Baculoviruses are virulent pathogens of a number of important insect pest species. In the host Trichoplusia ni, infection begins in the midgut when infectious virions of the occlusion-derived virus (ODV) phenotype enter and subsequently replicate in cells of the midgut epithelium. A second virion phenotype (budded virus [BV]) is produced there, and BV mediates systemic infection of the animal. Most prior detailed studies of baculovirus infections have focused on BV infections of cultured cells. In this study, we examined the transcriptional responses of the T. ni midgut to infection by ODV of the baculovirus AcMNPV and identified a variety of host genes that respond dramatically to viral infection. Understanding the transcriptional responses of the host midgut to viral infection is critically important for understanding the biphasic infection in the animal as a whole.
KEYWORDS: AcMNPV, baculovirus, Trichoplusia ni, differential expression, immune genes, midgut, transcriptome
ABSTRACT
Baculoviruses are large double-stranded DNA viruses that are virulent pathogens of certain insect species. In a natural host, Trichoplusia ni, infection by the model baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) begins when the occluded form of the virus disassembles in the midgut and virions infect midgut epithelial cells to establish the primary phase of the infection. To better understand the primary phase of the AcMNPV infection cycle, newly molted 5th-instar T. ni larvae were orally infected with AcMNPV occlusion bodies and the transcriptional responses of the T. ni midgut were analyzed at various times from 0 to 72 h postinfection, using transcriptome sequencing analysis and a T. ni reference genome. The numbers of differentially expressed host genes increased as the infection progressed, and we identified a total of 3,372 differentially expressed T. ni transcripts in the AcMNPV-infected midgut. Genes encoding orthologs of HMG176, atlastin, and CPH43 were among the most dramatically upregulated in response to AcMNPV infection. A number of cytochrome P450 genes were downregulated in response to infection. We also identified the effects of AcMNPV infection on a large variety of genes associated with innate immunity. This analysis provides an abundance of new and detailed information on host responses to baculovirus infection during the primary phase of the infection in the midgut and will be important for understanding how baculoviruses establish productive infections in the organism.
IMPORTANCE Baculoviruses are virulent pathogens of a number of important insect pest species. In the host Trichoplusia ni, infection begins in the midgut when infectious virions of the occlusion-derived virus (ODV) phenotype enter and subsequently replicate in cells of the midgut epithelium. A second virion phenotype (budded virus [BV]) is produced there, and BV mediates systemic infection of the animal. Most prior detailed studies of baculovirus infections have focused on BV infections of cultured cells. In this study, we examined the transcriptional responses of the T. ni midgut to infection by ODV of the baculovirus AcMNPV and identified a variety of host genes that respond dramatically to viral infection. Understanding the transcriptional responses of the host midgut to viral infection is critically important for understanding the biphasic infection in the animal as a whole.
INTRODUCTION
Baculoviruses are arthropod-specific viruses with circular double-stranded DNA genomes ranging from approximately 80 to 180 kbp that are packaged in rod-shaped nucleocapsids (1–3). Baculoviruses are used as biological control agents for agriculturally important insect pests in the order Lepidoptera (4). In general, baculovirus species have very narrow host ranges, often limited to one or a few closely related insect species (5). Autographa californica multiple nucleopolyhedrovirus (AcMNPV) is the type species of the family Baculoviridae and is the most intensively studied model of baculovirus biology. Unlike many other baculoviruses, AcMNPV has a wide host range, infecting at least 33 species of lepidopteran larvae in 10 families (1, 6). AcMNPV is highly pathogenic in the early-instar larvae of some species, yet it is less pathogenic in other species and when infecting later instars (7, 8).
