Abstract
Mammalian genomes are folded into tens of thousands of long-range looping interactions. The cause and effect relationship between looping and genome function is poorly understood, and the extent to which loops are dynamic on short time scales remains an unanswered question. Here we engineer a new class of synthetic architectural proteins for directed rearrangement of the 3-D genome using blue light. We target our light-activated-dynamic-looping (LADL) system to two genomic anchors with CRISPR guide RNAs and induce their spatial co-localization via light-induced heterodimerization of cryptochrome 2 and a dCas9-CIBN fusion protein. We apply LADL to redirect a stretch enhancer (SE) away from its endogenous Klf4 target gene and to the Zfp462 promoter. Using single molecule RNA FISH, we demonstrate that de novo formation of the Zfp462-SE loop correlates with a modest but significant increase in Zfp462 expression. LADL facilitates co-localization of genomic loci without exogenous chemical cofactors and will enable future efforts to engineer reversible and oscillatory loops on short time scales.
Introduction
The development of tools to manipulate 3-D genome folding on demand with spatiotemporal precision is of critical importance toward advancing studies in basic science, regenerative medicine, metabolic engineering, and synthetic biology. Mouse and human genomes are folded into more than 10,000 loops1, 2, but the functional role for individual and combinations of long-range connections on gene expression remains poorly understood. Published strategies for loop engineering involve synthetic transcription factors tethered to dCas93, 4 or zinc fingers5, 6, and synthetic looping factors have thus far been constitutively expressed or induced over long time scales by the presence of small molecules3–6. The paucity of tools to engineer genome folding on short time scales has prohibited our ability to understand the extent to which loops are dynamic and functionally contribute to the kinetics of transcriptional activation.
Results
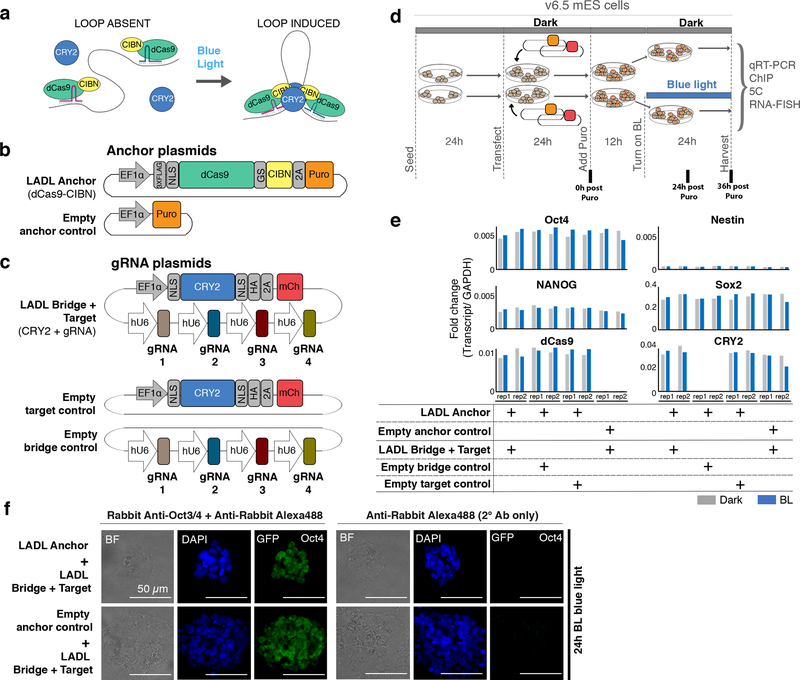
Here, we create synthetic architectural proteins with the capability of forming long-range physical contacts between multiple genomic loci on demand with blue light. We designed our light-activated dynamic looping (LADL) system in a modular manner with four key components (Figure 1a, Supplementary Figures 1–2, Supplementary Tables 1–6). First, we designed a synthetic architectural protein consisting of enzymatically inactive Cas9 (dCas9) tethered to a truncated CIBN protein derived from the CIB1 protein from Arabidopsis thaliana7 (Figure 1b). Second, we recruited the LADL Anchor (dCas9-CIBN) to two genomic target sites with sequence-specific CRISPR guide RNAs (gRNAs) (Figure 1c). We designed two gRNAs per anchoring genomic target site. Third, we hypothesized that the Photolyase-Homologous Region (PHR) domain of the CRY2 protein from Arabidopsis thaliana could serve as an inducible bridging factor due to its well-established ability to heterodimerize with CIBN in response to blue light on millisecond time scales in mammalian cells8, 9 (Figure 1c). Finally, we used blue light of wavelength 470nm as the loop inducing agent10. Because it is well-established that blue light illumination causes CIBN-CRY2 heterodimerization8, 9 and CRY2 oligomerization8, 11, 12, we hypothesized that LADL would spatially connect the two anchoring genomic fragments via a light-induced dCas9-CIBN and CRY2 bridge (Figure 1a). Thus, we designed LADL as a modular, four-component synthetic architectural protein system to spatially connect genomic loci in response to light via facile design of sequence-specific gRNAs.
