Abstract
Mangroves invade some very marginal habitats for woody plants—at the interface between land and sea. Since mangroves anchor tropical coastal communities globally, their origin, diversification and adaptation are of scientific significance, particularly at a time of global climate change. In this study, a combination of single-molecule long reads and the more conventional short reads are generated from Rhizophora apiculata for the de novo assembly of its genome to a near chromosome level. The longest scaffold, N50 and N90 for the R. apiculata genome, are 13.3 Mb, 5.4 Mb and 1.0 Mb, respectively. Short reads for the genomes and transcriptomes of eight related species are also generated. We find that the ancestor of Rhizophoreae experienced a whole-genome duplication ~70 Myrs ago, which is followed rather quickly by colonization and species diversification. Mangroves exhibit pan-exome modifications of amino acid (AA) usage as well as unusual AA substitutions among closely related species. The usage and substitution of AAs, unique among plants surveyed, is correlated with the rapid evolution of proteins in mangroves. A small subset of these substitutions is associated with mangroves’ highly specialized traits (vivipary and red bark) thought to be adaptive in the intertidal habitats. Despite the many adaptive features, mangroves are among the least genetically diverse plants, likely the result of continual habitat turnovers caused by repeated rises and falls of sea level in the geologically recent past. Mangrove genomes thus inform about their past evolutionary success as well as portend a possibly difficult future.
Keywords: mangrove, whole-genome sequencing, adaptive evolution, protein evolution, genetic diversity, sea-level changes
INTRODUCTION
One of the most productive and diverse environments for many life forms is at the interface between land and sea. Woody plants, however, are an exception to this species richness in intertidal zones. Globally, no more than 80 tree species have succeeded in invading intertidal zones to become mangroves, compared to over 10 000 that are found at the land–water interface in non-saline systems. (The term ‘mangrove’ refers to many independently evolved lineages of woody plants that occupy these land/saltwater interfaces.) Remarkably, the small number of mangrove species anchors tropical intertidal communities globally by providing key ecological services that include carbon sequestration [1], sediment accretion, seashore protection and ecosystem productivity [2].
How mangroves became adapted to the intertidal environments is thus a most interesting question. Mangroves differ from other plants living in hypersaline habitats [3,4] because their environments are stressful in multiple dimensions including high salinity, hypoxia, strong UV light and anaerobic soils [5]. All these stresses fluctuate daily as the tides ebb and flow. In the adapation, mangroves have evolved specialized traits that include viviparous embryonic development, aerial roots and high tannin content [2,6,7]. While there have been some attempts at understanding the molecular basis of these adaptations [8–11], a systematic investigation is hindered by the absence of genomic data in any mangrove species. This absence will be remedied in this study.
In this study, we sequenced the genomes and/or transcriptomes of nine species using the latest sequencing technology supplemented by the more conventional platforms. Among these species, seven are mangroves from the Rhizophoreae tribe, the most mangrove-rich taxon comprising 20 typical mangrove species. The remaining two are inland species most closely related to Rhizophoreae.
There is an urgency at this time for studying mangroves because of the impending sea-level rises. Plants of the tropical coasts are expected to be affected disproportionately and mangroves will likely bear the brunt of these environmental changes. Indeed, there have been several warnings for ‘a world without mangroves’ [12]. The availability of genome sequences may help to reveal the history of mangrove colonization and mechanisms of adaptation to intertidal zones. Perhaps most important of all, the genomic resources may help to spur research on these most interesting adaptations. Research activities in themselves could offer some needed protections for these fragile intertidal ecosystems.
MANGROVE GENOME SEQUENCE AND COMPOSITION
We obtained 16.2 Gb (gigabases) of single-molecule real-time long reads (SMRT; Supplementary Figs 1–3 and Supplementary Table 1, available as Supplementary Data at NSR online) from one mature plant of Rhizophora apiculata for de novo genome assembly. The final assembly contains 142 scaffolds of an aggregate size of 232 Mb, covering 85% of the R. apiculata genome (∼274 Mb; Supplementary Fig. 4, available as Supplementary Data at NSR online). The longest scaffold is 13.3 Mb long, N50 is 5.4 Mb and N90 is 1.0 Mb (Supplementary Table 2, available as Supplementary Data at NSR online). The 16 largest scaffolds cover half and the largest 48 scaffolds cover 90% of the genome. The 48 scaffolds are comparable in number to the Rhizophora chromosome count (2n = 36 [13]). This indicates that our assembly approaches the chromosome-level completeness.
To correct for long-read sequencing errors, we also generated 89.3 Gb of short paired-end reads, which are generally more accurate (Supplementary Note and Supplementary Table 3, available as Supplementary Data at NSR online). Nevertheless, had we used only the Illumina short reads, the N50 and N90 scaffolds would have decreased by 80% in length. Most crucially, we obtained contig N50 of 2.45 Mb using the SMRT assembly, whereas the short pair-end reads yielded an N50 of only 9.7 Kb (Supplementary Table 4, available as Supplementary Data at NSR online).
In order to study the evolution of mangroves that form the Rhizophoreae tribe, we further sequenced the genomes of R. mangle, R. stylosa and R. mucronata at lower depth (Supplementary Fig. 5 and Supplementary Table 5, available as Supplementary Data at NSR online). In the companion studies, we generated transcriptomes of Kandelia obovata, Bruguiera gymnorrhiza and Ceriops tagal, also from the Rhizophoreae tribe, as well as Pellacalyx yunnanesis and Carallia brachiata from the closest non-mangrove genera in the Rhizophoraceae family [14,15] (Supplementary Table 6, available as Supplementary Data at NSR online).
The quality of the assembly is reflected in the following statistics: 96% of the expressed sequences (Supplementary Note, available as Supplementary Data at NSR online) could be mapped to the genome; 93% of the core eukaryotic genes [16] are present and 99% of previously identified R. apiculata genes [17] could be uniquely mapped. Details of the procedures are given in the Supplementary Note and summarized in Supplementary Table 7 (both available as Supplementary Data at NSR online). The statistics indicate that these genomes are of the high quality necessary for advancing to the next stage of global mangrove research.
Using a combination of homology-based search and de novo prediction, we estimate that 29% of the R. apiculata genome consists of repetitive sequences (Supplementary Table 8, available as Supplementary Data at NSR online). The repetitive portions of the genome, comprising predominantly transposable element (TE) families, are drastically reduced in R. apiculata compared to the closely related non-mangrove plants (Lyu et al., unpublished data). By examining the long terminal repeats of many TEs, we conclude that the reduction is due to a lower rate of transposition, rather than a higher rate of TE loss. The underlying mechanisms of TE reductions are similar across independent mangrove lineages of Rhizophora, Avicennia and Sonneratia, the latter two being presented in the companion studies. The lower birth rate of TEs hints that active repression of TE jumping is a common strategy of mangrove genomes (see ‘Adaptation at the whole-genome level’ below). Decrease in TE numbers contributes substantially to the widespread genome-size reduction among true mangroves (Lyu et al., unpublished data).
