Abstract
In mycobacteria, D-alanine is an essential precursor for peptidoglycan biosynthesis. The only confirmed enzymatic pathway to form D-alanine is through the racemization of L-alanine by alanine racemase (Alr, EC 5.1.1.1). Nevertheless, the essentiality of Alr in Mycobacterium tuberculosis and Mycobacterium smegmatis for cell survivability in the absence of D-alanine has been a point of controversy with contradictory results reported in the literature. To address this issue, we examined the effects of alr inactivation on the cellular metabolism of M. smegmatis. The M. smegmatis alr insertion mutant TAM23 exhibited essentially identical growth to wild type mc2155 in the absence of D-alanine. NMR metabolomics revealed drastically distinct phenotypes between mc2155 and TAM23. A metabolic switch was observed for TAM23 as a function of supplemented D-alanine. In the absence of D-alanine, the metabolic response directed carbon through an unidentified transaminase to provide the essential D-alanine required for survival. The process is reversed when D-alanine is available, in which the D-alanine is directed to peptidoglycan biosynthesis. Our results provide further support for the hypothesis that Alr is not an essential function of M. smegmatis, and that specific Alr inhibitors will have no bactericidal action.
Keywords: NMR metabolomics, Mycobacterium smegmatis, Mycobacterium tuberculosis, Alanine racemase, D-alanine biosynthesis
Graphical Abstract
INTRODUCTION
Tuberculosis (TB) is a major global health problem and it remains the first worldwide cause of death from an infectious disease.1 Critical challenges to achieving the worldwide goals of disease control and elimination include the appearance of multiple and extensively drug resistant Mycobacterium tuberculosis strains, synergism with HIV driving co-infections in AIDS patients, and bacillary persistence in latently infected individuals.2 The cell wall of M. tuberculosis plays an important role in pathogenesis and its biosynthesis is a major source of targets for the design of antimicrobial agents and attenuated mutants.3–5 As in other eubacteria, peptidoglycan is a critical component of the cell wall backbone. D-alanine is an essential building block of peptidoglycan and is required for the formation of peptidoglycan crosslinks necessary to preserve cell integrity.3, 5–9
The main source of D-alanine in eubacteria is its conversion from L-alanine by alanine racemase (Alr), an enzyme encoded by the alr gene.3, 5, 8 The critical role of D-alanine in cell wall peptidoglycan biosynthesis has identified the biosynthesis of D-alanine as an important target for the generation of new anti-tuberculosis drugs. Also, attenuated mutants devoid of D-alanine biosynthesis are a potential source for vaccine candidates. Thus, alr essentiality and its role in mycobacterial physiology is core to the therapeutic value of targeting D-alanine biosynthesis. Studies on alr in the fast growing species Mycobacterium smegmatis, a useful model system for M. tuberculosis, rendered apparently contradictory results. In our initial reports, we found that the M. smegmatis alr insertion mutant grew without D-alanine.10–13 This data supported the existence of an alternative pathway of D-alanine biosynthesis in mycobacteria. However, contradictory results were reported as M. smegmatis and M. tuberculosis alr deletion mutants were unable to grow without D-alanine supplementation.14, 15 These results negate the existence of an alternative endogenous pathway of D-alanine biosynthesis in both M. smegmatis and M. tuberculosis. In effect, D-alanine provided by Alr, or an external source, were reported to be an absolute requirement for the growth of M. smegmatis and M. tuberculosis. A rational for the observed difference in the essentiality of M. smegmatis alr was attributed to the deletion mutant being a superior construct to the insertion mutant. This explanation was based on an inferred residual Alr activity despite evidence to the contrary.10–13 Furthermore, the argument ignores the fact that the insertion mutant was generated by the insertion of a drug marker that split alr approximately in the middle of the gene,11 a procedure similar to the creation of null mutant strains by transposon mutagenesis.16 Importantly, the deletion and insertion mutants were also selected and tested under different culture conditions that may impact D-alanine auxotrophy. Moreover, our recent studies on the mechanism of action of D-cycloserine (DCS) in both M. smegmatis and M. tuberculosis support our prior conclusions that Alr is non-essential and that D-alanine ligase is the lethal target of DCS in both mycobacterial species.10–13
Resolution of the controversy surrounding alr conditional essentiality in mycobacteria is important, not only in the context of the role of Alr, but also for its implications in the design of more effective drugs and vaccines targeting the endogenous synthesis of D-alanine in M. tuberculosis. Additionally, evidence of horizontal gene transfer in mycobacteria,17, 18 highlights the impact that the potential transfer of genes involved in an alternative pathway of D-alanine biosynthesis in a non-pathogenic species such as M. smegmatis might have on TB epidemiology and management. To address this issue, we undertook an NMR metabolomics study to determine whether D-alanine could be exchanged into the metabolic pool of the M. smegmatis alr insertion mutant TAM23. We report conclusive evidence for this exchange that suggest an alternative pathway of D-alanine biosynthesis in M. smegmatis.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains used in this study were M. smegmatis wild type mc215519 and alr mutant TAM23.11 For all experiments, strains were grown to an OD600 of 0.6 – 1.0 at 37 °C with shaking at 200 rpm in Middlebrook 7H9 complete media (MADC) supplemented with 0.2% (v/v) glycerol, 0.05% Tween 80, 0.01% cycloheximide, and ADC (0.5% bovine serum albumin (BSA) fraction V, 0.01 M glucose, and 0.015 M NaCl). Cycloheximide is a protein synthesis inhibitor, which was used to suppress the growth of yeast in the bacterial cell cultures. When necessary, cultures were supplemented with 50 mM D-alanine. For growth curve analysis, all cultures were inoculated to a starting OD600 of approximately 0.01 and the generation times were determined as previously described.20
Proteomics sample preparation and data collection.
All mc2155 and TAM23 cultures (50 mL) were grown to an OD600 of 0.8 – 1.1 in MADC media without D-alanine. The bacteria were removed, stored on ice for 10 min, and centrifuged at 3000 rpm at 4 °C for 15 min. The supernatant was removed from the cell pellet, filtered, and placed at −80 °C. Two washes with 30 mL ddH2O were performed, and the pellet was resuspended for a final volume of 1.0 mL in 50 mM ammonium bicarbonate containing 8 M urea and 1.5 mM phenyl-methyl-sulfonyl fluoride as a protease inhibitor. Cell lysis in the MP Biomedicals FastPrep-24 was performed 3 times for 20 s with a 5 min chill period between cycles. Two cycles of centrifugation were performed to remove cell debris. Protein (0.5 – 1 mg) was precipitated with cold acetone. Samples were vortexed and incubated overnight at −20 °C. Samples were then warmed to room temperature, pelleted by centrifugation at 14000g for 20 min, and the supernatant was discarded. After air-drying for 15 – 30 min, samples were subjected to MudPIT analysis at the University of Nebraska-Lincoln (UNL) Redox Biology Center Proteomics and Metabolomics Core Facility. A protease and trypsin digest was performed on the protein mixtures and then subjected to liquid chromatography electrospray mass spectrometry LC-ESI-MS. Protein identification was performed by a Mascot database search.21
NMR metabolomics sample preparation.
