Abstract
Despite impressive durable responses, immune checkpoint inhibitors do not provide a long-term benefit to the majority of patients with cancer. Understanding genomic correlates of response and resistance to checkpoint blockade may enhance benefits for patients with cancer by elucidating biomarkers for patient stratification and resistance mechanisms for therapeutic targeting. Here we review emerging genomic markers of checkpoint blockade response, including those related to neoantigens, antigen presentation, DNA repair, and oncogenic pathways. Compelling evidence also points to a role for T cell functionality, checkpoint regulators, chromatin modifiers, and copy-number alterations in mediating selective response to immune checkpoint blockade. Ultimately, efforts to contextualize genomic correlates of response into the larger understanding of tumor immune biology will build a foundation for the development of novel biomarkers and therapies to overcome resistance to checkpoint blockade.
Tumors have long been known to generate varying levels of immune response, a property termed immunogenicity. Early studies revealed that certain cancers maintained high levels of tumor-infiltrating lymphocytes (TILs), associated with improved prognosis1,2. TILs have been shown to be dysfunctional and to express multiple co-inhibitory or checkpoint receptors, such as cytotoxic T lymphocyte antigen-4 (CTLA-4, CD152), programmed cell death-1 (PD-1, CD279), T cell immunoglobulin and mucin-domain containing-3 (TIM-3), and lymphocyte-activation gene (LAG-3)3–5. The insight that blockade of these checkpoints can lead to reversal of TIL dysfunction with improved cytotoxicity and proliferative capacity of these cells has changed cancer treatment paradigms (Box 1). Immune checkpoint inhibitors, including monoclonal antibodies against PD-1 and CTLA-4, have generated durable responses across many tumor types6–13, leading to a number of Food and Drug Administration (FDA)-approved agents, with many others in the clinical trial pipeline. However, the majority of patients do not respond to checkpoint blockade, so predicting among patients the subset that will benefit from checkpoint inhibitors, either alone or in combination with other agents, remains a challenge.
Box 1 |. Tumor immunogenicity.
The foundation of anti-tumor immunity rests on the generation or reactivation of cytotoxic T cell responses. T cell activation is a highly coordinated and regulated activity, requiring initial stimulation through both the T cell receptor (TCR) and co-stimulatory molecules, such as CD28, a protein expressed on T cells that interacts with the ligands B7–1 (CD80) and B7–2 (CD86) on antigen-presenting cells. CTLA-4 competitively binds with high affinity to these ligands to limit initial co-stimulatory signals in lymph nodes184. Although murine models initially suggested that anti-CTLA-4 therapy also depleted regulatory T cells185–187, which constitutively express CTLA-4, recent human studies have shown conflicting results188,189. PD-1, in comparison, is induced following initial T cell activation to regulate T cells190. PD-1 binds the ligands PD-L1 and PD-L2 to attenuate TCR signaling191, thereby leading to decreased T cell proliferation, cytotoxicity, and cytokine production192.
Select tumors express high levels of the PD-1-binding ligand PD-L1, and initial trials of anti-PD-1 therapy found that PD-L1 expression, as detected by immunohistochemistry, correlated with response to therapy6,7,14, thereby leading the FDA to approve PD-L1 companion diagnostic tests for anti-PD-1/PD-L1 therapies in some cancers15. Additional studies of these histological markers showed that the association with response varied over time and by tumor type, with PD-L1 positivity distinguishing responders in some settings and not others12,16–18. Importantly, subsequent trials have demonstrated that a sizable portion of responses occurred in patients with PD-L1-negative tumors8,11,19–21. These findings have prompted efforts to identify other determinants of response to checkpoint blockade, including non-genomic factors, such as the gut microbiome, environmental influences, and metabolic pathways, reviewed elsewhere22–26.
Pivotal investigations of the cancer genome have uncovered oncogenic mutations underlying selective growth advantage, tumor suppressor inactivation, and tumorigenesis initiation, among others27–29. While such approaches have traditionally focused on genomic changes within tumor cells alone as a measure of responsiveness to small-molecule inhibitors or monoclonal antibodies, application of these strategies to immune checkpoint blockade requires consideration of tumor cells in conjunction with distinct immune cell populations and non-immune, non-tumor cells, such as stromal and endothelial cells. In this Review, we present a framework for emerging genomic correlates of clinical responses to immune checkpoint inhibitors (Fig. 1 and Table 1), discuss potential therapeutic applications of these findings, and outline crucial future studies needed to advance this field.
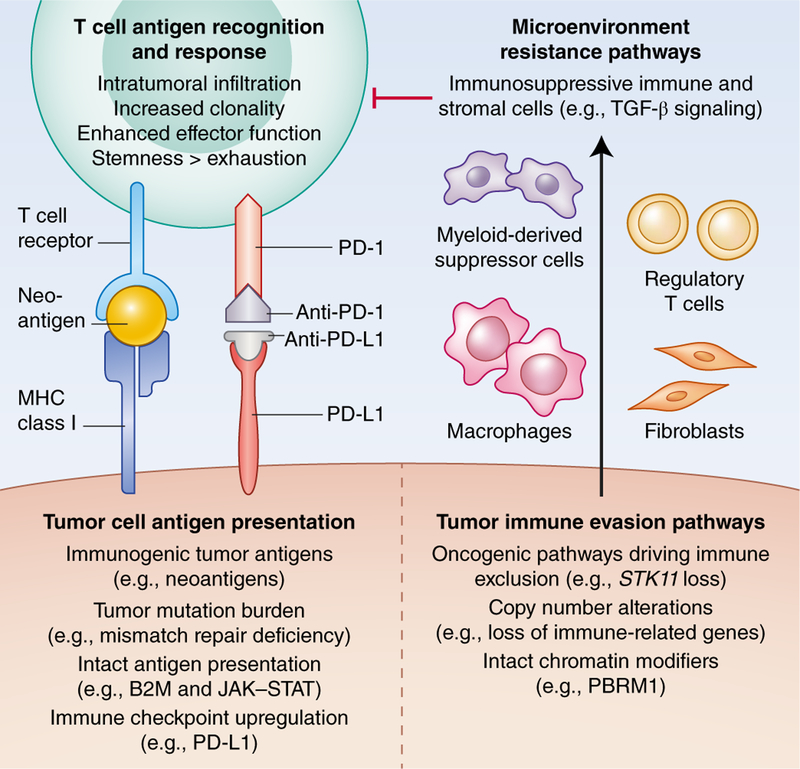
Fig. 1 |. Framework for genomic correlates of response to immune checkpoint blockade within the tumor immune microenvironment.
The left side outlines correlates of response, focusing on antigen presentation and recognition; the right side delineates resistance pathways that promote tumor immune evasion and induce immunosuppressive cells, which in turn inhibit the T cell-mediated anti-tumor response. Credit: Debbie Maizels/Springer Nature
Table 1 |.
Genomic correlates of response and resistance organized by primary location
| Primary location | Response category | Defining characteristics or examples |
|---|---|---|
| T cell | Intratumoral infiltration85,115,135–137,139 | Transcriptional signatures of cytotoxic lymphocytes infiltrating the tumor core |
| Enhanced effector function52,134,141 | Increased expression of PRF1, GZMA/B, CD8A, and IFNG | |
| Increased clonality14,144,41 | Ranging from 0 to 1, with 1 indicating a monoclonal population | |
| Greater sternness147,150 | Express chemokine receptor CXCR5 and transcription factor TCF7; lack TIM-3/CD39 | |
| Reduced exhaustion147,150 | Express co-inhibitory receptor TIM-3 and ectonucleotidase CD39; lack CXCR5/TCF7 | |
| Tumor cell (response mechanisms) | Tumor antigens31,32,34–40,54,57,65,67 | Neoantigens, viral antigens |
| Increased tumor mutation burden9,37,48 | Mismatch repair deficiency | |
| Immunogenic alterations159 | Inactivating mutations in SERPINB3 and SERPINB4 | |
| Mutational signatures39,53,108 | Smoking, ultraviolet light, alkylating agent therapy, APOBEC | |
| Genomic upregulation of PD-L1 (refs.50,92–94,97–100) |
PDL1 amplification and loss of CDK4, SPOP, and CMTM4 and CMTM6 | |
| Chromatin modifier loss152,154,157,158 | Inactivating mutations in PBRM1, ARID1A, and SMARCA4 | |
| Tumor cell (resistance mechanisms) | Tumor antigens68 | Cancer/testis antigens similar to self and less immunogenic |
| Deficient antigen presentation37,53 | Inactivating mutations in B2M, HLA, JAK/STAT, and IFN-γ response genes | |
| Oncogenicpathways45,113–115,117,118,124,125,129,130,133 | Inactivating STK11 and PTEN mutations, WNT/β-catenin, EGFR and KRAS mutations | |
| Immune evasion alterations141 | Increased expression of SERPINB9 | |
| CNAs144,160 | High levels of copy-number loss, chromosome arm and whole-chromosome CNAs | |
| Microenvironment | Immunosuppressive stromal cells115,123,126,140 | Transcriptional signatures of fibroblasts, endothelial cells, and TGF-β signaling |
| Immunosuppressive immune cells136,141 | Transcriptional signatures of myeloid-derived suppressor cells and regulatory T cells |
Tumor antigens and mutations
In order to avoid self-reactivity and the development of autoimmunity, the immune system discriminates self from non-self antigens. Tumors may express aberrant antigens that, in turn, may be recognized by the immune system. These include proteins normally expressed in immune-privileged sites, such as cancer/testis antigens; virally expressed proteins, as in the case of Epstein-Barr virus or human-papillomavirus-associated cancers; and proteins encoded by endogenous retroviruses, which are germline genomic elements resembling retroviruses that are transcriptionally repressed in normal cells but expressed in many tumor cells30–32. Furthermore, tumors contain the cumulative sum of thousands of mutations, a fraction of which alter the amino acid sequence of encoded proteins. These mutated proteins, unique to the tumor and not present in normal cells, are known as neoantigens25,30. Neoantigens are often specific to each tumor and are unlikely to be shared widely across tumors, even those with the same histology. If neoantigen epitopes are presented to the immune system, these epitopes generate an adaptive immune response selective for cancer cells that can be enhanced and strengthened by checkpoint blockade30,33.
Neoantigens and tumor mutational burden.
A diversity of neoantigens may be necessary but not sufficient for clinical responses to immune checkpoint inhibitors. Mouse models of sarcoma that respond to anti-CTLA-4 and/or anti-PD-1 therapy have shown reactivation of neoantigen-specific T cells (Box 1)34,35, and neoantigen-specific T cell reactivity has also been observed through tumor exome analysis in patients with advanced cancers who respond to checkpoint inhibitors36,37. The role of neoantigens in the response to immune checkpoint inhibitors has been further substantiated by studies showing that higher neoantigen load is associated with response to CTLA-4 and PD-1 blockade in patients with melanoma and non-small-cell lung cancer38–40, as well as analyses of acquired resistance in patients with non-small-cell lung cancer demonstrating loss of neoantigens through subclone elimination and chromosomal deletions in resistant tumors41.
