Abstract
Ligand-dependent Cre recombinases are pivotal tools for the generation of inducible somatic mutants. This method enables spatial and temporal control of gene activity through tamoxifen administration, providing new avenues for studying gene function and establishing animal models of human diseases. While this paved the way for developmental studies previously deemed impractical, the generation of tissue-specific transgenic mouse lines can be time-consuming and costly. Herein, we design a ‘smart’, biocompatible, and biodegradable nanoparticle system encapsulated with tamoxifen that is actively targeted to specific cell types in vivo through surface conjugation of antibodies. We demonstrate that these nanoparticles bind to cells of interest and activate Cre recombinase, resulting in tissue-specific Cre activation. This system provides a versatile, yet powerful approach to induce recombination in a ubiquitious Cre system for various biomedical applications and sets the stage for a time- and cost-effective strategy of generating new transgenic mouse lines.
Keywords: PLGA nanoparticles, 4-hydroxytamoxifen, Cre-mediated recombination, antibody surface conjugation, clonal analysis
INTRODUCTION
Within the past 50 years, nanoparticle (NP)-based carrier systems have emerged as promising vehicles for controlled drug delivery1,2. The carriers are designed to protect drugs from degradation, have a large surface to volume ratio, enhance tissue penetration, and importantly, control drug release. These systems were originally in the form of liposomes but the use of polymeric materials, particularly poly(lactic-co-glycolic acid) (PLGA) gained popularity in the early 1990s3. The appeal of PLGA polymers as a building-block for NP fabrication resides in their desirable properties, including: (1) biocompatibility and biodegradability, (2) FDA approval for parenteral delivery, (3) capability of fine-tuning drug release profiles, (4) protection of drug from degradation, and (5) ability of surface modifications to enhance stability or targeting2,4–6.
A strategy for improved targeting of NPs is through surface modification with ligands that recognize specific tissue types7,8. These targeting ligands play a pivotal role in navigating the NPs to desired cell types, where they can be recognized by direct ligand-receptor interaction. Examples of surface-bound ligands investigated include other polymers9–12, antibodies13,14, peptides15–17, and sugars18,19. Although surface modification with various ligands have been widely studied, only a few of these systems have surpassed research-bench testing to reach clinical development, with the majority in cancer medicine and imaging20. Major factors limiting the advancement of targeted NPs include an incomplete understanding of the fate of the NPs after delivery into the body, inability to load and deliver therapeutic levels of the intended drug, residual off-target effects, and the inefficiency of mass fabrication.
In this paper, we develop actively-targeted PLGA NPs that incorporates the CreER/LoxP system as a tool for tracing NP uptake and drug delivery at a cellular level. CreER/LoxP is an inducible, site-specific recombinase system comprised of the Cre recombinase enzyme fused to the estrogen receptor. This system allows for temporal control of gene expression through delivery of 4-hydroxytamoxifen (OH-Tam) to transgenic mice harboring the CreER gene. Through surface conjugation with antibodies, we provide qualitative and quantitative evidence of drug targeting at a cellular resolution using OH-Tam-loaded PLGA NPs administered to a mouse model harboring the CreER/LoxP reporter. This combined system provides a high-resolution approach to preferentially target cells of interest and for analyzing targeting efficiency. Tamoxifen-loading of targeted PLGA NPs provides an initial step for the development of a versatile, cost-effective, and timely methodology of generating transgenic mouse lines, potentially changing the current established approaches of the CreER/LoxP system.
METHODS
Generation of Transgenic Mouse Lines.
Rosa26CreER;tdTflox/+ mouse lines were obtained by crossing Rosa26-CreER mice with lineage reporter R26R-tdTomato mice. Rosa26CreER;R26VT2/GK3 mouse lines were obtained by crossing Rosa26-CreER mice with the multicolor reporter R26-”Rainbow” mice. Male and female mice ages 6–8 months were used for in vivo experiments unless otherwise specified. All animal studies were performed according to the guidelines of UCLA’s animal care and use committee and the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Studies performed are in accordance with humane treatment of the animals.
OH-Tam-loaded PLGA NP Fabrication.
NPs encapsulating OH-Tam were created using an emulsion-solvent evaporation technique. The “oil” phase of the emulsion was prepared by mixing 50 mg of PLGA and 4mg of 4-hydroxytamoxifenin 3ml of dichloromethane for 2 hours. 2 ml of this solution was then slowly poured into 8 ml of cold 1% polyvinyl alcohol (PVA) (in ultrapure water) solution. This solution was then sonicated on ice using a Fisher Scientific Model 500 ultrasonic dismembrator, with a microtip probe at 30% output for 120 seconds. The emulsion was stirred overnight (minimum of 12 hours) with a magnetic stir-bar at 700 rpm to allow the organic solvent to evaporate. The resulting NPs were washed of the PVA surfactant by 2 successive rinses in ddH20 (solution centrifugation at 14.5k rpm for 8 minutes followed by sonication of the pellet at 10% output for 10 seconds). With the final rinse, the NPs were resuspended in ddH20 at the desired concentration, frozen at -80°C for 24 hours, and lyophilized for 48 hours with the FreeZone 4.5 (Labconco, Kansas City, MO, United States).
Serum Detection of OH-Tam.
Serum quantification of OH-Tam was performed using a capillary high-performance liquid chromatography/tandem mass spectrometry (HPLC-MS) method. NPs loaded with OH-Tam were delivered via lateral tail vein and allowed to circulate for 1, 12, or 60 hours. Serum was collected and prepared for HPLC analysis. The area under the curve of the peak at a retention time of 2.37 minutes was quantified, and the concentration of OH-Tam was calculated using a generated standard curve.
Size and Surface Zeta-Potential Measurements.
To determine nanoparticle size and surface zeta-potential, lyophilized samples were suspended in PBS and dispensed into a cuvette (Malvern DTS0012) and inserted into a Zetasizer Nano ZS (Malvern, Worchestershire, UK) where the zeta-size, polydispersity index, and zeta-potential of triplicate samples was determined. Numerical results are presented as a zeta-average size or a zeta-potential ± standard deviation.
