Abstract
Historical mass extinction events had major impacts on biodiversity patterns. The most recent and intensively studied event is the Cretaceous – Paleogene (K-Pg) boundary (ca. 66 million years ago [MYA]). However, the factors that may have impacted diversification dynamics vary across lineages. We investigated the macroevolutionary dynamics with a specific focus on the impact of major historical events such as the K-Pg mass extinction event on two major subclasses – Lecanoromycetidae and Ostropomycetidae – of lichen-forming fungi and tested whether variation in the rate of diversification can be associated with the evolution of a specific trait state - macrolichen. Our results reveal accelerated diversification events in three families of morphologically complex lichen-forming fungi – Cladoniaceae, Parmeliaceae, and Peltigeraceae – which are from the subclass Lecanoromycetidae and mostly composed of macrolichens, those that form three dimensional structures. Our RTT plot result for the subclass Lecanoromycetidae also reveals accelerated diversification. Changes in diversification rates occurred around the transition between Mesozoic and Cenozoic eras and was likely related to the K-Pg mass extinction event. The phylogenetic positions for rate increases estimated based on marginal shift probability are, however, scattered from 100 to 40 MYA preventing us from making explicit inference. Although we reveal that the phenotypic state of macrolichens is associated with a higher diversification rate than microlichens, we also show that the evolution of macrolichens predated the K-Pg event. Furthermore, the association between macrolichens and increased diversification is not universal and can be explained, in part, by phylogenetic relatedness. By investigating the macroevolutionary dynamics of lichen-forming fungi our study provides a new empirical system suitable to test the effect of major historical event on shaping biodiversity patterns and to investigate why changes in biodiversity patterns are not in concordance across clades. Our results imply that multiple historical events during the transition from Mesozoic to Cenozoic eras, including the K-Pg mass extinction event, impacted the evolutionary dynamics in lichen-forming fungi. However, future studies focusing on individual lichen-forming fungal families are required to ascertain whether diversification rates are associated with growth form and certain geological events.
Subject terms: Speciation, Phylogenetics
Introduction
The five mass extinctions in earth’s history had major impacts on biodiversity, reshaping entire ecosystems and resulting in dramatic changes in the diversity of major clades1–4. The latest of these biotic crises, the Cretaceous – Paleogene (K-Pg) boundary 66 million years ago (MYA)5, is well known due to the extinction of non-avian dinosaurs6,7. There is a growing body of evidence that the composition of contemporary biodiversity has been significantly shaped by this last mass extinction event. The loss of entire clades during the K-Pg boundary created new ecological opportunities facilitating rapid diversification of surviving lineages2. For example, new ecological opportunities might be related to the extinction of predators and competitors, new open niches that were previously occupied or newly formed because of new environmental conditions, and the evolution of novel traits – biological (e.g., new phenotype) and ecological (e.g., symbiosis). Supporting evidence includes fossil data demonstrating mass extinction of all non-avian dinosaurs, but also mass extinction in other clades of terrestrial life, such as crocodyliforms, snakes and lizards, birds, mammals8–11. In addition, recent comparative phylogenetic studies have indicated that the K-Pg mass extinction event may have facilitated the subsequent rapid diversifications in modern day birds, their lice, and frogs12–14. These specific shifts have been proposed to facilitate the radiation of major clades of mammals, such as rodents15, which account for approximately 42% of extant mammalian diversity16, and of plant clades with duplicated genomes17.
The impact of the K-Pg boundary on diversification of organisms with scant fossil records, such as fungi and some photosynthetic organisms, is less well known. An increase of fungal spores in the fossil record following the mass extinction event and deforestation indicates increased frequency of decomposing fungal species18,19. In contrast, photosynthetic organisms, such as plants, are hypothesized to have undergone mass extinction events after the K-Pg boundary due to severe global climatic change20. However, the impact of this mass extinction on symbiotic fungi, such as lichen-forming fungi with poor fossil record21,22 has not yet been studied. Lichens are symbiotic systems containing at least a fungal host and a mutualistic algal/cyanobacterial partner, although lichen symbioses can also involve bacteria, accessory algae, and endolichenic fungi. Lichens are unique biological systems because the macroevolutionary dynamics of the free-living relative to the two main partners of the system – photosynthetic land plants and fungi – have been hypothesized to react differently to the K-Pg event18–20. It is of evolutionary interest to understand whether the long-term evolutionary dynamics is determined by the hosts that construct and fill specific ecological niches or the obligate symbionts that determine the survival of the hosts.
Investigating the interplay of historical events on macroevolutionary dynamics can provide further important insight into the unevenly distribution of species richness across the tree of life. The uneven distribution of species richness among clades across the tree of life can be explained by multiple factors leading to different rates of extinction and diversification. Increased rates of diversification in a clade are often explained by adaptive traits that evolve in a clade, so called key innovations23,24. For example, it has been demonstrated that speciation rate in color-polymorphic birds is significantly higher than that in other monomorphic bird lineages25. While attributing a single trait to the radiation of a lineage can be overly simplistic, identifying these putative key innovations, in tandem with other factors, can provide a better insight into overall diversification processes26. Lichen-forming fungi are unusual among fungi in that they form long-lived vegetative structures (thalli) to house the photosynthetic partners and these thalli can have different growth forms to resemble crusts (microlichens) or be more complex (macrolichens) being either leaf-like (foliose) or shrubby (fruticose)27 (Figs 1–3). A number of recent studies addressed the identification of key innovations in various groups of organisms, including in lichen-forming fungi28–38. The evolution of macrolichens may represent an innovation, because it allows lichens to explore additional niche spaces and to develop different structures that are not constrained to the surface of substrates. The impact of evolving a macrolichen phenotype is however unequivocal based on empirical data. For example, it has been shown that clades with macrolichens are more species-rich than clades forming primarily microlichens in some families of lichen-forming fungi, but also some clades containing only microlichens have been found to be hyperdiverse39–41.