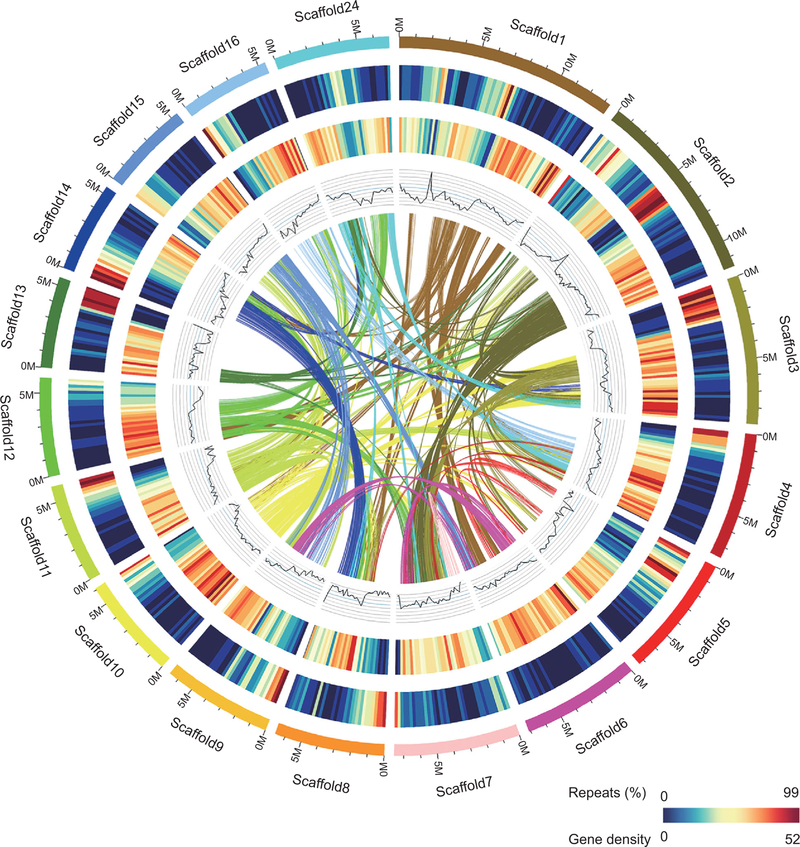
With repetitive elements masked, 26 640 protein-coding genes are predicted in the R. apiculata genome (Supplementary Tables 9 and 10, available as Supplementary Data at NSR online). By searching against public databases, we assigned these protein-coding genes to KEGG Ortholog terms and Gene Ontology terms (Supplementary Figs 6 and 7, and Supplementary Table 11, available as Supplementary Data at NSR online). We also predicted 2955 non-coding RNAs and 1783 transcription factors (Supplementary Tables 10 and 12, available as Supplementary Data at NSR online). A schematic representation of the genome is given in Fig. 1. Combining the R. apiculata genome with three well-annotated genomes of inland plants (Arabidopsis thaliana, Populus trichocarpa and Ricinus communis), we identified 17 806 gene families. Of these, 13 185 are found in R. apiculata and 10 054 families have at least one member from all four genomes (Supplementary Fig. 8, available as Supplementary Data at NSR online).
Figure 1.
Features of the R. apiculata genome. Each linking line in the center of the circle connects a pair of homologous genes. A cluster of such lines indicates a collinear block (see ‘Methods’ for details). Circular tracks present, from inner to outer, GC content (29.47–44.58% per 200 Kb), gene density (0–52 per 200 Kb) and percentage of repeats (0–99% per 200 Kb). The colored bars on the outer track demarcate the 17 scaffolds larger than 5 Mb.
WHOLE-GENOME DUPLICATION AND THE ORIGIN-DIVERSIFICATION OF RHIZOPHOREAE
Whole-genome duplication (WGD) is a feature of many taxa. WGD is, after all, an efficient way of expanding the genome [18]. We wish to know whether WGDs may have happened in Rhizophoreae. In particular, the timing of WGD in relation to geological events that permit the colonization of the intertidal zones has never been explored before.
We used MCScanX [19] (see ‘Methods’ and Supplementary Notes, available as Supplementary Data at NSR online) to align the R. apiculata genome to itself. We define ‘collinear blocks’ as regions of the genome that harbor at least five genes with homologs elsewhere in the genome and in the same order. We identified 377 such blocks that together cover 10 846 protein-coding genes (41% of all genes; Fig. 1). These genes are distributed among 4615 pairs, as well as some triplets/quadruplets, of genes. The extensive collinear blocks indicate at least one WGD event in the past.
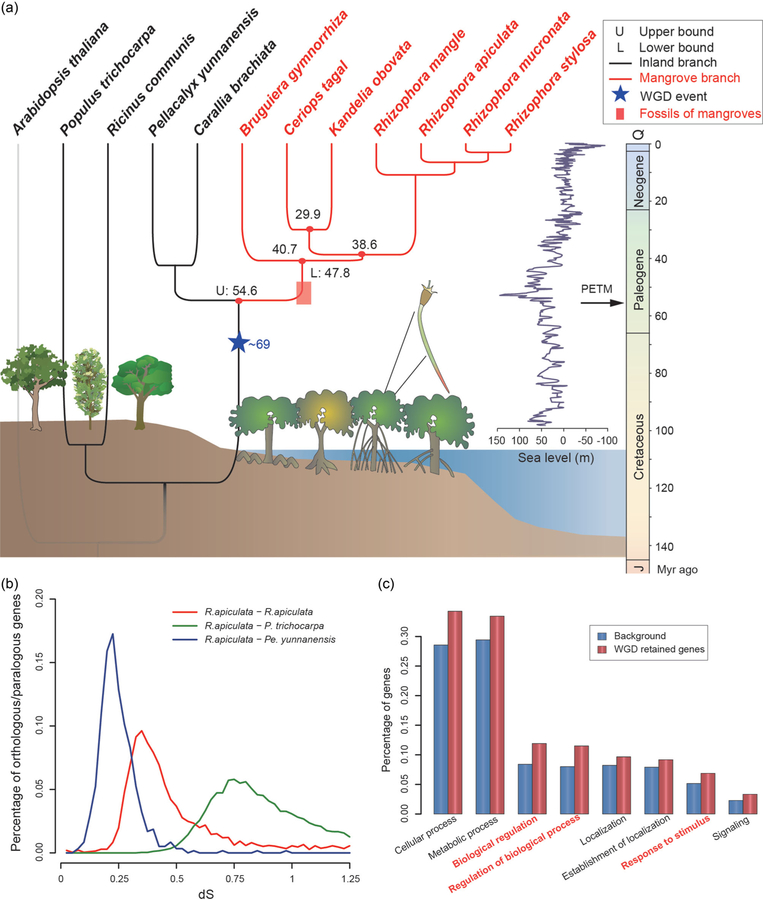
To estimate the timing of WGD, we calculate the synonymous divergence (dS) between paralogous genes of the same genome (Fig. 2b). The distribution of dS between paralogous genes is uni-modal, suggesting a single WGD in the ancestor of R. apiculata. Since the mean dS between paralogs (0.35, red line in Fig. 2b) is larger than that between R. apiculata and Ca. brachiate/Pe. yunnanessis (0.25) but smaller than that between R. apiculata and P. trichocarpa (0.75, green line; Supplementary Fig. 9, available as Supplementary Data at NSR online), the WGD likely happened between these two time points, as indicated by the star in Fig. 2a (see the Supplemental Note, available as Supplementary Data at NSR online, for details). This WGD event was expected and confirmed in related species (Supplementary Fig. 10, available as Supplementary Data at NSR online).
