Ewing sarcoma is characterized by a pathognomonic chromosomal translocation that generates the EWSR1-FLI1 chimeric transcription factor. The transcriptional targets of EWSR1-FLI1 that are essential for tumorigenicity are incompletely defined.
KEYWORDS: BNIP3L, CT antigen, cancer/testis antigen, EWSR1-FLI1, Ewing sarcoma, FATE1, RNF183
ABSTRACT
Ewing sarcoma is characterized by a pathognomonic chromosomal translocation that generates the EWSR1-FLI1 chimeric transcription factor. The transcriptional targets of EWSR1-FLI1 that are essential for tumorigenicity are incompletely defined. Here, we found that EWSR1-FLI1 modulates the expression of cancer/testis (CT) antigen genes, whose expression is biased to the testes but is also activated in cancer. Among these CT antigens, fetal and adult testis expressed 1 (FATE1) is most robustly induced. EWSR1-FLI1 associates with the GGAA repeats in the proximal promoter of FATE1, which exhibits accessible chromatin exclusively in mesenchymal progenitor cells (MPCs) and Ewing sarcoma cells. Expression of EWSR1-FLI1 in non-Ewing sarcoma cells and in MPCs enhances FATE1 mRNA and protein expression. Conversely, depletion of EWSR1-FLI1 in Ewing sarcoma cells leads to a loss of FATE1 expression. Importantly, we found that FATE1 is required for survival and anchorage-independent growth in Ewing sarcoma cells via attenuating the accumulation of BNIP3L, a BH3-only protein that is toxic when stabilized. This action appears to be mediated by the E3 ligase RNF183. We propose that engaging FATE1 function can permit the bypass of cell death mechanisms that would otherwise inhibit tumor progression.
INTRODUCTION
Ewing sarcoma is the second most common bone and soft tissue malignancy in children and adolescents. This cancer is characterized by one of several reciprocal chromosomal translocations that fuse a FET family protein with an ETS transcription factor (1–3). t(11;22)(q24;q12), which is by far the most common of these, fuses the amino terminal domain of EWSR1 to the DNA-binding domain of FLI1 (4). The resulting chimeric transcription factor, EWSR1-FLI1, associates with GGAA repeats in both the proximal promoters and distal enhancers of target genes to either anomalously activate or suppress their expression (5–8). This activity is considered to be the causative alteration in Ewing sarcoma. Suppression of EWSR1-FLI1 leads to a loss of proliferation and survival of tumor cells in vitro and in vivo (4). The depletion of EWSR1-FLI1 from Ewing sarcoma cells results in a gene expression profile reminiscent of that of mesenchymal progenitor cells (MPCs) (9). Furthermore, MPCs are among the few cell types that can be transformed by the EWSR1-FLI1 oncogene (10, 11). On the basis of these observations, MPCs are strongly considered to be the cell of origin for Ewing sarcoma, possibly due to the presence of a unique chromatin landscape that permits the disruption of normal mesenchymal differentiation programs by EWSR1-FLI1 (7, 12).
Current treatment of Ewing sarcoma consists of surgery, radiation, and systemic chemotherapy. In patients with local disease, the 5-year event-free survival (EFS) rate is 75% (13). These patients are at risk for long-term sequelae stemming from therapy. However, for patients with metastatic or relapsed disease, 5-year EFS rates drop dramatically to ∼20 and <15% in patients presenting with metastatic disease and relapsed disease, respectively (14, 15). Thus, all patients would benefit from more-effective and less-toxic treatment options. Several studies have shown that Ewing sarcoma is associated with very few other recurrent genetic alterations, limiting the availability of actionable targets (16, 17). Furthermore, the low mutation burden suggests limited production of neoantigens by tumor cells, likely restricting the efficacy of immunotherapeutic options.
Tumors frequently express genes that are otherwise restricted to the testis and trophoblasts (18–20). The corresponding gene products, collectively known as cancer/testis (CT) antigens, represent a class of tumor-associated antigens that are well-established targets for T-cell therapy and/or anticancer vaccines (21). For example, in synovial sarcoma, a soft tissue sarcoma of adolescents and young adults driven by a chimeric chromatin remodeler (SS18-SSX), 60% of patients exhibited an objective response to adoptive T cell therapy targeting the CT antigen, namely, NY-ESO-1 (22). A recent study reported that an anti-CT antigen vaccine was safe and well tolerated in Ewing sarcoma patients (23). Limited information exists regarding CT antigen expression in Ewing sarcoma. Studies performed to date have suggested that tumor-specific loss of CpG site methylation in CT antigen promoters is sufficient for their aberrant expression in tumors (24, 25). However, in addition to a permissive chromatin state, it is probable that transcriptional regulation is required to activate these genes.
Accumulating evidence now suggests that tumor-activated testis proteins are not merely bystanders in the oncogenic process but can also promote neoplastic behaviors (26–28). Recently, we reported that the CT antigen fetal and adult testis expressed 1 (FATE1) can attenuate apoptosis in multiple tumor types (28). FATE1 localizes to the mitochondria, where it can coordinate the action of E3 ligases to suppress BH3-only protein expression, thereby attenuating cell death. Importantly, elevated expression of FATE1 correlates with reduced survival in colon cancer. Here, we report that EWSR1-FLI1 directly activates the anomalous expression of FATE1 in Ewing sarcoma. FATE1 is essential for Ewing sarcoma survival through the destabilization of BNIP3L, a poorly characterized BH3-only protein that is toxic to Ewing sarcoma cells. This paper reports the first demonstration that chimeric transcription factors can directly activate CT antigens, with important implications for CT antigens as molecular targets or as the basis for Ewing sarcoma immunotherapeutic strategies.
RESULTS
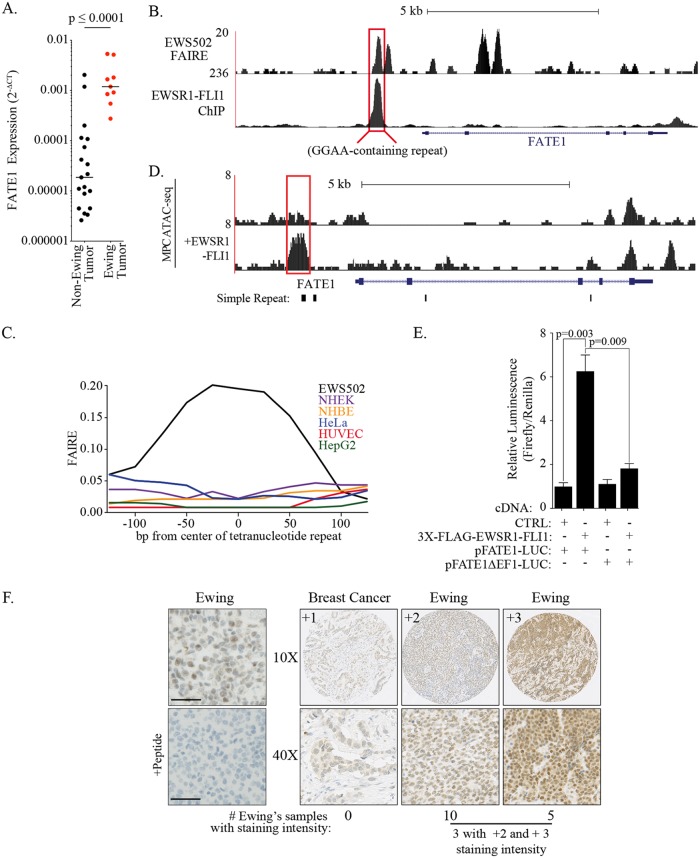
We have previously shown that a cancer/testis (CT) antigen, FATE1, is frequently expressed in tumors and is required for cell autonomous survival (28). As expected, a distributive sampling of FATE1 expression across a broad array of solid tumors returned unanimous presence calls. However, we observed elevated activation of FATE1 expression in a Ewing sarcoma-derived sample. Expansion of the original panel to include eight additional Ewing sarcoma cell lines revealed that the median level of FATE1 expression was 2 orders of magnitude higher in Ewing sarcoma cells than in other tumor types tested (Fig. 1A; see also Table 1). Given that EWSR1-FLI1 directly activates the anomalous expression of genes essential for tumorigenesis, we next asked whether FATE1 was a direct target of the chimeric transcription factor. Chromatin immunoprecipitation sequencing (ChIP-seq) analysis generated in Ewing sarcoma cell line EWS502 revealed strong EWSR1-FLI1 binding to a region 1,362 bp upstream of FATE1’s transcriptional start site (TSS) (Fig. 1B). This region included a microsatellite region containing 16 repeats of the sequence GGAA, the core of the canonical ETS binding motif (8). We then examined chromatin accessibility at this region using formaldehyde-assisted isolation of regulatory elements (FAIRE). This site demonstrated an increased signal consistent with enhanced chromatin accessibility, a condition which was not observed in normal and non-Ewing sarcoma tumor cell lines (Fig. 1B and C). Exploring a published MPC assay for transposase-accessible chromatin using sequencing (ATAC-seq) data to identify changes in nucleosome depletion that were dependent on EWSR1-FLI1, we observed that FATE1’s proximal promoter had an increased ATAC signal only after ectopic expression of EWSR1-FLI1 (Fig. 1D) (7). This finding suggests that the chromatin environment of the FATE1 promoter is permissive for EWSR1-FLI1-mediated activation during mesenchymal differentiation. Indeed, cotransfection of EWSR1-FLI1 with a luciferase reporter under the control of the FATE1 promoter demonstrated that the presence of the EWSR1-FLI1-targeted region was necessary and sufficient to activate gene expression (Fig. 1E). Importantly, immunohistochemical staining of a panel of Ewing sarcoma patient tumor samples revealed robust FATE1 protein expression (Fig. 1F). Expression exhibited both intratumoral heterogeneity and intertumoral heterogeneity, which is a common pattern for CT antigens (29). Taken together, these data suggest that EWSR1-FLI1 modulates the chromatin state to activate expression of FATE1 in Ewing sarcoma.
