−TEX) according to the manufacturer’s guidelines. TEX-treated and mock-treated samples were alcohol precipitated, suspended in DEPC-treated water, and then subjected to RNA fragmentation using the Ambion RNA fragmentation kit (catalog number AM8740). Fragmented RNA was subjected to RNA 5′-polyphosphatase (Epicenter) treatment, which was performed to facilitate the 5′-adapter ligation step. Small RNA libraries were generated by Macrogen using a TruSeq small RNA library kit (Illumina). One-hundred-base-pair paired-end read sequencing was performed using an Illumina HiSeq2000 sequencer.
sRNA-seq data analysis.
The raw sequencing reads were quality and adapter trimmed using Trimmomatic version 0.17 (54) with a minimum length of 90 bp. The trimmed reads were mapped on the Streptococcus pneumoniae D39 genome (RefSeq accession number NC_008533) and the D39 plasmid pDP1 sequence (RefSeq accession number NC_005022) using Bowtie2 version 2.0.0-beta7 (55). Custom Python scripts (DROOM), which are available at https://github.com/dhritis/DROOM, were used to generate read counts for the genes and 100-bp nonoverlapping intergenic regions of the genome. Predicted sRNAs were identified as peaks in expression relative to expression in neighboring portions of the genome (see Fig. S6 in the supplemental material). For this approach, a window of 1,000 bp in length was used. The test region was defined as a 100-bp interval located in the center of this window. To help eliminate edge effects, the test area was flanked on either side by 50 bp of unused sequence. The eight flanking 100-bp intervals (400 bp total to each side) covering the outer edges of the window served to determine background expression. A Z-score for the test interval was determined based on the mean and standard deviation observed for the flanking intervals. This analysis was performed genome-wide, and Z-scores were calculated at 50-bp sliding intervals across the length of the genome. The analysis was performed in a strand-specific manner using a combination of the mRNA and sRNA expression data and using a non-strand-specific method with sRNA expression data from only the wild-type samples (IU1781). We chose the sRNA non-strand-specific method for our final predictions, as it correlated best with previous experimental results. The test regions were ranked by Z-scores, and the top 200 regions were tested manually by examining the RNA-seq data in JBrowse or IGV (data visualization browsers), and the predicted sRNAs were experimentally validated using Northern blotting.
dRNA-seq data analysis.
The raw sequencing reads were quality and adapter trimmed using Cutadapt (56). The trimmed reads were mapped onto the Streptococcus pneumoniae D39 genome (RefSeq accession number NC_008533) and the D39 plasmid pDP1 sequence (RefSeq accession number NC_005022) using Bowtie2 version 2.0.6 (55). To compare read coverage between different experiments, coverage of single-end reads (for unpaired data) or paired-end fragments (for paired-end data) across the genome was calculated using BedTools (57). Paired-end data were prefiltered using Samtools (58) and custom scripts so that only properly paired reads of fragments of less than 600 nt were considered. Read coverage was normalized per million reads or paired-end fragments and averaged between replicates for coverage graphs. Coverage graphs were generated in R using a custom script and the R packages ggplot2, GenomicRanges (59), GenomicAlignments (59), GenomicFiles, and rtracklayer (60).
Antisense expression.
To determine genome-wide antisense expression, DESeq (version 1.9.12)-normalized reads per kilobase per million (RPKM) values were determined for both the sense and the antisense strands of all annotated ORFs of S. pneumoniae D39. Next, we determined the ratios of the RPKM values obtained for the antisense and the sense strands. If the ratio of antisense RPKM versus sense RPKM was >2-fold and the ORF was annotated to be present on the sense strand, then we designated that region to be a region of significant antisense expression.
Northern blot analysis.
RNA samples (2 μg) were either loaded onto 6% or 8% polyacrylamide gels containing 7 M urea or loaded onto 10% Criterion TBE-urea precast gels (Bio-Rad) and electrophoresed at 120 V or 70 V, respectively. Next, the RNA samples were transferred to a Zeta-Probe GT membrane (Bio-Rad) using a Trans-Blot SD semidry transfer apparatus (Bio-Rad) according to the manufacturer’s guidelines. Transferred RNA was UV cross-linked and hybridized overnight with radiolabeled probes or 100 ng/ml of 5′-biotinylated probes (Table S5) in ULTRAhyb hybridization buffer (Ambion) at 42°C as described previously (27, 61).
