Abstract
The development of biotechnology-based active pharmaceutical ingredients, such as GLP-1 analogs, brought changes in type 2 diabetes treatment options. For better therapeutic efficiency, these active pharmaceutical ingredients require appropriate administration, without the development of adverse effects or toxicity. Therefore, it is required to develop several quantification methods for GLP-1 analogs products, in order to achieve the therapeutic goals, among which ELISA and HPLC arise. These methods are developed, optimized and validated in order to determine GLP-1 analogs, not only in final formulation of the active pharmaceutical ingredient, but also during preclinical and clinical trials assessment. This review highlights the role of ELISA and HPLC methods that have been used during the assessment for GLP-1 analogs, especially for exenatide.
Keywords: Type 2 diabetes, Exenatide, Therapeutic drug monitoring, ELISA, HPLC
Graphical abstract
Highlights
-
•
Crucial role of exenatide's quantification during preclinical, clinical and biosimilaritystudies.
-
•
Description of ELISA and HPLC-MS main features and comparison for exenatide's quantification.
-
•
ELISA and HPLC-MS applicability in target proteomics.
-
•
Relevance of the validation quantification techniques.
1. Introduction
Type 2 diabetes (T2D) is the more common type of diabetes. It is a complex endocrine metabolic disease associated with obesity and a sedentary life style and it is considered to be, at present, a major health care problem in the world [1]. T2D arises from the reciprocal action between genetic and environmental causes that lead to an abnormal response of the skeletal muscle, liver and adipose tissue to insulin and the pancreatic ß-cells dysfunction [[1], [2], [3]]. The disease is characterized by the dysregulation of carbohydrates, lipids and proteins metabolism that outburst in insulin impaired secretion, insulin resistance or both [2].
Insulin resistance is considered to be the major “trademark” for T2D and its development is associated with tissue-specific inflammatory responses induced by pro-inflammatory cytokines and oxidative stress mediators [4]. Chronic exposure to the inflammatory and oxidative stress environment promotes the blockage of insulin receptor in the pancreatic ß-cell islets [5]. T2D begins to settle when the ability of the pancreatic ß-cells to segregate enough insulin to overcome insulin resistance in the tissues is compromised [4]. In T2D, due to insulin resistance, patients have high levels of blood glucose (hyperglycemia), which contribute to the oxidative stress, inflammatory pathways activation and microvascular conditions, which may evolve to macrovascular disease, augmenting the T2D's morbidity and mortality [1].
The key symptom that prones the development of T2D is hyperglycemia and it appears early in the settlement of the disease, known as prediabetes [2]. Patients with prediabetes can display modifications in three different markers: glycated hemoglobin (HbA1c) or fasting glucose (FG) or glucose tolerance (GT) [6]. Therefore, patients with prediabetes may have an enhancement in HbA1c or an impairment in the FG or GT levels [2].
T2D treatment is focused in achieving optimal control of blood glucose levels [2]. Oral therapies associated with changes in the life style, in combination, or not, with injectable therapy, are the core of T2D treatment. Even though, T2D is a multifactorial disease and its treatment responsiveness is highly variable accordingly to the complexity of the pathogenic process [1]. The therapies for T2D available in the market are biguanides, first generation sulfonylureas, second generation sulfonylureas, meglitinides, α-glucosidase inhibitors, thiazolidinediones, DPP-4 inhibitors [1,2], glucagon-like peptide-1 (GLP-1) receptor agonists and sodium-glucose co-transporter 2 (SGLT2) inhibitors. However, this review is focused on GLP-1 receptor agonists (GLP-1RA), more specifically on exenatide.
During GLP-1RA development process, as well as for other active pharmaceutical ingredients (API), preclinical studies are required to assess the API's pharmacokinetic and pharmacodynamic profiles and its safety and toxicity, in order to evaluate potential risks associated with the GLP-1RA and, consequently, identify and quickly act when a risk factor appears during the development in clinical trials onset [7]. Therefore, it is crucial to proceed to the monitoring of GLP-1RA during preclinical and clinical trials development, in order to predict the behavior of the API and to reduce possible erroneous information that may affect the therapeutic goal of GLP-1RA.
Nowadays the therapeutic pipeline for T2D is broad although during a long period this was not the reality [1]. Consequently, it is important that the patient's disease management includes a careful antidiabetic therapy selection, regarding the age, comorbidities, HbA1c levels and the adverse effects (AE) associated with each API [8]. Thus, it is important to minimize the AE of the API without compromising the required therapeutic concentration. Although the therapeutic concentration required for patient A may not be the same for patient B and, consequently, a posologic regimen may be accurate for one patient and fail in another, resulting in AE occurrence. Therefore, to avoid AE it is crucial to develop therapeutic drug monitoring (TDM) programs, allowing therapeutic individualization [8].
To develop and apply monitoring programs in preclinical and clinical trials and TDM programs for antidiabetic therapies, it is important to have quantification techniques allowing the evaluation of their pharmacokinetic and pharmacodynamic profiles, safety and toxicity, regarding the individualization of the therapy [9]. Since GLP-1RA are therapeutic peptides, their analytical quantification requires a specific, multiplexing and high throughput quantification technique [10]. The development of techniques for GLP-1RA quantification demands the development and optimization of sample pretreatment procedures, in order to decrease matrix complexity and to concentrate the API; however, it is a burdensome process.
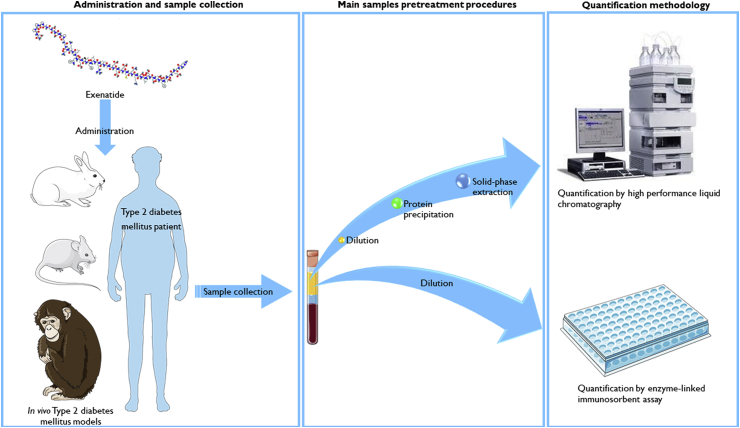
Fig. 1 outlines a scheme regarding the administration of exenatide, the sample collection from humans or laboratory animals, the pretreatment procedures that may be applied to the samples before quantification and the main bioanalytical methods applied for exenatide's quantification. Exenatide may be administered to humans, in clinical practice or in clinical trials assessment, and to laboratory animals, during preclinical assessment of new API formulations. As already emphasized, exenatide's monitoring is important and crucial; thus it requires sample collection (blood, tissue or API release medium). Before the quantification of exenatide through bioanalytical methodologies, collected samples need to be addressed properly through optimized pretreatment procedures, in order to avoid interferences, which may affect the quantification or promote damage to the analytical equipment, and to concentrate samples with low concentration of exenatide. Once the quantification is finished, clinical decisions may be taken regarding the best therapeutic outcome and preclinical development may continue towards clinical trials.
Fig. 1.
Exenatide's pathway since its administration to its quantification by high performance liquid chromatography or enzyme-linked immunosorbent assay, comprising the main pretreatment procedures applied to samples before each bioanalytical methodology.
The main aim of this review is to describe and critically analyze the quantification techniques applied to GLP-IRA, with focus on exenatide, including pretreatment procedures applied and recent advances regarding the detection of therapeutic proteins, such as exenatide, using mass spectrometry detectors, a leading marked change into the bioanalytical quantification.
2. GLP-1 receptor agonists
Discovered in 1987, GLP-1 is a 30 amino acid peptide and an incretin hormone released by intestinal L-cells after food intake and it has an important role in the glucose homeostasis [11,12]. After its release, GLP-1 binds to the GPL-1 receptor (GLP-1R) present in pancreas, brain, heart, kidney and gastrointestinal tract, activating it, promoting the increase in cAMP and intracellular calcium levels that lead to the glucose-dependent insulin exocytosis [11]. GLP-1 has antidiabetic functions important for T2D treatment amongst which are auto-limited insulinotropic effect that reduces the risk of hypoglycemia; regulation of postprandial glucose levels; stimulation of pancreatic ß-cells proliferation and neogenesis; inhibition of pancreatic ß-cells apoptosis; prevention of gastric emptying that promotes satiety and loss of body weight [11]. Although native GLP-1 has a short life-time due to its rapid degradation by dipeptidyl peptidase IV (DPP-IV), limiting its therapeutic application for T2D [11]. Therefore, the GLP-1RA were developed, in order to mimic the endogenous GLP-1 incretin action.
GLP-1RA are a therapeutic option for T2D and result in the increased demand for new T2D API, which has the ability to achieve optimal glycemic control and, at the same time, lower glucose plasma levels, to manage complications and to prevent the progression of the disease [11]. The GLP-1RA therapeutic group is heterogenous regarding pharmacology, administration, pharmacokinetics, tolerability and immunogenicity [13]. In order to extend the half-life of GLP-1 and decrease its renal elimination, strategies were undertaken to make GLP-1 resistant to DPP-IV degradation, through amino acid substitution at N-terminal positions or to allow the extension of GLP-1 actions, through binding the peptide to plasma albumin in order to augment its metabolic stability [11,14]. Regarding the strategy applied to the peptide, it is possible to classify the GLP-1RA into short-acting and long-acting [11].
A major difference between the short-acting and the long-acting GLP-1RA is that the short-acting peptides suffer fluctuations in plasma levels but the long-acting do not. Hence, it may lead to the supra-activation of the GLP-1R [14]. This difference is translated to different action mechanisms, efficacy and tolerability [14]. Therefore, the long-acting GLP-1RA are more prone to be accepted by patients, not only because they achieve better result in the same period, but also due to the lower injections frequency [14]. Although the decision of which API is more suitable for the patient is taken accordingly to his or her disease's profile.
2.1. Short-acting GLP-1RA
Exenatide is the synthetic form of exendin-4, a 39 amino acid peptide isolated from Gyla monster (Heloderma suspectum) saliva [11] with a molecular weight of 4186.6 Da [15] and natural resistance to DPP-IV degradation. It was the first approved GLP-1RA for human clinical application [12], as an adjunctive T2D therapy for patients who have not achieved the optimal glycemic control with oral therapies [16]. Although as a short-acting GLP-1RA with natural resistance to the proteolysis by DPP-IV, exenatide's half-life is 2.4 h, requiring two administrations per day [13], leading to a not simple treatment regimen, compromising the therapeutic adherence [17,18]. The other short-acting GLP-1RA is lixisenatide differing from exendin-4 at C-terminal amino acids, although it has a half-life of 3–4 h [14].