General procedures for the handling and preparation of M. smegmatis NMR samples for metabolomics analysis have been described elsewhere.22 Metabolomics samples for one-dimensional (1D) 1H NMR experiments were prepared from ten replicates of both M. smegmatis mc2155 and TAM23 cell cultures. M. smegmatis mc2155 and TAM23 strains were grown at 37 °C with shaking at 200 rpm in 50 mL of MADC media in 250 mL flask for 10 – 12 hours until an OD600 of 0.6 – 1.0. A second set of ten replicate cultures per strain were grown under the same condition with the exception that the TAM23 cultures were supplemented with 50 mM D-alanine after the OD600 reached approximately 0.6. All of the bacterial cultures were then allowed to grow for an additional 2 h. The cell cultures were placed on ice for 5 min and kept on ice throughout the entire process of metabolome extraction. Bacterial cells were harvested by centrifugation at 2000g at 4 °C for 15 min in 50 mL tubes. The cells were washed twice with 30 mL of ice-cold double distilled water. The cell pellets were re-suspended with 1.0 mL of double distilled water and transferred to a 2 mL vial containing 0.1 mm silica beads (Lysing Matrix B). The cells were then lysed for 40 s at 6 m/s using a MP Biomedicals FastPrep-24. The cell lysate was centrifuged for 10 min at 12 400g and 4 °C. The supernatant was extracted and then lyophilized and resuspended with 1.0 mL of 100% D2O containing 50 mM phosphate buffer (uncorrected pH = 7.2) with 50 µM 3-(trimethylsilyl) propionic-2,2,3,3-d4 acid sodium salt (TMSP). The samples were vortexed and then centrifuged. A 600 µL portion of the cell free extract was then transferred to an NMR tube. A total of 40 metabolomics samples were prepared for collecting one-dimensional (1D) 1H NMR spectra.
NMR metabolomics samples for two-dimensional (2D) 1H-13C HSQC experiments were prepared from triplicate M. smegmatis mc2155 and TAM23 cultures grown in 110 mL MADC media (250 mL flasks). The bacterial cultures were grown at 37 °C with shaking at 200 rpm for 10 – 12 h until an OD600 of approximately 0.6. The culture media was then supplemented with or without 100 μM [13C2]-D-alanine and the cultures were allowed to grow for an additional 2 h. The bacterial cells were processed and the metabolome was extracted following the same protocol described above.
NMR data collection.
The NMR data was collected and processed according to our previously described protocol.23 The NMR spectra were collected on a Bruker Avance DRX 500 MHz spectrometer equipped with a 5 mm triple-resonance cryogenic probe (1H, 13C, 15N) with a z-axis gradient, BACS-120 sample changer, and an automatic tuning and matching accessory was utilized for automated NMR data collection. 1D 1H NMR spectra were collected with Purge solvent saturation,24 a 1H 90° pulse of 8 μs, and a spectral width of 5482.5 Hz and 32k data points at 298 K. A total of 16 dummy scans and 128 scans were used with a recycle delay of 1.5 s for a total acquisition time of 10 minutes per spectrum.
A series of three time-zero 2D 1H-13C HSQC spectra (HSQC0) were collected using the pulse sequence described by Hu et al. for determination of absolute metabolite concentration changes.25, 26 The NMR spectra were collected with 2048 data points and a spectral width of 5000.0 Hz in the direct 1H dimension, and 64 data points and a spectral width of 17605.6 Hz in the indirect 13C dimension. A total of 16 dummy scans and 64 scans with a relaxation delay of 1.5 s were used to obtain each 2D 1H-13C HSQC0 spectrum with a total acquisition time of 6 hours.
Multivariate statistical analysis.
The 1D 1H NMR spectra were processed in our MVAPACK software suite (http://bionmr.unl.edu/mvapack.php).27 For principal component analysis (PCA), the 1D 1H NMR spectra were processed with a 1.0 Hz exponential apodization function, followed by Fourier transformation, automatic phasing and normalization with phase-scatter correction (PSC),28 and referenced to TMSP-D4 (0.0 ppm). The noise and solvent containing regions were manually removed, the spectrum was binned using an adaptive intelligent binning algorithm,29 and Pareto scaled. For orthogonal projections to latent structures discriminant analysis (OPLS-DA), the NMR data was processed using MVAPACK as described above. The table of integrals was center averaged and imported into SIMCA version 13.0 (Umetrics) to produce the S-plots. Supervised classification for each group was determined by cell type, with M. smegmatis mc2155 defined as the control (or assigned a value of 0) and M. smegmatis TAM23 was assigned a value of 1. Ellipses were generated for each group in the PCA scores plot using our PCA/PLS-DA utilities.30, 31 The ellipses correspond to the 95% confidence limits of a normal distribution for each group.
Metabolite identification and measurement of absolute concentration changes.
2D 1H-13C HSQC spectra were processed using NMRPipe.32 The spectra were Fourier transformed, manually phased, and baseline corrected. The processed 2D 1H-13C HSQC spectra were then analyzed using NMRView33 to assign chemical shifts, intensities, and volumes to each NMR resonance. Chemical shift lists were assigned to specific metabolites using the Human Metabolome Database,34 Madison Metabolomics Database,35 and the Platform for Riken Metabolomics.36 A chemical shift error-tolerance of 0.05 ppm and 0.40 ppm was used for 1H and 13C chemical shifts, respectively. The identification of metabolites and metabolomics pathways from the NMR metabolomics data were further verified using the Kyoto Encyclopedia of Genes and Genomes (KEGG)37 and MetaCyc38 databases.
Metabolite peak volumes from the HSQC0 experiment was calculated as previously described.25, 26 Briefly, peak volumes from a 2D 1H-13C HSQC experiment are dependent on the magnitude of J-coupling constants, relaxation processes, dynamics, and metabolite concentrations. The HSQC0 experiment is a series of three HSQC experiments with an increasing number of HSQC pulse sequence repetitions (i.e., HSQC1, HSQC2, HSQC3). In this manner, the HSQC peak volume will decrease proportional to the number of pulse sequence repetitions due to the impact of all factors except concentration. A natural log plot of peak volumes versus the increment number (1, 2, 3) allows for the extrapolation back to increment 0 or zero-time, which is proportional to the metabolite’s concentration. The calibration of metabolite peak volumes from the HSQC0 experiments with an absolute concentration was calculated with a standard calibration curve as previously described.23 Briefly, five different mixtures containing nine different 13C-labeled metabolites (2-keto-3-methylbutyrate, 2-ketobutyrate, acetate, alanine, fructose, glucose, glycine, pyruvate, succinate) with a range of known concentrations (5 to 300 μM) were used to collect a series of HSQC0 experiments. The HSQC0 peak volumes were determined for each metabolite at each concentration as described above. The HSQC0 peak volumes were then plotted versus the known metabolite concentrations to yield a standard calibration curve. The triplicate HSQC0 experiments were used to calculate averages and standard deviations for each metabolite concentration.
RESULTS
Growth curves of M. smegmatis alr mutants.