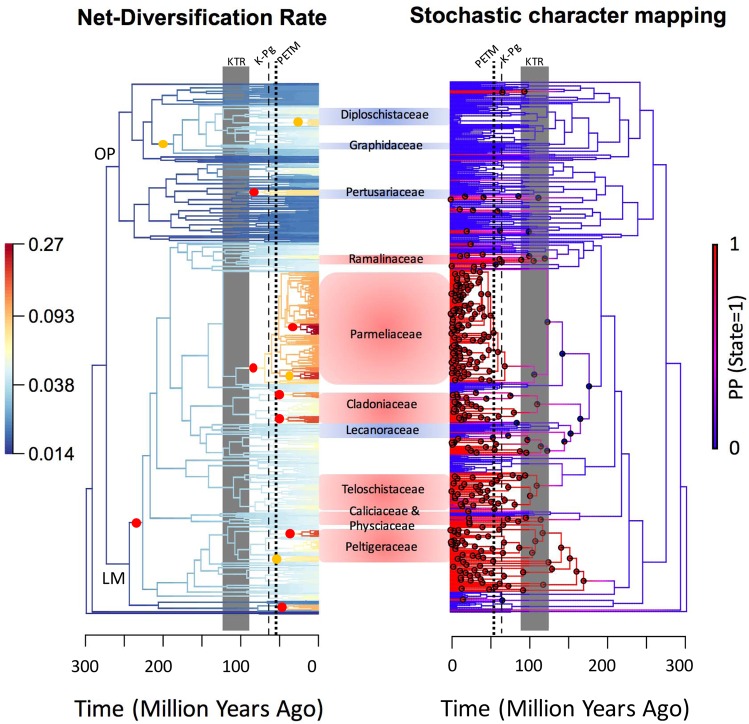
Figure 1.
Changes in diversification dynamics through time (left) and the evolutionary history of microlichen versus macrolichen (right). Branch color of the left figure depicts the net-diversification rate estimated from a BAMM analysis with a warmer color indicating higher rates. Color dots in the left panel indicate the positions where diversification rate has shifted (red dots indicate a marginal shift probability >0.5 and yellow dots >0.3). Branch color of the right figure depicts the probability of being state 1 (macrolichen), where a warmer color indicates a higher probability. Pie charts on nodes indicate the estimate trait states, where only nodes with >10% estimated probability of being macrolichens are shown. Dash lines in both figures indicate the K-Pg boundary (66 MYA) and the Paleocene-Eocene thermal maximum (PETM); the grey shaded areas indicate the period of rapid angiosperm diversification (KTR). LM = Lecanoromycetidae, OP = Ostropomycetidae.
Figure 3.
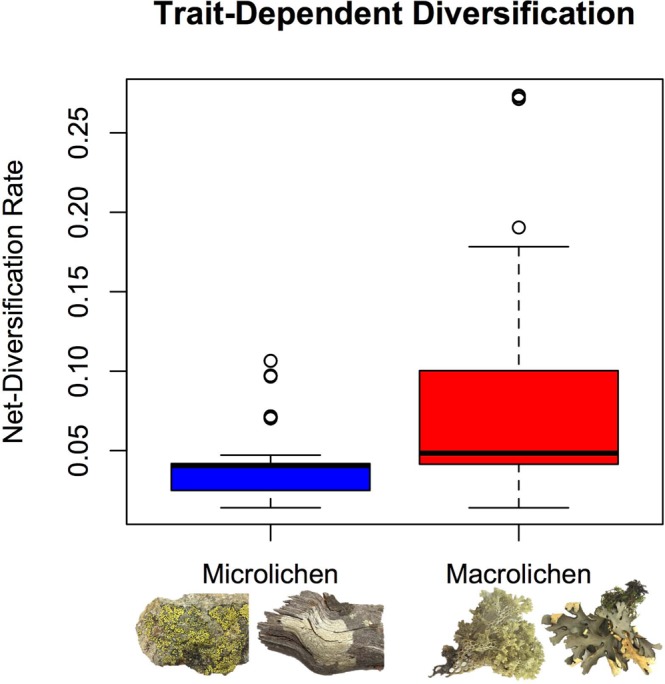
A boxplot for the estimated net-diversification rates for current microlichen (blue) and macrolichen (red) species.
Here we examine how major historical events and different diversification rates associated with a trait have shaped the diversity of lichen-forming fungi. Current evidence suggests that the lichenized life-style evolved in fungi during the Devonian22. Devonian lichen fossils already exhibit very complex structure, and there are other earlier fossils that might represent simplified/early-stages of lichenized life style. Hence, lichen-forming fungal clades have persisted through major historical events and are also species-rich and phenotypically diverse42. Given the impact of the K-Pg mass extinction event on terrestrial life, particularly on photosynthetic organisms20, it is plausible to assume that diversity of lichens, a mutualistic system of fungi and photosynthetic partners27, was also negatively affected by this event. On the other hand, should the K-Pg mass extinction event lead to extinction of epiphytic plant competitors, lichens that have similar morphology and ecological preference might have been benefited from the event and diversified to fill the newly open niches. Within a comparative phylogenetic context, we studied the association between the evolution of different trait states and major historical events and tested whether this putative evolutionary innovation may have accelerated diversification in lichen-forming fungi. Specifically, we investigated the macroevolutionary dynamics in the two major subclasses of lichen-forming fungi, Lecanoromycetidae and Ostropomycetidae, representing >90% of the species diversity in the class Lecanoromycetes (14900 estimated species). We used a recently reconstructed molecular phylogeny and classification of the two subclasses43 and a recently published monograph of the phylum Ascomycota that comprehensively reviewed and estimated the number of species per genus42. Our results provide a diversification history of lichen-forming fungi in general and also provide specific novel insights into how major historical events, such as the K-Pg mass extinction, might (or might not) impact diversification of mutualistic fungi.
Results and Discussion
Our current understanding of how historical events and evolutionary innovations might have affected variation in biodiversity patterns across geological periods and evolutionary lineages is largely derived from only a few well studied organismal groups, which can be incomplete and biased. Although the study of macroevolutionary dynamics has been revolutionized by molecular phylogenetic and comparative methods, these methods assume that the studies have complete taxon sampling or have been adjusted for missing samples. Therefore, studies have been restricted to vertebrates or some plant and insect groups that the species richness and the extant biodiversity pattern are better understood. Here, we extend our knowledge about the variation in diversification histories and patterns across different evolutionary lineages by studying two major clades of lichen-forming fungi. Our analysis of the macroevolutionary dynamics across lichen-forming fungi reveals multiple accelerated diversification events between 100 and 40 MYA, which encompasses the K-Pg boundary in several clades (based on estimated marginal shift probabilities; Fig. 1). Although most shifts in diversification rate occurred after the K-Pg boundary, some diversification shifts were also found to have occurred prior to the K-Pg boundary (Figs 1 and 2). Interestingly, our results suggest that no lineage of lichen-forming fungi within Lecanoromycetes experienced decelerated diversification. Accelerated diversification rates are particularly present in clades including macrolichens, and notably absent from most clades comprised microlichens (Figs 1 and 2). Significantly higher net-diversification rates are pronounced in clades consisting predominantly of macrolichens (Fig. 3). However, the hypothesis that such association can be explained by phylogenetic relatedness instead of the trait states per se cannot be ruled out (accessed with STRAPP44, Ps = 0.223, 0.187, and 0.222 using Spearman’s rho, Pearson correlation, and Mann-Whitney test, respectively). We note that in comparison to macrolichens, microlichen genera have not been subject to intensive molecular-based species delimitation and taxonomic studies, hence it cannot be ruled out that species diversity in microlichens might be underestimated. Future studies are needed to evaluate whether our reported pattern is an artifact due to inconsistent taxonomic efforts across lineages. We discuss how our finding sheds new lights on the effects of major historical events, ecological opportunities, and evolutionary innovations on biological diversification in the following sections.
