Abstract
Using non-reducing Western blotting to assess protein thiol redox state is challenging because most reduced and oxidised forms migrate at the same molecular weight and are, therefore, indistinguishable. While copper catalysed Click chemistry can be used to ligate a polyethylene glycol (PEG) moiety termed Click PEGylation to mass shift the reduced or oxidised form as desired, the potential for copper catalysed auto-oxidation is problematic. Here we define a catalyst-free trans-cyclooctene-methyltetrazine (TCO-Tz) inverse electron demand Diels Alder chemistry approach that affords rapid (k ~2000 M−1 s−1), selective and bio-orthogonal Click PEGylation. We used TCO-Tz Click PEGylation to investigate how fertilisation impacts reversible mitochondrial ATP synthase F1-Fo sub-unit alpha (ATP-α-F1) oxidation—an established molecular correlate of impaired enzyme activity—in Xenopus laevis. TCO-Tz Click PEGylation studies reveal substantial (~65%) reversible ATP-α-F1 oxidation at evolutionary conserved cysteine residues (i.e., C244 and C294) before and after fertilisation. A single thiol is, however, preferentially oxidised likely due to greater solvent exposure during the catalytic cycle. Selective reduction experiments show that: S-glutathionylation accounts for ~50–60% of the reversible oxidation observed, making it the dominant oxidative modification type. Intermolecular disulphide bonds may also contribute due to their relative stability. Substantial reversible ATP-α-F1 oxidation before and after fertilisation is biologically meaningful because it implies low mitochondrial F1-Fo ATP synthase activity. Catalyst-free TCO-Tz Click PEGylation is a valuable new tool to interrogate protein thiol redox state in health and disease.
Keywords: Mitochondria, ATP synthase, Fertilisation, Development, Redox signalling
Graphical abstract

Highlights
-
•
Catalyst-free TCO-Tz Click PEGylation can assess protein thiol redox state.
-
•
ATP-α-F1 is substantially oxidised before and after fertilisation.
-
•
S-glutathionylation is the dominant oxidative modification type.
-
•
A single thiol is preferentially oxidised due to greater solvent exposure.
-
•
Catalyst-free TCO-Tz Click PEGylation is a valuable new tool.
1. Introduction
Much mitochondrial redox biology from apoptosis and oxidative phosphorylation to uncoupling can be ascribed to matrix superoxide anion (O2.-) and hydrogen peroxide (H2O2) [[1], [2], [3], [4], [5], [6], [7], [8]]. Matrix O2.-/H2O2 may signal via intermediate thiyl radicals and sulfenic acids (SOH) [[9], [10], [11], [12]], species that establish a chemical route to covalent oxidative modifications capable of altering protein function (i.e., disulphide bonds, S-glutathionylation and S-nitrosation [[13], [14], [15], [16], [17], [18], [19], [20]]). The emerging consensus from redox proteomic studies is that: reversible protein thiol oxidation regulates respiratory chain complex activity (reviewed in Ref. [21]). To give a key example, S-nitrosation of a single cysteine in ND3 (Cys39) protects against ischemia-reperfusion injury induced O2.- production by locking complex I in a structurally inactive state [[22], [23], [24]]. Despite their many advantages, redox proteomic approaches are often unsuitable for addressing many hypothesis-driven questions [25,26]. Using non-reducing Western blotting to address hypothesis-driven questions is challenging because the reduced and oxidised forms of most protein thiols possess similar electrophoretic mobility. Functionalising maleimide with a polyethylene glycol (PEG) moiety can impart a mass shift to the reduced or oxidised form [27]. The bulky PEG moiety may, however, sterically impede thiol labelling [26].
Recently, van Leeuwen and colleagues [26] described a novel Click chemistry approach (termed Click PEGylation) that bypasses steric concerns by making the thiol labelling step PEG independent. To detect oxidised protein thiols they exploited copper (Cu+) catalysed Click chemistry [28] wherein alkyne-maleimide is ligated to a low molecular weight (e.g., 5 kDa) azide-PEG moiety to form a stable triazole. The oxidised form migrates at a higher molecular weight compared with the reduced form owing to the azide-PEG moiety induced redox mobility shift, which is detectable by Western blotting [26]. However, the need for a cytotoxic catalyst that requires additional reductants (e.g., l-ascorbic acid) and stabilisers (e.g., tris [(1-benzyl-1H-1,2,3,-triazol-4-yl)methyl] amine) is problematic. For example, Cu+ could adventitiously auto-oxidise samples [29]. One can omit Cu+ entirely using catalyst-free trans-cyclooctene-methyltetrazine (TCO-Tz) pair inverse electron demand Diels Alder chemistry [30,31]. The electron rich dienophile TCO reacts with the electron poor diene methyltetrazine to yield a conjugated pyridazine product via strained 4,5-dihydropyridazine and 1,4-dihydro-isomer intermediates [32]. TCO-Tz inverse electron demand Diels Alder chemistry is rapid (k ~2000 M−1 s−1), selective and bio-orthogonal [33]. The rate constant corresponds to a ~10-000 fold enhancement over Cu+ catalysed Click chemistry [32]. TCO-Tz Click PEGylation could help unravel how key biological phenomena (e.g., fertilisation) impact protein thiol redox state.
Fertilisation induces mitochondrial H2O2 release [[34], [35], [36]], but whether fertilisation impacts reversible protein thiol oxidation is an open question. This is important because thiol oxidation is a post-translational modification that can act as a negative regulator of mitochondrial ATP synthesis (see below). ATP demand appears to be comparatively low in the unfertilised egg before fertilisation initiates embryonic metabolism by increasing ATP expenditure [37]. ATP is required to support the biosynthetic demands of embryogenesis [38]. Existing knowledge suggests a mechanism of tuning ATP synthesis rate within the F1-Fo mitochondrial ATP synthase itself by reversibly oxidising the cognate ATP F1 alpha sub-unit (ATP-α-F1, reviewed in Refs. [[39], [40], [41]]). ATP-α-F1 is an integral component of a matrix facing multi sub-unit assembly responsible for chemiosmotic ATP synthesis [[42], [43], [44]]. In particular, arginine 373 helps stabilise transition states [45]. Reversible ATP-α-F1 oxidation was first demonstrated in 2006 by West and colleagues [46] and confirmed by redox proteomic studies showing that C244/294 are subject to S-glutathionylation and S-nitrosation [[47], [48], [49]]. Reversible ATP-α-F1 oxidation is an established molecular correlate of impaired catalysis [47]. Interestingly, ADP provides an instructive cue to restore catalytic activity by reversing thiol oxidation [48]. Inhibitory ATP-α-F1 oxidation may constrain ATP synthesis to preserve finite resources in the unfertilised egg before ADP provides a signal to increase mitochondrial ATP synthesis after fertilisation. We aimed to determine whether fertilisation alters ATP-α-F1 redox state in Xenopus laevis (X. laevis), an important developmental model [[50], [51], [52]], using TCO-Tz Click chemistry. Fertilisation induces H2O2 release in X. laevis mitochondria [34], making them an ideal model to test our experimental hypothesis that: reversible ATP-α-F1 oxidation is greater in the unfertilised egg than in the 1-cell embryo.
