Abstract
Little is known about the prevalence of drug-resistant mutations in HIV-1-positive individuals in Suzhou, China. To elucidate the transmitted drug resistance (TDR) and acquired drug resistance mutation (ADR) profiles, we collected blood specimens from 127 drug-naïve and 117 first-line drug-treated HIV-1-infected individuals sampled from 2014 to 2016 in Suzhou. We successfully amplified pol fragments from 100 drug-naïve and 20 drug-treated samples. We then determined the drug-resistant mutations to protease (PR) and reverse-transcriptase (RT) inhibitors according to the Stanford drug resistance database. Overall, 11 and 13 individuals had transmitted (drug-naïve group) and acquired (treated group) resistance mutations, respectively. Six transmitted drug-resistant mutations were found, including two mutations (L33F and L76V) in the protease region and four (K70N/E and V179D/E) in the RT region. Only L76V was a major mutation, and K70N/E and V179D/E are known to cause low-level resistance to RT inhibitors. All 13 treated participants who had major drug resistance mutations demonstrated intermediate to high resistance to efavirenz and nevirapine, and six had a treatment duration of less than three months. No major mutations to RT inhibitors were found, implying that the epidemic of transmitted resistance mutations was not significant in this area. Our results suggest that more frequent virus load and drug resistance mutation tests should be conducted for individuals receiving antiretroviral treatment, especially for newly treated patients. Our research provides insights into the occurrence of HIV-1 drug resistance in Suzhou and will help to optimize the treatment strategy for this population.
Electronic Supplementary Material
Supplementary material is available for this article at 10.1007/s12250-017-4002-y and is accessible for authorized users.
Keywords: HIV-1, drug resistance mutation, first-line regimen, China
Introduction
The concept of drug resistance has been well recognized and has become a serious concern in clinical bacteriology and virology. With respect to HIV-1, the emergence of antiviral drug resistance is inevitable, which can compromise the antiviral effect in HIV-1-infected individuals. Highly active antiretroviral therapy (HAART) has succeeded in reducing the morbidity and mortality of HIV-infected people, and has turned AIDS into a chronic and controllable disease (Cohen et al., 2013; Cohen et al., 2016). Despite this success, the widespread use of these drugs has led to antiviral drug resistance, which is becoming an issue of increasing concern attracting global attention (Stadeli and Richman, 2013). Antiretroviral drug resistance mutations can dominate both in the virus quasi-species from individuals under HAART as acquired drug resistance (ADR) mutations as well as in treatment-naïve individuals as transmitted drug resistance (TDR) mutations. The prevalence of TDR has been reported to be increasing in some regions, especially in more resource-limited settings (Gupta et al., 2012). In Europe and the United States, drug resistance mutations of HIV-1 were found in more than 60% of patients who had experienced antiretroviral treatment failure (Boyd and Cooper, 2007). Both ADR and TDR mutations can contribute to the clinical failure of treatment and lead to a suboptimal response to subsequent therapy (D’Aquila et al., 1995; Wittkop et al., 2011).
There were 501,000 reported individuals who were living with HIV-1/AIDS by the end of 2014 in China (UNAIDS, 2015). Like many other countries, China has spared no effort to propel HAART for HIV-1-infected individuals. Since the National Free Antiretroviral Treatment Program (NFATP) in China was established in 2003, there has been remarkable acceleration in the treatment population (Zhang et al., 2007; Chen et al., 2016). The total number of people who were receiving antiretroviral treatment was estimated at 295,000 until 2014. According to the Chinese National Free AIDS Antiretroviral Therapy Guidelines (China CDC, 2016), the first-line HAART regimens are mainly constituted by two nucleoside reverse transcriptase inhibitors (NRTIs) plus one of the non-nucleoside reverse transcriptase inhibitors (NNRTIs): [tenofovir (TDF) or azidothymidine (AZT)] + [lamivudine (3TC)] + [efavirenz (EFV) or nevirapine (NVP)]. In general, individuals that experience HAART failure are switched to the second-line regimen comprising TDF, 3TC, and the protease inhibitor (PI) lopinavir/ritonavir (LPV/r). Many investigations have been conducted to detect the presence of TDR or ADR mutations related to the therapy outcome to a certain degree (Li J et al, 2016). With respect to TDR, a previous study showed that although several common mutations were the cause of most of the transmitted and high-level resistance cases, the ratios differed according to geographic region (Rhee et al., 2015). In China, the TDR prevalence was reported to be initially low during 2004–2005 (Liao et al., 2010), but an increasing tendency was detected in recent years because of extensive HAART administration (Li H et al., 2016). After virological failure of first-line regimens, a small proportion of individuals reverted to second-line therapy (Wang et al., 2015), and this change might have left a signature on the protease of the HIV-1 sequence. Therefore, it is necessary to monitor the mutations both to protease and reverse transcriptase inhibitors (PIs and RTIs) when conducting a survey on drug resistance. It is common knowledge that drug resistance mutations are usually transmitted from treatment non-responding patients to treatment-naïve individuals, and some mutations can become epidemic among the latter group. Thus, analyzing the prevalence of ADR and TDR mutations will provide a clearer sense of the current situation of HIV-1 drug resistance and serve as a guideline for developing alternative treatment strategies to prevent such epidemics.