FIG 1.
FATE1 is a target of the oncogenic transcription factor EWSR1-FLI1. (A) FATE1 expression relative to RPL27 (2−ΔCT) in Ewing sarcoma and other tumor types. Ewing sarcoma cell lines: EWS502, EWS-894, RD-ES, SK-ES-1, MHH-ES-1, TC-32, SK-N-MC, TC-71, and CHLA-9. Other cancers: breast (BT-20, SUM159, and WHIM12), lung (SK-LC-5, H460, H1155, and H1299), melanoma (SK-MEL-37, SK-MEL-505, and SK-MEL-2), ovarian (ES-2, HEY, and SK-OV-3), colon (RKO, HT-29, and HCT116), and bone (SW-1353, U2OS, and Saos-2). P value calculated by two-tailed Mann-Whitney test. P ≤ 0.0001. (B) Track showing EWSR1-FLI1 signal (bottom) and FAIRE signal (top) in the proximal promoter of the FATE1 gene. (C) FAIRE signal within the proximal FATE1 promoter in EWS502 cells and indicated Encyclopedia of DNA Elements (ENCODE) cell lines. NHEK, normal human epidermal keratinocyte; NHBE, normal human bronchial epithelial; HUVEC, human umbilical vein endothelial cells. (D) ATAC-seq signal for mesenchymal progenitor cells (MPCs) with or without EWSR1-FLI1 (EF). (E) Luciferase activity was measured in HEK293T cells 48 hours after transfection with the indicated cDNAs and reporters. Bars represent means (n = 6) levels of firefly luminescence normalized to Renilla luminescence ± standard errors of the means (SEM). P values were calculated by one-tailed Student's t test. CTRL, control. (F) Tumor tissue sections stained with anti-FATE1. “+Peptide” indicates a peptide block. All specimens were from Ewing sarcoma tumors unless otherwise indicated. Bars, 50 μm.
TABLE 1.
FATE1 2−ΔCT in a panel of tumor-derived cell linesa
| Cell line | Tissue | 2−ΔCT value |
|---|---|---|
| TC-71 | Ewing | 0.0053340 |
| EWS502 | Ewing | 0.0051643 |
| BT-20 | Breast | 0.0020367 |
| RD-ES | Ewing | 0.0018127 |
| CHLA-9 | Ewing | 0.0016620 |
| SK-N-MC | Ewing | 0.0011911 |
| SW-1353 | Bone | 0.0011882 |
| TC-32 | Ewing | 0.0009143 |
| SK-ES-1 | Ewing | 0.0008477 |
| MHH-ES-1 | Ewing | 0.0005450 |
| EWS-894 | Ewing | 0.0002739 |
| SK-MEL-37 | Melanoma | 0.0002005 |
| SK-LC-5 | Lung | 0.0001136 |
| RKO | Colon | 0.0001079 |
| HT-29 | Colon | 0.0000743 |
| ES-2 | Ovary | 0.0000419 |
| HEY | Ovary | 0.0000340 |
| SUM159 | Breast | 0.0000217 |
| SK-MEL-505 | Melanoma | 0.0000186 |
| U20S | Bone | 0.0000120 |
| H460 | Lung | 0.0000111 |
| SK-MEL-2 | Melanoma | 0.0000099 |
| H1155 | Lung | 0.0000088 |
| Saos-2 | Bone | 0.0000045 |
| WHIM12 | Breast | 0.0000045 |
| SK-OV-3 | Ovary | 0.0000035 |
| HCT116 | Colon | 0.0000034 |
| H1299 | Lung | 0.0000026 |
Data represent distributions of 2−ΔCT values of cell lines and tissue origins used as described in the Fig. 1A legend.
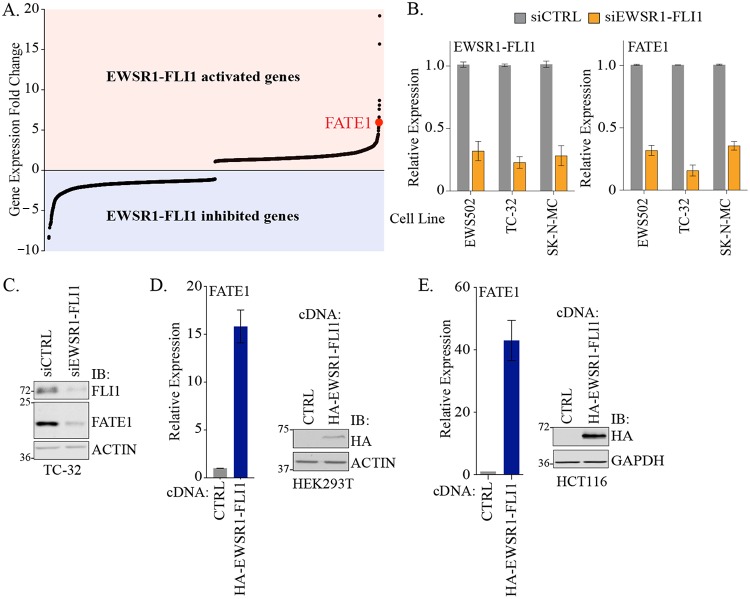
To directly assess the consequences of the presence of EWSR1-FLI1 for endogenous FATE1 gene expression, we analyzed a comparative genomics data set previously generated in EWS502 cells expressing short hairpin EWSR1-FLI1 (shEWSR1-FLI1) with or without concomitant ectopic expression of shRNA-insensitive EWSR1-FLI1 (8). In this analysis, FATE1 was the seventh most significantly differentially regulated gene (Fig. 2A). We further evaluated this finding using EWSR1-FLI1 small interfering RNA (siRNA) in EWS502 and two additional Ewing sarcoma-derived cell lines (Fig. 2B). Depletion of EWSR1-FLI1 in all cells reduced FATE1 mRNA accumulation (Fig. 2B). Furthermore, FATE1 protein levels were also diminished following loss of EWSR1-FLI1 (Fig. 2C). Conversely, transient expression of EWSR1-FLI1 activated FATE1 mRNA in non-Ewing sarcoma genetic backgrounds (HEK293T and HCT116 cells) 15-fold and 40-fold, respectively (Fig. 2D and E).
FIG 2.
EWSR1-FLI1 drives expression of FATE1. (A) Rank ordering of expression as assessed by whole-exome arrays (8). (B) mRNA expression of the indicated genes measured by qPCR following 48 h of siRNA transfection in the indicated cell lines. Bars represent means (n = 3) values relative to siCTRL ± standard deviations (SD). (C) At 96 h after transfection with the indicated siRNAs, whole-cell lysates (WCLs) were immunoblotted (IB) with the indicated antibodies. Molecular weight markers are indicated. (D) (Left) At 48 h after transfection with the indicated cDNA constructs, RNA was collected from HEK293T cells and used for qPCR analysis of FATE1 mRNA. Bars represent means (n = 3) ± SD. (Right) At 48 h after transfection, WCLs were collected and immunoblotted with the indicated antibodies. (E) (Left) At 48 h after transfection with the indicated cDNA constructs, RNA was collected from HCT116 cells and used for qPCR analysis of FATE1 mRNA. Bars represent means ± SD (n = 3). (Right) At 48 h after transfection, WCLs were collected and immunoblotted with the indicated antibodies.
