Not much is known about changes in host adaptation of avian influenza (AI) viruses in birds after long-term circulation in chickens or other terrestrial poultry. Although the origin of AI viruses affecting poultry is wild aquatic birds, the role of these birds in further dispersal of poultry-adapted AI viruses is not clear. Previously, we showed that HPAI viruses isolated early from poultry outbreaks could still infect and transmit well in mallards. In this study, we demonstrate that the Mexican H7N3 HPAI virus after four years of circulation in chickens replicates poorly and does not transmit in mallards but remains highly pathogenic in chickens. This information on changes in host adaptation is important for understanding the epidemiology of AI viruses and the role that wild waterfowl may play in disseminating viruses adapted to terrestrial poultry.
KEYWORDS: H7N3, avian influenza virus, chickens, highly pathogenic avian influenza, mallards, virus adaptation
ABSTRACT
Outbreaks of highly pathogenic avian influenza (HPAI) virus subtype H7N3 have been occurring in commercial chickens in Mexico since its first introduction in 2012. In order to determine changes in virus pathogenicity and adaptation in avian species, three H7N3 HPAI viruses from 2012, 2015, and 2016 were evaluated in chickens and mallards. All three viruses caused high mortality in chickens when given at medium to high doses and replicated similarly. No mortality or clinical signs and similar infectivity were observed in mallards inoculated with the 2012 and 2016 viruses. However, the 2012 H7N3 HPAI virus replicated well in mallards and transmitted to contacts, whereas the 2016 virus replicated poorly and did not transmit to contacts, which indicates that the 2016 virus is less adapted to mallards. In vitro, the 2016 virus grew slower and to lower titers than did the 2012 virus in duck fibroblast cells. Full-genome sequencing showed 115 amino acid differences between the 2012 and the 2016 viruses, with some of these changes previously associated with changes in replication in avian species, including hemagglutinin (HA) A125T, nucleoprotein (NP) M105V, and NP S377N. In conclusion, as the Mexican H7N3 HPAI virus has passaged through large populations of chickens in a span of several years and has retained its high pathogenicity for chickens, it has decreased in fitness in mallards, which could limit the potential spread of this HPAI virus by waterfowl.
IMPORTANCE Not much is known about changes in host adaptation of avian influenza (AI) viruses in birds after long-term circulation in chickens or other terrestrial poultry. Although the origin of AI viruses affecting poultry is wild aquatic birds, the role of these birds in further dispersal of poultry-adapted AI viruses is not clear. Previously, we showed that HPAI viruses isolated early from poultry outbreaks could still infect and transmit well in mallards. In this study, we demonstrate that the Mexican H7N3 HPAI virus after four years of circulation in chickens replicates poorly and does not transmit in mallards but remains highly pathogenic in chickens. This information on changes in host adaptation is important for understanding the epidemiology of AI viruses and the role that wild waterfowl may play in disseminating viruses adapted to terrestrial poultry.
INTRODUCTION
Avian influenza (AI) continues to be a threat to poultry worldwide. Recurring outbreaks of H5 and H7 highly pathogenic avian influenza (HPAI) viruses in chickens and other terrestrial poultry, and the continuing spread of H5Nx goose/Guangdong/1996 (Gs/GD) lineage viruses by wild migratory birds, underscore the need to better understand the pathogenesis and transmission of HPAI viruses in gallinaceous poultry and the role wild birds may play in initial introductions in an area and as continued vectors for farm-to-farm spread (1–3). Wild aquatic birds, especially of the orders Anseriformes (ducks, geese, and swans) and Charadriiformes (shorebirds, gulls, terns, and auks), are the natural reservoirs of AI viruses (4). AI viruses typically do not cause disease or mortality in these birds and are shed primarily through feces and spread by fecal contamination of the water or directly to other birds. Sporadically, these AI viruses can transmit to domestic birds producing subclinical infections, or occasionally, respiratory disease and drops in egg production (5). This phenotype of the virus is termed low-pathogenic avian influenza (LPAI) virus, based on low mortality produced by in vivo testing in chickens, and can be any combination of the 16 hemagglutinin (HA) and 9 neuraminidase (NA) virus subtypes (6). Some H5 and H7 LPAI viruses after circulating in gallinaceous poultry (chickens, turkeys, quail, etc.) have evolved to produce the HPAI virus pathotype. These HPAI viruses cause severe systemic disease and high mortality in gallinaceous poultry (7).
In June 2012, an outbreak of H7N3 HPAI was reported in commercial egg layer chickens in the state of Jalisco, Mexico. Since then, cases in poultry, principally chickens, have been repeatedly documented, mostly in the states of Jalisco, Guanajuato, and Puebla (http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=26143). Although the LPAI virus precursor has not been identified, the virus was determined to be closely related to wild aquatic bird isolates from North America (8). Thus, it is presumed that an LPAI virus from wild aquatic birds infected chickens and evolved into the HP form. In order to aid control, vaccination started in August 2012, and the initial vaccine strain, a 2006 H7N3 LPAI virus isolated from a cinnamon teal, was experimentally proven to confer protective immunity against the earlier virus (9, 10). Despite the use of vaccine, the H7N3 HPAI virus continues to circulate and evolve in chickens in Mexico (http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=26143).
Although wild aquatic birds are the genetic reservoirs of LPAI viruses, they are not considered genetic or long-term reservoirs of HPAI viruses (11). Experimental or natural infections and disease in domestic and wild ducks caused by HPAI viruses had been infrequently reported before the outbreaks of the Gs/GD lineage H5N1 HPAI viruses in Asia (5, 12, 13). In most experimental studies, ducks intranasally inoculated with non-Gs/GD H5 or H7 HPAI viruses showed mild or no clinical signs (14–20), but virus was isolated in some cases from tracheal and cloacal swabs (16, 19, 20) and recovered from the trachea, gut, liver, brain, and spleen (15). However, in a previous study, we demonstrated that mallards can become subclinically infected with HPAI viruses other than Gs/GD lineage H5N1 viruses and transmit to naive mallards under experimental conditions (21). Among these viruses, one of the early Mexican H7N3 HPAI chicken isolates from 2012, A/chicken/Jalisco/CPA1/2012 (H7N3), caused typical severe clinical signs of HPAI and high mortality in chickens (9) but no disease or mortality in mallards (21). The fact that mallards became infected and transmitted the early Mexican H7N3 virus suggests that the virus was still adapted to wild waterfowl. Long-term circulation of H7N3 HPAI viruses in gallinaceous poultry in Mexico since 2012 has provided ample opportunity to further adapt to gallinaceous species.
The presence of H7N3 HPAI virus in poultry in Mexico is of concern not only to Mexico’s poultry industry but also to neighboring countries. Spillover of poultry viruses into wild passerine birds was reported during the initial epizootic of H7N3 HPAI in June to October 2012 in Jalisco, Mexico, with infections detected in wild endemic great-tailed grackles (Quiscalus mexicanus) (22). The virus was also isolated from endemic clay-colored thrush (Turdus grayi) and plain chachalaca (Ortalis vetula) in a natural reserve in the State of Chiapas in 2015 (http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=17686). It is not known how common spillover into nonpoultry avian species happens with viruses that have circulated for long periods of time in gallinaceous poultry and if migratory waterfowl could become infected and spread these viruses. In this study, we determined the infectivity, pathogenesis, and transmission of Mexican H7N3 HPAI viruses in chicken and mallards, as representatives of the most common gallinaceous poultry and migratory waterfowl species affected by AI, to examine changes of virus adaptation between the initial isolate from 2012 and later isolates from 2015 and 2016.
RESULTS
Infectivity, transmission, and pathogenicity of the 2012, 2015, and 2016 H7N3 HPAI viruses in chickens.
The results for virus infectivity and transmission in chickens are shown in Table 1. Birds, both directly inoculated and contacts, were considered infected if they shed virus and/or seroconverted by 10 days postinoculation (dpi). All chickens inoculated with the 104 and 106 50% egg infectious dose (EID50) of any of the three viruses became infected and died by 3 days. Chickens infected with A/chicken/Puebla/CPA-28973/2015 (A/CK/PB/15) survived for longer than chickens infected with A/chicken/Jalisco/CPA1/2012 (A/CK/JL/12) and A/chicken/Puebla/CPA-03191-16-CENASA-95076/2016 (A/CK/PB/16) viruses. One chicken inoculated with the 102 dose of A/CK/PB/15 shed virus and died, but all other chickens in the 102 groups survived. The 50% bird infectious dose (BID50), although lowest for the A/CK/PB/15 virus, was similar for all three viruses (2.8 to 3.0 log10 BID50). Most chickens that became infected died without showing clinical signs (peracute disease). Ruffled feathers, lethargy, anorexia, prostration, swollen heads, green diarrhea, and cyanotic combs were observed in the rest. No differences in clinical signs were found between the groups infected with the three different viruses. The surviving chickens did not show evidence of clinical disease and were serologically negative.
TABLE 1.
Number of infected birds, 50% bird infectious dose, mortality, and seroconversion in chickens inoculated by the intrachoanal route or contact exposed with three H7N3 HPAI viruses from 2012, 2015, and 2016
| Virus | Inoculated birds (n = 5) |
Contact-exposed birds |
||||||
|---|---|---|---|---|---|---|---|---|
| Virus dose (log10 EID50) | No. of infected birds/total no. of birds inoculateda | BID50 (log10 EID50)b | Mortality (MDT)c | Serologyd | No. of infected birds/total no. of birds | Mortality (MDT)c | Serologyd | |
| A/CK/JL/12 | 2 | 0/5 | 3.0 | 0/5 | 0/5 | 0/3 | 0/3 | 0/3 |
| 4 | 5/5 | 5/5 (2) | NA | 1/3 | 1/3 (4) | 0/2 | ||
| 6 | 5/5 | 5/5 (2) | NA | 0/3 | 0/3 | 0/3 | ||
| A/CK/PB/15 | 2 | 1/5 | 2.8 | 1/5 (3) | 0/4 | 0/3 | 0/3 | 0/3 |
| 4 | 5/5 | 5/5 (2.8) | NA | 0/3 | 0/3 | 0/3 | ||
| 6 | 5/5 | 5/5 (2.8) | NA | 0/3 | 0/3 | 0/3 | ||
| A/CK/PB/16 | 2 | 0/5 | 3.0 | 0/5 | 0/5 | 0/3 | 0/3 | 0/3 |
| 4 | 5/5 | 5/5 (2.6) | NA | 0/3 | 0/3 | 0/3 | ||
| 6 | 5/5 | 5/5 (2) | NA | 0/3 | 0/3 | 0/3 | ||
Number of chickens infected/total number of birds inoculated; determined by qRRT-PCR and serology.
BID50, 50% bird infectious dose.
Number of dead birds/total number of birds inoculated (days postinoculation). MDT, mean death time (days post-virus exposure).
Number of birds with positive antibody titers/total number of birds that survived. NA, not applicable (because all died).
