Abstract
Neonicotinoids are effective insecticides used on many important arable and horticultural crops. They are nicotinic acetylcholine receptor agonists which disrupt the function of insect neurons and cause paralysis and death. In addition to direct mortality, there are numerous sublethal effects of low doses of neonicotinoids on bees. We hypothesize that some of these large array of effects could be a consequence of epigenetic changes in bees induced by neonicotinoids. We compared whole methylome (BS-seq) and RNA-seq libraries of the brains of buff-tailed bumblebee Bombus terrestris workers exposed to field-realistic doses of the neonicotinoid imidacloprid to libraries from control workers. We found numerous genes which show differential expression between neonicotinoid-treated bees and control bees, but no differentially methylated cytosines in any context. We found CpG methylation to be focused mainly in exons and associated with highly expressed genes. We discuss the implications of our results for future legislation.
Keywords: epigenetics, methylome, RNA-seq, BS-seq, social insects, pesticide
1. Introduction
Neonicotinoids are effective insecticides used on many important arable and horticultural crops, most frequently as seed dressing. They are systemic, meaning they are absorbed by the plant and transported to all tissues where they remain active for many weeks or months. This protects all parts of the plant, but also means that neonicotinoids are found in the nectar and pollen of flowering crops such as oilseed rape, and hence are consumed by bees [1]. It has also emerged that they are commonly found contaminating nectar and pollen of wild flowers growing on arable farmland, providing additional exposure of bees and other pollinators [1,2].
Neonicotinoids are nicotinic acetylcholine receptor agonists which disrupt the function of insect neurons and cause paralysis and death. In addition to direct mortality, laboratory and field studies have documented numerous sublethal effects of low doses of neonicotinoids on both honeybees and bumblebees (e.g. [3,4], reviewed in [5]). Sublethal effects at the individual level include reduced fecundity of queens, reduced fertility in males, impaired immune response, impaired navigation and learning, reduced pollen collection, and reduced food consumption. Collectively, these effects result in reduced colony growth and colony reproduction performance. The breadth of the effects of neonicotinoids on bees suggests that neonicotinoids have multiple modes of action beyond their designed direct impact on neurotransmission, for example their impact on immune signalling [6].
We hypothesize that some of these effects could be a consequence of epigenetic changes induced by neonicotinoids. Epigenetics is defined as the stable and heritable change in gene expression without any change in the DNA sequence [7]. Environmental contaminants have been found to affect the epigenetics of a diverse range of animal species from water fleas to polar bears [8] and include metals, endocrine disrupting compounds, air pollution, persistant organic pollutants, and pesticides [9], but much ecotoxicology research is centred on a direct link between exposure and response [8]. Epigenetic changes have the potential to weaken that link, with effects possibly manifesting much later in life or in subsequent generations. Thus if pesticide-induced epigenetic changes were shown to be heritable in bees, this would have implications for future ecological risk assessment.
In social insect research the role of DNA methylation, an epigenetic marker primarily involving the addition of a methyl group to a cytosine, has come under increasing scrutiny in recent years [10–20]. Methylation has also been implicated in important effects on the biology of bees, including the control of reproductive status [10,21] and memory [22], behaviours shown to be affected by neonicotinoids [23,24], although in the case of reproduction the link between methylation and social insect reproduction is controversial [16,18,25]. DNA methylation has been linked with alternative splicing in a number of insect species [11,14,17,26], and with histone modifications in the ant Camponotus floridanus [27]. In mammals, methylation on gene promoters leads to a reduction in gene expression. The effect of methylation on gene expression in insects is less well understood [28], though high levels of methylation have been associated with highly and stably expressed genes [29–31], while in honeybees hypomethylated genes are associated with caste-specific expression [16,32,33]. Gene expression differences due to neonicotinoid exposure have been found in honeyebee larval workers, adult workers, and queens [34–38].
In this study, we use whole-genome bisulfite sequencing (WGBS/BS-seq) and RNA-seq on brain tissue of neonicotinoid exposed and control Bombus terrestris workers in order to elucidate the effects of the neonicotinoid imidacloprid on the gene expression and methylation status of bumblebee workers.
2. Material and methods
(a). Beekeeping, experimental design, and brain dissection
Six colonies of Bombus terrestris audax were purchased from Agralan, UK. Each colony contained a queen and on average 10 workers and a small amount of brood. They were kept in wooden nest boxes and maintained under red light at 26°C and 60% humidity on a diet of 50% v/v glucose/fructose apiary solution (Meliose-Roquette, France) and pollen (Percie du set, France) [10]. Three colonies were used for the RNA-seq experiment and the other three for the BS-seq experiment (electronic supplementary material, figure S1).
Groups of five callow workers born on the same day were reared in Perspex boxes (18.5 cm × 12.5 cm × 6.5 cm). Boxes were then randomly assigned to control or treated groups. The control group was fed ad libitum with 50% v/v apiary solution for 6 days, whereas the treated group was fed ad libitum with a 10 ppb imidacloprid (Sigma-Aldrich) 50% v/v apiary solution, a field-realistic sublethal dose [39,40]. After a 6-day chronic exposure period [40], the bees were anaesthetized on ice at 4°C. The brains were dissected in phosphate buffered saline and immediately frozen in liquid nitrogen and stored at − 80°C. Their ovaries were checked for development to ensure that only non-reproductive workers were used [10,41].