Figure 2.
Dating the emergence of Rhizophoreae mangroves. (a) A phylogeny including Rhizophoreae mangroves (red lines) and their non-mangrove relatives (black lines). Arabidopsis thaliana (grey line) is the distant outgroup. The blue star indicates the time of whole-genome duplication (WGD) and the red box indicates the age of known fossils. The emergence of the Rhizophoreae mangroves is placed between an upper (U) and lower (L) bound as described in the main text. Historical sea-level changes are depicted in blue. Occurrence of the PETM is indicated by the arrow on the timeline. The cartoons of mangrove trees are contributed by Jane Thomas, Kris Beckert, Diana Kleine, Brianne Walsh, Dieter Tracey and Tracey Saxby (IAN Image Library, ian.umces.edu/imagelibrary/). (b) dS distributions between orthologs from pairs of species (blue and green lines) and between paralogs within R. apiculata (red line). (c) Prevalence of Gene Ontology terms among gene duplicates retained after WGD (red bars) compared to control genes that have no paralogs in collinear blocks (blue bars).
What then may be the timing of WGD in relation to mangrove emergence in the phylogeny? Did it occur before or after the origin of Rhizophoreae mangroves? To answer this question, we reconstruct the phylogeny of the 11 mangrove and non-mangrove species (Fig. 2a; Supplementary Table 6, available as Supplementary Data at NSR online). The tree topology was reconstructed using PhyML [20]. Ca. brachiata and Pe. yunnanensis are the non-mangrove members of the same family [21]. The divergence time is estimated by the MCMCTREE program from the PAML package [22,23] (see ‘Methods’) based on three dated events for calibration and confirmation. First, the root node of the common ancestor of Rhizophoraceae, Euphorbiaceae (Ri. communis) and Salicaceae (P. trichocarpa) has been placed in the interval of 105–120 Myr before present [24,25]. Second, a most recent fossil recognized as ancestral Rhizophora has been dated to the late Eocene (33.9–38 Mya) [26,27]. These two time points are used to constrain the estimation of substitution rates (Supplementary Fig. 11, available as Supplementary Data at NSR online). The third dated event is given by fossils of the ancestor of Rhizophoreae from the early Eocene (47.8–56 Myr ago) [27,28], which is used to corroborate the MCMCTREE estimates.
We estimate that the mangrove–non-mangrove divergence happened 54.6 Myr ago, while the most recent common ancestor of Rhizophoreae mangroves is estimated to be 40.7 Myr ago. The interval of 40.7–54.6 Myr corresponds well with the Eocene fossils of 47.8–56 Myr ago. The invasion hence took place in the small window of 47.8–54.6 Myr ago (Fig. 2a; Supplementary Figs 12 and 13, and Supplementary Tables 13 and 14, available as Supplementary Data at NSR online). Extrapolating from these time points, the WGD event is placed at 69 Myr ago (Fig. 2a; Supplementary Fig S14 and Supplementary Note, available as Supplementary Data at NSR online), slightly before the emergence and diversification of Rhizophoreae mangroves.
The invasion of the intertidal zones by Rhizophoreae appears to have coincided with a brief period of extreme global warming referred to as the Paleocene-Eocene Thermal Maximum (PETM) that occurred approximately 55.5 Myr ago [29] (Fig. 2a). Eustatic sea levels rose during PETM, likely submerging the angiosperms living at the margins of rainforests and forcing them to adapt to the new environment. Therefore, the emergence of mangroves may have been aided first by the genetic WGD event and then by suitable ecological conditions during the PETM.
It has been suggested that WGDs played important roles in the origin and diversification of many angiosperms [30]. The connection between genome duplication and evolutionary innovation seems particularly relevant in the emergence of mangroves. In addition to Rhizophoreae, two other major clades, Avicennia and Sonneratia, also experienced independent WGDs before their invasions of the intertidal zones (He et al., unpublished data).
A common pattern after WGD is that one of the two duplicated copies becomes lost shortly afterward [31]. Those genes that retain both copies are therefore of great interest. Across the genome, 2878 pairs are retained when we examined genes with the dS values in the 0.25–0.70 range. Genes with dS outside of this range may have unusual or complicated evolutionary dynamics and are excluded here. The retainers are enriched for ontology terms related to regulation and stress response (Fig. 2c; see also Supplementary Note, available as Supplementary Data at NSR online). The preferential retention of regulatory genes supports the evolutionary model of genome duplication [32], while the retention of stress response genes may pertain to the invasion of the intertidal zones.
ADAPTATION AT THE WHOLE-GENOME LEVEL
Inhabiting highly saline habitats relative to other woody plants, mangroves have to regulate intracellular salt levels to mitigate the effect of the environment [33]. However, daily sea-level fluctuations due to tides prevent an effective regulation. Indeed, it may take more than a week to reach an equilibrium when the salt concentration changes [9,33]. Thus, intracellular proteins have to adapt to an increase in environmental salinity.
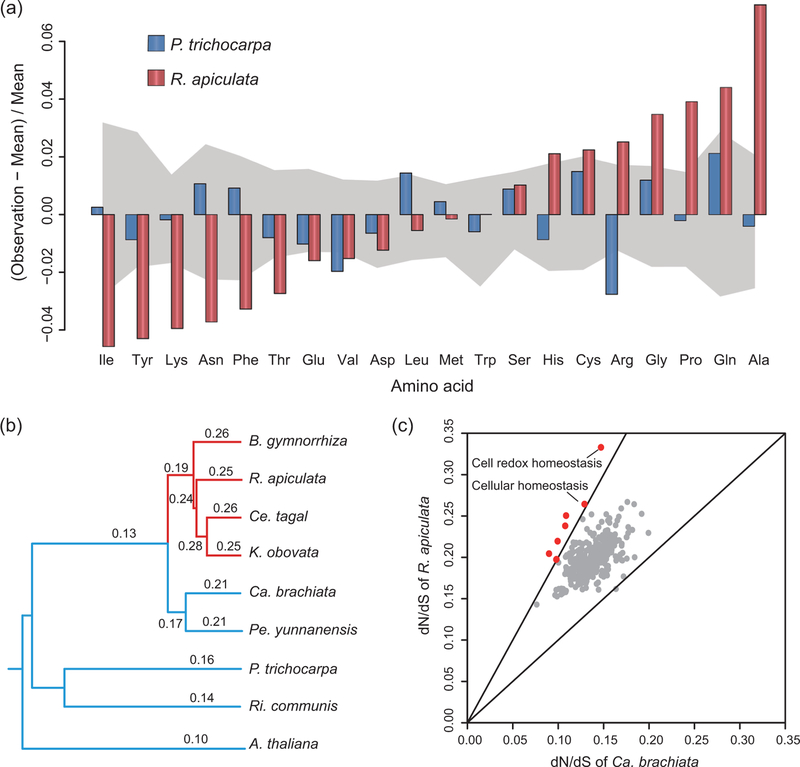
We surveyed amino acid (AA) compositions across all proteins among 50 plant species (Fig. 3a). The AA usages in R. apiculata (red bars) and P. trichocarpa (its closest non-mangrove relative with complete genome sequences, blue bars) are compared in the context of the other plant species. When the AAs are ranked from under-utilization to over-utilization, the divergence between this mangrove–non-mangrove pair is striking. In Rhizophora, two groups of AAs are found under- or over-utilized as shown on the opposite ends of Fig. 3a. In the more extreme cases, P. trichocarpa deviates from the mean in the opposite direction to R. apiculata (Asn, Arg and Ala). Overall, AA usage in R. apiculata deviates from the norm across the entire proteome.
