Abstract
Objective
Hypertrophic white adipose tissue (WAT) morphology is associated with insulin resistance and type 2 diabetes. The mechanisms governing hyperplastic versus hypertrophic WAT expansion are poorly understood. We assessed if epigenetic modifications in adipocytes are associated with hypertrophic adipose morphology. A subset of genes with differentially methylated CpG-sites (DMS) in the promoters was taken forward for functional evaluation.
Methods
The study included 126 women who underwent abdominal subcutaneous biopsy to determine adipose morphology. Global transcriptome profiling was performed on WAT from 113 of the women, and CpG methylome profiling on isolated adipocytes from 78 women. Small interfering RNAs (siRNA) knockdown in human mesenchymal stem cells (hMSCs) was used to assess influence of specific genes on lipid storage.
Results
A higher proportion of CpG-sites were methylated in hypertrophic compared to hyperplastic WAT. Methylation at 35,138 CpG-sites was found to correlate to adipose morphology. 2,102 of these CpG-sites were also differentially methylated in T2D; 98% showed directionally consistent change in methylation in WAT hypertrophy and T2D. We identified 2,508 DMS in 638 adipose morphology-associated genes where methylation correlated with gene expression. These genes were over-represented in gene sets relevant to WAT hypertrophy, such as insulin resistance, lipolysis, extracellular matrix organization, and innate immunity. siRNA knockdown of ADH1B, AZGP1, C14orf180, GYG2, HADH, PRKAR2B, PFKFB3, and AQP7 influenced lipid storage and metabolism.
Conclusion
CpG methylation could be influential in determining adipose morphology and thereby constitute a novel antidiabetic target. We identified C14orf180 as a novel regulator of adipocyte lipid storage and possibly differentiation.
Keywords: Adipocyte, Epigenetics, Adipogenesis, Insulin resistance
Abbreviations: DMS, differentially methylated CpG-sites; hMSCs, human mesenchymal stem cells; siRNA, small interfering RNAs; TSS, transcription start sites; WAT, white adipose tissue
Highlights
-
•
Hypertrophic adipose morphology display a distinct adipocyte CpG-methylome profile.
-
•
Adipose hypertrophy and type 2 diabetes display strong overlap in CpG-methylome profile.
-
•
C14orf180 is a novel regulator of adipocyte lipid storage and possibly adipogenesis.
1. Introduction
White adipose tissue (WAT) dysfunction is a hallmark of obesity and associated metabolic complications such as type 2 diabetes (T2D) [1]. WAT expands by increasing adipocyte number (hyperplasia) and size (hypertrophy). Their relative importance differs between individuals resulting in alternate adipose morphologies. Hypertrophic WAT is the pernicious morphology, which is associated with dyslipidemia, insulin resistance, and T2D [2], [3], [4], [5]. The increased incidence of metabolic disease observed with hypertrophic WAT may be explained by higher spontaneous lipolysis and release of pro-inflammatory adipokines [6], [7].
The mechanisms governing hyperplastic versus hypertrophic WAT expansion are poorly understood. Family history of diabetes is associated with enlarged adipocytes supporting a genetic impact on adipose morphology [8]. Furthermore, WAT hypertrophy has been linked to altered extracellular matrix function and impaired adipogenesis [9], [10]. The observation that generation of new adipocytes is constant during adulthood [11] gives support to the notion that long acting, e.g. epigenetic, mechanisms could participate in regulation of adipose morphology. One epigenetic mechanism by which cells are able to modulate their gene expression is DNA methylation at CpG dinucleotides [12]. In particular, tissue specific methylation in gene promoters inhibits gene expression [13]. Previous studies have compared CpG-methylation in WAT specimen between insulin resistant and sensitive, or T2D and non-diabetic subjects [14], [15]. However, a minority of cells in WAT constitutes adipocytes, the cell type defining adipose morphology [16].
To directly address the question if differentially methylated CpG-sites (DMS) in adipocytes are associated with adipose morphology and adipocyte function, we here report CpG-methylome profiling on abdominal subcutaneous adipocytes from a large cohort of women. CpG-methylation was related to adipose morphology and gene expression analyzed on the same biopsies. A subset of genes with DMS in the promoters were taken forward for functional evaluation by small interfering RNAs (siRNA) knockdown in adipose derived human mesenchymal stem cells (hMSCs) to assess influence on lipid metabolism and storage.
2. Methods
2.1. Patient examination
The study included 126 women recruited by local advertisement from the general adult population in the Stockholm (Sweden) area (Table 1); 113 women underwent gene expression and 78 CpG methylation profiling. The women displayed a large inter-individual variation in body mass index (BMI), and all were healthy. Those in the gene expression group have been described previously [17]. Participants were investigated at 8 AM after an overnight fast. Anthropometric and blood pressure measurements were performed followed by venous blood sampling. Fasting plasma glucose and lipids were analyzed at the hospital's routine chemistry laboratory. Serum insulin was measured by ELISA (Mercodia, Uppsala, Sweden). Homeostasis model assessment of insulin resistance (HOMA-IR) was computed according to Matthews et al. [18]. Following the clinical examination an abdominal subcutaneous WAT biopsy was obtained by needle aspiration as described [19]. The study was approved by the regional ethics board in Stockholm, and written informed consent was obtained from each subject. The work was carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans.
Table 1.
Clinical characteristics of studied women.
| CpG methylation in adipocytes |
Gene expression in WAT |
|||||
|---|---|---|---|---|---|---|
| Hypertrophy (n = 42) | Hyperplasia (n = 36) | Pb | Hypertrophy (n = 60) | Hyperplasia (n = 53) | Pb | |
| Age (y's) | 45 ± 13 | 39 ± 11 | 0.045 | 44 ± 12 | 41 ± 10 | 0.13 |
| BMI (kg/m2) | 33 ± 9 | 29 ± 9 | 0.021 | 34 ± 9 | 33 ± 10 | 0.43 |
| Systolic blood pressure (mm Hg) | 129 ± 15 | 124 ± 19 | 0.136 | 128 ± 17 | 127 ± 17 | 0.91 |
| Diastolic blood pressure (mm Hg) | 78 ± 7 | 75 ± 9 | 0.076 | 78 ± 8 | 77 ± 9 | 0.14 |
| fP-Glucose (mmol/L) | 5.17 ± 0.47 | 4.72 ± 0.40 | 8 × 10−5 | 5.32 ± 0.72 | 4.99 ± 0.53 | 0.011 |
| fS-Insulin (mU/l) | 11.99 ± 8.87 | 5.77 ± 3.99 | 3 × 10−5 | 12.79 ± 9.16 | 8.29 ± 6.28 | 0.0028 |
| HOMAIR | 2.82 ± 2.25 | 1.21 ± 0.85 | 8.6 × 10−6 | 3.09 ± 2.36 | 1.89 ± 1.59 | 0.0011 |
| Spontaneous lipolysis (μmol glycerol/2 h/g lipid) | 7.4 ± 6.7 | 3.3 ± 3.0 | 0.0004 | 7.0 ± 6.7 | 4.6 ± 5.0 | 0.0197 |
| Total Cholesterol, mmol/l | 5.16 ± 0.99 | 4.52 ± 0.84 | 0.0038 | 5.18 ± 0.85 | 4.51 ± 0.86 | 2.7 × 10−5 |
| fP-HDL Cholesterol, mmol/l | 1.36 ± 0.39 | 1.61 ± 0.40 | 0.004 | 1.36 ± 0.40 | 1.49 ± 0.39 | 0.048 |
| fP-Triglycerides (mmol/L) | 1.51 ± 0.85 | 0.92 ± 0.49 | 0.0004 | 1.57 ± 0.86 | 1.05 ± 0.61 | 0.0002 |
| Adipose morphology (delta)a | 145 ± 114 | −122 ± 99 | 4 × 10−14 | 143 ± 106 | −121 ± 93 | 6 × 10−20 |
Difference between average fat cell volume and expected volume for specific total body fat, see methods for detail.