For Northern blots probed with radiolabeled probes, T4 polynucleotide kinase (New England Biolabs) was used to end label 2 pmol of each synthetic DNA oligonucleotide probe with 1.67 pmol of [γ-32P]ATP (Perkin-Elmer) (6,000 Ci per mmol). Radiolabeled oligonucleotides were purified using Sephadex G-25 quick-spin columns (Roche). Labeled Northern blots were exposed to X-ray film to obtain an image and to a phosphor screen (Amersham) for 10 to 30 min for quantitation. The phosphor screen was scanned with a Typhoon 9200 variable-mode imager (Amersham), and quantitation of bands was performed with ImageQuant software (Molecular Dynamics). Blots that were hybridized with 5′-biotinylated DNA probes (100 ng/ml) were developed using the BrightStar BioDetect kit protocol (Ambion), imaged with a ChemiDoc MP imager (Bio-Rad), and quantified using Image Lab software version 5.2.1 (Bio-Rad).
Data availability.
Primary data from the RNA-seq and dRNA-seq analyses have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession numbers GSE124170 and GSE123437, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jiaqi Zheng, Tiffany Tsui, and other laboratory members for helpful discussions and input about this work.
This work was supported by McGovern Medical startup funds (to D.S., T.A.C., and N.R.D.L.) and NIGMS grants R01GM127715 and RO1GM128439 (M.E.W.).
The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00764-18.
REFERENCES
- 1.Caldelari I, Chao Y, Romby P, Vogel J. 2013. RNA-mediated regulation in pathogenic bacteria. Cold Spring Harb Perspect Med 3:a010298. doi: 10.1101/cshperspect.a010298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koo JT, Alleyne TM, Schiano CA, Jafari N, Lathem WW. 2011. Global discovery of small RNAs in Yersinia pseudotuberculosis identifies Yersinia-specific small, noncoding RNAs required for virulence. Proc Natl Acad Sci U S A 108:E709–E717. doi: 10.1073/pnas.1101655108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mann B, van Opijnen T, Wang J, Obert C, Wang Y-D, Carter R, McGoldrick DJ, Ridout G, Camilli A, Tuomanen EI, Rosch JW. 2012. Control of virulence by small RNAs in Streptococcus pneumoniae. PLoS Pathog 8:e1002788. doi: 10.1371/journal.ppat.1002788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori M-A, Soubigou G, Régnault B, Coppée J-Y, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 5.Sedlyarova N, Shamovsky I, Bharati BK, Epshtein V, Chen J, Gottesman S, Schroeder R, Nudler E. 2016. sRNA-mediated control of transcription termination in E. coli. Cell 167:111–121. doi: 10.1016/j.cell.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. 2002. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev 16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. 2009. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol 16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 8.Ramirez-Pena E, Trevino J, Liu Z, Perez N, Sumby P. 2010. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol Microbiol 78:1332–1347. doi: 10.1111/j.1365-2958.2010.07427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DebRoy S, Gebbie M, Ramesh A, Goodson JR, Cruz MR, van Hoof A, Winkler WC, Garsin DA. 2014. Riboswitches. A riboswitch-containing sRNA controls gene expression by sequestration of a response regulator. Science 345:937–940. doi: 10.1126/science.1255091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mellin JR, Koutero M, Dar D, Nahori MA, Sorek R, Cossart P. 2014. Riboswitches. Sequestration of a two-component response regulator by a riboswitch-regulated noncoding RNA. Science 345:940–943. doi: 10.1126/science.1255083. [DOI] [PubMed] [Google Scholar]
- 11.Trotochaud AE, Wassarman KM. 2005. A highly conserved 6S RNA structure is required for regulation of transcription. Nat Struct Mol Biol 12:313–319. doi: 10.1038/nsmb917. [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Lesnik EA, Hall TA, Sampath R, Griffey RH, Ecker DJ, Blyn LB. 2002. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems 65:157–177. doi: 10.1016/S0303-2647(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 13.Livny J, Brencic A, Lory S, Waldor MK. 2006. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res 34:3484–3493. doi: 10.1093/nar/gkl453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev 15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawano M, Reynolds AA, Miranda-Rios J, Storz G. 2005. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res 33:1040–1050. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. 2012. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J 31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melamed S, Peer A, Faigenbaum-Romm R, Gatt YE, Reiss N, Bar A, Altuvia Y, Argaman L, Margalit H. 2016. Global mapping of small RNA-target interactions in bacteria. Mol Cell 63:884–897. doi: 10.1016/j.molcel.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 19.Choi E, Han Y, Cho YJ, Nam D, Lee EJ. 2017. A trans-acting leader RNA from a Salmonella virulence gene. Proc Natl Acad Sci U S A 114:10232–10237. doi: 10.1073/pnas.1705437114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. 2009. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 21.Chao Y, Li L, Girodat D, Förstner KU, Said N, Corcoran C, Śmiga M, Papenfort K, Reinhardt R, Wieden H-J, Luisi BF, Vogel J. 2017. In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol Cell 65:39–51. doi: 10.1016/j.molcel.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chao Y, Vogel J. 2016. A 3′ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol Cell 61:352–363. doi: 10.1016/j.molcel.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 23.Lalaouna D, Carrier MC, Semsey S, Brouard JS, Wang J, Wade JT, Masse E. 2015. A 3′ external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise. Mol Cell 58:393–405. doi: 10.1016/j.molcel.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Smirnov A, Forstner KU, Holmqvist E, Otto A, Gunster R, Becher D, Reinhardt R, Vogel J. 2016. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc Natl Acad Sci U S A 113:11591–11596. doi: 10.1073/pnas.1609981113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henriques-Normark B, Tuomanen EI. 2013. The pneumococcus: epidemiology, microbiology, and pathogenesis. Cold Spring Harb Perspect Med 3:a010215. doi: 10.1101/cshperspect.a010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Opijnen T, Camilli A. 2012. A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res 22:2541–2551. doi: 10.1101/gr.137430.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsui HCT, Mukherjee D, Ray VA, Sham LT, Feig AL, Winkler ME. 2010. Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J Bacteriol 192:264–279. doi: 10.1128/JB.01204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slager J, Aprianto R, Veening J-W. 2018. Deep genome annotation of the opportunistic human pathogen Streptococcus pneumoniae D39. Nucleic Acids Res 46:9971–9989. doi: 10.1093/nar/gky725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halfmann A, Kovács M, Hakenbeck R, Brückner R. 2007. Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol Microbiol 66:110–126. doi: 10.1111/j.1365-2958.2007.05900.x. [DOI] [PubMed] [Google Scholar]
- 30.Acebo P, Martin-Galiano AJ, Navarro S, Zaballos A, Amblar M. 2012. Identification of 88 regulatory small RNAs in the TIGR4 strain of the human pathogen Streptococcus pneumoniae. RNA 18:530–546. doi: 10.1261/rna.027359.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar R, Shah P, Swiatlo E, Burgess SC, Lawrence ML, Nanduri B. 2010. Identification of novel non-coding small RNAs from Streptococcus pneumoniae TIGR4 using high-resolution genome tiling arrays. BMC Genomics 11:350. doi: 10.1186/1471-2164-11-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner GP, Kin K, Lynch VJ. 2013. A model based criterion for gene expression calls using RNA-seq data. Theory Biosci 132:159–164. doi: 10.1007/s12064-013-0178-3. [DOI] [PubMed] [Google Scholar]
- 33.Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery’s virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol 189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hör J, Gorski SA, Vogel J. 2018. Bacterial RNA biology on a genome scale. Mol Cell 70:785–799. doi: 10.1016/j.molcel.2017.12.023. [DOI] [PubMed] [Google Scholar]