The province of Jiangsu is a severe HIV-1 epidemic region, with over 10,000 HIV-1-infected cases reported by the end of 2014 (Qiu et al., 2014). Previous studies have elucidated the prevalence of TDR and ADR mutations in Jiangsu using samples collected from 2009 to 2012 (Zhou et al, 2016; Guo et al, 2015). However, the present status of drug resistance mutations in this area is not clear. Suzhou is one of the major cities in Jiangsu province, but there have been few investigations regarding HIV-1 drug resistance in the city. In this study, we collected 244 blood specimens that were consecutively sampled from 2014 to 2016 in Suzhou with the aim of elucidating the current HIV-1 drug resistance pattern and to offer suggestions for optimizing the treatment strategy.
Materials and Methods
Patients and samples
Plasma samples were collected from HIV-1-positive individuals that were enrolled at the Fifth People’s Hospital of Suzhou (an infectious disease hospital in Suzhou), China, during 2014–2016 after obtaining written informed consent. Basic epidemiological data such as gender, transmission route, CD4 cell count, viral load of some samples, and HIV-1/AIDS-related symptoms were recorded upon enrollment, samples without clear treatment history were excluded from the study.
RNA extraction, cDNA synthesis, nested-PCR, and sequencing of viral DNA
The plasma samples were collected and preserved in a −80 °C freezer until analysis. QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) was used to extract viral RNA from 140 µL of plasma according to the manufacturer protocol. cDNAs were synthesized from the total viral RNA using PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara, Japan). Finally, target gene amplification was performed using a nested PCR method to obtain two fragments: the protease (PR, HXB2: nucleotides 2014–3030) and reverse transcriptase (RT, nucleotides 2393–4672) 4672) regions in the pol gene. The PCR primers and conditions were the same as those reported previously (Zeng et al., 2014). The RNA extraction, cDNA synthesis, and PCR were operated in different rooms to prevent cross-contamination. The PCR products were sent to Sangon Biotechnology Co. for sequencing (ABI3730). The general primers for sequencing were designed as previously described (Li et al., 2014).
Sequence assembly, alignment, and quality control
Base electropherograms were visualized using Bioedit (Hall, 1999) and then the sequences were assembled by ContigExpress (Huang, 1992). Proper reference sequences were retrieved from the HIV-1 database (http://www.hiv.lanl.gov/) and aligned with the assembled sequences using the Muscle algorithm in Mega 6.0 (Tamura et al., 2013). All sequences were blasted for confirmation in case of potential contamination.
Phylogenetic analysis and genotyping of HIV-1 PR and RT sequences
First, all of the sequences were analyzed using the RIP tool in the HIV-1 database and REGA to identify their subtypes and recombination patterns, and then a maximum-likelihood phylogenetic tree was inferred using Mega 6.0 with 1000 bootstraps to confirm the subtypes and circulating recombination forms (CRFs). Unique recombination forms (URFs) were further bootscanned using Simplot (Lole et al., 1999).
Genotypic drug resistance analysis and prediction of drug susceptibility
Major and minor drug resistance sites in each sequence were determined according to the HIVdb program integrated in the HIV-1 drug resistance database (http://hivdb.stanford.edu/). The phenotype evaluation of drug resistance-related mutations to each antiviral drug was performed using the Stanford drug resistance algorithm. By assigning a “drug penalty score” for each mutation and adding it to a total score, five levels of drug resistance were inferred: susceptible, potential low-level resistance, low-level resistance, intermediate resistance, and highlevel resistance.
Results
Clinical characteristics of the participants
As shown in Table 1, among the 244 specimens collected, 237 were obtained from male subjects, and the majority (76.2%) became infected through having sex with a man (MSM). These HIV-1-positive individuals were distributed over a wide age range. We divided the patients into two groups based on whether or not they were receiving treatment: 127 were drug-naïve and 117 were treated with first-line drugs. Of the 117 treated subjects, 98 had a treatment duration of less than 6 months, and 19 individuals had been under treatment for more than 6 months.
Table 1.
Characteristics of all study participants in Suzhou, China
| Characteristic | Number | |
|---|---|---|
| gender | Male | 237 |
| Female | 7 | |
| Age | <30 | 107 |
| 30–50 | 103 | |
| >50 | 31 | |
| Unknown | 3 | |
| Risk factor | MSM | 186 |
| Heterosexual | 52 | |
| Unknown | 6 | |
| On HAART | Yes | 117 |
| No | 127 | |
| Drug duration | 0–6 months | 98 |
| 6–12 months | 11 | |
| >12 months | 8 | |
| Successfully sequenced | Drug treated | 20 |
| Drug naive | 100 | |
| Subtype | CRF01_AE | 78 |
| B | 1 | |
| C | 2 | |
| CRF07_BC | 25 | |
| CRF08_BC | 1 | |
| URF | 13 |
We obtained a total of 120 RT sequences, 100 from the drug-naïve subjects and 20 from the treated subjects. The PR gene was successfully amplified, yielding 87 sequences from the 244 samples, including 78 sequences from drug-naïve individuals and 9 from treated participants. The accession numbers of the sequences analyzed in this study were KY972127-KY972246.