In vitro Cre-Mediated Recombination Studies.
Induction of CreER-mediated recombination was studied in vitro in dermal fibroblasts from both the Rosa26CreER;tdTflox/+ and the Rosa26CreER;R26VT2/GK3 transgenic mice models. Briefly, the ear biopsies from mice from each transgenic model were performed, cut into small pieces, and digested with Liberase Blendzyme TH and TM in Medium 199 plus DNase I and polaxamer at 37°C for 1 hour. Cells were passed through a 70 μm cell strainer and centrifuged. The cells were resuspended in culture media (Dubecco’s Modified Eagle Medium with 10% FBS and 1% Penicillin/Streptomycin) and plated on 2-well chamber slides. After the cells were cultured to approximately 60–70% confluence, PLGA(50:50) nanoparticles (containing OH-Tam) were suspended in culture media at 100 μg/ml, and the nanoparticle/culture media suspension was incubated with the cells for 24 hours. At the end of the 24-hour incubation, the media was aspirated from the chamber, and the cells were fixed by first washing with cold PBS × 3, followed by cold 4% paraformaldehyde for 15 minutes, followed by repeat cold PBS rinse × 3. VectaShield DAPI mounting medium was then placed on each slide, and the slide was sealed with a coverslip. Imaging was performed under fluorescence microscopy (Leica Microsystems, Wetzlar, Germany), with 5 20x images recorded for each experimental condition. Quantification of cells in each image was performed using ImageJ image processing and analysis software (National Institutes of Health). Numerical results are presented as a percentage of positive to total cells in field of view ± standard deviation. To examine the extent of Cre recombination in response to increasing dose of PLGA nanoparticles containing OH-Tam, dermal fibroblasts isolated from Rosa26CreER;tdTflox/+ were treated with 1, 2, 5, and 8 μg of nanoparticles and fluorescence activated cell sorting (FACS) was performed after 24 hours to determine the percent of tdTomato+ fibroblasts.
In vivo Biodistribution Studies.
The in vivo biodistribution of OH-Tam-loaded NPs was studied by intravenous injection of generated NPs into Rosa26CreER;tdTflox/+ or Rosa26CreER;R26VT2/GK3 mice. Lyophilized NPs containing OH-Tam were suspended in PBS at a concentration of 1 mg/ml. 1mg (200 μg for 5 consecutive days) of nanoparticles were injected into the lateral tail vein of each mouse model, allowed to circulate for a total of 6 days (counting from the first injection). The mice were then euthanized by cervical dislocation after anesthesia with 5% isoflurane. For each mouse, the heart, one lung, one kidney, the liver, and the spleen were harvested. The organs were then fixed in 4% paraformaldehyde overnight at 4°C, followed by dehydration with sucrose solution for 24 hours. The organs were then mounted in OCT embedding compound, and frozen at -80°C for 48 hours. Sections of 6–8μm thick tissue were then cut with a cryostat and mounted on standard histological slides. Imaging was then performed under fluorescence microscopy (AF6000LX, Leica Microsystems, Wetzlar, Germany) as well as confocal microscopy (TCS SP5-STED, Leica Microsystems, Wetzlar, Germany).
Antibody Conjugation onto NP Surface.
NPs were “activated” for antibody conjugation by suspension for 30 min in 2-(N-morpholino) ethanesulfonic acid buffer (2 mg/ml) containing 1:1 ratio of EDC and sulfo-NHS. The solution was centrifuged and rinsed with phosphate buffered saline to remove excess reagents. 250 ug of anti-CD31 antibody (BD Biosciences, San Jose, CA, United States) was added to 1 mg of the “activated” NPs and incubated for 6 hours. Excess antibody was removed by PBS washes. NPs were suspended at a stock concentration of 10 mg/ml in 0.22um-sterile filtered PBS for tail vein delivery.
FACS Analysis of CD11b-NPs.
Targeting of bloodstream monocytes and neutrophils by anti-CD11b OH-Tam-loaded NPs was studied in the Rosa26CreER;tdTflox/+ transgenic mouse model. Briefly, anti-CD11b-conjugated NPs were prepared as described above. As a control, OH-Tam-loaded NPs that did not undergo any antibody conjugation were also prepared. For each of these conditions, 900 μg of conjugated NPs (300 μg for 3 consecutive days) were injected into the lateral tail vein of Rosa26CreER;tdTflox/+ transgenic mice. Triplicates were performed of each condition. After the fourth day, the mice were injected with heparin, and blood from each mouse was collected through retro-orbital capillary collection. For each mouse, 500 μL of blood was mixed with 10 mL RBC lysing buffer at room temperature for 5 minutes. The mixture was then centrifuged at 300g for 5 minutes, the aspirate removed, and the cell pellet gently resuspended and washed with cold PBS. This was again centrifuged at 300g for 5 minutes. The pellet was resuspended in 200 μL FACS buffer containing 1:50 AlexaFluor 647-conjugated anti-CD11b Ab. The antibody was allowed to incubate for 30 minutes at room temperature in the dark, after which excess antibody was washed off by adding 3ml of PBS and centrifuging at 300g for 3 minutes. The cells were resuspended in FACS buffer for analysis. Samples were acquired on a FACSAria Cell Sorter (BD Biosciences, San Jose, CA, United States), and data were analyzed with FlowJo software. Numerical results are presented as a percentage of tdTomato positive cells to CD11b positive cells.
Loading of Bioactive Molecules.