Figure 1. Concept, design, and implementation of the light-activated dynamic looping (LADL) system.
(a) Schematic of the LADL system. In the dark, the LADL anchoring protein dCas9-CIBN is recruited to two specific genomic fragments using guide RNAs (gRNAs). CRY2 is soluble and does not specifically associate with chromatin. Upon blue light illumination, CRY2 homodimerizes and CRY2 and dCas9-CIBN heterodimerize to form a bridge connecting the two gRNA-bound genomic fragments. We hypothesize that the bridge would loop out the intervening DNA. (b-c) Schematic of plasmid constructs encoding the (b) puromycin-selectable LADL Anchor and Empty anchor control and (c) the LADL Bridge + Target, the Empty target control, and the Empty bridge control. (d) Schematic timeline of seeding, transfection, puromycin selection, and blue light illumination of v6.5 mouse embryonic stem cells. (e) qRT-PCR analysis of Oct4, Nestin, Nanog, Sox2, dCas9, and CRY2 transcript levels in co-transfected mouse embryonic stem cells after 36 hours post-puromycin selection. Data from two independent experiments are shown. (f) Immunofluorescence staining for Oct4 in mouse embryonic stem cells co-transfected with the indicated plasmids. Scale bars, 50 μm. Images are representative of 3 independent experiments.
To determine the conditions in which blue light would induce a spatial chromatin contact, we first employed 24 hours of blue light exposure (Figure 1d). We built a light box to achieve 5 mW/cm2 intensity and 1-second pulses at 0.067 Hz as previously reported for optimal CRY2-CIBN heterodimerization10, 13 (detailed in Supplementary Methods). We confirmed that the light exposure conditions successfully induced CRY2 oligomerization (Supplementary Figure 3). Mouse embryonic stem (ES) cells were co-transfected to achieve one of four conditions: (1) LADL (Anchor (dCas9-CIBN) and Bridge+Target (CRY2+sgRNA) plasmids), (2) Empty anchor control (Empty anchor and Bridge+Target (CRY2+gRNA) plasmids), (3) Empty bridge control (Anchor (dCas9-CIBN) and Empty bridge (gRNA only) plasmids), or (4) Empty target control (Anchor (dCas9-CIBN) and (CRY2 only) plasmids). Overall plasmid mass and ratios were adjusted to optimize transfection efficiency (Supplementary Figure 4).
We exposed transfected cells to 24 hours of blue light or dark after puromycin selection (Figure 1d). ES cell densities were similar across conditions and exhibited morphology characteristic of the v6.5 feeder-dependent clone after passage onto gelatin (Supplementary Figure 5). All conditions showed equivalently high expression of pluripotency markers Oct4, Nanog, Sox2, and low levels of Nestin, suggesting that the pluripotent, self-renewing ES cell state was not compromised by transfection and light induction (Figure 1e–f, Supplementary Table 7). dCas9-CIBN and CRY2 transcripts were strongly expressed across all conditions transfected with vectors encoding the transgenes (Figure 1e). Moreover, equivalent levels were seen in dark and blue light exposure, thus ruling out artifacts caused by differential transgene levels between conditions. Together, these results demonstrate that the two plasmids encoding our 4-component synthetic architectural protein system were equivalently expressed and have minimal negative impact on ES cell morphology, viability, and pluripotent properties.