2. Methods
2.1. Materials and reagents
A complete list of the materials and reagents used is provided (see Supplementary Table 1).
2.1.1. Xenopus laevis
In-house bred X. laevis were maintained at the European Xenopus Resource Centre (EXRC) at 18 °C and fed daily on trout pellets (see https://xenopusresource.org/). Following ethical approval (#OLETHSHE1500), unfertilised eggs X. laevis were harvested. X. laevis embryos were dejellied and then harvested either immediately (designated as 15 min) or 90 min post in vitro fertilisation (IVF) [53]. IVF was performed according to standard X. laevis protocols [54]. Samples were stored at −80 °C until biochemical analysis. The 5 and 90 min time points were selected to capture redox changes as soon as practically possible after fertilisation and the first embryonic cell cycle, respectively [34]. To minimise the possibility that our results were attributable to any one female or outliers [55], samples were obtained from three adult females and unfertilised eggs/embryos were grouped into batches of five for biochemical analysis.
2.1.2. Click PEGOX protocol
Samples were homogenised in ice-cold lysis buffer (150 mM NaCl, 10 mM EDTA, 100 mM Tris, 1% Triton X-100, pH 7.2) supplemented with a protease inhibitor tablet (Sigma, UK, #11697498001) and 100 mM N-ethylmaleimide (NEM; Sigma, UK, #E3876). Homogenates were left to stand for 30 min on ice to enable NEM to alkylate reduced thiols, before being centrifuged at 14,000 g for 5 min at 4 °C. After performing a Bradford assay to determine protein content [56], samples were adjusted to 1000 μg/ml to normalise protein concentration. Samples were then passed through a 6000 kDa spin column (BioRad, UK, Micro Bio-Spin™ P-6 Gel Columns, #7326222) to remove excess NEM. The flow-through was treated with 5 mM Tris (2-carboxyethyl)phosphine hydrochloride (TCEP; Thermofisher Scientific, UK, #T2556) for 30 min on ice to reduce reversibly oxidised thiols. A spin column step was incorporated to prevent TCEP from interfering with subsequent thiol labelling [57]. Trans-cyclooctene (TCO)-3 polyethylene glycol (PEG)-maleimide (TCO-PEG3-NEM, Click Chemistry Tools, USA, #1002) was added (5 mM) to label newly reduced thiols for 30 min on ice. To initiate the Click reaction, 5 mM 6-methyltetrazine 5 kDa PEG (Tz-PEG5; Click Chemistry Tools, USA #1090) was added for 60 min at room temperature with gentle agitation. Samples were then passed through a 6000 kDa spin column to prevent excess PEG smearing gel bands by forming micelles with SDS [58]. To obtain a maximally oxidised control, samples were lysed with 50 mM diamide (Sigma, UK, #D3648) to oxidise thiols, centrifuged at 14,000 g for 5 min at 4 °C and the supernatant was passed through a 6000 kDa spin column to remove excess diamide. The TCO-Tz Click reaction was terminated by adding Laemmli buffer (4% SDS, 20% glycerol, 0.004% bromphenol blue and 0.125 M Tris, pH 6.8) supplemented with 100 mM 1,4-Dithiothreitol (DTT, VWR, UK, #443853 B), before samples were denatured at 80 °C for 5 min [59].
2.1.3. Click PEGRED protocol
Samples were homogenised in the presence of 100 mM TCO-PEG3-NEM. Homogenates were left to stand for 30 min on ice to enable TCO-PEG3-NEM to label reduced thiols, before being centrifuged at 14,000 g for 5 min at 4 °C. After determining protein content [56], samples were adjusted to 1000 μg/ml to normalise starting protein concentration. To initiate the Click reaction, 5 mM Tz-PEG5 was added for 60 min at room temperature with gentle agitation. Samples were then passed through a 6000 kDa spin column to remove excess Tz-PEG5. To obtain a maximally reduced control, samples were lysed with 50 mM TCEP to reduce reversibly oxidised thiols, centrifuged at 14,000 g for 5 min at 4 °C and the supernatant was passed through a 6000 kDa spin column to remove excess TCEP. The TCO-Tz Click reaction was terminated by adding Lammeli buffer supplemented with 100 mM DTT, before samples were denatured at 80 °C for 5 min [59].
2.1.4. Selective S-glutathionylation reduction protocol
The TCO-Tz Click PEGOX protocol was modified to selectively investigate S-glutathionylation by substituting TCEP with 8 U/ml recombinant human glutaredoxin 2 (GRX2) (#ab82665, Abcam, UK), 5 mM reduced l-glutathione (GSH) (#G4251, Sigma, UK), 4 U/ml Baker's yeast (S. cerevisiae) glutathione reductase (GR; #G3664, Sigma, UK) and 1 mM NADPH (#10107824001, Sigma, UK) for 30 min at room temperature with gentle agitation. Excess GSH and NADPH were removed by passing samples through a 6000 kDa spin column before the TCO-Tz Click reaction was performed as described above.
2.1.5. Redox mobility shift assay
Click reacted samples were resolved by molecular mass at 100 V for ~90–120 min on a pre-cast 4–15% gradient gel (BioRad, UK, #4561085) in running buffer (25 mM Tris, 190 mM glycine, 0.1% SDS, pH 8.3). Gels were transferred onto a low auto-fluorescence 0.45 μM PDVF (BioRad, UK, #1620261) membrane at 100 V for 60 min in transfer buffer (25 mM Tris, 190 mM glycine, 10% methanol, pH 8.3). Membranes were blocked with 5% non-fat dry milk (NFDM) in PBS for at least 60 min at room temperature. Membranes were incubated with an anti-ATP5A mouse monoclonal primary antibody (Abcam, UK, #ab14748) at a concentration of 1 μg/ml (in 3% NFDM TBST) overnight at 4 °C with gentle agitation [60,61]. A fluorescent pre-absorbed Alexa Flour(R) 750 mouse secondary antibody was used (dilution: 1:2000 in 3% NDFM TBST; Abcam, UK, #ab175741). Fluorescent signals were captured on an Analytik Jena scanner using the appropriate filters (excitation: 678–748 nm; emission: 767–807 nm). Bands were quantified on proprietary VisionWorks™ software. Consistent with previous research [27], the amount of Click PEGylated ATP-α-F1 (i.e., mass shifted) was compared to the amount of reduced protein to derive percentage ATP-α-F1 oxidation, which was calculated as: % oxidised protein = [oxidised protein (sum of mass shifted bands)/total protein] x 100. A substituted equation was used for the reduced protein: reduced protein = [sum of mass shifted bands/total signal] x 100. Internally normalising the analysis obviated the need for a loading control because it rendered the analysis within as opposed to between lanes. This was advantageous since many common loading controls (e.g., GAPDH [62]) possess solvent exposed thiols, which would confound parallel analysis.
2.1.6. Statistical analysis
Normal distribution was assessed using the D'Agostino-Pearson and Shapiro-Wilk normality tests. Both tests revealed that relevant data sets were normally distributed (i.e., alpha >0.05). Parametric testing was, therefore, justified. Click PEGOX and Click PEGRED data were analysed by a one way ANOVA to test differences between independent means with alpha <0.05. Selective S-glutathionylation experiments were analysed by an unpaired (i.e., independent means) Student's t-test with alpha <0.05. Differences in the contribution of the 5 and 10 kDa bands to the total mass shifted signal within a time point (e.g., 15 min post fertilisation) were analysed by a paired Student's t-test with alpha <0.05. Statistical analysis was performed on GraphPad Prism version 6 (https://www.graphpad.com/). Data are presented as Mean (M) and standard deviation (SD).