Figure 3.
Genome-wide signatures of adaptation in R. apiculata. (a) Amino acid (AA) usage in the R. apiculata genome vs. that of its closest relative, P. trichocarpa. The comparison between the two species is done in the context of 48 other species of plants, whose AA usages are shown in the shaded area by the quartile. It is apparent that the AA usage in R. apiculata, but not P. trichocarpa, deviates strongly from the norm for plants. (b) dN/dS ratio along each branch of the phylogenetic tree. Mangrove lineages are colored red. (c) dN/dS ratios among genes grouped by GO terms in R. apiculata vs. in Ca. brachiata. The lower line indicates equal ratios and the upper line indicates a two-fold increase in R. apiculata. GO terms above the upper line are marked in red.
It is most striking that other mangroves show the same trend in AA usage. In fact, R. apiculata has the least deviant AA usage among the three mangrove taxa that include Sonneratia alba and Avicennia marina (He et al., unpublished data). These two lineages, outside of the phylogeny in Fig. 2a, represent independent evolutionary events of mangrove emergence.
Given the unusual AA usage, we ask whether non-synonymous nucleotide substitutions might be more frequent in mangroves than in their relatives. This can be measured by the dN/dS ratio (ω) where dN and dS are, respectively, the number of non-synonymous and synonymous substitutions per site. In Fig. 3b, ω values along all lineages are calculated using the PAML suite of programs [22]. It shows that ω is elevated in the Rhizophoreae clade relative to its inland relatives. The ω values on mangrove branches are generally larger than 0.25, whereas they are typically smaller than 0.2 in non-mangrove lineages (Fig. 3b). Although a high ω ratio is indicative of selection, it is usually attributable to the relaxation of negative selection against deleterious AA substitutions, unless ω> 1. In other words, stronger positive selection driving a higher ω ratio cannot be distinguished from weaker selection that also permits a higher ω. Only positive selection leads to adaptation.
To distinguish between positive selection and relaxation of negative selection, we scanned the genome using the ‘branch-site method’ in PAML, which applies a likelihood ratio test to compare models that permit or forbid ω >1 [34]. We identified 209 genes that harbor codons with ω> 1. Of these, 19 are implicated in embryonic development of A. thaliana (Supplementary Table 15, available as Supplementary Data at NSR online). Three of these, EMB88 (embryo defective 88), EMB2768 and EMB3137, will be used in further analyses below.
Whole groups of genes may also show the sign of adaptive evolution. The average dN/dS ratios among genes grouped by ontology are given in Fig. 3c. Eight categories show a greater than two-fold increase in R. apiculata over its closest relative, Ca. brachiata (see Supplementary Table 16, available as Supplementary Data at NSR online, for detailed analyses), notably ‘cell redox homeostasis’ and ‘cellular homeostasis’. Because various stressors in the intertidal zone can break cellular homeostasis, especially redox homeostasis, the rapid evolution of these genes in R. apiculata deserves future investigation.
The unusual AA usage and high rate of non-synonymous changes can be observed in greater detail when we examine AA substitutions between R. apiculata and R. mangle. Previous studies have shown that AA substitutions among broad taxa follow a common, or nearly universal, pattern in which certain pairs of amino acids are rarely exchanged, even though their codons differ by only one bp [35,36]. Interestingly, these infrequent AA substitutions are unusually common between R. apiculata and R. mangle. Such patterns of AA substitution are also observed between closely related species from the Avicennia and Sonneratia genera (He et al., unpublished data). Thus, the dynamic pattern of AA substitutions, like the static pattern of AA usage reported in Fig. 3, may be quite general among mangroves.
SPECIALIZED ADAPTIVE TRAITS: VIVIPARY AND THE TANNIN CONTENT (THE RED BARK)
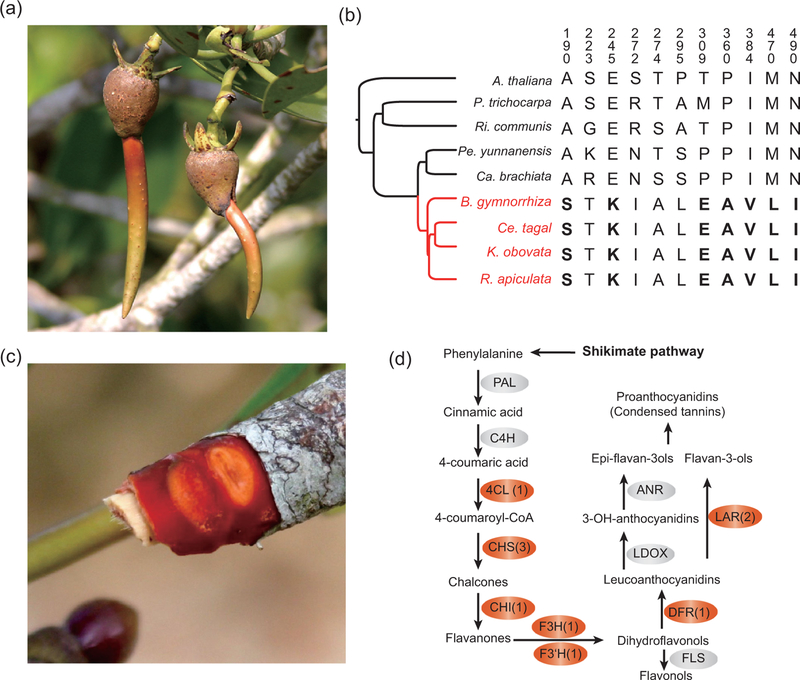
Two traits are particularly common in mangroves and rare in other woody plants: vivipary and the red-dish bark. Vivipary broadly means the ability of embryos to germinate while still attached to the parent (Fig. 4a). Previous works have suggested that viviparous embryos are protected from high salinity and other stresses during early development [2,37]. To identify candidate genes for this trait, we use a branch-site model implemented in PAML (modified A) [22], which focuses on a pre-determined set of genes to detect adaptive signals on a specific branch of phylogeny. We chose 255 genes from our orthologous gene set that are also found in the SeedGenes database [38]. These loci have been empirically confirmed to play a role in embryonic development in Arabidopsis. We focused on the branch spanning the interval of 47.8–54.6 Myr before present (Fig. 2a), which represents the most recent common ancestor of Rhizophoreae.