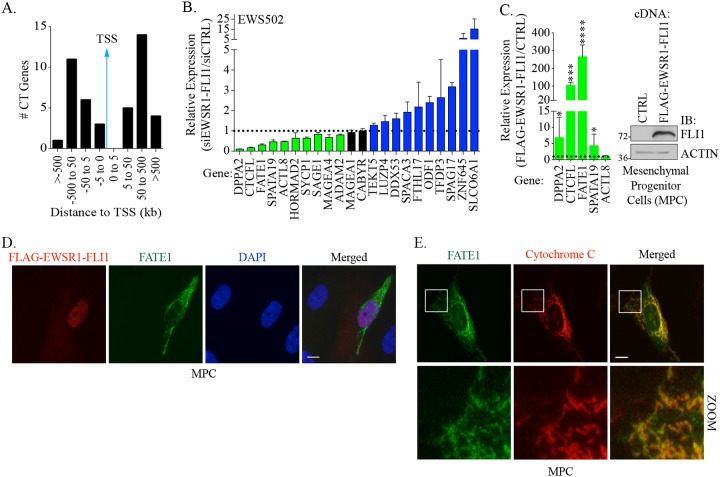
There are over 200 genes annotated as encoding CT antigens, and recent reports indicated that a number of these proteins can promote tumorigenic phenotypes and/or are targets for adoptive T cell transfer (30). To address whether FATE1 is unique among CT antigens as a target of EWSR1-FLI1, we intersected the EWSR1-FLI1-associated genes identified by ChIP-seq with the current compendium of CT antigens (8). This analysis identified 40 CT genes whose promoters were bound by EWSR1-FLI1 (Table 2). However, unlike FATE1, the majority of these binding sites were distal to the promoter regions of these genes (Fig. 3A). To examine the significance of EWSR1-FLI1 with respect to the expression of these genes, we depleted EWSR1-FLI1 in EWS502 cells and assessed the effects on their expression. Of the 22 CT antigens assayed, the expression levels of 10 (DPPA2, CTCFL, FATE1, SPATA19, ACTL8, HORMAD2, SYCP1, SAGE1, MAGEA4, and ADAM2) were reduced upon depletion of EWSR1-FLI1 (Fig. 3B). We next examined the effects of ectopic EWSR1-FLI1 expression on those CT antigens whose expression was reduced following EWSR1-FLI1 depletion. Here, we used MPCs, representing the putative cell of origin for Ewing sarcoma. Stable expression of EWSR1-FLI1 significantly induced expression of CTCFL and FATE1, with FATE1 exhibiting the highest level of induction (Fig. 3C). We next asked whether the increase in the level of FATE1 mRNA was associated with concomitant stabilization of protein. In control infected MPCs, we did not observe stabilization of FATE1 protein, indicating that its expression is low or undetectable in this cell type. However, we observed FATE1 protein stabilization in ∼10% of EWSR1-FLI1-expressing MPCs (Fig. 3D). In these cells, endogenous FATE1 localized to the mitochondria, its documented subcellular location (Fig. 3E) (28, 31). The heterogeneous stabilization of FATE1 protein in this setting suggests that additional, possibly posttranscriptional mechanisms regulate FATE1 protein stabilization in early stages of Ewing sarcoma.
TABLE 2.
Location of EWSR1-FLI1 binding sites in CT genesa
| CT gene | Chr. | Distance(s) from TSS (bp) |
|---|---|---|
| ACTL8 | 1 | 345,926 |
| ADAM2 | 8 | 129,615 |
| ADAM29b | 4 | 641,268; 798,778 |
| ANKRD45 | 1 | 57,520 |
| ARX | X | −136,105; −50,323; −8,193 |
| CABYR | 18 | −249,198 |
| CCDC33 | 15 | −7,491 |
| CEP290 | 12 | 592,754; 758,840 |
| CTCFL | 20 | 43,090 |
| CTNNA2 | 2 | 393,913 |
| DDX53 | X | −142,588 |
| DPPA2 | 3 | 128,585; 148,679 |
| ELOVL4 | 6 | 101,616 |
| FATE1 | X | −1,362; 7,391 |
| FTHL17 | X | −704,561; −123,766; 50,223 |
| GPATCH2 | 1 | 90; 59,101 |
| HORMAD2 | 22 | −51,019; 68,493 |
| LEMD1b | 1 | −73,547; −50,503; −22,986; −3,345; −1,951; 11,275 |
| LUZP4 | X | 169,138 |
| MAGEA1 | X | 59,575 |
| MAGEA4 | X | −51,623 |
| MORC1b | 3 | −69,790; −49,696; 84,884 |
| NOL4 | 18 | −955; 12,428; 29,016; 413,395 |
| ODF1 | 8 | −99,576; −75,881; 25,403 |
| PRSS54b | 16 | −36,475 |
| SAGE1 | X | −250,092 |
| SLCO6A1 | 5 | −238,290 |
| SPACA3 | 17 | −43,770; 161,106; 277,768; 281,678 |
| SPAG17 | 1 | −245,210; 11,187 |
| SPATA19 | 11 | 830,093 |
| SYCP1 | 1 | 26,743 |
| TDRD1b | 10 | −29,466 |
| TEKT5 | 16 | 80,004 |
| TEX15 | 8 | −101,981 |
| TFDP3 | X | −85,526; −13,585 |
| TMEFF1 | 9 | 95,481 |
| TMEFF2 | 2 | −805,941; −628,180; −588,256; −533,661; −244,625; −238,875; −183,539; −134,835; −115,893 |
| ZNF645 | X | 584,434 |
| NUF2 | 1 | 52,112; 208,417; 268,108; 388,760; 504,741; 547,070; 832,527 |
| OIP5 | 15 | 71,551 |
Data represent distributions of EWSR1-FLI1 binding sites in relation to cancer/testis antigen (CTA) gene transcription start sites determined by GREAT 3.0.0 analysis based on EWSR1-FLI1 ChIP-seq (8). Chr., chromosome.
mRNA was undetectable in EWS-502 by qPCR.
FIG 3.
EWSR1-FLI1 drives cancer/testis gene expression. (A) EWSR1-FLI1 binding sites near CT gene transcriptional start sites (TSSs) determined by GREAT 3.0.0 analysis based on EWSR1-FLI1 ChIP-seq (8). (B) EWS502 cells were transfected with siEWSR1-FLI1 for 48 h, and expression of the indicated mRNAs was measured by qPCR. Bars represent means (n = 3) of expression levels of the indicated genes relative to siCTRL ± SD. (C) (Left) At 96 h after infection with the indicated lentiviral constructs, RNA was collected from MPCs and used in qPCR analysis for the indicated genes. Bars represent means (n = 3) of expression levels of the indicated genes relative to CTRL cDNA (pLX304-Empty) ± SD. P values were calculated by an unpaired t test comparing data to CTRL values. ****, P ≤ 0.0001; ***, P ≤ 0.001; *, P ≤ 0.05. (Right) At 120 h after infection with the indicated lentiviral constructs, WCLs were collected and immunoblotted for the indicated antibodies. (D and E) At 96 h after infection with the indicated lentiviral constructs, MPCs were fixed and immunostained with the indicated antibodies. (D) Epifluorescent images were acquired with a 63× objective. (E) Cells were prepared as described for panel D and then confocally imaged with a 63× objective. Bars, 10 μm.
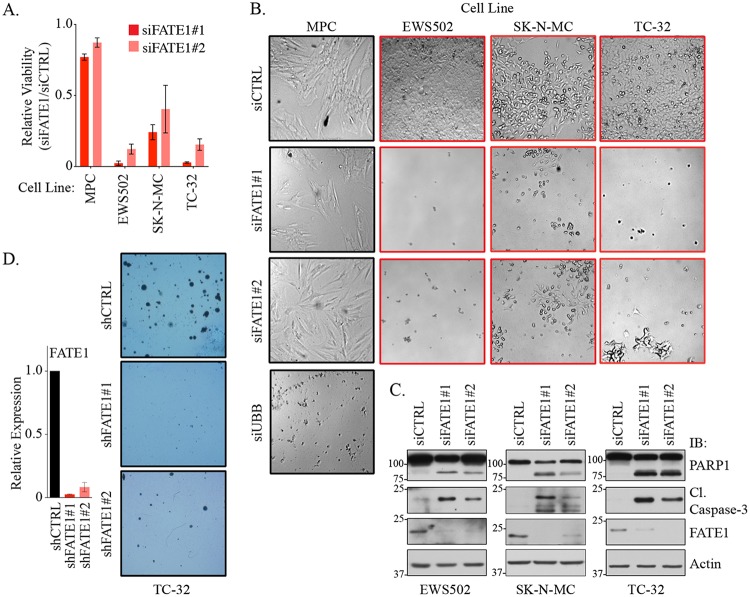
We next asked whether FATE1 is essential for Ewing sarcoma survival. In a panel of Ewing sarcoma-derived cell lines, depletion of FATE1 led to a substantial loss of viability, a phenotype not observed in MPCs (Fig. 4A and B). Furthermore, depletion of FATE1 in Ewing sarcoma cells led to accumulation of the apoptotic markers cleaved caspase-3 and cleaved poly(ADP-ribose) polymerase 1 (PARP1) (Fig. 4C). We also examined the effects of stable depletion of FATE1 on anchorage-independent growth. Using two independent shRNAs to stably deplete FATE1 in TC-32 cells, we observed a dramatic reduction of colony formation capacity (Fig. 4D). These data indicate that EWSR1-FLI1 can directly activate the expression of the CT antigen FATE1 as a means of supporting Ewing sarcoma tumor cell survival.