3. Results
3.1. Click PEGOX reveals substantial reversible ATP-α-F1 oxidation before and after fertilisation
We modified the Click PEGylation oxidation (Click PEGOX) method developed by Van Leeuwen et al. [26] for TCO-Tz Click chemistry by: (1) using NEM to irreversibly alkylate reduced thiols by Michael addition; (2) TCEP to reduce reversibly oxidised thiols; (3) TCO-PEG3-NEM to label newly reduced thiols by Michael addition; and (4) Tz-PEG5 treatment to initiate the inverse electron demand Diels Alder reaction that selectively mass shifts reversibly oxidised protein thiols [[31], [32], [33]] (see Fig. 1). The 3 PEG spacer was selected to improve TCO group solubility, flexibility and accessibility. ATP-α-F1 is an excellent TCO-Tz Click PEGylation candidate because it is abundant in unfertilised X. laevis eggs [63], both cysteine residues (i.e., C244 and C294) are conserved and reversible oxidation is biologically meaningful—it implies low mitochondrial ATP F1-Fo synthase activity [47]. C294 occurs earlier in the linear amino acid sequence in X. laevis due to a small deletion, but C294 is used throughout to allow phylogenetic comparisons (see Fig. 2). We used Click PEGOX to interrogate reversible ATP-α-F1 oxidation in the unfertilised egg and embryo at 15 and 90 min post fertilisation. The 15 and 90 min time-points were selected to capture redox changes as soon as practically possible after fertilisation and the first embryonic cell cycle, respectively [34]. Click PEGOX revealed that: ATP-α-F1 is substantially oxidised in the unfertilised egg (65.9 ± 8.4%), as well as, at 15 min (64 ± 5%) and 90 min (64.9 ± 11.3%) post fertilisation (see Fig. 3). A one way ANOVA revealed no significant differences between time-points (P = 0.8306). To put the result in perspective: the median reversible oxidation of cysteine residues in the mammalian proteome is in the order of 5–12% [64]. Diamide failed to increase the reversible oxidation observed. Instead, diamide decreased reversible oxidation, which could reflect irreversible oxidation to TCEP irreducible species (e.g., sulfinic acids). The doublet mass shift shows both C244 and C294 are reversibly oxidised. However, the percentage contribution of the 5 kDa band to the total mass shifted signal was significantly greater than the 10 kDa band within each time-point (see Table 1). Differential reversible oxidation shows that a single thiol is preferentially oxidised, making it the major contributor to the bulk signal. Contrary to our experimental hypothesis, ATP-α-F1 is subject to substantial (~65%) reversible oxidation before and after fertilisation in X. laevis.
Fig. 1.
Catalyst-free trans-cyclooctene-methyltetrazine (TCO-Tz) Click PEGylation schematic. The left side of the circle depicts the TCO-Tz Click PEGylation reduction (Click-PEGRED) protocol wherein reduced thiols are labelled with TCO-3 polyethylene glycol (PEG)-maleimide (TCO-PEG3-NEM, TPN); (2) before 6-methyltetrazine PEG 5 kDa (Tz-PEG5) is added to initiate the inverse electron demand Diels alder reaction and thereby mass shift reduced thiols. An optional Tris (2-carboxyethyl)phosphine hydrochloride (TCEP) reduction step to reduce reversibly oxidised thiols before labelling them with N-ethylmaleimide (NEM) is depicted. The right side of the circle depicts the TCO-Tz Click PEGylation oxidation (Click-PEGOX) protocol wherein (1) reduced thiols are labelled with NEM; (2) reversibly oxidised thiols are reduced with TCEP; (3) before being labelled with TCO-PEG3-NEM (TPN); and (4) 6 Tz-PEG5 is added to initiate the inverse electron demand Diels Alder reaction and thereby mass shift reversibly oxidised thiols. The insets on the far right depicts NEM mediated Michael addition (top), TPN mediated Michael addition to a reduced thiol (middle) and the inverse electron demand TCO-Tz Diels Alder reaction (bottom).
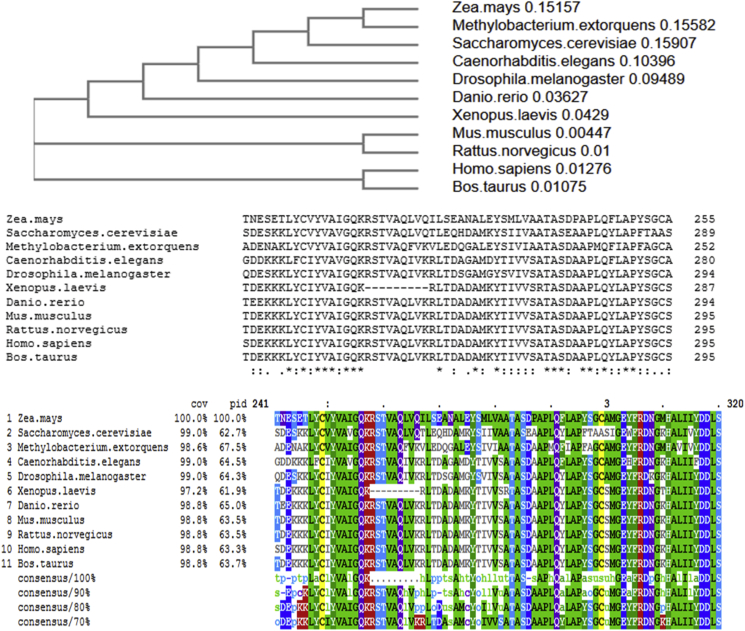
Fig. 2.
Cysteine residues in X. laevis ATP-α-F1are evolutionary conserved. Top. Phylogenetic tree of the common model organisms selected. Middle. Multiple sequence alignments of ATP-α-F1 derived from Clustal Omega (an open access online tool: https://www.ebi.ac.uk/Tools/msa/clustalo/). Bottom. Amino acid mark-up showing conserved cysteine residues in yellow. C294 occurs earlier in X. laevis due to a small deletion (see main text). Neighbouring amino acids surrounding C244 and C294 are highly conserved across phyla. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
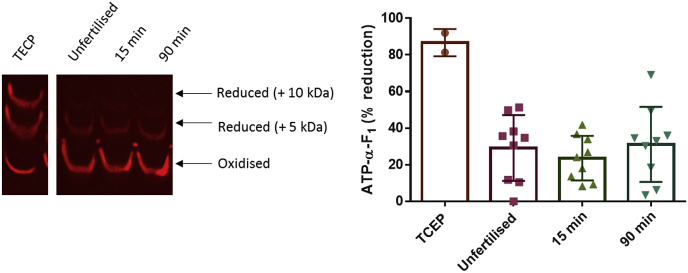
Fig. 3.