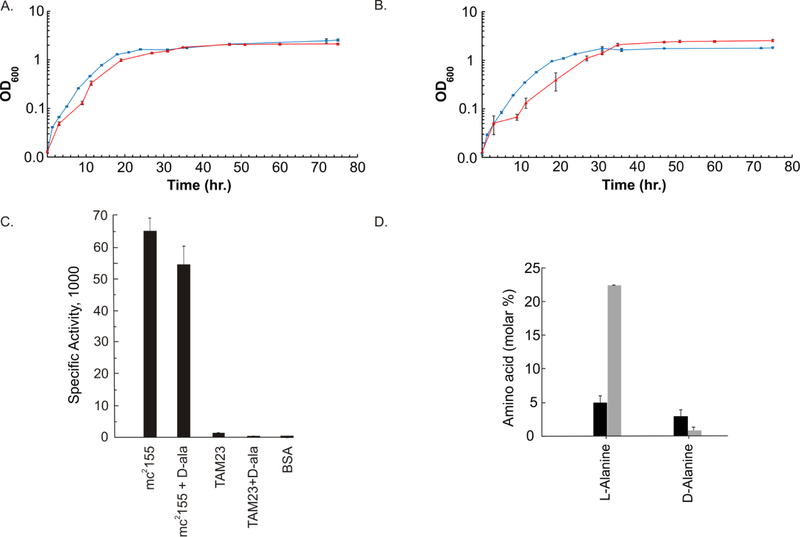
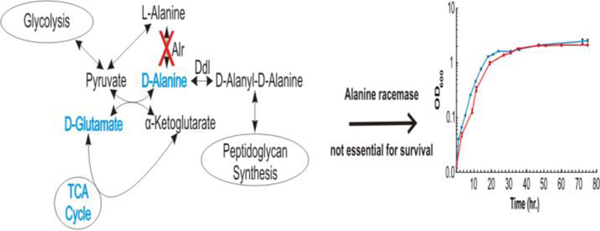
When grown in MADC, the wild type and the alr insertion mutant were still able to proliferate in the absence of D-alanine. In our original studies, we observed that TAM23 grew with a significant lag time when subcultured from an inoculum grown in media with D-alanine. However, in this study we subcultured from inocula prepared after the cells were passaged in media without D-alanine. Under these conditions, both strains yielded growth curves that followed similar trends (Fig. 1A). Strains mc2155 and TAM23 grew with a generation time of 208 ± 34 and 294 ± 29 min, had an initial lag phase of 5 and 12 h, and reached saturation densities (OD600) of 2.5 and 2.1, respectively. A similar result was obtained for both strains when the media was supplemented with D-alanine except for a moderately higher saturation density for TAM23 (Fig. 1B). Strains mc2155 and TAM23 had generation times of 310 ± 28 and 294 ± 33 min, lag phases of 4 and 6.5 h, and saturation densities of 1.7 and 2.54, respectively. When comparing the growth curves with and without D-alanine, TAM23 was able to grow more efficiently in the presence of D-alanine while the opposite was true for mc2155. Even though the slopes during the exponential phase of growth are significantly different (p-value < 0.001) when comparing TAM23 grown in no D-alanine to mc2155 grown with and without D-alanine, and TAM23 with D-alanine; there does not appear to be a biological difference. These results demonstrate that the alr gene is not essential for the survivability of M. smegmatis in the absence of D-alanine and that an alternative pathway for synthesizing D-alanine must exist.
Figure 1.
Growth curves for wild-type M. smegmatis mc2155 cells (blue) or alr mutant TAM23 cells (red) treated (A) without D-alanine or (B) with 50 mM D-alanine, respectively. Growth curves were collected in triplicate and the error bars were calculated using SEM. (C) Mean specific activities (µmoles of substrate consumed per min per mg protein) in M. smegmatis cell extracts. Data are as described previously by Chacon et al. 2002.11 Cells were grown to mid-exponential phase, harvested, and lysed. Cell-free extracts were assayed for Alr activity in triplicate. A bovine serum albumin (BSA) control was prepared at the same protein concentration as the cell extracts and assayed in triplicate. All error bars were calculated using standard deviations. (D) Intracellular steady-state concentrations of amino acids L- and D- alanine in mc2155 (black) and TAM23 (grey). Data reproduced from Chacon et al. 2009.10
Major proteomic changes in M. smegmatis alr mutants.
MudPIT analysis identified 904 proteins expressed in media without D-alanine. TAM23 protein overexpression was defined as a 10-fold or greater expression level relative to mc2155. Thirty-one proteins were identified as being overexpressed in TAM23 relative to mc2155 (Supplemental Table S1). Several enzymes directly involved in central carbon metabolism were affected by the alr mutation. Specifically, alanine dehydrogenase (ALD), and several alcohol dehydrogenases (ADHs), were all overexpressed in TAM23. The ADH enzymes exhibited an approximate 40-fold increase in TAM23 relative to mc2155. Similarly, ALD was upregulated by approximately 30-fold in TAM23.
Metabolic impact of media lacking D-alanine on M. smegmatis alr mutants.
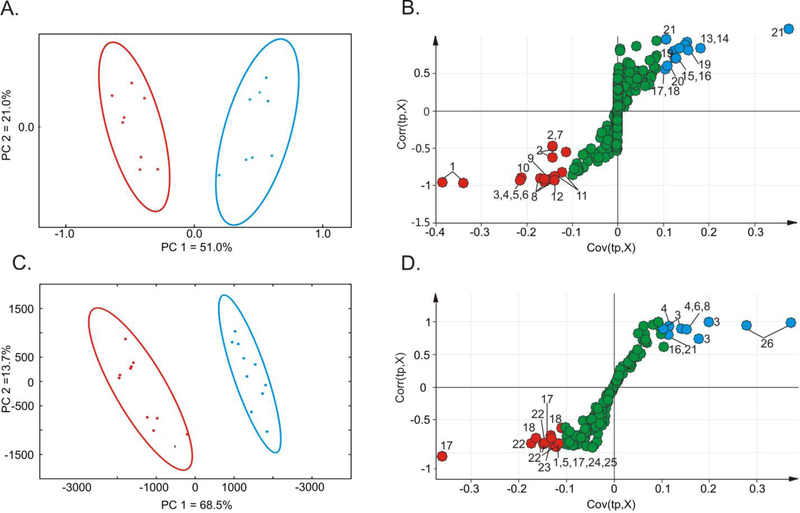
The set of 1D 1H NMR spectra were used to assess the global metabolome differences between wild type M. smegmatis mc2155 and the alanine racemase TAM23 insertion mutant (Fig. S1). The PCA scores plot (Fig. 2A) compares the metabolomes of M. smegmatis mc2155 cells and the TAM23 insertion mutant grown in MADC without D-alanine. The scores plot reduces the complex 1D 1H NMR spectrum that captures the state of the cellular metabolome into a single point in principal component space (PC1, PC2, etc.). Correspondingly, the principal components represent the largest variations between the individual 1D 1H NMR spectra. The scores plot indicates that PC1 and PC2 account for 51.0% and 21.0% of the variation in the NMR spectra, respectively. A total of 10 replicates for the wild type mc2155 and the alr TAM23 insertion mutant form two distinct clusters in the PCA scores plot. The unsupervised PCA model demonstrates a statistically significant difference between the metabolomes of M. smegmatis wild type mc2155 cells and the alr TAM23 insertion mutant.
Figure 2.
PCA scores plots generated from 1D 1H NMR spectra obtained from M. smegmatis TAM23 (red) and mc2155 (blue) cell lysates (A) without or (C) with media supplemented with D-alanine. OPLS-DA S-plots comparing the 1D 1H NMR spectra obtained from TAM23 and mc2155 cell lysates (B) without or (D) with media supplemented with D-alanine. Each point in the S-plot represents a specific bin of integrals for a chemical shift range of 0.25 ppm. The points at the extreme ends of the S-plots are major contributors to class distinction. The red points represent a decrease in the binned integrals for TAM23 compared to mc2155. The blue points represent an increase in the binned integrals for TAM23 compared to mc2155. Green points represent metabolites that are approximately equally in both strains. The labeled points were assigned to the following metabolites: 1) fatty acids, 2) ethanol, 3) glutamate, 4) oxaloacetate, 5) GABA, 6) succinate, 7) glycerol, 8) isoleucine, 9) valerate, 10) lactate, 11) N-acetylglucosamine-6-phosphate, 12) α-ketoisovalerate, 13) glycolate, 14) serine, 15) phosphoserine, 16) threonine, 17) lysine, 18) alanine, 19) glucarate, 20) ATP, 21) unidentified (UD), 22) trehalose, 23) valine, 24) ornithine, 25) α-ketoglutarate and 26) citrulline.