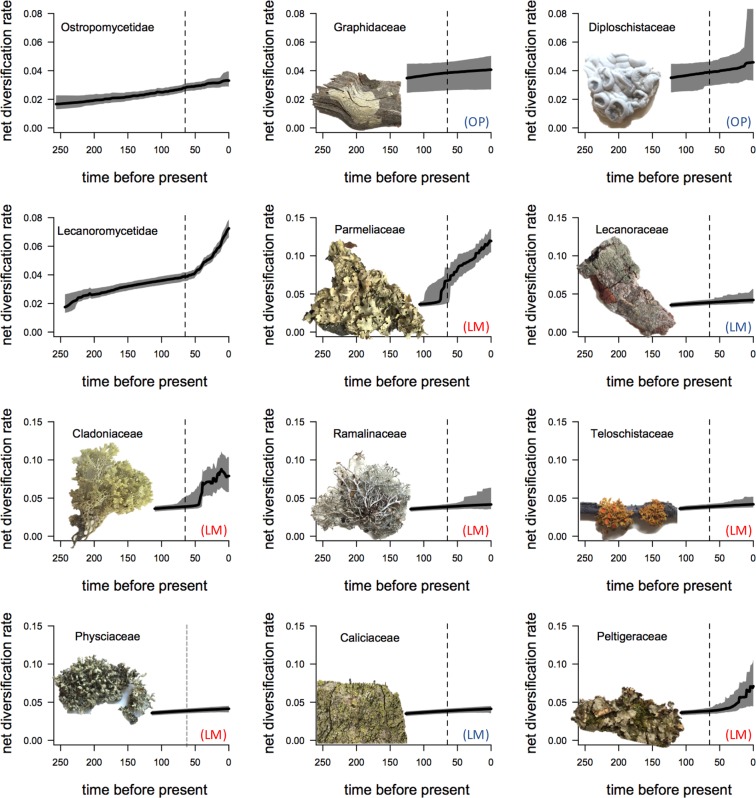
Figure 2.
Net-diversification rate through time plots of the two subclasses and families with more than 500 species in these two main subclasses of lichen-forming fungi. Solid lines are the mean net-diversification rates and the grey areas depict the 95% probability densities. Vertical dash lines indicate 66 MYA. LM = Lecanoromycetidae, OP = Ostropomycetidae. Red color = lineages that are mostly macrolichens, blue color = microlichen lineages.
Our finding implies that historical mass extinction events, and specifically the K-Pg event, might have impacted the diversification of lichen-forming fungi, as have been shown in other organismal groups1,2. However, in contrast to the predicted pattern of a decline in species richness that has been hypothesized for photosynthetic organisms20, the K-Pg extinction appears to have had positive effect on diversification in lichen-forming fungi. There are multiple instances of shifts in diversification rate after the K-Pg boundary, especially in the subclass Lecanoromycetidae (Figs 1 and 2). Furthermore, there is a sudden increase in diversification after the K-Pg boundary based on the estimated diversification rate through time in Lecanoromycetidae, whereas the estimated diversification rate increases gradually and continuously in subclass Ostropomycetidae (Fig. 2).
It is worth noting that many lichen-forming fungi can utilize different photosynthetic symbionts (both algae and cyanobacteria), and switches between different algal partners have been reported in empirical studies22,27,28. Therefore, mass extinction of other competitive evolutionary lineages such as other land plants might have led to the subsequent rapid diversification of the surviving lichen-forming fungal lineages that could develop new mutualistic relationships with the remaining algal/cyanobacterial partners. Nevertheless, given the uncertainty in phylogenetic dating and the geological time scale focused in our study, there were other major historical events and hypotheses that may also explain the observed pattern. For example, prior to the K-Pg event, the Cretaceous Terrestrial Revolution (KTR; from 125 to 80 MYA; Fig. 1) where angiosperms diversified rapidly could also account for the accelerated diversification in the epiphytic lichens45,46. The increase in diversification rate found in certain lichen-forming fungal lineages is not necessarily a result of mass extinction event of other lineages resulting in open niche space. The diversification and multiplication of angiosperms lineages might have provided new niches for epiphytic lichen radiation, e.g. novel durable and diverse substrates on vascular plants. Our study only provides a general time frame of lichen diversification and future studies are needed to specifically test the effect of different historical events and their interactions on diversification rate of lichen-forming fungi.
Our results imply that the evolution of macrolichens was an evolutionary innovation that helped certain lineages to rapidly diversify when new ecological opportunity emerged. Specifically, macrolichens might have diversified because of the new niches created by rapidly diversifying angiosperms during KTR; furthermore, those lichen-forming fungal lineages that survived the K-Pg mass extinction event might have rapidly filled the open niches resulted from the extinction of competitors. The three families showing patterns of acceleration in diversification rates based on the rate through time plots are from the subclass Lecanoromycetidae and form predominately macrolichens (Fig. 2). Such inference is further supported by the fact that the estimated net-diversification rate is significantly higher for macrolichens than microlichens (P < 0.001 based on t-test; Fig. 3).