Wilcoxon test was used to compare clinical and biochemical variables between groups. Values are mean ± SD.
2.2. WAT experiments
WAT specimens were subjected to collagenase treatment to obtain isolated adipocytes as described [20]. One portion of cells was immediately frozen in liquid nitrogen for subsequent RNA and DNA isolation. The mean weight and volume of remaining cells and spontaneous lipolysis were determined as described [21], [22]. A curve fit of the relationship between mean adipocyte volume and total fat mass was performed [23]. The difference between the measured and expected mean adipocyte volume at the corresponding total fat mass determines adipose morphology. If the measured adipocyte volume is larger than expected, WAT hypertrophy prevails, whereas the opposite is valid for hyperplasia. Thus, this measure of adipose morphology is independent of total fat mass.
2.3. Gene expression profiling
Global transcriptome profiling on WAT specimen was performed using Gene 1.0 or 1.1 ST Affymetrix arrays (Affymetrix, Inc., Santa Clara, CA). These array results have been reported earlier [17]. For significant phenotypic associations, we checked manually that the results were directionally consistent between array types.
Global transcriptome profiling on hMSCs was assessed using GeneChip® Human Transcriptome Array 2.0 (Affymetrix) according to the manufacturer's instructions. RNA was isolated as described in section 2.8. Pre-processing and normalization of arrays were carried out with Affymetrix Expression Console 4.0 (Affymetrix) using Gene Level SST-RMA.
2.4. DNA preparation
Genomic DNA was prepared from isolated adipocytes using the QiAamp DNA Mini kit (cat no. 51304, Qiagen, Hilden, Germany). DNA purity and quality was confirmed by A260/280 ratio >1.8 on a Nanodrop ND-1000 Spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA). DNA concentration was measured by Qubit (Life technologies, Stockholm, Sweden).
2.5. CpG methylation profiling
Global GpG-methylation profiling was performed on isolated adipocytes using the Infinium Human Methylation EPIC BeadChip (Illumina, San Diego, CA) as described in Appendix A. During pre-processing we removed probes overlapping SNPs, with unreliable measurement criteria (P ≥ 0.05), and 2,638 context-specific probes leaving 820,027 probes for further analysis. Methylation levels (beta values; β) were estimated as the ratio of signal intensity of the methylated alleles to the sum of methylated and unmethylated intensity signals. The β-values vary from 0 (no methylation) to 1 (100% methylation).
2.6. Microarray analyses
Qlucore Omics Explorer 3.2 (www.qlucore.com) was used for microarray analyses of clinical samples. Before phenotypic analysis, the 22,167 transcripts on the Gene 1.0 and 1.1 ST arrays were collapsed on to 20,506 unique transcripts by selecting probe-sets with highest average expression for each gene. Relationship between gene expression and adipose morphology was assessed by linear regression adjusting for array batch. We used Webgestalt (www.webgestalt.org, accessed 1/12-2018) and the KEGG and REACTOME databases to assess overrepresented gene sets among differentially expressed genes, as compared to all mRNA in the human genomes. Phenotypic analysis of EPIC arrays was limited to the 571,997 CpG-sites mapping to a unique site and linked to a transcript. Relationship between CpG-methylation beta-values and WAT morphology was assessed by linear regression adjusting for age. There was no batch effect between BeadChips according to PCA.
The Bioconductor R Limma package was used to identify differentially expressed genes between siC14orf180 and control transfected cells [24]. Gene set enrichment analysis (GSEA) was performed on these samples using the MSigDB Hallmark gene set collection [25], [26].
2.7. Cell culture experiments
hMSCs were isolated from abdominal subcutaneous WAT from a male donor (16 years old, BMI 24 kg/m2). hMSCs were cultured in 24 (1 × 105 cells/well) or 48-well (5 × 104 cells/well) plates and differentiated into adipocytes using described protocols [27]. For gene knockdown at day -1, hMSCs were transfected with small interfering RNAs (siRNA) targeting a candidate gene (Dharmacon™ OnTarget plus SMART pool or Dharmacon™ siGENOME SMART pool, VWR International, Sweden) or negative control siRNA (siNC) (Dharmacon™ siGENOME Control Pool) using DharmaFECT™ 3 transfection reagent (VWR International) as per manufacturer's instructions. For transfection at day 8 a Neon™ transfection system (MPK5000, Invitrogen, Göteborg, Sweden) was used as per manufacturer's instructions. Details of siRNAs used are given in Appendix B, Supplemental Table 1.
2.8. qRT-PCR
On day 9 of hMSCs differentiation, cells were harvested and RNA isolated using the RNeasy Mini Kit (217004, Qiagen). RNA concentration and purity was measured using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific). cDNA synthesis was performed using the iScript™ cDNA Synthesis Kit (1708891, Bio-Rad, Sundbyberg, Sweden) followed by SYBR green qRT-PCR analysis on an iCycler IQ (Bio-Rad). Primer sequences are given in Appendix B, Supplemental Table 2. Gene expression was normalized to β2M using the delta delta Ct method.
2.9. Lipid droplet quantification
On day 9 of hMSC differentiation cells were fixed in 4% paraformaldehyde for 10 min before washing in PBS. Lipid was stained using BODIPY™ (0.2 μg/mL) (Fisher Scientific, Göteborg, Sweden) and nucleus with Hoechst 33258 (10 mg/mL) (Fisher Scientific). Images were taken after total lipid per cell and total lipid droplets per cell were quantified on a CellInsight™ CX5 High Content Screening Platform (Fisher Scientific).