ATP-α-F1is subject to substantial reversible oxidation before and after fertilisation in X. laevis. Left. Western blot image showing reversibly oxidised (i.e., mass shifted 5 and 10 kDa bands) relative to reduced ATP-α-F1 (bottom band) before and after (15 and 90 min) fertilisation in X. laevis. Right. Percent reversible ATP-α-F1 oxidation before (n = 12) and after fertilisation (n = 12) quantified. Each n = a pool of 5 eggs/embryos. The positive control refers to a diamide (50 mM for 30 min) treated sample. A one way ANOVA revealed no significant differences between time-points (P = 0.8306).
Table 1.
Percentage contribution of the 5 and 10 kDa band signal to the total mass shift observed by experiment. Data are M and SD (±). Significance is derived from a paired within time-point Student's t-test. *Denotes a statistically significant difference between the 5 and 10 kDa band within sample time-point. A one way ANOVA was used to assess statistical significance between time-points based on the 5 kDa signal in experiment 1 and 2. In experiment 3, an unpaired Student's t-test was used. No statistically significant differences were observed between time-points.
| Time-point | 5 kDa (%) | 10 kDa (%) | Significance (P value) |
|---|---|---|---|
| Experiment 1: Oxidation | |||
| Unfertilised eggs | 64.4 ± 16.3 | 35.6 ± 16.3 | 0.0180* |
| Fertilised (15 min) | 60.4 ± 13.0 | 39.6 ± 12.3 | 0.0135* |
| Fertilised (90 min) | 64.3 ± 16.3 | 35.7 ± 19.4 | 0.0268* |
| Difference between time-points | n/a | n/a | 0.7921 |
| Experiment 2: Reduction | |||
| Unfertilised eggs | 74.7 ± 24.9 | 25.3 ± 24.9 | 0.0180* |
| Fertilised (15 min) | 80.9 ± 23.3 | 19.1 ± 23.2 | 0.0040* |
| Fertilised (90 min) | 71.2 ± 30.7 | 28.9 ± 30.7 | 0.0720 |
| Difference between time-points | n/a | n/a | 0.7362 |
| Experiment 3: S-glutathionylation | |||
| Unfertilised eggs | 68.5 ± 7.3 | 31.5 ± 7.3 | 0.0001* |
| Fertilised (90 min) | 74.0 ± 12.9 | 26.0 ± 12.9 | 0.0002* |
| Difference between time-points | n/a | n/a | 0.2509 |
3.2. Click PEGRED confirms substantial reversible ATP-α-F1 oxidation before and after fertilisation
If ATP-α-F1 is substantially oxidised before and after fertilisation, then mass shifting the reduced protein using a TCO-Tz Click PEG reduced (Click PEGRED) protocol should yield the inverse result. Put differently, if ATP-α-F1 is ~65% oxidised, then one would expect ~35% of the total protein to be mass shifted under Click PEGRED conditions. To determine percentage ATP-α-F1 reduction, we performed a Click PEGRED redox mobility shift assay by using TCO-PEG3-NEM to irreversibly alkylate reduced thiols by Michael addition and Tz-PEG5 to selectively mass shift reduced protein thiols (see Fig. 1). Click PEGRED showed that: ATP-α-F1 is ~20–30% reduced before (29.1 ± 17.9%) and after fertilisation (15min: 23.6. ± 12.2%; 90 min: 31.1 ± 20.5%; see Fig. 4) in X. laevis. A one way ANOVA revealed no significant differences between time-points (P = 0.6366). Analogous to the reversible oxidation results, band analysis confirms that one thiol was preferentially reduced as evidenced by the differential contribution of the 5 kDa and 10 kDa signals (see Table 1). The reduced signal, therefore, primarily reflects the redox state of a single thiol. Click PEGRED confirms substantial reversible ATP-α-F1 oxidation before and after fertilisation.
Fig. 4.
Trans-cyclooctene-methyltetrazine (TCO-Tz) Click PEGylation reduction (Click-PEGRED) confirms substantial reversible ATP-α-F1before and after fertilisation in X. laevis. Left. Western blot image showing reduced (i.e., mass shifted 5 and 10 kDa bands) relative to reversibly oxidised ATP-α-F1 (bottom band) before and after (15 and 90 min) fertilisation in X. laevis. Right. Percent ATP-α-F1 reduction before (n = 9) and after fertilisation (n = 9) in X. laevis quantified. Each n = a pool of 5 eggs/embryos. The positive control refers to a TCEP (50 mM for 30 min) treated sample. A one way ANOVA revealed no significant differences between time-points (P = 0.6366).
3.3. ATP-α-F1 is reversibly S-glutathionylated before and after fertilisation
To investigate reversible modification type, we substituted TCEP with recombinant glutaredoxin 2 (GRX2) to selectively reduce S-glutathionylated ATP-α-F1 (see Fig. 5). Given the lack of difference between the two post-fertilisation time-points, only the 90 min time-point was selected. Selective reduction experiments reveal that: ~30–40% of the total ATP-α-F1 protein is S-glutathionylated before (42.2 ± 12.6%) and after (36 ± 12%) fertilisation. The bottom band includes: reduced, reversibly oxidised (e.g., S-nitrosated) and irreversibly oxidised ATP-α-F1. The small (~6%) decrease in S-glutationylated ATP-α-F1 after fertilisation was statistically insignificant (P = 0.8901). Given ATP-α-F1 is ~65% reversibly oxidised before and after fertilisation, we estimate S-glutathionylation accounts for 50–60% of the total reversible oxidation observed, making it the dominant oxidative modification type. The mass shifted doublet shows both C244 and C294 are S-glutathionylated. Analogous to the reversible oxidation result, the percentage contribution of the 5 kDa band to the total mass shifted signal was significantly greater than the 10 kDa band within each time-point (see Table 1). A single thiol (potentially C244) is, therefore, preferentially S-glutathionylated likely due to greater solvent exposure during the catalytic cycle [65] (see Fig. 6). A selective chemical reduction strategy identifies S-gluthationylation as the dominant reversible oxidation type before and after fertilisation in X. laevis.
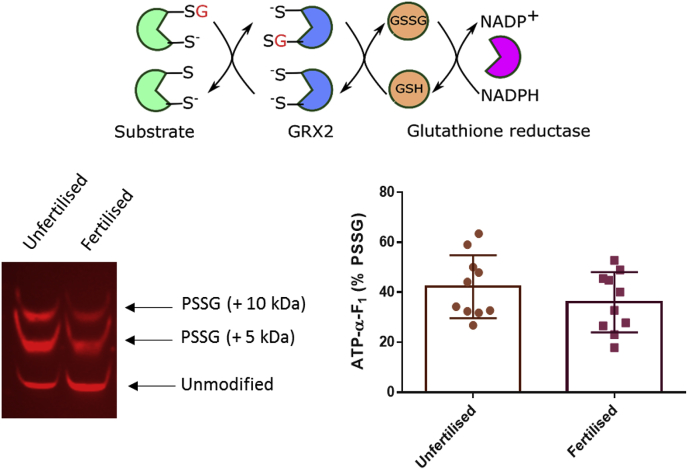
Fig. 5.