It is tempting to infer that the evolution of macrolichens, which may serve ecological functions like epiphytes, might prosper due to the extinction of competitors after the K-Pg boundary or because of the angiosperm radiation during KTR. It is, however, more difficult to determine how and why the macrolichen phenotype facilitated accelerated diversification rates. There are also patterns of accelerated diversification rate found in the Ostropomycetidae subclass, which consists of mainly microlichens (e.g., Pertusariaceae; Fig. 1). Some families predominately forming macrolichens in subclass Lecanoromycetidae did not undergo diversification rate increase. Furthermore, such association between macrolichen and a higher diversification rate disappeared when the phylogenetic relatedness among taxa were taken into consideration (STRAPP result). Note that, to test for trait-dependent hypothesis using STRAPP, it often requires a phylogeny with thousands of tips and a dozen of independent origins of the putative adaptive phenotype47. Most empirical data sets likely do not have the statistical power to detect trait-dependent diversification pattern using STRAPP even if the pattern exists. Additional comparative phylogenetic studies that focus on individual families, thus controlling for the phylogenetic lineage specific effect, will be required to explicitly test whether evolving the macrolichen growth form is an evolutionary innovation.
Macrolichens belong to young lineages nested within larger microlichen clades, and arose independently multiple times from microlichens at various points of time based on ancestral state reconstruction (Fig. 1). Specifically, both symmetric and asymmetric models of discrete character evolution fit the trait data and the phylogeny well (transition rate between states is 0.00136 for symmetric model with a log likelihood value of −132.8879; the transition rates are 0.00107 and 0.00149 from macrolichen to microlichen and vice versa, respectively with a log likelihood value of −132.6705; there is no statistical significance between the two models given the data, P = 0.49), so the result from the symmetric model is shown in Fig. 1. The early divergences occurred between microlichens, and it was not until ca. 200 MYA that macrolichens started to evolve from microlichens. Note that, macrolichens form a composite trait state that can be further divided into finer categories as growth forms – e.g., foliose and fruticose – and a diversity of different structure variations also exist within the growth forms27. It is likely that the different growth forms may represent different lineage-specific evolutionary innovations. This evolutionary innovation predated the evolutionary diversification of angiosperms and the K-Pg boundary in Lecanoromycetidae, but was synchronized with the evolutionary diversification of angiosperms in Ostropomycetidae (Fig. 1). Our result corroborates the finding from a previous study focusing on the evolutionary diversification among major Ascomycota lineages48. There is, however, substantial variation in the evolutionary history of microlichens versus macrolichens across lineages. As it predated both KTR event and the K-Pg boundary, the evolution of macrolichens in certain lineages may be pre-adaptive and led to subsequent accelerated diversification once provided with new ecological opportunities created by diversifying angiosperms during KTR and/or because of the extinction of competitors post K-Pg boundary20,44. However, this similar set of forces may not be operating in other lichen-forming fungal lineages as mentioned in the previous section – the effect is lineage-specific and can be confounded by phylogenetic relatedness.
Our results reveal that not all lineages could have seized the opportunity created by major historical events and underwent rapid diversification. Similar findings have also been reported in a recent study focusing on frogs, where only the evolutionary histories of three lineages unravel accelerated diversification after the K-Pg event. There are clearly lineage-specific patterns of macroevolutionary dynamics demonstrated in many macroevolutionary studies29, and our results further imply that there can be lineage-specific effects of evolving different trait states on variations of macroevolutionary dynamics. Specifically, the evolution of the macrolichen phenotype might be correlated with an increase in diversification rate, but only in certain lineages, after the K-Pg boundary, although the pattern is not universal and can be explained by phylogenetic relatedness. Our results imply that although the macroevolutionary dynamics in lichen-forming fungi, and other organisms in general, could be contingent to evolutionary innovations, other factors pertaining to biological variation among lineages, variation in the nature of the putative evolutionary innovation among lineages – e.g., different growth forms exist in macrolichens – and even stochastic events may mask the effect from the previous events. Furthermore, micro-evolutionary process was not taken into account in our macro-evolutionary analyses. The generation time for example may differ between lineages, and especially the disparity in growth rate between micro- and macrolichens could be an innate difference leading to distinct evolutionary rate across lineages. Additionally, as mentioned before the K-Pg mass extinction event was not the only major geohistorical or biological event that might have affected the rate of diversification during the transition from Mesozoic to Cenozoic eras. For example, the KTR event (125 to 80 MYA) is characterized by rapid diversification of angiosperms and mammals45,46. The rapid organismal diversification, particularly in angiosperms, during KTR has been hypothesized impacted the global biodiversity pattern and might have directly affected the diversification rates of other evolutionary lineages49. Furthermore, the Paleocene-Eocene thermal maximum (55 MYA; PETM) could have also impacted the pattern of evolutionary diversification50. Changes in temperature and precipitation patterns can significantly affect the distribution of lichen-forming fungi. Given the high uncertainty in phylogenetic dating and the estimated phylogenetic position of rate shifts scatter across 100 to 40 MYA, we cannot rule out the effect of environmental and climatic changes occurred during late Cretaceous or early Cenozoic on the variation in diversification dynamics of lichen-forming fungi. Nevertheless, our study augments the growing evidence for major effects on re-shaping terrestrial biodiversity pattern by both biological (e.g., KTR) and geohistorical e.g., (K-Pg and PETM) events.
Future comparative studies that include comprehensive sampling and family-specific study design may help to tease apart the relative effects of the major historical events – i.e., KTR, K-Pg, and PETM – and evolutionary innovation on variation in biodiversity pattern across geological times and evolutionary lineages. Comparative studies that have extensive taxon sampling within each family can also help to test whether the power to detect changes in macroevolutionary dynamics is not only determined by the abundance of taxon sampling and missing sample adjustment, but also by the phylogenetic depth – e.g., including different genera, or families – of the investigated phylogeny. For example, studies that focus on within family diversification often infer a major change in the diversification dynamics during the Miocene, while those that focus on a larger and deeper scale phylogenetic history, such as this study, reveal a significant effect around the Paleocene5. That is, the most apparent pattern that can be identified from a macroevolution comparative study may vary depending on the scope of phylogenetic sampling and geological time.
It is worthy of noting that the methods to infer shifts in diversification pattern and trait-dependent diversification based on reconstructed molecular phylogeny are still in its infancy. Limitations of methods used are widely debated and controversies do exist51,52. We too found an inconsistent result between our study and a previous study using the same method, BAMM specifically. Teloschistaceae was shown to have experienced significant diversification rate acceleration around 100 MYA40. Our result, which also places the common ancestor of Teloschistaceae around 100 MYA (Fig. 1), does not support a rate shift at the same node (marginal shift probability = 0.0013). This inconsistency may result because of different strategies of phylogenetic taxon sampling and geological time among studies. For example, more microlichens families have been included in the current study than in the previously mentioned study focusing on Teloschistaceae. Similarly, as pointed out in the previous paragraph, most empirical data sets may not have enough statistical power to detect trait-dependent diversification pattern using STRAPP. We want to emphasize that we do not reject the idea that there can be more shifts in diversification rate on the phylogeny, as BAMM has been shown to underestimate the number of rate shift, and that the diversification rate can be dependent upon trait states in lichen-forming fungi, even though they are highly lineage-specific. We only reveal that their effect on the variation of macroevolutionary dynamics do not result in statistically detectable differences using currently available data. The non-significant results can, for example, be due to the lack of statistical power, and the hypothesis of evolving macrolichens leading to accelerated diversification and a rate shift at the common ancestor for Teloschistaceae can still be real patterns that require additional investigations.