Baculoviruses, such as AcMNPV, that have been studied extensively produce two virion phenotypes: the occlusion-derived virus (ODV) and the budded virus (BV). ODV and BV are physically and functionally distinct, with BV being produced earlier by nucleocapsid budding from the surface of infected cells and ODV being produced later by nucleocapsid envelopment within the nucleus. ODV, which is subsequently occluded in a crystallized protein occlusion body (OB), is released into the environment and is orally infectious. When consumed by a susceptible host, OBs disassemble in the larval midgut, releasing ODV that subsequently enter midgut epithelial cells and initiate the primary phase of the infection in the animal. During the primary phase of the infection in the midgut, BV bud from the basal surfaces of midgut cells and BV subsequently infect most other tissues, initiating the secondary phase of the infection (reviewed in references 1 and 2). The majority of detailed studies of baculovirus infection and host responses have focused on infections of cultured cells by BV (representing the secondary phase of infection), yet the primary phase of the infection is critically important to the success of infection in the organism. In a prior study of AcMNPV infection of Heliothis virescens and Helicoverpa zea (representing permissive and semipermissive hosts, respectively), it was observed that even though susceptibility to mortal infection differed by >1,000-fold, detection of viral infection in the midgut appeared to be similar (9). Thus, the differences in host susceptibility appear to occur at a stage following viral entry into midgut cells, and the responses of the midgut cells to infection may be critical for the success of the primary infection and subsequent systemic infection. Differences in midgut intracellular responses to viral infection may lead to differences in the production of infectious progeny virions or differences in signaling or immune activation at the organismal level. Although midgut responses to AcMNPV infection are clearly important and likely determine the outcome of infection in the host, little detail is known about midgut responses, even in highly permissive host insects, such as Trichoplusia ni (the cabbage looper). To understand how midgut cells react to AcMNPV infection, we used a high-throughput sequencing approach to measure the effects of oral infection by AcMNPV ODV on global host gene expression in the Trichoplusia ni larval midgut. We previously performed a similar study of global host gene expression using a T. ni cell line (Tnms42) that was synchronously infected with AcMNPV BV (10, 11). BV infection of cultured Tnms42 cells is thought to most closely represent a model of events in the secondary phase of infection in the animal. Initiation of ODV infection of the midgut epithelium is substantially less efficient than synchronous infection of cultured cells by BV in the laboratory. High doses of ODV result in infection of only a subset of midgut epithelial cells in 5th-instar larvae. However, midgut cells that are infected by ODV may receive a relatively high dose of the virus since ODV virions contain multiple nucleocapsids. Because the midgut represents a first line of defense in oral infection by invading pathogens, the responses of midgut cells in the primary phase of infection may differ substantially from the responses of cultured cells to BV infection, which represent the secondary phase of infection.
Prior studies have shown that baculovirus infection triggers antiviral and other immune responses in cultured cells (1, 12, 13). In this study, we compared the transcriptomes of control (uninfected) and AcMNPV-infected T. ni midgut tissue and identified T. ni transcripts that were either up- or downregulated at each time point (0, 6, 12, 18, 24, 36, 48, and 72 h postinfection [p.i.]) following oral infection by ODV. Differential expression analysis resulted in the identification of a total of 3,372 transcripts with at least 4-fold changes (FC) in expression levels. Functional annotations of these differentially expressed (DE) transcripts revealed overall trends in important host cellular processes affected by virus infection. Of particular interest, AcMNPV infection dramatically increased the expression levels of several genes (HMG176, atlastin-2, cuticle CPH43, and E3 ubiquitin ligase SIAH) in the infected midgut, whereas the expression levels of several other genes (flippase, serine protease 33, and a variety of cytochrome P450 genes) were dramatically reduced. We also identified a small subset of T. ni transcripts that were differentially expressed across all time points postinfection. Thus, the global transcriptional analysis of the AcMNPV-infected T. ni midgut provides a detailed characterization of midgut-specific cellular responses to baculovirus infection and will be important for understanding the complex virus-host interactions in the primary phase of infection.
RESULTS AND DISCUSSION
Differential expression analysis of T. ni transcripts following AcMNPV infection.