HIV-1 subtype distribution through phylogenetic analysis
According to the RIP results (data not shown) and maximum-likelihood tree based on RT fragments (Figure 1), CRF01AE was found to be the most prominent strain (65.0%), followed by CRF07_BC (20.8%) (Table 1). We identified two subtype C sequences, which were genetically most closely related to the reference sequence from Africa. The unusual close genetic distance and clinical information confirmed that these sequences were a trans mitted pair. Furthermore, URFs accounted for a relatively high percentage (13/120, 10.8%) of the total sequences (Table 1). The URFs were not included in the phylogenetic tree and were analyzed separately by Simplot (Supplemental Figure S1). Bootscan analysis showed that their parental strains were CRF01_AE, subtype B and subtype C, and three of them showed the same breakpoints.
Figure 1.
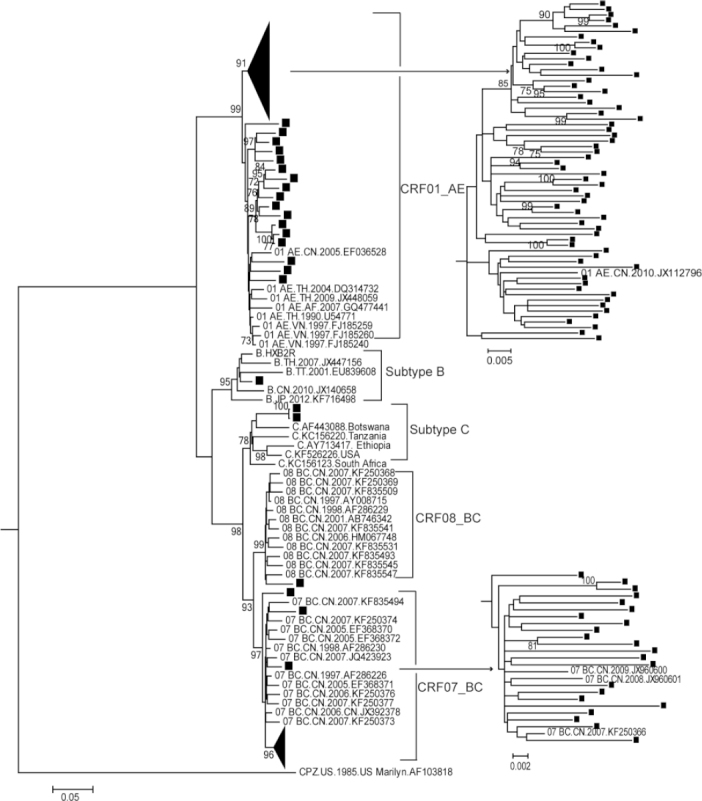
Maximum-likelihood phylogenetic tree of the RT gene region from HIV-1-positive individuals. One thousand bootstrap replicates were performed to confirm the reliability of the branching orders (bootstrap values over 70 are indicated at each node). The scale bar represents 5% nucleotide sequence divergence. Samples analyzed in this study are labeled with solid squares.
Drug resistance genotypes in the drug-naïve and treated groups
Among the 100 drug-naïve individuals, 11% (11/100) harbored six major or accessory drug resistance mutations (L33F, L76V, K70N/E, and V179D/E) in either the PR or RT region at four positions (Table 2). Only L76V was a major mutation in PR, whereas the others were accessory mutations. Furthermore, several “other mutations” were identified in both the PR and RT regions, including L10I/V, V11I, K20I, and A71V/T. A71V was the most prominent mutation identified, which mainly occurred in CRF07_BC in our cohort. In addition, V90I and V106I were identified, which are NNRTI-selected mutations with a very minimal effect on drug resistance.
Table 2.
Transmitted drug resistance mutations profile in drug-naïve subjects
| Mutation | Type | Mutation | Distribution | ||
|---|---|---|---|---|---|
| CRF01_AE | CRF07_BC | URFs | |||
| PI | Other | L10I | 1 | - | - |
| L10V | 2 | - | 1 | ||
| V11I | 1 | - | - | ||
| K20I | 1 | - | - | ||
| A71V | - | 6 | - | ||
| A71T | 1 | - | 1 | ||
| Accessory | L33F | 1 | - | - | |
| Major | L76V | 1 | - | - | |
| RTI | Other | V90I | - | 1 | - |
| V106I | 1 | - | - | ||
| NRTI | K70N | - | - | 1 | |
| K70E | 1 | - | - | ||
| NNRTI | V179D | 1 | - | ||
| V179E | 1 | - | 2 |
With regard to the 20 sequences from treated individuals, the first-line antiviral treatment regimens were as follows: 15 received 3TC+TDF+EFV, two received 3TC+ AZT+NVP, one received 3TC+AZT+EFV, one received 3TC+TDF+NVP, and one patient started on a regimen of 3TC+AZT+EFV that was then changed to 3TC+TDF+EFV; 13 harbored at least one major drug resistance mutation (Table 3). The most dominant mutation to NRTIs detected was M184V, which was found in 11 of the 13 samples, followed by D67N, which was found in four samples. The two frequently occurring mutations to NNRTIs were V106M and G190A, with a proportion of 7/13 and 4/13, respectively. All 13 participants showed relatively high resistance to EFV and NVP, and 12 had high resistance to 3TC, 5 individuals showed low or high resistance to TDF, only one (CNSZ70) was highly resistance to AZT contained in the regimen (Table 3).