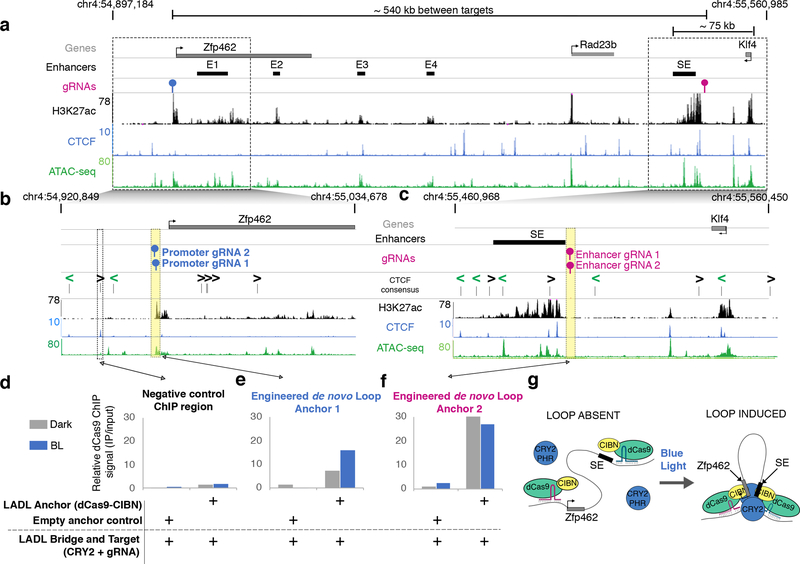
We chose an ~ 800 kb-sized locus around the Klf4 and Zfp462 genes as the genomic context for our LADL engineered loop. The Klf4 and Zfp462 genes are highly and lowly expressed in pluripotent ES cells, respectively, and are under the control of distal enhancer elements (Figure 2). As previously reported14, Zfp462 loops to at least four independent putative enhancers (E1, E2, E3, E4) marked by positive enrichment of the histone modification H3K27ac (Figure 2a, Supplementary Table 8). Klf4 forms an ~70 kb-sized long-range interaction with a large putative stretch enhancer (SE)14, 15. We reasoned that we could test LADL’s performance with a ‘Redirect and Reinforce’ strategy in which we spatially redirected the Klf4 SE away from Klf4 and to the Zfp462 promoter. To avoid disrupting endogenous transcription factors and architectural proteins, we designed LADL gRNAs directly adjacent to, but not overlapping, H3K27ac ChIP-seq and ATAC-seq signal marking the Klf4 SE and the Zfp462 promoter (Figure 2a–c, blue and magenta gRNA markers respectively, Supplementary Table 8). We used chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) to confirm recruitment of the LADL system to the specifically targeted genomic locations (Figure 2d–f, Supplementary Table 9). Using an anti-FLAG antibody, we demonstrated strong enrichment of FLAG-tagged dCas9-CIBN in the dark at both the Zfp462 promoter (Figure 2e) and the Klf4 SE (Figure 2f), but not a non-specific genomic region (Figure 2d). Importantly, this enrichment was not observed when the LADL Anchor was absent (Empty anchor control). Thus, the LADL Anchor can be effectively targeted to genomic loci adjacent to accessible chromatin using two gRNAs.
Figure 2. Chromatin binding of LADL Anchor (dCas9-CIBN) at the engineered sites increases after blue light exposure.
(a) Genome browser tracks overlaid at the Zfp462 and Klf4 genes and their target enhancers (chr4:54,897,184–55,560,985; mm9 reference genome). SE, the Klf4 stretch enhancer. E1, E2, E3, E4, the Zfp462 enhancers. (b-c) Zoomed-in genome browser tracks at the regions surrounding (b) the Zfp462 promoter and (c) the SE. (d-f) ChIP-qPCR data for the (d) negative control chromatin site, (e) engineered gRNAs at the Zfp462 promoter, and (f) engineered gRNAs at the Klf4 SE in co-transfected mouse embryonic stem cells after 24 hours of dark or blue light exposure. Data was acquired from one ChIP-qPCR experiment. (g) Model illustrating findings from the ChIP-qPCR data.
