Abstract
Background
At present, the schizophrenia diagnoses are based on the clinical symptoms and behaviors neglecting the laboratory test indicators.
Results
To better investigate the diagnostic potential of miRNAs for schizophrenia, we selected 14 candidate miRNAs and examined their expressions in the serums of 40 schizophrenia patients and 40 healthy controls by qRT-PCR. Ultimately three abnormally expressed microRNAs were identified, i.e., miR-34a-5p, miR-432-5p and miR-449a. Then, binary regression analysis was employed to combine 3 dysregulated miRNAs. ROC analysis revealed that the AUC of the combination of miR-432-5p + miR-449a in serums was 0.841 (95% CI: 0.791~0.887) with 90% sensitivity and 80% specificity. The AUC of the combination of miR-34a-5p + miR-432-5p + miR-449a in serums was 0.843 (95% CI: 0.791~0.887) with 90% sensitivity and 77.5% specificity. The results indicated that the combined model of miR-432-5p + miR-449a and miR-34a-5p + miR-432-5p + miR-449a have better prediction performances.
Conclusions
The study concludes that the two miRNAs combinations have the potential to be used as biomarkers for schizophrenia diagnoses. The finding may be conducive to overcoming the dilemmas faced by current schizophrenia diagnosis.
Keywords: Schizophrenia, MiRNAs, Biomarkers, Identification, Serum
Background
Schizophrenia is one of the common kinds of mental diseases that has a lifetime risk of nearly 1% [1]. Currently, schizophrenia diagnoses are based on the clinical assessment of symptoms and behavior abnormalities of patients. The laboratory tests of pathophysiological and biochemical indicators are not available to assist the clinical diagnoses of schizophrenia. MiRNAs are a class of short endogenous non-coding RNA, which participate in the post-transcriptional regulation of protein-coding genes [2, 3]. It has been found that miRNAs regulate many essential biological functions and processes [4, 5] and are involved in almost all the life processes [6]. The changes of miRNA expressions reflect the alteration of neuropsychiatric disorders in the genetic and biological aspects [7, 8], including schizophrenia [9–11].
Although the causes of schizophrenia is still unclear, abnormally expressed miRNAs were detected in brain tissues [12–14], whole blood [15, 16], serum [17], plasma [18, 19] and peripheral blood mononuclear cells (PBMCs) [20, 21] of schizophrenia patients. These miRNAs may be used as the biomarkers for schizophrenia diagnoses. In fact, the biomarkers based on blood diagnostics and treatment has drawn increasingly attention [21–23]. Lai et al. (2011) discovered 7 abnormally expressed miRNAs, i.e., miR-34a, miR-564, miR-449a, miR-548d, miR-432, miR-652 and miR-572 in PBMCs of schizophrenia patients [21]. Shi et al. (2012) verified that 5 miRNAs, i.e., miR-181b, miR-1308, miR-219-2-3p, miRNA-195 and let-7 g expressed aberrantly in the schizophrenia patient serums [17]. Sun et al. (2015) analyzed 9 schizophrenia-related miRNAs in plasma and PBMCs of schizophrenia patients and miRNA30e of plasma was confirmed as a biomarker for the schizophrenia diagnosis [19]. Wei et al. (2015) verified that miR-193a-3p and miR-130b could be used as biomarkers for the schizophrenia diagnosis [24]. Sun et al. (2015) identified that miR-30e, miR-34a, miR-346, miR-181b, and miR-7 in the plasma could be non-invasive schizophrenia diagnostic markers [18]. With 7 miRNAs, i.e., miR-34a, miR-564, miR-449a, miR-548d, miR-432, miR-652 and miR-572 were confirmed as potential biomarkers for schizophrenia, Lai et al. (2016) verified again that the 7 miRNA expressions did not change in the peripheral blood of schizophrenia patients that hospitalized for more than two months and whose symptoms were alleviated, so their expressions were stable as diagnostic biomarkers of schizophrenia [16]. Liu et al. (2017) indicated that the axis of EGR1-miR-30a-5p-NEUROD1 could be used as diagnosis biomarker in an acute psychotic state of schizophrenia patients [25]. Ma et al. (2018) found miR-22-3p, miR-137, and miR-92a-3p expressed aberrantly in schizophrenia patient’s peripheral blood and the combination of miR-22-3p + miR-137 + miR-92a-3p could be used as a schizophrenia diagnosis biomarker [26].