ATP-α-F1is reversibly S-glutathionylated before and after fertilisation in X. laevis. Top. A reaction scheme depicting the selected chemical reduction strategy for S-glutathionylated proteins. In this scheme, GRX2 reduces S-glutathionylated proteins using glutathione (GSH) derived electrons. Glutathione reductase (GR) is used to reduce oxidised glutathione (GSSG) using NADPH. Left. Western blot image showing the S-glutathionylated (i.e., mass shifted 5 and 10 kDa bands) relative to total ATP-α-F1 (bottom band) before and after (90 min) fertilisation in X. laevis. Right. Percent reversible ATP-α-F1 S-glutathionylation (i.e., PSSG) before (n = 10) and after fertilisation (n = 10) in X. laevis quantified. Each n = a pool of 5 eggs/embryos. An independent unpaired Student's t-test revealed no statistically significant difference in reversible S-glutathionylation between time-points (P = 0.8901).
Fig. 6.
Three dimensional molecular model of mitochondrial F1-FoATP synthase in state 3a derived fromRef. [65]in Bos taurus. State 3a corresponds to a catalytic rotary state of the F1 domain. Cysteine residues are highlighted in yellow, with C244 on the left and C294 on the right in each model display (left: surface; centre: space fill; right: ribbon). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Unravelling how fundamental biological phenomena impact redox homeostasis by Western blotting requires new techniques to monitor protein thiol redox state [26,27]. To do so, one must overcome a perennial technical difficulty: the reduced and oxidised forms of most protein thiols possess similar electrophoretic mobility [27]. Building on a recent Cu+ catalysed Click PEGylation method [26], we describe a novel catalyst-free TCO-Tz Click PEGylation approach to assess protein thiol redox state using Western blotting. Our approach enables one to estimate total reversible oxidation occupancy, differential occupancy between solvent exposed thiols, and establish reversible modification identity with selective reduction strategies (Table 2 lists key advantages and disadvantages of TCO-Tz Click PEGylation). TCO-Tz Click PEGylation may be a useful complementary technique for confirming redox proteomic findings [25]. For example, TCO-Tz Click PEGylation could be used to assess generator specific targets of mitochondrial O2.-/H2O2 (i.e., cytochrome c-1 for complex I derived O2.-/H2O2 produced by reverse electron transfer), which may enable investigators to infer the activity of a particular generator using an endogenous sentinel [2,66]. Click PEGylation relies on the primary antibody recognising the modified protein [26], but the bulky polymer itself and/or PEG-SDS micelles may obscure epitope recognition [27,58]. Potential solutions include using a polyclonal antibody and/or a smaller PEG polymer [26]. The latter strategy may be effective for small proteins (<30 kDa) with one or two solvent exposed thiols. TCO-Tz Click PEGylation creates new opportunities to interrogate protein thiol redox state in health and disease, which is significant since evolutionary conserved cysteine residues tend to be functionally important [64,67,68].
Table 2.
Key advantages and disadvantages of TCO-Tz Click PEGylation.
| Advantage | Disadvantage |
|---|---|
|
|
Underpinned by the pioneering work of Charles Manning Child [69] and reinvigorated by recent reports of fertilisation induced redox changes [[34], [35], [36], [37]], the redox regulation of fertility is re-emerging as a key area of interest (reviewed in Refs. [[70], [71], [72]]). Petrova and colleagues [36] show that fertilisation alters the reduced thiol proteome, but whether fertilisation impacts reversible protein thiol oxidation is unknown. Our TCO-Tz Click PEGylation studies advance current understanding by revealing substantial reversible ATP-α-F1 oxidation at C244/294 before and after fertilisation. This novel result rationalises further work to understand how fertilisation impacts reversible thiol oxidation across the mitochondrial proteome. Substantial reversible ATP-α-F1 oxidation implies low mitochondrial F1-Fo ATP synthase activity (i.e., low oligomycin sensitive respiration), consistent with suggestions that mitochondria in the unfertilised egg are functionally immature [73]. How reversible C244/294 oxidation impairs catalysis is unresolved, but may involve intermolecular disulphide bond formation between C294 and C103 of a proximal gamma F1 sub-unit (ATP-γ-F1) that impairs conformational flexibility [39,41]. Interestingly, intramolecular ATP-γ-F1 disulphide crosslinks hold chloroplast ATP synthase inactive in darkness to prevent ATP hydrolysis until daylight restores catalysis by triggering thioredoxin mediated reduction [44,74]. Perhaps, X. laevis exploit a similar strategy to prevent wasteful F1 mediated ATP hydrolysis. Recalcitrance of early X. laevis blastula to oligomycin [75], a canonical mitochondrial F1-Fo ATP synthase inhibitor, could reflect pre-existing inhibition by reversible oxidation. Persistent reversible ATP-α-F1 oxidation after fertilisation implies low matrix reductase activity and/or equilibrium between the rate of reduction and oxidation. Reductive systems (e.g., GRX2) may only be partially operative, consistent with relatively weak embryonic antioxidant defence [[76], [77], [78]]. We propose that: O2.-/H2O2 produced during oogenesis underlies the reversible oxidation observed as part of an ATP synthesis sensitive mitochondrial selection process [79]. Inhibitory ATP-α-F1 oxidation may constrain mitochondrial Fo-F1 ATP synthase activity until it is reversed to support metabolic differentiation later in development when the relevant reductive systems become operative [80].
Exploiting a GRX2 based selective reduction strategy, we show that: C244/294 are S-glutathionylated before and after fertilisation in X. laevis. Successful deglutathionylation suggests ATP-α-F1 may be a GRX2 substrate. ATP-α-F1 S-glutathionylation is consistent with redox proteomic studies demonstrating C244/294 S-glutathionylation in vitro in isolated mitochondrial and in vivo in canine hearts [[46], [47], [48]]. S-glutathionylation is the dominant oxidative modification type, accounting for ~50–60% of the reversible oxidation observed. The remaining 40–50% may comprise intermolecular disulphide bond formation since SOH and SNO species are relatively unstable [64,81]. Plausible chemical S-glutathionylation routes include: (1) un-catalysed addition of GSSG to a thiolate; (2) un-catalysed addition of GS− to a thiyl radical, SOH or SNO species; (3) GRX2 and/or glutathione S-transferase catalysed conjugation of PSSG to a substrate thiol (reviewed in Refs. [40,82,83]). S-glutathionylation may block intermolecular disulphide crosslinks with ATP-γ-F1 C103. S-glutathionylation still correlates with impaired enzyme activity [40], but may make it easier to reactivate the enzyme if disulphide crosslinks pose steric challenges and/or aggregate. S-glutathionylation may also protect C244/294 from irreversible oxidation to sulfinic and sulfonic acids [13]. Intriguingly, deglutathionylation of ATP-α-F1 and other proteins may contribute to the increase in the reduced glutathione pool that occurs after fertilisation [84,85]. Further studies are, however, required to investigate the scale of the embryonic S-glutathionylated proteome and whether deglutathionylation contributes to developmental fluxes in reduced glutathione availability. TCO-Tz Click PEGylation protocols can be readily adapted by substituting generic for selective reductants to unveil the dominant oxidative modification type.