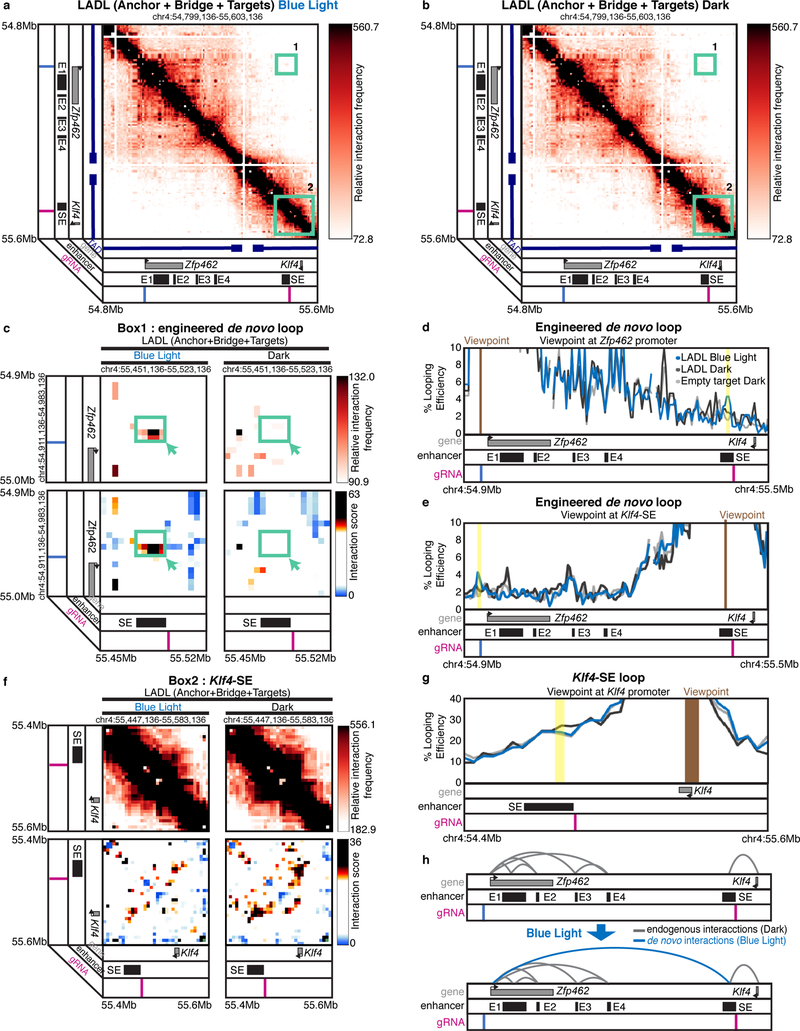
We next set out to determine if a spatial contact was induced by LADL in response to blue light. We hypothesized that an engineered long-range contact between our two targeted genomic fragments might alter dCas9-CIBN ChIP-qPCR signal due to indirect immunoprecipitation from the distal, spatially proximal fragment (Figure 2g). Indeed, we found that the intensity of dCas9-CIBN ChIP signal is altered after blue light illumination, increasing more than 2-fold at the Zfp462 promoter and slightly decreasing at the Klf4 SE (Figure 2e–f). We then directly assessed higher-order chromatin architecture with Chromosome-Conformation-Capture-Carbon-Copy (5C)14, 16–18 (Supplementary Tables 10–12). We generated a high- resolution map of long-range interactions for all genomic fragments in a ~3.5 Mb region around the Klf4 and Zfp462 genes in the conditions of (1) LADL (Anchor + Bridge + Target) after 24 hours of blue light, (2) LADL (Anchor + Bridge + Target) after 24 hours of dark, and (3) Empty target control (Anchor+Bridge only) after 24 hours of dark (Figure 3a–b, Supplementary Figure 6). Upon blue light illumination, a new long-range contact is gained between the SE and Zfp462 in mouse ES cells transfected with LADL vectors (Figure 3c–e, Supplementary Figure 7a). The engineered loop is specific to the LADL+blue light condition and not present in LADL+dark or Empty target+dark controls (Figure 3c–e, Supplementary Figure 7a). We reproduced the de novo Zfp462-Klf4 SE loop in LADL-transfected ES cells after 24 hours of blue light illumination at a lower intensity of 1.5 mW/cm2 (Supplementary Figure 7b) as well as in three more independent experiments at 5 mW/cm2 (Supplementary Figure 7c–f). An additional one-sided gRNA negative control (Anchor + Bridge + one-sided Target (CRY2+promoter-targeted sgRNA)) did not show looping signal (Supplementary Figure 7c, f). Classic 4C looping efficiency plots from the viewpoint of both gRNA anchors across five independent experiments confirmed that the median strength of the Zfp462-Klf4 SE interaction increased ~2.0–2.5-fold in the LADL+BL versus LADL+dark condition (Supplementary Figure 8). Together, these results demonstrate that LADL can form a new long-range interaction between two genomic fragments in a blue light-dependent manner.