As neoantigens are computationally predicted from somatic variant data, tumor mutational burden, measured as the number of nonsynonymous mutations per megabase sequenced, has therefore similarly been associated with improved responses to checkpoint blockade, initially in studies of melanoma and non-small-cell lung cancer, two of the tumor types with the highest mutation number38,39,42. Larger studies and phase III trials have confirmed that high tumor mutational burden correlates with enhanced checkpoint inhibitor responses and improved overall survival in certain tumor types, such as head and neck cancer43, urothelial carcinoma9, and lung cancer44–48, including small-cell lung cancer48. However, tumor mutational burden is an incomplete correlate of checkpoint blockade response in that mutation load distributions overlap considerably between responders and nonresponders38,39,42. Therefore, even the most ideal receiver operating charcteristic curve cutoffs for tumor mutational burden in melanoma and lung cancer have low statistical sensitivity and specificity for predicting clinical benefit from checkpoint inhibitors49. Furthermore, tumor mutational burden does not correlate with response in other tumor types, including Hodgkin’s lymphoma50–51, renal cell carcinoma52–53, and virally mediated tumors, such as polyomavirus-associated Merkel-cell carcinoma54–56, for which other factors likely drive responses to immune checkpoint inhibitors.
While a higher mutation and neoantigen load may reflect an increased likelihood of a T cell response being generated against that tumor, this has not necessarily correlated with the identification of circulating T cells that recognize the proposed neoepitope. Mouse models of sarcoma have demonstrated that anti-CTLA-4 and anti-PD-1 therapies reactivated neoantigen-specific CD8 + T cells to achieve tumor rejection34,35, and patients with cancers that responded to checkpoint inhibitors displayed neoantigen-specific T cell reactivity36,37,57. In both settings, the number of identified neoantigen-specific T cells was much lower than the predicted number of neo-epitopes. Whether this reflects underlying thymic deletion of tumor-reactive T cells with partial self-reactivity or a lack of sensitivity to identify the neoantigen-specific T cell population remains to be understood.
Neoantigen prediction.
Refining neoantigen prediction algorithms for neoepitopes may improve the ability to identify patients who are most likely to respond to checkpoint blockade. Initial techniques to identify the peptide sequences most likely to be presented by major histocompatibility complex (MHC) class I proteins relied on in silico binding assays followed by in vitro testing with T cell clones58,59. Subsequent efforts that were focused on large-scale mass-spectrometry approaches have led to the development of more advanced prediction algorithms60,61. Similar efforts to identify class II neoepitopes have lagged behind due to, at least in part, the longer amino acid sequences and less stringent binding affinity rules required for class II binding62. Recently, methods to detect class II epitopes have been developed using neural network analysis of data derived from proteomic experiments and single-cell T cell receptor (TCR) sequencing63. Improved prediction algorithms for both class I and II epitopes may enable a precision-medicine approach for neoantigen prioritization to be tested at an individual-patient level.
Neoantigen and mutation clonality.
The relative clonality of neoantigens and mutations, estimated by the portion of tumor cells harboring a mutation, has also been linked to response. In one study, clonal neoantigens, arising from founder mutations present in all or nearly all tumor cells, conferred sensitivity to PD-1 and CTLA-4 blockade in lung cancer and melanoma, while subclonal neoantigens, which arise from passenger mutations present in a subset of tumor cells, predominated in non-responders57. Similarly, clonal neoantigens and mutations occurred recurrently in responding tumors in a meta-analysis of 249 tumors, predominantly melanoma and non-small-cell lung cancer, that were treated with immune checkpoint blockade53. This correlation between neoantigen clonality and checkpoint blockade response may be explained by immu-noediting, in which immunogenic neoantigens trigger an antitumor immune response that then eliminates these neoantigens64. This phenomenon may thus restrict intratumor genetic heterogeneity and promote clonal dominance of nonimmunogenic neoantigens, an effect enhanced by checkpoint blockade in mouse models of cancer64. Neoantigen clonality may therefore identify tumors with less sophisticated immune evasion strategies than those of nonresponding tumors. Overall, neoantigen clonality needs additional validation before it can be used as a clinical biomarker.
Tumor antigen immunogenicity.
Furthermore, the similarity of neoantigens and other tumor antigens to non-self antigens, such as those expressed by viruses and other pathogens, may enhance immune recognition and checkpoint blockade response. Recent efforts have focused on quantifying the foreignness of neoantigens by comparing the neoantigen peptide sequences with those of immune epitopes that have been experimentally validated as targets of adaptive immune responses65,66. In one neoantigen fitness model, tumors with more immunogenic neoantigens, which had greater homology with infectious-disease-derived peptides, were defined as having lower fitness65,66. Patients with low-fitness melanoma and lung cancer tumors had longer survival after CTLA-4 and PD-1 blockade than patients with high-fitness tumors65. A similar concept may explain the improved responses to checkpoint blockade that were observed in virus-associated cancers, which have more immunogenic non-self antigens31,54. Likewise, tumors with aberrant expression of germline endogenous retroviruses, which are transcriptionally repressed in normal cells, have demonstrated greater immune cytolytic activity and more frequent responses to checkpoint blockade32,67. Altogether, the foreignness of tumor antigens may prove to be a stronger driver of checkpoint inhibitor response than the quantity of neoantigens.
Cancer/testis antigens.
Cancer/testis antigens, whose normal expression is restricted to developing reproductive tissues30,33, represent an alternative class of tumor antigens in that they consist of nonmutated self peptides that induce incomplete T cell tolerance and thus promote tumor immune evasion. Cancer/testis antigens are commonly expressed in many tumor types and have recently been implicated in resistance to immune checkpoint blockade. In metastatic melanoma, anti-CTLA-4 non-responders, but not anti-PD-1 non-responders harbored upregulated gene expression of a subcluster of melanoma-associated antigen (MAGE)-A cancer-germline antigens68. Notably, MAGE-A protein expression correlated with decreased expression of autophagy proteins, indicating that MAGE-A proteins may suppress autophagy to prevent T cell priming and avert immune recognition68. Whether other cancer/testis antigens similarly trigger immune tolerance and checkpoint blockade resistance remains to be determined.
Antigen presentation
HLA pathway alterations.
Anti-tumor immunity requires T cell recognition of the tumor epitope when presented on the MHC, a process known as antigen presentation. Human leukocyte antigen (HLA) genes HLA-A, HLA-B, and HLA-C encode the MHC class I proteins that present intracellular peptides on the cell surface to TCRs and require β−2 microglobulin (B2M) for stabilization on the cell surface (Fig. 2a)69. While B2M loss was initially observed in tumor immune escape after adoptive T cell therapies70, B2M mutations and loss of heterozygosity have more recently been demonstrated to be innate and acquired resistance mechanisms to CTLA-4 and PD-1 inhibitors in cohorts of patients with melanoma71, as well as in one or two patients who had melanoma, gastrointestinal cancer, or lung cancer37,72,73. These clinical observations are backed by preclinical experiments in which CRISPR-mediated knockout of B2m conferred resistance to PD-1 blockade in mouse models73,76. The heterogeneity of alterations in B2M and tumor types in which they are found suggest that this mechanism of resistance may prove to be more widespread as more studies are performed.
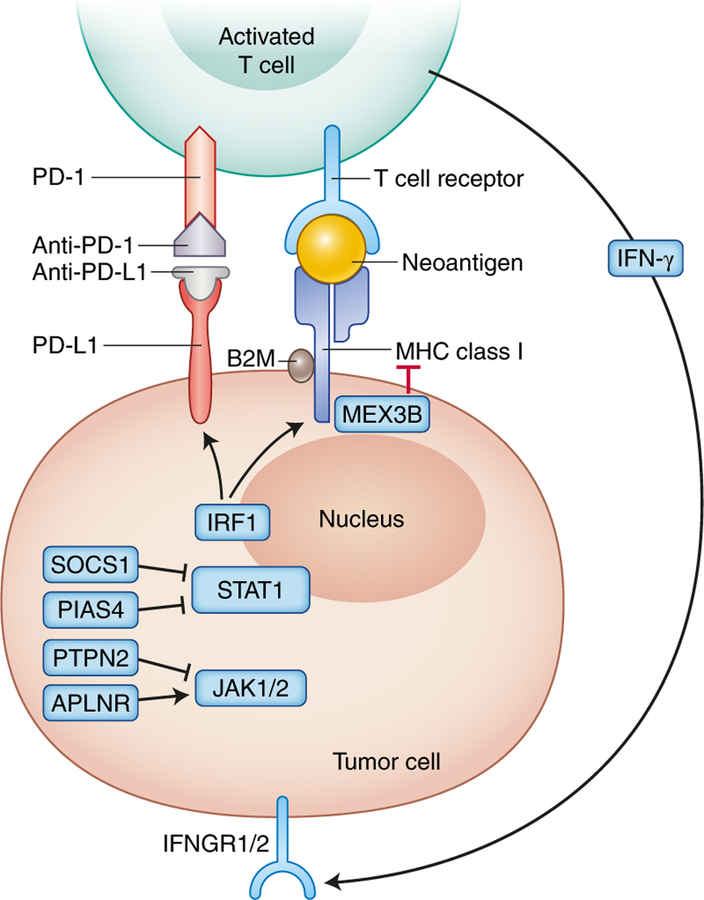
Fig. 2 |. Antigen presentation and genomic correlates of response to immune checkpoint blockade.
HLA genes HLA-A, HLA-B, and HLA-C encode the MHC class I proteins that present intracellular peptides on the cell surface to T cell receptors (TCRs) and require B2M for stabilization on the cell surface. IFN-γ, which can be released by activated T cells, binds IFNGR1/2 on tumors, which triggers JAK-STAT signaling that activates IFN response genes, including IRF1, which induces the transcription of other genes to increase the surface expression of PD-L1 and MHC molecules. Genomic mechanisms of checkpoint blockade response include upregulation of IFN-γ response gene expression85,86, loss of protein tyrosine phosphatase non-receptor type 2 (PTPN2)74, and germline HLA heterozygosity82. Conversely, genomic pathways of resistance include B2M loss37,71–73, somatic HLA loss of heterozygosity82, reduced HLA gene expression84, increased expression of MEX3B83, IFNGR1/2 or IRF1copy-number loss87, JAK1/2 inactivation72,88,89, APLNR (apelin receptor) loss76, and amplification of negative regulators of the IFN-γ pathway, including SOCS1 (suppressor of cytokine signaling 1) and PIAS4 (protein inhibitor of activated STAT4)87. Credit: Debbie Maizels/Springer Nature
Similarly, mutations within HLA genes, loss of diversity of HLA alleles, or alterations in HLA expression may diminish responses to checkpoint blockade. The extreme polymorphism of HLA genes has led to the concept that broader HLA diversity confers a survival advantage by promoting recognition by T cells of larger numbers of microbial and tumor antigens77,78. An individual’s HLA repertoire may exert a negative selection pressure on tumor mutations in that patient-specific MHC I and MHC II genotypes correlate with tumor mutations that produce neoantigens least likely to be presented, thus indicating immune evasion79,80. Likewise, the presence of HLA mutations directly correlated with neoantigen frequency and TIL quantity in colorectal tumors, suggesting that positive selection was occurring for HLA mutations in tumors under immune attack81. HLA loss of heterozygosity was similarly associated with a high burden of subclonal neoantigens that were predicted to bind to the lost HLA alleles, implying that there was enhanced immune evasion after the loss of heterozygosity event57.