Short-acting GLP-1RA have a shorter half-life, not affecting insulin postprandial secretion. However, they induce noticeable gastric emptying and promote similar glucagon secretion suppression as long-acting GLP-1RA. Therefore, they offer better postprandial hyperglycemia control and are useful to enhance or substitute rapid-acting insulin during meals [19]. Due to better effect on decreasing postprandial hyperglycemia, short-acting GLP-1RA are more prone to develop AE, particularly gastrointestinal AE [20].
Short-acting exenatide decreases HbA1c levels (mean 0.98%), fasting blood glucose (1.69 mmol/L) and body weight (1.5 kg) [13,21]. It has shown to be non-inferior to insulin glargine, in combination with metformin or sulfonylurea.
2.2. Long-acting GLP-1RA
The long-acting exenatide (exenatide-LA) may be administered by subcutaneous injection once weekly, due to its biodegradable microspheres formulation, which allows the slow release of exenatide through diffusion [14]. Exenatide-LA is not a different molecular entity from exenatide twice daily, although the differences in the formulation allow the slower metabolism of the API and, consequently, a longer half-life and a plasma concentration peak at 2 weeks after administration [22].
The other long-acting GPL-1RA are the products of different mechanisms applied to improve the half-life of the API. Liraglutide has a different amino acid at 34 position (Lys for Arg) and a linear fatty acid moiety bound to a glutamic acid, in order to improve its half-life (12–14 h) and its affinity to serum albumin, allowing an administration per day [13]. Albiglutide consists of two copies of a modified human GLP-1 coupled to recombinant albumin, resulting in DPP-IV proteolysis resistance, clearance reduction and extended half-life (6–8 days) [23]. Regarding dulaglutide, it is composed by two identical and DPP-IV resistant sulfidic chains coupled to a G4 modified immunoglobulin (IgG4), allowing the reduction of the clearance and the binding to the Fc receptors, in order to reduce antibodies production [14].
Exenatide-LA has the ability to reduce HbA1c (mean 1.4%), fasting blood glucose (mean −1.94 mmol/L) and the body weight (mean 2.5 kg), with sustained effects for 5 years [21]. It has shown to be higher than DPP-IV and not lower than metformin; however, it has shown inferiority regarding pioglitazone [13].
3. Preclinical trials and biosimilarity assessment monitoring
Preclinical trials enable the prevision of the API's action mechanism, its pharmacokinetic and pharmacodynamic profiles and its safe and non-toxic human dosages [7]. Therefore, preclinical trials are important regarding new API for T2D treatment. Preclinical investigation has been comprehensively researching exenatide's pharmacology, allowing the assessment of exenatide's ability to decrease glucose levels, its insulinotropic characteristics and longer period of activity [15]. Even though, the results may not always be parallel with the ones obtained during clinical trials. Through preclinical trials, it has been possible to assess exenatide's pharmacokinetic and pharmacodynamic profiles, allowing the elaboration of suppositions that may be corroborated during clinical trials. It is important to highlight that preclinical trials data combined with in vitro results regarding therapeutic peptides, such as exenatide, or other biotechnology-based API, enable the establishment of in vitro/in vivo correlations that are important to predict pharmacokinetic and pharmacodynamic profiles, enabling the assessment of the quantity of API that is available to exert therapeutic action [24].
However, the development of biotechnology-based API such as peptide-based API is expensive and, consequently, it affects negatively in the clinical practice [25]. Therefore, the development of biosimilars API is enhancing. A biosimilar API has resemblance to the biological medicine, even though it is not equal; it represents an advanced strategy for therapeutic application. Thus, it is highly important to evaluate the similarity between the biosimilar and the biological medicine during its development, in order to evaluate the existence or non-existence of significant clinical divergences [26].
Recently, there have been advances towards oral delivery systems for exenatide, due to poor therapeutic adherence to the available subcutaneous therapies for exenatide [27]. However, exenatide is submitted to enzymatic digestion and has poor intestinal permeability, leading to poor bioavailability [28]. Hence, researchers are investigating in in vitro studies and preclinical trials for oral delivery systems, in order to overcome exenatide's poor oral bioavailability. Amongst these systems, oral delivery systems with self-emulsifying properties, or methods combining an exenatide complex with ions of zinc and transferrin, or protamine-functionalized nanoparticles, may be highlighted [[27], [28], [29]]. Due to potential protective effects on cardiovascular system, researchers are investigating in in vitro studies and preclinical trials for biotechnology-based systems with exenatide, aiming its application in patients with myocardial ischemia [30]. Even though the results obtained so far are promising, it is still important to continue to study and investigate in vitro and preclinical studies, in order to obtain robust data, aiming the clinical trial scale-up and the market authorization by the authorities.
Monitoring during preclinical or biosimilar development is indispensable to provide qualitative and quantitative assessment of the pharmacokinetic and pharmacodynamic profiles and the safety and toxicity of new API. Therefore, it is required a sensible, sensitive, high-throughput and robust bioanalytical assay [31]. Only through monitoring it may achieve the efficacy, safety, toxicity and characterization information needed to, or not to, scale-up into clinical trials, in order to obtain authorities approval for its use in the clinical practice [31].
4. Therapeutic drug monitoring
Peptides are appearing as a new drug category in clinical therapy [9]. The interest in incretins has been growing since the last decade because of T2D treatment and the additional protective effects on central nervous and cardiovascular systems [32]. Thus, the GLP-1RA were, and still are, in investigation and, like other drugs, they have AE that compromise the quality of the patient's life and, consequently, reduce the treatment adherence [32]. GLP-1RA efficacy is dose dependent and the more common AE are nausea, vomiting, injection-site reactions and systemic allergic reactions [32]. However, there are concerns about their carcinogenic potential and capability to increase pancreatitis rate [32]. The gastrointestinal AE are dose dependent, normally transient, with a non-severe nature, limiting the administration at the utmost efficacious dosage, because the maximum tolerated doses are determined by the induction of nausea and vomiting [20,32]. Besides the above-descripted AE, GLP-1RA derived from exendin-4, the natural form of exenatide, promote the formation of antibodies, although they do not react with native GLP-1 and do not decrease the API efficacy [20]. Comparing exenatide's twice daily and long-acting formulations, significant HbA1c reductions were achieved with exenatide-LA, with lower incidence on gastrointestinal AE, however with higher incidence on injection-site AE and antibody formation [13].
In order to control GLP-1RA AE, it is important to establish TDM programs [33]. These programs are capable of determining the concentration of the API in blood, plasma, serum or tissues, allowing the optimization of the posologic regimen. There are few case reports regarding the TDM of exenatide; therefore, it is at the utmost importance to quantify of exenatide in biological matrices, in order to assess its pharmacokinetics and efficacy, monitoring the progression of the disease and predicting its toxicity. Even though, patients should be educated towards the symptoms of pancreatitis and the self-monitoring of blood glucose levels [34]. For the establishment of exenatide's TDM programs, it is necessary to develop and validate bioanalytical techniques that will allow a quick therapeutic intervention towards patient care optimization and the disease's control [35].
5. Quantification techniques
As above stated, exenatide quantification during clinical practice and preclinical and biosimilar assessment is crucial for the patient care and the formulation optimization, in order to achieve its efficacy and toxicity and to monitor the disease's progression [9,35]. In clinical practice laboratories, biosimilar API and therapeutic peptides, like exenatide, are submitted to quantitative analysis mostly by enzyme-linked immunosorbent assay (ELISA) or by high performance liquid chromatography (HPLC) [36].
ELISA is a classical analytical technique used for the quantification of peptides, including exenatide, due to its high sensitivity and throughput capacity. However, it is time consuming, associated with narrow linear ranges and promotes cross-reactivity between antibodies and similar structure compounds [9]. Some doubts have been raised about selectivity of ELISA, more precisely between a peptide and its metabolites or analogs and some trouble was found while quantifying a peptide at circulating concentrations [26,37]. Regarding the increase in the biosimilar API and therapeutic proteins pipeline, ELISA techniques have narrow multiplex ability during their development and the acquirement of antibodies for such new therapeutics is burdensome [38].
There has been a continuous evolution towards reaching more sensitive, reproducible, robust and multiplexing HPLC techniques for the quantification of exenatide, especially in what concerns mass spectrometry detection (MSD) [31]. The reason for this evolution is due to MSD ability to effectively identify and characterize proteins, even in complex matrices [39]. Thus, HPLC with MSD (HPLC-MSD) techniques allow the quantification of human endogenous hormones and theirs analogs, like exenatide, without a complex sample pretreatment procedure, allowing the quantification of multiple API [35,36]. However, it may occur ion suppression due to the electrospray, leading to loss of sensitivity and erroneous results. Applying HPLC-MSD techniques to the monitoring of biosimilar API and therapeutic proteins enables the quantification of bound and free forms of the API, whereas ELISA may only quantify the free form [40].
Table 1 allows a quick reading on the advantages and disadvantages of these two techniques, comparing ELISA, which were classically used for bioanalysis in general, and HPLC-MSD techniques, which are becoming the golden standard for bioanalysis during preclinical, clinical and biosimilarity assessment of therapeutic proteins [26,40].
Table 1.
Advantages and disadvantages of ELISA and HPLC-MSD techniques regarding the quantification of therapeutic proteins, such as exenatide, and biosimilar drugs during development and monitoring assessment.
| Technique | Advantages | Disadvantages |
|---|---|---|
| ELISA | ||
| HPLC-MSD |
|
5.1. Enzyme-linked immunosorbent assay (ELISA)
As stated above, until recently ELISA techniques were the golden choice for TDM programs and monitoring during preclinical or biosimilarity assessment, although since the evolution of HPLC-MSD techniques their use has been slowing down [26,40].
The fundamental basis of ELISA settles upon the analyte's separation through specific solvents and, frequently, it does not involve a sample pretreatment procedure; thus the endogenous components of the matrix are able to interact with the assay compounds or with the analyte [41]. This outcome is called matrix effect and it is a sterling disadvantage regarding ELISA techniques. Therefore, it is at the utmost importance to proceed to the identification and characterization of the interferents of the matrix that may affect the detection and quantification of exenatide [42].