An OPLS-DA model was generated from the metabolomics datasets to further characterize the mc2155 and TAM23 metabolome and to identify the discriminatory variations that maximize group separation.39 The OPLS-DA model without the addition of D-alanine yielded a reliable fit as evident by an R2X of 0.652, R2Y of 0.971, and a Q2 of 0.923 (Fig. S2A). Importantly, cross validation using CV-ANOVA produced a statistically significant p value of 5.17 × 10−6 indicating a valid OPLS-DA model. The group separation between the alanine racemase TAM23 mutant and the mc2155 wild type metabolomes is statistically significant, which is consistent with the group separation observed in the PCA scores plot. Furthermore, an S-plot generated from the OPLS-DA model identified the NMR spectral differences and, correspondingly, the major metabolome changes contributing to the group separations in the scores plot (Fig. 2B). The variables (chemical shifts) with a large modelled correlation [approximately 1 (correlated) or −1 (anticorrelated)] and large modeled co-variation ≥ 0.1 or ≤ −0.1 were identified as key contributors to group separations in the OPLS scores plot and are labeled on the S-plot. The metabolites from the S-plot identified as major contributors to group separation in the absence of supplemental D-alanine are summarized in Table 1.
Table 1.
Relative differences in metabolite concentrations between M. smegmatis wild-type and TAM23 alr mutant cells
| No D-alaninea,b |
50 mM D-alanine |
||
|---|---|---|---|
| Relative Increase |
Relative Decrease |
Relative Increase |
Relative Decrease |
| Alanine | α-ketoisovalerate | Citrulline | Alanine |
| ATP | Ethanol | Fatty acid | α-ketoglutarate |
| Fatty acid | GABA | Glutamate | GABA |
| Glucarate | Glutamate | Isoleucine | Lysine |
| Glycolate | Glycerol | Oxaloacetate | Ornithine |
| Lysine | Isoleucine | Succinate | Trehalose |
| Phosphoserine | Lactate | Threonine | Valine |
| Serine | N-acetyl-glucosamine-6-phosphate | ||
| Threonine | Oxaloacetate | ||
| Succinate | |||
| Valerate | |||
Observed increase or decrease in metabolite concentrations in M. smegmatis TAM23 alr mutant cells relative to wild type mc2155 cells. Metabolites were identified from the S-plots generated from the corresponding OPLS-DA model (see Fig. 2).
Metabolites in italics exhibited a reversal in concentration depending on whether media were supplemented with or without D-alanine.
Impact of D-alanine on the metabolome of M. smegmatis alr mutants.
The NMR metabolomics experiment was repeated using M. smegmatis mc2155 and TAM23 cells grown in MADC supplemented with 50 mM D-alanine. As observed previously, the PCA scores plot contained two distinct clusters indicating a difference between the mc2155 and TAM23 metabolomes (Fig. 2C). PC1 and PC2 account for 68.5% and 13.7% of the variation in the data, respectively.
The OPLS-DA scores plot exhibited a similar group separation between M. smegmatis mc2155 wild type cells and the TAM23 alr mutant (Fig. S2B). The OPLS-DA yielded a reliable fit as evident by R2X of 0.682, R2Y of 0.981 and a Q2 of 0.957. Importantly, cross validation using CV-ANOVA produced a statistically significant p value of 4.39 × 10−10 indicating a valid OPLS-DA model. The group separation between the alanine racemase TAM23 mutant and the mc2155 wild type metabolomes is statistically significant, which is consistent with the group separation observed in the PCA scores plot. The S-plot from OPLS-DA model was used to identify the chemical shifts (metabolites) that significantly contributed to the group separation in the OPLS-DA scores plot (Fig. 2D). The metabolites from the S-plot identified as major contributors to group separation in the presence of supplemental D-alanine are summarized in Table 1.
Quantitative analysis of metabolite changes in M. smegmatis alr mutants.
The NMR metabolomics experiment was repeated using M. smegmatis mc2155 and TAM23 cells grown in MADC supplemented with 100 µM of 13C-labeled [13C2]-D-alanine. Triplicate sets of 13C-labeled metabolite extracts were prepared per strain and then analyzed using time-zero 1H-13C HSQC (HSQC0) experiments.25, 26 A 2D 1H-13C HSQC NMR spectrum allows for easier identification of metabolites because of the reduced complexity and spectral overlap compared to a 1D 1H NMR spectrum. Only metabolites that originate from the supplemental 13C-D-alanine are observed in the 2D 1H-13C HSQC spectrum (Fig. S3). Each peak in the 2D 1H-13C represents a 13C-1H pair for a specific metabolite that were assigned by matching the chemical shifts against NMR metabolomics databases.34–36 A total of 38 metabolites were identified from the two M. smegmatis strains.
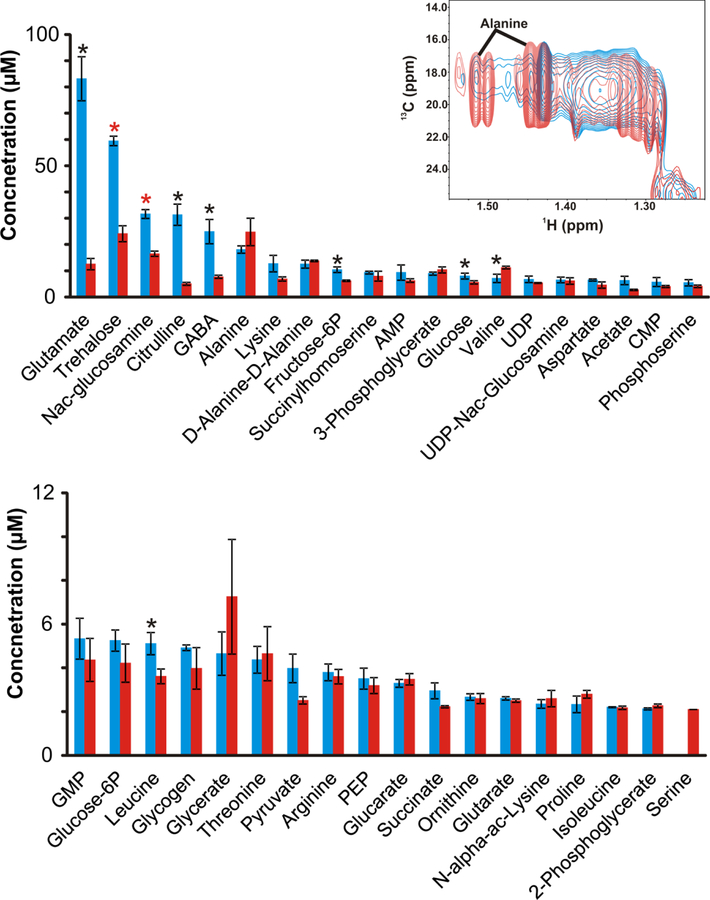
A concentration for each metabolite was determined by extrapolating peak volumes from a series of three 2D 1H-13C HSQC spectra at different time points to interpolate a time-zero peak volume. The spectra were collected in triplicate to determine an average peak volume and calculate a standard deviation. The average time-zero peak volume is directly proportional to a concentration based on a calibration curve using 9 13C-labeled compounds of known concentration.23 A plot of the absolute metabolite concentrations for the M. smegmatis mc2155 and TAM23 strains is shown in Fig. 3. Peptidoglycan precursors such as glutamate, lysine, and N-acetyl-glucosamine were significantly decreased in the alr insertion mutant. The concentration of trehalose, an essential precursor for trehalose-containing glycolipids, which allows the attachment of glycolipids to arabinogalactan in the cell wall was also decreased in the alr insertion mutant.40
Figure 3.