PLGA NPs loaded with OH-Tam and pMAXGFP were generated as follows. pMAXGFP was obtained commercially (Lonza) and vortexed with 0.4 mg of mannitol and 2 mg of NH4HCO3 until a clear solution forms (plasmid solution). Then 25 mg of PLGA and 2.5 mg of polyethyleinimine was dissolved along with 2 mg OH-Tam in 0.25 ml of dichloromethane (PLGA solution). The plasmid and PLGA solutions were sonicated using a tip sonicator for 60s. The resulting emulsion was mixed with 2.5 ml of 2% w/v of PVA aqueous solution for 120s to form a second emulsion. The mixture was magnetically stirred overnight at room temperature. NPs were collected by centrifugation at 13.5 krpm for 15 min and washed three times with deionized water to removed excess surfactant. 1% mannitol solution was added to disperse the NPs and the resulting solution lyophilized to generate porous PLGA NPs containing OH-Tam and pMAXGFP.
Clonal Analysis using the Rainbow System.
Rosa26CreER;R26VT2/GK3 neonates two days old were injected with 10 μg of CD31-conjugated OH-Tam NPs. Twenty-eight days post injection, hearts were harvested for clonal expansion analysis.
Statistical Analysis.
Student’s unpaired t-test and one-way ANOVA were used for statistical analyses. All data are presented as mean ± SEM. A probability value p ≤ 0.05 was considered statistically significant. All analyses were performed with GraphPad Prism 5.04 (San Diego, CA, United States).
RESULTS
Tamoxifen loading allows for high-resolution detection of NP uptake and drug delivery in an inducible CreER mouse model
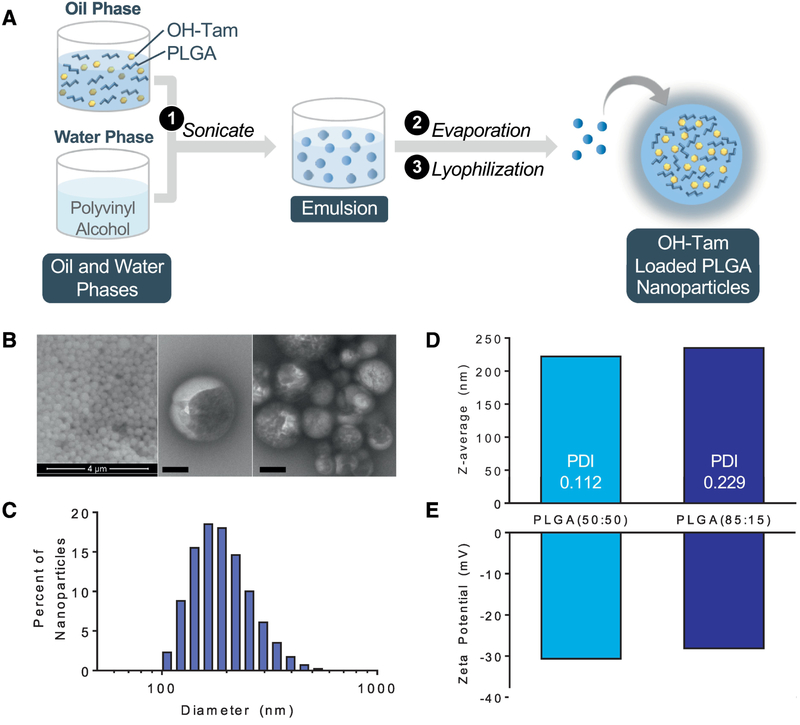
PLGA NPs were fabricated using an emulsion-evaporation technique (Figure 1A). Briefly, OHTam and PLGA were dissolved in dichloromethane and sonicated with a solution containing polyvinyl alcohol. OH-Tam loaded nanoparticles were extracted via evaporation and lyophilization. Scanning and transmission electron microscopy showed consistency of size and spherical shape (Figure 1B) and zetasizer analysis demonstrated particles ranging from 220–230 nm in diameter (Figure 1C, D). Surface zeta potential of the NPs was approximately -30mV, indicating stability and low risk of aggregation (Figure 1E).
Figure 1. Fabrication and characterization of of 4-hydroxytamoxifen loaded PLGA NPs.
(A) Schematic of NP generation using the emulsion-evaporation technique. (B) Scanning electron microscopy (left), transmission electron microscopy (middle, right) of PLGA NPs, bar = 200 nm. (C) Histogram of NP size distribution. (D) Average particle size, polydispersity, and (E) zeta-potential of OH-Tam-loaded PLGA(50:50) and PLGA(85:15) NPs.