Based on the above literature, we selected 14 candidate miRNAs that were reported as a possible biomarkers in the whole blood, plasma, PBMCs, or serums of schizophrenia patients. The 14 candidate miRNAs are miR-30e-5p, miR-130b-3p, miR-652-5p, miR-193a-3p, miR-181b-5p, miR-34a-5p, miR-346, miR-572, miR-7-5p, miR-449a, miR-564, miR-432-5p, miR-548d-3p, and miR-30a-5p. We used qRT-PCR to check the expressions of 14 miRNAs which expressed in the serums of 40 schizophrenia patients vs 40 healthy controls and further explored their diagnostic values and functions for schizophrenia.
Materials and methods
Participants
All the subjects of the study were diagnosed as schizophrenia patients by at least two experienced psychiatrists based on the criteria specified in the Diagnosis and Statistical Manual of Mental Disorders Fourth Edition (DSM-IV) and International Classification of Diseases 10 (ICD10). All the patients were treated with antipsychotic drugs including aripiprazole, risperidone, olanzapine, and clozapine. Control groups were recruited from volunteers. Blood donors and any individual who suffered or are suffering a mental illness and whose relatives suffered or are suffering with mental illness were excluded. Before conducting this study, we obtained the written consent from all the participants. The study was approved by the ethical committee of Tongliao Institute of Mental Health, Tongliao, China.
Peripheral blood collection
Independent peripheral blood samples (5 ml per participant) was obtained from 80 subjects in the morning on an empty stomach and were collected in EDTA anticoagulant tubes. After standing for half an hour on ice, the supernatant was aspirated. 10,000 g Centrifuged for 5 min and discard the precipitate. The supernatant that left was stored at − 80 °C until using for extraction of total RNA.
miRNA selection
We preliminarily selected 14 candidate miRNAs, i.e., miR-30e-5p, miR-130b-3p, miR-652-5p, miR-193a-3p, miR-181b-5p, miR-34a-5p, miR-346, miR-572, miR-7-5p, miR-449a, miR-564, miR-432-5p, miR-548d-3p, and miR-30a-5p in the whole blood, plasma, PBMCs or serums based on diagnostic potential confirmed by previous studies.
RNA extraction and miRNA quantification by QRT-PCR
Total RNAs were harvested using miRcute Serum/Plasma miRNA Isolation Kit (TIANGEN BIOTECH, China) and quantified using a NanoDrop 2000 (Thermo, USA) according to the manufacturer’s instructions. Total RNA (1 μg) from each sample was reverse-transcribed using the miRcute Plus miRNA First-Strand cDNA Kit (TIANGEN BIOTECH, China). Quantitative real-time PCR was performed with a 2 × SYBR qPCR Mix (Beijing Zoman Biotechnology Co., Ltd., China) to quantify the miRNA expression. In order to normalize miRNA expression, the study selected U6 small nuclear RNA as internal reference. Each sample was detected three times, and the 2-ΔΔCt method was used to analyze the expression data [27]. The primer information for the miRNAs and U6 is shown in Table 1.
Table 1.