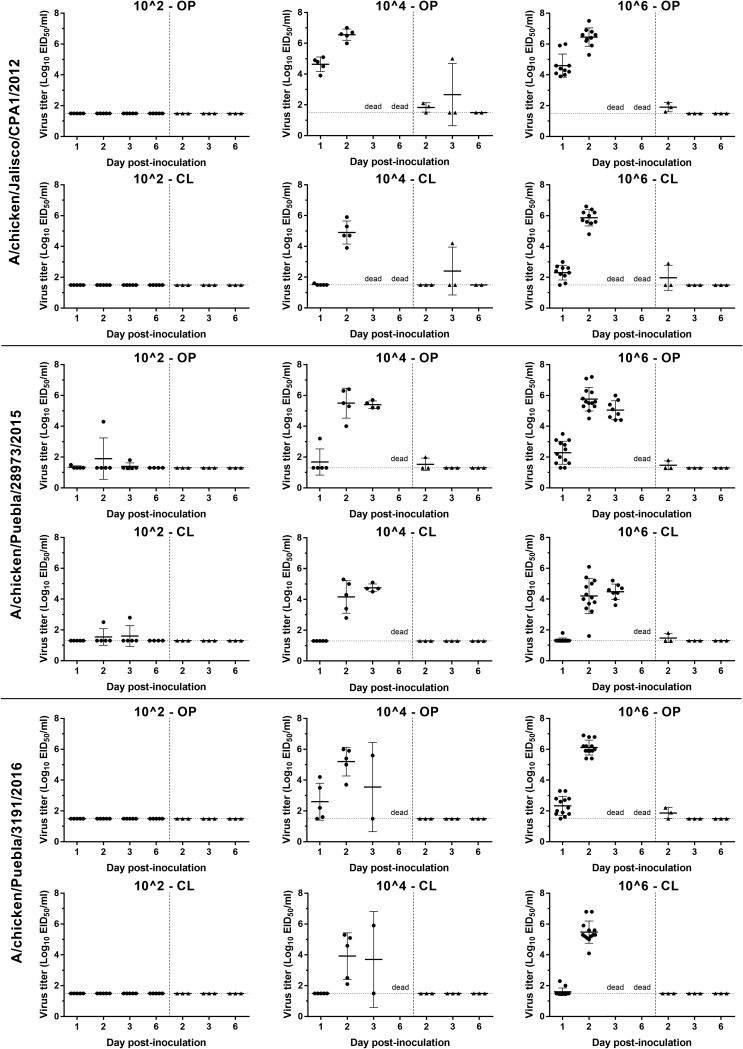
Moderate to high titers of all three viruses, as determined by viral RNA quantification, were shed by both the oropharyngeal (OP) (4.5 to 7.5 log10 EID50/ml) and cloacal (CL) (3.6 to 6.8 log10 EID50/ml) routes by the infected chickens at 2 dpi (Fig. 1). The OP titers were significantly higher than CL titers with all three viruses at the 106 dose. Among the chickens inoculated with the 106 dose of the three viruses, the mean virus titer of OP swabs in chickens infected with A/CK/JL/12 virus was higher than that in chickens infected with A/CK/PB/15 and A/CK/PB/16 viruses at 1 dpi (P < 0.0001) and with A/CK/PB/15 at 2 dpi (P = 0.0454). In groups inoculated with the 104 dose, the mean viral titer with A/CK/JL/12 virus was higher than with A/CK/PB/15 and A/CK/PB/16 viruses (P = 0.0006) at 1 dpi but only higher than that of A/CK/PB/16 at 2 dpi (P = 0.0477). In the CL swabs, the mean virus titer with A/CK/JL/12 was also significantly higher than that with A/CK/PB/15 and A/CK/PB/16 at 1 dpi (P < 0.0001) and with A/CK/PB/15 at 2 dpi (P = 0.0001).
FIG 1.
Scatter plot of oropharyngeal (OP) and cloacal (CL) virus shedding detected by qRRT-PCR from chickens inoculated with the 2012, 2015, and 2016 H7N3 HPAI viruses or exposed by direct contact. Virus titers are expressed as log10 with error bars. For statistical purposes, qRRT-PCR-negative samples were given a value of 0.1 log10 below the qRRT-PCR test limit of detection (1.5 log10 EID50/ml for the A/CK/JL/12 and A/CK/PB/16 viruses and 1.3 log10 EID50/ml for A/CK/PB/15 viruses). The circle (●) and triangle (▲) plots indicate viral shedding from inoculated and contact chickens, respectivelly.
Only one contact chicken in the group inoculated with the 104 dose of the A/CK/JL/12 virus became infected and died. Low levels of viral RNA were detected in swabs taken from a few contact chickens. However, as these birds did not show clinical signs and did not seroconvert, they were not considered infected.
The two birds necropsied at 2 dpi from each of the 106-dose groups had empty intestines and were dehydrated. Mild to moderate splenomegaly with parenchymal mottling, enlarged kidneys, catarrhal rhinitis, pale pancreas, and congested lungs were observed in all birds. Similar types and severities of histological lesions and virus antigen staining in tissues were present in all birds examined and were similar to lesions reported previously with A/CK/JL/12 in chickens (Table 2) (9). Moderate to severe multifocal necrosis was present in the parenchymal cells of many tissues but especially in the lung, heart, brain, liver, spleen, and adrenal gland, in some cases accompanied by mild to severe inflammation. Virus antigen, detected by immunohistochemical (IHC) analysis, was present in areas of necrosis and in infiltrating mononuclear cells in many organs, including the lung, brain, liver, adrenal gland, spleen, bursa, and thymus, as well as in parenchymal cells of some organs, including cardiac myocytes, Kupffer cells, hepatocytes, microglial cells and neurons, epithelium of air capillaries in the lung, kidney tubular epithelial and glomerular cells, bursa epithelium, and feather follicle epithelial cells (Table 2). Viral antigen staining in capillary endothelial cells was uncommon, restricted mainly to capillaries in the eyelid, comb, and air capillaries of the lungs. High virus titers were found in the lung, spleen, brain, heart, and muscle from all infected birds examined at 2 dpi (Table 3). Noninoculated control chickens showed no clinical signs or evidence of infection.
TABLE 2.
AI virus antigen staining in tissues from chickens inoculated by the intrachoanal route with H7N3 HPAI viruses
| Tissue | Result by virus (bird 1/bird 2)a |
||
|---|---|---|---|
| A/CK/JL/12 | A/CK/PB/15 | A/CK/PB/16 | |
| Nasal epithelium | +++/+++ | +++/+ | +++/+++ |
| Trachea | ++/+++ | ++/+++ | +++/+++ |
| Lung | +++/+++ | +++/++ | +++/+++ |
| Air sac | ++/+ | ++/+ | +/+ |
| Brain | ++/++ | +++/++ | +++/+++ |
| Heart | +++/+++ | +++/+++ | +++/+++ |
| Spleen | +++/+++ | +++/+++ | +++/+++ |
| Liver | ++/++ | +++/+ | +++/+++ |
| Kidney | ++/++ | +/+ | +++/+++ |
| Adrenal gland | +/+ | ++/+ | ++/++ |
| Skeletal muscle | ++/++ | ++/++ | ++/++ |
| Pancreas | −/− | +/+ | +++/+ |
| Cloacal bursa | +++/+++ | +++/++ | +++/++ |
| Cecal tonsils | +++/++ | +++/+ | ++/+++ |
| Thymus | +++/+++ | ++/++ | +/+ |
| Proventriculus | ++/++ | +/+ | +++/+++ |
The chickens were euthanized at 2 dpi. −, no positive cells; +, single-positive cells; ++, scattered groups of positive cells; +++, widespread positivity.
TABLE 3.
Comparison of AI virus titers in the lung, spleen, brain, muscle, and heart in chickens and mallards
| Virus by host | Virus titer (log10 EID50/g) for bird 1/bird 2 ina: |
||||
|---|---|---|---|---|---|
| Lung | Spleen | Brain | Heart | Muscle | |
| Chickens | |||||
| A/CK/JL/12 | 8.6/8.5 | 8.3/8.4 | 9.1/8.4 | 9.4/9.3 | 8.5/8.7 |
| A/CK/PB/15 | 7.5/6.8 | 7.5/6.9 | 8.8/8.7 | 8.3/8.3 | 8.2/7.6 |
| A/CK/PB/16 | 8.0/7.3 | 7.6/7.9 | 8.3/8.4 | 8.0/8.4 | 7.5/7.9 |
| Mallards | |||||
| A/CK/JL/12b | 3.5/3.7 | 2.6/4.3 | –/5.2 | ND | ND |
| A/CK/PB/16 | 2.1/– | 2.0/– | –/– | –/– | –/– |
| A/ML/MN/98 | 3.0/– | –/– | –/– | –/3.0 | –/– |
Tissues were taken from two birds per group at 2 dpi for chickens and 4 dpi for mallards. –, negative; ND, not done.
Data adopted from Pantin-Jackwood et al. (21).
In summary, although chickens infected with the 2012 H7N3 virus shed higher virus titers than did the 2015 and 2016 viruses, all three H7N3 HPAI viruses had similar infectivity (i.e., BID50) and pathogenicity and were poorly transmissible.
Infectivity, pathogenicity, and transmission of the 2012 and 2016 H7N3 HPAI viruses in mallards and comparison with a mallard-origin H6N2 LPAI virus.
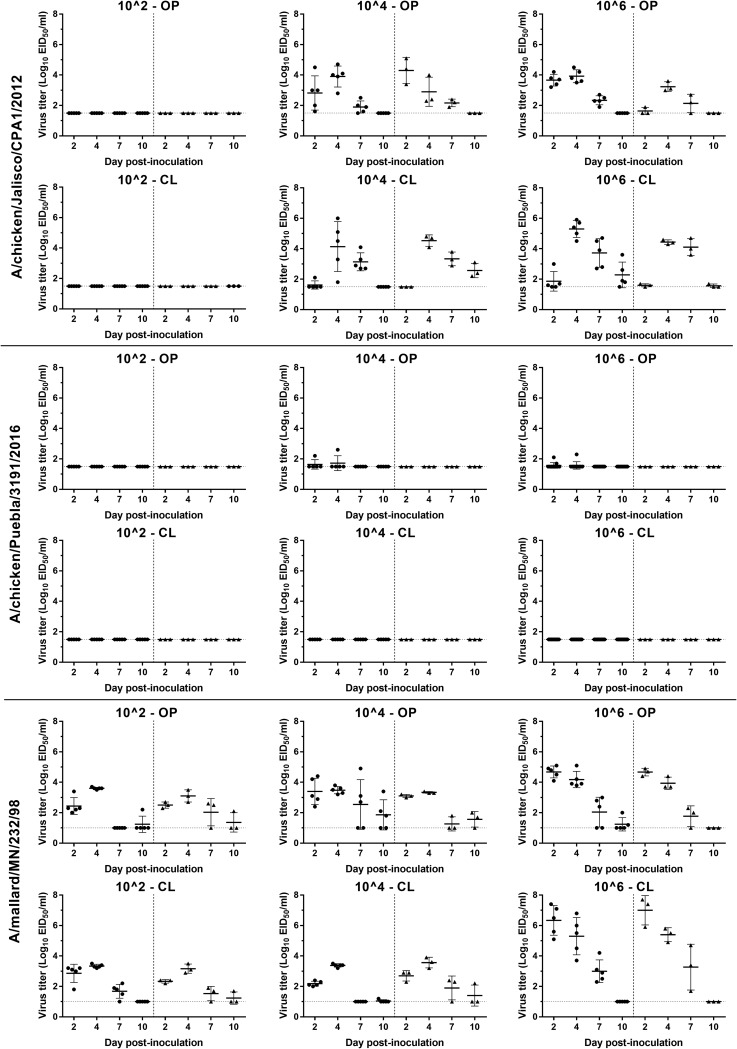
Since the results for the three H7N3 HPAI viruses were similar in chickens, the earlier and later viruses were examined in mallards. The results for virus infectivity and transmission in mallards of the 2012 and 2016 viruses and an H6N2 LPAI virus are shown in Table 4. No mortality was observed in any of the mallards inoculated with three viruses and the contacts regardless of the virus dose given. Based on virus shed and seroconversion, mallards inoculated with the 104 and 106 EID50 of A/CK/JL/12 virus, and all the contacts in these groups, became infected, which resulted in a BID50 of 3 log10 EID50. Seroconversion was observed in 2 to 5 mallards inoculated with all doses of A/CK/PB/16 virus, also resulting in a BID50 of 3 log10; however, no transmission to contacts occurred, and low or undetectable levels of virus was shed (Fig. 2). While viral shedding from the OP and CL routes was detected in all the mallards inoculated with 104 and 106 doses of A/CK/JL/12 virus and contacts, only one and three mallards inoculated with 104 and 106 doses of A/CK/JL/16 virus, respectively, shed virus from the OP route, with no viral shedding detected in contact mallards. No gross lesions were observed in the two mallards necropsied from the A/CK/PB/16 virus 106-dose group, and no microscopic lesions or virus staining was found in tissues collected from these birds. Noninoculated control mallards showed no clinical signs or evidence of infection.