5.1.1. ELISA methodology and incretins
For better consolidation of incretins' analytical quantification through ELISA, Table 2 briefly displays ELISA settings and protocol conditions for biological matrices and API formulation matrices [[43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54]].
Table 2.
ELISA experimental conditions and main settings for quantitative determination of incretins in biological and formulation matrices, in order to assess blood, plasmatic, serum and tissue concentrations, API pharmacokinetics or biotechnology entrapment efficiency.
| Incretin | Matrix | Sample pretreatment | LLOQ (pmol/L) | Ref |
|---|---|---|---|---|
| DPP-IV | Krill | Dilution | – | [43] |
| DPP-IV | Rat intestine | Homogenization | – | [44] |
| Exenatide | Human plasma | Dried blood spot | 23.89 | [45] |
| Exenatide | Microspheres | – | – | [46] |
| Exenatide | Microemulsion | – | – | [47] |
| Exenatide | Rat plasma | Dilution | 19.11a | [28,29,48] |
| Exenatide | Rat plasma | – | 119428.65a | [49] |
| Exenatide | Rat plasma | – | – | [50] |
| Exenatide | Rat serum | Dilution | 23885.73 | [51] |
| Exenatide | Rat serum | – | 19.11a | [52] |
| GLP-1 | Mice blood | – | – | [53] |
| GLP-1 | Mice intestinal cells | Dilution | 4.10a | [54] |
| Lithocholic acid exendin-4 derivatives | Rat plasma | Dilution | 19.11a | [12] |
Manufacturer values; LLOQ, lower limit of quantification.
As can be seen in Table 2, ELISA techniques are normally performed with commercially available kits regarding the manufacture instructions. Phoenix Pharmaceuticals, Inc™ provides the most used kit, although there are other ELISA kits also used in the literature provided by EMD Millipore™, Thermo Fisher Scientific™ and Sigma Aldrich®. The ELISA kit provided by Peninsula Laboratories International, Inc™ has a lower LLOQ than others (4.78 pmol/L) except when compared to the one used by Chepurny et al. [54]. Besides, it allows a more precise and accurate quantification and does not need a sample pretreatment procedure, taking less time to execute; still, obtained results may be less accurate due to a matrix effect occurrence.
In some cases, ELISA methodology may be developed without using a commercial kit, as already performed by some researchers (Table 2), allowing the selection of more suitable reagents, regarding particularly the analyte of interest. Even though it is a time-consuming procedure, in house developed ELISA methodology may be more fitted for exenatide quantification than the commercialized kit, obtaining more accurate results, due to the suitability of reagents and antibodies selected for the matrix and its endogenous composition, regarding exenatide's behavior in the matrix. However, ELISA's wide range may compromise the quantification of exenatide, because as a protein, exenatide does not have the same characteristics as its basic components, amino acids. The development of antibodies with perfect binding conditions for exenatide is burdensome. Therefore, ELISA methodology is not commonly applied in target proteomics [39].
5.1.2. How to address the matrix effect in ELISA?
In order to avoid or, at least, to diminish the matrix effect compromising ELISA sensitivity, selectivity and accuracy, it is important to select the fittest approach even though, nowadays the techniques applied for the quantification of exenatide are becoming more sensitive. There are two main approaches to this purpose, a sample pretreatment procedure and an exchange of the solvents or parameters of the assay.
Regarding the sample pretreatment approach, it may be applied a solvent that acts as a blocker for the interference, such as polyethylene glycol, protein A, G or L, although it may affect the concentration of exenatide [55]. The use of a low pH pretreatment procedure may enhance the selectivity of the ELISA for exenatide. However, it may induce its precipitation; hence the results obtained may be less accurate. The sample dilution as a pretreatment procedure is also widely exploited in order to handle matrix interference [12,28,29,43,48], but dilution procedures may affect negatively the sensitivity of the technique [56]. It is at the utmost importance to check whether there is any influence of the sample pretreatment procedure selected on the recovery of exenatide and the throughput capability of the assay [41].
Modifying the assay solvents or increasing their concentration, it is usually applied in order to address the interference compound that leads to the matrix effect. However, modifying an assay solvent may change the signal response of exenatide and, consequently, change its immunogenic response [57]. Augmenting incubation times allow extended interaction between exenatide and the solvents of the assay rather than the endogenous matrix interferences increasing the recovery of the API [56].
Although great efforts have been made to develop more refined ELISA techniques for bioanalysis of exenatide, HPLC-MSD techniques outperform them and will continue to improve, in order to acquire more selective, accurate, precise and reproducible methodologies for more quality and robust results.
5.2. High performance liquid chromatography (HPLC)
HPLC techniques have been used as the standard methodology for exenatide TDM programs, preclinical assessment and biotechnology-based API quality control. Thus, with the science evolution, HPLC-MSD is in its reaching, providing increased sensitivity and throughput capability for the quantification of high molecular weight API, like exenatide [31]. However, the demand for highly accurate and rugged HPLC-MSD techniques for the quantification of low molecular weight API is enhancing, especially in target proteomics [38,40].
Even though in this review we emphasize HPLC-MSD, Table 3 allows the assessment of various HPLC techniques developed so far [[58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73], [74], [75], [76], [77]], for incretins' quantification, assembling a brief compilation regarding matrix, sample volume, achieved LOD, HPLC technique, chromatographic column, mobile phase, elution mode, elution program and flow.
Table 3.
Liquid chromatography-based techniques for quantitative determination of incretins in biological matrices and API formulation matrices regarding TDM programs, preclinical assessment, pharmacokinetic studies, entrapment efficiency and quality control protocols.
| Incretin | Matrix | Sample volume (μL) | Technique | Detection | LOD (pmol/L) | Column | Mobile phase |
Flow (μL/min) | Ref. | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Organic phase | Aqueous phase | Elution | Composition | |||||||||
| DPP-IV antagonists | Krill | 100 | HPLC | QTOF-MS | – | C18 | MeOH | Water | Isocratic | – | 10.00 | [43] |
| DPP-IV antagonists | Hydrolysate | – | HPLC | MS/MS | – | C18 | ACN: TFA (B) | TFA (A) | Gradient | 0–60.00 min: 0–45% B | 200.00 | [58] |
| DPP-IV antagonists | BMPH | – | HPLC | UV | – | C18 | ACN (B) | 0.01% TFA (A) | Gradient | 0–54.00 min: 0–60% B | 1500.00 | [59] |
| Exenatide | Hydrogel | 500 | HPLC | – | – | C4 | 85 mmol PBS: ACN (61: 39, v/v) (B) | 85 mmol PBS: ACN (95: 5, v/v) (A) | Gradient | 0–6.00 min: 65%–72% B 6.00–33.00 min: 72% B | 1000.00 | [52,60] |
| Exenatide | Microspheres | 100 | HPLC | MS/MS | – | C18 | ACN: 0.01% TFA (B) | 0.01% TFA (A) | Gradient | 0–33.00 min: 30%–44% B | 1000.00 | [61] |
| Exenatide | Microspheres | – | HPLC | UV | – | C18 | 0.1% TFA in ACN | 0.1% TFA | Isocratic | – | – | [49] |
| Exenatide | Microspheres | – | HPLC | UV | – | Protein specific | 0.1% TFA in ACN | 0.1% TFA in 2% sodium sulfate | Isocratic | – | 800.00 | [48] |
| Exenatide | Microspheres | – | HPLC | UV | – | C18 | ACN (B) | Water with 0.1% TFA (A) | Gradient | 0–16.00 min: 20%–60% B | 1000.00 | [46,50] |
| Exenatide | Microspheres | – | HPLC | UV | – | C18 | 80% ACN with TFA (B) | 0.1% TFA (A) | Gradient | 0–10.00 min: 20%–80% B 10.00–15.00 min: 80% B | 1000.00 | [62] |
| Exenatide | Microspheres | – | HPLC | UV | – | C18 | ACN: 0.01% TFA (B) | 0.01% TFA (A) | Gradient | 0–20.00 min: 30%–44% B | 1000.00 | [63] |
| Exenatide | Microspheres | – | HPLC | UV | – | C18 | ACN (A) | 0.05 M KH2PO4 (B) | Gradient | 0–20.00 min: 27%–43% A 20.00–20.10 min: 43%-27% B 20.10–28.00 min: 27% B | 1000.00 | [64] |
| Exenatide | Nanoparticles | 2000 | HPLC | UV | – | C18 | 80% ACN with 0.1% TFA (B) | Water with 0.1% TFA (A) | Gradient | 0–10.00 min: 20%–80% B 10.00–15.00 min: 80% B | 1000.00 | [65] |
| Exenatide | Nanoparticles | 10000 | HPLC | UV | – | C18 | 0.2% H3PO4: ACN | 0.2% H3PO4 | – | – | 500.00 | [29] |
| Exenatide | Nanoparticles | 200 | HPLC | UV | 477.71 | C18 | 0.1% TFA in ACN (B) | 0.1% TFA (A) | Gradient | 0–20.00 min: 42%–74% B | 500.00 | [27] |
| Exenatide | Porcine skin | – | HPLC | UV | 126593.47 | C4 | 0.1 M KH2PO4 in MeOH: 0.2 M ClNaO₄*H₂O in MeOH | Water | Isocratic | – | – | [16] |
| Exenatide | Solution | – | UPLC | QTOF-MS | – | C18 | 0.1% FA in ACN (B) | 0.1% FA (A) | Gradient | 0–2.00 min: 5% B 2.00–40.00 min: 5%–60% B 40.00–42.00 min: 60%–95% B 42.00–45.00 min: 95% B 45.00–50.00 min: 5% B |
200.00 | [66] |
| Exenatide | Solution | – | HPLC | MS | – | C18 | ACN: 0.01% TFA (B) | 0.01% TFA (A) | Gradient | 0–30.00 min: 30%–44% B | 1000.00 | [67] |
| Exenatide and glucagon | Human plasma | 1000 | LC | HRMS | – | C18 | ACN in MeOH (B) | 0.1% FA (A) | Gradient | 0–3.50 min: 15%–55% B 3.50–4.25 min: 55% B 4.25–5.50 min: 15% B | 500.00 | [68] |
| Exendin-4 | Eluate | – | UPLC | MS | – | C18 | 5% water: 0.1% FA: 2 mM NH4HCO2 in ACN (B) | 5% ACN: 0.1% FA: 2 mM NH4HCO2 (A) | Gradient | 0–5.00 min: 0–40% B 5.00–10.00 min: 40% B | 350.00 | [69] |