In the clinic, broader HLA diversity enhanced immune recognition of tumor neoantigens and improved overall survival in individuals with cancer treated with immune checkpoint inhibitors82. Similarly, higher expression of MEX3B, a gene encoding an RNA-binding protein that downregulates HLA-A expression (Fig. 2a), was associated with primary resistance to anti-PD-1 therapy in mel-anoma83. Finally, in patients with melanoma who were treated with immune checkpoint inhibitors, loss of MHC class I protein expression on tumor cells correlated with transcriptional downregulation of HLA-A, HLA-B, HLA-C, and B2M and primary resistance to anti-CTLA-4 therapy, while MHC class II protein expression predicted response to anti-PD-1 therapy, again highlighting the importance of HLA expression in checkpoint blockade response84.
Interferon-γ pathway alterations.
Expression levels of HLA molecules and response to checkpoint blockade may in part be driven by signaling through the interferon (IFN)-γ receptor. As shown in Fig. 2b, IFN-γ, released by activated T cells upon recognizing neoantigens, binds IFN-γ receptors on tumors, which triggers Janus kinase 1 (JAK1) 1 and JAK2 and signal transducers and activators of transcription (STAT) signaling that activates IFN-related genes, including interferon regulatory factor 1 (IRF1), which induces the transcription of other genes to increase the surface expression of PD-L1 and MHC molecules. Gene expression from 220 anti-PD-1-treated patients with nine cancer types revealed a T-cell-inflamed signature that included upregulated IFN-γ response that was necessary but not always sufficient for clinical benefit from therapy85. Similarly, enriched expression of genes related to antigen presentation and the IFN-γ pathway was an early on-treatment signature of melanoma tumor response to anti-PD-1 therapy after prior progression on anti-CTLA-4 therapy86.
In addition to overall expression, copy-number alterations (CNAs) and mutations within the IFN-γ pathway have been identified as a mechanism of checkpoint inhibitor resistance. Of 12 melanoma tumors with primary resistance to anti-CTLA-4 therapy, 75% harbored genomic defects in the IFN-γ pathway, including copy-number loss of interferon-γ receptor 1 (IFNGR1), IFNGR2, IRF1, and JAK2, as well as amplification of IFN-γ pathway inhibitors, such as suppressor of cytokine signaling 1 (SOCS1) and protein inhibitor of activated STAT4 (PIAS4) (Fig. 2b)87. Likewise, significant enrichment of IFN-γ CNAs occurred in non-responding compared with responding tumors in a meta-analysis of 249 checkpoint-inhibitor-treated tumors53. Collectively, these results highlight the importance of antigen presentation and IFN-γ pathway genes in response to immune checkpoint blockade.
Mutations in the JAK-STAT pathway have similarly been identified in primary and acquired resistance to immune checkpoint inhibitors, albeit at a lower frequency than IFN-γ expression changes. JAK1 or JAK2 mutations have been reported in small numbers of patients with metastatic melanoma and MMR-deficient colon cancer that demonstrated primary or acquired resistance to anti-PD-1 therapy72,88,89. Although JAK1 and jAk2 are canonically thought to act downstream of the IFN-γ receptor, other JAK-STAT pathway components may be involved in PD-L1 induction, as demonstrated by genomic profiling of a lung adenocarcinoma tumor responding to anti-PD-L1 therapy, which revealed activating germline and somatic JAK3 mutations that were associated with increased PD-L1 expression in response to IFN-γ (ref.90). These clinical observations prompted preclinical CRISPR-Cas9 screens that aimed to identify regulators of the IFN-γ pathway74–76, which require validation in patients. These findings suggest that the full spectrum of factors mediating IFN-γ resistance is not fully understood and highlight this as an area to investigate for therapeutic targets to boost checkpoint blockade response.
PDL1 amplification and regulation
PDL1 amplification.
Amplification of the chromosome 9p24.1 locus, the region containing PDL1, PDL2, and JAK2, was first linked to increased expression of PD-L1 on the Reed-Sternberg cells of Hodgkin’s lymphoma91. Augmented JAK-STAT signaling and PD-L1 overexpression may contribute to impressive anti-PD-1 response rates of 69–87% in relapsed or refractory classic Hodgkin’s lymphoma50,92,93. PD-L1 overexpression has also been correlated with CNAs in 9p24.1 in primary central nervous system B cell lymphoma and primary testicular B cell lymphoma, with corresponding high responses to anti-PD-1 therapy94. Conversely, PD-L1 overexpression in T cell non-Hodgkin lymphoma has been linked to poor outcomes—specifically, rapid progression in three patients with adult T cell leukemia-lymphoma treated with PD-1 inhibitors95. These clinical observations may be explained by the role of PD-1 as a tumor suppressor in T cell lymphomas and particularly by the ability of PD-1 to block oncogenic TCR signaling96. Overall, these findings caution against employing PD-1 blockade in tumors driven by oncogenic T cell signaling.
The frequency of PDL1 amplification in solid tumors, in comparison, is reported to be much lower, with a prevalence of 0.7% in over 118,000 sequenced tumors, inclusive of 100 solid tumor histologic types97. Enrichment was seen in particular tumor types, including hepatocellular cholangiocarcinoma, head and neck squamous cell, nasopharyngeal carcinoma, undifferentiated soft-tissue sarcoma, kidney sarcomatoid, and thyroid anaplastic carcinoma, of which 12 of 13 patients with clinical annotation had co-amplification of JAK2 (ref.97). PDL1-amplified tumors did not always have high PD-L1 expression as determined by immunohistochemistry, echoing an earlier report in head and neck squamous cell tumors96. Within a clinically annotated cohort, 9 of 13 patients with identified PDL1 or PDL2 amplification were treated with PD-1/PD-L1 inhibitors either alone or in combination with CTLA-4 or an investigational agent and had an objective response rate of 67% and a median progression-free survival of 15 months97. This small cohort illustrates the challenges of widespread applicability given the low prevalence of PDL1 CNAs, emphasizing that this finding is preliminary
PD-L1 regulation.
Since PD-L1 associates with response to checkpoint blockade6,7, understanding PD-L1 regulation may reveal novel molecular mechanisms that could be targeted to overcome resistance to immune checkpoint inhibitors. Recent work has demonstrated that cyclin D-cyclin dependent kinase 4 (CDK4) and speckle-type POZ (pox virus and zinc finger) protein (SPOP) regulate PD-L1 protein abundance by promoting proteasome-mediated degradation of PD-L1 and that CDK4 and CDK6 (CDK4/6) inhibitors increased PD-L1 protein levels and enhanced response to PD-1 inhibitors in preclinical models of cancer98. In 97 primary human prostate cancers, 15 tumors with SPOP mutations had stronger PD-L1 staining and a lower number of TILs than tumors without SPOP mutations98. Additional PD-L1 regulators have been revealed by functional genomic screens in human cancer cell lines. Specifically, haploid genetic screens and CRISPR-Cas9 knockouts revealed that CMTM4 and CMTM6 (CMTM4/6), two closely related transmembrane proteins, reduced PD-L1 protein ubiquitination and enhanced T cell inhibition by PD-L1-expressing tumors99. Another similar CRISPR-Cas9 screen also identified CMTM6 as a regulator that prevents PD-L1 protein degradation and showed that CMTM6 depletion improved tumor-specific T cell activity in vitro and in vivo100. Together, these findings on PD-L1 regulators have important therapeutic implications in the enhancement of immune checkpoint blockade response. CDK4/6 inhibitors are already being investigated with PD-1/PD-L1 inhibitors in clinical trials, and CMTM4/6 first requires validation in human tumors.
DNA repair and mutational signatures
Mismatch repair deficiency and other DNA repair alterations.
Genomic analyses have also shown that mutational processes that directly alter DNA damage response and repair pathways influence response to immune checkpoint blockade101. Mismatch repair (MMR) deficiency has emerged as a robust indicator of response to immune checkpoint blockade37,40. Alterations in genes encoding MMR proteins, including MLH1, MSH2, MSH6, and PMS2, lead to uncorrected DNA replication errors and a large number of mutations, particularly small insertions and deletions in repetitive DNA sequences called microsatellites61. These genomic alterations often cause frameshift mutations that produce immunogenic neoantigens that are recognized and targeted by T cells102,103. Phase II clinical trials of single-agent PD-1 blockade in patients with MMR deficient tumors have demonstrated impressive response rates of 40–70% with durable progression-free and overall survival37,40. These results culminated in the first FDA approval of a cancer therapy based on a genomic biomarker regardless of tumor histology and site of origin. Notably, patients with primary resistance had similar tumor mutation burden as patients with responses, and responding patients had transient expansions of T cell clones specific for neoantigens that bound MHC proteins with > 100-fold higher affinity than wild-type peptides37, highlighting neoantigen immunogenicity rather than mutation burden as a potential driver of response to immune checkpoint blockade.
The success of MMR deficiency as a response predictor has raised the question of whether other DNA repair alterations are similarly associated with checkpoint blockade response. Tumors with inactivating mutations in POLE, encoding a DNA polymerase essential to nucleotide and base-excision repair, have demonstrated impressive responses to PD-1 inhibition in multiple case studies39,104–106. Another study of PD-1/PD-L1 inhibitors in metastatic urothelial carcinoma showed higher response rates in tumors with DNA damage response alterations compared with those without these alterations, and the highest response rates were in tumors with loss-of-function alterations as opposed to alterations of unknown signif-icance107. Importantly, statistical models that included DNA damage response alterations were superior at predicting survival compared with models accounting for mutation load107, indicating that alterations in DNA damage response pathways may impact response to immune checkpoint inhibitors more than mutation load.
Mutational signatures.
Beyond dysfunctional DNA repair, specific mutational signatures, which are characteristic patterns of mutation types generated by different mutational processes, have been linked to checkpoint blockade response. Among patients with lung cancer treated with checkpoint inhibitors, tumors harboring mutational signatures associated with smoking had a higher response rate than those without such smoking signatures39,53. Another cohort of patients with lung cancer who experienced durable clinical benefit from anti-PD-1 therapy displayed an enrichment of mutational signatures associated with apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC), a family of RNA and DNA editing proteins capable of promoting mutagenesis in cancer108. Likewise, patients with bladder cancer and head and neck squamous cell carcinoma demonstrated a significant association between APOBEC-related mutational signatures and response to immune checkpoint inhibitors53. Finally, patients with melanoma who were treated with checkpoint blockade were more likely to experience clinical benefit if their tumors harbored mutational signatures associated with ultraviolet light exposure or prior alkylating agent treatment as opposed to other dominant mutational signatures, and stratification by mutational signature eliminated the association of mutation burden with response, suggesting that mutation burden may not be an underlying driver of response in melanoma53. It will be of interest to understand whether these DNA-damaging processes generate more neoantigens or act by another mechanism to improve immunotherapy response.
Oncogenic signaling pathways
Certain cancer signaling pathways may influence response to immune checkpoint blockade by promoting immune evasion. Increasing evidence points to intratumoral immune exclusion as a dominant strategy employed by oncogenic pathways to limit response to immune checkpoint inhibitors109. Tumor immune microenvironments containing cytotoxic lymphocytes surrounding but not infiltrating the tumor core are termed immune-excluded or cold tumors, while those microenvironments with activated cytotoxic lymphocytes infiltrating the tumor core are referred to as immune-infiltrated or hot tumors110,111. In patients with metastatic melanoma, immune-excluded tumors demonstrated poorer responses to both CTLA-4 and PD-1 inhibitors than immune-infiltrated tumors14,112. Here, we discuss alterations in oncogenes and tumor-suppressor genes that contribute to intratumoral immune exclusion and checkpoint blockade resistance.