TABLE 4.
Number of infected birds, 50% bird infectious dose, mortality, and seroconversion in mallards inoculated via the intrachoanal route or contact exposed with two H7N3 HPAI viruses from 2012 and 2016, and a mallard origin H6N3 LPAI virus
| Virus | Inoculated birds (n = 5) |
Contact-exposed birds (n = 3) |
||||||
|---|---|---|---|---|---|---|---|---|
| Virus dose (log10 EID50) | No. of infected birdsa | BID50 (log10 EID50)b | Mortality | Serology (range of antibody titers, [log2])c | No. of infected birds | Mortality | Serology (range of antibody titers [log2])c | |
| A/CK/JL/12 (H7N3) | 2 | 0 | 3.0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 5 | 0 | 5 (4–5) | 3 | 0 | 3 (4–6) | ||
| 6 | 5 | 0 | 5 (4–5) | 3 | 0 | 3 (4–5) | ||
| A/CK/PB/16 (H7N3) | 2 | 2 | 3.0 | 0 | 2 (5) | 0 | 0 | 0 |
| 4 | 4 | 0 | 4 (4–6) | 0 | 0 | 0 | ||
| 6 | 5 | 0 | 5 (4–5) | 0 | 0 | 0 | ||
| A/ML/MN/98 (H6N2) | 2 | 5 | <2 | 0 | 1 (8) | 3 | 0 | 0 |
| 4 | 5 | 0 | 3 (7–9) | 3 | 0 | 0 | ||
| 6 | 5 | 0 | 5 (7–8) | 3 | 0 | 3 (7–8) | ||
Determined by qRRT-PCR and serology.
BID50, 50% bird infectious dose.
Number of birds with positive antibody titers.
FIG 2.
Scatter plot of oropharyngeal (OP) and cloacal (CL) virus shedding detected by qRRT-PCR in from mallards inoculated with the 2012 and 2016 H7N3 HPAI viruses and an H6N2 LPAI virus or exposed by direct contact. Virus titers are expressed as log10 with error bars. For statistical purposes, qRRT-PCR-negative samples were given a value of 0.1 log10 below the qRRT-PCR test limit of detection (1.5 log10 EID50/ml for A/CK/JL/12 and A/CK/PB/16 viruses and 1.0 log10 EID50/ml for A/ML/MN/98 virus). The circle (●) and triangle (▲) plots indicate viral shedding from inoculated and contacted chickens, respectively.
The mallard-origin H6N2 virus (A/ML/MN/98) was shed by all inoculated mallards by both the OP and CL routes regardless of the dose given. The BID50 was calculated to be less than 102 EID50. A/ML/MN/98 was transmitted to all the contacts based on viral shedding, but only contact mallards in the high-dose group seroconverted. No gross lesions were observed in the two necropsied mallards. Microscopic lesions consisted of mild lymphocytic rhinitis and tracheitis, and sporadic viral antigen staining was present in epithelial cells and infiltrating mononuclear cells of the nasal turbinates, intestine, and bursa of Fabricius.
The mean viral titers from OP and CL swabs were significantly lower in mallards inoculated with A/CK/PB/16 than with A/CK/JL/12 at all time points. In groups inoculated with the 106 dose, the mean virus titer with A/ML/MN/98 OP, 4.7 log10 EID50/ml; CL, 6.3 log10 EID50/ml) was higher than with the A/CK/JL/12 virus (OP, 3.7 log10 EID50/ml; CL, 1.9 log10 EID50/ml) in both OP (P = 0.0031) and CL (P < 0.0001) swabs at 2 dpi (Fig. 2). With the 104 dose, the mean virus titer from CL swabs with A/ML/MN/98 (2.2 log10 EID50/ml) was higher than with A/CK/JL/12 virus (1.6 log10 EID50/ml) at 2 dpi (P = 0.0059). The A/ML/MN/98 virus had faster onset of viral replication and higher earlier titers than did the 2012 H7N3 virus, but the A/CK/JL/12 virus was detected in cloacal swabs for longer than the A/ML/MN/98 virus in inoculated mallards.
The presence of viral RNA was examined in the tissues of the mallards necropsied at 4 dpi (Table 3). Because of limited mallard supply, we could not allocate mallards for pathogenicity testing with the A/CK/PB/12 virus. For comparison, we used the tissue viral titers of lung, spleen, and brain from a similar study we previously conducted following the exact methods used in this study (21). While the A/CK/PB/16 and A/ML/MI/98 viruses did not replicate or replicated poorly in the organs examined, the A/CK/JL/12 virus replicated to moderate titers in the lung, spleen, and brain.
In summary, the 2012 and 2016 H7N3 HPAI viruses showed similar infectivity and induced no mortality in mallards. However, while the 2016 H7N3 virus was barely shed and replicated poorly in tissues, the 2012 H7N3 virus replicated well in the respiratory and intestinal tract and internal organs and transmitted to contact mallards.
Comparison of viral growth in chicken and duck cells.
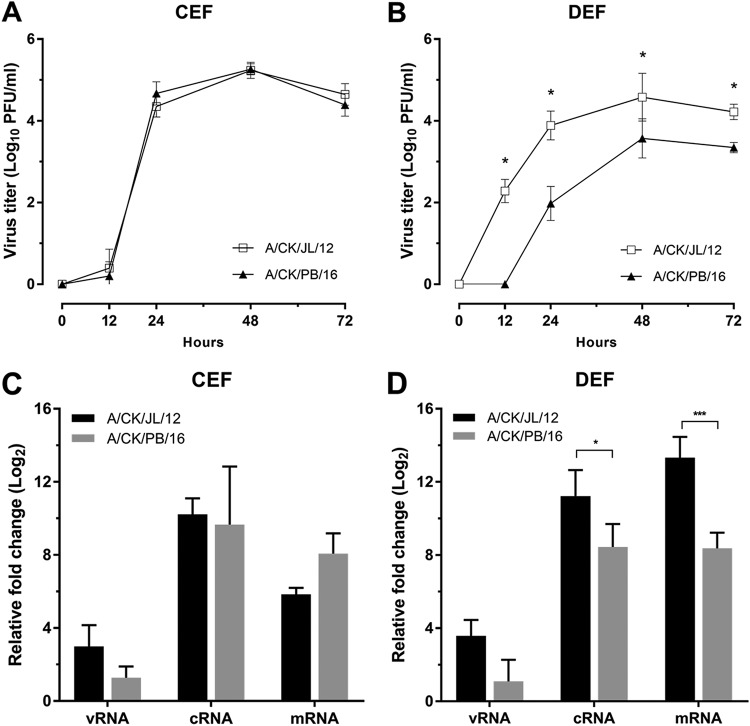
The growth of A/CK/JL/12 and A/CK/PB/16 viruses was examined in chicken and duck cells. In chicken embryo fibroblasts (CEF), both viruses grew to high titers (105.2 PFU/ml), without notable differences (Fig. 3A). This result is similar to the in vivo results in chickens. In contrast, in duck embryo fibroblasts (DEF), the A/CK/JL/12 virus grew to titers 1 to 2 logs higher than the A/CK/PB/16 virus at all time points (Fig. 3B), again similar to what was found in vivo in mallards.
FIG 3.
Viral growth kinetics and relative viral RNA quantification in chicken and duck cells. (A and B) CEF and DEF were inoculated with two chicken origin H7N3 HPAI viruses (A/CK/JL/12 and A/CK/PB/16 viruses) at a multiplicity of infection (MOI) of 0.001. The plaque titers from supernatant were determined at 12, 24, 48, and 72 h postinfection on MDCK cells, and each point represents the average of three independent replicates. The asterisk indicates the significance between the two H7N3 HPAI viruses (P < 0.05). (C and D) Total RNA was extracted from CEF and DEF lysates at 12 and 24 h postinfection. vRNA, cRNA, and mRNA were measured by strand-specific RRT-PCR. The fold increase was calculated by comparing the viral RNA detection at 24 hpi to that at 12 hpi for each type of RNA. Each value is the average from three independent experiments. Statistical significance was analyzed by using two-way ANOVA with Bonferroni multiple-comparison test. A P value of <0.05 was considered to be significant.
To compare the replication ability between A/CK/JL/12 and A/CK/PB/16 virus polymerases, we compared the expression of viral RNA (vRNA), cRNA, and mRNA in CEF and DEF lysates during exponential viral growth (from 12 to 24 hours postinoculation [hpi]) (Fig. 3C and D). Although the mean fold increase of mRNA in the A/CK/PB/16 virus was higher than in the A/CK/JL/12 virus, no significant difference in fold increase was found in all three types of RNA in CEF. However, the mean fold increases in cRNA and mRNA of the A/CK/JL/12 virus (cRNA, 11.22 ± 1.42; mRNA, 13.33 ± 1.13) were higher than those of the A/CK/JL/16 virus (cRNA, 8.44 ± 1.25; mRNA, 8.37 ± 0.86) in DEF, which correlated with the significant viral growth difference in the supernatant.
Sequence analysis.
The phylogenetic analysis showed that the all eight sequences of the initial H7N3 HPAI virus isolate from 2012 were genetically closely related to North American lineage wild bird isolates (see Fig. S1 in the supplemental material). After the first outbreak in 2012, and based on available sequences in the public database, all eight genes of H7N3 HPAI viruses evolved into two separate genetic clusters, which we designated clusters A and B. Consistent clustering of each gene suggests that the viruses evolved through genetic drift from initial outbreak viruses without gene reassortment. The nucleotide and amino acid similarities between clusters A and B are summarized in Table 5. The HA genes bifurcated into two clusters with extensive mutations (95.42% to 95.89%) compared to other segments, which was more weighted in the HA1 region (93.71% to 94.79%) than the HA2 region (97.44% to 98.04%). Sequence analysis also showed that the early H7N3 virus acquired a novel extended cleavage site, which probably originated from recombination with 28S rRNA from the avian host (23), and recent sequence analysis showed that the insertion has been maintained until 2016, with some amino acid changes (A/CK/JL/12, PENPKDRKSRHRRTR|GLF; cluster A, PENPKDRKNRHRRTR|GLF; cluster B, PENPKGKKSRHRKTR|GLF; and 2015 isolates, PENSKDMKSRHRKTR|GLF). In the polymerase segments, the PB1 gene showed the farthest nucleotide identity (95.33% to 95.68), which was not necessarily linked to the most distinct amino acid homology, the PA protein (96.93% to 97.35%).
TABLE 5.