Figure 3. LADL redirects and reinforces a long-range interaction between a stretch enhancer and a new target gene upon blue light illumination.
(a) Heatmap of long-range interactions around an ~800 kb genomic region encompassing the Klf4 and Zfp462 genes. SE, the Klf4 stretch enhancer. E1, E2, E3, E4, the Zfp462 enhancers. Mouse embryonic stem cells were co-transfected with LADL (Anchor + Bridge + Target) plasmids and then exposed to 24 hours blue light illumination. Box 1, the target de novo engineered loop between the pluripotency-specific Klf4 stretch enhancer and the Zfp462 promoter. Box 2, Klf4 interaction with its upstream, pluripotency-specific stretch enhancer (SE). Additional controls are shown in Supplementary Figure 6. Zoomed-in heatmaps of (c) Box 1 or (f) Box 2. Top, relative interaction frequency 5C signal. Bottom, distance-corrected interaction score 5C signal. (d, e, g) Classic 4C looping efficiency plots from the viewpoint of (d) the Zfp462 promoter targeted gRNA, (e) the stretch enhancer targeted gRNA, or (g) the Klf4 promoter. Additional negative controls and replicates are shown in Supplementary Figures 7–10. (h) Model of looping interaction reconfiguration in response to LADL and blue light illumination.
We next queried if endogenous chromatin loops were disrupted during the process of redirecting the Klf4 SE to Zfp462. In wild type mouse ES cells, the Klf4 gene forms a strong looping interaction with its target SE14, 15. We detected punctate looping signal in both LADL (Anchor + Bridge + Target) and the Empty target control (Anchor + Bridge only) in the dark (Figure 3f–g, Supplementary Figure 9a). The Klf4-SE loop remains largely intact, with slightly reduced contact frequency, in LADL transfected ES cells upon 24 hours of 5 mW/cm2 blue light illumination (Figure 3f–g, Supplementary Figure 9a). We observed a slight, but non-significant reduction in loop strength in the LADL + blue light condition compared to negative controls across n=4 total replicates at 5 mW/cm2 and one additional replicate at 1.5 mW/cm2 blue light exposure (Supplementary Figure 9b–f, Supplementary Figure 10). Importantly, our data suggests that the endogenous hub of enhancer-enhancer and enhancer-Zfp462 interactions14, 15 present in pluripotent ES cells was undisturbed across all conditions (Supplementary Figure 11). Together, these data demonstrate that the Klf4 SE can be redirected across a population of cells to Zfp462, with a slight disruption in endogenous Klf4-SE interactions and a negligible effect on endogenous Zfp462-enhancer interactions (Figure 3h).