Bar graph summarizing metabolite concentrations determined from 2D 1H-13C HSQC0 experiments. M. smegmatis mc2155 (blue) and TAM23 (red) cell cultures were supplemented with 100 µM 13C-labeled [13C2]-D-alanine. Statistical significance at the 95% confidence level is denoted by a black asterisk, and a red asterisk indicates a statistical significance after applying Bonferroni multiple hypothesis correction. (insert) Expanded view of an overlay of the wild type strain mc2155 (blue) and alr insertion mutant TAM23 (red) 2D 1H-13C HSQC0 spectra highlighting alanine concentration changes. The full spectral overlay is in supporting information (see Fig. S3).
DISCUSSION
Multiple resistant strains of TB are growing and spreading at a rapid pace; and the concurrent loss of antibiotic efficacy is predicted to cause upwards of 10 million deaths annually by 2050. Avoiding this potential pandemic requires developing new antibiotics that circumvent common mechanisms of resistance, which necessitates the identification of novel therapeutic targets. For antibiotics, therapeutic targets have commonly been an essential enzyme or protein. Alr is a pyridoxal 5’-phosphate (PLP) - containing enzyme which is thought to be necessary for the growth of mycobacteria.41–43 The racemization of L-alanine to D-alanine by Alr is the only confirmed enzymatic reaction in M. tuberculosis or M. smegmatis to produce the D form of alanine, which is an essential intermediate for peptidoglycan biosynthesis.8 Nevertheless, the essentiality of Alr in M. smegmatis and M. tuberculosis survivability and its significance as a therapeutic target have been highly controversial.
Chacon et al. previously observed that an alr knock-out mutant (TAM23) was able to grow under conditions without supplementing D-alanine into Middlebrook 7H9 or minimal medium.10, 11 The TAM23 strain did not exhibit any growth defects, but it was clear that the phenotypic changes between TAM23 and the wild type strain were pronounced.10, 11 Again, we did not observe any biologically significant differences when we compared the TAM23 growth curve, upon passage for growth in media without D-alanine, to the wild type mc2155 strain in the absence of D-alanine (Fig. 1A). In fact, we observed a higher saturation density for the TAM23 growth curve relative to mc2155 with the addition of D-alanine to the culture media (Fig. 1B). One potential interpretation is that the stress response TAM23 experiences due to the lack of both alr activity and D-alanine is partly relieved by supplemented D-alanine leading to a relative improvement in cell growth. Regardless, TAM23 does not exhibit any growth defects in the absence or presence of D-alanine strongly implying that the alr inactivation does not negatively impact cell survivability. This is in direct conflict with other studies that show Alr to be essential in mycobacteria.14, 15 These discrepancies may be attributed to contrasting mutant construction and cell growth conditions. TAM23 is an insertion mutant which still has the potential of producing a protein product with residual or defective activity. However, as discussed above, this possibility is extremely unlikely as the insertion mutation would be similar to a transposon insertion in the middle of the gene. Furthermore, we previously examined Alr activity in TAM23 and did not detect any activity above BSA background (Fig. 1C).10 The lack of Alr activity in TAM23 was also supported by examining the steady state concentrations of L- and D -alanine. L-alanine was observed to accumulate in TAM23, which is consistent with an inactive Alr (Fig. 1D). Conversely, the D-alanine production in TAM23 was below that of mc2155, but still above background levels. This is consistent with the alr mutant producing D-alanine through an alternative mechanism. Recently, we proposed an alternative metabolic pathway for the production of D-alanine in M. tuberculosis and M. smegmatis to compensate for an inactive Alr.12, 13 An alternative source of D-alanine may result from an unidentified transaminase converting pyruvate into D-alanine. This hypothesis resulted from a metabolomics investigation into the in vivo lethal target of DCS, in which Alr and D-alanine-D-alanine ligase (Ddl) had been previously identified as likely targets for DCS. We identified Ddl as the in vivo lethal target of DCS based on an observed decrease in the biosynthesis of D-alanyl-D-alanine that occurred simultaneously with a DCS induced inhibition on cell growth. We also previously observed the incorporation of 13C-carbons from D-alanine into glutamate was dependent on DCS concentration, which strongly suggests a transaminase compensating for an inactive Alr.
Herein, we further tested our hypothesis that Alr is nonessential for peptidoglycan biosynthesis and the survivability of M. smegmatis by examining the total proteomic and metabolic shifts that occurs in TAM23 relative to wild type mc2155 cells. Our proteomics analysis revealed that ALD and ADH enzymes were significantly overexpressed from Alr inactivation. Of particular note, ALD is involved in a metabolic recycling process, which includes an alternative pathway for the biosynthesis of D-alanine. ALD catalyzes the oxidative deamination of L-alanine to produce pyruvate and ammonia with concomitant formation of NADH.44 Thus, ALD may provide an alternative source of D-alanine via pyruvate, where pyruvate and D-glutamate are then converted into D-alanine and α-ketoglutarate by a transamination reaction. A D-amino acid aminotransferase (Dat) capable of catalyzing this transamination reaction has been previously observed in Bacillus subtilis, Listeria monocytogenes, and numerous other bacteria.45–50 While not verified, we previously identified a number of enzymes that may potentially perform the same function in M. smegmatis.51 In the absence of supplemental D-alanine, the overproduction of ALD in TAM23 increases the reducing potential (generation of NADH). This is compensated by the overproduction of ADH enzymes to regenerate NAD through the oxidation of NADH. Thus, this metabolic change is necessary to maintain redox balance when Alr is inactive.