Conclusions
By assembling publicly accessible sequence data that have been accumulated over the past decades from major lineages of the lichen-forming fungi, we provide a novel example from a mutualistic system that is biologically, ecologically, and evolutionarily distinct from the intensively investigated vertebrate (e.g., birds and frogs), plant (e.g., eudicots and ferns), and insect (e.g., beetles) systems and broaden our understanding on not only the origins of biodiversity, but also the possible causes of variation in biodiversity pattern. Studies that focus on different organismal groups and different geological time scales often come to different conclusions. This inconsistency can be due to the use of different data sets and methodological approaches, but can also unravel real variations in historical processes and contingencies among organismal groups, as was revealed in our study by comparing the diversification dynamics between the two-main lichen-forming fungal subclasses, Lecanoromycetidae and Ostropomycetidae. Both subclasses persisted throughout major historical events, but only certain families prospered and rapidly diversified when provided with new opportunities. The differences in reaction to historical events might be related to the evolution of specific traits, but the currently available data, as well as limited statistical models, prevent us from making definitive inferences. While there is no doubt that the methodological approaches will continue to improve and more and more data are becoming available, the best way to understand how lineage-specific factors may lead to variation in the diversification pattern is to study more of them.
Materials and Methods
Phylogeny, the estimation of sampling completeness, and trait data
Recent molecular phylogenetic studies have gradually built a broad picture of the diversification history in different families and classes of lichen-forming fungi40,41,53–56. We used a recent time-calibrated phylogeny of Lecanoromycetes that included samples from most families and genera of the subclasses Lecanoromycetidae and Ostropomycetidae (details in43 for all the subsequent analyses and also used the classification proposed in this paper. We note that phylogenies inferred from more comprehensively sampled data would likely provide higher statistical power and robust results. However, our analyses are based on the most comprehensive phylogeny currently available.
Bias in taxonomic completeness overall across the phylogeny and between different trait states are the main source of uncertainty in large-scale analysis of biodiversity patterns, particularly in poorly known groups, such as lichen-forming fungi. We acknowledge that there have been several taxonomic studies on lichen-forming fungi, and many cryptic species have been uncovered in recent years, suggesting the potential for many additional undescribed species that are disproportionally distributed across lineages57,58. While accounting for unknown species is a challenge in any analysis, we relied on a recent account of families and genera in Ascomycota42 to estimate sampling bias in different genera (Supplementary Materials; frag_genus.txt).
Our morphological trait data were based primarily on the work of Jaklitsch et al.42. Specifically, we included all types of foliose and fruticose lichens in the character state “macrolichen”, whereas crustose species and species with small squamules were categorized as “microlichen”. A table of trait states for each of the sampled taxa can be found in Supplementary Materials (Oslec_states.csv).
Macroevolutionary dynamics reconstruction
We estimated macroevolutionary rates and tested for their dependencies on morphological trait states using the program BAMM59. Our focus was on investigating support for lineage-specific and trait state differences in net diversification rate, given the difficulties in estimating extinction rates from molecular data60. One advantage of BAMM is that complex macroevolutionary mixture models are assessed with rate shifts across the tree, including accelerating and decelerating diversification rates. While trait-dependent diversification models are not directly fit to the tree, correlations between trait states and diversification rates can be assessed post hoc using STRAPP61. Therefore, we used BAMM to investigate evidence of accelerating or decelerating diversification through time and if the rate shifts were associated with critical geological events (e.g., the K-Pg boundary), and then to test for correlations between lichen growth forms (macro- versus microlichens) and net diversification rate.
Net-diversification, speciation and extinction rates through time based on43 were estimated using the program BAMM version 2.5. The initial values for speciation and extinction rates as well as the number of rate shift were estimated using the setBAMMpriors function in the R BAMMtools package59 and subsequently specified in the BAMM control file. A total of 2 × 108 generations of rjMCMC searches with samples stored every 1 × 105 generations were launched using the speciation-extinction mode. A total of 1000 post burnin samples (50%) were retained for the following analyses.
Using the posteriors generated from the rjMCMC searches, we aimed to (1) reconstruct the diversification history of Lecanoromycetidae and Ostropomycetidae, (2) test whether there is evidence of changes in diversification dynamics, particularly accelerating or decelerating diversification rate associated with major historical events, and (3) examine whether different growth forms in lichen-forming fungi are associated with different diversification rates. We visualized the reconstructed history of macroevolutionary dynamics with the plot.bammdata function from BAMMtools. We utilized marginal likelihood values to infer the phylogenetic position where diversification rate may have changed (accelerated/decelerated). We further plotted the diversification rate through time plots for each of the two subclasses and individual families that have a total species number greater than 500 (Table 1; family classification followed43 using the plotRateThroughTime function. We used the getTipRates function to extract the mean net-diversification rates at tips of the phylogeny, and then tested statistically whether species currently associated with different growth forms may exhibit different diversification rates. We also used the traitDependentBAMM [STRAPP61] function in BAMMtools to further investigate whether the net-diversification rate differs between species of different trait states while accounting for phylogenetic relatedness among taxa. The STRAPP analysis was performed with 1 × 104 iterations and Spearman’s rho as test statistics. We also applied Pearson correlation as test statistics and Mann-Whitney test to assess whether our results were robust.