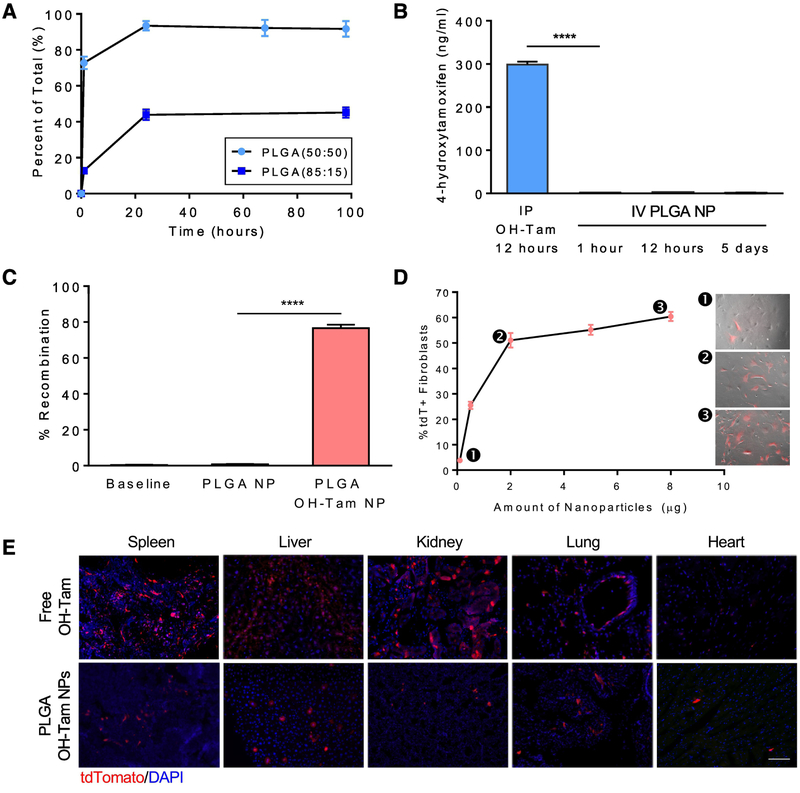
To assess nanoparticle uptake and drug delivery at a cellular level, we used the inducible CreER/LoxP system as a fluorescent readout. We generated a double transgenic mouse model, Rosa26CreER;tdTomatoflox/+, in which the presence of OH-Tam leads to Cre-mediated recombination and permanent labeling of cells with tdTomato (tdT) (Supplementary Figure 1A). Successful delivery of OH-Tam-loaded NPs to this mouse model would lead to the expression of the reporter protein, enabling accurate and sensitive confirmation of drug uptake and release. In vitro drug release profiles of OH-Tam-encapsulated NPs showed an initial burst of OH-Tam release within the first hour, with ~95% of the drug released within 24 hours (Figure 2A). Modulation of lactic:glycolic acid ratio of PLGA to 85:15 decreased the rate of drug release. Measurement of blood OH-Tam levels over the span of 5 days showed minimal drug release while in circulation (Figure 2B). To evaluate the effectiveness of the NPs in inducing Cre-recombination, OH-Tam-loaded NPs were added to cultures of dermal fibroblasts isolated from Rosa26CreER;tdTflox/+ and another independent inducible Cre/LoxP reporter mouse model, Rosa26CreER;R26VT2/GK3. Effective uptake of NPs and subsequent release of OH-Tam within Rosa26CreER;R26VT2/GK3 fibroblasts results in Cre recombinase activity and random expression of one of three fluorescent proteins, mCerulean, mCherry, or mOrange (Supplementary Figure 1B). As expected, tdT was abundantly expressed in Rosa26CreER;tdTflox/+ fibroblasts (Figure 2C and Supplementary Figure 1C) and a mosaic of colors were present in Rosa26CreER;R26VT2/GK3 fibroblasts (Supplementary Figure 1D). NP delivery to Rosa26CreER;tdTflox/+ fibroblasts showed a dose-dependent increase in Cre recombination (Figure 2D), as observed by an increase in the percent of tdT+ fibroblasts with greater amount of tamoxifen-loaded NPs. Additionally, as we increased the amount of OH-Tam in the encapsulated NPs, we observed an increase in tdT expression (Supplementary Figure 1E). We next evaluated whether in vivo delivery of OH-Tam-loaded NPs results in tissue recombination. Intravenous delivery of NPs loaded with OH-Tam showed high recombination rates within the liver and spleen, and only a few cells within the heart, kidney, and lung. The biodistribution profile of OH-Tam NPs is comparable to standard intraperitoneal (IP) injection of free OH-Tam although it appears that IP delivery resulted in more recombination events within the spleen, liver, and kidney compared to the lung and heart. This observation was consistent in both Rosa26CreER;tdTflox/+ (Figure 2E) and Rosa26CreER;R26VT2/GK3 (Supplementary Figure 1F) mouse models.
Figure 2. Cre-mediated recombination using PLGA NPs loaded with OH-Tam.
(A) Elution of OH-Tam from PLGA(50:50) and PLGA(85:15) NPs. (B) Comparison of blood OH-Tam levels between intraperitoneal injection of free OH-Tam and intravenous injection of OH-Tam encapsulated PLGA NPs over the span of 5 days. (C) In vitro quantification of recombination within Rosa26CreER;tdTF/+ fibroblasts upon delivery of PLGA NPs containing OH-Tam. (D) Dose-dependent increase in recombination within Rosa26CreER;tdTF/+ fibroblasts (as indicated by %tdT+ cells) analyzed by flow cytometry. Numbered insets depict merged brightfield and fluorescent images of cells prior to flow cytometry analysis. (E) Comparison of Cre-recombination in Rosa26CreER;tdTF/+ mice between intraperitoneal delivery of free OH-Tam vs tail vein delivery of OH-Tam loaded PLGA NPs, bar = 100 μm. **** p < 0.0001
Antibody surface-conjugation enhances targeted-delivery of NPs
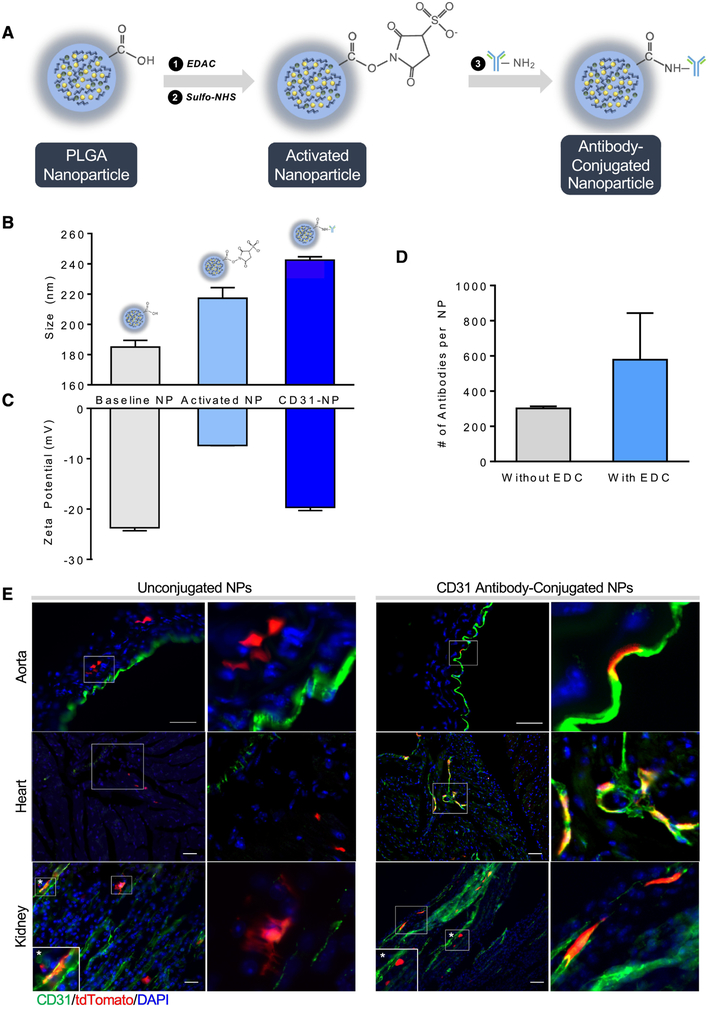
After validation of successful Cre recombination and delivery of biomolecules using PLGA NPs described above, we sought to actively target the NPs to specific cell types through NP surface conjugation of antibodies. The precise targeting of OH-Tam containing NPs to specific cell types that harbor Cre/LoxP transgene can offer an alternative strategy to generate transgenic mice, where NP delivery would lead to tissue specific Cre-mediated recombination. This approach would enhance NP recognition by a subset of cells that express specific antigens capable of recognizing and binding to the conjugated surface antibody. Antibody conjugation was achieved using 1-Ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDAC) linker chemistry (Figure 3A) and characterization of the conjugated NPs showed an increase in size (Figure 3B) and zeta potential (Figure 3C) consistent with antibody addition. The addition of EDAC crosslinkers almost doubled the amount of antibody binding to the surface of the NP compared to normal surface adsorption, thereby increasing efficiency of surface conjugation (Figure 3D).