Tumor suppressors and oncogenes.
Lung adenocarcinomas with inactivation of the tumor suppressor gene serine/threonine-protein kinase (STK11, also known as LKB1) have been shown to have a reduced number of TILs and worse responses to immune checkpoint inhibitors in multiple studies45,113–115. An emerging mechanism by which LKB1 alterations facilitate immune exclusion is methylation-induced suppression of stimulator of interferon genes (STING) expression that blocks cytoplasmic double-stranded DNA sensing, which otherwise induces type 1 interferons and T cell recruit-ment116. Furthermore, inactivation of LKB1 in a KRAS-driven genetically engineered mouse model of non-small-cell lung cancer was associated with increased interleukin-6 expression, enhanced recruitment of neutrophils, and decreased T cell infiltrate, phenomena that could be reversed with interleukin-6 blockade or neutrophil depletion113.
Loss of PTEN, another tumor suppressor, was similarly associated with increased immunosuppressive cytokines, decreased T cell tumor infiltration, and inferior responses to PD-1 inhibitors in patients with melanoma and xenograft models of melano-mas117, as well as in a patient with uterine leiomyosarcoma118. In addition, PTEN-deficient melanomas displayed reduced autophagy-related transcript and protein levels that led to resistance to T cell killing in vitro117. Importantly, autophagy has been reported to enhance tumor immunogenicity by recruiting dendritic cells, increasing cross-presentation and priming T cells119,120, an essential step in anti-tumor immunity in which dendritic cells introduce T cells to neoantigens and trigger cytotoxic differentiation121. Whether defective T cell priming contributes to immune exclusion and PD-1 resistance in PTEN-deficient tumors remains to be studied109.
Activating oncogenic alterations associated with intratumoral immune exclusion and checkpoint blockade resistance include WNT/β-catenin and transforming growth factor-β (TGF-β)122,123. β-catenin signaling has been shown to prevent T cell tumor infiltration by decreasing dendritic cell recruitment and T cell priming in autochthonous mouse melanoma models124,125. Conversely, multiple lines of evidence suggest that TGF-β signaling in fibroblasts creates a stromal barrier to physically exclude T cells from the tumor core in patients who did not respond to immune checkpoint inhibitors123,126. Hence, strategies to enhance checkpoint blockade response need to be tailored to the specific immune exclusion mechanisms employed by each oncogenic pathway.
Clonal driver alterations.
Certain clonal driver alterations also correlate with immune exclusion and checkpoint blockade response. Murine models of EGFR-mutated lung tumors showed higher levels of immunosuppressive growth factors and a lower ratio of cytotoxic (CD8+) to regulatory (Foxp3+) tumor-infiltrating T cells compared to normal lung127, and both human and murine EGFR-mutated lung tumors displayed lower numbers of infiltrating CD8 + T cells compared with KRAS-mutated lung tumors128. Notably, patients with EGFR-mutated non-small-cell lung cancer have demonstrated a low response rate to anti-PD-1/PD-L1 therapy45,129. Moreover, a large meta-analysis of patients with lung cancer who were randomized to anti-PD-1/PD-L1 therapy or chemotherapy found no overall survival benefit of checkpoint blockade in 271 patients with EGFR-mutation-positive lung cancers and a statistically significant treatment-EGFR mutation interaction in the overall cohort of more than 2,000 patients with known EGFR status130, suggesting that EGFR mutations are indeed a robust predictor of PD-1 blockade resistance.
Conversely, the relationship of KRAS with checkpoint blockade response is more variable. In the same meta-analysis, patients with KRAS-mutation-positive tumors had improved overall survival with anti-PD-1/PD-L1 therapy as compared to chemotherapy, but the interaction between mutation status and treatment effect was not significant for KRAS mutations130. Likewise, another smaller study found no enrichment of KRAS in lung cancers responding to PD-1/PD-L1 inhibitors45, and many studies have implicated KRAS in chemokine upregulation and immunosuppressive cell recruitment131,132. Therefore, the association of KRAS mutations with checkpoint inhibitor response likely depends on additional factors, such as tissue context and coexisting mutations. Specifically, KRAS-mutated pancreatic tumors have fewer infiltrating T cells than KRAS-mutated lung tumors132, and KRAS-mutated lung tumors with TP53 mutations have better responses to PD-1 inhibitors133, particularly when compared with KRAS-mutated lung tumors with LKB1 mutations114. Ultimately, the impact of tissue stroma and concurrent mutations on the immunologic effects of clonal driver mutations requires further clarification and must be considered in future biomarker-development efforts.
Transcriptional signatures and the tumor microenvironment
Transcriptional signatures linked to immune checkpoint inhibitor response encompass the broader tumor immune microenvironment and expose underlying pathways of response and resistance. These signatures have been derived from bulk RNA and thus represent an amalgamation of tumor cells, immune cells, and stromal cells. Gene expression analyses of clinical tumor samples have found that checkpoint blockade response correlates with T cell infiltration, cytotoxic function, antigen processing, checkpoint molecules, and decreased suppressor cells85,134–137, including immune checkpoint relationships derived from spontaneously regressing neuroblastoma tumors138. One important caveat is that all of the signatures presented here require confirmation by independent groups and prospective clinical validation.
Immune infiltration and stromal signaling.
From a functional standpoint, higher T effector and IFN-γ response gene expression, including of CD8A, EOMES, GZMA and GZMB, and IFNG, was strongly associated, in randomized phase II studies, with improved overall survival in patients with non-small-cell lung cancer treated with a PD-L1 inhibitor134 and improved progression-free survival in patients with metastatic renal cell carcinoma treated with a PD-L1 inhibitor with or without anti-vascular endothelial growth factor (VEGF) therapy52. More than likely, this gene signature points to the existing presence of an effector T cell response, poised to respond to anti-PD-1 therapy. Another immune infiltration transcriptional signature is a T-cell-inflamed gene expression profile that comprises 18 genes involved in antigen presentation, chemokine production, cytolytic activity, and T cell exhaustion85,139. This signature has been shown to predict clinical response to anti-PD-1 inhibition in over 300 patients across 22 tumor types115.
Transcriptional signatures associated with resistance to checkpoint immunotherapy have focused on stromal and vascular biology. The innate anti-PD-1 resistance signature (IPRES), derived from bulk RNA sequencing of tumor biopsies, was associated with primary resistance to PD-1 blockade in patients with melanoma and included enhanced expression of genes involved in stromal and vascular pathways, such as mesenchymal transition, cell adhesion, extracellular matrix remodeling, angiogenesis, and wound healing140. Similarly, resistance to anti-PD-1 inhibition in over 300 patients across 22 tumor types correlated with enriched expression of genes related to vascular endothelium, myeloid infiltrates, and stromal Wnt signaling115. These studies highlight the contribution of stromal and endothelial components of the tumor microenvironment to checkpoint blockade resistance, which requires further clarification in future work.
T cell dysfunction and exclusion.
Another transcriptional signature predictive of checkpoint blockade response integrated both T cell exclusion in cold tumors and T cell dysfunction in hot tumors in a computational framework termed tumor immune dysfunction and exclusion (TIDE) using gene expression and survival data from over 33,000 human tumor samples141. Whereas the TIDE T cell exclusion signature was derived from expression profiles of immunosuppressive cells, including cancer-associated fibroblasts, myeloid-derived suppressor cells, and tumor-associated macrophages, the TIDE T cell dysfunction signature employed Cox proportional hazards models to identify genes that interact with cytotoxic T lymphocyte function to influence survival, producing a signature enriched for immune inflammatory pathways and depleted of T cell activation pathways141. The combined overall TIDE signature predicted checkpoint immunotherapy response better than tumor mutation load and uncovered SERPINB9, a gene encoding a protein inactivator of granzyme B and normally expressed in immune cells and immune-privileged sites, as a novel mechanism underlying immune evasion in cancer cells141. Identifying additional mechanisms of resistance in immune-infiltrated tumors that have high levels of IFN-γ will facilitate preclinical investigation of combination agents aimed at overcoming acquired resistance to checkpoint blockade.
Unlike bulk RNA sequencing, single-cell RNA sequencing allows the detection of resistance mechanisms specific to tumor subpopulations, such as cancer cells, immune cells, and fibroblasts. One such single-cell RNA sequencing study in melanoma revealed a transcriptional signature in malignant cells associated with T cell exclusion and checkpoint immunotherapy resistance that consisted of reduced antigen presentation, IFN-γ signaling, complement response, and immune modulation pathways, as well as enriched expression of chromatin regulators, transcription factors, and cyclin-dependent kinases or targets, particularly CDK4 and CDK7 (ref.142). Interestingly, this malignant cell transcriptional signature, which outperformed other signatures in predicting response to PD-1 inhibitors, was more pronounced in cycling cells and repressed by CDK4/6 inhibitors142.
T cell functionality and heterogeneity
T cell clonality.
Efforts to define a functional T cell response to checkpoint blockade using genomic approaches initially focused on the clonality of the T cell response using T cell population RNA sequencing approaches. Assessments of T cell clonality traditionally relied on sequencing the β chain of the hypervariable complementarity-determining region 3 (CDR3) of the TCR to identify a unique T cell clone because the extreme polymorphism of this region is thought to be the primary determinant of antigen-recognition specificity143. Greater T cell clonality, ranging from 0 to 1, with 1 indicating a monoclonal population, suggests a more restricted T cell response to a small number of antigens, such as those within the tumor microenvironment. Increased T cell clonality has been correlated with improved response to PD-1 blockade in melanoma, with a higher abundance of pre-treatment clones identified after PD-1 blockade14,144. Although there was no difference in clonality among responders and non-responders to anti-CTLA-4 therapy, those patients who had received anti-CTLA-4 blockade before PD-1 therapy showed an increase in T cell clonality144.
In comparison, patients with non-small-cell lung cancer with acquired resistance to PD-1 blockade with or without CTLA-4 inhibition demonstrated diminished TCR clonality41. This decrease in clonal TCR frequency contrastsd with the clonal TCR expansion seen during initial response41. While this work highlights the overall diversity of the T cell response as a potential critical factor for checkpoint inhibitor response, this approach cannot match the TCR to its cognate neo-epitope. Instead, advances in single-cell RNA sequencing that identify the full-length TCR-α and TCR-β chains145, when combined with improved algorithms for prediction of neoantigens, may provide more definitive evidence about the relationship between T cell clonality and corresponding neoantigens in response to checkpoint blockade.
T cell exhaustion and stemness.