(b). BS-seq
(i). Genomic DNA extraction, sequencing, and mapping
Six libraries were prepared (three colonies, control, and treatment). For each colony, 10 boxes were reared (five control and five treatment). Each library was generated from 12 pooled brains of non-reproductive workers taken at random from the relevant boxes for a total of 72 brains. Genomic DNA was extracted, using QIAGEN QIAamp DNA Micro Kit following the manufacturer’s instruction. The concentration of genomic DNA was measured using a Qubitregistered dsDNA BR Assay Kit (ThermoFisher Scientific, USA) and Nanodrop. Sequencing was performed on a HiSeq 2000 machine (Illumina, Inc.) at the Beijing Genomics Institute (BGI), generating 100-bp paired-end reads.
Poor quality reads were removed using fastQC v.0.11.2 [42] and adapters trimmed using cutadapt v.1.11 [43] and trimmomatic v.0.36 [44]. Bismark v.0.18.1 [45] was used to align the reads to the Bter_1.0 genome (Refseq accession no. GCF_000214255.1 [46]), remove PCR artefacts, and extract methylation calls in CpG, CHH, and CHG contexts (where H represents adenine, thymine, or cytosine). The cytosine report files from Bismark and the B. terrestris annotation file (GCF_000214255.1) were combined using the sqldf library [47] in R v.3.4.0 [48] to generate the distribution of methylated Cs over genomic features. Cytosines with less than 10× coverage were excluded. For each cytosine, the proportion of methylation reads over total reads was calculated.
(ii). Methylation differences between treatments
Differential methylation analysis was performed using methylKit [49]. Bismark cytosine reports were filtered to exclude loci with extreme low or high coverage (less than 10 or greater than 500 reads) and those not covered in all samples. A mixture of binomial model (Cheng and Zhu, 2014 [50]) was used to make per-loci methylation status calls and only loci identified as methylated in at least one sample were tested. A logistic regression test was applied using overdispersion correction, controlling for colony as a covariate, and adjusting p-values for multiple testing using the SLIM method. A minimum change in methylation between treatments of 10% was used to filter results.
(c). RNA-seq
(i). RNA extraction and Illumina sequencing
Eighteen libraries were prepared (three colonies, three replicates per colony, two conditions). For each colony, six boxes were reared (three control and three treatment). Each library was generated from three pooled brains of non-reproductive workers taken from the relevant boxes, for a total of 54 brains. Total RNA was isolated using the GenElute Mammalian Total RNA Miniprep Kit. DNA and RNAase activity was eliminated using (Sigma-Aldrich DNase I treatment kit) following the manufacturer’s instruction. RNA concentration and integrity were determined by Bioanalyzer using the RNA Nano Kit (Agilent Technologies). From each sample, we isolated an average of 0.8 mg of RNA. Two samples appeared degraded and were not used. Nine control and seven treated samples were prepared and sequenced on HiSeq 200 (Illumina, Inc.) at Beijing Genomics Institute (BGI) and 100-bp paired-end reads were generated.
BGI removed adaptor sequences, contamination, and low-quality reads from raw data. Base calling and quality scoring of the raw reads were visualized using fastQC v.0.11.2 [42]. The clean reads for each sample were aligned to the reference genome Bter_1.0 genome (Refseq accession no. GCF_000214255.1 [46]) using Hisat2 v.2.0.4 [51] with default parameters. The output sam file was sorted and converted to a bam file using samtools [52]. Aligned reads were assembled and quantified using the assembler stringtie v.1.3.3b [53].
(ii). Differential gene expression analysis
A table of raw counts was generated using a Python script (https://github.com/gpertea/stringtie/blob/master/prepDE) and analysed using DESeq2 [54] in R v.3.4.0 [48] to estimate differentially expressed genes (DEGs) using an FDR-adjusted p-value threshold of 0.05 and controlling for colony effects. Genes with less than 10 reads were discarded from analysis. The normalized read counts were log 2 transformed. The quality of replicates was assessed by plotting read counts of samples against one another and assessing the dispersion and presence of any artefacts between samples [55]. A principal-component analysis was performed to visualize diversity between samples within treatment and between condition.
(d). Gene ontology term enrichment and Kyoto Encyclopedia of Genes and Genomes analysis
A list of gene ontology (GO) terms for the bumblebee were made by annotating the transcriptome using trinotate (default settings) [56] and blast2GO (against RefSeq) [57]. These lists were combined, using the pipeline implemented in [58] with a K-value of 1. A hypergeometric test was applied and significant GO terms identified after Benjamini-Hochberg (BH) correction (p corrected <0.05) [59] using GOstats [60], with all RNA features in the bumblebee genome used as a background (GCF_000214255.1). We filtered these to only those terms present in three or more DEGs and used REVIGO [61] to cluster and visualize enriched GO terms, selecting the whole UniProt database and SimRel semantic similarity measure.
The clusterprofiler R package (v.3.8.1) [62] identified differentially expressed genes associated with Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using the whole UniProt database. A hypergeometric test was applied and significant KEGG pathways were identified after BH correction (q-value <0.05) [58].