Nucleotide and amino acid similarities between H7N3 HPAI virus clusters A and B
| Segment | % identity |
|
|---|---|---|
| Nucleotide | Amino acid | |
| PB2 | 96.62–96.97 | 97.63–97.89 |
| PB1 | 95.33–95.68 | 97.23–97.62 |
| PA | 96.83–97.07 | 96.93–97.35 |
| HA | 95.42–95.89 | 95.00–96.07 |
| NP | 96.72–97.26 | 98.39–98.80 |
| NA | 96.52–96.94 | 95.74–96.80 |
| M | 97.25–98.17 | |
| M1 | 98.02–98.81 | |
| M2 | 94.85–95.88 | |
| NS | 96.72–97.26 | |
| NS1 | 93.91–96.09 | |
| NS2 | 98.35 | |
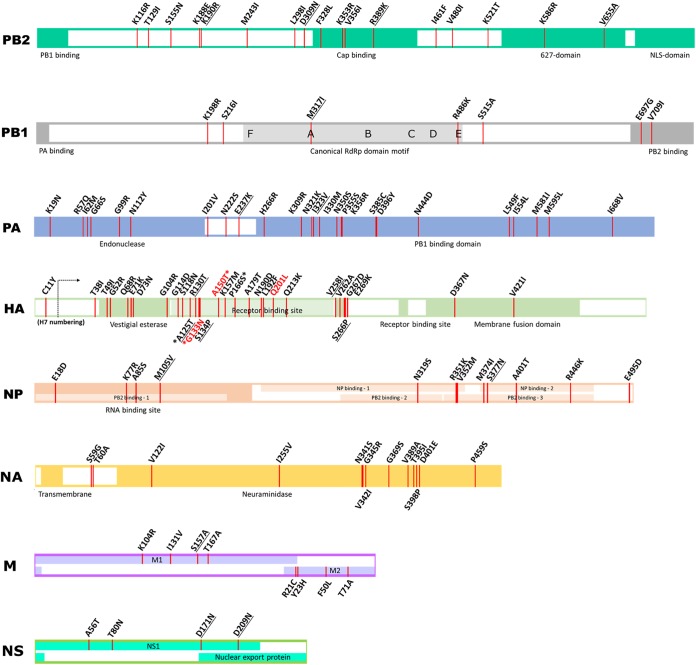
Full-genome sequence comparison between the A/CK/JL/12 and A/CK/PB/16 viruses showed 115 amino acid differences between the two viruses (PB2, n = 17; PB1, n = 7; PB1-F2, n = 1; PA, n = 25; HA, n = 29; NP, n = 12; neuraminidase [NA], n = 13; M1, n = 4; M2, n = 3; and NS1, n = 4). The amino acid changes are visualized in Fig. 4, with putative or functional domain annotation inferred from other studies (24–26). A total of 24 amino acid changes corresponding to those found in other studies associated with chicken or duck adaptation or virulence were identified and are summarized in Table 6 (27–40).
FIG 4.
Amino acid changes and functional domain annotation. Amino acid changes between the A/CK/JL/12 and A/CK/PB/16 viruses are marked at each segment in red lines. Specific amino acid changes underlined correspond to changes found in other studies associated with chicken or duck adaptation. Specific amino acid changes written in red indicate possible associations with vaccine escape mutations. Four amino acids marked with asterisks in the HA gene are linked to possible N-glycosylation acquisitions. RdRp, RNA-dependent RNA polymerase.
TABLE 6.
Amino acid changes associated with increase in virulence or adaptation of AI virus in chickens and ducks
| Protein | Amino acid substitution(s)a | Corresponding substitutions or sites in other studiesb | Remarks | Reference |
|---|---|---|---|---|
| NP | K77R | K77R | Introduction of wild bird H7N3 virus into domestic turkeys | Campitelli et al. (38) |
| M105V | M105V | |||
| PA | K19N | K19N (in quail) | H7N3 HPAI virus experimentally passaged in quail or turkeys | Giannecchini et al. (28) |
| H266R | R266H (in turkey) | |||
| M2 | S31N | S31N | Evolution of Gs/GD H5N1 HPAI virus after circulation in vaccinated commercial chickens | Arafa et al. (29) |
| PA | E237K | E237K | Increased pathogenicity of Gs/GD lineage H5N1 HPAI virus in chickens due to early destruction of innate immune response | Suzuki et al. (30) |
| NP | K77R | K77R | ||
| M105V | M105V | |||
| S377N | S377N | |||
| PB2 | D309N | N309D | Decreased mortality in chickens with Gs/GD lineage H5N1 HPAI reassortant viruses | Wasilenko et al. (31) |
| R389K | K389R | |||
| PB2 | V480I | V480I | Mexican H5N2 LPAI virus mutation to HPAI virus in chickens | Horimoto et al. (32) |
| PB1 | M317I | M317I | ||
| R486K | R486K | |||
| PA | G99R | G99R | ||
| NP | M105V | M105V | Amino acid change in Gs/GD lineage H5N1 HPAI virus increased polymerase activity and growth in chicken cells | Tada et al. (33) |
| HAc | A125T | A125T | H7N3 LPAI virus evolution in vaccinated poultry in Pakistan | Abbas et al. (34) |
| HAc | A125T | A/V/K125T | Amino acid sites positively selected after transmission of H7 viruses from wild to domestic birds | Lebarbenchon et al. (27) |
| R130T | 130 | |||
| S134P | 134 | |||
| V258I | 258 | |||
| S266P | 266 | |||
| PB1-F2 | E4G | E4G | Increased mortality in chickens with Gs/GD lineage H5N1 HPAI reassortant viruses | Bogs et al. (35) |
| NP | M105V | M105V | ||
| S377N | S377N | |||
| M2 | S82N | S82N | ||
| PB2 | R389K | K389R | Increased mortality in ducks with Gs/GD lineage H5N1 HPAI reassortant viruses | Song et al. (36) |
| PB2 | K190R | R190K | Increased mortality in ducks with Gs/GD lineage H5N1 HPAI reassortant viruses | Kajihara et al. (37) |
| PA | E237K | K237E | ||
| NP | S377N | N377S | ||
| NS1 | D209N | N209D | ||
| HAc | Q213K | K213Q | Changes in Gs/GD lineage H5 HPAI viruses associated with increased binding in duck intestines | Guo et al. (39) |
| NS1 | D209N | N209D | Decreased mean death time in ducks with Gs/GD lineage H5N1 HPAI reassortant viruses | Sarmento et al. (40) |
Amino acid changes from A/CK/JL/12 to A/CK/PB/16.
Amino acid changes specifically indicated in the publication or obtained from the published sequences.
Many amino acid changes in the polymerase complex proteins were observed between the 2012 and the 2016 viruses (Fig. 4 and S2), including changes in the PB2 and NP interaction site and RNA binding site of NP, endonuclease domain at the N terminus of PA, the C-terminal domain of PA, the contact area of the C terminus of PA, and the N terminus of PB1, the contact area of the C terminus of PB1 and the N terminus of PB2 the cap-binding site on PB2, and the 627-nuclear localization signal (627-NLS) domain of PB2. Some of the changes observed have been associated with changes related to adaptation or increased virulence in chickens or ducks (Table 6) (28, 30–33, 35–38).
Differences in HA amino acids and the acquisition of potential N-glycosylation sites of the A/CK/PB/16 virus compared to the A/CK/JL/12 virus are shown in Fig. 4 and 5A. The A/CK/PB/16 virus acquired four potential glycosylation sites at 123, 133, 149, and 164, all of which were localized in the globular head (H7 numbering). Overall, between 2012 and 2016, H7N3 Mexican strains obtained additional glycosylation sites in the globular head of the HA protein at the positions 123, 133, 149, and 164 (cluster A, 123, 133, 149, and 164; cluster B, 123 and 149). Some of the changes observed have been reported previously (Table 6) (27, 34, 39). Potential deglycosylation sites in the hypervariable region of the NA were found in the A/CK/PB/16 virus (Fig. 4 and 5B). Overall, between 2012 and 2016, the NA lost two potential glycosylation sites at positions 14 and 57 (cluster A, 57 and 2; cluster B, 14 and 57).
FIG 5.
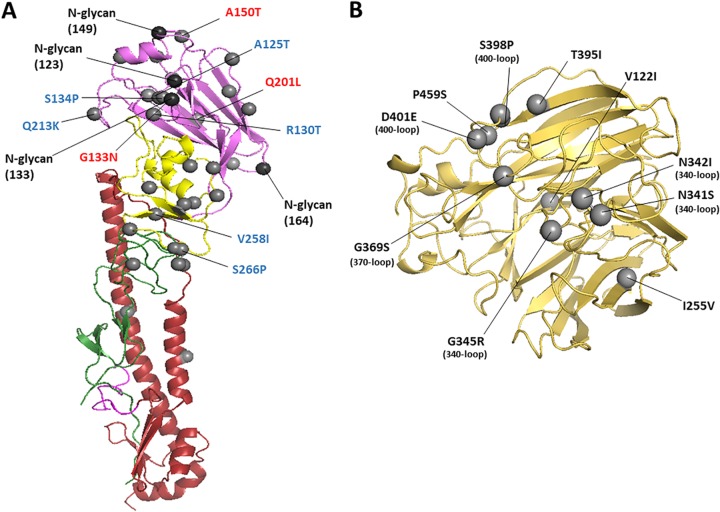
The hemagglutinin (HA) and neuraminidase (NA) structure of A/CK/PB/16 virus. The amino acid changes from A/CK/JL/12 to A/CK/PB/16 are shown as gray spheres using H7 numbering and N3 numbering. (A) Ribbon diagram of H7 HA. Only a monomer is shown. Potential vaccine escape mutations and residues related to chicken and duck adaptation are labeled in red and blue, respectively. The HA1 is depicted in green (F′ subdomain), yellow (vestigial esterase subdomain), and violet (receptor binding domain). The HA2 is shown in magenta (fusion peptide) and red (F subdomain). The amino acid changes from A/CK/JL/12 to A/CK/PB/16 are represented as gray spheres. The possible glycosylation sites are represented as black spheres. (B) Ribbon diagram of the head region of N3 NA. Only one monomer is shown. Amino acid changes located in the 340-, 370-, and 400-loop regions are marked in parentheses.
A lower number of changes were found in the M and NS genes; however, some of the changes have also been identified by others in association with increased adaptation in chickens or virulence in ducks (Table 6) (29, 35, 37, 40).
DISCUSSION
Surveillance of migratory waterfowl on the American continent has shown that the H7N3 subtype is predominant within H7 subtype viruses over all other subtype combinations (41). When it comes to domestic poultry, H7N3 LPAI virus was the second most frequently reported subtype and on four occasions mutated to HPAI virus by acquiring basic amino acids through homologous or nonhomologous recombination (23, 42). In 2002, an H7N3 HPAI outbreak at a broiler breeder farm in Chile was identified and later controlled by depopulation and strict biosecurity (43). H7N3 LPAI virus precursors were also suspected to be introduced to commercial poultry operations in Canada (2004 and 2007) and Jalisco, Mexico (2012), and evolved to cause outbreaks of H7N3 HPAI (8, 44, 45). Because all four HPAI viruses have different internal gene constellations and varied polybasic cleavage sites, each event represents a separate introduction of an LPAI virus from wild birds to domestic poultry. However, the similarity of viral genes between domestic and wild birds supported that virus flow from wild birds to domestic poultry (41). The LPAI poultry precursor for the Mexican H7N3 HPAI virus was not reported during the 2012 outbreak, so it is not clear for how long the LPAI virus circulated in poultry before evolving into the highly pathogenic form. After the initial outbreak of HPAI, with the accompanying severe disease in chickens, the implementation of a vaccine campaign reduced the severity of clinical disease and the incidence of cases, but the H7N3 HPAI virus has not been eradicated in poultry in Mexico.