Forward primers of miRNAs and primers of U6
| Name | Previous IDs | Accession number | Mature sequence | Sequence (from 5′to 3′) |
|---|---|---|---|---|
| hsa-miR-30e-5p | hsa-miR-30e | MIMAT0000692 | UGUAAACAUCCUUGACUGGAAG | TGTAAACATCCTTGACTGGA |
| hsa-miR-130b-3p | hsa-miR-130b | MIMAT0000691 | CAGUGCAAUGAUGAAAGGGCAU | CAGTGCAATGATGAAAGGGCAT |
| hsa-miR-193a-3p | hsa-miR-193a-3p | MIMAT0000459 | AACUGGCCUACAAAGUCCCAGU | AACTGGCCTACAAAGTCCCAGT |
| hsa-miR-181b-5p | hsa-miR-181b | MIMAT0000257 | AACAUUCAUUGCUGUCGGUGGGU | AACATTCATTGCTGTCGGTGGGTT |
| hsa-miR-34a-5p | hsa-miR-34a | MIMAT0000255 | UGGCAGUGUCUUAGCUGGUUGU | TGGCAGTGTCTTAGCTGGTTGTT |
| hsa-miR-346 | hsa-miR-346 | MIMAT0000773 | UGUCUGCCCGCAUGCCUGCCUCU | TGCCCGCATGCCTGCCTCT |
| hsa-miR-7-5p | hsa-miR-7 | MIMAT0000252 | UGGAAGACUAGUGAUUUUGUUGUU | TGGAAGACTAGTGATTTTGTT |
| hsa-miR-449a | hsa-miR-449 | MIMAT0001541 | UGGCAGUGUAUUGUUAGCUGGU | TGGCAGTGTATTGTTAGCTGGT |
| hsa-miR-564 | hsa-miR-564 | MIMAT0003228 | AGGCACGGUGUCAGCAGGC | AGGCACGGTGTCAGCAGGC |
| hsa-miR-432-5p | hsa-miR-432 | MIMAT0002814 | UCUUGGAGUAGGUCAUUGGGUGG | TCTTGGAGTAGGTCATTGGGTGG |
| hsa-miR-548d-3p | hsa-miR-548d | MIMAT0003323 | CAAAAACCACAGUUUCUUUUGC | GCCAAAAACCACAGTTTCTTTTGC |
| hsa-miR-572 | hsa-miR-572 | MIMAT0003237 | GUCCGCUCGGCGGUGGCCCA | CGCTCGGCGGTGGCCCA |
| hsa-miR-652-5p | hsa-miR-652 | MIMAT0022709 | CAACCCUAGGAGAGGGUGCCAUUCA | CAACCCTAGGAGAGGGTGCCATTCA |
| hsa-miR-30a-5p | hsa-miR-30a-5p | MIMAT0000087 | UGUAAACAUCCUCGACUGGAAG | CACTCTCATGTAAACATCCTCGAC |
| U6 snRNA (Forward) | NR_004394 | CTCGCTTCGGCAGCACA | ||
| U6 snRNA (Reverse) | AACGCTTCACGAATTTGCGT |
Statistical analyses
Data were analyzed using SPSS software version 20.0 (New York, USA), GraphPad Prism version 7.0 (California, USA) and MedCalc Statistical Software version 18.2.1 (MedCalc Software bvba, Ostend, Belgium; http://www.medcalc.org; 2018). Data (presented as mean ± SEM) were analyzed using the Mann-Whitney U test. Statistically significant level was set at P-values < 0.05. Using binary logistic regression to combine of 3 dysregulated miRNAs in the regression equation. The diagnostic performances of three miRNAs were evaluated by ROC curves.