FIG 4.
FATE1 supports Ewing sarcoma cell viability. (A) Relative viability levels of the indicated cell lines measured 120 h after transfection with the indicated siRNAs. Bars represent means (n = 3) of viability levels relative to siCTRL ± SD. (B) Bright-field images of cells assayed as described in the panel A legend and acquired with a 5× lens objective. (C) At 72 to 96 h after siRNA transfection, WCLs were collected from the indicated cell lines and immunoblotted with the indicated antibodies. (D) (Left) At 48 h after infection with the indicated lentiviral constructs, RNA was collected and used in qPCR for analysis of FATE1 mRNA expression. Bars represent means (n = 2) of expression levels of FATE1 relative to shCTRL ± range. (Right) At 48 h after infection with the indicated lentiviral constructs, cells were seeded in soft agar. After 3 weeks, colonies were stained and imaged.
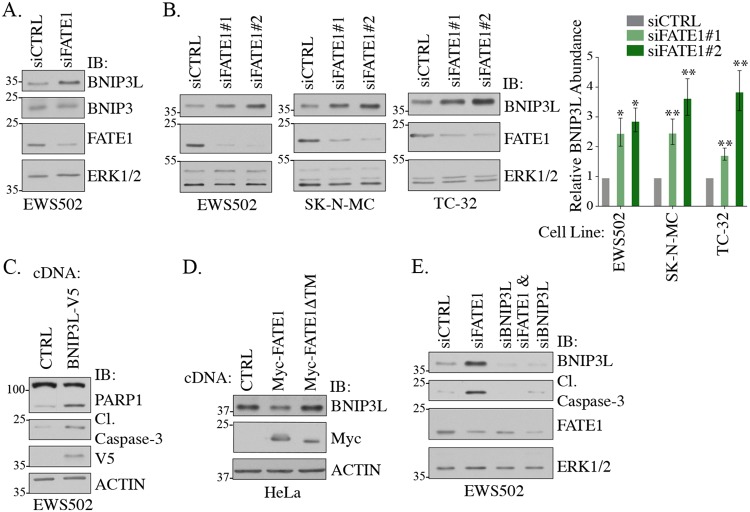
Previously, we demonstrated that FATE1 promotes stabilization of the BH3-only, proapoptotic protein BIK in colorectal, lung, breast, and cervical tumors. This activity was mediated by the FATE1-interacting partner RNF183, an endoplasmic reticulum (ER)-localized E3 ubiquitin ligase (28). We were unable to detect any appreciable levels of BIK protein in Ewing sarcoma cells, even in the presence of the proteasome inhibitor MG132 (data not shown). Furthermore, FATE1 depletion did not induce BIK accumulation, suggesting that FATE1 may have a context-selective mechanism in Ewing sarcoma (data not shown). FATE1 is reported to interact with two additional BH3 proteins, BNIP3 and BNIP3L (also known as NIX) (32, 33). These proteins are classified as atypical BH3-only proteins due to tryptophan substitutions in the highly conserved BCL2 consensus sequence (34). We evaluated accumulation of both of these proteins upon FATE1 depletion in Ewing sarcoma cells and observed stabilization of BNIP3L but not of BNIP3 (Fig. 5A). This phenotype was recapitulated in multiple Ewing sarcoma cell lines and with multiple independent siRNAs targeting FATE1 (Fig. 5B). BNIP3L is considered a tumor suppressor as its depletion increases tumorigenesis in vivo (35). Furthermore, BNIP3L is well documented to drive both apoptosis and mitophagy in multiple physiological systems (36). Given this, we hypothesized that FATE1 regulates BNIP3L to restrain cell death. Indeed, overexpression of BNIP3L in Ewing sarcoma cells promoted activation of cleaved caspase-3 and cleaved PARP (Fig. 5C). Furthermore, we found that overexpression of FATE1 is sufficient to destabilize endogenous BNIP3L protein accumulation (Fig. 5D). This phenotype required FATE1 localization to the mitochondria, as a mutant lacking the transmembrane domain, required for mitochondrial localization, did not reduce BNIP3L levels (Fig. 5D). Importantly, codepletion of BNIP3L with FATE1 rescued cleaved caspase-3, suggesting that death of Ewing sarcoma cells was mediated through BNIP3L stabilization (Fig. 5E).
FIG 5.
FATE1 destabilizes BNIP3L. (A and B) At 72 h after siRNA transfection, WCLs were collected from the indicated cell lines and immunoblotted with the indicated antibodies. Bars represent means (n = 3) of protein abundance levels relative to siCTRL ± SEM. (C) At 48 h after transfection with the indicated cDNA constructs, WCLs were collected from EWS502 cells and immunoblotted with the indicated antibodies. (D) At 48 h after transfection with the indicated cDNA constructs, WCLs were collected from HeLa cells and immunoblotted with the indicated antibodies. (E) At 72 h after siRNA transfection, WCLs were collected from EWS502 and immunoblotted with the indicated antibodies.
We next asked whether FATE1 associated with BNIP3L. We found that FATE1 interacted with BNIP3L in intact cells (Fig. 6A). On the basis of our previous findings determined with RNF183 and BIK, we next evaluated whether RNF183 was mediating BNIP3L turnover. We first validated that RNF183 protein is expressed in MPCs and a panel of Ewing sarcoma cell lines (Fig. 6B and C). Overexpression of wild-type RNF183, but not a ligase-dead (C13A/C59A) mutant, was sufficient to reduce BNIP3L protein stabilization (Fig. 6D). Moreover, addition of FATE1 and RNF183 together appeared to have an additive effect, reducing BNIP3L accumulation dramatically compared to overexpression of either protein alone (Fig. 6E). In agreement with a possible combinatorial effect, we found that BNIP3L association with RNF183 was enhanced in the presence of FATE1 (Fig. 6F). Taken together, these data indicate that BNIP3L expression is activated in Ewing sarcoma but must be tightly regulated for tumor survival. Given the relative rarity of FATE1 expression in MPCs, we hypothesize that during Ewing sarcoma tumor progression, cellular variants that engage FATE1 to promote BNIP3L turnover can overcome innate fail-safe barriers to transformation (Fig. 6G).
FIG 6.
FATE1 and RNF183 collaborate to promote BNIP3L turnover. (A) At 48 h after transfection in HEK293T, cell lysates were immunoprecipitated with c-Myc or V5 antibodies and immunoblotted with the indicated antibodies. (B) At 72 h after transfection with the indicated siRNAs, WCLs were collected from TC-32 cells and immunoblotted with the indicated antibodies. Arrows indicate RNF183 bands. (C) WCLs were collected from the indicated cell lines and immunoblotted for the indicated antibodies. (D) At 48 h after transfection with the indicated cDNA constructs, lysates were collected from HeLa cells and immunoblotted with the indicated antibodies. (E) At 48 h after transfection with the indicated cDNA constructs, lysates were collected from HeLa cells and immunoblotted with the indicated antibodies. (F) At 48 h after transfection in HEK293T cells, protein lysates were incubated with V5 antibodies and immunoblotted with the indicated antibodies. All samples contained pcDNA3-FLAG-Ub. (G) Proposed model for FATE1 contributing to Ewing sarcoma tumorigenesis.
DISCUSSION
EWSR1-FLI1 drives an anomalous gene expression program that blocks terminal differentiation and promotes proliferation, survival, and metastasis (7, 11, 37–39). Although many EWSR1-FLI1 targets have been identified through ChIP-seq and whole-genome expression analysis, only a small number of these have been validated and shown to have biological activity (7, 11, 37–42). Here, we defined the cancer/testis (CT) antigen FATE1 as a new EWSR1-FLI1 target that is essential for Ewing sarcoma survival. This finding defines CT antigens as representatives of an additional class of developmental genes aberrantly activated directly by EWSR1-FLI1. Previously, genes important for neuronal development (the NR0B1, NKX2.2, BCL11B, and SOX2 genes) have been identified as EWSR1-FLI1 targets and essential for tumorigenesis by promoting maintenance of an undifferentiated state (38, 40–42). Thus, our findings indicating that EWSR1-FLI1 can modulate expression of multiple CT antigens provide additional evidence to support the paradigm that EWSR1-FLI1 co-opts developmental programs to promote tumorigenesis.