Figure 3. Antibody conjugation improves targeting of NPs to specific cells.
(A) Schematic of the antibody conjugation process using EDC-NHS chemistry. (B) Size and (C) zeta potential changes before and after antibody conjugation. (D) Quantitation of the number of antibodies conjugated to the NP surface with and without EDC chemistry. (E) Comparison of NP uptake and Cre recombination in the aorta, kidney, and heart between unconjugated and anti-CD31 antibody conjugated NPs loaded with OH-Tam within Rosa26CreER;tdTF/+ mice, bar = 50 μm.Inset shows magnified view of boxed regions.
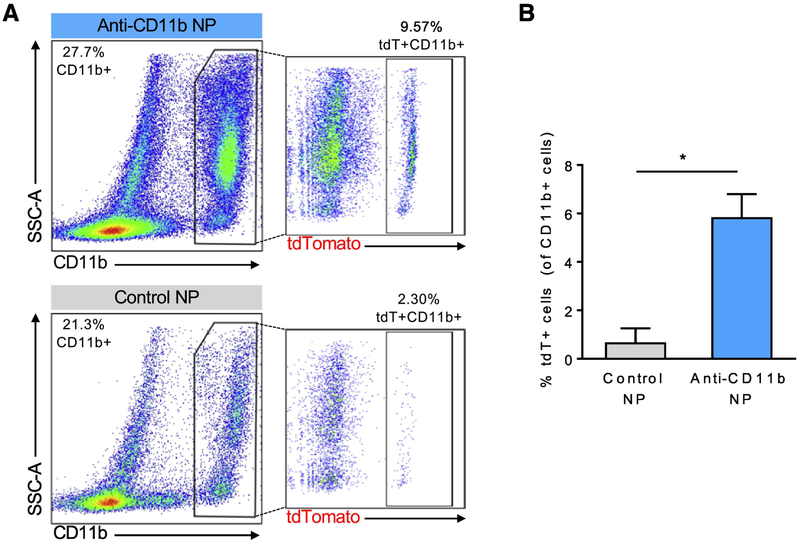
To test the specificity of NP uptake by cells of interest, we generated two different sets of targeted, OH-Tam-loaded NPs for delivery to (i) endothelial cells and (ii) monocytes/macrophages, using antibodies for CD31 and CD11b, respectively. First, we generated and delivered anti-CD31 antibody-conjugated NPs (CD31-NPs) to Rosa26CreER;tdTflox/+ mice. Immunofluorescence imaging showed co-localization of tdT (indicating successful recombination) in vascular endothelial cells (stained green for CD31), confirming specificity of targeting and OH-Tam release (Figure 3E). Compared to unconjugated NPs which resulted in single tdT+ cells randomly distributed within the aortas, kidneys, and hearts of the Rosa26CreER;tdTflox/+ mice, delivery of CD31-NPs led to preferential recombination events in endothelial cells. However, particularly in the kidney, we observed non-specific uptake with CD31-antibody conjugated NPs (starred inset). To further explore and quantify targeting efficiency, we conjugated anti-CD11b antibody to NPs containing OH-Tam as a way to target monocytes and macrophages within the blood. After injection of anti-CD11b-conjugated NPs (CD11b-NPs) loaded with OH-Tam, FACS analysis of blood showed a ~6-fold increase in tdT+ cells within the total CD11b+ population (Figure 4A,B) compared to untargeted PLGA nanoparticles. Again, we observed that while untargeted NPs loaded with OH-Tam resulted in cell labeling indiscriminately, CD11b-NPs led to more selective labeling of CD11b+ cells.
Figure 4. Targeting of monocytes/macrophages with CD11b-conjugated PLGA NPs.
(A)FACS analysis of peripheral blood after intravenous delivery of CD11b-conjugated, OH-Tam loaded PLGA NPs to Rosa26CreER;tdTF/+ mice. (B) Quantification of percent of recombined (tdT+) cells within CD11b+ cells, * p < 0.05.