2.10. Rate of lipolysis
Basal lipolysis was determined by measuring the concentration of glycerol in the media after three days incubation as previously described [28]. Briefly, at day 13 of differentiation, 50 μL of media was collected and added to 450 μl of resuspended ATP reagent (10 μL purified glycerokinase (Sigma–Aldrich, Stockholm, Sweden) and 10 μL ATP Reagent SL (Biotherma, Handen, Sweden) in 0.05 M Tris buffer). The decay in luminescence over 1 min was measured and compared with a glycerol standard curve to determine concentration. Fresh media not exposed to cells was used as blank and subtracted from sample results.
2.11. De novo lipogenesis
Incorporation of [1-14C]-acetic acid (Perkin Elmer, Stockholm, Sweden) into hMSCs was analyzed as previously described [29]. On day 13 of differentiation cells were insulin starved for 18 h in DMEM media containing additives as described [27] and 1 μM glucose. Following starvation, cells were incubated in DMEM media containing 2 mM glucose, 10 mM HEPES, 0.2% BSA, 2 mM acetate and 1 μCi [1-14C]-acetic acid with or without 100 nM insulin. Cells were lysed in 0.1% SDS and radioactivity in lysate determined on a scintillation counter.
2.12. Statistical analyses
Wilcoxon test was used to compare clinical and biochemical variables between groups. Student's t test (2 sided) was used to compare the global average proportion of methylated CpG-sites between the two clinical groups, and the average difference in methylation of DMS between adipose morphology and T2D. Relationship between CpG-methylation and gene expression was assessed by Pearson correlation. Chi-square test was used to compare distribution of DMS between groups. A one-way ANOVA test with a Dunnett's multiple comparison test was used to test for significance between treatment and control samples in cell culture experiments.
3. Results
3.1. Clinical characteristics of study participants
Seventy-eight women underwent CpG-methylation profiling on isolated adipocytes, and 113 women underwent transcriptome profiling on WAT; 65 study subjects overlapped between these analyses (Table 1). Subjects were grouped based on adipose morphology. WAT hypertrophy was associated with a pernicious metabolic profile with higher plasma fasting glucose, fasting insulin, and HOMA-IR, as well as total cholesterol and triglycerides. Adipocyte spontaneous lipolysis was also higher in the subgroup with WAT hypertrophy. In the CpG-methylation cohort age and BMI were slightly higher in the subjects with WAT hypertrophy as compared to those with WAT hyperplasia, whereas no difference in these parameters was observed between WAT hypertrophy and hyperplasia in the transcriptome cohort.
3.2. Transcriptome profile in relation to WAT morphology
Global transcriptome analysis on WAT identified 1,131 genes whose expression significantly correlated with adipose morphology (FDR ≤0.01; Appendix B, Supplemental Table 3). The 1,131 genes were enriched for gene sets relevant to WAT hypertrophy including Regulation of lipolysis rate, Insulin resistance, Adipocytokine signaling, Extracellular matrix organization, and Neutrofil degranulation (FDR <0.15; Appendix B, Supplemental Table 4). The genes assigned to each pathway and correlated to adipose morphology are presented in Appendix B, Supplemental Table 5. Examples of genes in metabolic pathways inversely correlated with adipose morphology include IRS2, a central player in insulin signaling (FDR <0.01, R = −0.35). Expression of genes in pro-inflammatory responses such as IL1RN (FDR <0.01, R = 0.39), CCL22 (FDR <0.01, R = 0.35), and CCR1 (FDR <0.01, R = 0.34) correlated positively with adipose morphology.
3.3. CpG-methylation landscape in relation to WAT morphology
We assessed methylation at 820,026 distinct CpG-sites in relation to adipose morphology. There was a borderline significant overall difference in CpG-methylation between women with hypertrophic versus hyperplastic WAT (Table 2, P = 0.058). However, in and around the transcription start sites (TSS) of genes [TSS 1500, TSS 200, 5’ untranslated region (UTR) and 1st Exon] there was a significantly higher proportion of methylated CpG-sites in hypertrophic compared to hyperplastic WAT. Significant differences were not observed for other gene regions examined. In relation to CpG islands, significant alterations in the methylation state between the adipose morphologies occurred within CpG islands and surrounding shores.
Table 2.
Global CpG methylome signature in relation to adipose morphology.a
| Global average proportion of methylated CpG sites |
Genome distribution of differentially methylated CpG sites |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of CpG- sites | hypertrophy | hyperplasia | difference | Pb | No. of CpG sites | No. of DMS | % of all CpG sites | % of all DMS | |
| In relation to gene regionc | |||||||||
| TSS 1500 | 99,132 | 0.399 ± 0.011 | 0.392 ± 0.008 | 0.007 | 0.008 | 99,132 | 5591 | 0.173 | 0.159 |
| TSS 200 | 62,009 | 0.186 ± 0.005 | 0.182 ± 0.005 | 0.004 | 0.004 | 62,009 | 2015 | 0.108 | 0.057 |
| 5′ UTR | 68,096 | 0.449 ± 0.012 | 0.443 ± 0.009 | 0.006 | 0.018 | 68,096 | 4594 | 0.119 | 0.131 |
| 1st Exon | 24,866 | 0.217 ± 0.006 | 0.214 ± 0.005 | 0.003 | 0.008 | 24,866 | 833 | 0.043 | 0.024 |
| Body | 292,753 | 0.662 ± 0.018 | 0.656 ± 0.012 | 0.006 | 0.100 | 292,753 | 20,491 | 0.512 | 0.583 |
| Exon Bnd | 5440 | 0.785 ± 0.021 | 0.780 ± 0.014 | 0.005 | 0.241 | 5440 | 270 | 0.010 | 0.008 |
| 3′ UTR | 19,701 | 0.702 ± 0.019 | 0.694 ± 0.014 | 0.008 | 0.056 | 19,701 | 1344 | 0.034 | 0.038 |
| In relation to CpG Islandc | |||||||||
| N Shelf | 30,283 | 0.701 ± 0.019 | 0.696 ± 0.013 | 0.006 | 0.147 | 18,646 | 1352 | 0.033 | 0.038 |
| N Shore | 79,745 | 0.446 ± 0.013 | 0.439 ± 0.009 | 0.007 | 0.008 | 64,165 | 4381 | 0.112 | 0.125 |
| CpG Island | 158,706 | 0.184 ± 0.006 | 0.182 ± 0.005 | 0.003 | 0.025 | 133,745 | 2296 | 0.234 | 0.065 |
| S Shore | 68,056 | 0.44 ± 0.013 | 0.432 ± 0.009 | 0.007 | 0.006 | 55,930 | 3901 | 0.098 | 0.111 |
| S Shelf | 28,054 | 0.709 ± 0.019 | 0.703 ± 0.013 | 0.005 | 0.155 | 16,998 | 1232 | 0.030 | 0.035 |
| Open sea | 455,182 | 0.694 ± 0.019 | 0.688 ± 0.014 | 0.006 | 0.103 | 282,513 | 21,976 | 0.494 | 0.625 |
| All | 820,026 | 0.551 ± 0.015 | 0.545 ± 0.011 | 0.006 | 0.058 | 571,997 | 35,138 | 1 | 1 |
Values are mean ± SD.