3. Results
(a). Methylation analysis
The overall sequence alignment rate was (). The proportion of methylated cytosine reads calculated by Bismark were for CpGs, for CHGs, for CHHs, and for CNs or CHNs ((H = A, C or T). While insect methylation levels are often low [15] these methylation levels are lower even than in the honeybee, Apis mellifera, estimated at approximately at the genome level using similar metrics [63,64]. In a CpG context, across all samples, of loci with a minimum coverage of 10 reads were considered methylated by the mixture of binomial model. The distribution of CpG methylation shows a mild bimodal distribution with the vast majority of sites being not or only modestly methylated and a few fully methylated (electronic supplementary material, figure S2a). Methylated CpGs are more abundant in coding regions (sevenfold) and exons (fivefold) than introns (figure 1a). Non-CpG per-loci methylation levels were reported as less than 0.001% by the mixture of binomial model. This, in conjunction with the uniformity of non-CpG methylation across genomic features (figure 1b,c), led to the conclusion that such levels were indistinguishable from error and as such were excluded from subsequent analysis.
Figure 1.
Methylated Cs distribution. Average proportion of methylation reads ±s.d. per CpG (a), CHG (b), and CHH (c) positions over genomic features. Control samples in black and neonicotinoid-treated samples in grey.
(i). Methylation differences between control and neonicotinoid-treated samples
In total, 4 424 986 loci were analysed using the mixture of binomial model, which subsequently identified 6080 sites to test. No differentially methylated loci were identified using logistic regression at a q-value of 0.05 or 0.1. MethylKit includes an option to pool replicates into single control/treatment samples and use Fisher’s exact test; using this approach we identified a small number of differentially methylated CpGs at q-value <0.1, including loci within histone-lysine N-methyltransferase 2C, histone acetyltransferase p300, CXXC1 (a transcriptional activator that binds to unmethylated CpGs), and genes involved with axon formation (electronic supplementary material, diff_meth_fisher).
(b). Expression analysis
Alignment rate to the genome was 93.6% (92.1–94.1) and after filtering a total of 10 772 genes were analysed. All libraries from the same treatment showed low variation in their gene expression patterns (electronic supplementary material, figures S3 and S4).
(i). Differential expression
A total of 405 genes were differentially expressed: 192 genes upregulated and 213 downregulated in neonicotinoid samples compared to controls (see electronic supplementary material, differentially_expressed_genes). Four cytochrome P450 (CYP) genes were differentially expressed, two upregulated and two downregulated. Upregulated genes in neonicotinoid-treated bees also include apyrase that hydrolyzes ATP to AMP, the neuropeptide receptor pyrokinin-1 receptor, and ionotropic receptor 25a that is involved in circadian clock resetting in Drosophila [65]. Downregulated genes include neurexin, involved in synaptic formation and maintenance, peptide methionine sulfoxide reductase, involved in repair of oxidation-damaged proteins, and a number of genes related to photoreceptor function. Three genes belonging to the homeotic box gene (Hox) family were downregulated in neonicotinoid-treated bees. lethal(2)essential for life (Efl21) displayed the highest downregulation. We found 105 enriched biological process GO terms (BH corrected p < 0.05) associated with differential gene expression (electronic supplementary material, expression_GO), subsequently clustered using REVIGO to 58 terms (electronic supplementary material, figure S5). Many of the most significantly enriched terms were associated with energy reserve metabolism. Also enriched were terms associated with apoptotic processes, apoptotic cell clearance, immune effector processes, cell death, and response to chemical stimulus. No KEGG pathways were over-represented for differentially expressed genes (q < 0.05).
(c). DNA methylation: expression correlation
We calculated the average percentage of methylated reads per gene for the most differentially expressed genes (log2 fold-change greater than 0.5 or less than −0.5) and non-differentially expressed genes (figure 2), fitting a generalized linear model (GLM) with a quasi-binomial error distribution with treatment (control versus neonicotinoid) and expression state (DEG versus non-DEG) as independent variables. There were no significant interactions between the independent variables (interaction model versus main effects only model: χ2 = −0.014, d.f. = 1, p = 0.82). For CpGs, non-differentially expressed genes had more methylation than differentially expressed genes (z1,19673 = 4.641, p < 0.001). There was no significant treatment effect on methylation levels (z1,19673 = −0.772, p = 0.692).
Figure 2.
Average percentage of methylated CpG per gene. Differentially expressed genes (DEG) and non-differentially expressed genes (non-DEG) are plotted separately. Dots represent genes.