The increase in ammonia production from an elevated ALD activity did not lead to an observable toxicity for M. smegmatis. In fact, ammonia is a preferred source of nitrogen for M. smegmatis, and is a diffusible substance that participates in many metabolic processes and, consequently, is under tight regulatory control.52–54 There is always a balance of oxidative deaminations and reductive aminations. Further, there are outer membrane proteins that participate in the influx and efflux of ammonia. As a result, M. smegmatis is readily able to adapt to large variations in nitrogen (ammonia) availability through a variety of cellular processes. Similarly, the increase in ADH activity to regenerate NAD would be expected to increase the conversion of an acetaldehyde to an alcohol, which is then excreted from the cell. This is also consistent with the prior observation that M. smegmatis ADH has a preference to function as an aldehyde reductase and convert acetaldehydes into alcohols.55
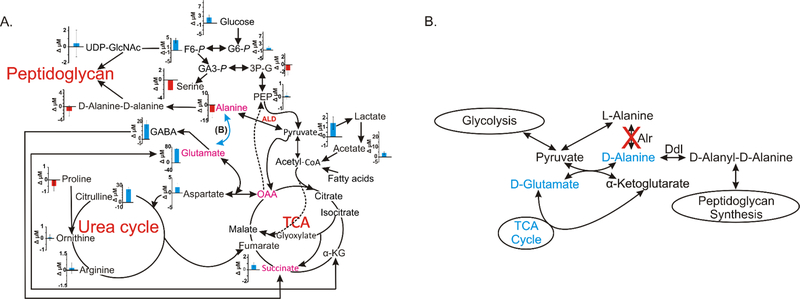
From a 1D 1H NMR metabolomics study we observed statistically significant differences between the mc2155 and TAM23 metabolomes based on the distinct clusters in the PCA scores plot (Fig. 2A). The metabolite changes identified from the resulting S-plot (Fig. 2B) are listed in Table 1. As previously observed, the difference in the mc2155 and TAM23 metabolomes was eliminated when TAM23 was complemented with the wild type alr gene.12 Conversely, supplementing the culture media with D-alanine had no effect, the mc2155 and TAM23 global metabolomes are still significantly different (Figs. 2C and 2D and Table 1). The pronounced difference in the mc2155 and TAM23 metabolomes and the inability of supplemental D-alanine to reverse these metabolic differences eliminates the hypothesis that TAM23 is able to survive in the absence of D-alanine due to residual or defective Alr activity. Instead, the metabolic differences persist because a clear switch in the metabolic state of TAM23 ensues when TAM23 is cultured in the presence or absence of D-alanine. TAM23 cells enter an anabolic state when D-alanine is not readily available. Intriguingly, when D-alanine is available to TAM23, we observe a complete reversal in the flow of carbon through some metabolic processes. In effect, the addition of D-alanine circumvents the need for a metabolic response to replace the missing D-alanine due to the alr inactivation. Instead, TAM23 simply directs the D-alanine into peptidoglycan biosynthesis, which then allows the cell to pursue normal metabolic processes. Nevertheless, TAM23 still exhibits a distinct metabolism compared to mc2155. For instance, fatty acids are increased in TAM23 relative to mc2155, presumably to redirect fatty acids to increase cell wall integrity. The metabolites that exhibited a reversal in cellular concentrations based on the availability of D-alanine are colored in the metabolic pathways in Fig. 4. Indeed, supplementation with D-alanine induced a metabolic switch for TAM23 as revealed by the 1D 1H NMR metabolomics experiments (Fig. 4B).
Figure 4.
(A) Metabolic network map displays the 2D 1H-13C-HSQC time zero absolute metabolite concentration differences between M. smegmatis wild type mc2155 and alr insertion mutant TAM23 grown in MADC media and treated at mid exponential phase with [13C2]-D-alanine. A positive blue bar indicates a concentration increase in mc2155 relative to TAM23. A negative red bar indicates a concentration decrease in mc2155 relative to TAM23. The plotted concentrations are on a µM scale and the error bars represent standard deviations. Metabolites highlighted in magenta exhibited a reversal in concentration depending on whether media were supplemented with or without D-alanine from the 1D 1H NMR experiments and Table 1. L-alanine dehydrogenase (ALD), highlighted in red, was overexpressed in TAM23 relative to mc2155. The dark arrows identify the primary carbon flux pathway, blue arrows represent a hypothetical pathway for D-alanine biosynthesis that is depicted in detail in (B), and the dotted lines represent established alternative routes. (B) Metabolic network summarizing the 1D 1H NMR data and the L-D amino acid pools (see Fig. 1D and Table 1) comparing mc2155 with TAM23 from cultures without supplemental D-alanine. Metabolites colored blue were observed to increase in mc2155 relative to TAM23. Diagram adapted from Halouska et al. 2014.51
To further evaluate the metabolic response due to alr inactivation, we traced the distribution of 13C-carbon derived from [13C2]-D-alanine throughout the TAM23 and mc2155 metabolome using 2D 1H-13C HSQC NMR experiments (Figs. 3 and 4A). In M. tuberculosis and M. smegmatis, D-alanine can be directly converted into L-alanine (by Alr), into D-alanyl-D-alanine (by Ddl), into pyruvate (by an unidentified transaminase), or into alanyl-poly(glycerolphosphate)/D-alanine--poly(phosphoribitol). As expected, alr inactivation causes more of the supplemented D-alanine to be biosynthesized into D-alanyl-D-alanine in TAM23 relative to mc2155. More notably, a drastic decline is observed in the conversion of D-alanine into glutamate and pyruvate along with an accumulation of [13C2]-D-alanine in TAM23. This is consistent with an inactive Alr in TAM23 that prevents the D-alanine carbon from entering into other metabolic pathways besides peptidoglycan biosynthesis. In fact, a likely pathway is the conversion of D-alanine into pyruvate by the unidentified transaminase, which, apparently, is significantly less efficient than Alr in directing the supplemental D-alanine into central carbon metabolism. Conversely, the supplied D-alanine is not essential to peptidoglycan biosynthesis in mc2155 since Alr is still active. Instead, D-alanine can be used as an alternative carbon source by converting it into L-alanine, which is then diverted into other metabolic processes. We observed an increase in the conversion of [13C2]-D-alanine into glucose, fructose-6-phosphatate, pyruvate, glutamate, GABA, succinate aspartate and citrulline for mc2155 (Figs. 3 and 4A). This indicates an increase in the flow of carbon through glycolysis, the TCA cycle and urea cycle with a simultaneous decrease from peptidoglycan biosynthesis. Notably, the 1D 1H NMR experiments indicate an increase in the cellular concentration of citrulline, glutamate, and succinate for TAM23 with supplemental D-alanine. This apparent discrepancy highlights the fact that the different NMR experiments actually measure two distinctly different metabolic changes. The 1D 1H NMR experiments measure the total pool of these metabolites within the cell culture regardless of the carbon source. Conversely, the 2D 1H-13C HSQC experiment is only measuring the amount of citrulline, glutamate, and succinate derived from the supplied [13C2]-D-alanine. The observed changes in the concentrations of GABA and glutamate for TAM23 may also be indicative of a stress response resulting from an inactive Alr in our mutant strain especially in conditions without D-alanine.55 The decrease in GABA and glutamate derived from [13C2]-D-alanine are consistent with D-alanine being shunted into other critically needed metabolites, where other carbon sources may be used for GABA and glutamate production.
CONCLUSION
Taken together, these new data uniformly agree with the hypothesis of an inactive Alr in TAM23 and an alternative transaminase as a source of D-alanine for TAM23 (Fig. 4). Our results further confirm that Alr is not an essential enzyme in M. smegmatis. Since central metabolic pathways are conserved in mycobacteria, these results also suggest that Alr is not essential in M. tuberculosis and that Alr inhibitors will not have a bactericidal action.
Supplementary Material
Table S1. Proteins exhibiting a significant change in expression levels between TAM23 and mc2155 cells cultured in media without D-alanine.
Figure S1. Representative examples of 1D 1H NMR spectra of M. smegmatis metabolite extracts.
Figure S2. OPLS-DA model generated from the 1D 1H NMR spectra obtained from M. smegmatis metabolite extracts.
Figure S3. Overlay of 2D 1H-13C HSQC spectra generated from M. smegmatis metabolite extracts.