EWSR1-FLI1 expression is tolerated but is not oncogenic in MPCs; thus, additional alterations are required for proliferation and survival (11). It is notable that only ∼10% of EWSR1-FLI1-expressing MPCs also express FATE1. As immunohistochemistry (IHC) staining results indicated that FATE1 expression is prevalent in most Ewing tumors, we hypothesize that FATE1-expressing cells may gain a selective advantage early during tumorigenesis. On the basis of our studies, we hypothesize that BNIP3L destabilization is critical for proliferation and survival of cells following EWSR1-FLI1 expression. Indeed, BNIP3L is documented to promote cell death following exposure to hypoxia in a variety of pathological settings (36, 43–45). In addition, during cardiac hypertrophy, Gαq signaling induces expression of BNIP3L, which promotes cell death (46). Alternatively, FATE1-mediated BNIP3L turnover may be essential to deflect MPC differentiation programs. EWSR1-FLI1 repression of mesenchymal differentiation programs is a key step in the pathogenesis of Ewing sarcoma (7, 12, 39). Previous reports indicated that BNIP3L promotes autophagy during differentiation of MPCs during osteogenesis and adipogenesis (47, 48). Thus, it is possible that BNIP3L destruction is essential to disrupt the autophagic and/or mitophagic balance that must be maintained for appropriate differentiation. In summary, we hypothesize that EWSR1-FLI1 expression may engage a BNIP3L-mediated tumor-suppressive response. When BNIP3L is destabilized through the activity of FATE1 and RNF183, this barrier is removed, thereby permitting uncontrolled proliferation and survival in the presence of EWSR1-FLI1.
The mechanisms that activate the expression of CT antigens are poorly defined. While DNA demethylation is thought to contribute significantly to expression in cancer cells and is exploited clinically, the specific factors that activate transcription following demethylation have been poorly described (25, 49, 50). Here, we report EWSR1-FLI1 as the first chimeric oncogenic transcription factor to directly activate CT antigens. EWSR1-FLI1 associates with GGAA repeats, which are also bound by the ETS family of transcription factors. Importantly, the founding member of the CT antigen family, MAGEA1, was reported to contain multiple ETS binding sites in its promoter (51). ETS transcription factors are frequently upregulated through either genomic alterations or signaling cascades that promote their activity (52). Thus, our findings suggest a generalizable concept that ETS transcription factors may promote oncogenesis in part through the activation of CT antigens.
MATERIALS AND METHODS
Cell lines.
H1299 and H1155 cells were obtained from John Minna (UT Southwestern [UTSW]). RKO and HT-29 cells were obtained from Jerry Shay (UTSW). TC-32, TC-71, and CHLA-9 cells were obtained from the Children’s Oncology Group. SK-MEL-2 cells were obtained from the National Cancer Institute. SK-LC-5 and SK-MEL-37 were obtained from Lloyd Old (Ludwig Institute). SUM159 cells were obtained from Asterand. HEY and ES-2 cells were obtained from Rolf Brekken (UTSW). Human mesenchymal progenitor (hMPro) cells were obtained from Aruna Biomedical. HCT116 cells were obtained from Cyrus Vaziri (University of North Carolina at Chapel Hill [UNC]). WHIM12 cells were obtained from Charles Perou (UNC). SK-MEL-505 and U2OS cells were obtained from Michael White (UTSW). EWS502, EWS894, RD-ES, SK-ES-1, MHH-ES-1, and SK-N-MC cells were obtained from Ian Davis (UNC) in 2012. BT-20, SW-1353, H460, Saos-2, and SK-OV-3 cells were purchased from ATCC. HEK293T cells were obtained from Gary Johnson (UNC) in 2010. All cell lines were cultured in the medium recommended by the provider. Authentication by short-tandem-repeat analysis was performed on H1299, H1155, RKO, HT-29, TC-32, TC-71, CHLA-9, SK-MEL-2, SK-LC-5, SK-MEL-37, SUM159, HEY, ES-2, HCT116, WHIM12, SK-MEL-505, U2OS, EWS502, EWS894, RD-ES, SK-ES-1, MHH-ES-1, SK-N-MC, BT-20, SW-1353, H460, Saos-2, and SK-OV-3 cells prior to initiation of studies. Cell lines were periodically evaluated for mycoplasma contamination by DAPI (4′,6-diamidino-2-phenylindole) staining for the presence of DNA in the cytoplasm. All cell lines were cryopreserved in liquid nitrogen, and fresh stocks were used routinely throughout the study.
siRNA transfections.
Small interfering RNA (siRNA) pools were obtained from GE Healthcare (M-015068 [FATE1#1], M-011815 [BNIP3L], and MU-007134 [RNF183#1; siGENOME siRNA]), Thermo Fisher (a pool of s40185, s40186, and s228434 for FATE1#2), or Sigma-Aldrich (a pool of SASI_Hs01_00131347, SASI_Hs01_00131348, SASI_Hs01_00131352, and SASI_Hs01_00131354 for RNF183#2). Control (siCTRL) siRNAs were either nontargeting siRNAs from GE Healthcare (D-001206; siGENOME siRNA) or a pool of 4 siRNAs targeting DLNB14 from GE Healthcare (MU-027240; siGENOME siRNA) (53). A siRNA oligonucleotide targeting the type I EWSR1-FLI1 fusion was previously described (37). A siRNA targeting UBB (M-013382-01; GE Healthcare) was used to monitor transfection efficiency, as it causes toxicity. CellTiter-Glo (CTG) values for UBB transfection had to result in a viability loss of >90% compared to the control transfected cells for inclusion (28).
Reagents.
CellTiter-Glo (CTG), a Dual-Glo luciferase assay system, and FuGENE 6 were from Promega. Lipofectamine RNAiMAX, Lipofectamine 2000, and Opti-MEM were from Thermo Fisher. Giemsa stain and Sequa-brene were from Sigma.
Antibodies.
Antibodies used for immunoblotting were as follows: from Santa Cruz Biotechnology, GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (sc-51907; 1:1,000), hemagglutinin (HA) (sc-805; 1:500), actin (sc-8432 [1:2,500] and sc-47778 [1:5,000]), c-Myc (9E10 [sc-40; 1:2,500] and A-14 [sc-789; 1:1,000]), and extracellular signal-regulated kinase 1 and 2 (ERK1/2) (sc-93; 1:1,000); from Sigma-Aldrich, FATE1 (HPA034604; 1:1,000), RNF183 (SAB2106627; 1:1,000), and HA (clone 3F10; 1:1,000); from Cell Signaling Technology, cleaved caspase-3 (9661; 1:500), PARP (9532; 1:1,000), BNIP3L (12396; 1:1,000), and V5 (13202; 1:2,500); from Abcam, FATE1 (ab111486; 1:1,000); from BD Biosciences, FLI1 (554266; 1:500) and BNIP3 (10433; 1:1,000). Antibodies for immunofluorescence were as follows: from Sigma-Aldrich, FATE1 (HPA034604; 1:100) and FLAG (F1804; 1:100); from Abcam, cytochrome c (556432; 1:200). Antibodies for immunoprecipitation were as follows: from Santa Cruz Biotechnology, c-Myc (9E10) (sc-40); from Invitrogen, V5 (46-0705).
Expression plasmids.
pCMV-HA-EWSR1-FLI1 (HA-EWSR1-FLI1) was generated by cloning HA-EWSR1-FLI1 from pLentiLox5.5 between SalI and NotI of pCMV-HA (Clontech). pCMV-HA was used as a control. 3×FLAG-EWSR1-FLI1 was obtained from James Amatruda (UTSW) and inserted into pLX304 via the Gateway LR Clonase II reaction. The control plasmid, pLX304-HcRed, was generated in a similar manner using pENTR221-HcRed and pLX304 (Addgene plasmid; catalog no. 25890). PLX304-BNIP3L-V5 was generated via the Gateway LR Clonase II reaction from pDNR221-BNIP3L. Myc-FATE1 cDNA was from pCMV-Myc-FATE. Myc-FATE1ΔTM was produced by amplifying the 486-base-pair fragment on the 5-prime end of the FATE gene and then inserting it into pCMV-Myc vector. Full-length RNF183 cDNA was obtained in pLX304 and cloned into pCMV-HA (Clontech) between SalI and NotI. HA-RNF183-C13A/C59A was generated using site-directed mutagenesis. pcDNA3-FLAG-Ub was obtained from Yue Xiong at UNC. pFATE1-Luc was produced by amplifying a 1,606-base-pair fragment from bp −1634 to −29 upstream of the FATE1 translational start site from HCT116 genomic DNA with KpnI and HindIII extensions. The insertion was ligated into pTA-Luc (Clontech) placing the FATE1 promoter insertion directly upstream of the luc gene from Photinus pyralis. pFATE1ΔEF1-Luc was produced by amplifying the 1,268 bp on the 3-prime end of the FATE1 promoter and then inserting it into pTA-Luc as described for pFATE1-Luc. pRL-CMV expressing Renilla luciferase was a gift from Deborah Chapman, University of Pittsburgh; and pCMV-dr8.91 and pCMV-VSV-G were gifts from Michael White (UTSW). psPAX2 and pMD2.G lentiviral constructs were purchased from Addgene. For shRNA experiments, pLKO.1 vectors from The RNA Consortium (TRC) expressing FATE1-targeted shRNAs (A7 [TRCN0000130192] and A9 [TRCN0000146597]) were obtained from the UNC Lentiviral core, and a nontargeting shRNA in pLKO.1 (shSCR) was used as a control (Addgene).