Table 3.
Acquired drug resistance mutations in treated subjects and the predicted phenotypes to NRTIs and NNRTIs
| Drug Resistance Mutations | Treatment Duration (months) |
ART start date | Sample date | Predicted Phenotypes | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient ID | NRTI | NNRTI | Regimen | Viral load (date) | NRTI AZT 3TC TDF | NNRTI EFVNVP | ||||||
| CNSZ12 | D67NM184V | K101EG3190A | 3TC+AZT+NVP | 20 | 2011/12/3 | 2013/8/22 | 1.1*105(2013/10/21); 1.3*105(2013/11/22) | S | H | S | H | H |
| 4*104(2013/11/12); | ||||||||||||
| CNSZ16 | M184V | Y181I | 3TC+TDF+NVP | 10 | 2012/10/20 | 2013/8/28 |
3.9*105(2013/12/20) 1.2*105(2014/1/21); 6.6*105(2014/2/21) |
S | H | S | I | H |
| 3.14*104(2015/8/21) | ||||||||||||
| CNSZ28 | K65RY115F M184V | L100IK103E | 3TC+EFV+TDF | 2 | 2015/10/8 | 2015/12/3 | Undetectable (2016/10.18) | S | H | H | H | H |
| CNSZ32 | M184V | V106MG190A | 3TC+EFV+TDF | 3 | 2015/10/10 | 2016/1/13 | 1.49*105(2015/7/27) | S | H | S | H | H |
| CNSZ70 | M41LE44DD67N M184VT215Y | K101HG190A N348I | 3TC+AZT+NVP | 48 | 2011/7/1 | 2015/8/25 | 5.52*102(2015/9/4); 8.55*103(2015/11/30) | H | H | L | I | H |
| CNSZ83 | K65R | V106MW179D | 3TC+EFV+TDF | 3 | 2015/9/9 | 2015/12/2 | <5.0*102(2015/9/25); 3*105(2016/8/16) | S | I | H | H | H |
| <5.0*102(2015/10/16); | ||||||||||||
| CNSZ128 | M184V | V106MV179E | 3TC+EFV+TDF | 1 | 2015/11/9 | 2015/12/7 | 5.55*102(2016/2/22) <5*102(2016/4/8) | S | H | S | H | H |
| CNSZ134 | Y188L | 3TC+EFV+TDF | 1 | 2015/11/11 | 2015/12/9 | 3.4*105(2015/12/18) | s | S | S | H | H | |
| CNSZ149 | D67NK70E M184V | V106MV179D | 3TC+EFV+TDF | 11 | 2015/2/9 | 2016/1/7 | 1.5*105(2015/10/18); 2.8*105(2016/1/11) | s | H | L | H | H |
| 1.6*105(2015/8/21); | ||||||||||||
| CNSZ161 | T69NM184I | V90IK103N V106IM230L | 3TC+EFV+TDF | 3 | 2015/9/30 | 2016/1/5 | 5.2*104(2015/10/30) 6.4*105(2016/1/29); 6.6*104(2016/9/13) | s | H | S | H | H |
| CNSZ235 | D67NM184V | V106MV179D | 3TC+AZT+EFV 3TC+TDF+EFV | 16 | 2014/10/28 | 2016/3/28 | 2.1*105(2015/10/27); 2.8*104(2016/1/19) | s | H | S | H | H |
| CNSZ265 | M184V | V106MV179D | 3TC+TDF+EFV | 8 | 2015/8/19 | 2016/4/20 | 6.3*102(2015/8/21); 7.1*105(2016/7/26) | s | H | S | H | H |
| CNSZ365 | D67NK70R M184V | V106MG190A F227V | 3TC+TDF+EFV | 13 | 2015/7/1 | 2016/8/26 | 2.6*105(2016/6/7); 5.0*105(2016/8/19) | s | H | L | H | H |
Twelve of the 13 participants (except for patient CNSZ32) that harbored major drug resistance mutations had records on viral loads after the ART start date and sample date (Table 3). Ten had viral loads >1000 IU/mL after the sample dates, and two had a viral load < 500 IU/mL despite the presence of the identified drug resistance mutations.