5. Conclusion
We describe a novel catalyst-free TCO-Tz Click PEGylation approach to assess protein thiol redox state by mass shifting the reduced or oxidised form as desired. Knowledge of how fertilisation—a key biological phenomenon—impacts matrix protein thiol redox state is fragmentary. To advance current understanding, we used TCO-Tz Click PEGylation to study the mitochondrial F1-Fo ATP synthase by assessing the redox state of the essential cognate sub-unit ATP-α-F1 in X. laevis. Three key novel findings are that: (1) ATP-α-F1 is substantially reversibly oxidised at evolutionary conserved cysteine residues (i.e., C244/C294) before and after fertilisation; and (2) S-glutathionylation is the dominant oxidative modification type; (3) a single thiol is preferentially oxidised likely due to greater solvent exposure during the catalytic cycle. Substantial reversible ATP-α-F1 oxidation before and after fertilisation is biologically meaningful because it implies low mitochondrial F1-Fo ATP synthase activity. Catalyst-free TCO-Tz Click PEGylation is a valuable new tool to interrogate protein thiol redox state in health and disease.
Acknowledgements
J.N.C., E.J.F., and H.H. gratefully acknowledge Highlands and Islands Enterprise (HMS 9353763) funding. The EXRC is funded by the Wellcome Trust (212942/Z/18/Z) and BBSRC (BB/R014841/1).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101258.
Author contributions
Conceptualization, J.N.C. & H·H.; Methodology, J.N·C.; Investigation, J.N·C., E.J.F., M.T.V.M., & A.N.; Writing – Original Draft, J.N·C; Writing – Review & Editing, ALL; Funding Acquisition, J.N·C., H·H., and M.G.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brand M.D. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic. Biol. Med. 2016;100:14–31. doi: 10.1016/j.freeradbiomed.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Collins Y., Chouchani E.T., James A.M., Menger K.E., Cocheme H.M., Murphy M.P. Mitochondrial redox signalling at a glance. J. Cell Sci. 2012;125:1837. doi: 10.1242/jcs.098475. [DOI] [PubMed] [Google Scholar]
- 4.Echtay K.S., Roussel D., St-Pierre J., Jekabsons M.B., Cadenas S., Stuart J.A., Harper J.A., Roebuck S.J., Morrison A., Pickering S., Clapham J.C., Brand M.D. Superoxide activates mitochondrial uncoupling proteins. Nature. 2002;415:96–99. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- 5.Giorgio M., Trinei M., Migliaccio E., Pelicci P. Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals? Nat. Rev. Mol. Cell Biol. 2007;8:722–728. doi: 10.1038/nrm2240. [DOI] [PubMed] [Google Scholar]
- 6.Cobley J.N., Fiorello M.-L., Bailey D.M. 13 reasons why the brain is susceptible to oxidative stress. Redox Biol. 2018;15:490–503. doi: 10.1016/j.redox.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mailloux R.J. Teaching the fundamentals of electron transfer reactions in mitochondria and the production and detection of reactive oxygen species. Redox Biol. 2015;4:381–398. doi: 10.1016/j.redox.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sena L.A., Chandel N.S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell. 2012;48:158–166. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winterbourn C.C., Metodiewa D. Reactivity of biologically important thiol compounds with superoxide and hydrogen peroxide. Free Radic. Biol. Med. 1999;27:322–328. doi: 10.1016/s0891-5849(99)00051-9. [DOI] [PubMed] [Google Scholar]
- 10.Winterbourn C.C., Hampton M.B. Thiol chemistry and specificity in redox signaling. Free Radic. Biol. Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Stöcker S., Van Laer K., Mijuskovic A., Dick T.P. The conundrum of hydrogen peroxide signaling and the emerging role of peroxiredoxins as redox relay hubs. Antioxidants Redox Signal. 2018;28:558–573. doi: 10.1089/ars.2017.7162. [DOI] [PubMed] [Google Scholar]
- 12.Lo Conte M., Carroll K.S. The redox biochemistry of protein sulfenylation and. J. Biol. Chem. 2013;288:26480–26488. doi: 10.1074/jbc.R113.467738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy M.P. Mitochondrial thiols in antioxidant protection and redox signaling: distinct roles for glutathionylation and other thiol modifications. Antioxidants Redox Signal. 2012;16:476–495. doi: 10.1089/ars.2011.4289. [DOI] [PubMed] [Google Scholar]
- 14.Holmström K.M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 15.Yang J., Carroll K.S., Liebler D.C. The expanding landscape of the thiol redox proteome. Mol. Cell. Proteom. 2016;15:1–11. doi: 10.1074/mcp.O115.056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hess D.T., Matsumoto A., Kim S.-O.O., Marshall H.E., Stamler J.S. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 17.Mailloux R.J., Treberg J.R. Protein S-glutathionlyation links energy metabolism to redox signaling in mitochondria. Redox Biol. 2016;8:110–118. doi: 10.1016/j.redox.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forman H.J., Maiorino M., Ursini F. Signaling functions of reactive oxygen species. Biochemistry. 2010;49:835–842. doi: 10.1021/bi9020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parvez S., Long M.J.C., Poganik J.R., Aye Y. Redox signaling by reactive electrophiles and oxidants. Chem. Rev. 2018;118:8798–8888. doi: 10.1021/acs.chemrev.7b00698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulsen C.E., Carroll K.S. Cysteine-mediated redox Signaling : chemistry , biology , and tools for discovery. Chem. Rev. 2013;133:4633–4679. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dröse S., Brandt U., Wittig I. Mitochondrial respiratory chain complexes as sources and targets of thiol-based redox-regulation. Biochim. Biophys. Acta Protein Proteonomics. 2014;1844:1344–1354. doi: 10.1016/j.bbapap.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Galkin A., Meyer B., Wittig I., Karas M., Schägger H., Vinogradov A., Brandt U. Identification of the mitochondrial ND3 subunit as a structural component involved in the active/deactive enzyme transition of respiratory complex I. J. Biol. Chem. 2008;283:20907–20913. doi: 10.1074/jbc.M803190200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chouchani E.T., Pell V.R., Gaude E., Aksentijević D., Sundier S.Y., Robb E.L., Logan A., Nadtochiy S.M., Emily N.J., Smith A.C., Eyassu F., Shirley R., Hu C., Dare A.J., James A.M., Rogatti S., Hartley R.C., Eaton S., Ana S.H., Brookes P.S., Davidson S.M., Duchen M.R., Saeb-parsy K. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515:431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chouchani E.T., Methner C., Nadtochiy S.M., Logan A., Victoria R., Ding S., James A.M., Cochemé H.M., Reinhold J., Lilley K.S., Partridge L., Fearnley I.M., Robinson A.J., Hartley R.C., Smith R.A.J., Krieg T., Brookes P.S., Murphy M.P. Cardioprotection by S-nitrosation of a cysteine switch on mitochondrial complex I. Nat. Med. 2013;19:753–759. doi: 10.1038/nm.