Results
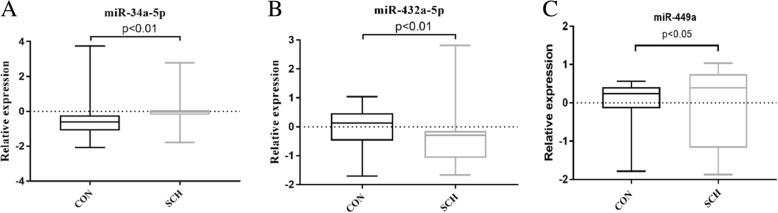
Baseline demographic characteristics of the patient and control groups are shown in Table 2. No significant difference is found in age and gender between the two groups (all p > 0.05). QRT-PCR was performed to compare the expression differences of the 14 candidate miRNAs between patients and controls. The expressions of miR-34a-5p, miR-432-5p and miR-449a in schizophrenia patients were significantly different from those of the healthy control. Relative levels of the 3 significantly altered miRNAs can be seen in Fig. 1.
Table 2.
The demographics of all subjects
| Patients | Healthy controls | F/χ2 | p | |
|---|---|---|---|---|
| Age (years) | 30.35 ± 6.367 | 27.9 ± 7.873 | 2.342 | 0.232 |
| Gender (M/F) | 22/18 | 19/21 | 0.45 | 0.502 |
Fig. 1.
Discrimination of 3 miRNAs levels in serums of SCH and CON. The y axis indicates log102-ΔΔCt relative expression of aberrant miRNAs of quantitative real-time PCR. The expression of 3 miRNAs of 14 miRNAs was significantly different in 40 SCH than in 40 CON. (a) miR-34a, (b) miR- 432-5p, (c) miR-449a. The plots were constructed using GraphPad Prism 7 software and statistical difference was analyzed by Mann-Whitney U test. CON, healthy controls; SCH, schizophrenia patients
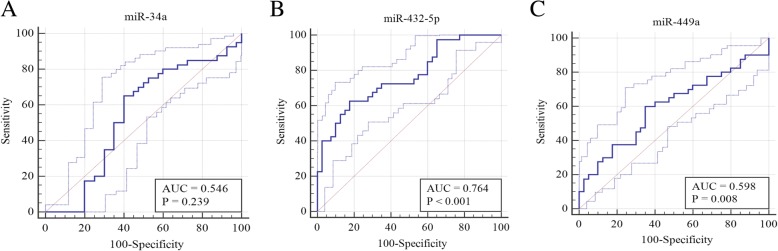
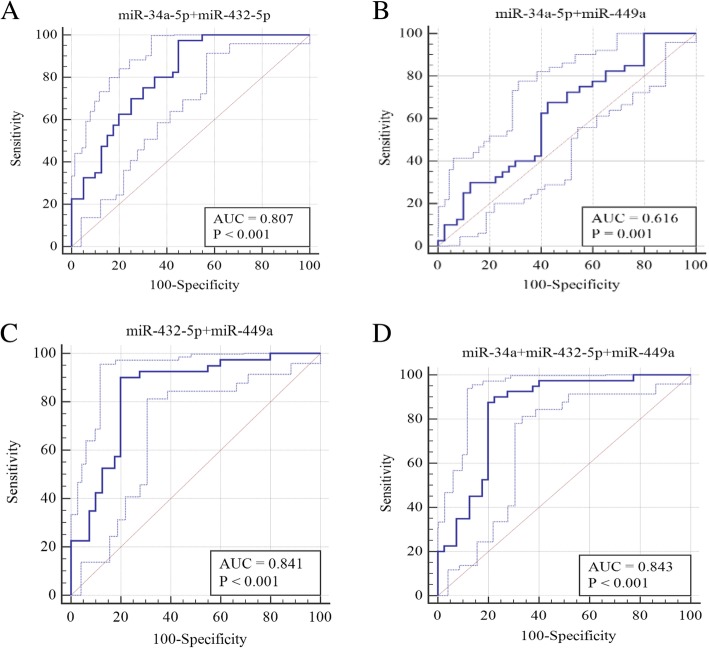
ROC curves of the 3 miRNAs were constructed to further evaluate their diagnostic values. As shown in Fig. 2 and Table 3, miR-432a-5p has the best discriminatory performance with an AUC of 0.764 (95% CI: 0.706~0.817), 62.5% sensitivity, 82.5% specificity and 0.45 Youden index. The ROC curves demonstrated that the combination of miR-34a-5p + miR-432-5p, miR-432-5p + miR-449a, miR-34a-5p + miR-432-5p + miR-449a had a better discriminatory performance with a respective AUC of 0.807 (95%CI: 0.752~0.855), 0.841 (95%CI: 0.789~0.885) and 0.843 (95%CI: 0.791~0.887) (See Fig. 3 and Table 3). The combination of miR-34a-5p + miR-432-5p has 97.5% sensitivity, 55% specificity and 0.525 Youden index J; the combination of miR-432-5p + miR-449a has 90% sensitivity, 80% specificity and 0.7 Youden index J; the combination of miR-34a-5p + miR-432-5p + miR-449a has 90% sensitivity, 77.5% specificity and 0.675 Youden index J (See Table 3). In terms of the indicators such as sensitivity, specificity and Yoden index, the combination of miR-432-5p + miR-449a and the combination of miR-34a-5p + miR-432-5p + miR-449a display a better discriminatory performance.