Figure 4.
Genomic basis of phenotypic change. (a) Viviparous seedlings of Rhizophora. The embryos developed with the hypocotyls growing out of fruits before being detached from mother plant. (b) Amino acid changes in SAE2. Changes in boldface are inferred to be under positive selection in the ancestral Rhizophoreae using the branch-site test (see text). (c) On a twig of R. apiculata, oxidized tannin is responsible for the red inner bark. (d) Flavonoid biosynthesis pathway governing tannin production in plants (see Supplementary Table 18, available as Supplementary Data at NSR online, for full enzyme names). Enzymes catalysing each reaction are listed next to the arrows. Red boxes highlight genes differentially expressed under increased salt concentration. For enzymes coded by more than one gene, the number of differentially expressed copies is given in the parentheses.
Five genes are identified by the branch-site test (Supplementary Table 17, available as Supplementary Data at NSR online). The most dramatic gene is SAE2 (SUMO-activating enzyme 2), which carries 11 Rhizophoreae-specific AA substitutions (Fig. 4b) [39]. Seven of these changes show signs of positive selection, including a site predicted to be functionally critical (predicted by PROVEAN [40]). Most importantly, our recent study found SAE2 to have experienced convergent evolution in three independently evolved mangrove clades [41]. Two other genes, ent-kaurene synthase (KS; Supplementary Fig. 15, available as Supplementary Data at NSR online) and GA3β-hydroxylase (GA3ox), are also candidate loci for vivipary (Supplementary Note, available as Supplementary Data at NSR online). Curiously, both have been duplicated in tandem in mangroves.
The second specialized trait in Rhizophoreae mangroves is the characteristic red bark that earns mangroves the nickname of ‘red trees’ in some languages. The red color is caused by a high concentration of tannins, a collection of flavonoid polymers, in several tissues (Fig. 4c) [42,43]. Tannin and other polyphenols have antioxidant activities and play a role in photoprotection, pathogen and herbivory resistance as well as salt tolerance [43–45]. We searched for adaptive signals in a set of genes involved in salt tolerance and flavonoid biosynthesis by analysing transcriptome profiles under different salt concentrations (see Supplementary Note and Supplementary Figs 16–18, available as Supplementary Data at NSR online).
Among the 34 genes coding for key enzymes in the flavonoid biosynthesis pathway (Supplementary Table 18, available as Supplementary Data at NSR online), 10 are differentially expressed under salt stress in R. apiculata (Supplementary Table 19, available as Supplementary Data at NSR online) and are highlighted in Fig. 4d. An interesting gene is Dihydroflavonol reductase B [DFR(B)], which is often lost in other taxa but is expressed at an elevated level in Rhizophora in high-salt environments (Supplementary Fig. 19, available as Supplementary Data at NSR online). DFR(B) differs from other members of the DFR family by 23 AAs, all of which are in the NAD(P)-binding domain.
Vivipary and tannin concentration are only two of the most conspicuous traits in mangroves. When recent expansions of gene families are analysed, it is evident that many have evolved to cope with the various stresses in these inhospitable environments. Using a maximum-likelihood method implemented in the CAFE software [46], we identify 112 gene families that have expanded in R. apiculata during recent evolution (Supplementary Figs 20 and 21, available as Supplementary Data at NSR online). The expanded families are enriched mostly for pathways involved in plant–pathogen interaction and biosynthesis of secondary metabolites (Supplementary Table 20, available as Supplementary Data at NSR online). We also find 2963 (11% of the total) R. apiculata genes that have been duplicated in tandem. Many of these genes are in the category of ‘response to chemical stimulus’ (Supplementary Table 21, available as Supplementary Data at NSR online).
DISCUSSION
The extensive set of high-quality genomic sequences provides a glimpse into the emergence, diversification and adaptation of Rhizophoreae, the largest monophyletic group of mangroves.
Rhizophoreae mangroves originated from inland ancestors about 50 Myr ago during PETM and after a WGD event. We suggest that the WGD event provided the genetic material and the PETM provided the suitable ecological conditions for this emergence. The genome-wide AA usage modification and acceleration of substitution rates, together with positive selection on AA sequence or copy number of genes underlying specific traits, contributed to the adaptation and diversification of this most successful mangrove clade.
The greater significance of the genomic sequences of mangroves lies in the future research possibilities. In addition to viviparous embryo and high tannin content, Rhizophoreae mangroves have other specialized traits, such as the aerial roots and cuticular waxes, the molecular bases of which have never been investigated. At the population level, Rhizophoreae is much more prone to undergo speciation than all other mangroves. This tendency could be due to both its genetic architecture and ecological conditions [47,48]. Furthermore, the independent evolution of mangroves makes them ideal candidates for studying convergent evolution. A recent analysis [41] provides a glimpse of the possible extent of molecular convergence among mangrove clades.
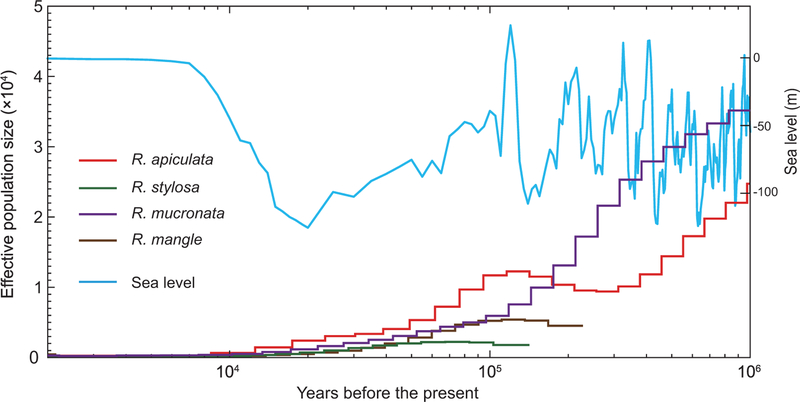
Despite their prominent global presence on the tropical coasts, mangroves should not be considered abundant in the genetic sense. All four Rhizophora species have extremely low genetic diversity: 3.1–5.5 × 10−4 per bp (Supplementary Note, available as Supplementary Data at NSR online). For a confirmation, we use the pairwise sequentially Markovian coalescent analysis (PSMC) [49] to infer the recent demographic histories of the Rhizophora species based on their whole-genome sequences. The model suggests a decrease in effective population size (Ne) starting about 100 000 years ago. Interestingly, this drop coincides with a dramatic change in global sea level (Fig. 5; Supplementary Fig. 22, available as Supplementary Data at NSR online). Although sea levels started to rise in the last 20 000 years, and mangrove populations expanded to colonize the newly available habitat, genetic diversity has yet to recover from the earlier reduction in effective population size. This trend is observable across all species.