| Exendin-4 | Hydrolysate | – | HPLC | MS | – | C18 | ACN: 0.1% TFA (B) | – | Gradient | 0–35.00 min: 0–85% B | 1000.00 | [70] |
| Exendin-4 | Monkey plasma | 150 | UHPLC | MS/MS | 2388.56 | C18 | ACN (B) | 0.1% FA (A) | Gradient | 0–2.50 min: 25%–60% B 2.50–2.60 min: 60% B 2.60 min: 25% B | 1000.00 | [71] |
| Exendin-4 | Solution | – | HPLC | UV | – | C5 | ACN: water (70: 30, v/v) with 10 mM TFA (B) | 10 mM TFA A | Gradient | 0–2.00 min: 35% B 2.00–9.00 min: 35%–100% B | 2000.00 | [72] |
| Exendin-4 | Solution | – | HPLC | UV | – | C18 | ACN | 0.1% TFA | Isocratic | – | 500.00 | [73] |
| Exendin-4 analog | Rat retina | 10 | HPLC | FL | – | C18 | MeOH (B) | 0.1 M KH2PO4: 35% MeOH: 2% THF (A) | Gradient | 0–12.00 min: 0–40% B 12.00–17.00 min: 40% B 17.00–22.00 min: 0% B | 1000.00 | [74] |
| Exendin | Human plasma | 100 | UHPLC | MS/MS | 385.78 | C18 | 0.1% FA in ACN (B) | 0.1% FA (A) | Gradient | 0–12.00 min: 20%–65% B | 0.45 | [36] |
| 12.00–15.00 min: 85% B | 0.70 | |||||||||||
| 15.00–21.00 min: 20% B | 0.45 | |||||||||||
| GIP (1‐42) GIP (3‐42) |
Human plasma | 1000 | IM-LC | MS | 5.54 5.24 |
C18 | MeOH: water (90: 10, v/v) with 0.1% FA (A) | MeOH: water (10: 90, v/v) with 0.1% FA (B) | Gradient | 0–10.00 min: 100% B | 40.00 | [75] |
| 10.00–15.00 min: 100%-30% B | 40.00 | |||||||||||
| GLP-1 (7‐36) GLP-1 (9‐36) |
5.84 | 15.00–30.00 min: 100% A | 20.00 | |||||||||
| 30.00–35.00 min: 100% B | 40.00 | |||||||||||
| GLP-1 | Solution | 10 | HPLC | UV | – | C18 | ACN: 0.1% TFA (B) | Water: ACN: 0.1% TFA (A) | Gradient | 0–60.00 min: 40%–100% B | 500.00 | [76] |
| GLP-1 | GLP-1/HSA | – | HPLC | UV | – | Protein specific | 0.15 M sodium chloride | 0.02 M sodium phosphate | – | – | 800.00 | [53] |
| GLP-1 analogs | Mice plasma | 100 | LC | MS/MS | 10.00 | C18 | ACN with 0.1% FA (B) | Water: ACN (90: 10, v/v) (A) |
Gradient | 0–0.20 min: 5%–20% B 0.20–1.70 min: 20%–45% B 1.70–1.80 min: 40%–100% B 1.80–2.40 min: 100% B 2.40–3.00 min: 5% B |
500.00 | [9] |
| GLP-1 analogs | Mice serum | – | 2DLC | MS | – | C4 | ACN with 0.2% FA, 0.05% TFA and 10% 2-propanol (B) | Water with 0.2% FA, 0.05% TFA and 10% 2-propanol (A) | Gradient | 0–39.00 min: 20% B 39.00–79.00 min: 20%–90% B |
50.00 | [35] |
| Glucagon | Human plasma | 400 | UHPLC | MS/MS (SRM) |
7.18 | C18 | 0.2% FA in ACN (A) | 0.2% FA (B) | Gradient | 0–2.00 min: 22%–32% A 2.00–3.00min: 95% A 3.00–7.00 min: 22% A |
800.00 | [77] |
| Glucagon | Human plasma | 500 | IA-LC | MS/MS | 0.78 | C18 | 0.1% FA in ACN (B) | 0.1% FA (A) | Gradient | 0–2.00 min: 2% B | 15.00 | [37] |
| GLP-1 | 0.79 0.84 |
2.00–6.00 min: 20%–50% B 6.00–9.00 min: 50%–95% B 9.0–011.00 min: 95% B 11.00–13.50 min: 2% B |
4.00 | |||||||||
| Insulin | Human plasma | 500 | HPLC | MS/MS | 16.49 | C4 | 0.1% FA in ACN (B) | 0.1% FA (A) | Gradient | 0–2.50 min: 15%–40% B | 200.00 | [26] |
| – | 500.00 | |||||||||||
| Insulin Lixisenatide Exenatide Liraglutide Semaglutide |
Rat, minipig and human subcutaneous tissue | – | LC | HRMS (DDM) |
– | C18 | 0.1% FA in ACN (B) | 0.1% FA (A) | Gradient | 0–0.50 min: 0.5% B 0.50–5.00 min: 80% B 5.00–6.00 min: 80% B 6.00–6.10 min: 0.5%B 6.10–7.00 min: 0.5% B |
400.00 | [24] |
| Human plasma | 50 | – | ||||||||||
ACN, acetonitrile. BMPH, barbel muscle protein hydrolysate. ClNaO4*H2O, sodium perchlorate monohydrate. FA, formic acid. GIP, glucose-dependent insulinotropic peptide. GLP-1/HAS, glucagon-like peptide 1/human serum albumin fusion protein. HRMS, high resolution mass spectrometry. HRMS (DDM), high-resolution mass spectrometry data-dependent monitoring. H3PO4, phosphoric acid. IA-LC, immunoaffinity liquid chromatography. IM-LC, immunoprecipitation liquid chromatography. KH2PO4, monopotassium phosphate. LOD, limit of detection. MeOH, methanol. MS/MS, tandem mass spectrometry. MS/MS (SRM), tandem mass spectrometry selected reaction monitoring. NH4HCO2, ammonium formate. PBS, phosphate buffer solution. PDA, photodiode array. QTOF-MS, quadrupole time of flight mass spectrometry. TFA. trifluoracetic acid. THF, tetrahydrofuran. UPLC, ultra-performance liquid chromatography. 2DLC, 2-dimensional liquid chromatography.
5.2.1. Chromatographic column
During the development of a liquid chromatography-based technique, it is important to select the chromatographic column, in order to assess the consistency of the results within columns from different lots and the carryover effect, selecting a pretreatment approach suitable for the quantification of the API [26]. Although it is also crucial to take into consideration the physical and chemical properties of the API.
Exenatide has a high molecular weight and an isoelectric point of 4.96, even though the API comprise amino acids with cationic properties, conferring cationic characteristics to the drug [65]. As a peptide, exenatide promotes chromatographic peak tailing and carryover effect due to its hydrophobicity [78]. However, the carryover effect may be avoided or, at least, decreased, through a wash run between analytical runs, in order to clean-up remaining residues from the sample previously injected [26]. Regarding the peak tailing, it is fundamental to proceed with a sample pretreatment approach that enables clean samples and the extraction of exenatide without compromising its quantification, in order to obtain sharp and symmetric chromatographic peaks.
Despite the great diversity of available chromatographic columns, reversed-phase C18 columns are the golden choice columns for exenatide quantification [43,46,58,59,64,66,76,77]. Reversed-phase C18 columns have high porosity and surface, allowing a fast elution with shorter run times; however, a fast elution may reduce the peak resolution and the interaction between exenatide and the stationary phase [79]. To overcome the lack of peak resolution, some researchers use reversed-phase C4 columns [52,60], enhancing the stationary phase hydrophobicity, selectivity, allowing to achieve a symmetric chromatographic peak. In lower extension, chromatographic columns specific for proteins or reversed-phase C5 columns are also used for exenatide quantification [72], in order to obtain sharp and symmetrical chromatographic peaks, avoiding the peak tailing effect.
5.2.2. Mobile phase
During the optimization of the mobile phase, it is important to consider the proportion of the organic phase and the final pH [79]. Since exenatide is hydrophilic, the mobile phase used for its elution must be constituted by a small percentage of water, enhancing the interaction between exenatide and the stationary phase and the peak resolution. Regarding Table 3, it is possible to analyze that, generally, researchers use a small proportion of aqueous phase and alkaline pH conditions, since the elution of exenatide with acidic mobile phases is not suitable, due to its cationic behavior [16].
However, the selection of the mobile phase needs also to take into consideration how exenatide will be detected, because in the case of mass spectrometry detection, the mobile phase affects the sensitivity and efficiency of the detection [80]. In some cases, the addition of volatile solvents is demanding, in order to enhance the sensitivity and the signal of the chromatographic peak [81].
Despite the wide variety concerning the solvents applied as mobile phase for the optimal quantification of exenatide, water and ACN with FA or TFA are the most selected mobile phase constituents [49,58,59,[61], [62], [63],67,68], achieving symmetrical peaks shape, with residue peak tailing and high signal.
5.2.3. Detection
5.2.3.1. Ultraviolet detection (UVD)
Since the emergence of HPLC techniques, as a reliable bioanalytical instrument for the quantification of exenatide and other incretins in biological and biotechnology-based matrices, the chromatographic instrumentation is evolving, allowing the quality enhancement of the detectors available.
Despite this evolution, many authors rely on the UVD for exenatide and incretins quantification [46,[48], [49], [50],53,59,62,72,73,76]. UVD can identify exenatide with efficacy and sensitivity, although it is expensive and difficult for the gradient elution of exenatide, due to the mobile phase refraction index shift [46]. However, nowadays the detection golden choice, in general, is the MSD.
5.2.3.2. Mass spectrometry detection (MSD)
Mass spectrometers quantify ionized molecule mass-to-charge quotient (m/z) through ion capture and individualized ions intensity measurement [39]. Due to the substantial improvement in the MSD equipment, it is now possible to obtain robust, reproducible and accurate quantification results regarding target proteomics, including therapeutic and recombinant proteins and therapeutic recombinant monoclonal antibodies [38]. The MSD allows the quantification and identification of exenatide, associating exenatide's molecular structure with its retention time, allowing the identification of possible isomers, metabolites and analogs [31,46].
The sensitivity of MSD is dependent on exenatide ionization efficiency, which is dependent on its physical and chemical properties and on the matrix interferences [79]. The ionization efficiency is affected by the polarity and the size of the analyte, since exenatide is a polar and large protein, the ionization is not completely affected, and thus the ion suppression effect does not augment significantly. Table 3 displays a wide application array of MSD for the quantification of exenatide in biological and biotechnology-based matrices.