Fig. 2.
ROC analysis for the miR-34a, miR-432-5p, and miR-449a
(a) miR-34a (b) miR-432-5p (c) miR-449a.
Table 3.
Diagnostic accuracy based on serum miRNA levels
| miRNAs | AUC | 95%CI | P-value | P-Bonferroni | Sensitivity | Specificity | Youden index J |
|---|---|---|---|---|---|---|---|
| miR-34a-5p | 0.546 | 0.480~0.610 | 0.2392 | 3.3488 | 65% | 60% | 0.25 |
| miR-432-5p | 0.764 | 0.706~0.817 | < 0.0001 | < 0.01 | 62.50% | 82.50% | 0.45 |
| miR-449a | 0.598 | 0.533~0.661 | 0.008 | 0.112 | 60.00% | 65.00% | 0.25 |
| miR-34a + miR-432-5p | 0.807 | 0.752~0.855 | < 0.0001 | < 0.01 | 97.50% | 55.00% | 0.525 |
| miR-34a + miR-449a | 0.616 | 0.552~0.678 | 0.0014 | 0.0196 | 67.50% | 57.50% | 0.25 |
| miR-432-5p + miR-449a | 0.841 | 0.789~0.885 | < 0.0001 | < 0.01 | 90.00% | 80.00% | 0.7 |
| miR-34a + miR-432-5p + miR-449a | 0.843 | 0.791~0.887 | < 0.0001 | < 0.01 | 90.00% | 77.50% | 0.675 |
Fig. 3.
ROC curves for diagnostic models of the combination of miR-34a-5p + miR-432, miR-34a-5p + miR-449a, miR-432-5p + miR-449a, miR-34a-5p + miR-432-5p + miR-449a
(a) The combination of miR-34a-5p + miR-432 (b) The combination of miR-34a-5p + miR-449a (c) The combination of miR-432-5p + miR-449a. (d) The combination of miR-34a-5p + miR-432-5p + miR-449a.
Discussion
The current study selected 14 candidate miRNAs and examined their expressions in the serums to explore their diagnostic value for schizophrenia. The qRT-PCR result showed that the expressions of miR-34a-5p, miR-432-5p, and miR-449a have difference in schizophrenia patients. The AUC of the combinations of miR-432-5p + miR-449a and miR-34a-5p + miR432-5p + miR-449a were greater compared with any single miRNA, indicating that the two combination of miRNAs can be used as biomarkers for schizophrenia.