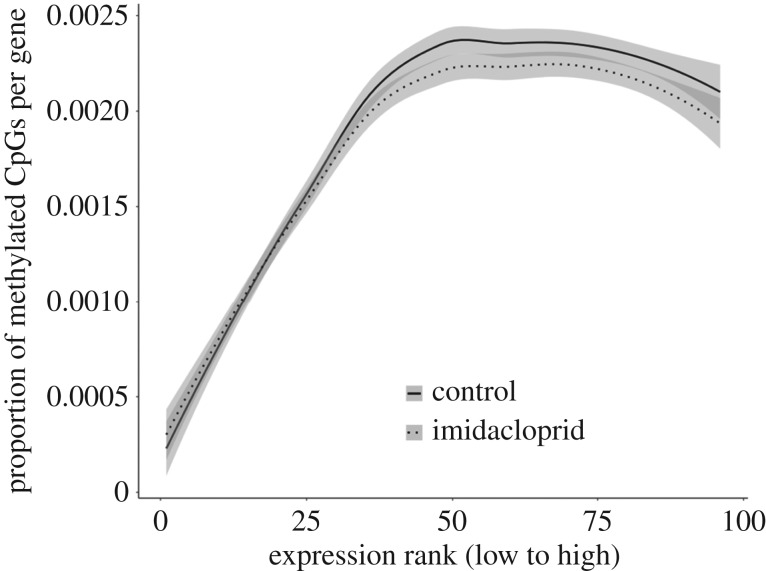
To have a more fine-scale understanding of the correlation between methylation and expression, we plotted mean proportion of methylation per gene against ranked expression level (log10 fragments per kilobase of exon model per million reads mapped (fpkm) per gene) in 100 bins (from low to high) (figure 3) fitting a linear model with treatment and expression level as independent variables. There was no significant interaction between the effects of expression and treatment on methylation (interaction model versus main effects only model: F1,189 = 1.0347, p = 0.3104). We found a significant association between expression and methylation (F1,189 = 281.654, p = <2 × 10−16). Neonicotinoid-treated bees had comparable levels of CpG methylation to control bees (F1,189 = 1.8125, p = 0.1798).
Figure 3.
The proportion of methylated CpGs is plotted against gene expression rank. One hundred ‘bins’ of progressively increasing level of expression were generated and genes with a similar level of expression have been grouped in the same bin. Solid lines represent control samples and dotted lines neonicotinoid-treated samples. The grey shading represents 95% confidence intervals.
4. Discussion
We found numerous genes which show differential expression between bees treated with field-realistic doses of the neonicotinoid imidacloprid and control bees. We found CpG methylation to be focused in exons, and high CpG methylation was associated with highly expressed genes, but no differentially methylated loci were detected between treatments. Non-differentially expressed genes had higher methylation levels than differentially expressed genes.
Four cytochrome P450 (CYP) genes were identified as differentially expressed, in line with other studies assessing the impact of insecticides on honeybees [37,38,66–68]. Two were upregulated (CYP6k1 and 4c3) and two downregulated (28d1 and 9e2). CYP6, 9, and 28 genes are linked to xenobiotic metabolism and resistance to insecticides [69] and CYP6 genes specifically have been found to be upregulated in honeybees after treatment with sublethal doses of the neonicotinoid thiamethoxam [67], as has CYP4C1 after treatment with the neonicotinoid clothianidin [37]. The CYP9Q subfamily was recently shown to be responsible for bee sensitivity to neonicotinoids [70].
The identification of DEGs associated with synaptic transmission (electronic supplementary material, expression_GO) is to be expected, given that we used brain tissue and given the known target effects of neonicotinoids. The identification of a downregulated neurexin gene aligns with the results of Shi et al. [67]. The effect seen here on metabolic pathways has also been found in honeybees, with GO term enrichment for catabolic carbohydrate and lipid metabolism [37]. These authors suggested that due to the intensive energy demands of the brain, negative effects on metabolic pathways could affect brain function and therefore behaviour. During the review period, a further study was published examining gene expression changes in B. terrestris after exposure to neonicotinoids, again showing changes in carbohydrate and lipid metabolism [71]. Efl21, the most downregulated gene identified, has been found to be involved in foraging behaviour in bees [72], a potential genetic link to the findings of Mommaerts et al. [73]. Impaired foraging has implications for pollination, reproduction, and overall colony survival. Downregulation of carbohydrate metabolism pathways has also been shown in honeybee larvae [38,68]. Also downregulated were three hox genes. This may be indicative of an impaired immune system, as hox genes have been found to play a role in invertebrate innate immune responses [74,75]. Hox genes have been found to be downregulated in response to insecticide treatment in honeybees [34]. The bumblebee visual system may also be impacted by imidacloprid treatment, given the downregulation of genes such as protein scarlet, protein glass, and ninaC.
No differentially methylated loci between control and treatment were identified using a logistic regression model, and we suggest that if acute neonicotinoid exposure does alter methylation status in B. terrestris it is subtle and the data reported here may be underpowered to detect it due to low per-sample coverage. A small number of differentially methylated loci were identified by pooling replicates and using Fisher’s exact test (electronic supplementary material, diff_meth_fisher), but unlike logistic regression, this approach cannot control for covariates and the results should be treated with caution. Using this approach, a CpG loci in CXXC-type zinc finger protein 1 was identified as hypermethylated in neonicotinoid-treated bees; this gene was also upregulated in that group. In mammals, CXXC1 is a transcriptional activator that binds to unmethylated CpGs to regulate gene expression [76]. Other loci identified by pooling were located within histone acetyltransferase p300 and histone-lysine N-methyltransferase 2C. These findings raise the possibility that neonicotinoids may have a more detectable effect on methylation and subsequent gene expression over a longer period through a cascade of epigenetic processes. A study on the effects of imidacloprid on bumblebees found no effect on mortality or reproduction over 11 weeks using 10 ppb when workers were not required to forage for food, while 20 ppb affected mortality and foraging was impaired at both doses [73]. It may, therefore, be that a higher dose or longer exposure time might have a detectable impact on CpG methylation, and further work investigating chronic rather than acute exposure to imidacloprid at different doses would be valuable. Also worthy of investigation is the potential effect on epigenetic processes other than DNA methylation, such as histone modification, which has been found to have a similar, but non-redundant, association with gene expression in the ant Camponotus floridanus [27].