Analyses of global average proportion of methylated CpG sites in relation to CpG Islands were performed in all 820,026 interrogated CpG sites. Analyses in relation to gene region and of DMS were limited to 571,997 CpG sites linked to genes.
Student's t test (2 sided) was used to compare global average proportion of methylated CpG sites between the two clinical groups.
Annotation is based on Illumina annotation of EPIC arrays; within 1500 basepairs of transcriptional start site (TSS 1500); within 200 basepairs of transcriptional start site (TSS 200); untranslated region (UTR), exon boundary (Exon Bnd); upstream of CpG island (N); downstream CpG island (S); unrelated to CpG island (Open sea).
Methylation at 35,138 CpG-sites were found to significantly correlate to adipose morphology at a FDR ≤0.01 (Appendix B, Supplemental Table 6). Table 2 displays the distribution of the DMS in relation to gene regions and CpG islands. The genome distribution of the DMS associated with adipose morphology significantly differed from expected (P < 0.0001). An enrichment of DMS were found within the gene body and 5’ UTR, and an underrepresentation of such sites upstream of the transcriptional start site and within the first exon. There was also a significant over-representation of DMS at the CpG island shores and open sea regions, and under-representation in CpG islands.
3.4. DMS common to WAT hypertrophy and T2D
We next compared the 35,138 DMS associated with adipose morphology in the present study with previously reported DMS associated with T2D [15]. Among 15,627 DMS in WAT associated with T2D, 2,102 CpG-sites were associated with WAT morphology of which 2,061 (98%) showed directionally consistent change in methylation in WAT hypertrophy and T2D (Appendix B, Supplemental Table 6). Differential methylation at DMS was overall more pronounced in isolated adipocytes in relation to adipose morphology as compared to in WAT in relation to T2D. The average difference in beta-value of all DMS displaying higher methylation in hypertrophic WAT and T2D was 0.068 and 0.04, respectively (P 5.8 × 10−282). The average difference in beta-value of all DMS displaying lower methylation in hypertrophic WAT and T2D was −0.050 and −0.045, respectively (P 5.5 × 10−9).
3.5. Identification of differentially methylated genes associated with adipose morphology
To identify epigenetically regulated genes of potential importance for adipose morphology we overlapped the 35,138 DMS with the 1,131 differentially expressed genes and hereby identified 2,508 DMS in 638 genes where methylation correlated with expression of the corresponding gene with an r2 ≥ 0.06 (Appendix B, Supplemental Table 7). The 638 genes were over-represented in gene sets relevant to WAT hypertrophy, such as Insulin resistance (e.g. AKT2, PIK3CG, SLC2A4, and IRS2), Regulation of lipolysis in adipocytes (e.g. ADRB1, ADRB2, ABHD5), Extracellular matrix organization (e.g. collagens, cathepsins, and integrins), and Innate Immune System (Appendix B; Supplemental Table 8).
3.6. Functional evaluation of adipose morphology candidate genes
11 of 638 genes (Table 3) were selected for functional evaluation based on the following filtering criteria [1]: ≥1 CpG-site in the promoter region that positively correlated with adipocyte hypertrophy [2], inverse correlation between CpG methylation and gene expression in the clinical cohort, and [3] expression enriched in adipocytes. For the latter assessment, we used the FANTOM5 project database (http://fantom.gsc.riken.jp/) which interrogates the expression level of genes in various primary cell types including our adipose derived hMSCs across differentiation. The 11 selected genes displayed the highest expression and largest fold increase during differentiation into adipocytes, as well as high expression in mature adipocytes compared with other tissue and cells.
Table 3.
Summary of selected gene displaying correlation between promoter CpG methylation, expression. and adipose morphology.a
| Gene | CpGb | Adipose morphology |
Gene expression vs CpG methylatione | effect of siRNA on hMSCsf |
Concordant DMS in T2D | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| vs CpG methylationc |
vs gene expressiond |
total lipid/cell | no. lipid droplets/cell | ADIPOQ | FABP4 | basal lipolysis | insulin stimulated lipogenesis | ||||||
| FDR | r | FDR | r | r2 | |||||||||
| ADH1B | cg05641529 | 0.001 | 0.5 | 0.0001 | −0.48 | 0.25 | down | down | down | blunted | |||
| cg24368912 | 0.0034 | 0.43 | 0.38 | yes | |||||||||
| cg07059704 | 0.0009 | 0.5 | 0.41 | ||||||||||
| AQP7 | cg16447312 | 0.0012 | 0.48 | 0.0069 | −0.34 | 0.18 | up | down | blunted | ||||
| AZGP1 | cg08008696 | 0.0013 | 0.48 | 0.0008 | −0.41 | 0.4 | down | down | down | down | blunted | ||
| cg26429636 | 0.0009 | 0.51 | 0.47 | ||||||||||
| cg13241889 | 0.0025 | 0.44 | 0.45 | ||||||||||
| C14orf180 | cg16063716 | 0.001 | 0.5 | 0.0022 | −0.37 | 0.22 | down | down | down | down | down | blunted | yes |
| cg21198755 | 0.0023 | 0.45 | 0.16 | ||||||||||
| cg02868468 | 0.0019 | 0.46 | 0.16 | ||||||||||
| cg12981387 | 0.0011 | 0.49 | 0.14 | ||||||||||
| cg00497078 | 0.0011 | 0.49 | 0.13 | ||||||||||
| cg14803282 | 0.0016 | 0.47 | 0.2 | ||||||||||
| cg15665105 | 0.0014 | 0.47 | 0.18 | ||||||||||
| cg24102702 | 0.0071 | 0.4 | 0.09 | ||||||||||
| cg14735905 | 0.0026 | 0.44 | 0.08 | ||||||||||
| cg18484736 | 0.0063 | 0.4 | 0.11 | ||||||||||
| cg04220416 | 0.0022 | 0.45 | 0.06 | ||||||||||
| CAMK1 | cg01396022 | 0.0096 | 0.38 | 0.001 | −0.40 | 0.07 | down | ||||||
| cg10836985 | 0.0008 | 0.52 | 0.15 | ||||||||||
| cg08237614 | 0.0045 | 0.42 | 0.16 | ||||||||||
| GYG2 | cg02075301 | 0.0034 | 0.43 | 0.009 | −0.33 | 0.34 | down | down | up | ||||
| HADH | cg15330017 | 0.0024 | 0.44 | 0.0003 | −0.44 | 0.29 | down | down | |||||
| cg01573501 | 0.0079 | 0.39 | 0.23 | ||||||||||
| PC | cg10486891 | 0.0044 | 0.42 | 0.0088 | −0.33 | 0.11 | down | ||||||
| cg04301544 | 0.0093 | 0.38 | 0.1 | ||||||||||
| PFKFB3 | cg22224625 | 0.0066 | 0.4 | 0.0008 | −0.41 | 0.15 | down | down | down | blunted | |||
| cg06002198 | 0.0082 | 0.39 | 0.16 | ||||||||||
| cg03889890 | 0.0046 | 0.42 | 0.24 | ||||||||||
| cg22692545 | 0.0074 | 0.39 | 0.07 | ||||||||||
| cg10405605 | 0.0091 | 0.39 | 0.16 | ||||||||||
| PRKAR2B | cg21250978 | 0.0012 | 0.48 | 0.0022 | −0.37 | 0.46 | down | down | up | yes | |||
| cg24538401 | 0.0018 | 0.46 | 0.43 | ||||||||||
| RXRG | cg22052044 | 0.0063 | 0.4 | 0.0018 | −0.38 | 0.18 | |||||||
| cg02364825 | 0.0031 | 0.43 | 0.14 | ||||||||||
| cg26419494 | 0.0065 | 0.4 | 0.1 | ||||||||||
Adipose morphology was assessed as difference between average fat cell volume and expected volume for specific total body fat. see methods for detail. For selection of genes see results section.