The study confirmed that miR-34a, miR-432, and miR-449a expressed significant changes in serums of schizophrenia patients, suggesting that they may play an important role in the development of schizophrenia. MiR-34a has some well-documented involvements in neurogenesis and neural differentiation [28, 29]. The change of the expression of miR-34a has been found in brain tissues of patients with psychiatric disorder [30, 31]. Schipper et al. (2007) found that the expression of miR-34a significantly up-regulated in the PBMCs of patients with Alzheimer’s disease [32]. The expression level of miR-432 was observed to alter in postmortem cerebellar cortex from autism patients [33]. The miR-449 expression level was found deregulated in cerebrospinal fluid of AD patient brains [34]. Wu et al. (2014) verified that miR-449 and miR-34b/c are very important in normal brain development [35]. Using the results of genome-wide association analysis (GWAS) of schizophrenia, Hauberg et al. predict miRNAs target genes and performed regression and enrichment analysis to explore the regulation of miRNAs on schizophrenia risk genes, they listed 10 conserved miRNAs in the enrichment analysis, including miR-34 ac-5p and miR-449a, and indicated these miRNAs play an important role in schizophrenia [36]. In addition, several target genes of hsa-miR-34a were found using the predictions of TargetScan, such as GREM2, TANC2, CAMSAP1, RGMB, CALN1, RTN4RL1 and FKBP1B are related to the development and function of neuron [16]. The study showed that miR-34a-5p, miR-432-5p, and miR-449a expressed aberrantly in the serums of schizophrenia patients. Before then, Sun et al. (2015) verified the abnormal expression of miR-34a existed in PBMCs of schizophrenia patients [19]. Sun et al. (2015) found again the expressions of miR-34a and miR-432 exhibited the difference in plasma [18]. Yu et al. (2014) verified that the expression level of miR-432 were significantly down-regulated in PBMCs of schizophrenia patients before treatment compared with healthy controls [23]. Lai et al. (2016) verified that the miR-34a, miR-432 and miR-449a did not change in the PBMCs of patients that hospitalized for more than two months and whose symptoms were alleviated [16]. They believe the hospitalization and symptom alleviation could not affected the expressions of circulating miRNAs, so they were sufficiently stable and detectable biomarkers in peripheral blood [16]. In addition, Lai et al. (2014) investigated the expression changes of blood-based miRNA from preterm infants to adulthood, and found miR-34a, miR-432, and miR-449a were expressed consistently from infancy to adulthood [37].
The limitations of the study includes the lack of distinction among psychotic symptoms and the limited number of subjects. All the selected patients in the current study were treated with antipsychotic drugs. We can not exclude the possibility of the expressions of change in these miRNAs before and after the treatment.
In conclusion, we check the expressions of 14 candidate miRNAs in the serums of 80 subjects and found that miR-34a-5p, miR432-5p, and miR-449a expressed aberrantly in schizophrenia patients. The AUC value indicated that the combination of miR-432-5p + miR-449a and all three miRNAs could be used as biomarkers for schizophrenia. We propose that two novel schizophrenia diagnostic model presented here can be treated as biomarkers for schizophrenia and they are useful for overcoming the limitations faced by current schizophrenia diagnoses.
Acknowledgements
We are deeply grateful to all participants.
Abbreviations
- AUC
Area under the curve
- DSM-IV
Diagnosis and Statistical Manual of Mental Disorders Fourth Edition
- ICD10
International Classification of Diseases 10
- MiRNAs
MicroRNAs
- QRT-PCR
Quantitative real-time polymerase chain reaction
- ROC
Receiver operating characteristics
Authors’ contributions
KH, HS, HL, YS designed the study. KH, CG, MG, SW did RT-qPCR experiment and analyzed the data of the study. ST, QZ, and HS managed sample collection. KH, YS and CG wrote the first draft of the manuscript. All authors contributed to and approved the final manuscript.
Funding
This study was supported by the Natural Science Foundations of China (81501151, 81660234), the Natural Science Foundation of Inner Mongolia (2017MS08165, 2017MS(LH)0801).
Availability of data and materials
The original datasets are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Inner Mongolia University for Nationalities and Tongliao Institute of Mental Health, Tongliao, China, and it is also in accordance with the principles of the Declaration of Helsinki. Before conducting this study, we obtained the written consent from all participants.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kuanjun He and Chuang Guo contributed equally to this work.