However, new MS instrumentation with increased accuracy, reproducibility, sensitivity, throughput and multiplexing capabilities for the detection of high molecular weight proteins, such as exenatide, is continuously emerging, similar to the interest in target proteomics. MSD coupled to LC-based techniques, such as triple quadrupole (QqQ) mass spectrometers and quadrupole time of flight (QTOF) mass spectrometers, have the ability to, simultaneously, separate and detect exenatide in complex matrices [40,82]. However, not every equipment has these abilities.
Some MSD instruments applied in target proteomics only allow the detection of exenatide in complex matrices after its extraction from the matrix, such as matrix assisted laser desorption/ionization (MALDI) and surface enhanced laser desorption/ionization (SELDI) [39]. MALDI and SELDI are usually associated with TOF mass analyzer systems and, normally, they do not promote the fragmentation of exenatide, allowing its fingerprint analysis; however, these MSD instruments lack interlaboratory reproducibility [83]. SELDI arises from MALDI due to the ability to increase the sample surface area that is exposed to the radiation, enhancing the methodology's specificity [84].
High-resolution mass spectrometers (HRMS) have been replacing QqQ mass spectrometers due to the ability to reach inferior sensitivity and dynamic ranges, with higher resolution. Therefore, target proteomics associated with HRMS is becoming refined, enabling the selection of three main approaches for detection of the exenatide in biological and biotechnology-based formulations matrices: data-independent acquisition (DIA), selected reaction monitoring (SRM) or selected ion monitoring (SIM) or multiple reaction monitoring (MRM) and parallel reaction monitoring (PRM) [31,40].
DIA is an MS method that is characterized by its high throughput and multiplexing abilities [85]. It allows gathering information of the sample, independently of the selected ion precursor, without the predetermination of the target index; hence, it enables the acquisition of information for a wide precursor's range [86]. Although due to the wide range for ion precursors, the accuracy of the DIA is diminished ensuring the obtained results and enhancing the possibility of false positive results [38]. However, associated with QTOF MSD, DIA decreases the range for ion precursors, identifying specific peptides with fast acquisition of information [87]. Applied to proteomics, DIA yields results with reproducibility, linearity, sensitivity and accuracy without time-consuming method development and it has the advantage of, not only quantifying, but also identifying possible biomarkers, allowing a multiplexing quantification and the simultaneous recognition of peptides [38].
SRM is intrinsic to QqQ MSD and it is a multiplexing method with high sensitivity and selectivity, which enables the precise detection of proteins in complex matrices at very low concentration ranges (pg/mL), due to its capability to enhance the method resolution and reduce the background interference [40,88]. It is applied in biological and biotechnology-based formulation matrices, due to its advantages regarding the multiplexing capability, specific detection, fast development methodology, feasibility and reliability for the quantification of proteins and their isomers or metabolites in complex matrices. However, it requires a time-consuming optimization and its sensitivity is dependent on the fragmentation efficiency; thus high collision energy increases low abundance ions, diminishing methodology's LLOQ [89]. SIM operating mode enables exenatide quantification with enhanced resolution; however, in biological samples it does not achieve high selectivity, due to the lack of ability to reduce the background noise [40,89]. Samples containing exenatide submitted to QqQ MSD in MRM mode achieve high selectivity, demonstrating capability to distinguish exenatide from the endogenous matrix components [71].
QTOF MSD techniques operating in the PRM may fragment and isolate the ion precursor of the peptide in the same way of SRM; however, PRM is able to obtain the full MS scan for all the ion precursors [90]. PRM provides superior sensitivity, enabling the discrimination of the peptide's signal from the eluted endogenous matrix interferences [91]. The multiplexing ability and the sensitivity of PRM are proportionally inversed. Despite its ability to achieve lower LLOQ, which is, in cases of low peptide concentration samples, such as plasma samples containing exenatide, an important feature, it requires more time to reach the same quality performance of SRM [38,92,93]. Even though PRM is still settling regarding the bioanalysis, it has already demonstrated to achieve high quality results, enabling the discrimination between both matrix interferences and modifications in the peptide [38].
5.3. Sample pretreatment procedure for HPLC quantification
To obtain samples with higher concentrations without endogenous components inherent to biological samples, it is required the development of sample pretreatment procedures to achieve such goals. However, they are dependent of the detection system used for the quantification [80].
Regarding the possible interferences of the matrix in exenatide and target proteomics MSD, it is at the utmost importance to implement pretreatment procedures before its injection in the chromatographic system, in order to reduce the ion suppression and to enhance the method sensitivity. Therefore, to decrease the matrix complexicity it is required the application of a bottom-up aproach, which proceeds to the extraction, fragmentation, cleavage and sepation of abundant interferent proteins, in order to obtain cleaner samples for quantification [39].
Table 4 assembles the main sample pretreatment procedures developed for biological and biotechnology-based formulation matrices for incretins' quantification by HPLC.
Table 4.
Pretreatment procedures applied to biological and API formulation samples for incretins quantification by liquid chromatography-based techniques during preclinical assessment, TDM programs and quality control.
| Incretin | Matrix | Sample pretreatment | Ref |
|---|---|---|---|
| Exenatide | Microspheres | Dilution | [62] |
| Dilution | [64] | ||
| Precipitation | |||
| Exenatide | Nanoparticles | Precipitation | [29] |
| Exenatide | Solution | Dilution | [66] |
| Exenatide and glucagon | Human plasma | Protein precipitation | [68] |
| Dilution | |||
| Exendin-4 | Imaging eluate | Dilution | [69] |
| SPE | |||
| Monkey plasma | SPE | [71] | |
| Rat retina | Dilution | [74] | |
| Derivatization | |||
| Exendin ([9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39]) | Human plasma | Dilution | [36] |
| SPE | |||
| Solution | Dilution | [66] | |
| GIP and GLP-1 | Human plasma | Immunoprecipitation | [75] |
| Dilution | |||
| Glucagon | Human plasma | Protein precipitation | [77] |
| SPE | |||
| Dilution | |||
| Glucagon GLP-1 ([7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36]) GLP-1 ([9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36]) | Human plasma | Dilution | [37] |
| GLP-1 analogs | Human plasma and rat subcutaneous tissue | Protein precipitation | [24] |
| GLP-1 analogs | Mice plasma | Dilution | [9] |
| Insulin | Human plasma | Immunoaffinity purification | [26] |
GIP, glucose-dependent insulinotropic peptide. GLP-1, glucagon-like peptide-1. SPE, solid-phase extraction.
5.3.1. Dilution
The dilution of a biological sample requires the addition of an organic reagent, in order to obtain a less concentrated sample, avoiding the clogging of the chromatographic system or the column damage, due to the entrapment of exenatide or matrix endogenous components [80]. When the detection is made by mass spectrometry, the dilution procedure allows the decrease of the surface tension [94].
Regarding exenatide sample pretreatment procedures, dilution is a usual procedure step, especially when mass spectrometry is the selected detection system, in order to obtain wider surface area for the ionization and to avoid the matrix effect [94].
5.3.2. Protein precipitation
In the majority of biological samples, including blood, plasma, urine and milk, proteins are an endogenous component. Therefore. proteins are a source of matrix interferents that, when not dealt with, may promote the column collapse or clogging of the chromatographic system.
Protein precipitation is an old, simple and commonly used sample pretreatment procedure that allows the reduction of sample complexity. However, it only removes proteins, allowing the remaining of phospholipids, highly present in plasma samples [10,94,95].
Usually it is performed through the addition of precipitant agents followed by centrifugation and it may be performed alone or in combination with other sample pretreatment procedure. Although due to the fact that exenatide is a peptide, protein precipitation procedures are not suitable, due to the fact that the API may precipitate with the sample endogenous proteins, resulting in erroneous results.
Biological matrices containing exenatide are frequently submitted to protein precipitation [24,67,76]. The precipitant agent of choice is ACN due to the ability to precipitate proteins without degrading the sample. Although it may not promote the precipitation of all the proteins in the sample, enabling the damage of the chromatographic system.
In proteomics, it is common to proceed to the enzymatic cleavage of abundant proteins that are not of interest, promoting the fragmentation of complex proteins into smaller peptides, obtaining samples more prone to be quantified by MSD [39]. Trypsin grants a specific and predictable enzymatic cleavage; however, it is possible that the digestion does not occur as predicted, resulting in peptides with similar molecular weight, turning the identification of the fragmented peptides burdensome, due to the convergence of the mass spectra [96]. Nevertheless, when samples are separated by HPLC-MSD, the convergence of the mass spectra is avoided, because peptides with the same molecular weight have different hydrophobic behavior, eluting at different retention times [39].
Therefore, the combination of protein precipitation procedures with other sample pretreatment procedures allows the acquisition of cleaner samples. When combined with solid-phase extraction, it is possible to remove lipids from the sample, including phospholipids, enhancing its efficiency [94,95].
5.3.3. Solid-phase extraction (SPE)
SPE is the prime procedure for sample pretreatment and it is a reliable, efficient and selective procedure, which allows better phospholipids and proteins clean-up, without affecting the drug's concentration [94,95]. It enables the removal of matrix endogenous interferences, diminishes the matrix effect, and extracts and concentrates exenatide in biological samples, without requiring large sample or reagent volumes [80,94].
It is important to highlight the importance of the correct selection of the SPE's sorbent, in order to enhance the selectivity, efficiency and adsorption capability. Therefore, the cartridge's type, elution and extraction reagents and samples' pH are dependent on the chemical characteristics of the analyte, more specifically exenatide [80].
Focusing on exenatide samples pretreatment procedure, it is possible to remark that SPE is a frequently applied methodology, especially using Oasis® MCX cartridges. These cartridges allow a satisfactory recovery of exenatide at neutral pH [80] and a greater sample clean-up, with the retention of phospholipids and proteins, avoiding the matrix effect. The sorbent used in Oasis® MCX cartridges is a combination of a silica chemically bonded sorbent and a carbon and polymer-based sorbents, which are characterized by the lack of chemical and physical stability and low capacity, without the possibility to be reusable [97]. Therefore, there are under development of new sorbents that will allow the extraction and concentration [97] of biological samples. Hopefully, this new sorbent class will be quickly applied to exenatide biological samples, enabling faster clean-up of the sample, without affecting the concentration levels of exenatide.