We found patterns of CpG methylation to be in line with other insect species. It is mainly focused in exons [15], and high CpG methylation was associated with highly expressed genes (figure 3) [11,13,16,18,29,31], and non-differentially expressed genes showed higher levels of methylation [13,14,16,77]. As well as inducing no changes in methylation at individual loci, neonicotinoids appear to have no effect on overall levels of CpG methylation (figures 2 and 3). This failure to identify methylation differences between experimental groups is consistent with findings of robust methylation between castes in various insects [78] but contrasts with studies finding differences resulting from removal of maternal care [11], or within castes with differing reproductive status [33].
Non-CpG methylation plays a role in gene silencing in flowering plants [79] and to a lesser extent, in mammals [80]. In this study, while we identified a very small number of loci showing methylation in CHG/CHH contexts we could not exclude the possibility that much of it was noise, as bisulfite sequencing is prone to false positives from sources such as incomplete bisulfite conversion, miscalled bases, and SNPs. Overall, we conclude that there is no notable methylation of non-CpG cytosines in B. terrestris, as with the honeybee [17] and Nasonia vitripennis [31]. In contrast to the preponderance of CpG methylation in exons, we found that CHH and CHG methylation was uniformly spread throughout genes (figure 1) a pattern which would be consistent with the idea that there is no significant methylation in these contexts.
Recently, it has become clear that epigenetics can play a role in the interplay between man-made chemicals and natural ecosystems, and their constituent species [9]. Hymenopteran insects (ants, bees, and wasps) are ideal models to study this. They are both strongly affected by man-made chemicals and are important emerging models for epigenetics, with a number of species with relatively small genomes showing a confirmed role for methylation in their biology [81–84].
However, on the evidence of this study, imidacloprid does not appear to have epigenetic effects, at least through DNA methylation. This finding is important in the context of future legislation for pesticide control, as it is evidence suggesting a potential lack of transgenerational effects on B. terrestris with the use of imidacloprid.
Supplementary Material
Acknowledgements
Thanks to Dr Swidbert Ott and Prof. Dave Goulson for discussions.
Data accessibility
All sequencing data related to this project can be found under NCBI BioProject PRJNA524132.
Authors' contributions
E.B.M., E.R., and P.S.A.B designed the study. P.S.A.B. carried out the experiments. P.S.A.B., B.J.H., M.P., A.R.C.J., and H.M. analysed the data. M.P., P.S.A.B., and E.B.M. wrote the initial draft. All authors were involved in redrafting.
Competing interests
We declare we have no competing interests.
Funding
E.B.M., B.J.H., and M.P. were funded by NERCgrant NE/N010019/1. P.S.A.B. was supported by a scholarship from the Human Capacity Development Program (Koya University, Iraq). H.M. was supported by a NERC CENTA DTP studentship. A.R.C.J. was supported by a BBSRC MIBTP DTP studentship. This research used the ALICE High Performance Computing Facility at the University of Leicester.
References
- 1.Botías C, David A, Horwood J, Abdul-Sada A, Nicholls E, Hill E, Goulson D. 2015. Neonicotinoid residues in wildflowers, a potential route of chronic exposure for bees. Environ. Sci. Technol. 49, 12 731–12 740. ( 10.1021/acs.est.5b03459) [DOI] [PubMed] [Google Scholar]
- 2.David A, Botías C, Abdul-Sada A, Nicholls E, Rotheray EL, Hill EM, Goulson D. 2016. Widespread contamination of wildflower and bee-collected pollen with complex mixtures of neonicotinoids and fungicides commonly applied to crops. Environ. Int. 88, 169–178. ( 10.1016/j.envint.2015.12.011) [DOI] [PubMed] [Google Scholar]
- 3.Rundlöf M. et al. 2015. Seed coating with a neonicotinoid insecticide negatively affects wild bees. Nature 521, 77–80. ( 10.1038/nature14420) [DOI] [PubMed] [Google Scholar]
- 4.Whitehorn PR, O’Connor S, Wackers FL, Goulson D. 2012. Neonicotinoid pesticide reduces bumble bee colony growth and queen production. Science 336, 351–352. ( 10.1126/science.1215025) [DOI] [PubMed] [Google Scholar]
- 5.Pisa LW. et al. 2015. Effects of neonicotinoids and fipronil on non-target invertebrates. Environ. Sci. Pollut. Res. 22, 68–102. ( 10.1007/s11356-014-3471-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prisco GD, Cavaliere V, Annoscia D, Varricchio P, Caprio E, Nazzi F, Gargiulo G, Pennacchio F. 2013. Neonicotinoid clothianidin adversely affects insect immunity and promotes replication of a viral pathogen in honey bees. Proc. Natl Acad. Sci. USA 110, 18 466–18 471. ( 10.1073/pnas.1314923110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldberg AD, Allis CD, Bernstein E. 2007. Epigenetics: a landscape takes shape. Cell 128, 635–638. ( 10.1016/j.cell.2007.02.006) [DOI] [PubMed] [Google Scholar]
- 8.Head JA. 2014. Patterns of DNA methylation in animals: an ecotoxicological perspective. Integr. Comp. Biol. 54, 77–86. ( 10.1093/icb/icu025) [DOI] [PubMed] [Google Scholar]
- 9.Vandegehuchte MB, Janssen CR. 2014. Epigenetics in an ecotoxicological context. Mutat. Res./Genet. Toxicol. Environ. Mutagen. 764–765, 36–45. ( 10.1016/j.mrgentox.2013.08.008) [DOI] [PubMed] [Google Scholar]