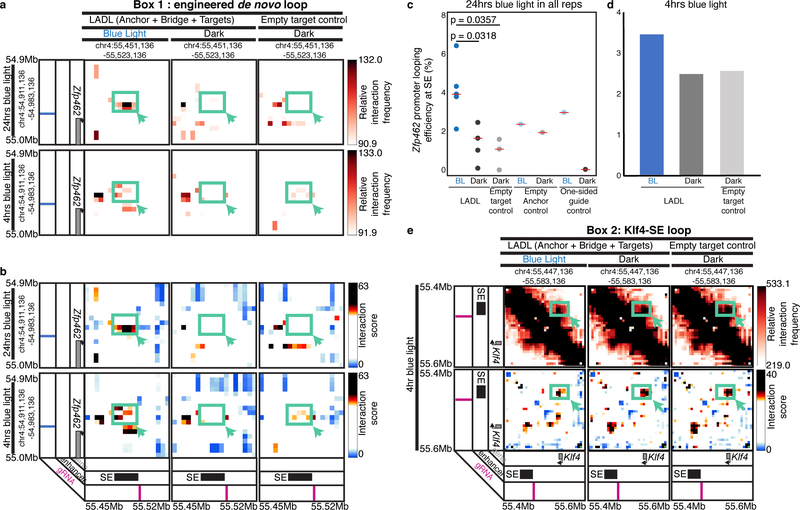
To gain insight into the time scale upon which the LADL-induced loops are formed, we mapped chromatin architecture in LADL engineered ES cells with 5C after varying the time scale of blue light exposure (Supplementary Figure 12). We observed the de novo engineered loop between Zfp462 and the Klf4 SE after as little as 4 hours blue light illumination, but it was slightly shifted spatially compared to the LADL-induced loop observed at 24 hours blue light (Figure 4a–b, Supplementary Figure 8a). Classic 4C looping efficiency plots confirmed that the engineered Zfp462-Klf4 SE loop showed an increase in interaction frequency between LADL blue light and dark conditions after 4 hours light exposure, however 24 hours blue light exposure led to stronger loop induction (Figure 4c–d). Consistent with our observations at 24 hours, the Klf4-SE interaction was only slightly reduced in LADL-engineered ES cells after 4 hours 5 mW/cm2 blue light compared to the LADL+dark and Empty target+dark conditions (Figure 4e). Together, these results indicate that LADL can enable the formation of long-range looping interactions on-demand in as early as 4 hours after application of the induction stimulus. Chemical induction of looping is reported to occur on the time scale of 24 hours or more3, thus LADL provides an advance in shortening the time scale of loop induction.
Figure 4. The LADL-induced engineered loop occurs as early as 4 hours after blue light illumination.
(a-b) Zoomed in heatmaps corresponding to Box 1 from Figure 3a–b for (a) relative interaction frequency 5C signal and (b) distance-corrected interaction score 5C signal. (c-d) Percent looping efficiency for the Zfp462 promoter and Klf4 SE interaction after (c) 24 hours (n=5 independent experiments) and (d) 4 hours (n=1 experiment) of blue light illumination. Red bars, medians of each condition. P-values computed using the unpaired two-sided Mann Whitney U test. (e) Zoomed in heatmaps corresponding to Box2 from Figure 3a–b for (top) relative interaction frequency 5C signal and (bottom) distance-corrected interaction score 5C signal.
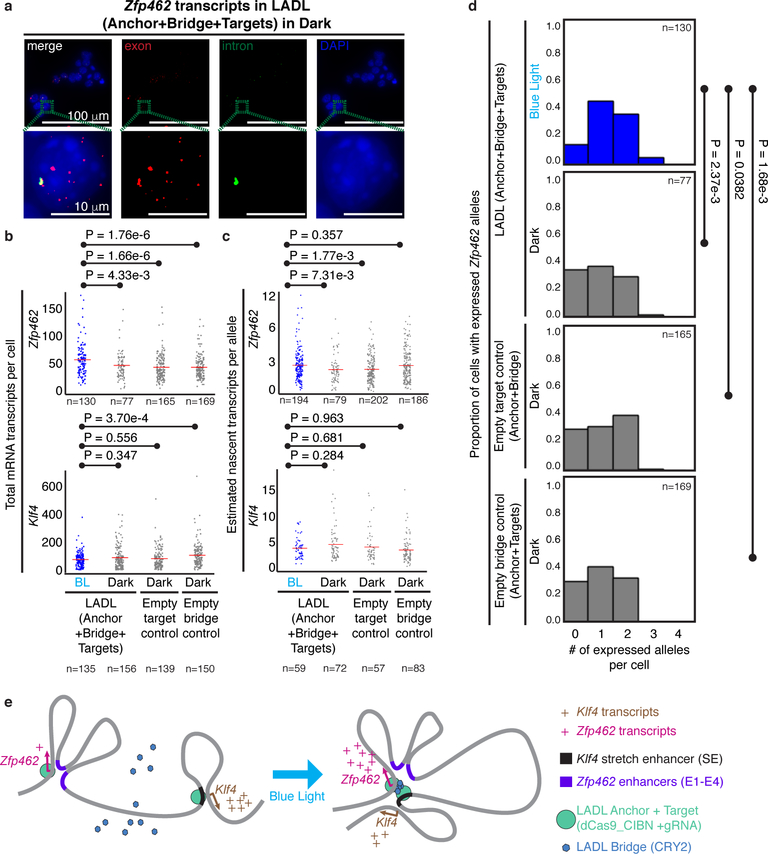
To understand the possible functional role of the de novo engineered loop, we next measured the effects of LADL-engineered looping interactions on gene expression. We performed single molecule RNA FISH to assess Zfp462 and Klf4 expression changes on a single cell basis after 24 hours blue light illumination (Figure 5a, Supplementary Table 13). The mean number of total Zfp462 mRNA transcripts per cell (53.47; 95% CI: 48.54 < μ Zfp462_LADL+BL < 58.40) was significantly higher in LADL+BL compared to LADL+dark (43.68; 95% CI: 38.24 < μZfp462_LADL+dark < 49.11), Empty target control+dark (40.14; 95% CI: 36.69 < μZfp462_Empty target control+dark < 43.59), or Empty bridge control+dark (40.33; 95% CI: 37.24 < μZfp462_Empty bridge control+dark < 43.41) (Figure 5b). Moreover, the mean of estimated nascent Zfp462 transcripts per allele (2.62; 95% CI: 2.43 < μZfp462_LADL+BL < 2.81) was also significantly higher in LADL+BL compared to LADL+dark (2.25; 95% CI: 1.99 < μZfp462_LADL+dark < 2.51) and Empty target