Figure 5.
Rhizophora mangrove demography. Historical effective population size (Ne, y-axis) changes going back in time (x-axis). The changes in Ne are inferred using the PSMC method [48], which relies on the varying level of genetic diversity in different DNA segments across the genome as a basis for the inference of historical Ne changes. In this graph, the generation time (g) is set to 20 years and the mutation rate (μ) is 1.6 × 10−8/bp/generation. Historical sea-level fluctuations are plotted for comparison (blue line) [50].
While mangroves and the tropical intertidal ecosystems appear vibrant at present, genetic data suggest that this may not have been the case in recent geological times. Sea-level changes may have taken their toll until recently, when the levels became relatively stable in the last 7000 years. With sea levels projected to rise, mangrove populations could conceivably recede to levels even lower than those indicated by their low extant genetic diversity, especially when we factor in human-driven disturbance of their habitats.
The analyses and research resources provided by this study are significant because they will enable modern evolutionary, ecological and genomic research to expand to mangroves. The transition from inland to intertidal zones is an important model of adaptation and species proliferation. The genome-wide changes in AA usage are but one example of adaptation in this transition. Further active research on mangroves will also be crucial for the understanding and appreciation of the tropical coastal ecosystems anchored by these ‘red trees’. Since a large fraction of Earth’s human population lives near them, a sense of urgency should be very appropriate.
ONLINE METHODS
Genome sequencing and assembly.
Tissues from one mature individual of Rhizophora apiculata. (Qinglan Harbor, Hainan, China (19°37′N, 110°48′E)) were collected for DNA extraction. Genomic DNA was extracted from leaves using the CTAB method [51] and total RNA was extracted from leaves, roots, flowers and stems using the modified CTAB method [52]. Short-reads libraries were constructed following the TruSeq DNA Sample Preparation Guide. Ten libraries with different DNA fragment sizes were sequenced using Illumina Hiseq 2000 platform. 20 kb Single Molecule Real Time (SMRT) long-read library were prepared following PacBio SMRTbell 20 kb Template Preparation BluePippin Size Selection protocal and were sequenced using Biosciences RS II platform. The sequencing data of R. mangle, R. mucronata and R. stylosa were produced in the same way as the short-reads libraries. The transcriptome of R. apiculata and other five species in the Rhizophoraceae family (Kandelia obovata, Bruguiera gymnorrhiza, Ceriops tagal, Pellacalyx yunnanensis and Carallia brachiata) was sequenced on the Illumina Hiseq 2000 platform with insert size of 300 bp.
Before assembling, PCR duplication, adaptor contamination and low-quality reads were filtered out. The SMRT long reads and Illumina short reads were combined to assemble a draft genome. The de novo assembled genome based on the SMRT long reads was produced using four programs: falcon (https://github.com/PacificBiosciences/FALCON/), DBG2OLC [53], smartdenovo (https://github.com/ruanjue/smartdenovo) and wtdbg (https://github.com/ruanjue/wtdbg). The result obtained with smartdenovo was used as the final assembly because of its superior quality. Genome polishing was performed using Quiver [54] to further improve site-specific consensus accuracy. Illumina reads were then mapped to the polished genome assembly by BWA [55]. SNPs as well as small indels were called and corrected by SAMTOOLS [56] and in-house scripts. Finally, gap-filling were performed on the scaffolds with SSPACE 3.0 [57] using 10 Kb mate-pair sequences with the key parameters set as: -x 1 -m 50 -o 10 -z 200 -p 1.
The sequences of the transcriptome of R. apiculata, 458 core eukaryotic genes (CEGMA) [16] and 79 randomly selected genes from our previous work were used to evaluate the genome coverage and structural accuracy of the genome assembly (Supplementary Note).
The three re-sequenced congeneric genomes were mapped to the de novo assembled R. apiculata genome for comparison. Transcriptomes of the other five species (K. obovata, B. gymnorrhiza, Ce. tagal, Pe. yunnanensis and Ca. brachiata) were assembled and annotated using a common procedure [14] (see also in Supplementary Table 6, available as Supplementary Data at NSR online).
Genome annotation.
Three approaches were used to predict protein coding genes: homolog-based, de novo and transcriptome based prediction. Repeat sequences were masked throughout the genome using RepeatMasker (version 3.2.9) [58] and the RepBase library (version 16.08) [59] before further analysis. Homologous proteins from five known whole-genome sequences: Oryza sativa, Mimulus guttatus, Sesamum indicum, Populus trichocarpa and Eucalyptus grandis, were aligned to the repeat-masked R. apiculata genome using exonerate (v1.1.1) [60] for homolog-based prediction. Gene structures were generated using Genewise (version 2.2.0) [61]. The Augustus (version 3.2.2) [62] and GeneMark-ET (version 4.29) [63] algorithms were used to predict protein coding genes ab initio. Thirdly, RNA-seq reads were mapped to the genome using Tophat (version v2.1.1) [64], and gene models from spliced transcripts were identified using cufflinks (version v2.2.1) [65]. Finally, the three sets of predicted genes were combined using EVidenceModeler (EVM) [66] to generate a weighted and non-redundant consensus set of gene structures.
To annotate the functions of genes, coding sequences were aligned against the SwissProt, TrEMBL [67] and NCBI non-redundant protein databases using BLAST (v2.2.6) with an e-value threshold of 1 × 10−5. Gene Ontology (GO) annotation was obtained by aligning against the Pfam database [68] using HMMER2GO (https://github.com/sestaton/HMMER2GO). The protein sequences were also searched against the KEGG database [69] for KO (KEGG Orthology) assignments and pathway annotation.
Phylogenetic analyses and time dating.
The de novo genome of R. apiculata, the short-read sequences of R. mangle, R. mucronata and R. stylosa, the published genomes of P. trichocarpa and Ri. communis and the transcriptome data from five other species of Rhizophoraceae were used to reconstruct phylogenetic trees as well as estimate divergence times.
Orthologous genes were identified using the OrthoMCL software [70]. Phylogenetic trees were built using PhyML [20]. The species-divergent times were estimated using the program MCMC-TREE from the PAML 4.8 package [22] with the HKY85+gamma model, assuming an independent rate for each branch.
Collinearity analysis.
To detect the signature of whole-genome duplication, self-alignment was performed on protein sequences of R. apiculata using BLASTp (with an e-value cutoff of 1 × 10−5, identity ≥40%), followed by identification of syntenic blocks using MCScanX [19]. Collinear blocks having at least five paired homologous genes were accepted as duplicated blocks in this study. Genome distribution of the collinear blocks was visualized using the Circos software (v0.65) [71]. The time of WGD events was estimated following methods described in the Supplementary Note, available as Supplementary Data at NSR online.
Gene family analysis.