Combined delivery of OH-Tam and Proteins/Plasmid DNA
To demonstrate the use of OH-Tam as a means of detecting effective NP uptake and drug delivery, we performed proof-of-concept applications with delivery of FITC-albumin protein and a GFP DNA plasmid. We utilized a water-in-oil-in water double emulsion method (Supplemental Figure 2A) to generate these two sets of PLGA NPs. For protein delivery, PLGA and OH-Tam were dissolved together to form the oil phase and an aqueous solution of FITC-BSA comprised the first water phase. After sonication of these two phases to form a primary emulsion, another water phase consisting of PVA was used to generate a secondary emulsion from which the NPs were extracted through evaporation and lyophilization. Delivery of these NPs to cultured dermal fibroblasts from Rosa26CreER;tdTflox/+ showed aggregates of FITC-albumin within recombined cells (Supplemental Figure 2B). These in vitro observations were corroborated by in vivo tail vein administration of the NPs showing only FITC-BSA within recombined cells (Supplemental Figure 2C). These results confirmed NP uptake and concomitant release of both OH-Tam to mediate Cre-recombination and FITC-albumin into the cytoplasm of the same cell.
We achieved similar results with delivery of pMAXGFP (Lonza), a 3.5 kbp plasmid driven by a CMV promoter, which expresses an enhanced green fluorescent protein (eGFP). In vivo delivery of NPs containing OH-Tam and pMAXGFP into Rosa26CreER;tdTflox/+ mice revealed successful uptake of both pMAXGFP and OH-Tam within the same cells, based on GFP and tdTomato co-expression (Supplemental Figure 3D).
Clonal expansion analysis using our targeted PLGA system
In addition to serving as a readout of effective uptake and drug release, another benefit with the loading of OH-Tam into PLGA NPs is its potential application in developmental biology, particularly for clonal expansion analysis. Our developed NPs provide an ideal platform for clonal analysis as we can examine any cell type of interest by modulating the surface antibody and adjusting the extent of recombination through dose-titration of NPs. To test this application, we delivered a limiting amount of CD31-NPs containing OH-Tam to Rosa26CreER;R26VT2/GK3 neonates to induce rare recombination events. When the neonates reached 30 days old, we collected tissues for clonal expansion analysis (Supplemental Figure 3A). In this model, endothelial cell clones are expected to randomly express one of three fluorescent proteins, mCerulean, mCherry, or mOrange. Each clone of cells would be distinguishable by the fluorescent protein they express, and clonal analysis can be achieved by counting the total number of cells from each clone. Recombination was observed in endothelial cells as confirmed with CD31 staining. Clusters of cells were identified and expressed the same reporter protein, indicating clonal expansion of a parental cell that recombined after NP uptake and passed on the genetic labeling to its progeny (Supplemental Figure 3B).
DISCUSSION
Nanoparticles have long been utilized as a platform for targeted and controlled drug delivery. Methods to directly track nanoparticle targeting and distribution rely on imaging techniques such as fluorescence microscopy, optical imaging, or magnetic resonance imaging. However, they rarely address the success of drug delivery itself. The inducible Cre/LoxP system has been a powerful tool in developmental and molecular biology, with temporal and spatial control of gene recombination by administration of tamoxifen and utilization of tissue-specific promoters, respectively. We have combined the precision and versatility of PLGA nanoparticles as a drug delivery platform with the sensitive and robust CreER/LoxP construct to develop a system that accurately reports successful uptake and drug release. This capability is vital for determining efficiency and specificity of drug delivery, which are current limitations with NP-based carriers.
We used PLGA to design our NP system for several reasons, including their biocompatibility, versatility in modulating their physico-chemical properties, and their FDA approval for clinical use. As the constituent building blocks of PLGA are monomers of lactic acid and glycolic acid, its degradation products are therefore physiological molecules naturally processed during the citric acid cycle. The double-emulsion method for NP fabrication allows for the encapsulation of both hydrophilic as well as hydrophobic compounds, increasing its versatility for drug delivery. We validated this strategy by co-delivery of DNA plasmids along with OH-Tam. This approach could be applied to a variety of biological systems, in which a reporter gene will permanently mark cells that successfully received DNA plasmids. Additionally, this method of fabrication allows for fine-tuning of payload encapsulation, enabling adjustment in the amount of OH-Tam or drug delivered in a specified dose of NP. Utilizing this method, we provide proof-of-concept for delivery of both macromolecules as well as plasmid DNA.
An important extension to efficient and specific reporting of drug delivery is the applicability of our system in the generation of tissue-specific transgenic mice for biomedical studies. Alteration of mouse genome by gene-targeting approaches has greatly facilitated studies in developmental and cellular biology. Additionally, transgenic mouse models can be utilized to turn on a reporter gene to track individual cells for lineage tracing. A major technical advance in this field has been the development of models that allow for control of the timing, cell-type, and tissue-specificity of gene activation or repression. However, the generation of tissue-specific CreER/LoxP mouse models is a rigorous process that can be costly and time-consuming. Therefore, the development of a system that combines a generic CreER/LoxP mouse with ‘smart’, OH-Tam-loaded NPs capable of targeting specific cell types would convert this generic mouse into a tissue-specific transgenic mouse. This is advantageous for several reasons. First, it circumvents the need to generate and maintain different transgenic mice with cell-type specific promoters. Second, it allows for the use of a single mouse model, reducing possible genetic and phenotype variability. Finally, by using nanoparticles with several different ligands, it may be possible to induce recombination within more than one cell type of interest. We have shown that by utilizing an actively-targeted approach via surface conjugation of antibodies, we can improve delivery of biomolecules to specific cells within the body. Our study sets the stage for a timely, cost-effective, and novel application of NPs in the potential generation of transgenic mouse models. We also envision their applicability in improving CRISPR/Cas9 technology for targeted delivery of Cas9 mRNA, guide RNA, or donor DNA to induce gene editing in specific cell types.