The objective of the present study was to compare the pathobiology of early (2012) and more recent (2015 and 2016) Mexican H7N3 HPAI viruses in chickens and mallards to determine if the continuing circulation of these viruses in gallinaceous poultry has affected the fitness of the virus in migratory waterfowl, using the mallard as a model for waterfowl and using chickens as a model for gallinaceous poultry. We define virus adaptation as the ability of a virus to efficiently infect, replicate, and transmit in a host. This includes strategies developed by viruses to improve fitness and survival within the host. No differences in infectivity in chickens, as defined by BID50, were found between the 2012, 2015, and 2016 viruses, and all three viruses caused 100% mortality in infected birds. Gross and microscopic lesions, virus tissue tropism, and amount of virus staining were similar between the three viruses; however, some differences in the patterns of virus replication and shed were observed, with earlier virus detection and 1-log10-higher titers at peak of shedding in OP and CL swabs in chickens infected with the A/CK/JL/12 virus. With the exception of one chicken from the A/CK/JL/12 virus 104-dose group, no virus transmission to contacts was observed despite the BID50 of all three viruses being lower than the suggested dose needed for virus transmissibility in chickens (<4.7 log10 EID50) (46). The one contact chicken in the group that received a 104 dose of A/CK/JL/12 virus became infected and died at 4 dpi, but chickens in the group that received the high dose survived. As the virus shed by inoculated chickens was comparable between doses of 106 and 104, it is inferred that there was no significant difference in virus exposure. Therefore, this discrepancy is likely caused by differences in susceptibility in individual birds. The transmission results obtained in this study in chickens are similar to what we have found with other HPAI viruses examined under the same experimental conditions, in which, in spite of the virus causing severe disease and being shed in high titers, little or no transmission is observed (47–50). However, LPAI viruses that are known to be well adapted to chickens, including the Asian H9N2 and H7N9 viruses, readily transmitted in chickens in our model (51). Results from other research groups show that the transmissibility of AI viruses in chickens, including HPAI viruses, varies depending on the virus strain and how long the virus has circulated in chickens (14, 52, 53). It is not clear why HPAI viruses do not transmit well under experimental conditions when they easily transmit under field conditions. One explanation might be the relatively short period of contact exposure due to the acuteness of the disease during experimental HPAI infection. Also, under field conditions, other factors could increase virus transmissibility in chickens, such as breed and age of the birds, stress, higher density of birds, different husbandry conditions, exposure to other pathogens, and extended viral shedding periods under improper vaccine immune status. Further studies are needed to better understand the transmission of AI viruses in birds.
In order to address the possible role of waterfowl in the spread of H7N3 HPAI viruses, the infectious dose and transmissibility of the 2012 and 2016 H7N3 HPAI viruses were examined in mallards and compared to those of a mallard origin LPAI virus (A/ML/MI/98). No mortality or clinical signs were observed in any of the experimental groups. The BID50 for A/ML/MI/98 virus was <2 log10 EID50, and the virus transmitted to all contacts, indicating as expected that this virus is well adapted to mallards. For the 2012 Mexican HPAI virus (A/CK/JL/12), the BID50 was 3 log10 EID50, and the virus transmitted to all contacts in the groups infected with the medium and high doses. Although the BID50 was the same for the more recent 2016 virus, virus shed was minimal, and the virus did not transmit to contacts, indicating that this virus was less adapted to mallards. Generally, when a gallinaceous poultry-adapted virus infects ducks, the virus retains the respiratory-tropic replication pattern (54, 55). The early Mexican H7N3 virus did not seem to be fully gallinaceous poultry adapted based on virus being shed by mallards by the CL route. In contrast, very low OP viral titers and no CL virus shed were found in mallards infected with A/CK/PB/16 virus, suggesting that long-term circulation in terrestrial poultry affected the ability of the virus to replicate in the respiratory and enteric tracts of mallards, and systemically. This decreased in virus fitness in mallards was also observed with HPAI viruses from previous outbreaks, such as the 1983 Pennsylvania H5N2 and the 1994 Mexican H5N2 HPAI outbreaks (21). The mallards inoculated with A/ML/MN/98 shed virus by both the OP and CL routes but primarily through the CL route, which is typical in wild ducks (21). The contact mallards in the group that received a medium dose of A/ML/MN/98 virus exhibited OP and CL virus shed with no detectable seroconversion, which also has been observed when ducks are exposed to low-LPAI-virus titers (56, 57). Antibody titers could have also been detected if serology was done beyond 10 dpi, but in our experience, most birds seroconvert by this time point. On the other hand, contact mallards in the group that received a medium dose of A/CK/JL/12 shed virus and seroconverted, even though the A/CK/JL/12-inoculated mallards shed an amount of virus comparable to that of mallards inoculated with the same dose of A/ML/MN/98 virus. It is assumed that, because HPAI virus replication is not limited to the respiratory and intestinal tracts, the systemic virus replication was able to induce seroconversion.
In order to confirm that the host species affects the replication of A/CK/JL/12 and A/CK/PB/16 viruses, we compared the growth kinetics of the two viruses in CEF and DEF. Consistent with the results of the experimental infection in mallards and chickens, we found that A/CK/JL/12 virus replicated at 1.6 times higher titers than did A/CK/PB/16 in DEF, but the two viruses replicated equally in CEF. These observed differences in virus replication in chicken and duck cells might be a reflection of the differences in amino acid homology in the polymerase proteins of the two viruses (PB2, 97.5%; PB1, 96.8%; PA, 97.1%). Several amino acid changes (PB2 R389K and V655A, PB1 M317I, and PA I323V) in the functional domains were observed between the polymerase genes of the A/CK/JL/12 and A/CK/PB/16 viruses, similar to what reported in other studies (31, 37, 58), thus contributing to the higher replication of A/CK/JL/12 virus in DEF. Changes in the other virus genes, including the HA (94.8% homology) and NA (97.2% homology), could contribute to virus receptor binding and cell release, which could also explain the differences observed in viral growth in CEF and DEF.
Although there are not many studies examining the adaptation of AI viruses in different bird species, 24 of the changes we found have been previously reported to be changes associated with chicken or duck virus adaptation or virulence (Table 6) (27–40). Among the 14 references, five studies analyzed sequences of cases of virus transmission from wild bird to domestic poultry and circulation in domestic poultry in vaccinated or unvaccinated flocks (27, 29, 32, 34, 38). Six studies examined pathogenicity in chickens and ducks by using reassortant viruses (30, 31, 35–37, 40). Two studies determined amino acid changes that increased polymerase activity in chicken cells and receptor binding properties in chicken and duck cells (33, 39). Another study showed that experimental passages of wild duck influenza virus in quail or turkey resulted in amino acid changes possibly associated with adaptation (28). In our studies, we found 29 and 13 amino acid differences in the HA and NA proteins, respectively, between A/CK/JL/12 and A/CK/PB/16 viruses. Three amino acid changes in the HA receptor binding domain (G133N, A151T, and Q201L) were consistent with mutations found in H7N3 LPAI viruses from poultry in Italy following heterologous vaccination (59). The addition and subtraction of N-glycosylation sites in the HA and NA genes could be a consequence of vaccine immune pressure and host-specific viral replication fitness (60–62). In the NA protein, six out of 13 amino acid changes were located in the 340-, 370-, and 400-loop regions, where secondary a sialic acid-binding site is adjacent to the active neuraminidase region (63, 64). The heterogenicity of the loop regions was also found to affect in vitro replication but was not necessarily associated with changes in viral replication in ducks (65). Two changes in NP, M105V and S377N, have been repeatedly found in several independent studies as being associated with increased pathogenicity in chickens and adaptation in chickens and turkeys (30, 33, 35, 37, 38). Some studies report that deletions in either NA or NS are associated with an increase in pathogenicity in avian species (28, 38, 66). However, no deletions were observed in the Mexican H7N3 viruses. As more studies similar to those mentioned above are conducted, a better understanding on the molecular markers of AI virus adaptation in different avian species will be achieved.
The phylogenetic analysis of the H7N3 HPAI viruses showed that after 2012, the viruses evolved into separate genetic clusters. Since there were not enough sequence data available for 2012 to 2014, it is assumed that the A/CK/PB/15 virus clustered independently as a transitional virus on the way of bifurcation. Both the A/CK/PB/15 and A/CK/PB/16 viruses have further evolved from the A/CK/JL/12 virus. The A/CK/PB/15 virus did not cluster with A/CK/PB/16 virus, and only seven of the changes observed in the A/CK/PB/16 virus compared to A/CK/JL/12 virus were also found in A/CK/PB/15 (PB2 K190R and V655A; PB1 M317I; HA A125T, S134P, and V258I; and NP S377N). In this study, we did not examine A/CK/PB/15 virus in mallards, so we do not know if it is less fit in mallards than A/CK/JL/12; however, of the 93 amino acid changes in A/CK/PB/15, 39 matched amino acid changes in the A/CK/PB/16 virus. For both viruses, it is not clear which changes are the result of normal accumulation of mutations during virus replication and vaccine immune pressure, or are related to changes in viral fitness in terrestrial poultry. Additional studies using reverse genetics would be required to elucidate which gene or amino acid substitutions contribute to the changes in host adaptation.
Historically, after the introduction into poultry, AI viruses, including HPAI viruses, rarely reinfect wild birds, with the exception of the H5Nx HPAI Gs/GD lineage viruses, which have undergone unique circumstances by circulating in both domestic chickens and ducks. Long-term surveillance in North America did not detect reverse transmission, i.e., from domestic to wild birds, for H7 viruses (both LPAI and HPAI viruses) (67), but most of the poultry outbreaks were short-lived, with the virus eradicated in a timely matter, the exception being the Mexico H7N3 HPAI virus. In this study, we showed that H7N3 HPAI virus after several years of circulation in poultry has changed to replicate and transmit less in mallards while maintaining similar pathogenicity in chickens. These results might explain why no spillover of H7N3 HPAI virus to migratory waterfowl has been reported and that the outbreaks have been confined to Mexico. The unusual extensive mutations with acquisition of possible glycosylation sites on HA suggests that the H7N3 HPAI virus has evolved to evade vaccine immunity (58). By comparing the mutations observed to ones previously reported in other studies, it is clear that host adaptation of AI viruses can be achieved by changes in more than one viral gene. Further studies are needed to continue elucidating the phenotypic markers associated with AI virus adaptation in different bird species.
MATERIALS AND METHODS
Viruses.
The following Mexican H7N3 HPAI viruses were used as challenge viruses: A/chicken/Jalisco/CPA1/2012 (A/CK/JL/12), A/chicken/Puebla/CPA-28973/2015 (A/CK/PB/15), and A/chicken/Puebla/CPA-03191-16-CENASA-95076/2016 (A/CK/PB/16) (courtesy of Joaquín B. Delgadillo, Igor Romero, and Mario Solís Hernández, Servicio Nacional de Sanidad, Inocuidad y Calidad Agroalimentaria [SENASICA], Mexico). An LPAI virus, H6N2 A/mallard/MN/232/1998 (A/ML/MN/98) from the Southeast Poultry Research Laboratory (SEPRL) repository, was also used for bird challenge. The viruses were propagated and titrated in specific-pathogen-free (SPF) embryonating chicken eggs (ECE) using standard methods (68). Stocks were diluted to the target dose with brain heart infusion (BHI) broth (Becton, Dickinson and Company, Sparks, MD). The experiments were performed in biosecurity level 3 enhanced facilities in accordance with procedures approved by the U.S. National Poultry Research Center (USNPRC) Institutional Biosecurity Committee.
Animals and housing.
Three-week-old SPF white Leghorn chickens (Gallus gallus domesticus) were obtained from the USNPRC in-house flocks. Mallard ducks (Anas platyrhynchos) were provided at 1 day of age by a commercial hatchery and held for 2 weeks at the USNPRC. Serum samples were collected from 15 chickens and 15 ducks to confirm that the birds were serologically negative to AI virus by blocking enzyme-linked immunosorbent assay (ELISA; FlockCheck avian influenza multiscreen antibody test; Idexx Laboratories, Westbrook, ME, USA). Each experimental group was housed in self-contained isolation units ventilated under negative pressure with inlet and exhaust high-efficiency particulate air (HEPA)-filtered ventilation. Feed and water were provided with ad libitum access. This study and associated procedures were reviewed and approved by the USNPRC Institutional Animal Care and Use Committee.
Experimental design and sampling.