OrthoMCL software [70] was used to identify orthologous and paralogous groups of genes from four genomes (R. apiculata, A. thaliana, Ri. communis and P. trichocarpa). For genes with alternative splicing, the longest transcript was selected to represent the gene. All proteins from these four species were merged to perform an all-vs.-all alignment using BLASTp with an e-value cutoff of 1 × 10−10. The alignments were fed into a standalone OrthoMCL program with the default MCL inflation parameter (2.0). In the next step, CAFÉ [46] took the gene family sizes as input and used a stochastic birth and death process to model the evolution of gene family sizes across a given phylogenetic tree and detected expanded or contracted gene families with P-values < 0.05.
Heterozygosity and demographic history.
Using the aligner bowtie2 [72], clean short reads from R. apiculata (insert size: 200 bp, 300 bp, 400 bp and 600 bp), R. mangle (insert size: 300 bp), R. mucronata (insert size: 300 bp) and R. stylosa (insert size: 300 bp) were mapped to the assembled reference genome to identify the single nucleotide polymorphism sites (SNPs). Several filters were applied to ensure the accuracy of SNP calling: (i) removing potential PCR-duplicated, single-end mapped and improperly paired mapped reads; (ii) only sites having adequate sequencing depth (20– 200× for R. apiculata, 15–80× for R. mangle, R. mucronata and R. stylosa) were used; (iii) the called heterozygous sites had to have minor allele frequency larger than 0.15. More than 99.9% of heterozygous sites were retained according to the binomial function, assuming that the two alleles are equally sequenced, which indicated a good quality of the SNP data set. Heterozygosity was estimated as the number of identified heterozygous sites divided by the total number of sites meeting our depth criteria.
We used a pairwise sequentially Markovian coalescent (PSMC) analysis [49] to infer the history of population size with the parameters ‘-N25 -t500 -r5 - p “4+25*2+4+6”.’ The generation time was set as 20 years, and the mutation rate for each species was set to a previously estimated value (1.6 × 10−8).
Supplementary Material
ACKNOWLEDGEMENT
We thank X. He, J. Lu, J. Liu and F. He for insightful comments.
FUNDING
This work was supported by the National Natural Science Foundation of China (91331202, 41130208 and 31600182), the 985 Project (33000–18821105), the National Postdoctoral Program for Innovative Talents (BX201700300), the National Key Research and Development Plan (2017FY100705), the China Postdoctoral Science Foundation (2014M552264 and 2015T80931), the Fundamental Research Funds for the Central Universities (17lgpy99) and the Chang Hungta Science Foundation of Sun Yat-Sen University.
Footnotes
SUPPLEMENTARY DATA
Supplementary Data are available at NSR online.
ACCESSION CODES
Genome assembly of R. apiculata and raw reads of R. mucronata, R. stylosa and R. mangle has been deposited in the European Nucleotide Archive (ENA) under project accession numbers PRJEB8423, PRJEB20990, PRJEB20992 and PRJEB21001.
REFERENCES
- 1.Donato DC, Kauffman JB and Murdiyarso D et al. Mangroves among the most carbon-rich forests in the tropics. Nat Geo-science 2011; 4: 293–7. [Google Scholar]
- 2.Tomlinson P The Botany of Mangroves Cambridge: Cambridge University Press, 1986. [Google Scholar]
- 3.Ma T, Wang J and Zhou G et al. Genomic insights into salt adaptation in a desert poplar. Nat Commun 2013; 4: 2797. [DOI] [PubMed] [Google Scholar]
- 4.Olsen JL, Rouze P and Verhelst B et al. The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 2016; 530: 331–5. [DOI] [PubMed] [Google Scholar]
- 5.Giri C, Ochieng E and Tieszen LL et al. Status and distribution of mangrove forests of the world using earth observation satellite data. Global Ecol Biogeogr 2011; 20: 154–9. [Google Scholar]
- 6.Ball MC. Ecophysiology of mangroves. Trees 1988; 2: 129–42. [Google Scholar]
- 7.Parida AK and Jha B. Salt tolerance mechanisms in mangroves: a review. Trees 2010; 24: 199–217. [Google Scholar]
- 8.Fu X, Huang Y and Deng S et al. Construction of a SSH library of Aegiceras corniculatum under salt stress and expression analysis of four transcripts. Plant Sci 2005; 169: 147–54. [Google Scholar]
- 9.Liang S, Fang L and Zhou R et al. Transcriptional homeostasis of a mangrove species, Ceriops tagal, in saline environments, as revealed by microarray analysis. PloS one 2012; 7: e36499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, Yang S and Li J et al. Transcriptome analysis of the holly mangrove acanthus ilicifolius and its terrestrial relative, acanthus leucostachyus, provides insights into adaptation to intertidal zones. BMC Genomics 2015; 16: 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, He Z and Xu S et al. Transcriptome analyses provide insights into the phylogeny and adaptive evolution of the mangrove fern genus Acrostichum. Sci Rep 2016; 6: 35634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duke NC, Meynecke J-O and Dittmann S et al. A world without mangroves? Science 2007; 317: 41–2. [DOI] [PubMed] [Google Scholar]
- 13.Sidhu SS. Further studies on the cytology of mangrove species of India. Caryologia 1968; 21: 353–7. [Google Scholar]
- 14.Yang Y, Yang S and Li J et al. De novo assembly of the transcriptomes of two yellow mangroves, Ceriops tagal and C. zippeliana, and one of their terrestrial relatives, Pellacalyx yunnanensis. Mar Genomics 2015; 23: 33–6. [DOI] [PubMed] [Google Scholar]
- 15.Guo W, Wu H and Zhang Z et al. Comparative analysis of transcriptomes in rhizophoraceae provides insights into the origin and adaptive evolution of mangrove plants in intertidal environments. Front Plant Sci 2017; 8: 795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parra G, Bradnam K and Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 2007; 23: 1061–7. [DOI] [PubMed] [Google Scholar]
- 17.Guo Z, Huang Y and Chen Y et al. Genetic discontinuities in a dominant mangrove Rhizophora apiculata (Rhizophoraceae) in the Indo-Malesian region. J Biogeogr 2016; 43: 1856–68. [Google Scholar]
- 18.McGrath CL and Lynch M. Evolutionary significance of whole-genome duplication. In: Soltis PS and Soltis DE, (eds). Polyploidy and Genome Evolution Berlin, Heidelberg: Springer Berlin Heidelberg; 2012: 1–20. [Google Scholar]
- 19.Wang Y, Tang H and DeBarry JD et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 2012; 40: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guindon S, Dufayard JF and Lefort V et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 2010; 59: 307–21. [DOI] [PubMed] [Google Scholar]
- 21.Schwarzbach AE and Ricklefs RE. Systematic affinities of Rhizophoraceae and Anisophylleaceae, and intergeneric relationships within Rhizophoraceae, based on chloroplast DNA, nuclear ribosomal DNA, and morphology. Am J Bot 2000; 87: 547–64. [PubMed] [Google Scholar]
- 22.Yang Z PAML 4. phylogenetic analysis by maximum likelihood. Mol Biol Evol 2007; 24: 1586–91. [DOI] [PubMed] [Google Scholar]
- 23.Yang Z and Rannala B. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol Biol Evol 2006; 23: 212–26. [DOI] [PubMed] [Google Scholar]
- 24.Xi Z, Ruhfel BR and Schaefer H et al. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc Natl Acad Sci USA 2012; 109: 17519–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis CC, Webb CO and Wurdack KJ et al. Explosive radiation of malpighiales supports a mid-cretaceous origin of modern tropical rain forests. Am Nat 2005; 165: E36–65. [DOI] [PubMed] [Google Scholar]