Discussion
The goal of our study was to elucidate the drug resistance mutation profiles of HIV-1-infected therapy-naïve and treated individuals in Suzhou, China. Most of the HIV-1-positive participants enrolled in the study became infected through MSM, which is in line with a recent report showing a marked uptrend of people living with HIV-1 transmitted through MSM in China (UNAIDS, 2015). Drug resistance analysis showed that the prevalence of the TDR mutations to PIs and RTIs was 2% (2/100) and 9% (9/100), respectively, this overall ratio of individuals harboring at least one mutation to PIs or RTIs (11%) is slightly higher than that reported in previous studies conducted in China (3.8% and 4.5%) (Liao et al., 2010; Zhao et al., 2011). Major mutations were detected in most of the samples for which the HIV-1 fragments were successfully amplified, and are known to cause severe resistance to 3TC, EFV, and NVP, which were included in the regimens of these participants.
As for the TDRs, there was one major mutation (L76V) and one accessory mutation (L33F) to PIs identified. These mutations may represent the imprints of HIV-1 PIs in China. L76V reduces the susceptibility to indinavir, Lopinavir, darunavir, and fosamprenavir. L33F is an accessory drug resistance mutation in the PR region and, when combined with other accessory and major mutations, it can reduce drug susceptibility to each PI or compensate for the fitness costs caused by major mutations. The rest were polymorphic per se, which were also found to be related to HIV drug pressure, and were designated as “other mutations”. Among these, the most dominant mutation to PIs was A71V, which mainly occurs in CRF07B_C in China (the protease region of CRF07BC actually belongs to subtype B). After searching the related literature, we found that A71V has been frequently found in the untreated population in China (Li et al., 2014), and the ratio was higher than the frequency of natural polymorphism at this position. Given the fact that PIs are not in the list of first-line antiretroviral drugs provided by the National ART Program and their use is relatively limited in China at present, this phenomenon can be explained by the epidemic of a drug-resistant strain in China that originated from Europe or America. Mutations to RTIs were detected at two positions (K70N/E and V179D/E). The mutations K70N/E tend to be selected in patients receiving TDF-containing regimens and may cause low resistance to NRTIs (TDF and 3TC). V179D is a polymorphic accessory mutation selected by exposure to NNRTIs (EFV and NVP); V179E is non-polymorphic and has a similar effect as V179D.
Our surveillance of TDRs in Suzhou was somewhat consistent with a former study in some aspects (Guo et al., 2015), which found a total TDR frequency of 2% in Jiangsu when using the sample collected from 2009 to 2011, and only two mutations to RTIs (V179D and K101E) were found. It is interesting that we also detected the V179D mutation in our sample, indicating that this mutation may persist and be transmitted in this area. Many other TDR survey studies in China have reported major TDR mutation clusters (Zhao et al., 2015); however, no such major mutations were detected in our study. This might be due to the fact that the drug-naïve cases in the present study were not newly infected, so that major mutations (if any) would have had enough time to be outcompeted by the wild type because of their fitness cost when the drug pressure was removed.
By contrast, of the 117 participants who were receiving treatment, only 20 HIV-1 sequences were amplified. Upon treatment, the virus can quickly evolve to a resistance state, as evident in some of the newly treated subjects. Concretely speaking, among the 98 treated individuals that had been under treatment for less than 6 months, six showed major drug resistance mutations. All cases showed higher-level resistance to NNRTIs, which reflects the fact that this class of drugs has a relatively lower genetic barrier to resistance so that even one mutation can be sufficient to confer high-level resistance (Richman et al., 1994). In the 13 cases with major mutations, 10 had a viral load >1000 IU/mL, indicating that the mutations might be the cause of ineffective viral load suppression. In fact, eight of these individuals (CNSZ16, CNSZ70, CNSZ128, CNSZ149, CNSZ161, CNSZ235, CNSZ265, and CNSZ365) reverted to second-line regimens according to follow-up visit records from the hospital (data not shown). Only one patient (CNSZ70) showed high resistance to all drugs in the regimen; the thymidine analog mutations M41L and T215Y detected in this individual can confer high-level resistance to AZT when combined. The other three cases were still susceptible or showed a low resistance level to one of the drugs (AZT or TDF) in their regimens. A single mutation that conferred resistance to the NRTI (M184V) and NNRTI (Y181I) in subject CNSZ16 was the cause of treatment failure. Indeed, a clinical study showed that in individuals with virological failure, the virus sometimes developed resistance to some but not all drugs in the regimen (Wainberg et al., 2005). Of note, six of the 13 individuals with a treatment duration of no more than three months harbored major drug resistance mutation. These results suggest that drug adherence, which was related to tolerance of the drug side effects, might be a critical problem in the early treatment process.
The main limitation of the present study is that we did not have complete viral load information for all subjects because of the scarcity of the sample; however, we could deduce that the viral suppression was probably effective among most of the treated individuals given the low successful amplification ratio in this group, which likely reflects a low virus copy number of HIV-1 RNA since most individuals (83.8%, 98/117) receiving antiretroviral drugs were newly treated. Furthermore, the amplification rate (100/127) in the drug-naïve group was much higher than that of the treated group, which indicates that this distinct discrepancy was due to the difference in the viral load.