6. Validation of quantification techniques
To guarantee the reliability of the results obtained, it is necessary to perform a validation procedure of the analytical technique, in order to reach the drug's safety and efficacy parameters. Therefore, the European Medicines Agency (EMA) and the U. S. Food and Drug Administration (FDA) elaborated guidelines with recommendations for the correct validation of bioanalytical methods [98,99].
In most of the cases, it is required the full validation of the technique developed to exhibit that it is reliable for the quantification of the analyte's concentration in the biological matrix [98]. Thus, there are some parameters that may not be withdrawn from the validation procedure, such as selectivity, sensibility, accuracy, precision, lower limit of quantification (LLOQ), calibration range, matrix effects, reproducibility, stability and incurred sample reanalysis [98,99]. All the previous parameters are outlined on both guidelines as well as how to proceed during their evaluation.
The above stated parameters need to be confirmed for both ligand-binding assays, such as ELISA techniques, and HPLC methodology.
7. Conclusion
Exenatide is a drug for T2D treatment that, in therapeutic concentrations, it can inhibit the pancreatic ß-cells apoptosis and prevent the gastric emptying, promoting satiety and loss of body weight. The development of exenatide's TDM programs is important, in order to achieve optimal individual therapeutic concentrations; thus a suitable and validated quantification technique is needed. Notwithstanding, such quantification techniques are also required for preclinical and clinical assessment of incretins, as well as for the assessment of biotechnology-based API and in proteomics, in order to obtain their safety, toxicity and pharmacokinetic and pharmacodynamic profiles.
The main characteristics of ELISA and HPLC techniques developed so far for the quantification of exenatide were described and critically analyzed. ELISA are, generally, supplied as validated commercial kit. However, when the analytes, antibodies or matrices suffer a modification, it is recommended to validate the technique. It is possible to develop and validate ELISA protocols based on those available in the market, creating individualized and more suitable techniques for the aim of the study. On the other hand, HPLC techniques are used often as quality control for exenatide, but they may also be used to quantify exenatide in biological and biotechnology-based API matrices, requiring development, optimization and validation procedures. There have been advances towards higher specificity and sensitivity HPLC–MSD analysis of therapeutic peptides and in proteomics, due to the rapid development of the method. HPLC-MSD has been invaluable for the analysis of exenatide in several matrices. It achieves wider concentration ranges, enables the quantification and identification of the active peptide and its metabolites or analogs, and it provides structural specific molecular details for exenatide and its metabolites and analogs, unlikely to address when using ELISA techniques alone. Despite the advances in HPLC-MSD technology in proteomics, there are still challenges concerning the complexity towards the proteomic itself and the urgent need of the development of statistical methods specific for proteomic application.
Columns mostly used for exenatide's quantification are C18, in spite of the existence of columns specific for proteins. Amongst the detection, MSD is the most wanted due to its ability to quantify and identify exenatide in biological and biotechnology-based matrices. Although it is an expensive detection, when compared with the UVD, and it is not always available in the laboratories. In these cases, it is usual to develop and validate HPLC techniques for UVD and then apply them to MSD, only needing a partial validation in order to assure the reliability of the obtained results.
When dealing with biological matrices and biotechnology-based matrices, sample pretreatment techniques may be required to achieve the high sensitivity demanded by exenatide quantification, not only by reducing background interference, but also by enriching exenatide prior to analysis. Therefore, it is important to highlight the need to incorporate a SPE procedure into the sample pretreatment method, in order to obtain cleaner samples and to avoid protein precipitation procedures, due to the possibility of precipitating exenatide, obtaining erroneous results or damaging the chromatographic equipment.
A big challenge is to detect exenatide by UVD with similar sensitivity and efficiency of MSD, using a small sample volume, and a simple mobile phase, in the shorter time possible with a lower LLOQ. Not all laboratories have access to MSD, because it is an expensive equipment, needing more qualified personal and suitable sample pretreatment procedures. Thus, in some cases, laboratories developed the methodology in UVD as it was MSD and then hired a company to perform MSD. When there is this need, it is important to develop a sample pretreatment procedure, as well as the chromatographic technique, which may be easily adapted to the MSD. Therefore, samples need to be cleaner, in order to avoid the matrix effects and the impairment of the ionization procedure.
Target proteomics has been becoming an essential instrument for trustworthy quantification of therapeutic and recombinant proteins and biotechnology-based API, due to its features of multiplexing ability, reproducibility and specificity. However, it remains the lack of sensitivity to quantify proteins with lower abundance in complex samples, without compromising the samples. Therefore, it is still required to continue the development of MSD instruments, in order to achieve effective, sensitive and high-throughput methodologies, without the need for complex sample pretreatment procedures that will be able to be applied in API's preclinical, clinical and biosimilar assessment and quality control of biotechnology-based API. Thus, this paper can be a support material for the development of new HPLC techniques for the quantification of exenatide in biological or biotechnology-based matrices.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
References
- 1.Tahrani A.A., Barnett A.H., Bailey C.J. Pharmacology and therapeutic implications of current drugs for type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2016;12:566–592. doi: 10.1038/nrendo.2016.86. [DOI] [PubMed] [Google Scholar]
- 2.DeFronzo R.A., Ferrannini E., Groop L. Type 2 diabetes mellitus. Nat. Rev. Dis. Primers. 2015;1:15019. doi: 10.1038/nrdp.2015.19. [DOI] [PubMed] [Google Scholar]
- 3.Jonietz E. Pathology: cause and effect. Nature. 2012;485:S10–S11. doi: 10.1038/485s10a. [DOI] [PubMed] [Google Scholar]
- 4.Rehman K., Akash M.S. Mechanisms of inflammatory responses and development of insulin resistance: how are they interlinked? J. Biomed. Sci. 2016;23:87. doi: 10.1186/s12929-016-0303-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akash M.S.H., Shen Q., Rehman K. Interleukin-1 receptor antagonist: a new therapy for type 2 diabetes mellitus. J. Pharmacol. Sci. 2012;101:1647–1658. doi: 10.1002/jps.23057. [DOI] [PubMed] [Google Scholar]
- 6.DeFronzo R.A. From the triumvirate to the ominous octet: a new paradigm for the treatment of type 2 diabetes mellitus. Diabetes. 2009;58:773–795. doi: 10.2337/db09-9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sim D.S., Kauser K. In vivo target validation using biological molecules in drug development. In: Nielsch U., Fuhrmann U., Jaroch S., editors. New Approaches to Drug Discovery. Springer International Publishing; 2016. pp. 59–70. [Google Scholar]
- 8.R. Drab S. Glucagon-like peptide-1 receptor agonists for type 2 diabetes: a clinical update of safety and efficacy. Curr. Diabetes Rev. 2016;12:403–413. doi: 10.2174/1573399812666151223093841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H., Xin B., Caporuscio C. Bioanalytical strategies for developing highly sensitive liquid chromatography/tandem mass spectrometry based methods for the peptide GLP-1 agonists in support of discovery PK/PD studies. Mass Spectrom. 2011;25:3427–3435. doi: 10.1002/rcm.5241. [DOI] [PubMed] [Google Scholar]
- 10.Dittrich J., Becker S., Hecht M. Sample preparation strategies for targeted proteomics via proteotypic peptides in human blood using liquid chromatography tandem mass spectrometry. Proteonomics Clin. Appl. 2015;9:5–16. doi: 10.1002/prca.201400121. [DOI] [PubMed] [Google Scholar]
- 11.Manandhar B., Ahn J.M. Glucagon-like peptide-1 (GLP-1) analogs: recent advances, new possibilities, and therapeutic implications. J. Med. Chem. 2015;58:1020–1037. doi: 10.1021/jm500810s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chae S.Y., Jin C.H., Shin J.H. Biochemical, pharmaceutical and therapeutic properties of long-acting lithocholic acid derivatized exendin-4 analogs. J. Contr. Release. 2010;142:206–213. doi: 10.1016/j.jconrel.2009.10.025. [DOI] [PubMed] [Google Scholar]
- 13.Lovshin J.A., Drucker D.J. Incretin-based therapies for type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2009;5:262–269. doi: 10.1038/nrendo.2009.48. [DOI] [PubMed] [Google Scholar]
- 14.Meier J.J. GLP-1 receptor agonists for individualized treatment of type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2012;8:728–742. doi: 10.1038/nrendo.2012.140. [DOI] [PubMed] [Google Scholar]
- 15.Parkes D.G., Mace K.F., Trautmann M.E. Discovery and development of exenatide: the first antidiabetic agent to leverage the multiple benefits of the incretin hormone, GLP-1. Exp. Opin. Drug Discov. 2013;8:219–244. doi: 10.1517/17460441.2013.741580. [DOI] [PubMed] [Google Scholar]
- 16.Bachhav Y.G., Kalia Y.N. Development and validation of a rapid high-performance liquid chromatography method for the quantification of exenatide. Biomed. Chromatogr. 2011;25:838–842. doi: 10.1002/bmc.1526. [DOI] [PubMed] [Google Scholar]
- 17.Syed Y.Y., McCormack P.L. Exenatide extended-release: an updated review of its use in type 2 diabetes mellitus. Drugs. 2015;75:1141–1152. doi: 10.1007/s40265-015-0420-z. [DOI] [PubMed] [Google Scholar]