Contributor Information
Kuanjun He, Email: hekuanjun666@126.com.
Lin He, Email: helinhelin@gmail.com.
Yongyong Shi, Email: shiyongyong@gmail.com.
References
- 1.Mayilyan KR, Weinberger DR, Sim RB. The complement system in schizophrenia. Drug News Perspect. 2008;21(4):200–210. doi: 10.1358/dnp.2008.21.4.1213349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 4.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Bartel David P. MicroRNAs. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Miller BH, Wahlestedt C. MicroRNA dysregulation in psychiatric disease. Brain Res. 2010;1338:89–99. doi: 10.1016/j.brainres.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun E, Shi Y. MicroRNAs: small molecules with big roles in neurodevelopment and diseases. Exp Neurol. 2015;268:46–53. doi: 10.1016/j.expneurol.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8(2):R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, et al. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008;17(8):1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- 11.Mellios N, Galdzicka M, Ginns E, Baker SP, Rogaev E, Xu J, et al. Gender-specific reduction of estrogen-sensitive small RNA, miR-30b, in subjects with schizophrenia. Schizophr Bull. 2012;38(3):433–443. doi: 10.1093/schbul/sbq091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ. Upregulation of dicer and MicroRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol Psychiatry. 2011;69(2):180–187. doi: 10.1016/j.biopsych.2010.09.030. [DOI] [PubMed] [Google Scholar]
- 13.Moreau MP, Bruse SE, David-Rus R, Buyske S, Brzustowicz LM. Altered MicroRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol Psychiatry. 2011;69(2):188–193. doi: 10.1016/j.biopsych.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mor Eyal, Kano Shin-Ichi, Colantuoni Carlo, Sawa Akira, Navon Ruth, Shomron Noam. MicroRNA-382 expression is elevated in the olfactory neuroepithelium of schizophrenia patients. Neurobiology of Disease. 2013;55:1–10. doi: 10.1016/j.nbd.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Morena MT, Eitson JL, Dozmorov IM, Belkaya S, Hoover AR, Anguiano E, et al. Signature MicroRNA expression patterns identified in humans with 22q11.2 deletion/DiGeorge syndrome. Clin Immunol. 2013;147(1):11–22. doi: 10.1016/j.clim.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai C, Lee S, Scarr E, Yu Y, Lin Y, Liu C, et al. Aberrant expression of microRNAs as biomarker for schizophrenia: from acute state to partial remission and from peripheral blood to cortical tissue. Transl Psychiatry. 2016;6(1):e717. doi: 10.1038/tp.2015.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi W, Du J, Qi Y, Liang G, Wang T, Li S, et al. Aberrant expression of serum miRNAs in schizophrenia. J Psychiatr Res. 2012;46(2):198–204. doi: 10.1016/j.jpsychires.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 18.Sun X, Zhang J, Niu W, Guo W, Song H, Li H, et al. A preliminary analysis of microRNA as potential clinical biomarker for schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2015;168(3):170–178. doi: 10.1002/ajmg.b.32292. [DOI] [PubMed] [Google Scholar]
- 19.Sun X, Lu J, Zhang L, Song H, Zhao L, Fan H, et al. Aberrant microRNA expression in peripheral plasma and mononuclear cells as specific blood-based biomarkers in schizophrenia patients. J Clin Neurosci. 2015;22(3):570–574. doi: 10.1016/j.jocn.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 20.Gardiner E, Beveridge NJ, Wu JQ, Carr V, Scott RJ, Tooney PA, et al. Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol Psychiatry. 2011;17(8):827–840. doi: 10.1038/mp.