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cobley J.N., Sakellariou G.K., Husi H., Mcdonagh B. Proteomic strategies to unravel age-related redox signalling defects in skeletal muscle. Free Radic. Biol. Med. 2019;132:24–32. doi: 10.1016/j.freeradbiomed.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 26.van Leeuwen L.A.G., Hinchy E.C., Murphy M.P., Robb E.L., Cochemé H.M. Click-PEGylation – a mobility shift approach to assess the redox state of cysteines in candidate proteins. Free Radic. Biol. Med. 2017;108:374–382. doi: 10.1016/j.freeradbiomed.2017.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burgoyne J.R., Oviosu O., Eaton P. The PEG-switch assay: a fast semi-quantitative method to determine protein reversible cysteine oxidation. J. Pharmacol. Toxicol. Methods. 2013;68:297–301. doi: 10.1016/j.vascn.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Rostovtsev V., Green L., Fokin V., Sharpless K. A stepwise huisgen cycloaddition process catalyzed by copper ( I ): regioselective ligation of azides and terminal alkynes. Angew. Chem. Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 29.Halliwell B., Gutteridge J.M.C. fifth ed. Oxford University Press; 2015. Free Radicals in Biology & Medicine. [Google Scholar]
- 30.Jewett J., Bertozzi C. Cu-free click cycloaddition reactions in chemical biology. Chem. Soc. Rev. 2010;39:1272–1279. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diels O., Alder K. Synthesen in der hydroaromatischen Reihe. Justus Liebigs Ann. Chem. 1928;460:98–122. [Google Scholar]
- 32.Oliveira B.L., Guo Z., Bernardes G.J.L. Inverse electron demand Diels-Alder reactions in chemical biology. Chem. Soc. Rev. 2017;46:4895–4950. doi: 10.1039/c7cs00184c. [DOI] [PubMed] [Google Scholar]
- 33.Blackman M.L., Royzen M., Fox J.M. Tetrazine Ligation : fast bioconjugation based on inverse-electron-demand Diels - alder reactivity. J. Am. Chem. Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han Y., Ishibashi S., Iglesias-Gonzalez J., Chen Y., Love N.R., Amaya E. Ca2+-Induced mitochondrial ROS regulate the early embryonic cell cycle. Cell Rep. 2018;22:218–231. doi: 10.1016/j.celrep.2017.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dumollard R., Carroll J., Duchen M.R., Campbell K., Swann K. Mitochondrial function and redox state in mammalian embryos. Semin. Cell Dev. Biol. 2009;20:346–353. doi: 10.1016/j.semcdb.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 36.Petrova B., Liu K., Tian C., Kitaoka M., Freinkman E., Yang J., Orr-Weaver T.L. Dynamic redox balance directs the oocyte-to-embryo transition via developmentally controlled reactive cysteine changes. Proc. Natl. Acad. Sci. Unit. States Am. 2018;115:201807918. doi: 10.1073/pnas.1807918115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dumollard R. Sperm-triggered [Ca2+] oscillations and Ca2+ homeostasis in the mouse egg have an absolute requirement for mitochondrial ATP production. Development. 2004;131:3057–3067. doi: 10.1242/dev.01181. [DOI] [PubMed] [Google Scholar]
- 38.Vastag L., Jorgensen P., Peshkin L., Wei R., Rabinowitz J.D., Kirschner M.W. Remodeling of the metabolome during early frog development. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaludercic N., Giorgio V. The dual function of reactive oxygen/nitrogen species in bioenergetics and cell death: the role of ATP synthase. Oxid. Med. Cell. Longev. 2016 doi: 10.1155/2016/3869610. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mailloux R.J., Willmore W.G. S-glutathionylation reactions in mitochondrial function and disease. Front. Cell Dev. Biol. 2014;2:1–17. doi: 10.3389/fcell.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S.B., Murray C.I., Chung H.S., Van Eyk J.E. Redox regulation of mitochondrial ATP synthase. Trends Cardiovasc. Med. 2013;23:18. doi: 10.1016/j.tcm.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abrahams J.P., Leslie A.G.W., Lutter R., Walker J.E. Structure at 2 . 8 A resolution of F1 · ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 43.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 44.Walker J.E., Walker S.J. Keilin memorial lecture keilin memorial lecture the ATP synthase : the understood , the uncertain and the unknown. Biochem. Soc. Trans. 2013;41:1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- 45.Menz R.I., Walker J.E., Leslie A.G.W. Structure of bovine mitochondrial F 1 -ATPase with nucleotide bound to all three catalytic Sites : implications for the mechanism of rotary catalysis. Cell. 2001;106:331–341. doi: 10.1016/s0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- 46.West M.B., Hill B.G., Xuan Y.T., Bhatnagar A. Protein glutathiolation by nitric oxide: an intracellular mechanism regulating redox protein modification. FASEB J. 2006;20:1715–1717. doi: 10.1096/fj.06-5843fje. [DOI] [PubMed] [Google Scholar]
- 47.Wang S.B., Foster D.B., Rucker J., O'Rourke B., Kass D.A., Van Eyk J.E. Redox regulation of mitochondrial ATP synthase: implications for cardiac resynchronization therapy. Circ. Res. 2011;109:750–757. doi: 10.1161/CIRCRESAHA.111.246124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia J., Han D., Sancheti H., Yap L.P., Kaplowitz N., Cadenas E. Regulation of mitochondrial glutathione redox status and protein glutathionylation by respiratory substrates. J. Biol. Chem. 2010;285:39646–39654. doi: 10.1074/jbc.M110.164160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun J., Nguyen T., Aponte A.M., Menazza S., Kohr M.J., Roth D.M., Patel H.H., Murphy E., Steenbergen C. Ischaemic preconditioning preferentially increases protein S-nitrosylation in subsarcolemmal mitochondria. Cardiovasc. Res. 2015;106:227–236. doi: 10.1093/cvr/cvv044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Session A.M., Uno Y., Kwon T., a Chapman J., Toyoda A., Takahashi S., Fukui A., Hikosaka A., Suzuki A., Kondo M., Van Heeringen S.J., Quigley I., Heinz S., Ogino H., Ochi H., Hellsten U., Lyons J.B., Simakov O., Putnam N., Stites J., Wallingford J.B., Ito Y., Asashima M., Ueno N., Matsuda Y., Veenstra G.J.C. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538:1–15. doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harland R.M., Grainger R.M. Xenopus research: metamorphosed by genetics and genomics. Trends Genet. 2011;27:507–515. doi: 10.1016/j.tig.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sidlauskaite E., Gibson J.W., Megson I.L., Whitfield P.D., Tovmasyan A., Batinic-Haberle I., Murphy M.P., Moult P.R., Cobley J.N. Mitochondrial ROS cause motor deficits induced by synaptic inactivity: implications for synapse pruning. Redox Biol. 2018;16:344–351. doi: 10.1016/j.redox.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guille M. Microinjection into Xenopus oocytes and embryos. Mol. Methods Dev. Biol. 1999;127:111–123. doi: 10.1385/1-59259-678-9:111. [DOI] [PubMed] [Google Scholar]
- 54.Sive H.L., Grainger R.M., Harland R.M. first ed. Cold Spring Harbor Laboratory Press; New York: 2000. Early Development of Xenopus laevis: A Laboratory Manual. [Google Scholar]