- 18.Karagiannis T., Liakos A., Bekiari E. Efficacy and safety of once-weekly glucagon-like peptide 1 receptor agonists for the management of type 2 diabetes: a systematic review and meta-analysis of randomized controlled trials. Diabetes Obes. Metab. 2015;17:1065–1074. doi: 10.1111/dom.12541. [DOI] [PubMed] [Google Scholar]
- 19.Guo X.H. The value of short- and long-acting glucagon-like peptide-1 agonists in the management of type 2 diabetes mellitus: experience with exenatide. Curr. Med. Res. Opin. 2016;32:61–76. doi: 10.1185/03007995.2015.1103214. [DOI] [PubMed] [Google Scholar]
- 20.Gallwitz B. Novel therapeutic approaches in diabetes. In: Stettler C., Christ E., Diem P., editors. Novelties in Diabetes. 2016. pp. 43–56. [DOI] [PubMed] [Google Scholar]
- 21.Blevins T., Pullman J., Malloy J. DURATION-5: exenatide once weekly resulted in greater improvements in glycemic control compared with exenatide twice daily in patients with type 2 diabetes. J. Clin. Endocrinol. Metab. 2011;96:1301–1310. doi: 10.1210/jc.2010-2081. [DOI] [PubMed] [Google Scholar]
- 22.Ostergaard L., Frandsen C.S., Madsbad S. Treatment potential of the GLP-1 receptor agonists in type 2 diabetes mellitus: a review. Expert Rev. Clin. Pharmacol. 2016;9:241–265. doi: 10.1586/17512433.2016.1121808. [DOI] [PubMed] [Google Scholar]
- 23.Baggio L.L., Huang Q., Brown T.J. A recombinant human glucagon-like peptide (GLP)-1-albumin protein (Albugon) mimics peptidergic activation of GLP-1 receptor-dependent pathways coupled with satiety, gastrointestinal motility, and glucose homeostasis. Diabetes. 2004;53:2492–2500. doi: 10.2337/diabetes.53.9.2492. [DOI] [PubMed] [Google Scholar]
- 24.Esposito S., de Leonibus M.L., Ingenito R. A liquid chromatography high-resolution mass spectrometry in vitro assay to assess metabolism at the injection site of subcutaneously administered therapeutic peptides. J. Pharm. Biomed. Anal. 2018;159:449–458. doi: 10.1016/j.jpba.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 25.Afonso J., de Sousa H.T., Rosa I. Therapeutic drug monitoring of CT-P13: a comparison of four different immunoassays. Therap. Adv. Gastroenterol. 2017;10:661–671. doi: 10.1177/1756283X17722915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y., Sun L., Anderson M. Insulin glargine and its two active metabolites: a sensitive (16 pM) and robust simultaneous hybrid assay coupling immunoaffinity purification with LC-MS/MS to support biosimilar clinical studies. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017;1063:50–59. doi: 10.1016/j.jchromb.2017.08.018. [DOI] [PubMed] [Google Scholar]
- 27.Menzel C., Holzeisen T., Laffleur F. In vivo evaluation of an oral self-emulsifying drug delivery system (SEDDS) for exenatide. J. Contr. Release. 2018;277:165–172. doi: 10.1016/j.jconrel.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L., Shi Y., Song Y. The use of low molecular weight protamine to enhance oral absorption of exenatide. Int. J. Pharm. 2018;547:265–273. doi: 10.1016/j.ijpharm.2018.05.055. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L., Shi Y., Song Y. Tf ligand-receptor-mediated exenatide-Zn2+ complex oral-delivery system for penetration enhancement of exenatide. J. Drug Target. 2018;26:931–940. doi: 10.1080/1061186X.2018.1455839. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., Qian P., Zhou H. Pharmacological signatures of the exenatide nanoparticles complex against myocardial ischemia reperfusion injury. Kidney Blood Press. Res. 2018;43:1273–1284. doi: 10.1159/000492409. [DOI] [PubMed] [Google Scholar]
- 31.Liu S., Gao W.J., Wang Y. Comprehensive N-glycan profiling of cetuximab biosimilar candidate by NP-HPLC and MALDI-MS. PLoS One. 2017;12:e0170013. doi: 10.1371/journal.pone.0170013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Consoli A., Formoso G. Potential side effects to GLP-1 agonists: understanding their safety and tolerability. Expert Opin. Drug Saf. 2015;14:207–218. doi: 10.1517/14740338.2015.987122. [DOI] [PubMed] [Google Scholar]
- 33.Yamada T., Ishiguro N., Oku K. Successful colistin treatment of multidrug-resistant Pseudomonas aeruginosa infection using a rapid method for determination of colistin in plasma: usefulness of therapeutic drug monitoring. Biol. Pharm. Bull. 2015;38:1430–1433. doi: 10.1248/bpb.b15-00323. [DOI] [PubMed] [Google Scholar]
- 34.Prasad-Reddy L., Isaacs D. A clinical review of GLP-1 receptor agonists: efficacy and safety in diabetes and beyond. Drugs Context. 2015;4:212283. doi: 10.7573/dic.212283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy R.E., Kinhikar A.G., Shields M.J. Combined use of immunoassay and two-dimensional liquid chromatography mass spectrometry for the detection and identification of metabolites from biotherapeutic pharmacokinetic samples. J. Pharmaceut. Biomed. Anal. 2010;53:221–227. doi: 10.1016/j.jpba.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 36.Lasaosa M., Patel P., Givler S. A liquid chromatography-mass spectrometry assay for quantification of Exendin 9-39 in human plasma. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2014;947–948:186–191. doi: 10.1016/j.jchromb.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee A.Y.H., Chappell D.L., Bak M.J. Multiplexed quantification of proglucagon-derived peptides by immunoaffinity enrichment and tandem mass spectrometry after a meal tolerance test. Clin. Chem. 2016;62:227–235. doi: 10.1373/clinchem.2015.244251. [DOI] [PubMed] [Google Scholar]
- 38.Shi T., Song E., Nie S. Advances in targeted proteomics and applications to biomedical research. Proteomics. 2016;16:2160–2182. doi: 10.1002/pmic.201500449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karpievitch Y.V., Polpitiya A.D., Anderson G.A. Liquid chromatography mass spectrometry-based proteomics: biological and technological aspects. Ann. Appl. Stat. 2010;4:1797–1823. doi: 10.1214/10-AOAS341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Todoroki K., Yamada T., Mizuno H. Current mass spectrometric tools for the bioanalyses of therapeutic monoclonal antibodies and antibody-drug conjugates. Anal. Sci. 2018;34:397–406. doi: 10.2116/analsci.17R003. [DOI] [PubMed] [Google Scholar]
- 41.Gorovits B., McNally J., Fiorotti C. Protein-based matrix interferences in ligand-binding assays. Bioanalysis. 2014;6:1131–1140. doi: 10.4155/bio.14.56. [DOI] [PubMed] [Google Scholar]
- 42.Partridge M.A., Pham J., Dziadiv O. Minimizing target interference in PK immunoassays: new approaches for low-pH-sample treatment. Bioanalysis. 2013;5:1897–1910. doi: 10.4155/bio.13.128. [DOI] [PubMed] [Google Scholar]
- 43.Ji W., Zhang C., Ji H. Purification, identification and molecular mechanism of two dipeptidyl peptidase IV (DPP-IV) inhibitory peptides from Antarctic krill (Euphausia superba) protein hydrolysate. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017;1064:56–61. doi: 10.1016/j.jchromb.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 44.Srivastava S., Shree P., Tripathi Y.B. Active phytochemicals of Pueraria tuberosa for DPP-IV inhibition: in silico and experimental approach. J. Diabetes Metab. Disord. 2017;16 doi: 10.1186/s40200-017-0328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Y.Q., Khetarpal R., Zhang Y. Combination of ELISA and dried blood spot technique for the quantification of large molecules using exenatide as a model. J. Pharmacol. Toxicol. Methods. 2011;64:124–128. doi: 10.1016/j.vascn.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 46.Qi F., Wu J., Fan Q. Preparation of uniform-sized exenatide-loaded PLGA microspheres as long-effective release system with high encapsulation efficiency and bio-stability. Colloids Surfaces B Biointerfaces. 2013;112:492–498. doi: 10.1016/j.colsurfb.2013.08.048. [DOI] [PubMed] [Google Scholar]
- 47.Li X., Huang Y., Huang Z. Enhancing stability of exenatide-containing pressurized metered-dose inhaler via reverse microemulsion system. AAPS PharmSciTech. 2018;19:2499–2508. doi: 10.1208/s12249-018-1026-z. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y., Sun T., Zhang Y. Exenatide loaded PLGA microspheres for long-acting antidiabetic therapy: preparation, characterization, pharmacokinetics and pharmacodynamics. RSC Adv. 2016;6:37452–37462. [Google Scholar]
- 49.Kwak H.H., Shim W.S., Hwang S. Pharmacokinetics and efficacy of a biweekly dosage formulation of exenatide in Zucker diabetic fatty (ZDF) rats. Pharm. Res. 2009;26:2504–2512. doi: 10.1007/s11095-009-9966-3. [DOI] [PubMed] [Google Scholar]
- 50.Qi F., Wu J., Yang T. Mechanistic studies for monodisperse exenatide-loaded PLGA microspheres prepared by different methods based on SPG membrane emulsification. Acta Biomater. 2014;10:4247–4256. doi: 10.1016/j.actbio.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 51.Li X.G., Li L., Zhou X. Pharmacokinetic/pharmacodynamic studies on exenatide in diabetic rats. Acta Pharmacol. Sin. 2012;33:1379–1386. doi: 10.1038/aps.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu L., Li K., Liu X. In vitro and in vivo evaluation of a once-weekly formulation of an antidiabetic peptide drug exenatide in an injectable thermogel. J. Pharmacol. Sci. 2013;102:4140–4149. doi: 10.1002/jps.23735. [DOI] [PubMed] [Google Scholar]
- 53.Chen J., Bai G., Cao Y. One-step purification of a fusion protein of glucagon-like peptide-1 and human serum albumin expressed in Pichia pastoris by an immunomagnetic separation technique. Biosci. Biotechnol. Biochem. 2007;71:2655–2662. doi: 10.1271/bbb.70190. [DOI] [PubMed] [Google Scholar]
- 54.Chepurny O.G., Leech C.A., Tomanik M. Synthetic small molecule GLP-1 secretagogues prepared by means of a three-component indole annulation strategy. Sci. Rep. 2016;6 doi: 10.1038/srep28934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bjerner J., Olsen K.H., Bormer O.P. Human heterophilic antibodies display specificity for murine IgG subclasses. Clin. Biochem. 2005;38:465–472. doi: 10.1016/j.clinbiochem.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 56.Hansen T.K., Fisker S., Hansen B. Impact of GHBP interference on estimates of GH and GH pharmacokinetics. Clin. Endocrinol. 2002;57:779–786. doi: 10.1046/j.1365-2265.2002.01668.x. [DOI] [PubMed] [Google Scholar]