No major TDR mutations to RTIs were found in the study. K70N/E and V179D/E can cause low-level resistance to RTIs, and V179D/E do not appear to reduce the virological response to a first-line regimen (Mackie et al., 2013). Based on the facts above, we might infer that TDR in this area is not a serious problem, and that a genotypic drug resistance test would not be absolutely necessary before initiating HAART. By contrast, given the high ratio of drug resistance mutations detected in the treatment-experienced individuals, more frequent drug resistance tests should be performed for this population. Based on these results, it is reasonable to expect that a large proportion of HIV-1-infected individuals under ART will eventually revert to a second-line drug in the future (usually with the addition of the PI LPV/r). When this occurs, it is necessary to take the baseline drug resistance mutations to PIs into account for determining an optimal treatment strategy. In our study, the most common TDR mutations to PIs were polymorphic mutations, whose main functions are to increase the virus replication capacity (caused by major mutations) instead of increasing drug resistance. Major mutations have been shown to persist within the host for a very long time (Gandhi et al., 2003; Little et al., 2008; Barbour et al., 2004), but they finally often revert to the wild type because of the fitness cost. However, this might not be the case for polymorphic mutations because they do not impair but rather rescue the fitness of the virus under drug pressure. A previous study (Theys et al., 2012) showed that even in the absence of drug pressure, these treatment-associated polymorphisms in the PR region were estimated to increase virus fitness. The high prevalence of PR-related mutations detected in our study may have affected the replication capacity, especially the mutations L10I/V and A71V/T, which were also reported in many other studies (Han et al., 2007; Ye et al., 2012; Li et al., 2014). Therefore, further research is warranted to understand the impact of these mutations.
In summary, our survey provides an overview of the current TDR and ADR mutation profile in Suzhou, China. No major TDR mutations to a first-line regimen were observed, whereas ADR mutations were found in some newly treated individuals. Our results point to the importance of continuous genotypic drug resistance mutation monitoring in the treated HIV-1-infected population, especially for those that are newly treated (treatment duration < 3 months). The findings in this study might help to guide ART strategies for this region and China at large.
Electronic supplementary material
Drug resistance mutation profiles of the drug-naïve and first-line regimen-treated HIV-1-infected population of Suzhou, China
Acknowledgments
This study was supported by grants from the Natural Science Foundation of Jiangsu Province (BL2013017) and the Suzhou Science and Technology Bureau (SYS201156) to Dr. Feng Qian, the Suzhou Health and Family Planning Commission (LCZX201413) to Ming Li, and the Key National Science and Technology Program in the Thirteen Five-Year Plan Period of China (2017ZX10201102-007-002).
Author Contributions
TL, FQ, TY, RY, YG, CZ and BS conceived the experiments; WX, LZ, JH, HW, YZ, YW, XL, SG, ZT, XL, WZ, PX, HC, WL and ML collected the samples; TL and YC performed the experiments; TL analyzed the data; TL, FQ and BS wrote and finalized the manuscript. All authors have read and approved the final manuscript.
Compliance with Ethics Guidelines
The authors declare that there is no conflict of interest. The study was approved by the Ethics Committees of the Fifth People’s Hospital of Suzhou and Wuhan Institute of Virology. Informed consent was obtained from all participants in this study.
Footnotes
These authors contributed equally to this work.
Contributor Information
Chuanwu Zhu, Phone: +86-521-65180193, FAX: +86-512-65291020, Email: zhuchw@126.com.
Binlian Sun, Phone: +86-27-87198040, FAX: +86-27-87198560, Email: sunbl@wh.iov.cn.
References
- Barbour JD, Hecht FM, Wrin T, Liegler TJ, Ramstead CA, Busch MP, Segal MR, Petropoulos CJ, Grant RM. Persistence of primary drug resistance among recently HIV-1 infected adults. AIDS. 2004;18:1683–1689. doi: 10.1097/01.aids.0000131391.91468.ff. [DOI] [PubMed] [Google Scholar]
- Boyd MA, Cooper DA. Second-line combination antiretroviral therapy in resource-limited settings: facing the challenges through clinical research. AIDS. 2007;21(4):S55–S63. doi: 10.1097/01.aids.0000279707.01557.b2. [DOI] [PubMed] [Google Scholar]
- Chen X, Ye M, Zhou YH, Liu FL, Duo L, Li H, Pang W, Zheng YT. High seroprevalence of toxoplasma gondii and HIV-1 co-infection among drug users in Yunnan province, southwest China. Sci China Life Sci. 2015;59:857–859. doi: 10.1007/s11427-015-0358-y. [DOI] [PubMed] [Google Scholar]
- Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, Hakim JG, Kumwenda J, Grinsztejn B, Pilotto JH, Godbole SV, Chariyalertsak S, Santos BR, Mayer KH, Hoffman IF, Eshleman SH, Piwowar-Manning E, Cottle L, Zhang XC, Makhema J, Mills LA, Panchia R, Faesen S, Eron J, Gallant J, Havlir D, Swindells S, Elharrar V, Burns D, Taha TE, Nielsen-Saines K, Celentano DD, Essex M, Hudelson SE, Redd AD, Fleming TR. Antiretroviral Therapy for the Prevention of HIV-1 Transmission. N Engl J Med. 2016;375:830–839. doi: 10.1056/NEJMoa1600693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MS, Smith MK, Muessig KE, Hallett TB, Powers KA, Kashuba AD. Antiretroviral treatment of HIV-1 prevents transmission of HIV-1: where do we go from here? Lancet. 2013;382:1515–1524. doi: 10.1016/S0140-6736(13)61998-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aquila RT, Johnson VA, Welles SL, Japour AJ, Kuritzkes DR, DeGruttola V, Reichelderfer PS, Coombs RW, Crumpacker CS, Kahn JO, Richman DD. Zidovudine resistance and HIV-1 disease progression during antiretroviral therapy. AIDS Clinical Trials Group Protocol 116B/117 Team and the Virology Committee Resistance Working Group. Ann Intern Med. 1995;122:401–408. doi: 10.7326/0003-4819-122-6-199503150-00001. [DOI] [PubMed] [Google Scholar]
- Guo H, Xu X, Hu H, Zhou Y, Yang H, Qiu T, Fu G, Huan X. Low prevalence of the transmitted HIV-1 drug resistance among newly diagnosed HIV-1 individuals in Jiangsu Province, China during 2009–2011. BMC Public Health. 2015;15:120. doi: 10.1186/s12889-015-1489-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Jordan MR, Sultan BJ, Hill A, Davis DH, Gregson J, Sawyer AW, Hamers RL, Ndembi N, Pillay D, Bertagnolio S. Global trends in antiretroviral resistance in treatmentnaive individuals with HIV after rollout of antiretroviral treatment in resource-limited settings: a global collaborative study and meta-regression analysis. Lancet. 2012;380:1250–1258. doi: 10.1016/S0140-6736(12)61038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids SympSer 1999. 1999;41:95–98. [Google Scholar]
- Han X, Zhang M, Dai D, Wang Y, Zhang Z, Liu J, Geng W, Jiang Y, Takebe Y, Shang H. Genotypic resistance mutations to antiretroviral drugs in treatment-naive HIV/AIDS patients living in Liaoning Province, China: baseline prevalence and subtype-specific difference. AIDS Res Hum Retroviruses. 2007;23:357–364. doi: 10.1089/aid.2006.0094. [DOI] [PubMed] [Google Scholar]
- Huang X. A contig assembly program based on sensitive detection of fragment overlaps. Genomics. 1992;14:18–25. doi: 10.1016/S0888-7543(05)80277-0. [DOI] [PubMed] [Google Scholar]
- Li H, Chang S, Han Y, Zhuang D, Li L, Liu Y, Liu S, Bao Z, Zhang W, Song H, Li T, Li J. The prevalence of drug resistance among treatment-naive HIV-1-infected individuals in China during pre-and post- 2004. BMC Infect Dis. 2016;16:605. doi: 10.1186/s12879-016-1928-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li L, Yang S, Li J, Zhang M, Yang C, Liu J, Li H. Identification and characterization of two human immunodeficiency virus type 1 unique recombinant forms from Yunnan, China. Virol Sin. 2016;31:184–187. doi: 10.1007/s12250-015-3671-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Sun B, Zeng H, Sun Z, Sun G, Yang R. Relatively high prevalence of drug resistance among antiretroviral-naive patients from Henan, Central China. AIDS Res Hum Retroviruses. 2014;30:160–164. doi: 10.1089/aid.2013.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L, Xing H, Shang H, Li J, Zhong P, Kang L, Cheng H, Si X, Jiang S, Li X, Shao Y. The prevalence of transmitted antiretroviral drug resistance in treatment-naive HIV-infected indi-viduals in China. J Acquir Immune Defic Syndr. 2010;53(1):S10–S14. doi: 10.1097/QAI.0b013e3181c7d363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Fulllength human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie NE, Dunn DT, Dolling D, Garvey L, Harrison L, Fearnhill E, Tilston P, Sabin C, Geretti AM. The impact of HIV-1 reverse transcriptase polymorphisms on responses to first-line nonnucleoside reverse transcriptase inhibitor-based therapy in HIV-1-infected adults. Aids. 2013;27:2245–2253. doi: 10.1097/QAD.0b013e3283636179. [DOI] [PubMed] [Google Scholar]
- Chinese Center for Disease Control and Prevention (China CDC). 2016. Chinese National Free AIDS Antiretroviral Therapy Manual. (In Chinese)
- Qiu T, Ding P, Liu X, Xu J, Guo H, Fu G, Xu X, Huan X. Epidemiological characteristics of HIV/AIDS patients newly received highly active antiretroviral therapy during 2005–2013 in Jiangsu province. Chin J Epidemiol. 2014;35:1320–1323. [PubMed] [Google Scholar]