All CpG sites listed below are located within 1500 basepairs from transcription start site.
Relationship between CpG-methylation, as assessed by beta-values, and adipose morphology was assessed by linear regression adjusting for age.
Relationship between gene expression and adipose morphology was assessed by linear regression adjusting for array batch.
Pearson r.2.
Genes in the leftmost column were knocked down by siRNA and effect on lipid accumulation and gene expression assessed as described in method section.
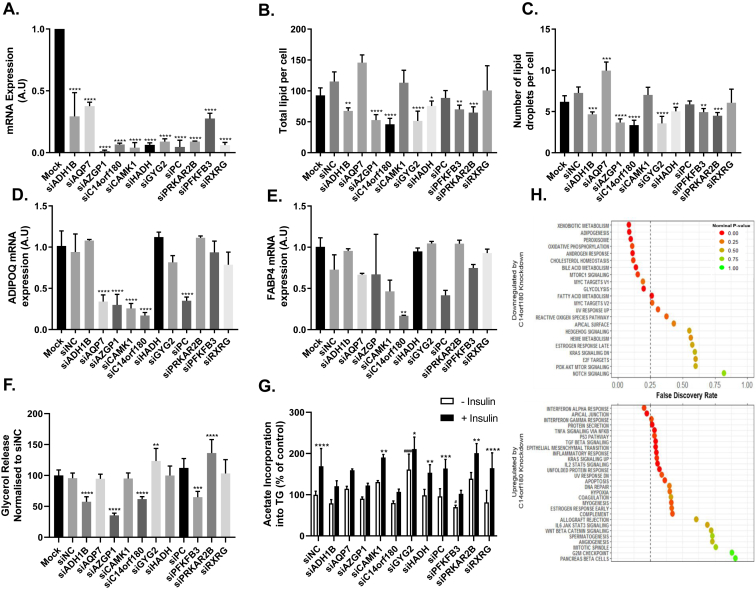
To examine if the 11 adipose morphology associated genes influenced adipocyte function, their expression was knocked down in hMSCs at the early (day -1) and late (day 8) stage of differentiation into adipocytes. Each siRNA was able to markedly and significantly knockdown its target gene nine days after transfection and into differentiation (Figure 1A). To assess target genes influence on adipogenesis, we quantified lipid content in cells after knockdown. Knockdown of seven genes (ADH1B, AZGP1, C14orf180, GYG2, HADH, PFKFB3, PRKAR2B) decreased both the total amount of lipid and number of lipid droplets per cell (Figure 1B,C), whereas knockdown of AQP7 increased number of lipid droplets per cell (Figure 1C). Knockdown of C14orf180 was accompanied by decreased expression of the established adipocyte markers ADIPOQ and FABP4 (Figure 1D,E) [30], [31]. Knockdown of four more genes decreased ADIPOQ levels (AQP7, AZGP1, CAMPK1, PC), of which knockdown of AZGP1 showed consistent inhibitory impact on ADIPOQ mRNA and lipids per cell. No additional gene influenced FABP4 mRNA levels. Late knockdowns (day 8) of six genes influenced lipolysis; knockdown of ADH1B, AZGP1, C14orf180, and PFKFB3 had a consistent inhibitory effect on lipid storage and lipolysis, whereas knockdown of GYG2 and PRKAR2B increased lipolysis (Figure 1F). Knockdown of five genes blunted the response of lipogenesis to insulin; results for four genes were consistent with influence on lipid storage (ADH1B, AZGP1, C14orf180, and PFKFB3) whereas knockdown of AQP7 stimulated lipid storage but caused blunted the response to insulin (Figure 1G). Gene knockdown had modest effect on basal lipogenesis (Figure 1G). siRNA knockdown of RXR had no effect on any investigated phenotype.
Figure 1.
Impact of candidate gene knockdown in adipose derived human mesenchymal stem cells (hMSCs) on lipids per cell, gene expression and lipid turnover. siRNAs targeting the indicated genes or siNegative Control (siNC) were transfected on Day -1 (A–E and H) or on Day 8 (F–G) of differentiation alongside a Mock transfection. For Day -1 knockdown, cells were lysed or fixed and mRNA or lipid was quantified on day 9 of differentiation. For day 8 knockdown, glycerol in the media or lipid synthesized in the cells was measured at day 13. A. mRNA quantification of the siRNA target gene compared with mock in transfected cells. B. Total lipid per cell or C. Total lipid droplets per cell were quantified after lipid staining and normalized per nucleus. n = 3 per group. D. ADIPOQ or E. FABP4 mRNA quantification after target gene knockdown as indicated on the x-axis. n = 3–7 per group. F. Basal lipolysis measured as glycerol release into the media n = 8. G. De novo lipogenesis over 3 h with or without the addition of insulin. Lipogenesis was measured via radiolabeled acetate incorporation into the cell. H. Global transcriptome profiles on siC14orf180 or siNC transfected samples were subjected to Gene Set Enrichment Analysis (GSEA). The upper panel shows gene sets downregulated and the lower panel gene sets upregulated by siC14orf180 knockdown compared to siNC. Statistical analysis carried out for B-F was target siRNA versus siNC. Statistical analysis for G carried out was (−) insulin versus (+) insulin significance denoted by * and siNC (−) insulin versus target siRNA (−) insulin significance denoted by #. One-way ANOVA with Dunnetts post hoc test was carried out for all experiments shown. *P ≤ 0.05, **P ≤ 0.01,***P ≤ 0.001, ****P ≤ 0.0001, #P ≤ 0.05, ###P ≤ 0.001.