Table 1.
| Subclass | Order | Family | Number of Species |
|---|---|---|---|
| Lecanoromycetidae | Lecanorales | Parmeliaceae | 2760* |
| Gypsoplacaceae | 1 | ||
| Tephromelataceae | 50 | ||
| Lecanoraceae | 780 | ||
| Pilocarpaceae | 380 | ||
| Malmideaceae | 53 | ||
| Cladoniaceae | 810 | ||
| Ramalinaceae | 675 | ||
| Biatoraceae | 150 | ||
| Psoraceae | 55 | ||
| Sphaerophoraceae | 38 | ||
| Catillariaceae | 175 | ||
| Scoliciosporaceae | 16 | ||
| Psilolechiaceae | 4 | ||
| Teloschistales | Teloschistaceae | 700 | |
| Megalosporaceae | 38 | ||
| Brigantiaeaceae | 50 | ||
| Caliciales | Physciaceae | 580 | |
| Caliciaceae | 630 | ||
| Peltigerales | Peltigeraceae | 611 | |
| Massalongiaceae | 5 | ||
| Vahliellaceae | 8 | ||
| Koeberiaceae | 8 | ||
| Collemataceae | 190 | ||
| Placynthiaceae | 30 | ||
| Pannariaceae | 400 | ||
| Coccocarpiaceae | 38 | ||
| Lecideales | Lecideaceae | 200 | |
| Rhizocarpales | Rhizocarpaceae | 230 | |
| Sporastatiales | Sporastatiaceae | 5 | |
| Ostropomycetidae | Graphidales | Diploschistaceae | 600 |
| Thelotremataceae | 250 | ||
| Graphidaceae | 1250 | ||
| Fissurinaceae | 160 | ||
| Gomphillaceae | 420 | ||
| Gyalectales | Porinaceae | 360 | |
| Coenogoniaceae | 90 | ||
| Sagiolechiaceae | 4 | ||
| Gyalectaceae | 90 | ||
| Phlyctidaceae | 20 | ||
| Odontotrematales | Odontotremataceae | 22 | |
| “Ostropales” | Asconditella clades | 14 | |
| Stictidaceae | 210 | ||
| Ascarosporinaceae | 5 | ||
| Thelenellales | Thelenellaceae | 65 | |
| Ochrolechiales | Ochrolechiaceae | 60 | |
| Varicellariaceae | 7 | ||
| Variolariaceae | 5 | ||
| Microcaliciaceae | 4 | ||
| Megasporaceae | 240 | ||
| Pertusariales | Pertusariaceae | 400 | |
| Agyriaceae | 5 | ||
| Icmadophilaceae | 55 | ||
| Coccotremataceae | 26 | ||
| Baeomycetales | Trapeliaceae | 117 | |
| Xylographaceae | 33 | ||
| Arctomiaceae | 12 | ||
| Baeomycetaceae | 16 | ||
| Cameroniaceae | 2 | ||
| Protothelenellaceae | 14 | ||
| Arthrorhaphidaceae | 14 | ||
| Hymeneliaceae | 28 |
*For families that contain more than 500 species the species numbers are shown in bold.
The evolution of macro- and micro-lichens
To reconstruct the evolutionary history of macrolichens and microlichens, we fit discrete models (binary states) of character evolution using the ace function in the ape package62. Likelihood ratio tests were used to compare models that assume either equal or asymmetric rate of transition between trait states. The ancestral states at each node of the empirical tree were estimated using the selected model. To visualize the evolution of lichen growth forms, we further performed 100 stochastic character mappings on the empirical tree using the make.simmap function and plotted a summary of state probabilities for each branch with the densityMap function in the phytools package63.
Supplementary information
Acknowledgements
Financial support by the Negaunee Foundation is greatly acknowledged.
Author Contributions
H.T.L. conceived the study; J.-P.H. and H.T.L. designed the work; M.P.N. gathered the data set; J.-P.H. performed the analyses with inputs from H.T.L.; J.-P.H., E.K., S.D.L., M.P.N. and H.T.L. wrote the manuscript. All authors read and approved the final manuscript.
Data Availability
The studied phylogeny was retrieved from43. The sampling fraction and trait state files for BAMM analyses were appended as Supplementary Materials.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44881-1.
References
- 1.Erwin DH. Lessons from the Past: Biotic Recoveries from Mass Extinctions. Proc. Natl. Acad. Sci. USA. 2001;98:5399–5403. doi: 10.1073/pnas.091092698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erwin DH. The End and the Beginning: Recoveries from Mass Extinctions. Trends Ecol. Evol. 1998;13:344–349. doi: 10.1016/S0169-5347(98)01436-0. [DOI] [PubMed] [Google Scholar]
- 3.Knoll AH, Bambach RK, Canfield DE, Grotzinger JP. Comparative Earth History and Late Permian Mass Extinction. Science. 1996;273:452–457. doi: 10.1126/science.273.5274.452. [DOI] [PubMed] [Google Scholar]
- 4.Jablonski D. Background and Mass Extinctions - the Alternation of Macroevolutionary Regimes. Science. 1986;231:129–133. doi: 10.1126/science.231.4734.129. [DOI] [PubMed] [Google Scholar]
- 5.Renne PR, et al. Time Scales of Critical Events around the Cretaceous-Paleogene Boundary. Science. 2013;339:684–687. doi: 10.1126/science.1230492. [DOI] [PubMed] [Google Scholar]
- 6.Sereno PC. The Evolution of Dinosaurs. Science. 1999;284:2137–2147. doi: 10.1126/science.284.5423.2137. [DOI] [PubMed] [Google Scholar]
- 7.Padian K, Chiappe LM. The Origin and Early Evolution of Birds. Biol. Rev. 1998;73:1–42. doi: 10.1017/S0006323197005100. [DOI] [Google Scholar]
- 8.Longrich NR, Bhullar B-AS, Gauthier JA. Mass Extinction of Lizards and Snakes at the Cretaceous-Paleogene Boundary. Proc. Natl. Acad. Sci. USA. 2012;109:21396–21401. doi: 10.1073/pnas.1211526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Longrich NR, Tokaryk T, Field DJ. Mass Extinction of Birds at the Cretaceous-Paleogene (K-Pg) Boundary. Proc. Natl. Acad. Sci. USA. 2011;108:15253–15257. doi: 10.1073/pnas.1110395108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Leary MA, et al. The Placental Mammal Ancestor and the Post-K-Pg Radiation of Placentals. Science. 2013;339:662–667. doi: 10.1126/science.1229237. [DOI] [PubMed] [Google Scholar]
- 11.Kellner AWA, Pinheiro AEP, Campos DA. A New Sebecid from the Paleogene of Brazil and the Crocodyliform Radiation after the K-Pg Boundary. Plos One. 2014;9:1. doi: 10.1371/journal.pone.0081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claramunt S, Cracraft J. A New Time Tree Reveals Earth History’s Imprint on the Evolution of Modern Birds. Sci. Adv. 2015;1:e1501005–e1501005. doi: 10.1126/sciadv.1501005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Y-J, et al. Phylogenomics Reveals Rapid, Simultaneous Diversification of Three Major Clades of Gondwanan Frogs at the Cretaceous-Paleogene Boundary. Proc. Natl. Acad. Sci. USA. 2017;114:e5864–e5870. doi: 10.1073/pnas.1704632114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith VS, et al. Multiple Lineages of Lice Pass through the K-Pg Boundary. Biol. Lett. 2011;7:782–785. doi: 10.1098/rsbl.2011.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu S, et al. Molecular and Paleontological Evidence for a Post-Cretaceous Origin of Rodents. Plos One. 2012;7:10. doi: 10.1371/annotation/75d95a55-a58d-4831-8de4-d0f935fe512d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson, D. E. & Reeder, D. M. Mammal Species of the World. A Taxonomic and Geographic Reference. (Baltimore, Maryland, Johns Hopkins University Press, 2005).