However, there are limitations with our current system that need to be addressed before it can be fully applied to biological systems. First, targeting to specific cell types was not achieved at a high efficiency, since there is non-specific uptake of nanoparticles, especially within the reticuloendothelial system. We observed recombination events in the liver and spleen resulting from high trafficking of the nanoparticles within these organs. Second, it is well established that there is rapid protein adsorption onto surfaces of circulating nanomaterials, known as the protein corona21–23. The coating of nanoparticles with circulating proteins and peptides may adversely affect the targeting of the particles to the cells of interest and may modify NP size, surface charge, surface composition, and functionality24. And finally, in order to utilize this system to generate tissue-specific transgenic mice, surface antibodies to the tissue of interest must be available for nanoparticle surface modification. While further studies are needed to refine the targeting efficiency, particularly within solid organs, the versatility and sensitivity of our targeted PLGA NP system provides it with the potential to significantly improve the cost and time of biomedical research. Together, our results demonstrate that NP surface antibody conjugation improves recognition and uptake by cells that express specific ligands to the surface receptor. In combination with OH-Tam loading, this system holds promise for future application of inducing Cre-recombination in specific cell types to generate transgenic models and perform developmental studies such as clonal expansion analysis.
Supplementary Material
Supplemental Figure 1. Induction of Cre recombination using OH-Tam-loaded PLGA NPs. Schematic of the inducible Cre-LoxP technology in (A) Rosa26CreER;tdTF/+ and (B) Rosa26CreER;R26VT2/GK3 mice. Fluorescence microscopy of dermal fibroblasts isolated from(C) Rosa26CreER;tdTF/+ and (D) Rosa26CreER;R26VT2/GK3 mice cultured with OH-Tam loaded NPs showing efficient recombination and expression of reporter proteins. (E) Dermal fibroblasts from Rosa26CreER;tdTF/+ mice treated with NPs loaded with varying percentage (w/w) of OH-Tam compared to free OH-Tam treatment. (F) Comparison of recombination in various organs of Rosa26CreER;R26VT2/GK3 mice receiving either intraperitoneal injection of free OH-Tam or intravenous administration of OH-Tam encapsulated PLGA NPs.
Supplemental Figure 2. Combined delivery of OH-Tam and Proteins/Plasmid DNA. (A) Schematic of FITC-albumin NP fabrication. (B) Mouse dermal fibroblasts isolated from Rosa26CreER;tdTF/+ mice treated with NPs loaded with OH-Tam and FITC-albumin showing aggregates of FITC-albumin within recombined cells, bar = 100 um. (C) In vivo delivery of OH Tam and FITC-BSA encapsulated PLGA NPs in Rosa26CreER;tdTF/+ mice. Images show recombination in hepatocytes containing FITC-albumin aggregates, bar = 5 μm. (D) Tail vein delivery of PLGA NPs containing 4-OHT and pMAXGFP in Rosa26CreER;tdTF/+ mice. Liver section showing recombined hepatocytes expressing both tdT (red) and GFP (green), bar = 50 μm.
Supplemental Figure 3. Clonal expansion analysis within Rainbow hearts. (A) Schematic of NP delivery. Neonatal day 2 pups were injected intraperitoneally with CD31-NPs loaded with OH-Tam and hearts harvested 3 weeks post-injection for clonal analysis. (B) Cell clones (red) within Rosa26CreER;R26VT2/GK3 neonatal hearts identified as CD31+ from immunohistochemistry (green), bar = 50 μm.
SOURCES OF FUNDING
This work was supported in part by grants from the National Institutes of Health (NIH) DP2 HL127728 (R.A.), California Institute of Regenerative Medicine (CIRM) New Faculty Physician Scientist Award (RN3–06378) (R.A.), Eli & Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA Research Award (R.A.), and the Sarnoff Cardiovascular Research Foundation (N.B.N.).
Abbreviations:
- FACS
fluorescence activated cell sorting
- HPLC-MS
high-performance liquid chromatography/tandem mass spectrometry
- NP
nanoparticle
- OH-Tam
4-hydroxytamoxifen
- PLGA
poly(lactic-co-glycolic acid
- PVA
polyvinyl alcohol
- tdT
tandem dimer Tomato
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure: All authors report no disclosures.
REFERENCES
- 1.Acharya S, Sahoo SK. PLGA nanoparticles containing various anticancer agents and tumour delivery by EPR effect. Advanced Drug Delivery Reviews. 2011;63(3):170–183. [DOI] [PubMed] [Google Scholar]
- 2.Peres C, Matos AI, Conniot J, et al. Poly(lactic acid)-based particulate systems are promising tools for immune modulation. Acta Biomaterialia. 2017;48(Supplement C):41–57. [DOI] [PubMed] [Google Scholar]
- 3.Kamaly N, Xiao Z, Valencia PM, Radovic-Moreno AF, Farokhzad OC. Targeted polymeric therapeutic nanoparticles: design, development and clinical translation. Chemical Society Reviews. 2012;41(7):2971–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Locatelli E, Comes-Franchini M. Biodegradable PLGA-b-PEG polymeric nanoparticles: Synthesis, properties, and nanomedical applications as drug delivery system. Vol 142012. [Google Scholar]
- 5.Ratzinger G, Agrawal P, Körner W, et al. Surface modification of PLGA nanospheres with Gd-DTPA and Gd-DOTA for high-relaxivity MRI contrast agents. Biomaterials. 2010;31(33):8716–8723. [DOI] [PubMed] [Google Scholar]
- 6.Panyam J, Zhou W-Z, Prabha S, Sahoo SK, Labhasetwar V. Rapid endo-lysosomal escape of poly(dl-lactide-co-glycolide) nanoparticles: implications for drug and gene delivery. The FASEB Journal. 2002;16(10):1217–1226. [DOI] [PubMed] [Google Scholar]