Luciferase assays.
Luciferase (pFATE1-LUC or pFATE1ΔEF1-LUC; 100 ng) and Renilla (pRL-CMV; 2 ng) reporters were cotransfected with 100 ng of the indicated cDNAs into HEK293T cells using FuGENE 6. At 48 h later, firefly and Renilla luciferase activities were measured using a Dual-Glo luciferase assay system modified to use 30 μl of both the Dual-Glo luciferase and Stop and Glo reagents per well.
Immunohistochemistry.
Analysis was performed on a Dako Autostainer Link 48 system. Briefly, the slides were baked for 20 min at 60°C and then deparaffinized and hydrated before the antigen retrieval step. Heat-induced antigen retrieval was performed at high pH for 20 min in a Dako PT Link system. The tissue was incubated with a peroxidase block, and then an antibody incubation (1:500 dilution) was performed for 20 min. The tissue microarrays (TMA) were composed of Ewing sarcoma selected from The University of North Carolina Surgical Pathology and St. Jude Children's Research Hospital archives under an Institutional Review Board (IRB)-approved protocol. Hematoxylin and eosin (H&E)-stained slides were reviewed, and representative areas of tumor were marked for coring. TMA blocks, containing triplicate 0.6-mm cores per case, were constructed. TMA and cell line array (CLA) blocks were cut into 4-μm and 5-μm sections, respectively, and placed on positively charged glass slides. Ewing sarcoma TMA were stained according to the protocol described above, except that a 1:250 dilution of anti-FATE (Sigma) was used. Samples from a Ewing sarcoma case were also obtained from UTSW under an IRB-approved protocol. This section was stained at 1:500. For the peptide block, a 5:1 ratio of peptide to antibody was used.
Transfections.
siRNAs were reverse transfected with Lipofectamine RNAiMAX. HCT116 cells were transfected in six-well dishes using 6 μl of Lipofectamine 2000 incubated with 2 μg/per plasmid DNA. HeLa and HEK293T cells were transfected in six-well dishes using 5 μl of FuGENE 6 incubated with 1 μg total DNA.
Immunoprecipitation.
HEK293T cells were transfected in 60-mm-diameter dishes using calcium phosphate incubated with 3 to 3.5 μg of plasmid DNA. Cells where lysed on ice for 30 min in nondenaturing lysis buffer (NDLB) containing the following ingredients: 50 mM HEPES [pH 7.4], 1.0% Triton X-100, 0.5% sodium deoxycholate, 150 mM NaCl, 1 mM NaVO4, 25 mM β-glycerophosphate, 1 mM EDTA, 1 mM EGTA, 1 μg ml−1 pepstatin, 2 μg ml−1 leupeptin, 2 μg ml−1 aprotinin, and 10 μM bestatin. Lysates were clarified at 12,000 × g for 10 min and then precleared with protein A/G beads for 2 h at 4°C. A total of 5% of each clarified lysate was set aside as an input loading control, and the remainder was immunoprecipitated for 30 min at 4°C with antibodies and protein A/G beads. Beads were washed once in high-salt (500 mM NaCl) NDLB and three times in NDLB and were then resuspended and boiled in 2× Laemmli sample buffer.
Lentiviral transduction.
Lentivirus was produced through cotransfection of HEK293T cells with 3 to 5 μg of viral expression vector, 3 μg of pCMV-dr8.91 or psPAX2 packing vector, and 300 ng of pCMV-VSV-G or 2 μg of pMD2.G envelope vector. Virus-conditioned medium was harvested, passed through 0.45-μm-pore-size filters, and then used to infect target cells in the presence of 10 μg/ml Sequa-brene.
ChIP, ATAC, and FAIRE data sets.
Whole-exome-expression EWS502 ChIP and FAIRE data sets were generated previously and deposited under GEO accession number GSE31838 (8). All FAIRE data were normalized for read depth for each experiment. MPC ATAC-seq data (GEO accession number GSE61951) were taken from reference 7.
Soft agar assays.
At 48 h after lentiviral-mediated transduction of shRNAs, 8.0 × 103 TC-32 cells were suspended in 1 ml of 0.35% Bacto agar mixed in RPMI medium containing 10% fetal calf serum (FCS) over a 1-ml base layer of 0.7% Bacto agar in the same media. A 1-ml volume of medium was added to the top of each well and exchanged twice weekly with RPMI medium containing 20% FCS for 3 weeks. At the end of 3 weeks, colonies were stained overnight with Giemsa stain and 5 distinct fields in each well were manually counted.
Gene expression.
RNA was isolated using a mammalian total RNA isolation kit (Sigma), and the concentration of RNA obtained using a NanoDrop spectrophotometer (Thermo Fisher). Equal microgram volumes of RNA were reverse transcribed for each experimental condition by the use of a High-Capacity cDNA reverse transfection kit with random hexamer primers (Thermo Fisher). The resulting cDNA was used for expression analysis performed with an Applied Biosystems real-time PCR system with TaqMan real-time PCR (Thermo Fisher) or Solaris (GE Healthcare) quantitative PCR (qPCR) probes. The TaqMan qPCR probes were as follows: FATE1 (Hs03044961_g1), EWSR1-FLI1 (Hs03024497_ft), DPPA2 (Hs00819993_g1), CTCFL (Hs00540740_m1), SPATA19 (Hs00545367_m1), ACTL8 (Hs00380546_m1), HORMAD2 (Hs00381066_m1), SYCP1 (Hs00172654_m1), SAGE1 (Hs04188949_g1), ADAM2 (Hs00155182_m1), MAGEA1 (Hs00607097_m1), MAGEA4 (Hs01025554_m1), TEXT5 (Hs01025979_m1), DDX53 (Hs00704566_s1), SPACA3 (Hs00420105_m1), FTHL17 (Hs01853260_g1), ODF1 (Hs00225767_m1), SPAG17 (Hs00400873_m1), ZNF645 (Hs00537405_s1), and SLC06A1 (Hs00542846_m1). The Solaris qPCR probes were as follows: CABYR (AX-012436-00-0100), LUZP4 (AX-021108-00-0100), and TFDP3 (AX-013860-00-0200). RPL27 was used as an internal loading assay for all expression assays (TaqMan: Hs03044961_g1). Relative expression values were calculated using the comparative threshold cycle (2−ΔΔCT) method in all experiments aside from those performed in the experiments whose results are shown in Fig. 1A (54).
Immunoblotting.
Whole-cell lysates (WCLs) were prepared in 2× Laemmli sample buffer and resolved using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Gels were transferred to Immobilon polyvinylidene difluoride (PVDF) or low-fluorescence PVDF (Millipore) membranes, blocked in either Tris-buffered saline containing 0.1% Tween 20 (TBST) and 5% nonfat dry milk or Odyssey blocking buffer (Li-COR Biosciences). This was followed by incubation with the indicated primary antibodies overnight in 5% nonfat dry milk, 5% bovine serum albumin (BSA), or Odyssey blocking buffer (Li-COR Biosciences) mixed in TBST. After washes in TBST, appropriate horseradish peroxidase (HRP)-coupled secondary antibodies (Jackson Immunoresearch) or IRDye-conjugated antibodies (Li-COR Bioscience) were used for chemiluminescence or fluorescence detection (Odyssey), respectively. Chemiluminescence (total − background) was calculated using the ImageJ integrated density function.
Immunofluorescence.
Cells plated on glass coverslips were fixed with 3.7% formaldehyde–phosphate-buffered saline (PBS) for 10 min. The coverslips were then washed once in PBS and permeabilized with 0.5% Triton X-100–PBS for 10 min. Cells were blocked in 1% bovine serum albumin (BSA)–0.1% Tween 20–1× PBS (PBTA) for 15 min and then incubated with primary antibodies for 1 h at room temperature followed by three washes in PBTA. Coverslips were then incubated with Alexa Fluor-conjugated secondary antibodies (Thermo Fisher) for 30 min followed by 3 washes in PBTA and a wash in H2O. Cells were then mounted with Prolong Gold antifade reagent with DAPI (Thermo Fisher). Images were acquired on using a Leica CTR6000B inverted microscope equipped with a Leica DFC300FX color charge-coupled-device (CCD) camera or a Zeiss LSM510 confocal microscope.
Viability assays.