Similar experiments were conducted with each bird species to evaluate the 50% bird infectious dose (BID50), pathogenicity, and transmissibility of the viruses. A/CK/JL/12, A/CK/PB/15, and A/CK/PB/16 were examined in chickens. A control group using a chicken-adapted virus was not included since our previous studies have shown efficient transmission of chicken-adapted viruses with the model used (51). Based on the results in chickens, A/CK/JL/12 and A/CK/PB/16 were chosen to be examined in mallards. A control group for a mallard-adapted virus, the A/ML/MN/98 LPAI virus, was included. To evaluate the BID50, birds were divided into three groups of five birds, and birds from each group were inoculated intranasally (by the choanal cleft) with 102, 104, or 106 50% egg infectious doses (EID50) in 0.1 ml of the respective viruses. The inoculum titers were verified by back titration in ECE. Sham birds were inoculated intranasally with 0.1 ml of sterile allantoic fluid diluted 1:300 in brain heart infusion broth. To evaluate the transmissibility of the viruses, 3 noninoculated hatch mates were added to each dose group at 1 day postinoculation (dpi). Oropharyngeal (OP) and cloacal (CL) swabs were collected from chickens at 1, 2, 3, and 6 dpi and from mallards at 2, 4, 7, 10 dpi; these schedules were based on our previous studies (47). Swabs were placed in 1.0 ml of BHI with penicillin (2,000 units/ml; Sigma-Aldrich), gentamicin (200 μg/ml; Sigma-Aldrich), and amphotericin B (5 μg/ml; Sigma-Aldrich), and stored at −80°C. At 10 dpi, survivors were bled to evaluate seroconversion and euthanized.
For each species, two additional birds challenged with the 106 EID50 of the viruses were euthanized and necropsied at 2 dpi (chickens) and 4 dpi (mallards) to evaluate gross and microscopic lesions, with the exception of A/CK/JL/12, which was previously characterized in mallards (21). The timing of the necropsies is based on our previous study (9). A full set of tissues were collected from each bird and fixed in 10% neutral-buffered formalin solution, paraffin embedded, sectioned, and stained with hematoxylin and eosin for histopathologic evaluation. Duplicate sections were stained by immunohistochemical (IHC) methods to determine influenza viral antigen distribution in tissues (69). Portions of the lung, heart, brain, muscle, and spleen were also collected and stored at −80°C for subsequent virus detection and quantification. Serum samples were collected from all surviving birds at the end of each experiment to evaluate infection status by determining antibody levels by hemagglutination inhibition (HI) assays using standard methods and homologous antigen (70). The virus infectious dose was calculated by the Reed-Muench method (71), using the criteria that birds were considered infected if they shed detectable levels of virus at any time and/or were positive for antibody at the end of the study.
Viral RNA quantification in swabs and tissues.
OP and CL swabs and tissues were processed for quantitative real-time reverse transcription-PCR (qRRT-PCR) to quantify viral RNA. RNA was extracted using MagMAX-96 AI/ND viral RNA isolation kit (Ambion, Inc.), following the manufacturer’s instructions. qRRT-PCRs targeting the influenza virus M gene (72) were conducted using the AgPath-ID one-step RT-PCR kit (Ambion, Austin, TX) and the ABI 7500 fast real-time PCR system (Applied Biosystems, Carlsbad, CA). Total RNA was extracted from tissues using TRIzol LS reagent (Invitrogen, Carlsbad, CA) and the Qiagen RNeasy minikit (Qiagen Corp., Valencia, CA). Virus titers in tissue samples were determined by weighing, homogenizing, and diluting tissues in BHI to a 10% (wt/vol) concentration. In order to further standardize the amount of nonspecific RNA from the tissue, the resulting RNA extracts were quantified using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific), following the manufacturer’s instructions, and accordingly diluted with phosphate-buffered saline to obtain 50 ng/μl. For virus quantification, standard curves were established with RNA extracted from dilutions of the same titrated stock of the challenge virus. This methodology has been used as a standard protocol among many published veterinary influenza studies and has demonstrated a high correlation between qRRT-PCR results and infectious titers (73). Results were reported as EID50/ml or EID50/g equivalents, and the lower limit of detection was set based on each standard curve. For statistical purposes, qRRT-PCR-negative samples were given a value of 0.1 log10 below the qRRT-PCR test limit of detection.
Viral growth kinetics and relative cellular virus RNA.
In order to compare in vitro growth of H7N3 HPAI viruses, 12-well plates were seeded with chicken embryo fibroblasts (CEF; ATCC CRL-12203) or duck embryo fibroblasts (DEF; ATCC CCL-141) and allowed to grow until confluent monolayers were obtained. One monolayer from each type of cells was trypsinized in 0.25% trypsin-EDTA and counted in a hemocytometer. Titrated viral stocks of A/CK/JL/12 and A/CK/PB/16 viruses were diluted and used to infect cells in triplicate at a multiplicity of infection of 0.001. Briefly, viruses were allowed to adsorb for 60 min, and then the virus inoculum was aspirated and cells were washed once with sterile PBS. Supernatants were collected at 12, 24, 48, and 72 h postinfection (hpi) and titrated by plaque assay on fresh Madin-Darby canine kidney (MDCK) monolayers (74).
For relative quantification of viral, complementary, and mRNA (vRNA, cRNA, and mRNA) of the AI viruses, virus-infected CEF and DEF were lysed at 12 and 24 hpi using the TRIzol reagent (Life Technologies, Carlsbad, CA). Total RNA extraction was performed using the RNeasy minikit (Qiagen), according to the manufacturer’s instructions. Strand-specific real-time RT-PCR was utilized to distinguish the three different forms of virus RNA, as described previously (75), with primer modifications. Briefly, cDNAs complementary to the three types of influenza virus RNA were synthesized using SuperScript IV reverse transcriptase with saturated trehalose (Sigma-Aldrich). As a trigger of viral RNA polymerase switching from a transcriptase to a replicase (76), real-time PCR targeting the NP gene of influenza was performed with PowerUp SYBR Green mastermix (Thermo Fisher Scientific). The replicating virus RNA was normalized with a housekeeping gene expression (glyceraldehyde 3-phosphate dehydrogenase [GAPDH]) (77, 78). The primer sets used are found in Table 7. The fold increase was calculated by comparing virus RNA detection between 12 hpi and 24 hpi for each RNA type.
TABLE 7.
Reverse transcription and real-time PCR primer sets used for vRNA, cRNA, and mRNA quantification
| Target | Purpose | Primer name | Sequence (5′–3′) |
|---|---|---|---|
| vRNA | Reverse transcription | vRNAtag_CPAseg5_675F | GGCCGTCATGGTGGCGAATAAATGGACGGAGGACAAGAATTGC |
| Real-time PCR | vRNAtag | GGCCGTCATGGTGGCGAAT | |
| CPAseg5_822R | CTCAGAATGAGAGCAGACCGTGCA | ||
| cRNA | Reverse transcription | cRNAtag_CPAseg5_1541R | GCTAGCTTCAGCTAGGCATCAGTAGAAACAAGGGTATTTTTCTTT |
| Real-time PCR | cRNAtag | GCTAGCTTCAGCTAGGCATC | |
| CPAseg5_1466F | CGATCGTGCCTTCCTTTG | ||
| mRNA | Reverse transcription | mRNAtag_CPAseg5_dTR | CCAGATCGTTCGAGTCGTTTTTTTTTTTTTTTTTCTTTAATTGTC |
| Real-time PCR | mRNAtag | CCAGATCGTTCGAGTCGT | |
| CPAseg5_1466F | CGATCGTGCCTTCCTTTG | ||
| Chicken GAPDH | Normalization | CGAPD_F | CCTCTCTGGCAAAGTCCAAG |
| CGAPDH_R | CATCTGCCCATTTGATGTT | ||
| Duck GAPDH | Normalization | DGAPDH_F | ATGTTCGTGATGGGTGTGAA |
| DGAPDH_R | CTGTCTTCGTGTGTGGCTGT |
Statistical analysis.
Statistical differences in the mean hemagglutinin inhibition (HI) titers and mean virus shedding titers were analyzed using the Tukey one-way analysis of variance (ANOVA) with the GraphPad Prism 7 software. Viral growth kinetics and virus RNA quantification were determined using two-way ANOVA with a Bonferroni multiple-comparison test. A P value of <0.05 was considered to be significant.
Sequence analysis.
Full-length-genome sequencing of the 2012, 2015, and 2016 H7N3 HPAI viruses was conducted using next-generation sequencing. All eight DNA segments were synthesized and amplified by reverse transcription-PCR (RT-PCR) using the OneTaq one-step RT-PCR kit (New England BioLabs) (79). The Nextera XT DNA sample preparation kit (Illumina, USA) and 5 μl of 0.2 ng/μl of double-stranded (ds) cDNA were used to generate multiplexed paired-end sequencing libraries, according to the manufacturer’s instructions. The library pool was loaded into the flow cell of the 500-cycle MiSeq reagent kit v2 (Illumina). The barcoded multiplexed library sequencing (2 × 250 bp) was performed on a MiSeq platform (Illumina). The sequences were reconstituted by de novo and directed assembly using the Geneious 10.0.9 software. All eight sequences from A/CK/JL/12 were identical to the sequences from GenBank (accession numbers JX317626, JX397993, and JX465631 to JX465636). All the Mexican H7N3 HPAI and the ancestral wild bird sequences with high homology to initial H7N3 (A/CK/JL/12) were collected from GenBank and included to construct the phylogenetic trees. The maximum likelihood tree of each gene was estimated by RAxML (80) using the general time-reversible model of nucleotide substitution. Bootstrap support values were generated by using 1,000 rapid bootstrap replicates. Bootstrap values of >70% are shown at the branch nodes. Genotypes were designated with a bootstrap value of >70%. In order to identify genetic changes associated with the changes observed in virus adaptation, complete coding regions of each segment of the 2012 (A/CK/JL/12) and 2016 (A/CK/PB/16) H7N3 viruses were aligned and used for amino acid change identification. The coding sequences discriminating single-nucleotide polymorphisms (SNPs) were classified as either nonsynonymous or synonymous based on whether or not they correspond to differences in encoded amino acid sequences. Amino acid changes were compared to changes found in other studies examining chicken or duck adaptation or virulence of AI viruses. For changes in the HA gene, alignments were conducted to compare with other HA subtypes following the recommended numbering scheme (81).
Molecular characterization of the hemagglutinin, neuraminidase, and polymerase proteins: residues and protein structure.
The molecular characterization of A/CK/JL/12 and A/CK/PB/16 proteins was performed using different methodologies. The A/CK/JL/12 and A/CK/PB/16 protein sequences were used for alignment and residue analysis in the Lasergene 12 using Clustal W MegAlign software (DNAStar, Madison, WI). The potential N-glycosylation sites for HA and NA were predicted using the NetNGlyc server 1.0 (82). The A/CK/PB/16 HA (PDB ID 3M5G), NA (PDB ID 4HZV), and polymerase fragment (endonuclease and PA-Cter-PB1-Nter [PDB IDs 2ZN1 and 3CM8], PB1-Cter/PB2-Nter [PDB ID 3A1G], PB2-cap, and 627-NLS domains [PDB ID 2VY6] structures were modeled using the known structures of influenza A in the SWISS-MODEL server (28–30). The three-dimensional (3D) molecular structures were visualized using the PyMOL molecular graphics system (version 2.0; Schrödinger, LLC).
Data availability.
The A/CK/PB/15 (accession numbers MK027368 to MK027375) and A/CK/PB/16 (accession numbers MK027376 to MK027383) sequences have been deposited in GenBank.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate the technical assistance provided by Scott Lee and Nikolai Lee and the animal care provided by Roger Brock, Keith Crawford, and Gerald Damron in conducting these studies.
This research was supported by the ARS Project 6040-32000-066-00D, the Center of Research in Influenza Pathogenesis (CRIS), an NIAID-funded Center of Excellence in Influenza Research and Surveillance (CEIRS) contract (HHSN272201400008C), and the ARS/APHIS interagency agreement 60-6040-6-007.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the USDA or the NIH. The mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00543-19.