Based on work in mouse models of chronic viral infections, a nuanced view of T cell functionality has emerged with the recognition that the ‘exhausted’ CD8 + PD-1 + T cell population itself is heterogenous, and a subset of the T cells respond to PD-1 blockade146,147. These populations may be more broadly broken into two classes: a subset that is considered to be ‘progenitors’ or ‘stem-cell-like’, which expresses the chemokine receptor CXCR5 + and the transcription factor TCF7 (also known as TCF1), lacks the co-inhibitory receptor TIM-3, and is capable of responding to checkpoint blockade and giving rise to effector T cell progeny; and a ‘terminally exhausted’ subset that lacks CXCR5 or TCF7 but expresses TIM-3 and does not proliferate in response to PD-1/PD-L1 blockade147,148. The stem-like population of Cd8 + PD-1 + CXCR5 + TIM-3− T cells has been identified in lung cancer tumors via multiparameter flow cytometry and is similar to a subset of CD8 + T cells in melanoma identified by single-cell RNA sequencing approaches149. More recently, this CD8 + TCF7+T cell population, also expressing the IL-7 receptor, has been found to be enriched in melanoma tumors that respond to checkpoint blockade, which was confirmed in an independent cohort150. A CD8 + T cell population expressing TIM-3 and CD39, an ectonucleotidase involved in adenosine signaling, was enriched in non-responders150. While this work focused on the CD8 + T cell compartment, additional analyses of single-cell data will likely further illuminate the role that heterogeneity plays in other T cell subsets, including regulatory T cells, CD4+helper and effector cells, and tissue-resident T cells, as well as the myeloid cell compartment, and the contributions of these immune cell subtypes to responsiveness to immunotherapy.
Chromatin modifier genes
SWI-SNF chromatin remodeling complexes regulate genomic architecture151 and impact response to checkpoint inhibitors. Inactivating mutations in the PBRM1 gene, which encodes a subunit of the PBAF SWI-SNF chromatin remodeling complex, conferred susceptibility to dual PD-1 and CTLA-4 inhibition in mouse melanoma tumors otherwise resistant to checkpoint blockade, and PBRM1-deficient cells showed enhanced chromatin accessibility and significantly enriched expression of IFN-γ response genes, including genes encoding chemokines that recruit effector T cells, resulting in increased intratumoral cytotoxic CD8+ T cells152. Moreover, PBRM1-deficient tumors in the Cancer Genome Atlas co-associated with expression of genes related to stimulated 3’ antisense retroviral coding sequences (SPARCS), a subclass of endogenous retroviruses that triggers innate immunity by generating double-stranded RNA153. PBRM1 loss additionally correlated with checkpoint inhibitor response in patients with metastatic renal cell carcinoma154. Notably, the benefit conferred by PBRM1 loss was predominantly observed in patients who had been previously treated, the majority of whom received VEGF inhibitors, and not in patients receiving anti-PD-1/PD-L1 as first-line treatment154. Prior analyses of patients with metastatic renal cell carcinoma demonstrated that PBRM1 mutations correlated with improved response to first-line VEGF inhibitors155,156. A subsequent randomized phase II study of patients with treatment-naive metastatic renal cell carcinoma also found that PBRM1 mutations were associated with improved responses in patients for whom first-line treatment was a VEGF inhibitor, but not in patients for whom it was a PD-L1 inhibitor with or without a monoclonal anti-VEGF antibody52. Altogether, these works indicate that prior anti-angiogenic therapy influences the association between PBRM1 status and response to anti-PD-1/PD-L1 therapy, underscoring the need for further investigation of this pathway and the conclusion that PBRM1 mutations should not presently be advanced as a clinical biomarker.
AT-rich interactive domain-containing protein 1A (ARID1A) and SWI-SNF related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 4 (SMARCA4), two other genes encoding SWI-SNF chromatin remodeling complex subunits, have similarly been linked to response to checkpoint inhibitors. In mice with ARID1A-deficient ovarian tumors, PD-L1 inhibition led to reduced tumor burden and prolonged survival compared with controls157. This study found that ARID1Aloss and impaired ARID1A binding to the MMR protein MSH2 similarly reduced MMR and increased mutation frequency, leading the authors to hypothesize that ARID1A loss may increase response to checkpoint blockade through MMR deficiency157. Another study reported that four patients with ovarian small-cell carcinoma, a low-mutation cancer driven by loss-of-function SMARCA4 mutations, had impressive responses to anti-PD-1 therapy158. Taken together, these studies demonstrate a role for chromatin-modifier genes in response to checkpoint inhibitors and highlight the need to confirm these findings and investigate the relationship between other epigenetic changes and checkpoint blockade response.
Additional recurrent genetic alterations
Other recurrent genetic alterations have been linked to response to immune checkpoint blockade, including SERPINB3 and SERPINB4 (SERPINB3/4), genes encoding serine protease inhibitors involved in apoptosis and autoimmunity159, and CNAs. In two independent melanoma cohorts, it was found that SERPINB3/4 mutations were associated with clinical benefit and longer survival after anti-CTLA-4 therapy159. Inactivating mutations in SERPINB3/4 cause proteins to misfold and form inflammatory aggregates capable of triggering autoimmune disease, leading to the hypothesis that SERPINB3/4 mutations initiate a broad tumor immune response reactivated by checkpoint inhibitors159.
In contrast, CNAs have been connected to resistance to checkpoint blockade. A high number of somatic CNAs in patients with melanoma treated with anti-CTLA-4 therapy correlated with worse survival, and over 5,000 tumors of 12 cancer types with high levels of whole-chromosome and chromosome-arm CNAs exhibited reduced expression of cytolytic immune cell genes, specifically CD247, CD2, CD3E, GZMH, NKG7, PRF1, and GZMK160. Similarly, an analysis of patients with melanoma treated with checkpoint inhibitors found that non-responding tumors had a higher burden of copy-number loss and reduced expression of immune-related genes, including cytolytic markers, HLA molecules, IFN-γ pathway genes, chemokines, and adhesion molecules144. In sum, these studies indicate that tumor aneuploidy may be associated with decreased tumor immune infiltrates and resistance to checkpoint blockade, suggesting that CNAs may represent a survival strategy for tumors, allowing them to escape strong anti-tumor immune responses. The mechanism underlying this association may be related to lost immunogenic neoantigens enhancing immune evasion or another pathway that has yet to be discovered.
Toward clinical translation
The aforementioned genomic correlates of immune checkpoint response may serve as a blueprint around which to develop targeted therapies to overcome immune checkpoint inhibitor resistance. This section presents examples of potential therapeutic applications sparked by these genomic observations (Table 2). Numerous combination regimens with immune checkpoint blockade exist beyond those related to genomic alterations and are reviewed comprehensively elsewhere23,161,162.
Table 2 |.
Trials targeting genomic correlates of response to immune checkpoint blockade
| Intervention | Trial number | Patient population | Phase | Setting | Status |
|---|---|---|---|---|---|
| Neoantigen vaccines | |||||
| NeoVax + anti-CTLA-4 ipilimumab | NCT02950766 | Stage III or low-volume stage IV RCC | I | Adjuvant/1st line | Not yet recruiting |
| NeoVax + anti-PD-1 nivolumab ± anti-CTLA-4 ipilimumab | NCT03422094 | Newly diagnosed, unmethylated glioblastoma | I | 1st line | Recruiting |
| NeoVax + ant-CD20 rituxumab | NCT03361852 | Follicular lymphoma | I | 1st line | Not yet recruiting |
| NeoVax + alkylating agent cyclophosphamide | NCT03219450 | Chronic lymphocytic leukemia | I | 1st line | Not yet recruiting |
| Interferon therapies | |||||
| Interferon-γ-1β + anti-PD-1 pembrolizumab | NCT03063632 | Mycosis fungoides and Sézary syndrome | II | Refractory | Recruiting |
| Interferon-γ + anti-PD-1 nivolumab | NCT02614456 | Advanced solid tumors | I | 2nd line | Recruiting |
| JAK1 inhibitor itacitinib or PI3Kδ inhibitor + anti-PD-1 pembrolizumab | NCT02646748 | Advanced solid tumors | I/II | Refractory | Recruiting |
| JAK2 inhibitor ruxolitinib + anti-PD-1 pembrolizumab | NCT03012230 | Metastatic triple-negative breast cancer | I | 2nd line | Recruiting |
| JAK1 inhibitor itacitinib + anti-PD-1 pembrolizumab | NCT03425006 | Metastatic PD-L1+ NSCLC | II | 1st line | Recruiting |
| STING agonist MIW815 + anti-PD-1 spartalizumab | NCT03172936 | Advanced solid tumors or lymphomas | I | Any line | Recruiting |
| CDK4/6 inhibitors | |||||
| Fulvestrant ± CDK4/6 inhibitor palbociclib ± anti-PD-L1 avelumab | NCT03147287 | Metastatic HR+HER2− breast cancer | II | 2nd-3rd line | Recruiting |
| Tamoxifen ± CDK4/6 inhibitor palbociclib followed by anti-PD-LI avelumab | NCT03573648 | Stage II or III HR+ breast cancer | II | Neoadjuvant | Recruiting |
| CDK4/6 inhibitor ribociclib + anti-PD-1 spartalizumab (+ fulvestrant if breast) | NCT03294694 | Metastatic ovarian cancer or HR+HER2− breast cancer | I | Any line | Recruiting |
| CDK4/6 inhibitor abemaciclib + anti-PD-1 pembrolizumab | NCT02779751 | Metastatic NSCLC or HR+HER2− breast cancer | I | 1st-3rd line | Recruiting |
| Anti-PD-L1 avelumab, cetuximab, + CDK4/6 inhibitor palbociclib | NCT03498378 | Recurrent/metastatic headand neck squamous cell carcinoma | I | Any line | Recruiting |
| Anti-TGF-β therapies | |||||
| Anti-TGF-β NIS793 + anti-PD-1 spartalizumab | NCT02947165 | Advanced solid tumors | I | Refractory | Recruiting |
| TGF-β receptor 1 inhibitor galunisertib + anti-PD-LI durvalumab | NCT02734160 | Metastatic pancreatic cancer | I | 3rd line | Recruiting |
| TGF-β receptor 1 inhibitor galunisertib + ant-PD-1 nivolumab | NCT02423343 | HCC or recurrent NSCLC | I/II | 2nd line | Recruiting |
| Anti-PD-L1/TGF-β trap M7824 | NCT02517398 | Metastatic or locally advanced solid tumors | I | Refractory | Recruiting |
| Anti-PD-LI/TGF-β trap M7824 + treatment of physician’s choice | NCT03620201 | Stage II-III HER2+ breast cancer | I | Neoadjuvant | Recruiting |
| Anti-PD-LI/TGF-β trap M7824 versus anti-PD-1 pembrolizumab | NCT03631706 | PD-L1+ NSCLC | II | 1st line | Recruiting |
HCC, hepatocellular carcinoma; HER2, human epidermal growth factor receptor 2; HR+, hormone receptor positive; NSCLC, non-small-cell lung cancer; Refractory, failed prior standard therapy (including subject refusal or intolerance); RCC, renal cell carcinoma
Neoantigen vaccines.
One of the most advanced areas in immunogenomics therapeutic development is personalized cancer vaccines. These efforts utilize next-generation-sequencing-based analyses to identify and target predicted neoantigens with patient-specific vaccines that stimulate an immune response tailored to each individual’s tumor mutational profile163. Two recent studies have demonstrated that personalized neoantigen vaccines based on individual tumor whole-exome and transcriptome sequencing augmented existing T cell responses and activated new T cell responses in patients with advanced or newly metastatic mela-noma164,165. In addition, three patients with radiologically detected metastatic lesions at the start of neoantigen vaccination, who subsequently progressed, experienced complete responses with the addition of anti-PD-1 therapy164,165. These neoantigen vaccines are currently being examined in multiple clinical trials in other cancers with lower neoantigen burden, including renal cell carcinoma (NCT02950766), glioblastoma (NCT03422094), follicular lymphoma (NCT03361852), and chronic lymphocytic leukemia (NCT03219450) with and without immune checkpoint inhibitors. Whether the enhanced T cell priming and activation effects of neoantigen vaccines can overcome checkpoint blockade resistance has yet to be established.