- 17.Lohaus R, Van de Peer Y. Of Dups and Dinos: Evolution at the K/Pg Boundary. Curr. Opin. Plant Biol. 2016;30:62–69. doi: 10.1016/j.pbi.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Vajda V, McLoughlin S. Fungal Proliferation at the Cretaceous-Tertiary Boundary. Science. 2004;303:1489–1489. doi: 10.1126/science.1093807. [DOI] [PubMed] [Google Scholar]
- 19.Vajda V, Raine JI, Hollis CJ. Indication of Global Deforestation at the Creataceous-Tertiary Boundary by New Zealand Fern Spike. Science. 2001;294:1700–1702. doi: 10.1126/science.1064706. [DOI] [PubMed] [Google Scholar]
- 20.Alvarez LW, Alvarez W, Asaro F, Michel HV. Extraterrestrial Cause for the Cretaceous - Tertiary Extinction - Experimental Results and Theoretical Interpretation. Science. 1980;208:1095–1108. doi: 10.1126/science.208.4448.1095. [DOI] [PubMed] [Google Scholar]
- 21.Honegger R, Edwards D, Axe L. The Earliest Records of Internally Stratified Cyanobacterial and Algal Lichens from the Lower Devonian of the Welsh Borderland. New Phytol. 2013;197:264–275. doi: 10.1111/nph.12009. [DOI] [PubMed] [Google Scholar]
- 22.Lumbsch, H. T. & Rikkinen, J. Evolution of Lichens. In: The Fungal Community: Its Organization and Role in the Ecosystem. Pp 53–62, Edited by Dighton J., White J. F. & Boca Raton, F. L. (CRC Press, 2017).
- 23.Simpson, G. G. The Major Features of Evolution. (New York, Columbia University Press, 1953).
- 24.Hodges SA, Arnold ML. Spurring Plant Diversification: Are Floral Nectar Spurs a Key Innovation? Proc. Roy. Soc. London B-Biol. Sci. 1995;262:343–348. doi: 10.1098/rspb.1995.0215. [DOI] [Google Scholar]
- 25.Hugall AF, Stuart-Fox D. Accelerated Speciation in Colour-Polymorphic Birds. Nature. 2012;485:631. doi: 10.1038/nature11050. [DOI] [PubMed] [Google Scholar]
- 26.Donoghue MJ, Sanderson MJ. Confluence, Synnovation, and Depauperons in Plant Diversification. New Phytol. 2015;207:260–274. doi: 10.1111/nph.13367. [DOI] [PubMed] [Google Scholar]
- 27.Nash, T. H. Lichen Biology, 2nd ed. (Cambridge, UK, Cambridge University Press, 2008).
- 28.Kraichak E, Luecking R, Lumbsch HT. A Unique Trait Associated with Increased Divesification in a Hyperdiverse Family of Tropical Lichen-Forming Fungi. Internatl. J. Plant Sci. 2015;176:597–606. doi: 10.1086/682061. [DOI] [Google Scholar]
- 29.Alfaro ME, et al. Nine Exceptional Radiations Plus High Turnover Explain Species Diversity in Jawed Vertebrates. Proc. Natl. Acad. Sci. USA. 2009;106:13410–13414. doi: 10.1073/pnas.0811087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maddison WP, Midford PE, Otto SP. Estimating a Binary Character’s Effect on Speciation and Extinction. Syst. Biol. 2007;56:701–710. doi: 10.1080/10635150701607033. [DOI] [PubMed] [Google Scholar]
- 31.Morlon H. Phylogenetic Approaches for Studying Diversification. Ecol. Lett. 2014;17:508–525. doi: 10.1111/ele.12251. [DOI] [PubMed] [Google Scholar]
- 32.Rabosky DL. Automatic Detection of Key Innovations, Rate Shifts, and Diversity-Dependence on Phylogenetic Trees. Plos One. 2014;9:e89543. doi: 10.1371/journal.pone.0089543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Werner, G. D. A., Cornwell, W. K., Sprent, J. I., Kattge, J. & Kiers, E. T. A Single Evolutionary Innovation Drives the Deep Evolution of Symbiotic N-2-Fixation in Angiosperms. Nature Comm. 5 (2014). [DOI] [PMC free article] [PubMed]
- 34.Silvestro D, Zizka G, Schulte K. Disentangling the Effects of Key Innovations on the Diversification of Bromelioideae (Bromeliaceae) Evolution. 2014;68:163–175. doi: 10.1111/evo.12236. [DOI] [PubMed] [Google Scholar]
- 35.Rainford JL, Hofreiter M, Nicholson DB, Mayhew PJ. Phylogenetic Distribution of Extant Richness Suggests Metamorphosis Is a Key Innovation Driving Diversification in Insects. Plos One. 2014;9:10. doi: 10.1371/journal.pone.0109085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng J, Smith SD. How Traits Shape Trees: New Approaches for Detecting Character State-Dependent Lineage Diversification. J. Evol. Biol. 2014;27:2035–2045. doi: 10.1111/jeb.12460. [DOI] [PubMed] [Google Scholar]
- 37.de Vos JM, Hughes CE, Schneeweiss GM, Moore BR, Conti E. Heterostyly Accelerates Diversification Via Reduced Extinction in Primroses. Proc. Roy. Soc. B-Biol. Sci. 2014;281:1784. doi: 10.1098/rspb.2014.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia-Porta J, Ord TJ. Key Innovations and Island Colonization as Engines of Evolutionary Diversification: A Comparative Test with the Australasian Diplodactyloid Geckos. J. Evol. Biol. 2013;26:2662–2680. doi: 10.1111/jeb.12261. [DOI] [PubMed] [Google Scholar]
- 39.Divakar PK, Kauff F, Crespo A, Leavitt SD, Lumbsch HT. Understanding Phenotypical Character Evolution in Parmelioid Lichenized Fungi (Parmeliaceae, Ascomycota) Plos One. 2013;8:e83115. doi: 10.1371/journal.pone.0083115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaya E, et al. The Adaptive Radiation of Lichen-Forming Teloschistaceae Is Associated with Sunscreening Pigments and a Bark-to-Rock Substrate Shift. Proc. Natl. Acad. Sci. USA. 2015;112:11600–11605. doi: 10.1073/pnas.1507072112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kraichak E, et al. A Tale of Two Hyper-Diversities: Diversification Dynamics of the Two Largest Families of Lichenized Fungi. Sci. Rep. 2015;5:e10028. doi: 10.1038/srep10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaklitsch, W. M., Baral, H. O., Lücking, R. & Lumbsch, H. T. Ascomycota. In: Syllabus of Plant Families - Adolf Engler’s Syllabus Der Pflanzenfamilien. Pp 1–150. Edited by Frey W., vol. 1/2, 13 ed. Stuttgart, Gebr. Borntraeger Verlagsbuchhandlung (2016).