REFERENCES
- 1.Alexander DJ. 2000. A review of avian influenza in different bird species. Vet Microbiol 74:3–13. doi: 10.1016/S0378-1135(00)00160-7. [DOI] [PubMed] [Google Scholar]
- 2.Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus AD, Fouchier RA. 2006. Global patterns of influenza a virus in wild birds. Science 312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 3.Spackman E. 2009. The ecology of avian influenza virus in wild birds: what does this mean for poultry? Poult Sci 88:847–850. doi: 10.3382/ps.2008-00336. [DOI] [PubMed] [Google Scholar]
- 4.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pantin-Jackwood MJ, Swayne DE. 2009. Pathogenesis and pathobiology of avian influenza virus infection in birds. Rev Sci Tech 28:113–136. doi: 10.20506/rst.28.1.1869. [DOI] [PubMed] [Google Scholar]
- 6.Swayne DE, Suarez DL. 2000. Highly pathogenic avian influenza. Rev Sci Tech 19:463–482. doi: 10.20506/rst.19.2.1230. [DOI] [PubMed] [Google Scholar]
- 7.Swayne DE, Pantin-Jackwood MJ. 2008. Pathobiology of avian influenza virus infections in birds and mammals, p 87–122. In Swayne DE. (ed), Avian influenza. Blackwell Publishing, Ames, IA. [Google Scholar]
- 8.Lu L, Lycett SJ, Leigh Brown AJ. 2014. Determining the phylogenetic and phylogeographic origin of highly pathogenic avian influenza (H7N3) in Mexico. PLoS One 9:e107330. doi: 10.1371/journal.pone.0107330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapczynski DR, Pantin-Jackwood M, Guzman SG, Ricardez Y, Spackman E, Bertran K, Suarez DL, Swayne DE. 2013. Characterization of the 2012 highly pathogenic avian influenza H7N3 virus isolated from poultry in an outbreak in Mexico: pathobiology and vaccine protection. J Virol 87:9086–9096. doi: 10.1128/JVI.00666-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertran K, Sá e Silva M, Pantin-Jackwood MJ, Swayne DE. 2013. Protection against H7N3 high pathogenicity avian influenza in chickens immunized with a recombinant fowlpox and an inactivated avian influenza vaccines. Vaccine 31:3572–3576. doi: 10.1016/j.vaccine.2013.05.039. [DOI] [PubMed] [Google Scholar]
- 11.Röhm C, Horimoto T, Kawaoka Y, Suss J, Webster RG. 1995. Do hemagglutinin genes of highly pathogenic avian influenza viruses constitute unique phylogenetic lineages? Virology 209:664–670. doi: 10.1006/viro.1995.1301. [DOI] [PubMed] [Google Scholar]
- 12.Alexander DJ, Allan WH, Parsons DG, Parsons G. 1978. The pathogenicity of four avian influenza viruses for fowls, turkeys and ducks. Res Vet Sci 24:242–247. doi: 10.1016/S0034-5288(18)33080-7. [DOI] [PubMed] [Google Scholar]
- 13.Capua I, Mutinelli F. 2001. Mortality in Muscovy ducks (Cairina moschata) and domestic geese (Anser anser var. domestica) associated with natural infection with a highly pathogenic avian influenza virus of H7N1 subtype. Avian Pathol 30:179–183. doi: 10.1080/03079450120044597. [DOI] [PubMed] [Google Scholar]
- 14.Alexander DJ, Parsons G, Manvell RJ. 1986. Experimental assesment of the pathogenicity of eight avian influenza A viruses of H5 subtype for chickens, turkeys, ducks and quail. Avian Pathol 15:647–662. doi: 10.1080/03079458608436328. [DOI] [PubMed] [Google Scholar]
- 15.Wood GW, Parsons G, Alexander DJ. 1995. Replication of influenza A viruses of high and low pathogenicity for chickens at different sites in chickens and ducks following intranasal inoculation. Avian Pathol 24:545–551. doi: 10.1080/03079459508419093. [DOI] [PubMed] [Google Scholar]
- 16.Westbury HA, Turner AJ, Kovesdy L. 1979. The pathogenicity of three Auatralian fowl plague viruses for chickens, turkeys and ducks. Vet Microbiol 4:223–234. doi: 10.1016/0378-1135(79)90058-0. [DOI] [Google Scholar]
- 17.Wood JM, Webster RG, Nettles VF. 1985. Host range of A/Chicken/Pennsylvania/83 (H5N2) influenza virus. Avian Dis 29:198–207. doi: 10.2307/1590708. [DOI] [PubMed] [Google Scholar]
- 18.Slemons RD, Easterday BC. 1972. Host response differences among 5 avian species to an influenzavirus–A-turkey-Ontario-7732-66 (Hav5N?). Bull World Health Organ 47:521–525. [PMC free article] [PubMed] [Google Scholar]
- 19.Sá e Silva M, Mathieu-Benson C, Kwon YK, Pantin-Jackwood M, Swayne DE. 2011. Experimental infection with low and high pathogenicity H7N3 Chilean avian influenza viruses in Chiloe wigeon (Anas sibilatrix) and cinnamon teal (Anas cyanoptera). Avian Dis 55:459–461. doi: 10.1637/9665-012011-Reg.1. [DOI] [PubMed] [Google Scholar]
- 20.van der Goot JA, van Boven M, Koch G, de Jong MC. 2007. Variable effect of vaccination against highly pathogenic avian influenza (H7N7) virus on disease and transmission in pheasants and teals. Vaccine 25:8318–8325. doi: 10.1016/j.vaccine.2007.09.048. [DOI] [PubMed] [Google Scholar]
- 21.Pantin-Jackwood MJ, Costa-Hurtado M, Shepherd E, DeJesus E, Smith D, Spackman E, Kapczynski DR, Suarez DL, Stallknecht D, Swayne DE. 2016. Pathogenicity and transmission of H5 and H7 highly pathogenic avian influenza viruses in mallards. J Virol 90:9967–9982. doi: 10.1128/JVI.01165-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navarro-López R, Vazquez-Mendoza LF, Villarreal Chavez CL, Casaubon Y, Marquez Ruiz MA. 2014. Highly pathogenic avian influenza A/H7N3 in great-tailed grackles (Quiscalus mexicanus) in the Altos de Jalisco region of Mexico. JMM Case Rep 1:e001461. doi: 10.1099/jmmcr.0.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurer-Stroh S, Lee RT, Gunalan V, Eisenhaber F. 2013. The highly pathogenic H7N3 avian influenza strain from July 2012 in Mexico acquired an extended cleavage site through recombination with host 28S rRNA. Virol J 10:139. doi: 10.1186/1743-422X-10-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pflug A, Lukarska M, Resa-Infante P, Reich S, Cusack S. 2017. Structural insights into RNA synthesis by the influenza virus transcription-replication machine. Virus Res 234:103–117. doi: 10.1016/j.virusres.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 25.Boivin S, Cusack S, Ruigrok RW, Hart DJ. 2010. Influenza A virus polymerase: structural insights into replication and host adaptation mechanisms. J Biol Chem 285:28411–28417. doi: 10.1074/jbc.R110.117531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sriwilaijaroen N, Suzuki Y. 2012. Molecular basis of the structure and function of H1 hemagglutinin of influenza virus. Proc Jpn Acad Ser B Phys Biol Sci 88:226–249. doi: 10.2183/pjab.88.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lebarbenchon C, Stallknecht DE. 2011. Host shifts and molecular evolution of H7 avian influenza virus hemagglutinin. Virol J 8:328. doi: 10.1186/1743-422X-8-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giannecchini S, Clausi V, Di Trani L, Falcone E, Terregino C, Toffan A, Cilloni F, Matrosovich M, Gambaryan AS, Bovin NV, Delogu M, Capua I, Donatelli I, Azzi A. 2010. Molecular adaptation of an H7N3 wild duck influenza virus following experimental multiple passages in quail and turkey. Virology 408:167–173. doi: 10.1016/j.virol.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Arafa A, Suarez D, Kholosy SG, Hassan MK, Nasef S, Selim A, Dauphin G, Kim M, Yilma J, Swayne D, Aly MM. 2012. Evolution of highly pathogenic avian influenza H5N1 viruses in Egypt indicating progressive adaptation. Arch Virol 157:1931–1947. doi: 10.1007/s00705-012-1385-9. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki K, Okada H, Itoh T, Tada T, Mase M, Nakamura K, Kubo M, Tsukamoto K. 2009. Association of increased pathogenicity of Asian H5N1 highly pathogenic avian influenza viruses in chickens with highly efficient viral replication accompanied by early destruction of innate immune responses. J Virol 83:7475–7486. doi: 10.1128/JVI.01434-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wasilenko JL, Lee CW, Sarmento L, Spackman E, Kapczynski DR, Suarez DL, Pantin-Jackwood MJ. 2008. NP, PB1, and PB2 viral genes contribute to altered replication of H5N1 avian influenza viruses in chickens. J Virol 82:4544–4553. doi: 10.1128/JVI.02642-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horimoto T, Rivera E, Pearson J, Senne D, Krauss S, Kawaoka Y, Webster RG. 1995. Origin and molecular changes associated with emergence of a highly pathogenic H5N2 influenza virus in Mexico. Virology 213:223–230. doi: 10.1006/viro.1995.1562. [DOI] [PubMed] [Google Scholar]
- 33.Tada T, Suzuki K, Sakurai Y, Kubo M, Okada H, Itoh T, Tsukamoto K. 2011. NP body domain and PB2 contribute to increased virulence of H5N1 highly pathogenic avian influenza viruses in chickens. J Virol 85:1834–1846. doi: 10.1128/JVI.01648-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbas MA, Spackman E, Swayne DE, Ahmed Z, Sarmento L, Siddique N, Naeem K, Hameed A, Rehmani S. 2010. Sequence and phylogenetic analysis of H7N3 avian influenza viruses isolated from poultry in Pakistan 1995–2004. Virol J 7:137. doi: 10.1186/1743-422X-7-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bogs J, Veits J, Gohrbandt S, Hundt J, Stech O, Breithaupt A, Teifke JP, Mettenleiter TC, Stech J. 2010. Highly pathogenic H5N1 influenza viruses carry virulence determinants beyond the polybasic hemagglutinin cleavage site. PLoS One 5:e11826. doi: 10.1371/journal.pone.0011826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song J, Feng H, Xu J, Zhao D, Shi J, Li Y, Deng G, Jiang Y, Li X, Zhu P, Guan Y, Bu Z, Kawaoka Y, Chen H. 2011. The PA protein directly contributes to the virulence of H5N1 avian influenza viruses in domestic ducks. J Virol 85:2180–2188. doi: 10.1128/JVI.01975-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kajihara M, Sakoda Y, Soda K, Minari K, Okamatsu M, Takada A, Kida H. 2013. The PB2, PA, HA, NP, and NS genes of a highly pathogenic avian influenza virus A/whooper swan/Mongolia/3/2005 (H5N1) are responsible for pathogenicity in ducks. Virol J 10:45. doi: 10.1186/1743-422X-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campitelli L, Mogavero E, De Marco MA, Delogu M, Puzelli S, Frezza F, Facchini M, Chiapponi C, Foni E, Cordioli P, Webby R, Barigazzi G, Webster RG, Donatelli I. 2004. Interspecies transmission of an H7N3 influenza virus from wild birds to intensively reared domestic poultry in Italy. Virology 323:24–36. doi: 10.1016/j.virol.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 39.Guo H, de Vries E, McBride R, Dekkers J, Peng W, Bouwman KM, Nycholat C, Verheije MH, Paulson JC, van Kuppeveld FJ, de Haan CA. 2017. Highly pathogenic influenza A(H5Nx) viruses with altered H5 receptor-binding specificity. Emerg Infect Dis 23:220–231. doi: 10.3201/eid2302.161072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarmento L, Wasilenko J, Pantin-Jackwood M. 2010. The effects of NS gene exchange on the pathogenicity of H5N1 HPAI viruses in ducks. Avian Dis 54:532–537. doi: 10.1637/8917-050409-Reg.1. [DOI] [PubMed] [Google Scholar]