ACKNOWLEDGMENTS
We thank Dr. Renu Nandakumar and Dr. Nandakumar Madayiputhiya at the Proteomic and Metabolomics Core facility for providing the mudPIT proteomics data. This manuscript was supported in part by funds from the National Institute of Health (AI087668, P20 RR-17675, P30 GM103335), the USDA NIFA Station Animal Health Project (NEB 39–162), the American Heart Association (0860033Z), the UNL School of Veterinary Medicine and Biomedical Sciences, and the Nebraska Research Council. S. Halouska and R. J. Fenton were partially supported by O. Chacon’s NIH grant (R21 AI087561) to standardize NMR techniques included in this publication. K.K. was supported by the Undergraduate Creative Activities and Research Experience (UCARE) program at UNL. The research was performed in facilities renovated with support from the National Institutes of Health (RR015468–01).
Footnotes
ASSOCIATED CONTENT
Supporting information
The Supporting Information is available free of charge on the ACS Publications website at DOI:
The authors declare no competing financial interest.
REFERENCES
- (1).Global Tuberculosis Control Report; Who [World Health Organization]: 2015. [Google Scholar]
- (2).Gengenbacher M; Kaufmann SHE, Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol. Rev 2012, 36 (3), 514–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Belanger AE; Porter JC; Hatfull GF, Genetic analysis of peptidoglycan biosynthesis in mycobacteria: characterization of a ddlA mutant of Mycobacterium smegmatis. J. Bacteriol 2000, 182 (23), 6854–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Moat AG; Foster JW; Spector MP, Cell Structure and Function. In Microbial Physiology, John Wiley & Sons, Inc: 2003; pp 277–349. [Google Scholar]
- (5).Brennan PJ, Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2003, 83 (1–3), 91–7. [DOI] [PubMed] [Google Scholar]
- (6).Neuhaus FC, Selective inhibition of enzymes utilizing alanine in the biosynthesis of peptidoglycan. Antimicrob. Agents Chemother 1968, 1967 (7), 304–13. [PubMed] [Google Scholar]
- (7).Lambert MP; Neuhaus FC, Factors affecting the level of alanine racemase in Escherichia coli. J. Bacteriol 1972, 109 (3), 1156–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Walsh CT, Enzymes in the D-alanine branch of bacterial cell wall peptidoglycan assembly. J. Biol. Chem 1989, 264 (5), 2393–6. [PubMed] [Google Scholar]
- (9).van Heijenoort J, Biosynthesis of bacterial peptidoglycan unit. . In Bacterial Cell Wall, 1st ed.; Ghuysen JM; Hakenbeck R, Eds. Elsevier Medical Press: Amsterdam, 1994; Vol. 27, pp 39–54. [Google Scholar]
- (10).Chacon O; Bermudez LE; Zinniel DK; Chahal HK; Fenton RJ; Feng Z; Hanford K; Adams LG; Barletta RG, Impairment of D-alanine biosynthesis in Mycobacterium smegmatis determines decreased intracellular survival in human macrophages. Microbiology 2009, 155 (Pt 5), 1440–50. [DOI] [PubMed] [Google Scholar]
- (11).Chacon O; Feng Z; Harris NB; Caceres NE; Adams LG; Barletta RG, Mycobacterium smegmatis D-Alanine Racemase Mutants Are Not Dependent on D-Alanine for Growth. Antimicrobial agents and chemotherapy 2002, 46 (1), 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Halouska S; Chacon O; Fenton RJ; Zinniel DK; Barletta RG; Powers R, Use of NMR metabolomics to analyze the targets of D-cycloserine in mycobacteria: role of D-alanine racemase. Journal of proteome research 2007, 6 (12), 4608–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Halouska S; Fenton RJ; Zinniel DK; Marshall DD; Barletta RG; Powers R, Metabolomics analysis identifies d-Alanine-d-Alanine ligase as the primary lethal target of d-Cycloserine in mycobacteria. J Proteome Res 2014, 13 (2), 1065–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Milligan DL; Tran SL; Strych U; Cook GM; Krause KL, The alanine racemase of Mycobacterium smegmatis is essential for growth in the absence of D-alanine. J. Bacteriol 2007, 189 (22), 8381–8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Awasthy D; Bharath S; Subbulakshmi V; Sharma U, Alanine racemase mutants of Mycobacterium tuberculosis require D-alanine for growth and are defective for survival in macrophages and mice. Microbiology (Reading, U. K.) 2012, 158 (2), 319–327. [DOI] [PubMed] [Google Scholar]
- (16).Sassetti CM; Boyd DH; Rubin EJ, Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 2003, 48 (1), 77–84. [DOI] [PubMed] [Google Scholar]
- (17).Krzywinska E; Krzywinski J; Schorey JS, Naturally occurring horizontal gene transfer and homologous recombination in Mycobacterium. Microbiology (Reading, U. K.) 2004, 150 (6), 1707–1712. [DOI] [PubMed] [Google Scholar]
- (18).Veyrier F; Pletzer D; Turenne C; Behr MA, Phylogenetic detection of horizontal gene transfer during the step-wise genesis of Mycobacterium tuberculosis. BMC Evol. Biol 2009, 9, No pp. given. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Snapper SB; Melton RE; Mustafa S; Kieser T; Jacobs WR Jr., Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol 1990, 4 (11), 1911–9. [DOI] [PubMed] [Google Scholar]
- (20).Miller JH, Determination of Viable Cell Counts: Bacterial Growth Curves. In Experiments in Molecular Genetics, Cold Spring Harbor Laboratory: Cold Spring Harbor, N.Y, 1972; pp 31–36. [Google Scholar]
- (21).Perkins DN; Pappin DJC; Creasy DM; Cottrell JS, Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20 (18), 3551–3567. [DOI] [PubMed] [Google Scholar]
- (22).Zinniel DK; Fenton RJ; Halouska S; Powers R; Barletta RG, Sample Preparation of Mycobacterium tuberculosis Extracts for Nuclear Magnetic Resonance (NMR) Metabolomic Studies. Journal of Visualized Experiments 2012, 67, e3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Halouska S; Zhang B; Gaupp R; Lei S; Snell E; Fenton RJ; Barletta RG; Somerville GA; Powers R, Revisiting Protocols for the NMR Analysis of Bacterial Metabolomes. Journal of Integrated OMICS 2013, 2 (3), 120–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Simpson AJ; Brown SA, Purge NMR: effective and easy solvent suppression. J. Magn. Reson 2005, 175 (2), 340–346. [DOI] [PubMed] [Google Scholar]
- (25).Hu KF; Westler WM; Markley JL, Simultaneous Quantification and Identification of Individual Chemicals in Metabolite Mixtures by Two-Dimensional Extrapolated Time-Zero H-1-C-13 HSQC (HSQC(0)). Journal of the American Chemical Society 2011, 133 (6), 1662–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Hu K; Ellinger JJ; Chylla RA; Markley JL, Measurement of absolute concentrations of individual compounds in metabolite mixtures by gradient-selective time-zero 1H-13C HSQC with two concentration references and fast maximum likelihood reconstruction analysis. Anal Chem 2011, 83 (24), 9352–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Worley B; Powers R, MVAPACK: A Complete Data Handling Package for NMR Metabolomics. ACS Chem. Biol 2014, 9 (5), 1138–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Worley B; Powers R, Simultaneous phase and scatter correction for NMR datasets. Chemom. Intell. Lab. Syst 2014, 131, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Worley B; Powers R, Generalized adaptive intelligent binning of multiway data. Chemom. Intell. Lab. Syst 2015, 146, 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Worley B; Halouska S; Powers R, Utilities for quantifying separation in PCA/PLS-DA scores plots. Anal Biochem 2013, 433 (2), 102–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Werth MT; Halouska S; Shortridge MD; Zhang B; Powers R, Analysis of Metabolomic PCA Data using Tree Diagrams. Analytical Biochemistry 2010, 399 (1), 56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Delaglio F; Grzesiek S; Vuister GW; Zhu G; Pfeifer J; Bax A, NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR 1995, 6 (3), 277–93. [DOI] [PubMed] [Google Scholar]