3.7. C14orf180 is critical for adipogenic gene expression
Among the candidate genes for adipose morphology from the siRNA experiment, only C14orf180 significantly blunted all examined phenotypes. As little is known about the function of C14orf180, we carried out global transcriptome analysis on hMSCs transfected with siC14orf180 or siNC at day -1 of differentiation. 1,116 genes were differentially expressed between these groups at a FDR ≤0.01 (Appendix B, Supplemental Table 9). C14orf180 knockdown downregulated expression of genes in pathways such as Adipogenesis and Oxidative phosphorylation and upregulated genes in the interferon-α pathway (FDR <0.25) (Figure 1H, Appendix C, Supplemental Figure 1). Specific genes downregulated in siC14orf180 treated cells include the master regulators of adipogenesis PPARG and CEBPa, as well genes essential for lipid metabolism (LIPE, LPL, FABP4, PLIN2, and DGAT1) (Appendix B, Supplemental Table 9).
4. Discussion
In this study, we present results linking adipocyte epigenetic profile to adipose morphology. In a genome-wide analysis, we demonstrate a modest but significant increase in proportion of methylated CpG-sites in the 5’ regions of genes, as well as in CpG islands and surrounding shore regions in adipocytes from women with hypertrophic WAT. Among DMS overlapping between WAT hypertrophy and T2D, 98% display concordant change in CpG-methylation between these clinical conditions. 638 genes display correlation between promoter CpG-methylation, expression, and adipose morphology, including genes related to insulin resistance, regulation of lipolysis, and extracellular matrix organization. By siRNA knockdown we identify, among differentially methylated genes, novel regulators of adipocyte lipid storage including C14orf180.
The factors regulating adipose morphology, in particular diabetes-predisposing hypertrophy, are poorly understood. In the present study, adipocyte DMS associated with adipose hypertrophy display strong overlap with those observed in T2D further strengthening the causality between adipose morphology and diabetes. The measure of adipose morphology applied herein is adjusted for, and thus not related to, total body fat [23]. Bariatric surgery induces a hypercellular WAT phenotype with small adipocytes, which is associated with a global decrease in proportion of methylated CpG-sites in adipocytes [32], [33]. This is concordant with the finding in the present study where hypertrophic WAT is associated with an increase in global CpG-methylation in 5’ regions of genes, and gives support to the notion that adipocyte DNA methylation is causally linked to cell size. Life style interventions with physical exercise and altered dietary fatty acid composition have been shown to influence WAT CpG-methylation [34], [35], but the impact on the adipocyte epigenome or adipose morphology has, to our knowledge, not been investigated.
Most previous epigenome profiling on WAT have studied tissue specimen. Adipocytes constitute a minority of cells in WAT, which also comprise fibroblasts, endothelial, immune, and precursor cells [36]. In the present study, we examined only the mature adipocytes and, in this subpopulation of cells in WAT, identified numerous genes displaying correlation between promoter CpG-methylation, expression, and adipose morphology. DMS associated with WAT hypertrophy were over-represented outside CpG islands, regions where methylation is thought to be more dynamic and tissue specific, as compared to CpG islands [12], [13]. CpG-sites in the vicinity of the transcription start site are of particular importance for cell function since their methylation prevent initiation of transcription [12]. Inverse correlation between adipocyte promoter CpG-methylation and WAT gene expression in vivo can be validated in vitro, i.e. methylation at such CpG-sites inhibits gene expression [37]. This supports the notion that at least some of the genes reported here and related to e.g. insulin resistance, regulation of lipolysis, and extracellular matrix organization are linked to adipocyte function and WAT hypertrophy. A subset of genes with DMS was taken forward for functional evaluation by siRNA knockdown in hMSCs and were shown to influence lipid storage and turnover thus representing new potential regulators of adipose morphology (Table 3):
-
-
C14orf180 has been reported to encode a plasma-membrane protein with unknown function displaying enriched expression in WAT [38]. We report that knockdown of C14orf180 inhibits lipid storage in hMSCs. C14orf180 deficiency downregulated expression of key genes in adipogenesis including PPARG and CEBPA, as well as numerous genes involved in lipid metabolism. The interferon-α pathway, which was upregulated upon C14orf180 knockdown, has previously been shown to inhibit adipogenesis [39]. Thus, we identified C14orf180 as a novel regulator of adipocyte lipid storage and possibly differentiation. C14orf180 displays numerous DMS in the promoter region, one of which overlaps with T2D (Table 3). Together these findings support the notion that epigenetic regulation of C14orf180 contributes to diabetogenic hypertrophic WAT.
-
-
AZGP1 encodes the adipokine ZAG1, which has been reported to reduce body weight and expression of lipogenic enzymes in WAT of rodents [40]. The latter is in contrast to our results in which knockdown of AZGP1 inhibited lipid storage and blunted insulin stimulated lipogenesis. The discrepant results could be due either to reduced lipid storage in the present study being secondary to reduced adipogenesis, as supported by reduced ADIPOQ expression, or species specific effects of AZGP1.
-
-
PFKFB3 inhibition impairs basal and insulin stimulated lipogenesis, and gene knockdown may inhibit adipocyte lipid storage as observed in the present study [41].
-
-
PRKAR2B is required for adipogenesis and low levels are associated with increased lipolysis [42], [43]. Consistent with these findings, in the present study, PRKAR2B knockdown inhibited lipid accumulation and increased basal lipolysis. DMS in the PRKAR2B promoter region overlap between WAT hypertrophy and T2D thus supporting the notion that epigenetic regulation of PRKAR2B predisposes to hypertrophic WAT and T2D.
-
-
Genetic variants in ADH1B affect body fat [44] consistent with ADH1B knockdown inhibiting lipid storage.
-
-
HADH is involved in beta oxidation and GYG2 controls glycogen storage. Our findings that HADH and GYG2 knockdown inhibits lipid storage adds novel metabolic function to these genes. GYG2 knockdown also increased basal lipogenesis perhaps resulting from decreased free glycogen levels which has been shown to increase glucose uptake in adipocytes [45].
-
-
AQP7 was the only gene whose knockdown increased lipid storage in hMSCs. This effect is consistent with AQP7 encoding an adipocyte membrane protein responsible for glycerol transport, a substrate and product of lipid metabolism [46].
WAT hypertrophy has been shown to be caused by impaired recruitment of new adipocytes and epigenetic factors control adipogenesis [10], [47]. We do not have samples to directly study the precursor cell epigenome in clinical cohorts. Therefore, it is unclear if DMS adipocytes reported here are present in precursor cells. However, it has been reported that DMS between lean and obese are largely maintained between human adipose derived stem cells and mature adipocytes [48]. The impaired lipid storage observed after candidate gene knockdown could thus be the result of impaired adipogenesis.