control+dark (2.26; 95% CI: 2.1 < μZfp462_Empty target control+dark < 2.42) (Figure 5c). Total Zfp462 mRNA transcripts per cell were reproducibly upregulated upon LADL-induced loop formation in two out of the three RNA FISH experiments (Supplementary Figure 13a, c). Moreover, estimated nascent Zfp462 transcripts per allele were reproducibly upregulated upon LADL-induced loop formation in three out of three experiments (Supplementary Figure 13b, d). We also queried Klf4 expression and observed that total mRNA and estimated nascent transcript levels were highly variable across experiments, consistent with the slight but non-significant reduction in loop formation in LADL+BL versus negative controls (Figure 5b–c, Supplementary Figure 13a–d). Overall, our data are consistent with our working model that forced looping between the Klf4 SE and the Zfp462 promoter correlates with a modest increase in total mRNA and nascent transcripts of Zfp462 per cell.
Figure 5. Functional effect of the LADL-engineered de novo loop on endogenous gene expression.
(a) Full field (top row) and zoomed microscopy images (bottom row) from single molecule RNA fluorescence in situ hybridization (FISH) analysis of LADL-engineered mouse embryonic stem cells in dark. Quantitative image analysis of Zfp462 and Klf4 transcripts from LADL engineered mouse ES cells after 24 hours of blue light (detailed in Supplementary Methods). Scale bars, 100 μm. Images are representative of n=3 independent experiments. (b-c) Strip charts representing (b) the total number of mRNA transcripts per cell and (c) the estimated number of nascent transcripts per allele for Zfp462 (upper row) and Klf4 (lower row). Red bars, means of each condition. (d) Histograms represent the proportion of cells with a specific number of actively expressing Zfp462 alleles. P-values computed using the unpaired one-tailed Mann Whitney U test. Sample sizes represent (b, d) number of cells or (c) number of active transcription alleles. Additional replicates from independent experiments are shown in Supplementary Figure 13. (e) Schematic diagram illustrating the link between LADL engineered de novo loop formation and changes in endogenous gene expression.
We also hypothesized that an increase in loop frequency might lead to an increase in the number of transcriptionally active alleles within a population that simultaneously express the target gene. Using single molecule RNA FISH, we observed that the number of alleles per cell actively transcribing Zfp462 was significantly increased in LADL+BL (1.32; 95% CI: 1.19 < μZfp462_LADL+dark < 1.46) compared to LADL+dark (0.97; 95% CI: 0.79 < μZfp462_LADL+dark < 1.16), Empty target control+dark (1.12; 95% CI: 0.99 < μZfp462_Empty target control+BL < 1.24), or Empty bridge control+dark (1.02; 95% CI: 0.91 < μZfp462_Empty bridge control+BL < 1.14) (Figure 5d). The increase in Zfp462 expressing alleles in LADL+BL was reproducible in two out of three RNA FISH experiments (Supplementary Figure 13e–f). Our data suggest that LADL-induced formation of the de novo Zfp462-Klf4 SE loop can result in an increase in the proportion of alleles expressing Zfp462 (Figure 5e, Supplementary Figure 13).