- Rhee S-Y, Blanco JL, Jordan MR, Taylor J, Lemey P, Varghese V, Hamers RL, Bertagnolio S, de Wit TFR, Aghokeng AF, et al. Geographic and Temporal Trends in the Molecular Epidemiology and Genetic Mechanisms of Transmitted HIV-1 Drug Resistance: An Individual-Patient- and Sequence-Level Meta-Analysis. PLoS Med. 2015;12:e1001810. doi: 10.1371/journal.pmed.1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman DD, Havlir D, Corbeil J, Looney D, Ignacio C, Spector SA, Sullivan J, Cheeseman S, Barringer K, Pauletti D, et al. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J Virol. 1994;68:1660–1666. doi: 10.1128/jvi.68.3.1660-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi RT, Wurcel A, Rosenberg ES, Johnston MN, Hellmann N, Bates M, Hirsch MS, Walker BD. Progressive reversion of human immunodeficiency virus type 1 resistance mutations in vivo after transmission of a multiply drug-resistant virus. Clin Infect Dis. 2003;37:1693–1698. doi: 10.1086/379773. [DOI] [PubMed] [Google Scholar]
- Little SJ, Frost SD, Wong JK, Smith DM, Pond SL, Ignacio CC, Parkin NT, Petropoulos CJ, Richman DD. Persistence of transmitted drug resistance among subjects with primary human. J Virol. 2008;82:5510–5518. doi: 10.1128/JVI.02579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadeli KM, Richman DD. Rates of emergence of HIV drug resistance in resource-limited settings: a systematic review. Antivir Ther. 2013;18:115–123. doi: 10.3851/IMP2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theys K, Deforche K, Vercauteren J, Libin P, van de Vijver DAMC, Albert J, Åsjö B, Balotta C, Bruckova M, Camacho RJ, et al. Treatment-associated polymorphisms in protease are significantly associated with higher viral load and lower CD4 count in newly diagnosed drug-naive HIV-1 infected patients. 2012. p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNAIDS. 2015. 2015 China AIDS Response Progress Report. Available: http://www.unaids.org/sites/default/files/country/documents/CHN_narrative_report_2015.pdf
- Wainberg MA, Brenner BG, Turner D. Changing patterns in the selection of viral mutations among patients receiving nucleoside and nucleotide drug combinations directed against human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 2005;49:1671–1678. doi: 10.1128/AAC.49.5.1671-1678.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang Z, Liu J, Yue Y, Yang S, Huang H, He C, Liao L, Xing H, Ruan Y, Shao Y. Efficacy and HIV drug resistance profile of second-line ART among patients having received long-term first-line regimens in rural China. Sci Rep. 2015;5:14823. doi: 10.1038/srep14823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkop L, Günthard HF, de Wolf F, Dunn D, Cozzi-Lepri A, de Luca A, Kücherer C, Obel N, von Wyl V, Masquelier B, et al. Effect of transmitted drug resistance on virological and immunological response to initial combination antiretroviral therapy for HIV (EuroCoord-CHAIN joint project): a European multicohort study. The Lancet Infectious Diseases. 2011;11:363–371. doi: 10.1016/S1473-3099(11)70032-9. [DOI] [PubMed] [Google Scholar]
- Ye JR, Lu HY, Wang WS, Guo L, Xin RL, Yu SQ, Wu TC, Zeng Y, He X. The prevalence of drug resistance mutations among treatment-naive HIV-infected individuals in Beijing, China. AIDS Res Hum Retroviruses. 2012;28:418–423. doi: 10.1089/aid.2011.0097. [DOI] [PubMed] [Google Scholar]
- Zeng H, Sun B, Li L, Li Y, Liu Y, Xiao Y, Jiang Y, Yang R. Reconstituting the epidemic history of mono lineage of HIV-1 CRF01_AE in Guizhou province, Southern China. Infection, Genetics and Evolution. 2014;26:139–145. doi: 10.1016/j.meegid.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Zhang F, Haberer JE, Wang Y, Zhao Y, Ma Y, Zhao D, Yu L, Goosby EP. The Chinese free antiretroviral treatment program: challenges and responses. Aids. 2007;21(8):S143–S148. doi: 10.1097/01.aids.0000304710.10036.2b. [DOI] [PubMed] [Google Scholar]
- Zhao B, Han X, Dai D, Liu J, Ding H, Xu J, Chu Z, Bice T, Diao Y, Shang H. New trends of primary drug resistance among HIV type 1-infected men who have sex with men in Liaoning Province, China. AIDS Res Hum Retroviruses. 2011;27:1047–1053. doi: 10.1089/aid.2010.0119. [DOI] [PubMed] [Google Scholar]
- Zhao B, Han X, Xu J, Hu Q, Chu Z, Zhang J, Lu L, Wang Z, Fu J, Chen X, Yan H, Zhuang M, Wang L, Sun A, Zhang CM, Shang H. Increase of RT-related transmitted drug resistance in non-CRF01_AE among HIV type 1-infected men who have sex with men in the 7 cities of China. J Acquir Immune Defic Syndr. 2015;68:250–255. doi: 10.1097/QAI.0000000000000467. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Lu J, Wang J, Yan H, Li J, Xu X, Zhang Z, Qiu T, Ding P, Fu G, Huan X, Hu H. Prevalence of HIV Antiretroviral Drug Resistance and Its Impacts on HIV-1 Virological Failures in Jiangsu, China: A Cross-Sectional Study. BioMed Research International. 2016;2016:1752437. doi: 10.1155/2016/1752437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Drug resistance mutation profiles of the drug-naïve and first-line regimen-treated HIV-1-infected population of Suzhou, China