- 10.Amarasinghe HE, Clayton CI, Mallon EB. 2014. Methylation and worker reproduction in the bumble-bee (Bombus terrestris). Proc. R. Soc. B 281, 20132502 ( 10.1098/rspb.2013.2502) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arsenault SV, Hunt BG, Rehan SM. 2018. The effect of maternal care on gene expression and DNA methylation in a subsocial bee. Nat. Commun. 9(1), 3468 ( 10.1038/s41467-018-05903-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foret S, Kucharski R, Pittelkow Y, Lockett GA, Maleszka R. 2009. Epigenetic regulation of the honey bee transcriptome: unravelling the nature of methylated genes. BMC Genomics 10, 472 ( 10.1186/1471-2164-10-472) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glastad KM, Hunt BG, Goodisman MAD. 2013. Evidence of a conserved functional role for DNA methylation in termites. Insect Mol. Biol. 22, 143–154. ( 10.1111/imb.2013.22.issue-2) [DOI] [PubMed] [Google Scholar]
- 14.Glastad KM, Gokhale K, Liebig J, Goodisman MAD. 2016. The caste- and sex-specific DNA methylome of the termite Zootermopsis nevadensis. Sci. Rep. 6, 37110 ( 10.1038/srep37110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glastad KM. et al. 2017. Variation in DNA methylation is not consistently reflected by sociality in hymenoptera. Genome Biol. Evol. 9, 1687–1698. ( 10.1093/gbe/evx128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Libbrecht R, Oxley PR, Keller L, Kronauer DJC. 2016. Robust DNA methylation in the clonal raider ant brain. Curr. Biol. 26, 391–395. ( 10.1016/j.cub.2015.12.040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. 2010. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 8, e1000506 ( 10.1371/journal.pbio.1000506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patalano S. et al. 2015. Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies. Proc. Natl Acad. Sci. USA 112, 13 970–13 975. ( 10.1073/pnas.1515937112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rehan SM, Glastad KM, Lawson SP, Hunt BG. 2016. The genome and methylome of a subsocial small carpenter bee, Ceratina calcarata. Genome Biol. Evol. 8, 1401–1410. ( 10.1093/gbe/evw079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Standage DS, Berens AJ, Glastad KM, Severin AJ, Brendel VP, Toth AL. 2016. Genome, transcriptome and methylome sequencing of a primitively eusocial wasp reveal a greatly reduced DNA methylation system in a social insect. Mol. Ecol. 25, 1769–1784. ( 10.1111/mec.2016.25.issue-8) [DOI] [PubMed] [Google Scholar]
- 21.Kucharski R, Maleszka J, Foret S, Maleszka R. 2008. Nutritional control of reproductive status in honeybees via DNA methylation. Science 319, 1827–1830. ( 10.1126/science.1153069) [DOI] [PubMed] [Google Scholar]
- 22.Biergans SD, Jones JC, Treiber N, Galizia CG, Szyszka P. 2012. DNA methylation mediates the discriminatory power of associative long-term memory in honeybees. PLoS ONE 7, e39349 ( 10.1371/journal.pone.0039349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanley DA, Smith KE, Raine NE. 2015. Bumblebee learning and memory is impaired by chronic exposure to a neonicotinoid pesticide. Sci. Rep. 5, 16508 ( 10.1038/srep16508) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams GR, Troxler A, Retschnig G, Roth K, Yañez O, Shutler D, Neumann P, Gauthier L. 2015. Neonicotinoid pesticides severely affect honey bee queens. Sci. Rep. 5, 14621 ( 10.1038/srep14621) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herb BR, Shook MS, Fields CJ, Robinson GE. 2018. Defense against territorial intrusion is associated with DNA methylation changes in the honey bee brain. BMC Genomics 19, 216 ( 10.1186/s12864-018-4594-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li-Byarlay H. et al. 2013. RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee. Proc. Natl Acad. Sci. USA 110, 12 750–12 755. ( 10.1073/pnas.1310735110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glastad KM, Hunt BG, Goodisman MAD. 2015. DNA methylation and chromatin organization in insects: insights from the ant Camponotus floridanus. Genome Biol. Evol. 7, 931–942. ( 10.1093/gbe/evv039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pegoraro M, Marshall H, Lonsdale ZN, Mallon EB. 2017. Do social insects support Haig’s kin theory for the evolution of genomic imprinting? Epigenetics 12, 725–742. ( 10.1080/15592294.2017.1348445) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonasio R. et al. 2012. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr. Biol. 22, 1755–1764. ( 10.1016/j.cub.2012.07.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foret S, Kucharski R, Pellegrini M, Feng S, Jacobsen SE, Robinson GE, Maleszka R. 2012. DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. Proc. Natl Acad. Sci. USA 109, 4968–4973. ( 10.1073/pnas.1202392109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Wheeler D, Avery A, Rago A, Choi J-H, Colbourne JK, Clark AG, Werren JH. 2013. Function and evolution of DNA methylation in Nasonia vitripennis. PLoS Genet. 9, e1003872 ( 10.1371/journal.pgen.1003872) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elango N, Hunt BG, Goodisman MAD, Yi SV. 2009. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc. Natl Acad. Sci. USA 106, 11 206–11 211. ( 10.1073/pnas.0900301106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall H, Lonsdale ZN, Mallon EB. 2019. Methylation and gene expression differences between reproductive castes of bumblebee workers. bioRxiv, p. 517698.