- 35.Saliba A-E, Santos SC, Vogel J. 2017. New RNA-seq approaches for the study of bacterial pathogens. Curr Opin Microbiol 35:78–87. doi: 10.1016/j.mib.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Colgan AM, Cameron ADS, Kröger C. 2017. If it transcribes, we can sequence it: mining the complexities of host-pathogen-environment interactions using RNA-seq. Curr Opin Microbiol 36:37–46. doi: 10.1016/j.mib.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 37.García-Del Portillo F, Pucciarelli MG. 2017. RNA-Seq unveils new attributes of the heterogeneous Salmonella-host cell communication. RNA Biol 14:429–435. doi: 10.1080/15476286.2016.1276148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Broach WH, Weiss A, Shaw LN. 2016. Transcriptomic analysis of staphylococcal sRNAs: insights into species-specific adaption and the evolution of pathogenesis. Microb Genom 2:e000065. doi: 10.1099/mgen.0.000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gómez-Lozano M, Marvig RL, Molina-Santiago C, Tribelli PM, Ramos J-L, Molin S. 2015. Diversity of small RNAs expressed in Pseudomonas species. Environ Microbiol Rep 7:227–236. doi: 10.1111/1758-2229.12233. [DOI] [PubMed] [Google Scholar]
- 40.Zengel JM, Lindahl L. 1994. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog Nucleic Acid Res Mol Biol 47:331–370. doi: 10.1016/S0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
- 41.Johnsen M, Christensen T, Dennis PP, Fiil NP. 1982. Autogenous control: ribosomal protein L10-L12 complex binds to the leader sequence of its mRNA. EMBO J 1:999–1004. doi: 10.1002/j.1460-2075.1982.tb01284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henkin TM. 2008. Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev 22:3383–3390. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ontiveros-Palacios N, Smith AM, Grundy FJ, Soberon M, Henkin TM, Miranda-Ríos J. 2008. Molecular basis of gene regulation by the THI-box riboswitch. Mol Microbiol 67:793–803. doi: 10.1111/j.1365-2958.2007.06088.x. [DOI] [PubMed] [Google Scholar]
- 44.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res 30:3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winkler WC, Cohen-Chalamish S, Breaker RR. 2002. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci U S A 99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henkin TM. 2014. The T box riboswitch: a novel regulatory RNA that utilizes tRNA as its ligand. Biochim Biophys Acta 1839:959–963. doi: 10.1016/j.bbagrm.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnorpfeil A, Kranz M, Kovács M, Kirsch C, Gartmann J, Brunner I, Bittmann S, Brückner R. 2013. Target evaluation of the non-coding csRNAs reveals a link of the two-component regulatory system CiaRH to competence control in Streptococcus pneumoniae R6. Mol Microbiol 89:334–349. doi: 10.1111/mmi.12277. [DOI] [PubMed] [Google Scholar]
- 48.Guo MS, Updegrove TB, Gogol EB, Shabalina SA, Gross CA, Storz G. 2014. MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev 28:1620–1634. doi: 10.1101/gad.243485.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyakoshi M, Chao Y, Vogel J. 2015. Regulatory small RNAs from the 3′ regions of bacterial mRNAs. Curr Opin Microbiol 24:132–139. doi: 10.1016/j.mib.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 50.Giudice E, Macé K, Gillet R. 2014. trans-translation exposed: understanding the structures and functions of tmRNA-SmpB. Front Microbiol 5:113. doi: 10.3389/fmicb.2014.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wassarman KM, Storz G. 2000. 6S RNA regulates E. coli RNA polymerase activity. Cell 101:613–623. doi: 10.1016/S0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 52.Kazmierczak KM, Wayne KJ, Rechtsteiner A, Winkler ME. 2009. Roles of relSpn in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39. Mol Microbiol 72:590–611. doi: 10.1111/j.1365-2958.2009.06669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramos-Montañez S, Tsui H-CT, Wayne KJ, Morris JL, Peters LE, Zhang F, Kazmierczak KM, Sham L-T, Winkler ME. 2007. Polymorphism and regulation of the spxB (pyruvate oxidase) virulence factor gene by a CBS-HotDog domain protein (SpxR) in serotype 2 Streptococcus pneumoniae. Mol Microbiol 67:729–746. doi: 10.1111/j.1365-2958.2007.06082.x. [DOI] [PubMed] [Google Scholar]
- 54.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B. 2012. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40:W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lawrence M, Gentleman R, Carey V. 2009. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics 25:1841–1842. doi: 10.1093/bioinformatics/btp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sinha D, Matz L, Cameron T, De Lay NR. 2018. Poly(A) polymerase is required for RyhB sRNA stability and function in Escherichia coli. RNA 24:1496–1511. doi: 10.1261/rna.067181.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Primary data from the RNA-seq and dRNA-seq analyses have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession numbers GSE124170 and GSE123437, respectively.