- (33).Johnson BA, Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods in molecular biology 2004, 278, 313–52. [DOI] [PubMed] [Google Scholar]
- (34).Wishart DS; Jewison T; Guo AC; Wilson M; Knox C; Liu Y; Djoumbou Y; Mandal R; Aziat F; Dong E; Bouatra S; Sinelnikov I; Arndt D; Xia J; Liu P; Yallou F; Bjorndahl T; Perez-Pineiro R; Eisner R; Allen F; Neveu V; Greiner R; Scalbert A, HMDB 3.0--The Human Metabolome Database in 2013. Nucleic acids research 2013, 41 (Database issue), D801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Cui Q; Lewis IA; Hegeman AD; Anderson ME; Li J; Schulte CF; Westler WM; Eghbalnia HR; Sussman MR; Markley JL, Metabolite identification via the Madison Metabolomics Consortium Database. Nature biotechnology 2008, 26 (2), 162–4. [DOI] [PubMed] [Google Scholar]
- (36).Akiyama K; Chikayama E; Yuasa H; Shimada Y; Tohge T; Shinozaki K; Hirai MY; Sakurai T; Kikuchi J; Saito K, PRIMe: a Web site that assembles tools for metabolomics and transcriptomics. In silico biology 2008, 8 (3–4), 339–45. [PubMed] [Google Scholar]
- (37).Okuda S; Yamada T; Hamajima M; Itoh M; Katayama T; Bork P; Goto S; Kanehisa M, KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res 2008, 36 (Web Server issue), W423–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Caspi R; Foerster H; Fulcher CA; Kaipa P; Krummenacker M; Latendresse M; Paley S; Rhee SY; Shearer AG; Tissier C; Walk TC; Zhang P; Karp PD, The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic acids research 2008, 36 (Database issue), D623–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Bylesjo M; Rantalainen M; Cloarec O; Nicholson JK; Holmes E; Trygg J, OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. Journal of Chemometrics 2006, 20 (8–10), 341–351. [Google Scholar]
- (40).Woodruff PJ; Carlson BL; Siridechadilok B; Pratt MR; Senaratne RH; Mougous JD; Riley LW; Williams SJ; Bertozzi CR, Trehalose is required for growth of Mycobacterium smegmatis. J Biol Chem 2004, 279 (28), 28835–43. [DOI] [PubMed] [Google Scholar]
- (41).Copie V; Faraci WS; Walsh CT; Griffin RG, Inhibition of alanine racemase by alanine phosphonate: detection of an imine linkage to pyridoxal 5’-phosphate in the enzyme-inhibitor complex by solid-state 15N nuclear magnetic resonance. Biochemistry 1988, 27 (14), 4966–70. [DOI] [PubMed] [Google Scholar]
- (42).Lambert MP; Neuhaus FC, Mechanism of D-cycloserine action: alanine racemase from Escherichia coli W. J Bacteriol 1972, 110 (3), 978–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Silverman RB, The potential use of mechanism-based enzyme inactivators in medicine. J Enzyme Inhib 1988, 2 (2), 73–90. [DOI] [PubMed] [Google Scholar]
- (44).Feng Z; Caceres NE; Sarath G; Barletta RG, Mycobacterium smegmatis L-alanine dehydrogenase (Ald) is required for proficient utilization of alanine as a sole nitrogen source and sustained anaerobic growth. J. Bacteriol 2002, 184 (18), 5001–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Thompson RJ; Bouwer HGA; Portnoy DA; Frankel FR, Pathogenicity and immunogenicity of a Listeria monocytogenes strain that requires D-alanine for growth. Infect. Immun 1998, 66 (8), 3552–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Kwak M-S; Lee S-G; Jeong S-C; Suh S-H; Lee J-H; Jeon Y-J; Kim Y-H; Sung M-H, Screening and taxonomic characterization of D-amino acid aminotransferase-producing thermophiles. Sanop Misaengmul Hakhoechi 1999, 27 (3), 184–190. [Google Scholar]
- (47).Taylor PP; Fotheringham IG, Nucleotide sequence of the Bacillus licheniformis ATCC 10716 dat gene and comparison of the predicted amino acid sequence with those of other bacterial species. Biochim. Biophys. Acta, Gene Struct. Expression 1997, 1350 (1), 38–40. [DOI] [PubMed] [Google Scholar]
- (48).Berger BJ; English S; Chan G; Knodel MH, Methionine regeneration and aminotransferases in Bacillus subtilis, Bacillus cereus, and Bacillus anthracis. J. Bacteriol 2003, 185 (8), 2418–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Kobayashi J; Shimizu Y; Mutaguchi Y; Doi K; Ohshima T, Characterization of D-amino acid aminotransferase from Lactobacillus salivarius. J. Mol. Catal. B: Enzym 2013, 94, 15–22. [Google Scholar]
- (50).Lee S-G; Hong S-P; Song JJ; Kim S-J; Kwak M-S; Sung M-H, Functional and structural characterization of thermostable D-amino acid aminotransferases from Geobacillus spp. Appl. Environ. Microbiol 2006, 72 (2), 1588–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Halouska S; Fenton RJ; Zinniel DK; Marshall DD; Barletta RG; Powers R, Metabolomics analysis identifies D-Alanine-D-Alanine ligase as the primary lethal target of D-cycloserine in mycobacteria. J. Proteome Res 2014, 13 (2), 1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Amon J; Braeu T; Grimrath A; Haenssler E; Hasselt K; Hoeller M; Jessberger N; Ott L; Szoekoel J; Titgemeyer F; Burkovski A, Nitrogen control in Mycobacterium smegmatis: nitrogen-dependent expression of ammonium transport and assimilation proteins depends on the OmpR-type regulator GlnR. J. Bacteriol 2008, 190 (21), 7108–7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Jenkins VA; Barton GR; Robertson BD; Williams KJ, Genome wide analysis of the complete GlnR nitrogen-response regulon in Mycobacterium smegmatis. BMC Genomics 2013, 14, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Williams KJ; Jenkins VA; Barton GR; Bryant WA; Krishnan N; Robertson BD, Deciphering the metabolic response of Mycobacterium tuberculosis to nitrogen stress. Mol. Microbiol 2015, 97 (6), 1142–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Galamba A; Soetaert K; Buyssens P; Monnaie D; Jacobs P; Content J, Molecular and biochemical characterisation of Mycobacterium smegmatis alcohol dehydrogenase C. FEMS Microbiol Lett 2001, 196 (1), 51–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Proteins exhibiting a significant change in expression levels between TAM23 and mc2155 cells cultured in media without D-alanine.
Figure S1. Representative examples of 1D 1H NMR spectra of M. smegmatis metabolite extracts.
Figure S2. OPLS-DA model generated from the 1D 1H NMR spectra obtained from M. smegmatis metabolite extracts.
Figure S3. Overlay of 2D 1H-13C HSQC spectra generated from M. smegmatis metabolite extracts.