Interferon therapies.
Another approach to addressing checkpoint blockade resistance focuses on therapies that boost anti-tumor immunity through IFN activation. IFN-γ-responsive genes involved in antigen presentation, cytokine signaling, cytotoxic activity, and adaptive immunity have been shown to correlate with response to checkpoint inhibitors85,115. Phase I/II trials investigating combinations of IFN-γ and checkpoint inhibitors are underway in advanced solid tumors and refractory lymphomas (NCT02614456, NCT03063632). The IFN-γ receptor depends on the JAK-STAT signal transduction pathway to regulate transcription of IFN-γ-inducible genes166, so JAK inhibitors may curtail the efficacy of checkpoint inhibitors. In support of this concept, a phase I clinical trial in advanced solid tumors showed that the JAK1 inhibitor itacitinib combined with the PD-1 inhibitor pembrolizumab reduced peripheral T cell activation and did not affect intratumoral CD8 + T cell or T regulatory cell levels, leading to lower-than-expected response rates167. Alternatively, T cell priming may depend on IFN-β activation by the STING protein complex acting as an endoplasmic reticulum receptor that propagates cytosolic double-stranded DNA sensing to trigger an anti-tumor immune response113,168. Although many STING agonists are under clinical development in combination with checkpoint inhibitors (NCT02675439, NCT03172936, NCT03010176), the single-agent activity of these therapies has been limited169,170. Compelling evidence indicates that STING expression is repressed by hyper-methylation in LKB1-deficient lung cancers, perhaps explaining the limited efficacy of current STING agonists and raising the question of whether combinations of hypomethylating agents and antibody-STING agonist conjugates would enhance responses to checkpoint inhibitors116.
CDK4/6 inhibitors.
Another promising therapeutic strategy to overcome checkpoint inhibitor resistance is boosting T cell infiltration of tumors by blocking immune exclusion pathways. Targeting CDK4, a defining component of the malignant cell transcriptional resistance signature discovered in single-cell sequencing analyses of melanoma142, embodies an emerging tactic to transform immune-excluded tumors into immune-infiltrated tumors. In mouse models of cancer, human breast cancer cell lines, and clinical breast cancer biopsies, CDK4/6 inhibitors have been shown to promote anti-tumor immunity by reducing regulatory T cell proliferation, enhancing effector T cell activation, and improving antigen presentation through decreased methylation of endogenous retro-viruses171–173. Moreover, mouse models of colon, lung, and breast cancer have shown that CDK4/6 inhibitors augment responses to PD-1 blockade171–173, and preliminary clinical data suggest that CDK4/6 inhibition improves responses to PD-1 blockade in hormone-receptor-positive metastatic breast cancer as compared with historical response rates for either as a monotherapy174. Numerous phase I/II trials combining CDK4/6 inhibitors with checkpoint immunotherapy are ongoing across tumor types, including breast, ovarian, lung, and head and neck cancers (NCT03147287, NCT03294694, NCT02779751, NCT03498378).
Anti-TGF-β therapies.
Finally, TGF-β signaling in fibroblasts constitutes another critical mechanism of immune exclusion and resistance to checkpoint inhibitors126, as demonstrated by the finding that combined blockade of TGF-β and PD-L1 reduced stromal TGF-β signaling and augmented T cell tumor infiltration and tumor regression in a mouse model of immune-excluded mammary carcinoma123. Likewise, in mouse models of colorectal adenocarcinomas, TGF-β inhibition conferred susceptibility to anti-PD-L1 therapy with augmented infiltrating lymphocytes and type 1 T helper cell activation that led to metastasis eradication175. On the basis of this evidence, combined TGF-β and PD-1/PD-L1 inhibition is under evaluation in phase I/II trials in advanced solid tumors (NCT02947165, NCT02734160, NCT02423343). Moreover, the bifunctional fusion protein M7824, consisting of a PD-L1 monoclonal antibody linked to TGF-β receptor II, which traps all three TGF-β isoforms, displayed more effective tumor suppression than single-agent therapy in syngeneic mouse models, including long-term survival and anti-tumor immunity176. The dose-escalation component of a phase I study (NCT02517398) investigating this agent demonstrated a manageable safety profile and some durable responses177, although whether this agent is more effective than PD-L1 inhibition alone has yet to be established. Table 2 displays ongoing clinical trials of therapeutic mechanisms targeting these genomic correlates of response to improve efficacy of immune checkpoint inhibitors.
Clinical trial innovation.
Innovation in trial design, patient recruitment, and clinical endpoints will help determine the best therapies informed by genomic data to combine with checkpoint blockade to overcome resistance. Basket trials organized around a specific molecular alteration constitute an important strategy to test immunotherapy combinations across tumor types in biomarker-selected patients with the highest probability of benefit (Fig. 3a)178. Adaptive trials may similarly augment enrollment into subgroups with promising response rates by using interim analyses to make sample size modifications that ensure statistical power in the subgroup of interest (Fig. 3b)179. Novel trial designs for combination dose-escalation studies include run-in periods that allow sequential combination after monotherapy within the same patient, zig-zag schedules that alternate agents for dose increases, and bifurcated arms with monotherapy escalation and combination escalation at a lower dose180. Beyond trial designs, patient recruitment can be improved with automated tools that prompt clinicians to consider patient-specific trials matched to actionable genomic alterations at key clinical and research time points, including when scans or labs indicate progressive disease, when tumor genetic testing is completed, and when new trials open178. To address the nonlinear dose-response and dose-toxicity kinetics of checkpoint inhibitors, early-phase trials should explore novel biomarker-driven response criteria and surrogate endpoints, such as circulating tumor DNA and target receptor occupancy, while avoiding additive dosing without efficacy benefits181,182. Likewise, new endpoints for later-stage studies, such as immune-related response criteria and durable response rate, require continued investigation180. Most importantly, in light of the rapidly ballooning quantity of immuno-oncology trials, academic institutions and pharmaceutical companies need to coordinate trial efforts and sharing of correlative science data to prevent redundancy and prioritize the most promising checkpoint inhibitor combinations161.
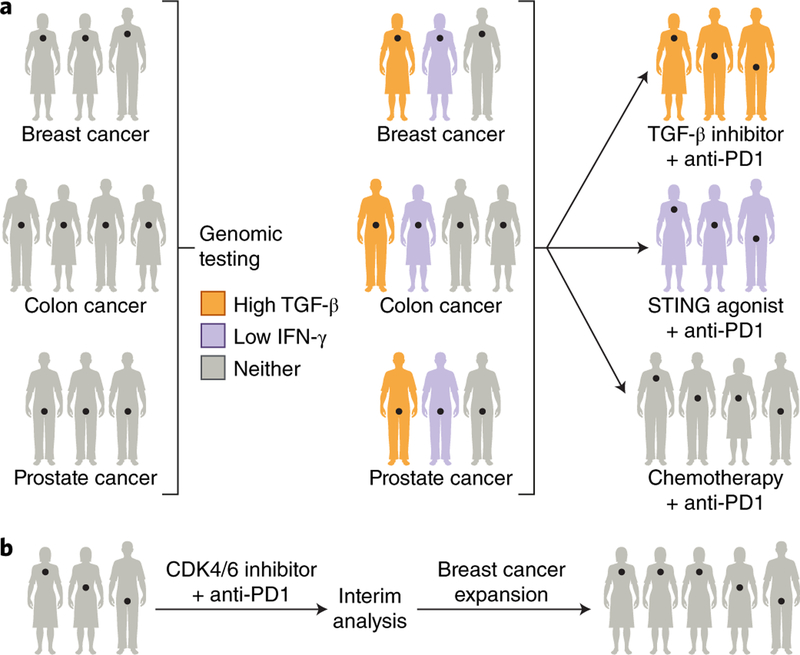
Fig. 3 |. Trial designs to overcome checkpoint blockade resistance.
a, A basket trial matches therapeutic agents based on genomic alterations irrespective of tumor site of origin. In the illustrated example, breast, colorectal, and prostate tumors are tested for TGF-β and IFN-γ transcriptomic signatures. Those patients whose tumors possess high TGF-β or low IFN-γ signatures are treated with TGF-β inhibitors or STING agonists, respectively, in addition to anti-PD-1 therapy, while tumors without genomic targets are treated with chemotherapy and anti-PD-1 therapy. b, An adaptive trial design employs interim analyses to make modifications, such as expansion of patient cohorts with more promising responses, illustrated in the figure as an expansion of the breast cancer cohort. Credit: Debbie Maizels/Springer Nature.
Future directions
Much work remains to develop these correlates of checkpoint blockade response into reliable biomarkers that can guide treatment decisions and therapeutic development. First and foremost, many of these concepts need to be validated in functional preclinical models and prospective clinical cohorts. Moreover, the search for additional genomic correlates requires clinically annotated patient cohorts with sample sizes into the hundreds and even thousands to detect rare response-associated variants53. This effort necessitates populations with greater diversity of race, ethnicity, age, and tumor histology183. Future discovery efforts should focus on detailed genomic and clinical characterization coupled with thorough investigation of less explored areas, including epigenomics, proteomics, and metabolomics, to fully characterize the relationship of host immunity, tumor biology, and the microbiome to checkpoint inhibitor response. Comprehensive clinical databases of patients treated with checkpoint inhibitors must be effectively integrated with preclinical models and screens. Ultimately, systems biology and bioinformatic approaches are needed to coordinate this vast array of information, to assess the relative importance of each component, and to develop risk scores to capture the complexity of multiple driving alterations influencing response. Altogether, these efforts to contex-tualize genomic correlates into the larger tumor-immune biology will build a deeper quantitative and conceptual understanding of response and resistance to checkpoint blockade to better promote the development of rational biomarkers and therapies.
Acknowledgements
This work was supported by BroadIgnite (E.M.V.A.), BroadNext10 (E.M.V.A., D.M.), NIH R01CA227388 (E.M.V.A.), NIH K08CA188615 (E.M.V.A.), and NIH U01CA233100 (E.M.V.A). This work was also supported by the Center for Immuno-Oncology at the Dana- Farber Cancer Institute and a Stand Up To Cancer-American Cancer Society Lung Cancer Dream Team Translational Research Grant (SU2C-AACR-DT17–15). Stand Up To Cancer (SU2C) is a program of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C.