- 34.Aufauvre J, Misme-Aucouturier B, Vigués B, Texier C, Delbac F, Blot N. 2014. Transcriptome analyses of the honeybee response to Nosema ceranae and insecticides. PLoS ONE 9, e91686 ( 10.1371/journal.pone.0091686) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaimanee V, Evans JD, Chen Y, Jackson C, Pettis JS. 2016. Sperm viability and gene expression in honey bee queens (Apis mellifera) following exposure to the neonicotinoid insecticide Imidacloprid and the organophosphate acaricide coumaphos. J. Insect. Physiol. 89, 1–8. ( 10.1016/j.jinsphys.2016.03.004) [DOI] [PubMed] [Google Scholar]
- 36.Christen V, Mittner F, Fent K. 2016. Molecular effects of neonicotinoids in honey bees (Apis mellifera). Environ. Sci. Technol. 50, 4071–4081. ( 10.1021/acs.est.6b00678) [DOI] [PubMed] [Google Scholar]
- 37.Christen V, Schirrmann M, Frey JE, Fent K. 2018. Global transcriptomic effects of environmentally relevant concentrations of the Neonicotinoids Clothianidin, Imidacloprid, and Thiamethoxam in the brain of honey bees (Apis mellifera). Environ. Sci. Technol. 52, 7534–7544. ( 10.1021/acs.est.8b01801) [DOI] [PubMed] [Google Scholar]
- 38.Derecka K. et al. 2013. Transient exposure to low levels of insecticide affects metabolic networks of honeybee larvae. PLoS ONE 8, e68191 ( 10.1371/journal.pone.0068191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blacquiére T, Smagghe G, van Gestel CAM, Mommaerts V. 2012. Neonicotinoids in bees: a review on concentrations, side-effects and risk assessment. Ecotoxicology 21, 973–992. ( 10.1007/s10646-012-0863-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cresswell JE. 2011. A meta-analysis of experiments testing the effects of a neonicotinoid insecticide (Imidacloprid) on honey bees. Ecotoxicology 20, 149–157. ( 10.1007/s10646-010-0566-0) [DOI] [PubMed] [Google Scholar]
- 41.Harrison MC, Hammond RL, Mallon EB. 2015. Reproductive workers show queenlike gene expression in an intermediately eusocial insect, the buff-tailed bumble bee Bombus terrestris. Mol. Ecol. 24, 3043–3063. ( 10.1111/mec.13215) [DOI] [PubMed] [Google Scholar]
- 42.Andrews S. 2010. FastQC a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 43.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. ( 10.14806/ej.17.1) [DOI] [Google Scholar]
- 44.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572. ( 10.1093/bioinformatics/btr167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sadd BM. et al. 2015. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 16, 76 ( 10.1186/s13059-015-0623-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grothendieck G. 2017. sqldf: Manipulate R Data Frames Using SQL. R package version 0.4-11.
- 48.R Core Team. 2014. R: A language and environment for statistical computing. Vienna, Austria: R foundation for statistical computing.
- 49.Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE. 2012. MethylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87 ( 10.1186/gb-2012-13-10-r87) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng L, Zhu Y. 2014. A classification approach for DNA methylation profiling with bisulfite next-generation sequencing data. Bioinformatics (Oxford, England), 30(2), 172–179. [DOI] [PubMed] [Google Scholar]
- 51.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. ( 10.1038/nmeth.3317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H. et al. 2009. The sequence alignment/map format and samtools. Bioinformatics 25, 2078–2079. ( 10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. ( 10.1038/nbt.3122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 ( 10.1186/s13059-014-0550-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rich C. et al. 2018. Cell type identity determines transcriptomic immune responses in Arabidopsis thaliana roots. bioRxiv, p. 302448.