- 26.Muller J Fossil pollen records of extant angiosperms. Bot Rev 1981; 47: 1–142. [Google Scholar]
- 27.Graham A Paleobotanical evidence and molecular data in reconstructing the historical phytogeography of rhizophoraceae. Ann MO Bot Gard 2006; 93: 325–34. [Google Scholar]
- 28.Collinson ME. Fossil plants of the London Clay Palaeontological Association; 1983. [Google Scholar]
- 29.Handley L, Crouch EM and Pancost RD. A New Zealand record of sea level rise and environmental change during the paleocene–eocene thermal maximum. Palaeogeogr, Palaeoclimatol, Palaeoecol 2011; 305: 185–200. [Google Scholar]
- 30.De Bodt S, Maere S and Van de Peer Y. Genome duplication and the origin of angiosperms. Trends Ecol Evol 2005; 20: 591–7. [DOI] [PubMed] [Google Scholar]
- 31.Freeling M Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol 2009; 60: 433–53. [DOI] [PubMed] [Google Scholar]
- 32.Maere S, De Bodt S and Raes J et al. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA 2005; 102: 5454–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kura-Hotta M, Mimura M and Tsujimura T et al. High salt-treatment-induced Na+ extrusion and low salt-treatment-induced Na+ accumulation in suspension-cultured cells of the mangrove plant, Bruguiera sexangula. Plant, Cell Environ 2001; 24: 1105–12. [Google Scholar]
- 34.Zhang J, Nielsen R and Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol 2005; 22: 2472–9. [DOI] [PubMed] [Google Scholar]
- 35.Tang H, Wyckoff GJ and Lu J et al. A universal evolutionary index for amino acid changes. Mol Biol Evol 2004; 21: 1548–56. [DOI] [PubMed] [Google Scholar]
- 36.Lu J, Tang T and Tang H et al. The accumulation of deleterious mutations in rice genomes: a hypothesis on the cost of domestication. Trends Genet 2006; 22: 126–31. [DOI] [PubMed] [Google Scholar]
- 37.Elmqvist T and Cox PA. The evolution of vivipary in flowering plants. Oikos 1996: 3–9.
- 38.Meinke D, Muralla R and Sweeney C et al. Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci 2008; 13: 483–91. [DOI] [PubMed] [Google Scholar]
- 39.Saracco SA, Miller MJ and Kurepa J et al. Genetic analysis of SUMOylation in arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 2007; 145: 119–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi Y and Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015; 31: 2745–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu S, He Z and Guo Z et al. Genome-wide convergence during evolution of mangroves from woody plants. Mol Biol Evol 2017; 34: 1008–15. [DOI] [PubMed] [Google Scholar]
- 42.Hernes PJ and Hedges JI. Tannin signatures of barks, needles, leaves, cones, and wood at the molecular level. Geochim Cosmochim Acta 2004; 68: 1293–1307. [Google Scholar]
- 43.Wang Y, Zhu H and Tam NFY. Polyphenols, tannins and antioxidant activities of eight true mangrove plant species in South China. Plant Soil 2014; 374: 549–63. [Google Scholar]
- 44.Hsu F-L, Nonaka G-I and Nishioka I. Tannins and related compounds. XXXI. Isolation and characterization of proanthocyanidins in Kandelia candel (L.) Druce. Chem Pharm Bull 1985; 33: 3142–52. [Google Scholar]
- 45.Kandil FE, Grace MH and Seigler DS et al. Polyphenolics in Rhizophora mangle L. leaves and their changes during leaf development and senescence. Trees 2004; 18: 518–28. [Google Scholar]
- 46.De Bie T, Cristianini N and Demuth JP et al. CAFE: a computational tool for the study of gene family evolution. Bioinformatics 2006; 22: 1269–71. [DOI] [PubMed] [Google Scholar]
- 47.Wu C-I and Ting C-T. Genes and speciation. Nat Rev Genet 2004; 5: 114–22. [DOI] [PubMed] [Google Scholar]
- 48.Yang M, He Z and Shi S et al. Can genomic data alone tell us whether speciation happened with gene flow? Mol Ecol 2017; 26: 2845–9. [DOI] [PubMed] [Google Scholar]
- 49.Li H and Durbin R. Inference of human population history from individual whole-genome sequences. Nature 2011; 475: 493–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller KG, Kominz MA and Browning JV et al. The Phanerozoic record of global sea-level change. Science 2005; 310: 1293–8. [DOI] [PubMed] [Google Scholar]
- 51.Doyle J and Doyle J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 1987; 19: 11–5. [Google Scholar]
- 52.Yang G, Zhou R and Tang T et al. Simple and efficient isolation of high-quality total RNA from Hibiscus tiliaceus, a mangrove associate and its relatives. Prep Biochem Biotechnol 2008; 38: 257–64. [DOI] [PubMed] [Google Scholar]
- 53.Ye C, Hill CM and Wu S et al. DBG2OLC: Efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci Rep 2016; 6: 31900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chin C-S, Alexander DH and Marks P et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 2013; 10: 563–9. [DOI] [PubMed] [Google Scholar]
- 55.Li H and Durbin R. Fast and accurate short read alignment with Burrows– Wheeler transform. Bioinformatics 2009; 25: 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, Handsaker B and Wysoker A et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25: 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boetzer M, Henkel CV and Jansen HJ et al. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011; 27: 578–9. [DOI] [PubMed] [Google Scholar]
- 58.Chen N Using repeatmasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics 2004: 4.10.11–14.10.14. [DOI] [PubMed]
- 59.Jurka J, Kapitonov VV and Pavlicek A et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 2005; 110: 462–7. [DOI] [PubMed] [Google Scholar]
- 60.Slater GS and Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 2005; 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Birney E, Clamp M and Durbin R. GeneWise and Genomewise. Genome Res 2004; 14: 988–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stanke M, Keller O and Gunduz I et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res 2006; 34: W435–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lomsadze A, Burns PD and Borodovsky M. Integration of mapped RNA-Seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res 2014: gku557. [DOI] [PMC free article] [PubMed]
- 64.Trapnell C, Pachter L and Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009; 25: 1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trapnell C, Williams BA and Pertea G et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28: 511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haas BJ, Salzberg SL and Zhu W et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol 2008; 9: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boeckmann B, Bairoch A and Apweiler R et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 2003; 31: 365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Finn RD, Coggill P and Eberhardt RY et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 2016; 44: D279–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanehisa M and Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 2000; 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li L, Stoeckert CJ Jr and Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 2003; 13: 2178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krzywinski M, Schein J and Birol I et al. Circos: an information aesthetic for comparative genomics. Genome Res 2009; 19: 1639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Langmead B and Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9: 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.