Footnotes
Competing interests
T.E.K. and K.P.B. declare no competing interests. E.M.V.A. holds consulting roles with Tango Therapeutics, Invitae, Genome Medical, Illumina, Foresite Capital, and Dynamo, receives research support from Bristol-Myers Squibb and Novartis, owns equity in Tango Therapeutics, Genome Medical, Syapse, and Microsoft, received travel reimbursement from Roche/Genentech, and is on an institutional patent for chromatin mutations and immunotherapy response.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Foley EJ Antigenic properties of methylcholanthrene-induced tumors in mice of the strain of origin. Cancer Res 13, 835–837 (1953). [PubMed] [Google Scholar]
- 2.Smith HJ Antigenicity of carcinogen-induced and spontaneous tumours in inbred mice. Br. J. Cancer 20, 831–837 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walunas TL et al. CTLA-4 can function as a negative regulator of T cell activation. Immunity 1, 405–413 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Fourcade J et al. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J. Exp. Med 207, 2175–2186 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsuzaki J et al. Tumor-infiltrating NY-ESO-1-specific CD8+ T cells are negatively regulated by LAG-3 and PD-1 in human ovarian cancer. Proc. Natl Acad. Sci. USA 107, 7875–7880 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topalian SL et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med 366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garon EB et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med 372, 2018–2028 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Motzer RJ et al. Nivolumab for metastatic renal cell carcinoma: results of a randomized phase II trial. J. Clin. Oncol 33, 1430–1437 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenberg JE et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 387, 1909–1920 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolchok JD et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med 377, 1345–1356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Khoueiry AB et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 389, 2492–2502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim ST et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med 24, 1449–1458 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Ribas A & Wolchok JD Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tumeh PC et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen AR & Siu LL PD-L1 testing in cancer: challenges in companion diagnostic development. JAMA Oncol 2, 15–16 2016). [DOI] [PubMed] [Google Scholar]
- 16.Carbognin L et al. Differential activity of nivolumab, pembrolizumab and MPDL3280A according to the tumor expression of programmed death-ligand-1 (PD-L1): sensitivity analysis of trials in melanoma, lung and genitourinary cancers. PLoS One 10, e0130142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antonia SJ et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol 17, 883–895 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Sun C, Mezzadra R & Schumacher TN Regulation and function of the PD-L1 checkpoint. Immunity 48, 434–452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma P & Allison JP The future of immune checkpoint therapy. Science 348, 56–61 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Brahmer J et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med 373, 123–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Socinski MA et al. Atezolizumab for first-line treatment of metastatic nonsquamous NSCLC. N. Engl. J. Med 378, 2288–2301 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A & Wargo JA The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell 33, 570–580 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma P, Hu-Lieskovan S, Wargo JA & Ribas A Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Syn NL, Teng MWL, Mok TSK & Soo RA De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol 18, e731–e741 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Chen DS & Mellman I Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Leone RD & Emens LA Targeting adenosine for cancer immunotherapy. J. Immunother. Cancer 6, 57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stratton MR, Campbell PJ & Futreal PA The cancer genome. Nature 458, 719–724 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vogelstein B et al. Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey MH et al. Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385 e318 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schumacher TN & Schreiber RD Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Seiwert TY et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol 17, 956–965 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Rooney MS, Shukla SA, Wu CJ, Getz G & Hacohen N Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yarchoan M, Johnson BA III, Lutz ER, Laheru DA & Jaffee EM Targeting neoantigens to augment antitumour immunity. Nat. Rev. Cancer 17, 209–222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gubin MM et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577–581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fehlings M et al. Checkpoint blockade immunotherapy reshapes the high-dimensional phenotypic heterogeneity of murine intratumoural neoantigen-specific CD8+ T cells. Nat. Commun 8, 562 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Rooij N et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J. Clin. Oncol 31, e439–e442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le DT et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Allen EM et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rizvi NA et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le DT et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med 372, 2509–2520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anagnostou V et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov 7, 264–276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snyder A et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med 371, 2189–2199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seiwert TY et al. Biomarkers predictive of response to pembrolizumab in head and neck cancer (HNSCC). Cancer Res 73, LB–339 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hellmann MD et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell 33, 843–852 e844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rizvi H et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J. Clin. Oncol 36, 633–641 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hellmann MD et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N. Engl. J. Med 378, 2093–2104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carbone DP et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N. Engl. J. Med 376, 2415–2426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hellmann MD et al. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell 33, 853–861 e854 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colli LM et al. Burden of nonsynonymous mutations among TCGA cancers and candidate immune checkpoint inhibitor responses. Cancer Res 76, 3767–3772 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen R et al. Phase II study of the efficacy and safety of pembrolizumab for relapsed/refractory classic hodgkin lymphoma. J. Clin. Oncol 35, 2125–2132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Armand P et al. Nivolumab for relapsed/refractory classic hodgkin lymphoma after failure of autologous hematopoietic cell transplantation: extended follow-up of the multicohort single-arm phase II CheckMate 205 trial. J. Clin. Oncol 36, 1428–1439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McDermott DF et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat. Med 24, 749–757 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miao D et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat. Genet 50, 1271–1281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nghiem PT et al. PD-1 blockade with pembrolizumab in advanced merkel-cell carcinoma. N. Engl. J. Med 374, 2542–2552 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goh G et al. Mutational landscape of MCPyV-positive and MCPyV-negative Merkel cell carcinomas with implications for immunotherapy. Oncotarget 7, 3403–3415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaufman HL et al. Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: a multicentre, single-group, open-label, phase 2 trial. Lancet Oncol 17, 1374–1385 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McGranahan N et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robbins PF et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat. Med 19, 747–752 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tran E et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 350, 1387–1390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yadav M et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515, 572–576 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Abelin JG et al. Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity 46, 315–326 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nielsen M, Lund O, Buus S & Lundegaard C MHC class II epitope predictive algorithms. Immunology 130, 319–328 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Graham DB et al. Antigen discovery and specification of immunodominance hierarchies for MHCII-restricted epitopes. Nat. Med 24, 1762–1772 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Milo I et al. The immune system profoundly restricts intratumor genetic heterogeneity. Sci. Immunol 3, 30470696 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Łuksza M et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 551, 517–520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Balachandran VP et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, 512–516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Panda A et al. Endogenous retrovirus expression is associated with response to immune checkpoint blockade in clear cell renal cell carcinoma. JCI Insight 3, 30135306 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shukla SA et al. Cancer-germline antigen expression discriminates clinical outcome to CTLA-4 blockade. Cell 173, 624–633 e628 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sidney J, Peters B, Frahm N, Brander C & Sette A HLA class I supertypes: a revised and updated classification. BMC Immunol 9, 1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Restifo NP et al. Loss of functional beta 2-microglobulin in metastatic melanomas from five patients receiving immunotherapy. J. Natl Cancer Inst 88, 100–108 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sade-Feldman M et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat. Commun 8, 1136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zaretsky JM et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med 375, 819–829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gettinger S et al. Impaired HLA class I antigen processing and presentation as a mechanism of acquired resistance to immune checkpoint inhibitors in lung cancer. Cancer Discov 7, 1420–1435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Manguso RT et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kearney CJ et al. Tumor immune evasion arises through loss of TNF sensitivity. Sci. Immunol 3, 29776993 (2018). [DOI] [PubMed] [Google Scholar]
- 76.Patel SJ et al. Identification of essential genes for cancer immunotherapy. Nature 548, 537–542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Parham P & Ohta T Population biology of antigen presentation by MHC class I molecules. Science 272, 67–74 (1996). [DOI] [PubMed] [Google Scholar]
- 78.Messaoudi I, Guevara Patiño JA, Dyall R, LeMaoult J & Nikolich-Zugich J Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science 298, 1797–1800 (2002). [DOI] [PubMed] [Google Scholar]
- 79.Marty R et al. MHC-I genotype restricts the oncogenic mutational landscape. Cell 171, 1272–1283 e1215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marty R, Thompson WK, Salem RM, Zanetti M & Carter H Evolutionary pressure against MHC class II binding cancer mutations. Cell 175, 416–428 e413 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Giannakis M et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep 15, 857–865 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chowell D et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 359, 582–587 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang L et al. The RNA-binding protein MEX3B mediates resistance to cancer immunotherapy by downregulating HLA-A expression. Clin. Cancer Res 24, 3366–3376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rodig SJ et al. MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci. Transl. Med 10, 30021886 (2018). [DOI] [PubMed] [Google Scholar]
- 85.Ayers M et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest 127, 2930–2940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen PL et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov 6, 827–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gao J et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 167, 397–404 e399 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sucker A et al. Acquired IFNy resistance impairs anti-tumor immunity and gives rise to T-cell-resistant melanoma lesions. Nat. Commun 8, 15440 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shin DS et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov 7, 188–201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Van Allen EM et al. Long-term benefit of PD-l1 blockade in lung cancer associated with JAK3 activation. Cancer. Immunol. Res 3, 855–863 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roemer MG et al. PD-L1 and PD-L2 genetic alterations define classical hodgkin lymphoma and predict outcome. J. Clin. Oncol 34, 2690–2697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ansell SM et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N. Engl. J. Med 372, 311–319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Green MR et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 116, 3268–3277 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nayak L et al. PD-1 blockade with nivolumab in relapsed/refractory primary central nervous system and testicular lymphoma. Blood 129, 3071–3073 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ratner L, Waldmann TA, Janakiram M & Brammer JE Rapid progression of Adult T-cell leukemia-lymphoma after PD-1 inhibitor therapy. N. Engl. J. Med 378, 1947–1948 (2018). [DOI] [PubMed] [Google Scholar]
- 96.Wartewig T et al. PD-1 is a haploinsufficient suppressor of T cell lymphomagenesis. Nature 552, 121–125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Goodman AM et al. Prevalence of PDL1 amplification and preliminary response to immune checkpoint blockade in solid tumors. JAMA. Oncol 4, 1237–1244 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang J et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 553, 91–95 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mezzadra R et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 549, 106–110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Burr ML et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 549, 101–105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mouw KW, Goldberg MS, Konstantinopoulos PA & D’Andrea AD DNA damage and repair biomarkers of immunotherapy Response. Cancer Discov 7, 675–693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Linnebacher M, Wienck A, Boeck I & Klar E Identification of an MSI-H tumor-specific cytotoxic T cell epitope generated by the (−1) frame of U79260(FT0). J. Biomed. Biotechnol 2010, 841451 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Germano G et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature 552, 116–120 (2017). [DOI] [PubMed] [Google Scholar]