5. Conclusion
In conclusion, results presented herein support the notion that differential CpG-methylation in adipocytes is linked to hypertrophic WAT morphology predisposing to T2D. Several genes which appear to be under epigenetic control and influence adipocyte lipid storage and metabolism were identified.
Acknowledgements
The microarray hybridizations were done at BEA (www.bea.ki.se). This study was supported by the Strategic Research Program in Diabetes at Karolinska Institutet, Centre for Innovative Medicine (CIMED), Swedish Research Council, Novo Nordisk Foundation, Novo Nordisk A/S, Stockholm County Council, and Diabetesfonden. The sponsors took no part in the collection, analysis, and interpretation of data, in the writing of the report, or in the decision to submit the article for publication.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.04.009.
Conflict of interest
None declared.
Appendix ASupplementary data
The following are the Supplementary data to this article:
Multimedia component 1
Multimedia component 2
References
- 1.Kahn S.E., Hull R.L., Utzschneider K.M. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444(7121):840–846. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- 2.Veilleux A., Caron-Jobin M., Noel S., Laberge P.Y., Tchernof A. Visceral adipocyte hypertrophy is associated with dyslipidemia independent of body composition and fat distribution in women. Diabetes. 2011;60(5):1504–1511. doi: 10.2337/db10-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guilherme A., Virbasius J.V., Puri V., Czech M.P. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nature Reviews Molecular Cell Biology. 2008;9(5):367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weyer C., Foley J.E., Bogardus C., Tataranni P.A., Pratley R.E. Enlarged subcutaneous abdominal adipocyte size, but not obesity itself, predicts type II diabetes independent of insulin resistance. Diabetologia. 2000;43(12):1498–1506. doi: 10.1007/s001250051560. [DOI] [PubMed] [Google Scholar]
- 5.Lonn M., Mehlig K., Bengtsson C., Lissner L. Adipocyte size predicts incidence of type 2 diabetes in women. The FASEB Journal. 2010;24(1):326–331. doi: 10.1096/fj.09-133058. [DOI] [PubMed] [Google Scholar]
- 6.Gao H., Mejhert N., Fretz J.A., Arner E., Lorente-Cebrian S., Ehrlund A. Early B cell factor 1 regulates adipocyte morphology and lipolysis in white adipose tissue. Cell Metabolism. 2014;19(6):981–992. doi: 10.1016/j.cmet.2014.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanda H., Tateya S., Tamori Y., Kotani K., Hiasa K., Kitazawa R. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. Journal of Clinical Investigation. 2006;116(6):1494–1505. doi: 10.1172/JCI26498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahlman I., Ryden M., Arner P. Family history of diabetes is associated with enhanced adipose lipolysis: evidence for the implication of epigenetic factors. Diabetes & Metabolism. 2018;44(2):155–159. doi: 10.1016/j.diabet.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 9.Minchin J.E., Dahlman I., Harvey C.J., Mejhert N., Singh M.K., Epstein J.A. Plexin D1 determines body fat distribution by regulating the type V collagen microenvironment in visceral adipose tissue. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(14):4363–4368. doi: 10.1073/pnas.1416412112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arner E., Westermark P.O., Spalding K.L., Britton T., Ryden M., Frisen J. Adipocyte turnover: relevance to human adipose tissue morphology. Diabetes. 2010;59(1):105–109. doi: 10.2337/db09-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spalding K.L., Arner E., Westermark P.O., Bernard S., Buchholz B.A., Bergmann O. Dynamics of fat cell turnover in humans. Nature. 2008;453(7196):783–787. doi: 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- 12.Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature Reviews Genetics. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 13.Irizarry R.A., Ladd-Acosta C., Wen B., Wu Z., Montano C., Onyango P. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nature Genetics. 2009;41(2):178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arner P., Sahlqvist A.S., Sinha I., Xu H., Yao X., Waterworth D. The epigenetic signature of systemic insulin resistance in obese women. Diabetologia. 2016;59(11):2393–2405. doi: 10.1007/s00125-016-4074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nilsson E., Jansson P.A., Perfilyev A., Volkov P., Pedersen M., Svensson M.K. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes. 2014;63(9):2962–2976. doi: 10.2337/db13-1459. [DOI] [PubMed] [Google Scholar]
- 16.Ehrlund A., Acosta J.R., Bjork C., Heden P., Douagi I., Arner P. The cell-type specific transcriptome in human adipose tissue and influence of obesity on adipocyte progenitors. Scientific Data. 2017;4:170164. doi: 10.1038/sdata.2017.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dahlman I., Ryden M., Brodin D., Grallert H., Strawbridge R.J., Arner P. Numerous genes in loci associated with body fat distribution are linked to adipose function. Diabetes. 2016;65(2):433–437. doi: 10.2337/db15-0828. [DOI] [PubMed] [Google Scholar]
- 18.Matthews D.R., Hosker J.P., Rudenski A.S., Naylor B.A., Treacher D.F., Turner R.C. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 19.Kolaczynski J.W., Morales L.M., Moore J.H., Jr., Considine R.V., Pietrzkowski Z., Noto P.F. A new technique for biopsy of human abdominal fat under local anaesthesia with Lidocaine. International Journal of Obesity and Related Metabolic Disorders. 1994;18(3):161–166. [PubMed] [Google Scholar]
- 20.Rodbell M., Krishna G. Preparation of isolated fat cells and fat cell "ghosts"; methods for assaying adenylate cyclase activity and levels of cyclic AMP. Methods in Enzymology. 1974;31:103–114. doi: 10.1016/0076-6879(74)31010-5. [DOI] [PubMed] [Google Scholar]
- 21.Lofgren P., Hoffstedt J., Naslund E., Wiren M., Arner P. Prospective and controlled studies of the actions of insulin and catecholamine in fat cells of obese women following weight reduction. Diabetologia. 2005;48(11):2334–2342. doi: 10.1007/s00125-005-1961-6. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch J., Gallian E. Methods for the determination of adipose cell size in man and animals. The Journal of Lipid Research. 1968;9(1):110–119. [PubMed] [Google Scholar]
- 23.Andersson D.P., Arner E., Hogling D.E., Ryden M., Arner P. Abdominal subcutaneous adipose tissue cellularity in men and women. International Journal of Obesity. 2017;41(10):1564–1569. doi: 10.1038/ijo.2017.148. [DOI] [PubMed] [Google Scholar]