- 41.Krauss S, Stucker KM, Schobel SA, Danner A, Friedman K, Knowles JP, Kayali G, Niles LJ, Dey AD, Raven G, Pryor P, Lin X, Das SR, Stockwell TB, Wentworth DE, Webster RG. 2015. Long-term surveillance of H7 influenza viruses in American wild aquatic birds: are the H7N3 influenza viruses in wild birds the precursors of highly pathogenic strains in domestic poultry? Emerg Microbes Infect 4:e35. doi: 10.1038/emi.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suarez DL, Senne DA, Banks J, Brown IH, Essen SC, Lee CW, Manvell RJ, Mathieu-Benson C, Moreno V, Pedersen JC, Panigrahy B, Rojas H, Spackman E, Alexander DJ. 2004. Recombination resulting in virulence shift in avian influenza outbreak, Chile. Emerg Infect Dis 10:693–699. doi: 10.3201/eid1004.030396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Max V, Herrera J, Moreira R, Rojas H. 2007. Avian influenza in Chile: a successful experience. Avian Dis 51:363–365. doi: 10.1637/7631-042806R1.1. [DOI] [PubMed] [Google Scholar]
- 44.Pasick J, Handel K, Robinson J, Copps J, Ridd D, Hills K, Kehler H, Cottam-Birt C, Neufeld J, Berhane Y, Czub S. 2005. Intersegmental recombination between the haemagglutinin and matrix genes was responsible for the emergence of a highly pathogenic H7N3 avian influenza virus in British Columbia. J Gen Virol 86:727–731. doi: 10.1099/vir.0.80478-0. [DOI] [PubMed] [Google Scholar]
- 45.Pasick J, Berhane Y, Hooper-McGrevy K. 2009. Avian influenza: the Canadian experience. Rev Sci Tech 28:349–358. doi: 10.20506/rst.28.1.1875. [DOI] [PubMed] [Google Scholar]
- 46.Swayne DE, Slemons RD. 2008. Using mean infectious dose of wild duck- and poultry-origin high- and low-pathogenicity avian infleunza viruses as one measure of infectivity and adaptation to poultry. Avian Dis 52:455–460. doi: 10.1637/8229-012508-Reg.1. [DOI] [PubMed] [Google Scholar]
- 47.DeJesus E, Costa-Hurtado M, Smith D, Lee DH, Spackman E, Kapczynski DR, Torchetti MK, Killian ML, Suarez DL, Swayne DE, Pantin-Jackwood MJ. 2016. Changes in adaptation of H5N2 highly pathogenic avian influenza H5 clade 2.3.4.4 viruses in chickens and mallards. Virology 499:52–64. doi: 10.1016/j.virol.2016.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bertran K, Swayne DE, Pantin-Jackwood MJ, Kapczynski DR, Spackman E, Suarez DL. 2016. Lack of chicken adaptation of newly emergent Eurasian H5N8 and reassortant H5N2 high pathogenicity avian influenza viruses in the U.S. is consistent with restricted poultry outbreaks in the Pacific flyway during 2014–2015. Virology 494:190–197. doi: 10.1016/j.virol.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 49.Bertran K, Lee DH, Criado MF, Smith D, Swayne DE, Pantin-Jackwood MJ. 2018. Pathobiology of Tennessee 2017 H7N9 low and high pathogenicity avian influenza viruses in commercial broiler breeders and specific pathogen free layer chickens. Vet Res 49:82. doi: 10.1186/s13567-018-0576-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pantin-Jackwood MJ, Stephens CB, Bertran K, Swayne DE, Spackman E. 2017. The pathogenesis of H7N8 low and highly pathogenic avian influenza viruses from the United States 2016 outbreak in chickens, turkeys and mallards. PLoS One 12:e0177265. doi: 10.1371/journal.pone.0177265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spackman E, Pantin-Jackwood M, Swayne DE, Suarez DL, Kapczynski DR. 2015. Impact of route of exposure and challenge dose on the pathogenesis of H7N9 low pathogenicity avian influenza virus in chickens. Virology 477:72–81. doi: 10.1016/j.virol.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan R, Cui J, Zhang S, Cao L, Liu X, Kang Y, Song Y, Gong L, Jiao P, Liao M. 2014. Pathogenicity and transmission of H5N1 avian influenza viruses in different birds. Vet Microbiol 168:50–59. doi: 10.1016/j.vetmic.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 53.van der Goot JA, de Jong MC, Koch G, Van Boven M. 2003. Comparison of the transmission characteristics of low and high pathogenicity avian influenza A virus (H5N2). Epidemiol Infect 131:1003–1013. doi: 10.1017/S0950268803001067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spackman E, Gelb J Jr, Preskenis LA, Ladman BS, Pope CR, Pantin-Jackwood MJ, McKinley ET. 2010. The pathogenesis of low pathogenicity H7 avian influenza viruses in chickens, ducks and turkeys. Virol J 7:331. doi: 10.1186/1743-422X-7-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin Y, Yu D, Ren H, Yin Z, Huang Z, Hu M, Li B, Zhou W, Yue J, Liang L. 2014. Phylogeography of avian influenza A H9N2 in China. BMC Genomics 15:1110. doi: 10.1186/1471-2164-15-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kida H, Yanagawa R, Matsuoka Y. 1980. Duck influenza lacking evidence of disease signs and immune response. Infect Immun 30:547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferreira HL, Vangeluwe D, Van Borm S, Poncin O, Dumont N, Ozhelvaci O, Munir M, van den Berg T, Lambrecht B. 2015. Differential viral fitness between H1N1 and H3N8 avian influenza viruses isolated from mallards (Anas platyrhynchos). Avian Dis 59:498–507. doi: 10.1637/11074-033015-Reg. [DOI] [PubMed] [Google Scholar]
- 58.Criado MF, Bertran K, Lee D-H, Killmaster L, Stephens C, Spackman E, Sa e Silva M, Atkins E, Mebatsion T, Widener J, Pritchard N, King H, Swayne DE. 2019. Efficacy of novel recombinant fowlpox vaccine against recent Mexican H7N3 highly pathogenic avian influenza virus. Vaccine 37:2232–2243. doi: 10.1016/j.vaccine.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 59.Beato MS, Xu Y, Long LP, Capua I, Wan XF. 2014. Antigenic and genetic evolution of low-pathogenicity avian influenza viruses of subtype H7N3 following heterologous vaccination. Clin Vaccine Immunol 21:603–612. doi: 10.1128/CVI.00647-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wagner R, Matrosovich M, Klenk HD. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol 12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 61.Hervé PL, Lorin V, Jouvion G, Da Costa B, Escriou N. 2015. Addition of N-glycosylation sites on the globular head of the H5 hemagglutinin induces the escape of highly pathogenic avian influenza A H5N1 viruses from vaccine-induced immunity. Virology 486:134–145. doi: 10.1016/j.virol.2015.08.033. [DOI] [PubMed] [Google Scholar]
- 62.Alvarado-Facundo E, Vassell R, Schmeisser F, Weir JP, Weiss CD, Wang W. 2016. Glycosylation of residue 141 of subtype H7 influenza A hemagglutinin (HA) affects HA-pseudovirus infectivity and sensitivity to site A neutralizing antibodies. PLoS One 11:e0149149. doi: 10.1371/journal.pone.0149149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yen HL, Hoffmann E, Taylor G, Scholtissek C, Monto AS, Webster RG, Govorkova EA. 2006. Importance of neuraminidase active-site residues to the neuraminidase inhibitor resistance of influenza viruses. J Virol 80:8787–8795. doi: 10.1128/JVI.00477-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shtyrya YA, Mochalova LV, Bovin NV. 2009. Influenza virus neuraminidase: structure and function. Acta Naturae 1:26–32. [PMC free article] [PubMed] [Google Scholar]
- 65.Kobasa D, Rodgers ME, Wells K, Kawaoka Y. 1997. Neuraminidase hemadsorption activity, conserved in avian influenza A viruses, does not influence viral replication in ducks. J Virol 71:6706–6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y, Chen S, Zhang X, Fu Q, Zhang Z, Shi S, Zhu Y, Gu M, Peng D, Liu X. 2014. A 20-amino-acid deletion in the neuraminidase stalk and a five-amino-acid deletion in the NS1 protein both contribute to the pathogenicity of H5N1 avian influenza viruses in mallard ducks. PLoS One 9:e95539. doi: 10.1371/journal.pone.0095539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dugan VG, Chen R, Spiro DJ, Sengamalay N, Zaborsky J, Ghedin E, Nolting J, Swayne DE, Runstadler JA, Happ GM, Senne DA, Wang R, Slemons RD, Holmes EC, Taubenberger JK. 2008. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog 4:e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spackman E, Killian ML. 2014. Avian influenza virus isolation, propagation, and titration in embryonated chicken eggs. Methods Mol Biol 1161:125–140. doi: 10.1007/978-1-4939-0758-8_12. [DOI] [PubMed] [Google Scholar]
- 69.Pantin-Jackwood MJ. 2014. Immunohistochemical staining of influenza virus in tissues. Methods Mol Biol 1161:51–58. doi: 10.1007/978-1-4939-0758-8_5. [DOI] [PubMed] [Google Scholar]
- 70.OIE. 2015. Avian influenza (infection with avian influenza viruses), chapter 2.3.4, p 821–843. In Manual for diagnostic tests and vaccines for terrestrial animals. World Organisation for Animal Health, Paris, France: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.03.04_AI.pdf. [Google Scholar]
- 71.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Hyg 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 72.Spackman E, Senne DA, Myers TJ, Bulaga LL, Garber LP, Perdue ML, Lohman K, Daum LT, Suarez DL. 2002. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J Clin Microbiol 40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee CW, Suarez DL. 2004. Application of real-time RT-PCR for the quantitation and competitive replication study of H5 and H7 subtype avian influenza virus. J Virol Methods 119:151–158. doi: 10.1016/j.jviromet.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 74.Gaush CR, Smith TF. 1968. Replication and plaque assay of influenza virus in an established line of canine kidney cells. Appl Microbiol 16:588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kawakami E, Watanabe T, Fujii K, Goto H, Watanabe S, Noda T, Kawaoka Y. 2011. Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA. J Virol Methods 173:1–6. doi: 10.1016/j.jviromet.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Portela A, Digard P. 2002. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J Gen Virol 83:723–734. doi: 10.1099/0022-1317-83-4-723. [DOI] [PubMed] [Google Scholar]
- 77.Adams SC, Xing Z, Li J, Cardona CJ. 2009. Immune-related gene expression in response to H11N9 low pathogenic avian influenza virus infection in chicken and Pekin duck peripheral blood mononuclear cells. Mol Immunol 46:1744–1749. doi: 10.1016/j.molimm.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 78.Cui Z, Hu J, He L, Li Q, Gu M, Wang X, Hu S, Liu H, Liu W, Liu X, Liu X. 2014. Differential immune response of mallard duck peripheral blood mononuclear cells to two highly pathogenic avian influenza H5N1 viruses with distinct pathogenicity in mallard ducks. Arch Virol 159:339–343. doi: 10.1007/s00705-013-1820-6. [DOI] [PubMed] [Google Scholar]
- 79.Chrzastek K, Lee DH, Smith D, Sharma P, Suarez DL, Pantin-Jackwood M, Kapczynski DR. 2017. Use of sequence-independent, single-primer-amplification (SISPA) for rapid detection, identification, and characterization of avian RNA viruses. Virology 509:159–166. doi: 10.1016/j.virol.2017.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Burke DF, Smith DJ. 2014. A recommended numbering scheme for influenza A HA subtypes. PLoS One 9:e112302. doi: 10.1371/journal.pone.0112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gupta R, Jung E, Brunak S. 2004. Prediction of N-glycosylation sites in human proteins. http://www.cbs.dtu.dk/services/NetNGlyc/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The A/CK/PB/15 (accession numbers MK027368 to MK027375) and A/CK/PB/16 (accession numbers MK027376 to MK027383) sequences have been deposited in GenBank.