Cells were reverse transfected in 96-well format with 50 to 75 nM siRNA using 0.1 to 0.3 μl of RNAiMAX diluted in Opti-MEM. The CellTiter-Glo assay was used to quantitate levels of cellular ATP modified to use 15 μl of the assay reaction mixture for each well. Luminescence was read using a Pherastar Plus plate reader (BMG Labtech). Bright-field images of cells were acquired on a Leica CTR6000B inverted microscope.
ACKNOWLEDGMENTS
Z.R.G. was supported by NIH Mechanisms of Diseases and Translational Science (MoDTS) training grant T32GM109776-02 and NSF GRFP training grant 2016219621. Z.A.G. was supported by pharmacology training grant NIH T32GM007062. P.T. was supported by general medicine training grants T32GM007040-37 and F30CA183464. I.D. was supported in part by the NCI (R01CA166447 and P30CA016086), the Corn-Hammond Fund for Childhood Cancer Research, the V Foundation for Cancer Research, and the Rita Allen Foundation. A.W.W. was supported in part by Stand Up to Cancer (IRG1211), the National Cancer Institute (CA196905), and grants from the One Million 4 Anna Foundation, Pipers Legacy, and St. Baldrick’s Foundation. The Simmons Cancer Center Support Grant (5P30 CA142543-05) supported shared resources used in this study at UTSW.
We have no conflicts of interest to disclose.
REFERENCES
- 1.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G. 1992. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 2.Aman P, Panagopoulos I, Lassen C, Fioretos T, Mencinger M, Toresson H, Hoglund M, Forster A, Rabbitts TH, Ron D, Mandahl N, Mitelman F. 1996. Expression patterns of the human sarcoma-associated genes FUS and EWS and the genomic structure of FUS. Genomics 37:1–8. doi: 10.1006/geno.1996.0513. [DOI] [PubMed] [Google Scholar]
- 3.Bailly RA, Bosselut R, Zucman J, Cormier F, Delattre O, Roussel M, Thomas G, Ghysdael J. 1994. DNA-binding and transcriptional activation properties of the EWS-FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol Cell Biol 14:3230–3241. doi: 10.1128/MCB.14.5.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lessnick SL, Ladanyi M. 2012. Molecular pathogenesis of Ewing sarcoma: new therapeutic and transcriptional targets. Annu Rev Pathol 7:145–159. doi: 10.1146/annurev-pathol-011110-130237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeva V, Surdez D, Guillon N, Tirode F, Fejes AP, Delattre O, Barillot E. 2010. De novo motif identification improves the accuracy of predicting transcription factor binding sites in ChIP-Seq data analysis. Nucleic Acids Res 38:e126. doi: 10.1093/nar/gkq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guillon N, Tirode F, Boeva V, Zynovyev A, Barillot E, Delattre O. 2009. The oncogenic EWS-FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PLoS One 4:e4932. doi: 10.1371/journal.pone.0004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suva ML, Rossetti NE, Boonseng WE, Oksuz O, Cook EB, Formey A, Patel A, Gymrek M, Thapar V, Deshpande V, Ting DT, Hornicek FJ, Nielsen GP, Stamenkovic I, Aryee MJ, Bernstein BE, Rivera MN. 2014. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 26:668–681. doi: 10.1016/j.ccell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel M, Simon JM, Iglesia MD, Wu SB, McFadden AW, Lieb JD, Davis IJ. 2012. Tumor-specific retargeting of an oncogenic transcription factor chimera results in dysregulation of chromatin and transcription. Genome Res 22:259–270. doi: 10.1101/gr.125666.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. 2007. Mesenchymal stem cell features of Ewing tumors. Cancer Cell 11:421–429. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 10.Riggi N, Cironi L, Provero P, Suva ML, Kaloulis K, Garcia-Echeverria C, Hoffmann F, Trumpp A, Stamenkovic I. 2005. Development of Ewing's sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Res 65:11459–11468. doi: 10.1158/0008-5472.CAN-05-1696. [DOI] [PubMed] [Google Scholar]
- 11.Riggi N, Suva ML, Suva D, Cironi L, Provero P, Tercier S, Joseph JM, Stehle JC, Baumer K, Kindler V, Stamenkovic I. 2008. EWS-FLI-1 expression triggers a Ewing's sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res 68:2176–2185. doi: 10.1158/0008-5472.CAN-07-1761. [DOI] [PubMed] [Google Scholar]
- 12.Gomez NC, Hepperla AJ, Dumitru R, Simon JM, Fang F, Davis IJ. 2016. Widespread chromatin accessibility at repetitive elements links stem cells with human cancer. Cell Rep 17:1607–1620. doi: 10.1016/j.celrep.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Womer RB, West DC, Krailo MD, Dickman PS, Pawel BR, Grier HE, Marcus K, Sailer S, Healey JH, Dormans JP, Weiss AR. 2012. Randomized controlled trial of interval-compressed chemotherapy for the treatment of localized Ewing sarcoma: a report from the Children's Oncology Group. J Clin Oncol 30:4148–4154. doi: 10.1200/JCO.2011.41.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bacci G, Ferrari S, Longhi A, Donati D, De Paolis M, Forni C, Versari M, Setola E, Briccoli A, Barbieri E. 2003. Therapy and survival after recurrence of Ewing's tumors: the Rizzoli experience in 195 patients treated with adjuvant and neoadjuvant chemotherapy from 1979 to 1997. Ann Oncol 14:1654–1659. doi: 10.1093/annonc/mdg457. [DOI] [PubMed] [Google Scholar]
- 15.McTiernan AM, Cassoni AM, Driver D, Michelagnoli MP, Kilby AM, Whelan JS. 2006. Improving outcomes after relapse in Ewing's sarcoma: analysis of 114 patients from a single institution. Sarcoma 2006:1. doi: 10.1155/SRCM/2006/83548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, Kiezun A, Carter SL, Shukla SA, Mehta SS, Thorner AR, de Torres C, Lavarino C, Sunol M, McKenna A, Sivachenko A, Cibulskis K, Lawrence MS, Stojanov P, Rosenberg M, Ambrogio L, Auclair D, Seepo S, Blumenstiel B, DeFelice M, Imaz-Rosshandler I, Schwarz-Cruz YCA, Rivera MN, Rodriguez-Galindo C, Fleming MD, Golub TR, Getz G, Mora J, Stegmaier K. 2014. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov 4:1326–1341. doi: 10.1158/2159-8290.CD-13-1037. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, Kiezun A, Hammerman PS, McKenna A, Drier Y, Zou L, Ramos AH, Pugh TJ, Stransky N, Helman E, Kim J, Sougnez C, Ambrogio L, Nickerson E, Shefler E, Cortes ML, Auclair D, Saksena G, Voet D, Noble M, DiCara D, Lin P, Lichtenstein L, Heiman DI, Fennell T, Imielinski M, Hernandez B, Hodis E, Baca S, Dulak AM, Lohr J, Landau DA, Wu CJ, Melendez-Zajgla J, Hidalgo-Miranda A, Koren A, McCarroll SA, Mora J, Lee RS, Crompton B, Onofrio R, et al. 2013. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scanlan MJ, Simpson AJ, Old LJ. 2004. The cancer/testis genes: review, standardization, and commentary. Cancer Immun 4:1. [PubMed] [Google Scholar]
- 19.Gibbs ZA, Whitehurst AW. 2018. Emerging contributions of cancer/testis antigens to neoplastic behaviors. Trends Cancer 4:701–712. doi: 10.1016/j.trecan.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitehurst AW. 2014. Cause and consequence of cancer/testis antigen activation in cancer. Annu Rev Pharmacol Toxicol 54:251–272. doi: 10.1146/annurev-pharmtox-011112-140326. [DOI] [PubMed] [Google Scholar]
- 21.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. 2014. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer 14:135–146. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 22.Robbins PF, Morgan RA, Feldman SA, Yang JC, Sherry RM, Dudley ME, Wunderlich JR, Nahvi AV, Helman LJ, Mackall CL, Kammula US, Hughes MS, Restifo NP, Raffeld M, Lee CC, Levy CL, Li YF, El-Gamil M, Schwarz SL, Laurencot C, Rosenberg SA. 2011. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol 29:917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnadas DK, Shusterman S, Bai F, Diller L, Sullivan JE, Cheerva AC, George RE, Lucas KG. 2015. A phase I trial combining decitabine/dendritic cell vaccine targeting MAGE-A1, MAGE-A3 and NY-ESO-1 for children with relapsed or therapy-refractory neuroblastoma and sarcoma. Cancer Immunol Immunother 64:1251–1260. doi: 10.1007/s00262-015-1731-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sigalotti L, Fratta E, Coral S, Tanzarella S, Danielli R, Colizzi F, Fonsatti E, Traversari C, Altomonte M, Maio M. 2004. Intratumor heterogeneity of cancer/testis antigens expression in human cutaneous melanoma is methylation-regulated and functionally reverted by 5-aza-2'-deoxycytidine. Cancer Res 64:9167–9171. doi: 10.1158/0008-5472.CAN-04-1442. [DOI] [PubMed] [Google Scholar]