Discussion
Overall, we present LADL as a new synthetic architectural protein system that is capable of forming inducible long-range looping interactions in response to light. Our new ‘3-D optoepigenetic tools’ to engineer chromatin topology will be useful in the future to: (1) facilitate loop activation and reversibility on rapid time scales, (2) enable the previously unachievable ability to oscillate spatial contacts, and (3) overcome signal-to-noise issues in population-based genomics assays by synchronizing chromatin topology across a large population of cells in response to blue light illumination. Although the looping strength achieved in this first variation of LADL was modest, we can further optimize looping strength in the future by adjusting light intensity, gRNA numbers, Cry2 bridge size, or other light-inducible dimerization systems. At the single locus investigated here, we see that a 2–2.5 fold increase in loop strength correlates with a modest 1.2–1.3-fold increase in gene expression. It will be important to use LADL and other 3D genome engineering tools in the future to obtain a truly quantitative understanding of the relationship between loop strength, enhancer strength, and gene expression levels. We also see opportunities to use LADL to form possible phase separated nuclear bodies or hubs of multi-way chromatin contacts. Should 3-D genome engineering prove useful for correcting chromatin misfolding patterns in disease, LADL will open up future opportunities for spatial targeting of specific cell types in vivo for dynamic looping and control of gene expression on short time scales.
Supplementary Material
Acknowledgements
We thank members of the Cremins lab for helpful discussions. Jennifer E. Phillips-Cremins is a New York Stem Cell Foundation – Robertson Investigator and an Alfred P. Sloan Foundation Fellow. This research was supported by The New York Stem Cell Foundation (J.E.P.C), the Alfred P. Sloan Foundation (J.E.P.C), the NIH Director’s New Innovator Award from the National Institute of Mental Health (1DP2MH11024701; J.E.P.C), a 4D Nucleome Common Fund grant (1U01HL12999801; J.E.P.C), a joint NSF-NIGMS grant to support research at the interface of the biological and mathematical sciences (1562665; J.E.P.C), and a National Science Foundation Graduate Research Fellowship under Grant No. DGE-1321851 (J.A.B.).
Footnotes
Draft Competing interests
The authors declare no competing interests.
Reference
- 1.Phanstiel DH et al. Static and Dynamic DNA Loops form AP-1-Bound Activation Hubs during Macrophage Development. Mol Cell 67, 1037–1048 e1036 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rao SS et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan SL et al. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nature Communications 8, 15993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hao N, Shearwin KE & Dodd IB Programmable DNA looping using engineered bivalent dCas9 complexes. Nature Communications 8, 1628 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng W et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149, 1233–1244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng W et al. Reactivation of Developmentally Silenced Globin Genes by Forced Chromatin Looping. Cell 158, 849–860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu H et al. Photoexcited CRY2 Interacts with CIB1 to Regulate Transcription and Floral Initiation in Arabidopsis. 322, 1535–1539 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS & Schaffer DV Optogenetic protein clustering and signaling activation in mammalian cells. Nature Methods 10, 249 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Kennedy MJ et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods 7, 973–975 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konermann S et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozkan-Dagliyan I et al. Formation of Arabidopsis Cryptochrome 2 Photobodies in Mammalian Nuclei: APPLICATION AS AN OPTOGENETIC DNA DAMAGE CHECKPOINT SWITCH. 288, 23244–23251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Che DL, Duan L, Zhang K & Cui B The Dual Characteristics of Light-Induced Cryptochrome 2, Homo-oligomerization and Heterodimerization, for Optogenetic Manipulation in Mammalian Cells. ACS Synth Biol 4, 1124–1135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polstein LR & Gersbach CA A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol 11, 198–200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beagan JA et al. Local Genome Topology Can Exhibit an Incompletely Rewired 3D-Folding State during Somatic Cell Reprogramming. Cell Stem Cell 18, 611–624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beagan JA et al. YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dostie J et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res 16, 1299–1309 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips-Cremins JE et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153, 1281–1295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JH et al. 5C-ID: Increased resolution Chromosome-Conformation-Capture-Carbon-Copy with in situ 3C and double alternating primer design. Methods 142, 39–46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.