- 55.Kilkenny C., Browne W.J., Cuthill I.C., Emerson M., Altman D.G. Improving bioscience research Reporting : the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8:6–10. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 57.Tyagarajan K., Pretzer E., Wiktorowicz J.E. Thiol-reactive dyes for fluorescence labeling of proteomic samples. Electrophoresis. 2003;24:2348–2358. doi: 10.1002/elps.200305478. [DOI] [PubMed] [Google Scholar]
- 58.Zheng C.Y., Ma G., Su Z. Native PAGE eliminates the problem of PEG-SDS interaction in SDS-PAGE and provides an alternative to HPLC in characterization of protein PEGylation. Electrophoresis. 2007;28:2801–2807. doi: 10.1002/elps.200600807. [DOI] [PubMed] [Google Scholar]
- 59.Laemmli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 60.Cobley J.N., Bartlett J.D., Kayani A., Murray S.W., Louhelainen J., Donovan T., Waldron S., Gregson W., Burniston J.G., Morton J.P., Close G.L. PGC-1a transcriptional response and mitochondrial adaptation to acute exercise is maintained in skeletal muscle of sedentary elderly males. Biogerontology. 2012;13:621–631. doi: 10.1007/s10522-012-9408-1. [DOI] [PubMed] [Google Scholar]
- 61.Cobley J.N., Sakellariou G.K., Owens D.J., Murray S., Waldron S., Gregson W., Fraser W.D., Burniston J.G., Iwanejko L.A., McArdle A., Morton J.P., Jackson M.J., Close G.L. Lifelong training preserves some redox-regulated adaptive responses after an acute exercise stimulus in aged human skeletal muscle. Free Radic. Biol. Med. 2014;70:23–32. doi: 10.1016/j.freeradbiomed.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 62.Peralta D., Bronowska A.K., Morgan B., Dóka É., Van Laer K., Nagy P., Gräter F., Dick T.P. A proton relay enhances H2O2 sensitivity of GAPDH to facilitate metabolic adaptation. Nat. Chem. Biol. 2015;11:156–163. doi: 10.1038/nchembio.1720. [DOI] [PubMed] [Google Scholar]
- 63.Wühr M., Freeman R.M., Presler M., Horb M.E., Peshkin L., Gygi S.P., Kirschner M.W. Deep proteomics of the Xenopus laevis egg using an mRNA-derived reference database. Curr. Biol. 2014;24:1467–1475. doi: 10.1016/j.cub.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Go Y.-M., Chandler J.D., Jones D.P. The cysteine proteome. Free Radic. Biol. Med. 2015;84:227–245. doi: 10.1016/j.freeradbiomed.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou A., Rohou A., Schep D.G., V Bason J., Montgomery M.G., Walker J.E., Grigorieff N. Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. Elife. 2015;4 doi: 10.7554/eLife.10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bleier L., Wittig I., Heide H., Steger M., Brandt U., Drose S. Generator-specific targets of mitochondrial reactive oxygen species. Free Radic. Biol. Med. 2015;78:1–10. doi: 10.1016/j.freeradbiomed.2014.10.511. [DOI] [PubMed] [Google Scholar]
- 67.Marino S.M., Gladyshev V.N. Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J. Mol. Biol. 2010;404:902–916. doi: 10.1016/j.jmb.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weerapana E., Wang C., Simon G.M., Richter F., Khare S., Dillon M.B.D., Bachovchin D.A., Mowen K., Baker D., Cravatt B.F. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Child C.M. Chicago University Press; Chicago: 1947. Patterns and Problems in Development. [Google Scholar]
- 70.Rampon C., Volovitch M., Joliot A., Vriz S. Hydrogen peroxide and redox regulation of developments. Antioxidants. 2018;7:159. doi: 10.3390/antiox7110159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blackstone N.W. Multicellular redox regulation: integrating organismal biology and redox chemistry. Bioessays. 2006;28:72–77. doi: 10.1002/bies.20337. [DOI] [PubMed] [Google Scholar]
- 72.Blackstone N.W. Redox control and the evolution of multicellularity. Bioessays. 2000;22:947–953. doi: 10.1002/1521-1878(200010)22:10<947::AID-BIES10>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 73.Allen J.F. Separate sexes and the mitochondrial theory of ageing. J. Theor. Biol. 1996;180:135–140. doi: 10.1006/jtbi.1996.0089. [DOI] [PubMed] [Google Scholar]
- 74.Nalins C.M., Mccarty R.E. Role of a disulfide bond in the y subunit in activation of the ATPase of chloroplast coupling factor 1 ” P- J. Biol. Chem. 1984;259:7275–7280. [PubMed] [Google Scholar]
- 75.Gibeaux R., Acker R., Kitaoka M., Georgiou G., van Kruijsbergen I., Ford B., Marcotte E.M., Nomura D.K., Kwon T., Veenstra G.J.C., Heald R. Paternal chromosome loss and metabolic crisis contribute to hybrid inviability in Xenopus. Nature. 2018;553:337–341. doi: 10.1038/nature25188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bell K.F.S., Al-Mubarak B., Martel M.-A., McKay S., Wheelan N., Hasel P., Márkus N.M., Baxter P., Deighton R.F., Serio A., Bilican B., Chowdhry S., Meakin P.J., Ashford M.L.J., Wyllie D.J.A., Scannevin R.H., Chandran S., Hayes J.D., Hardingham G.E. Neuronal development is promoted by weakened intrinsic antioxidant defences due to epigenetic repression of Nrf2. Nat. Commun. 2015;6:7066. doi: 10.1038/ncomms8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dennery P.A. Oxidative stress in development: nature or nurture? Free Radic. Biol. Med. 2010;49:1147–1151. doi: 10.1016/j.freeradbiomed.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 78.Cobley J.N. Synapse pruning: mitochondrial ROS with their hands on shears. Bioessays. 2018;40 doi: 10.1002/bies.201800031. [DOI] [PubMed] [Google Scholar]
- 79.Lieber T., Jeedigunta S., Palozzi J., Lehmann R., Hurd T. Removal of deleterious mtDNA in the germline. Nature. 2019 doi: 10.1038/s41586-019-1213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Agathocleous M., Love N.K., Randlett O., Harris J.J., Liu J., Murray A.J., Harris W.A. Metabolic differentiation in the embryonic retina. Nat. Cell Biol. 2012;14:859–864. doi: 10.1038/ncb2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wolhuter K., Eaton P. How widespread is stable protein S-nitrosylation as an end-effector of protein regulation? Free Radic. Biol. Med. 2017;109:156–166. doi: 10.1016/j.freeradbiomed.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 82.Hurd T.R., Costa N.J., Dahm C.C., Beer S.M., Brown S.E., Filipovska A., Murphy M.P. Glutathionylation of mitochondrial proteins. Antioxidants Redox Signal. 2005;7:999–1010. doi: 10.1089/ars.2005.7.999. [DOI] [PubMed] [Google Scholar]
- 83.Mieyal J.J., Gallogly M.M., Qanungo S., Sabens E., Shelton M. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxidants Redox Signal. 2008;10:1943–1988. doi: 10.1089/ars.2008.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hansen J.M., Harris C. Glutathione during embryonic development. Biochim. Biophys. Acta Gen. Subj. 2015;1850:1527–1542. doi: 10.1016/j.bbagen.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 85.Timme-Laragy A.R., Goldstone J.V., Imhoff B.R., Stegeman J.J., Hahn M.E., Hansen J.M. Glutathione redox dynamics and expression of glutathione-related genes in the developing embryo. Free Radic. Biol. Med. 2013;65:89–101. doi: 10.1016/j.freeradbiomed.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.