- 24.Smyth G.K., Michaud J., Scott H.S. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21(9):2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Systems. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao H., Volat F., Sandhow L., Galitzky J., Nguyen T., Esteve D. CD36 is a marker of human adipocyte progenitors with pronounced adipogenic and triglyceride accumulation potential. Stem Cells. 2017;35(7):1799–1814. doi: 10.1002/stem.2635. [DOI] [PubMed] [Google Scholar]
- 28.Hellmer J., Arner P., Lundin A. Automatic luminometric kinetic assay of glycerol for lipolysis studies. Analytical Biochemistry. 1989;177(1):132–137. doi: 10.1016/0003-2697(89)90027-4. [DOI] [PubMed] [Google Scholar]
- 29.Barquissau V., Beuzelin D., Pisani D.F., Beranger G.E., Mairal A., Montagner A. White-to-brite conversion in human adipocytes promotes metabolic reprogramming towards fatty acid anabolic and catabolic pathways. Molecular Metabolism. 2016;5(5):352–365. doi: 10.1016/j.molmet.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hotamisligil G.S., Bernlohr D.A. Metabolic functions of FABPs--mechanisms and therapeutic implications. Nature Reviews Endocrinology. 2015;11(10):592–605. doi: 10.1038/nrendo.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sowers J.R. Endocrine functions of adipose tissue: focus on adiponectin. Clinical Cornerstone. 2008;9(1):32–38. doi: 10.1016/s1098-3597(08)60026-5. discussion 9-40. [DOI] [PubMed] [Google Scholar]
- 32.Hoffstedt J., Andersson D.P., Eriksson Hogling D., Theorell J., Naslund E., Thorell A. Long-term protective changes in adipose tissue after gastric bypass. Diabetes Care. 2017;40(1):77–84. doi: 10.2337/dc16-1072. [DOI] [PubMed] [Google Scholar]
- 33.Dahlman I., Sinha I., Gao H., Brodin D., Thorell A., Ryden M. The fat cell epigenetic signature in post-obese women is characterized by global hypomethylation and differential DNA methylation of adipogenesis genes. International Journal of Obesity. 2015;39(6):910–919. doi: 10.1038/ijo.2015.31. [DOI] [PubMed] [Google Scholar]
- 34.Ronn T., Volkov P., Davegardh C., Dayeh T., Hall E., Olsson A.H. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genetics. 2013;9(6):e1003572. doi: 10.1371/journal.pgen.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perfilyev A., Dahlman I., Gillberg L., Rosqvist F., Iggman D., Volkov P. Impact of polyunsaturated and saturated fat overfeeding on the DNA-methylation pattern in human adipose tissue: a randomized controlled trial. American Journal of Clinical Nutrition. 2017;105(4):991–1000. doi: 10.3945/ajcn.116.143164. [DOI] [PubMed] [Google Scholar]
- 36.Eto H., Suga H., Matsumoto D., Inoue K., Aoi N., Kato H. Characterization of structure and cellular components of aspirated and excised adipose tissue. Plastic and Reconstructive Surgery. 2009;124(4):1087–1097. doi: 10.1097/PRS.0b013e3181b5a3f1. [DOI] [PubMed] [Google Scholar]
- 37.Bialesova L., Kulyte A., Petrus P., Sinha I., Laurencikiene J., Zhao C. Epigenetic regulation of PLIN 1 in obese women and its relation to lipolysis. Scientific Reports. 2017;7(1):10152. doi: 10.1038/s41598-017-09232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang R., Yao F., Gao F., Abou-Samra A.B. Nrac, a novel nutritionally-regulated adipose and cardiac-enriched gene. PLoS One. 2012;7(9):e46254. doi: 10.1371/journal.pone.0046254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee K., Um S.H., Rhee D.K., Pyo S. Interferon-alpha inhibits adipogenesis via regulation of JAK/STAT1 signaling. Biochimica et Biophysica Acta. 2016;1860(11 Pt A):2416–2427. doi: 10.1016/j.bbagen.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Gong F.Y., Zhang S.J., Deng J.Y., Zhu H.J., Pan H., Li N.S. Zinc-alpha2-glycoprotein is involved in regulation of body weight through inhibition of lipogenic enzymes in adipose tissue. International Journal of Obesity. 2009;33(9):1023–1030. doi: 10.1038/ijo.2009.141. [DOI] [PubMed] [Google Scholar]
- 41.Trefely S., Khoo P.S., Krycer J.R., Chaudhuri R., Fazakerley D.J., Parker B.L. Kinome screen identifies PFKFB3 and glucose metabolism as important regulators of the insulin/insulin-like growth factor (IGF)-1 signaling pathway. Journal of Biological Chemistry. 2015;290(43):25834–25846. doi: 10.1074/jbc.M115.658815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peverelli E., Ermetici F., Corbetta S., Gozzini E., Avagliano L., Zappa M.A. PKA regulatory subunit R2B is required for murine and human adipocyte differentiation. Endocr Connect. 2013;2(4):196–207. doi: 10.1530/EC-13-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arner P., Andersson D.P., Backdahl J., Dahlman I., Ryden M. Weight gain and impaired glucose metabolism in women are predicted by inefficient subcutaneous fat cell lipolysis. Cell Metabolism. 2018;28(1):45–54 e3. doi: 10.1016/j.cmet.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Winnier D.A., Fourcaudot M., Norton L., Abdul-Ghani M.A., Hu S.L., Farook V.S. Transcriptomic identification of ADH1B as a novel candidate gene for obesity and insulin resistance in human adipose tissue in Mexican Americans from the Veterans Administration Genetic Epidemiology Study (VAGES) PLoS One. 2015;10(4):e0119941. doi: 10.1371/journal.pone.0119941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Markan K.R., Jurczak M.J., Brady M.J. Stranger in a strange land: roles of glycogen turnover in adipose tissue metabolism. Molecular and Cellular Endocrinology. 2010;318(1–2):54–60. doi: 10.1016/j.mce.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kishida K., Kuriyama H., Funahashi T., Shimomura I., Kihara S., Ouchi N. Aquaporin adipose, a putative glycerol channel in adipocytes. Journal of Biological Chemistry. 2000;275(27):20896–20902. doi: 10.1074/jbc.M001119200. [DOI] [PubMed] [Google Scholar]
- 47.Longo M., Raciti G.A., Zatterale F., Parrillo L., Desiderio A., Spinelli R. Epigenetic modifications of the Zfp/ZNF423 gene control murine adipogenic commitment and are dysregulated in human hypertrophic obesity. Diabetologia. 2018;61(2):369–380. doi: 10.1007/s00125-017-4471-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ejarque M., Ceperuelo-Mallafre V., Serena C., Maymo-Masip E., Duran X., Diaz-Ramos A. Adipose tissue mitochondrial dysfunction in human obesity is linked to a specific DNA methylation signature in adipose-derived stem cells. International Journal of Obesity. 2018 doi: 10.1038/s41366-018-0219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 1
Multimedia component 2