- 25.De Smet C, Lurquin C, Lethe B, Martelange V, Boon T. 1999. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol 19:7327–7335. doi: 10.1128/MCB.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitehurst AW, Xie Y, Purinton SC, Cappell KM, Swanik JT, Larson B, Girard L, Schorge JO, White MA. 2010. Tumor antigen acrosin binding protein normalizes mitotic spindle function to promote cancer cell proliferation. Cancer Res 70:7652–7661. doi: 10.1158/0008-5472.CAN-10-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cappell KM, Sinnott R, Taus P, Maxfield K, Scarbrough M, Whitehurst AW. 2012. Multiple cancer testes antigens function to support tumor cell mitotic fidelity. Mol Cell Biol 32:4131–4140. doi: 10.1128/MCB.00686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maxfield KE, Taus PJ, Corcoran K, Wooten J, Macion J, Zhou Y, Borromeo M, Kollipara RK, Yan J, Xie Y, Xie XJ, Whitehurst AW. 2015. Comprehensive functional characterization of cancer-testis antigens defines obligate participation in multiple hallmarks of cancer. Nat Commun 6:8840. doi: 10.1038/ncomms9840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jungbluth AA, Stockert E, Chen YT, Kolb D, Iversen K, Coplan K, Williamson B, Altorki N, Busam KJ, Old LJ. 2000. Monoclonal antibody MA454 reveals a heterogeneous expression pattern of MAGE-1 antigen in formalin-fixed paraffin embedded lung tumours. Br J Cancer 83:493–497. doi: 10.1054/bjoc.2000.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almeida LG, Sakabe NJ, deOliveira AR, Silva MC, Mundstein AS, Cohen T, Chen YT, Chua R, Gurung S, Gnjatic S, Jungbluth AA, Caballero OL, Bairoch A, Kiesler E, White SL, Simpson AJ, Old LJ, Camargo AA, Vasconcelos AT. 2009. CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res 37:D816–D819. doi: 10.1093/nar/gkn673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doghman-Bouguerra M, Granatiero V, Sbiera S, Sbiera I, Lacas-Gervais S, Brau F, Fassnacht M, Rizzuto R, Lalli E. 2016. FATE1 antagonizes calcium- and drug-induced apoptosis by uncoupling ER and mitochondria. EMBO Rep 17:1264–1280. doi: 10.15252/embr.201541504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu H, Tardivo L, Tam S, Weiner E, Gebreab F, Fan C, Svrzikapa N, Hirozane-Kishikawa T, Rietman E, Yang X, Sahalie J, Salehi-Ashtiani K, Hao T, Cusick ME, Hill DE, Roth FP, Braun P, Vidal M. 2011. Next-generation sequencing to generate interactome datasets. Nat Methods 8:478–480. doi: 10.1038/nmeth.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. 2005. Towards a proteome-scale map of the human protein-protein interaction network. Nature 437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 34.Aouacheria A, Brunet F, Gouy M. 2005. Phylogenomics of life-or-death switches in multicellular animals: Bcl-2, BH3-Only, and BNip families of apoptotic regulators. Mol Biol Evol 22:2395–2416. doi: 10.1093/molbev/msi234. [DOI] [PubMed] [Google Scholar]
- 35.Fei P, Wang W, Kim SH, Wang S, Burns TF, Sax JK, Buzzai M, Dicker DT, McKenna WG, Bernhard EJ, El-Deiry WS. 2004. Bnip3L is induced by p53 under hypoxia, and its knockdown promotes tumor growth. Cancer Cell 6:597–609. doi: 10.1016/j.ccr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Ney PA. 2009. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ 16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prieur A, Tirode F, Cohen P, Delattre O. 2004. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol Cell Biol 24:7275–7283. doi: 10.1128/MCB.24.16.7275-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riggi N, Suva ML, De Vito C, Provero P, Stehle JC, Baumer K, Cironi L, Janiszewska M, Petricevic T, Suva D, Tercier S, Joseph JM, Guillou L, Stamenkovic I. 2010. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev 24:916–932. doi: 10.1101/gad.1899710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torchia EC, Jaishankar S, Baker SJ. 2003. Ewing tumor fusion proteins block the differentiation of pluripotent marrow stromal cells. Cancer Res 63:3464–3468. [PubMed] [Google Scholar]
- 40.Smith R, Owen LA, Trem DJ, Wong JS, Whangbo JS, Golub TR, Lessnick SL. 2006. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell 9:405–416. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Wiles ET, Lui-Sargent B, Bell R, Lessnick SL. 2013. BCL11B is up-regulated by EWS/FLI and contributes to the transformed phenotype in Ewing sarcoma. PLoS One 8:e59369. doi: 10.1371/journal.pone.0059369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kinsey M, Smith R, Lessnick SL. 2006. NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing's sarcoma. Mol Cancer Res 4:851–859. doi: 10.1158/1541-7786.MCR-06-0090. [DOI] [PubMed] [Google Scholar]
- 43.Bruick RK. 2000. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci U S A 97:9082–9087. doi: 10.1073/pnas.97.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sowter HM, Ratcliffe PJ, Watson P, Greenberg AH, Harris AL. 2001. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res 61:6669–6673. [PubMed] [Google Scholar]
- 45.Regula KM, Ens K, Kirshenbaum LA. 2002. Inducible expression of BNIP3 provokes mitochondrial defects and hypoxia-mediated cell death of ventricular myocytes. Circ Res 91:226–231. doi: 10.1161/01.RES.0000029232.42227.16. [DOI] [PubMed] [Google Scholar]
- 46.Yussman MG, Toyokawa T, Odley A, Lynch RA, Wu G, Colbert MC, Aronow BJ, Lorenz JN, Dorn GW II.. 2002. Mitochondrial death protein Nix is induced in cardiac hypertrophy and triggers apoptotic cardiomyopathy. Nat Med 8:725–730. doi: 10.1038/nm719. [DOI] [PubMed] [Google Scholar]
- 47.Song BQ, Chi Y, Li X, Du WJ, Han ZB, Tian JJ, Li JJ, Chen F, Wu HH, Han LX, Lu SH, Zheng YZ, Han ZC. 2015. Inhibition of Notch signaling promotes the adipogenic differentiation of mesenchymal stem cells through autophagy activation and PTEN-PI3K/AKT/mTOR pathway. Cell Physiol Biochem 36:1991–2002. doi: 10.1159/000430167. [DOI] [PubMed] [Google Scholar]
- 48.Sbrana FV, Cortini M, Avnet S, Perut F, Columbaro M, De Milito A, Baldini N. 2016. The role of autophagy in the maintenance of stemness and differentiation of mesenchymal stem cells. Stem Cell Rev 12:621–633. doi: 10.1007/s12015-016-9690-4. [DOI] [PubMed] [Google Scholar]
- 49.Srivastava P, Paluch BE, Matsuzaki J, James SR, Collamat-Lai G, Blagitko-Dorfs N, Ford LA, Naqash R, Lübbert M, Karpf AR, Nemeth MJ, Griffiths EA. 2016. Induction of cancer testis antigen expression in circulating acute myeloid leukemia blasts following hypomethylating agent monotherapy. Oncotarget 7:12840–12856. doi: 10.18632/oncotarget.7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Griffiths EA, Srivastava P, Matsuzaki J, Brumberger Z, Wang ES, Kocent J, Miller A, Roloff GW, Wong HY, Paluch BE, Lutgen-Dunckley LG, Martens BL, Odunsi K, Karpf AR, Hourigan CS, Nemeth MJ. 2018. NY-ESO-1 vaccination in combination with decitabine induces antigen-specific T-lymphocyte responses in patients with myelodysplastic syndrome. Clin Cancer Res 24:1019–1029. doi: 10.1158/1078-0432.CCR-17-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Smet C, Courtois SJ, Faraoni I, Lurquin C, Szikora JP, De Backer O, Boon T. 1995. Involvement of two Ets binding sites in the transcriptional activation of the MAGE1 gene. Immunogenetics 42:282–290. doi: 10.1007/BF00176446. [DOI] [PubMed] [Google Scholar]
- 52.Sizemore GM, Pitarresi JR, Balakrishnan S, Ostrowski MC. 2017. The ETS family of oncogenic transcription factors in solid tumours. Nat Rev Cancer 17:337–351. doi: 10.1038/nrc.2017.20. [DOI] [PubMed] [Google Scholar]
- 53.Whitehurst AW, Bodemann BO, Cardenas J, Ferguson D, Girard L, Peyton M, Minna JD, Michnoff C, Hao W, Roth MG, Xie XJ, White MA. 2007. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature 446:815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- 54.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. [DOI] [PubMed] [Google Scholar]