- 104.Bouffet E et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J. Clin. Oncol 34, 2206–2211 (2016). [DOI] [PubMed] [Google Scholar]
- 105.Johanns TM et al. Immunogenomics of hypermutated glioblastoma: a patient with Germline POLE deficiency treated with checkpoint blockade Immunotherapy. Cancer Discov 6, 1230–1236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bhangoo MS et al. Tumor mutational burden guides therapy in a treatment refractory POLE-mutant uterine carcinosarcoma. Oncologist 23, 518–523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Teo MY et al. Alterations in DNA damage response and repair genes as potential marker of clinical benefit from PD-1/PD-L1 blockade in advanced urothelial cancers. J. Clin. Oncol 36, 1685–1694 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang S, Jia M, He Z & Liu XS APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer. Oncogene 37, 3924–3936 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Spranger S & Gajewski TF Impact of oncogenic pathways on evasion of antitumour immune responses. Nat. Rev. Cancer 18, 139–147 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Binnewies M et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med 24, 541–550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gajewski TF, Schreiber H & Fu YX Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol 14, 1014–1022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ji RR et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol. Immunother 61, 1019–1031 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Koyama S et al. STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress T-cell activity in the lung tumor microenvironment. Cancer Res 76, 999–1008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Skoulidis F et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov 8, 822–835 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cristescu R et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362, 30309915 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kitajima S et al. Suppression of STING associated with LKB1 loss in KRAS-driven lung cancer. Cancer Discov 9, 34–45 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Peng W et al. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov 6, 202–216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.George S et al. Loss of PTEN is associated with resistance to anti-PD-1 checkpoint blockade therapy in metastatic uterine leiomyosarcoma. Immunity 46, 197–204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Michaud M et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 334, 1573–1577 (2011). [DOI] [PubMed] [Google Scholar]
- 120.Page DB et al. Glimpse into the future: harnessing autophagy to promote anti-tumor immunity with the DRibbles vaccine. J. Immunother. Cancer 4, 25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Borst J, Ahrends T, Bąbała N, Melief CJM & Kastenmüller W CD4+ T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol 18, 635–647 (2018). [DOI] [PubMed] [Google Scholar]
- 122.Jimenez-Sanchez A et al. Heterogeneous tumor-immune microenvironments among differentially growing metastases in an ovarian cancer patient. Cell 170, 927–938 e920 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mariathasan S et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Spranger S, Bao R & Gajewski TF Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 523, 231–235 (2015). [DOI] [PubMed] [Google Scholar]
- 125.Spranger S, Dai D, Horton B & Gajewski TF Tumor-residing Batf3 dendritic cells are required for effector t cell trafficking and adoptive t cell therapy. Cancer Cell 31, 711–723 e714 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chakravarthy A, Khan L, Bensler NP, Bose P & De Carvalho DD TGF-β-associated extracellular matrix genes link cancer-associated fibroblasts to immune evasion and immunotherapy failure. Nat. Commun 9, 4692 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Akbay EA et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov 3, 1355–1363 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Busch SE et al. Lung cancer subtypes generate unique immune Responses. J. Immunol 197, 4493–4503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gainor JF et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: a retrospective analysis. Clin. Cancer Res 22, 4585–4593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee CK et al. Clinical and molecular characteristics associated with survival among patients treated with checkpoint inhibitors for advanced non-small cell lung carcinoma: a systematic review and meta-analysis. JAMA Oncol 4, 210–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Anderson KG, Stromnes IM & Greenberg PD Obstacles posed by the tumor microenvironment to T cell activity: a case for synergistic therapies. Cancer Cell 31, 311–325 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cullis J, Das S & Bar-Sagi D Kras and tumor immunity: friend or foe?. Cold Spring Harb. Perspect. Med 8, 29229670 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dong ZY et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin. Cancer Res 23, 3012–3024 2017). [DOI] [PubMed] [Google Scholar]
- 134.Fehrenbacher L et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 387, 1837–1846 (2016). [DOI] [PubMed] [Google Scholar]
- 135.Şenbabaoğlu Y et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol 17, 231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Charoentong P et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Reports 18, 248–262 (2017). [DOI] [PubMed] [Google Scholar]
- 137.Prat A et al. Immune-related gene expression profiling after PD-1 blockade in non-small cell lung carcinoma, head and neck squamous cell carcinoma, and melanoma. Cancer Res 77, 3540–3550 (2017). [DOI] [PubMed] [Google Scholar]
- 138.Auslander N et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat. Med 24, 1545–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Danaher P et al. Pan-cancer adaptive immune resistance as defined by the Tumor Inflammation Signature (TIS): results from The Cancer Genome Atlas (TCGA). J. Immunother. Cancer 6, 63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hugo W et al. Genomic and transcriptomic features of response to Anti-PD-1 therapy in metastatic melanoma. Cell 165, 35–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jiang P et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med 24, 1550–1558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jerby-Arnon L et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell 175, 984–997 e924 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xu JL & Davis MM Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity 13, 37–45 (2000). [DOI] [PubMed] [Google Scholar]
- 144.Roh W et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci. Transl. Med 9, eaah3560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Stubbington MJT et al. T cell fate and clonality inference from single-cell transcriptomes. Nat. Methods 13, 329–332 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Blackburn SD et al. Coregulation of CD8+T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol 10, 29–37 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Im SJ et al. Defining CD8+T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Utzschneider DT et al. T cell factor 1-expressing memory-like CD8+ T cells sustain the immune response to chronic viral infections. Immunity 45, 415–427 (2016). [DOI] [PubMed] [Google Scholar]
- 149.Brummelman J et al. High-dimensional single cell analysis identifies stem-like cytotoxic CD8+ T cells infiltrating human tumors. J. Exp. Med 215, 2520–2535 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sade-Feldman M et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175, 998–1013 e1020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kadoch C & Crabtree GR Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci. Adv 1, e1500447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pan D et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 359, 770–775 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cañadas I et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat. Med 24, 1143–1150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Miao D et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 359, 801–806 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Carlo MI et al. Genomic alterations and outcomes with VEGF-targeted therapy in patients with clear cell renal cell carcinoma. Kidney. Cancer 1, 49–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Voss MH et al. Integrated biomarker analysis for 412 renal cell cancer (RCC) patients (pts) treated on the phase 3 COMPARZ trial: correlating common mutation events in PBRM1 and BAP1 with angiogenesis expression signatures and outcomes on tyrosine kinase inhibitor (TKI) therapy. J. Clin. Oncol 35, 4523–4523 (2017). [Google Scholar]
- 157.Shen J et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat. Med 24, 556–562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jelinic P et al. Immune-active microenvironment in small cell carcinoma of the ovary, hypercalcemic type: rationale for immune checkpoint blockade. J. Natl Cancer Inst 110, 787–790 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Riaz N et al. Recurrent SERPINB3 and SERPINB4 mutations in patients who respond to anti-CTLA4 immunotherapy. Nat. Genet 48, 1327–1329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Davoli T, Uno H, Wooten EC & Elledge SJ Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355, 28104840 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tang J, Shalabi A & Hubbard-Lucey VM Comprehensive analysis of the clinical immuno-oncology landscape. Ann. Oncol 29, 84–91 (2018). [DOI] [PubMed] [Google Scholar]
- 162.Patel SA & Minn AJ Combination cancer therapy with immune checkpoint blockade: mechanisms and strategies. Immunity 48, 417–433 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu XS & Mardis ER Applications of Immunogenomics to Cancer. Cell 168, 600–612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sahin U et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017). [DOI] [PubMed] [Google Scholar]
- 165.Ott PA et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ikeda H, Old LJ & Schreiber RD The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev 13, 95–109 (2002). [DOI] [PubMed] [Google Scholar]
- 167.Kirkwood JM et al. Effect of JAK/STAT or PI3Kδ plus PD-1 inhibition on the tumor microenvironment: biomarker results from a phase Ib study in patients with advanced solid tumors. Cancer. Res 78, abstr. CT176 (2018). [Google Scholar]
- 168.Corrales L et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep 11, 1018–1030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Meric-Bernstam F et al. Phase I dose-finding study of MIW815 (ADU-S100), an intratumoral STING agonist, in patients with advanced solid tumors or lymphomas. J. Immunother. Cancer 6, 114 (2018).30400835 [Google Scholar]
- 170.Harrington KJ et al. Preliminary results of the first-in-human (FIH) study of MK-1454, an agonist of stimulator of interferon genes (STING), as monotherapy or in combination with with pembrolizumab (pembro) in patients with advanced solid tumors or lymphomas. Ann. Oncol 29, LBA15 (2018). [Google Scholar]
- 171.Goel S et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 548, 471–475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Schaer DA et al. The CDK4/6 inhibitor abemaciclib induces a T cell inflamed tumor microenvironment and enhances the efficacy of PD-L1 checkpoint blockade. Cell Rep 22, 2978–2994 (2018). [DOI] [PubMed] [Google Scholar]
- 173.Deng J et al. CDK4/6 inhibition augments antitumor immunity by enhancing T-cell activation. Cancer Discov 8, 216–233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tolaney SM et al. Updated efficacy, safety, & PD-L1 status of patients with HR+, HER2− metastatic breast cancer administered abemaciclib plus pembrolizumab. J. Clin. Oncol 36, 1059 (2018). [Google Scholar]
- 175.Tauriello DVF et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 554, 538–543 (2018). [DOI] [PubMed] [Google Scholar]
- 176.Lan Y et al. Enhanced preclinical antitumor activity of M7824, a bifunctional fusion protein simultaneously targeting PD-L1 and TGF-β. Sci. Transl. Med 10, 29343622 (2018). [DOI] [PubMed] [Google Scholar]
- 177.Strauss J et al. Phase I trial of M7824 (MSB0011359C), a bifunctional fusion protein targeting PD-L1 and TGFβ, in advanced solid tumors. Clin. Cancer Res 24, 1287–1295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tao JJ, Schram AM & Hyman DM Basket studies: redefining clinical trials in the era of genome-driven oncology. Annu. Rev. Med 69, 319–331 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Thorlund K, Haggstrom J, Park JJ & Mills EJ Key design considerations for adaptive clinical trials: a primer for clinicians. Br. Med. J 360, k698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ott PA, Hodi FS, Kaufman HL, Wigginton JM & Wolchok JD Combination immunotherapy: a road map. J. Immunother. Cancer 5, 16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Emens LA et al. Cancer immunotherapy: opportunities and challenges in the rapidly evolving clinical landscape. Eur. J. Cancer 81, 116–129 (2017). [DOI] [PubMed] [Google Scholar]
- 182.Anagnostou V et al. Immuno-oncology trial endpoints: capturing clinically meaningful activity. Clin. Cancer Res 23, 4959–4969 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Landry LG, Ali N, Williams DR, Rehm HL & Bonham VL Lack of diversity in genomic databases is a barrier to translating precision medicine research into practice. Health Aff. (Millwood) 37, 780–785 (2018). [DOI] [PubMed] [Google Scholar]
- 184.Chambers CA, Kuhns MS, Egen JG & Allison JP CTLA-4-mediated inhibition in regulation of T cell responses: mechanisms and manipulation in tumor immunotherapy. Annu. Rev. Immunol 19, 565–594 (2001). [DOI] [PubMed] [Google Scholar]
- 185.Simpson TR et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J. Exp. Med 210, 1695–1710 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Selby MJ et al. Anti-CTLA-4 antibodies of IgG2a isotype enhance antitumor activity through reduction of intratumoral regulatory T cells. Cancer. Immunol. Res 1, 32–42 (2013). [DOI] [PubMed] [Google Scholar]
- 187.Wei SC et al. Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell 170, 1120–1133 e1117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Romano E et al. Ipilimumab-dependent cell-mediated cytotoxicity of regulatory T cells ex vivo by nonclassical monocytes in melanoma patients. Proc. Natl Acad. Sci. USA 112, 6140–6145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sharma A et al. Anti-CTLA-4 immunotherapy does not deplete F0XP3+ regulatory T cells (Tregs) in human cancers. Clin. Cancer Res 25, 1233–1238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sharpe AH & Pauken KE The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol 18, 153–167 (2018). [DOI] [PubMed] [Google Scholar]
- 191.Chemnitz JM, Parry RV, Nichols KE, June CH & Riley JL SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J. Immunol 173, 945–954 (2004). [DOI] [PubMed] [Google Scholar]
- 192.Wei F et al. Strength of PD-1 signaling differentially affects T-cell effector functions. Proc. Natl Acad. Sci. USA 110, E2480–E2489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]