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai CY, Yu SL, Hsieh MH, Chen CH, Chen HY, Wen CC, et al. MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS One. 2011;6(6):e21635. doi: 10.1371/journal.pone.0021635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang F, Xu Y, Shugart YY, Yue W, Qi G, Yuan G, et al. Converging evidence implicates the abnormal microRNA system in schizophrenia. Schizophr Bull. 2015;41(3):728–735. doi: 10.1093/schbul/sbu148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu H, Wu J, Zhang H, Zhang G, Sui J, Tong W, et al. Alterations of miR-132 are novel diagnostic biomarkers in peripheral blood of schizophrenia patients. Prog Neuro-Psychopharmacol Biol Psychiatry. 2015;63:23–29. doi: 10.1016/j.pnpbp.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Wei H, Yuan Y, Liu S, Wang C, Yang F, Lu Z, et al. Detection of circulating miRNA levels in schizophrenia. Am J Psychiatry. 2015;172(11):1141–1147. doi: 10.1176/appi.ajp.2015.14030273. [DOI] [PubMed] [Google Scholar]
- 25.Liu S, Zhang F, Shugart YY, Yang L, Li X, Liu Z, et al. The early growth response protein 1-miR-30a-5p-neurogenic differentiation factor 1 axis as a novel biomarker for schizophrenia diagnosis and treatment monitoring. Transl Psychiatry. 2017;7(1):e998. doi: 10.1038/tp.2016.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma J, Shang S, Wang J, Zhang T, Nie F, Song X, et al. Identification of miR-22-3p, miR-92a-3p, and miR-137 in peripheral blood as biomarker for schizophrenia. Psychiatry Res. 2018;265:70–76. doi: 10.1016/j.psychres.2018.03.080. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Jauhari A, Singh T, Singh P, Parmar D, Yadav S. Regulation of miR-34 family in neuronal development. Mol Neurobiol. 2018;55(2):936–945. doi: 10.1007/s12035-016-0359-4. [DOI] [PubMed] [Google Scholar]
- 29.Chua C, Tang BL. miR-34a in neurophysiology and neuropathology. J Mol Neurosci. 2019;67(2):235–246. doi: 10.1007/s12031-018-1231-y. [DOI] [PubMed] [Google Scholar]
- 30.Kim AH, Reimers M, Maher B, Williamson V, Mcmichael O, Mcclay JL, et al. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res. 2010;124(1–3):183–191. doi: 10.1016/j.schres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azevedo JA, Carter BS, Meng F, Turner DL, Dai M, Schatzberg AF, et al. The microRNA network is altered in anterior cingulate cortex of patients with unipolar and bipolar depression. J Psychiatr Res. 2016;82:58–67. doi: 10.1016/j.jpsychires.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schipper HM, Maes OC, Chertkow HM, Wang E. MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul Syst Bio. 2007;1:263–274. doi: 10.4137/grsb.s361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abu-Elneel K, Liu T, Gazzaniga FS, Nishimura Y, Wall DP, Geschwind DH, et al. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics. 2008;9(3):153–161. doi: 10.1007/s10048-008-0133-5. [DOI] [PubMed] [Google Scholar]
- 34.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14(1):27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 35.Wu J, Bao J, Kim M, Yuan S, Tang C, Zheng H, et al. Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc Natl Acad Sci U S A. 2014;111(28):E2851–E2857. doi: 10.1073/pnas.1407777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hauberg ME, Roussos P, Grove J, Borglum AD, Mattheisen M. Analyzing the role of MicroRNAs in schizophrenia in the context of common genetic risk variants. JAMA Psychiatry. 2016;73(4):369–377. doi: 10.1001/jamapsychiatry.2015.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai CY, Wu YT, Yu SL, Yu YH, Lee SY, Liu CM, et al. Modulated expression of human peripheral blood microRNAs from infancy to adulthood and its role in aging. Aging Cell. 2014;13(4):679–689. doi: 10.1111/acel.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original datasets are available from the corresponding author on reasonable request.