- 57.Hennig C., Rink L., Kirchner H. Evidence for presence of lgG4 anti-immunoglobulin autoantibodies in all human beings. Lancet. 2000;355:1617–1618. doi: 10.1016/s0140-6736(00)02223-6. [DOI] [PubMed] [Google Scholar]
- 58.Silveira S.T., Martinez-Maqueda D., Recio I. Dipeptidyl peptidase-IV inhibitory peptides generated by tryptic hydrolysis of a whey protein concentrate rich in β-lactoglobulin. Food Chem. 2013;141:1072–1077. doi: 10.1016/j.foodchem.2013.03.056. [DOI] [PubMed] [Google Scholar]
- 59.Sila A., Alvarez O.M., Haddar A. Purification, identification and structural modelling of DPP-IV inhibiting peptides from barbel protein hydrolysate. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2016;1008:260–269. doi: 10.1016/j.jchromb.2015.11.054. [DOI] [PubMed] [Google Scholar]
- 60.Li K., Yu L., Liu X. A long-acting formulation of a polypeptide drug exenatide in treatment of diabetes using an injectable block copolymer hydrogel. Biomaterials. 2013;34:2834–2842. doi: 10.1016/j.biomaterials.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 61.Liang R., Zhang R., Li X. Stability of exenatide in poly(D,L-lactide-co-glycolide) solutions: a simplified investigation on the peptide degradation by the polymer. Eur. J. Pharm. Sci. 2013;50:502–510. doi: 10.1016/j.ejps.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 62.Yang H.J., Park I.S., Na K. Biocompatible microspheres based on acetylated polysaccharide prepared from water-in-oil-in-water (W-1/O/W-2) double-emulsion method for delivery of type II diabetic drug (exenatide) Colloids Surf. A Physicochem. Eng. Asp. 2009;340:115–120. [Google Scholar]
- 63.Liang R., Li X., Shi Y. Effect of water on exenatide acylation in poly(lactide-co-glycolide) microspheres. Int. J. Pharm. 2013;454:344–353. doi: 10.1016/j.ijpharm.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 64.Wang P., Li Y., Jiang M. Effects of the multilayer structures on Exenatide release and bioactivity in microsphere/thermosensitive hydrogel system. Colloids Surf. B Biointerfaces. 2018;171:85–93. doi: 10.1016/j.colsurfb.2018.04.063. [DOI] [PubMed] [Google Scholar]
- 65.Kim J.Y., Lee H., Oh K.S. Multilayer nanoparticles for sustained delivery of exenatide to treat type 2 diabetes mellitus. Biomaterials. 2013;34:8444–8449. doi: 10.1016/j.biomaterials.2013.07.040. [DOI] [PubMed] [Google Scholar]
- 66.Liu S., Moulton K.R., Auclair J.R. Mildly acidic conditions eliminate deamidation artifact during proteolysis: digestion with endoprotease Glu-C at pH 4.5. Amino Acids. 2016;48:1059–1067. doi: 10.1007/s00726-015-2166-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang R., Li X., Zhang R. Acylation of exenatide by glycolic acid and its anti-diabetic activities in db/db mice. Pharm. Res. 2014;31:1958–1966. doi: 10.1007/s11095-014-1298-2. [DOI] [PubMed] [Google Scholar]
- 68.Morin L.P., Mess J.N., Garofolo F. Large-molecule quantification: sensitivity and selectivity head-to-head comparison of triple quadrupole with Q-TOF. Bioanalysis. 2013;5:1181–1193. doi: 10.4155/bio.13.87. [DOI] [PubMed] [Google Scholar]
- 69.Kiesewetter D.O., Guo N., Guo J. Evaluation of an [(18)F]AlF-NOTA analog of exendin-4 for imaging of GLP-1 receptor in insulinoma. Theranostics. 2012;2:999–1009. doi: 10.7150/thno.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng J., Zhu Y., Yan Q. Recombination and functional studies of a dual-action peptide for diabetes. J. Drug Target. 2013;21:443–449. doi: 10.3109/1061186X.2012.761225. [DOI] [PubMed] [Google Scholar]
- 71.Kehler J.R., Bowen C.L., Boram S.L. Application of DBS for quantitative assessment of the peptide Exendin-4; comparison of plasma and DBS method by UHPLC-MS/MS. Bioanalysis. 2010;2:1461–1468. doi: 10.4155/bio.10.108. [DOI] [PubMed] [Google Scholar]
- 72.Selvaraju R.K., Velikyan I., Johansson L. In vivo imaging of the glucagon like peptide 1 receptor in the pancreas with 68Ga-labeled DO3A-exendin-4. J. Nucl. Med. 2013;54:1458–1463. doi: 10.2967/jnumed.112.114066. [DOI] [PubMed] [Google Scholar]
- 73.Zheng X., Li Y., Fu G. Application of novel peptide (Pp1) improving the half-life of exendin-4 in vivo. Peptides. 2011;32:964–970. doi: 10.1016/j.peptides.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y., Zhang J., Wang Q. Intravitreal injection of exendin-4 analogue protects retinal cells in early diabetic rats. Invest. Ophthalmol. Vis. Sci. 2011;52:278–285. doi: 10.1167/iovs.09-4727. [DOI] [PubMed] [Google Scholar]
- 75.Wolf R., Hoffmann T., Rosche F. Simultaneous determination of incretin hormones and their truncated forms from human plasma by immunoprecipitation and liquid chromatography-mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2004;803:91–99. doi: 10.1016/j.jchromb.2003.11.044. [DOI] [PubMed] [Google Scholar]
- 76.Zheng X., Li Y., Li X. Peptide complex containing GLP-1 exhibited long-acting properties in the treatment of type 2 diabetes. Diabetes Res. Clin. Pract. 2011;93:410–420. doi: 10.1016/j.diabres.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 77.Howard J.W., Kay R.G., Tan T. Development of a high-throughput UHPLC-MS/MS (SRM) method for the quantitation of endogenous glucagon from human plasma. Bioanalysis. 2014;6:3295–3309. doi: 10.4155/bio.14.226. [DOI] [PubMed] [Google Scholar]
- 78.Chambers E.E., Legido-Quigley C., Smith N. Development of a fast method for direct analysis of intact synthetic insulins in human plasma: the large peptide challenge. Bioanalysis. 2013;5:65–81. doi: 10.4155/bio.12.290. [DOI] [PubMed] [Google Scholar]
- 79.Bicker J., Fortuna A., Alves G. Liquid chromatographic methods for the quantification of catecholamines and their metabolites in several biological samples-A review. Anal. Chim. Acta. 2013;768:12–34. doi: 10.1016/j.aca.2012.12.030. [DOI] [PubMed] [Google Scholar]
- 80.Kim C., Ryu H.D., Chung E.G. A review of analytical procedures for the simultaneous determination of medically important veterinary antibiotics in environmental water: sample preparation, liquid chromatography, and mass spectrometry. J. Environ. Manag. 2018;217:629–645. doi: 10.1016/j.jenvman.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 81.Wei H.M., Tao Y.F., Chen D.M. Development and validation of a multi-residue screening method for veterinary drugs, their metabolites and pesticides in meat using liquid chromatography-tandem mass spectrometry. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2015;32:686–701. doi: 10.1080/19440049.2015.1008588. [DOI] [PubMed] [Google Scholar]
- 82.Nesvizhskii A.I., Vitek O., Aebersold R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods. 2007;4:787–797. doi: 10.1038/nmeth1088. [DOI] [PubMed] [Google Scholar]
- 83.Baggerly K.A., Morris J.S., Coombes K.R. Reproducibility of SELDI-TOF protein patterns in serum: comparing datasets from different experiments. Bioinformatics. 2004;20:777–785. doi: 10.1093/bioinformatics/btg484. [DOI] [PubMed] [Google Scholar]
- 84.Petricoin E.F., Ardekani A.M., Hitt B.A. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 85.Sajic T., Liu Y., Aebersold R. Using data-independent, high-resolution mass spectrometry in protein biomarker research: perspectives and clinical applications. Proteonomics Clin. Appl. 2015;9:307–321. doi: 10.1002/prca.201400117. [DOI] [PubMed] [Google Scholar]
- 86.Gillet L.C., Navarro P., Tate S. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.O111.016717. O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Egertson J.D., Kuehn A., Merrihew G.E. Multiplexed MS/MS for improved data-independent acquisition. Nat. Methods. 2013;10:744–746. doi: 10.1038/nmeth.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shi T., Su D., Liu T. Advancing the sensitivity of selected reaction monitoring-based targeted quantitative proteomics. Proteomics. 2012;12:1074–1092. doi: 10.1002/pmic.201100436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dillen L., Cools W., Vereyken L. Comparison of triple quadrupole and high-resolution TOF-MS for quantification of peptides. Bioanalysis. 2012;4:565–579. doi: 10.4155/bio.12.3. [DOI] [PubMed] [Google Scholar]
- 90.Schilling B., MacLean B., Held J.M. Multiplexed, scheduled, high-resolution parallel reaction monitoring on a full scan QqTOF instrument with integrated data-dependent and targeted mass spectrometric workflows. Anal. Chem. 2015;87:10222–10229. doi: 10.1021/acs.analchem.5b02983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Domon B., Gallien S. Recent advances in targeted proteomics for clinical applications. Proteonomics Clin. Appl. 2015;9:423–431. doi: 10.1002/prca.201400136. [DOI] [PubMed] [Google Scholar]
- 92.Gallien S., Duriez E., Demeure K. Selectivity of LC-MS/MS analysis: implication for proteomics experiments. J. Proteom. 2013;81:148–158. doi: 10.1016/j.jprot.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 93.Majovsky P., Naumann C., Lee C.W. Targeted proteomics analysis of protein degradation in plant signaling on an LTQ-orbitrap mass spectrometer. J. Proteome Res. 2014;13:4246–4258. doi: 10.1021/pr500164j. [DOI] [PubMed] [Google Scholar]
- 94.Carmical J., Brown S. The impact of phospholipids and phospholipid removal on bioanalytical method performance. Biomed. Chromatogr. 2016;30:710–720. doi: 10.1002/bmc.3686. [DOI] [PubMed] [Google Scholar]
- 95.Lopes B.R., Barreiro J.C., Cass Q.B. Bioanalytical challenge: a review of environmental and pharmaceuticals contaminants in human milk. J. Pharm. Biomed. Anal. 2016;130:318–325. doi: 10.1016/j.jpba.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 96.Tubaon R.M., Haddad P.R., Quirino J.P. Sample clean-up strategies for ESI mass spectrometry applications in bottom-up proteomics: trends from 2012 to 2016. Proteomics. 2017;17 doi: 10.1002/pmic.201700011. [DOI] [PubMed] [Google Scholar]
- 97.Hashemi B., Zohrabi P., Shamsipur M. Recent developments and applications of different sorbents for SPE and SPME from biological samples. Talanta. 2018;187:337–347. doi: 10.1016/j.talanta.2018.05.053. [DOI] [PubMed] [Google Scholar]
- 98.European Medicines Agency . 2011. Guideline on Bioanalytical Method Validation. [DOI] [PubMed] [Google Scholar]
- 99.Food & Drug Administration . 2018. Guidance for Industry Bioanalytical Method Validation. [Google Scholar]