- 7.Kocbek P, Obermajer N, Cegnar M, Kos J, Kristl J. Targeting cancer cells using PLGA nanoparticles surface modified with monoclonal antibody. Journal of Controlled Release. 2007;120(1):18–26. [DOI] [PubMed] [Google Scholar]
- 8.Hans ML, Lowman AM. Biodegradable nanoparticles for drug delivery and targeting. Current Opinion in Solid State and Materials Science. 2002;6(4):319–327. [Google Scholar]
- 9.Gref R, Minamitake Y, Peracchia MT, Trubetskoy V, Torchilin V, Langer R. Biodegradable long-circulating polymeric nanospheres. Science. 1994;263(5153):1600. [DOI] [PubMed] [Google Scholar]
- 10.Li Y-P, Pei Y-Y, Zhang X-Y, et al. PEGylated PLGA nanoparticles as protein carriers: synthesis, preparation and biodistribution in rats. Journal of Controlled Release. 2001;71(2):203–211. [DOI] [PubMed] [Google Scholar]
- 11.Avgoustakis K, Beletsi A, Panagi Z, et al. Effect of copolymer composition on the physicochemical characteristics, in vitro stability, and biodistribution of PLGA–mPEG nanoparticles. International Journal of Pharmaceutics. 2003;259(1):115–127. [DOI] [PubMed] [Google Scholar]
- 12.F Zambaux M, Bonneaux F, Gref R, Dellacherie E, Vigneron C. MPEO-PLA nanoparticles: Effect of MPEO content on some of their surface properties. Vol 441999. [DOI] [PubMed] [Google Scholar]
- 13.Karra N, Nassar T, Ripin A, Schwob O, Borlak J, Benita S. Antibody Conjugated PLGA Nanoparticles for Targeted Delivery of Paclitaxel Palmitate: Efficacy and Biofate in a Lung Cancer Mouse Model. Small. 2013(9):4221–4236. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Lee C-H, Park J, et al. Antibody conjugated magnetic PLGA nanoparticles for diagnosis and treatment of breast cancer. Journal of Materials Chemistry. 2007;17(26):2695–2699. [Google Scholar]
- 15.Zhang N, Chittapuso C, Ampassavate C, Siahaan TJ, Berkland C. PLGA Nanoparticle-Peptide Conjugate Effectively Targets Intercellular Cell-Adhesion Molecule-1. Bioconjugate chemistry. 2008;19(1):145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasconcelos A, Vega E, Pérez Y, Gómara MJ, García ML, Haro I. Conjugation of cell-penetrating peptides with poly(lactic-co-glycolic acid)-polyethylene glycol nanoparticles improves ocular drug delivery. International Journal of Nanomedicine. 2015;10:609–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathew A, Fukuda T, Nagaoka Y, et al. Curcumin Loaded-PLGA Nanoparticles Conjugated with Tet-1 Peptide for Potential Use in Alzheimer’s Disease. PLoS ONE. 2012;7(3):e32616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain SK, Gupta Y, Ramalingam L, et al. Lactose-Conjugated PLGA Nanoparticles for Enhanced Delivery of Rifampicin to the Lung for Effective Treatment of Pulmonary Tuberculosis. PDA Journal of Pharmaceutical Science and Technology. 2010;64(3):278–287. [PubMed] [Google Scholar]
- 19.Yin Y, Chen D, Qiao M, Lu Z, Hu H. Preparation and evaluation of lectin-conjugated PLGA nanoparticles for oral delivery of thymopentin. Journal of Controlled Release. 2006;116(3):337–345. [DOI] [PubMed] [Google Scholar]
- 20.Anselmo AC, Mitragotri S. Nanoparticles in the clinic. Bioengineering & Translational Medicine. 2016;1(1):10–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch I, Dawson KA. Protein-nanoparticle interactions. Nano Today. 2008;3(1):40–47. [Google Scholar]
- 22.Cedervall T, Lynch I, Lindman S, et al. Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proceedings of the National Academy of Sciences. 2007;104(7):2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monopoli MP, Åberg C, Salvati A, Dawson KA. Biomolecular coronas provide the biological identity of nanosized materials. Nature Nanotechnology. 2012;7:779. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen VH, Lee B-J. Protein corona: a new approach for nanomedicine design. International Journal of Nanomedicine. 2017;12:3137–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Induction of Cre recombination using OH-Tam-loaded PLGA NPs. Schematic of the inducible Cre-LoxP technology in (A) Rosa26CreER;tdTF/+ and (B) Rosa26CreER;R26VT2/GK3 mice. Fluorescence microscopy of dermal fibroblasts isolated from(C) Rosa26CreER;tdTF/+ and (D) Rosa26CreER;R26VT2/GK3 mice cultured with OH-Tam loaded NPs showing efficient recombination and expression of reporter proteins. (E) Dermal fibroblasts from Rosa26CreER;tdTF/+ mice treated with NPs loaded with varying percentage (w/w) of OH-Tam compared to free OH-Tam treatment. (F) Comparison of recombination in various organs of Rosa26CreER;R26VT2/GK3 mice receiving either intraperitoneal injection of free OH-Tam or intravenous administration of OH-Tam encapsulated PLGA NPs.
Supplemental Figure 2. Combined delivery of OH-Tam and Proteins/Plasmid DNA. (A) Schematic of FITC-albumin NP fabrication. (B) Mouse dermal fibroblasts isolated from Rosa26CreER;tdTF/+ mice treated with NPs loaded with OH-Tam and FITC-albumin showing aggregates of FITC-albumin within recombined cells, bar = 100 um. (C) In vivo delivery of OH Tam and FITC-BSA encapsulated PLGA NPs in Rosa26CreER;tdTF/+ mice. Images show recombination in hepatocytes containing FITC-albumin aggregates, bar = 5 μm. (D) Tail vein delivery of PLGA NPs containing 4-OHT and pMAXGFP in Rosa26CreER;tdTF/+ mice. Liver section showing recombined hepatocytes expressing both tdT (red) and GFP (green), bar = 50 μm.
Supplemental Figure 3. Clonal expansion analysis within Rainbow hearts. (A) Schematic of NP delivery. Neonatal day 2 pups were injected intraperitoneally with CD31-NPs loaded with OH-Tam and hearts harvested 3 weeks post-injection for clonal analysis. (B) Cell clones (red) within Rosa26CreER;R26VT2/GK3 neonatal hearts identified as CD31+ from immunohistochemistry (green), bar = 50 μm.