- 56.Hébert FO, Grambauer S, Barber I, Landry CR, Aubin-Horth N. 2016. Transcriptome sequences spanning key developmental states as a resource for the study of the cestode Schistocephalus solidus, a threespine stickleback parasite. GigaScience 5, 24 ( 10.1186/s13742-016-0128-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2go: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. ( 10.1093/bioinformatics/bti610) [DOI] [PubMed] [Google Scholar]
- 58.Amar D, et al. 2014. Evaluation and integration of functional annotation pipelines for newly sequenced organisms: the potato genome as a test case. BMC Plant Biology 14(1), 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodological) 57, 289–300. [Google Scholar]
- 60.Falcon S, Gentleman R. 2007. Using GOstats to test gene lists for GO term association. Bioinformatics 23, 257–258. ( 10.1093/bioinformatics/btl567) [DOI] [PubMed] [Google Scholar]
- 61.Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800 ( 10.1371/journal.pone.0021800) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu G, Wang L-G, Han Y, He Q-Y. 2012. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A J. Integr. Biol. 16, 284–287. ( 10.1089/omi.2011.0118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bewick AJ, Vogel KJ, Moore AJ, Schmitz RJ. 2017. Evolution of DNA methylation across insects. Mol. Biol. Evol. 34, 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feng S. et al. 2010. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl Acad. Sci. USA 107, 8689–8694. ( 10.1073/pnas.1002720107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen C. et al. 2015. Drosophila ionotropic receptor 25a mediates circadian clock resetting by temperature. Nature 527, 516–520. ( 10.1038/nature16148) [DOI] [PubMed] [Google Scholar]
- 66.Li Z. et al. 2017. Differential physiological effects of neonicotinoid insecticides on honey bees: a comparison between Apis mellifera and Apis cerana. Pestic. Biochem. Physiol. 140, 1–8. ( 10.1016/j.pestbp.2017.06.010) [DOI] [PubMed] [Google Scholar]
- 67.Shi T-F, Wang Y-F, Liu F, Qi L, Yu L-S. 2017. Sublethal effects of the neonicotinoid insecticide Thiamethoxam on the transcriptome of the honey bees (Hymenoptera: Apidae). J. Econ. Entomol. 110, 2283–2289. ( 10.1093/jee/tox262) [DOI] [PubMed] [Google Scholar]
- 68.Wu M-C, Chang Y-W, Lu K-H, Yang E-C. 2017. Gene expression changes in honey bees induced by sublethal imidacloprid exposure during the larval stage. Insect. Biochem. Mol. Biol. 88, 12–20. ( 10.1016/j.ibmb.2017.06.016) [DOI] [PubMed] [Google Scholar]
- 69.Feyereisen R. 2006. Evolution of insect P450. Biochem. Soc. Trans. 34, 1252–1255. ( 10.1042/BST0341252) [DOI] [PubMed] [Google Scholar]
- 70.Manjon C. et al. 2018. Unravelling the molecular determinants of bee sensitivity to neonicotinoid insecticides. Curr. Biol. 28, 1137–1143.e5. ( 10.1016/j.cub.2018.02.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Colgan TJ, Fletcher IK, Arce AN, Gill RJ, Rodrigues AR, Stolle E, Chittka L, Wurm Y. 2019. Caste- and pesticide-specific effects of neonicotinoid pesticide exposure on gene expression in bumblebees. Mol. Ecol. 28, 1964–1974. ( 10.1111/mec.2019.28.issue-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hernández LG. et al. 2012. Worker honeybee brain proteome. J. Proteome Res. 11, 1485–1493. ( 10.1021/pr2007818) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mommaerts V, Reynders S, Boulet J, Besard L, Sterk G, Smagghe G. 2009. Risk assessment for side-effects of neonicotinoids against bumblebees with and without impairing foraging behavior. Ecotoxicology 19, 207 ( 10.1007/s10646-009-0406-2) [DOI] [PubMed] [Google Scholar]
- 74.Irazoqui JE, Ng A, Xavier RJ, Ausubel FM. 2008. Role for beta-catenin and HOX transcription factors in Caenorhabditis elegans and mammalian host epithelial-pathogen interactions. Proc. Natl Acad. Sci. USA 105, 17 469–17 474. ( 10.1073/pnas.0809527105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Uvell H, Engström Y. 2007. A multilayered defense against infection: combinatorial control of insect immune genes. Trends Genet. 23, 342–349. ( 10.1016/j.tig.2007.05.003) [DOI] [PubMed] [Google Scholar]
- 76.Shin Voo K, Carlone DL, Jacobsen BM, Flodin A, Skalnik DG. 2000. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol. Cell. Biol. 20, 2108–2121. ( 10.1128/MCB.20.6.2108-2121.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sarda S, Zeng J, Hunt BG, Yi SV. 2012. The evolution of invertebrate gene body methylation. Mol. Biol. Evol. 29, 1907–1916. ( 10.1093/molbev/mss062) [DOI] [PubMed] [Google Scholar]
- 78.Hunt BG, Brisson JA, Yi SV, Goodisman MAD. 2010. Functional conservation of DNA methylation in the pea Aphid and the honeybee. Genome Biol. Evol. 2, 719–728. ( 10.1093/gbe/evq057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE. 2014. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21, 64–72. ( 10.1038/nsmb.2735) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dyachenko OV, Schevchuk TV, Kretzner L, Buryanov YI, Smith SS. 2010. Human non-CG methylation. Epigenetics 5, 569–572. ( 10.4161/epi.5.7.12702) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Glastad KM, Hunt BG, Yi SV, Goodisman MAD. 2011. DNA methylation in insects: on the brink of the epigenomic era. Insect Mol. Biol. 20, 553–565. ( 10.1111/imb.2011.20.issue-5) [DOI] [PubMed] [Google Scholar]
- 82.Weiner SA, Toth AL. 2012 Epigenetics in social insects: a new direction for understanding the evolution of castes. Genetics Research International. 2012, Article ID 609810.
- 83.Welch M, Lister R. 2014. Epigenomics and the control of fate, form and function in social insects. Curr. Opin. Insect Sci. 1, 31–38. ( 10.1016/j.cois.2014.04.005) [DOI] [PubMed] [Google Scholar]
- 84.Yan H, Simola DF, Bonasio R, Liebig J, Berger SL, Reinberg D. 2014. Eusocial insects as emerging models for behavioural epigenetics. Nat. Rev. Genet. 15, 677–688. ( 10.1038/nrg3787) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data related to this project can be found under NCBI BioProject PRJNA524132.