- 43.Kraichak E, Huang J-P, Nelsen MP, Leavitt SD, Lumbsch HT. A Revised Classification of Orders and Families in the Two Major Subclasses of Lecanoromycetes (Ascomycota) Based on a Temporal Approach. Bot. J. Linn. Soc. 2018;188:233–249. [Google Scholar]
- 44.Rabosky DL, Huang H. A Robust Semi-Parametric Test for Detecting Trait-Dependent Diversification. Syst. Biol. 2015;65:181–193. doi: 10.1093/sysbio/syv066. [DOI] [PubMed] [Google Scholar]
- 45.Meredith RW, et al. Impacts of the Cretaceous Terrestrial Revolution and Kpg Extinction on Mammal Diversification. Science. 2011;334:521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- 46.Dilcher DL. Toward a New Synthesis: Major Evolutionary Trends in the Angiosperm Fossil Record. Proc. Natl. Acad. Sci. USA. 2000;97:7030–7036. doi: 10.1073/pnas.97.13.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooper N, Thomas GH, FitzJohn RG. Shedding Light on the ‘Dark Side’ of Phylogenetic Comparative Methods. Methods Ecol. Evol. 2016;7:693–699. doi: 10.1111/2041-210X.12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prieto M, Wedin M. Dating the Diversification of the Major Lineages of Ascomycota (Fungi) Plos One. 2013;8:e65576. doi: 10.1371/journal.pone.0065576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feldberg K, et al. Epiphytic Leafy Liverworts Diversified in Angiosperm-Dominated Forests. Sci. Rep. 2014;4:5974. doi: 10.1038/srep05974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gingerich PD. Environment and Evolution through the Paleocene-Eocene Thermal Maximum. Trends Ecol. Evol. 2006;21:246–253. doi: 10.1016/j.tree.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 51.Moore BR, Höhna S, May MR, Rannala B, Huelsenbeck JP. Critically Evaluating the Theory and Performance of Bayesian Analysis of Macroevolutionary Mixtures. Proc. Natil. Acad. Sci. 2016;113:9569–9574. doi: 10.1073/pnas.1518659113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rabosky DL, Mitchell JS, Chang J. Is Bamm Flawed? Theoretical and Practical Concerns in the Analysis of Multi-Rate Diversification Models. Syst. Biol. 2017;66:477–498. doi: 10.1093/sysbio/syx037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Divakar PK, et al. Evolution of Complex Symbiotic Relationships in a Morphologically Derived Family of Lichen-Forming Fungi. New Phytol. 2015;208:1217–1226. doi: 10.1111/nph.13553. [DOI] [PubMed] [Google Scholar]
- 54.Divakar PK, et al. Using a Temporal Phylogenetic Method to Harmonize Family- and Genus-Level Classification in the Largest Clade of Lichen-Forming Fungi. Fungal Divers. 2017;84:101–117. doi: 10.1007/s13225-017-0379-z. [DOI] [Google Scholar]
- 55.Miadlikowska J, et al. Multigene Phylogenetic Synthesis for 1307 Fungi Representing 1139 Infrageneric Taxa, 312 Genera and 66 Families of the Class Lecanoromycetes (Ascomycota) Mol. Phylogenet. Evol. 2014;79:132–168. doi: 10.1016/j.ympev.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lutzoni F, et al. Assembling the Fungal Tree of Life: Progress, Classification, and Evolution of Subcellular Traits. Am. J. Bot. 2004;91:1446–1480. doi: 10.3732/ajb.91.10.1446. [DOI] [PubMed] [Google Scholar]
- 57.Crespo A, Lumbsch HT. Cryptic Species in Lichen-Forming Fungi. IMA Fungus. 2010;1:167–170. doi: 10.5598/imafungus.2010.01.02.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lumbsch HT, Leavitt SD. Goodbye Morphology? A Paradigm Shift in the Delimitation of Species in Lichenized Fungi. Fungal Divers. 2011;50:59–72. doi: 10.1007/s13225-011-0123-z. [DOI] [Google Scholar]
- 59.Rabosky DL, et al. Bammtools: An R Package for the Analysis of Evolutionary Dynamics on Phylogenetic Trees. Methods Ecol. Evol. 2014;5:701–707. doi: 10.1111/2041-210X.12199. [DOI] [Google Scholar]
- 60.Rabosky DL. Extinction Rates Should Not Be Estimated from Molecular Phylogenies. Evolution. 2010;64:1816–1824. doi: 10.1111/j.1558-5646.2009.00926.x. [DOI] [PubMed] [Google Scholar]
- 61.Rabosky DL, Huang H. A Robust Semi-Parametric Test for Detecting Trait-Dependent Diversification. Syst. Biol. 2016;65:181–193. doi: 10.1093/sysbio/syv066. [DOI] [PubMed] [Google Scholar]
- 62.Paradis E, Claude J, Strimmer K. Bioinformatics. 2004. Ape: Analyses of Phylogenetics and Evolution in R Language; pp. 289–290. [DOI] [PubMed] [Google Scholar]
- 63.Revell LJ. Phytools: An R Package for Phylogenetic Comparative Biology (and Other Things) Methods Ecol. Evol. 2012;3:217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The studied phylogeny was retrieved from43. The sampling fraction and trait state files for BAMM analyses were appended as Supplementary Materials.