To analyze the midgut cell responses to AcMNPV infection, developmentally synchronized and newly molted 5th-instar T. ni larvae were orally infected with wild-type (WT) AcMNPV OBs as described previously (14). Each larva was orally inoculated with 7 × 104 OBs, a dose determined empirically to be the minimum approximate dose resulting in maximal numbers of infected midgut cells. At selected time points postinfection (0, 6, 12, 18, 24, 36, 48, and 72 h p.i.), the midguts were excised and pooled (6 larval midguts per replicate, 3 replicates per time point) and used for isolation of poly(A) mRNA and construction of strand-specific RNA sequencing (ssRNA-Seq) libraries for Illumina sequencing. From each replicate sample, we mapped cleaned reads to the T. ni reference genome (14,384 genes) (15). Previously, we also estimated viral reads in each T. ni midgut sample by similarly mapping cleaned reads to the AcMNPV genome (156 genes) (NCBI accession no. NC_001623 and BioProject accession no. PRJNA484772) (14). The percentage of total reads mapping to T. ni genes declined gradually as the infection proceeded (Fig. 1 and Table 1), with the largest decline in T. ni read counts being observed between 36 and 72 h p.i., when the percentage of viral reads simultaneously increased the most dramatically (Fig. 1). To identify changes in T. ni transcript levels that resulted from AcMNPV infection, we estimated the expression levels of each T. ni gene by calculating reads per kilobase of transcript per million mapped reads (RPKM) (see Table S1 in the supplemental material) and performed differential expression analysis between AcMNPV-infected and uninfected midguts at each of the selected time points postinfection. We considered only transcripts with at least a 4-fold change (4-FC) in transcript levels (between the infected and uninfected samples) to be differentially expressed (DE) transcripts. Using this threshold, we identified a total of 3,372 DE T. ni transcripts in the AcMNPV-infected midgut (Fig. 2 and Table S2A and B). At early time points (0, 6, 12 h p.i.), few DE transcripts were detected (67 DE transcripts total), while substantial numbers of DE transcripts were identified at later times: 18 h p.i. (424 transcripts), 24 h p.i. (82 transcripts), 36 h p.i. (414 transcripts), 48 h p.i. (475 transcripts), and 72 h p.i. (1910 transcripts) (Table S2A and B). In general and as expected, DE transcript numbers increased as the infection proceeded. We further subclassified DE transcripts (4-FC and P < 0.05) based on the degree of up- or downregulation at each time point. For these calculations, RPKM values of <1 were treated as 0. Therefore, T. ni transcripts with RPKM values of <1 in one of the midgut samples (uninfected or infected) were classified as either induced or suppressed, respectively, in response to AcMNPV infection (Table S2A). For example, a transcript with an RPKM value of <1 in the uninfected midgut and ≥1 in the infected midgut at a time point was classified as induced, while a transcript with an RPKM value of ≥1 in the uninfected midgut and <1 in the infected midgut was classified as suppressed. If transcript RPKM values were ≥1 in both infected and uninfected midguts (and they differed by ≥4-fold), they were classified as either upregulated or downregulated (Table S2B). Using these criteria, we identified a total of 920 (induced), 1,419 (upregulated), 578 (suppressed), and 455 (downregulated) transcripts in the midgut (Fig. 2).
FIG 1.
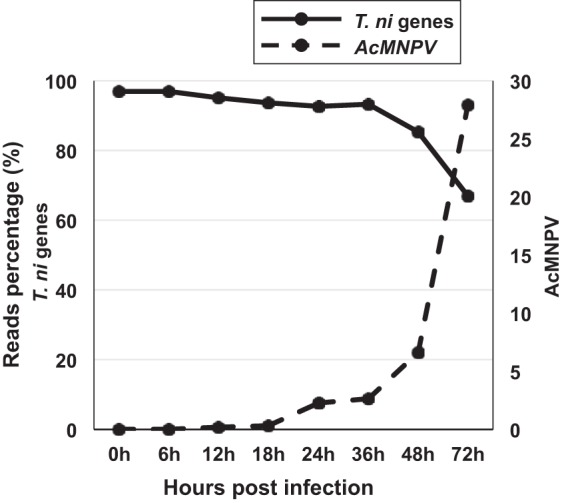
T. ni and AcMNPV mRNA reads as a percentage of total mRNA reads at each time point from 0 to 72 h p.i. The percentages of AcMNPV mRNA reads in the infected midgut are indicated by the dashed line (right y axis), and the percentage of T. ni mRNA reads in the infected midgut are shown as a solid line (left y axis). T. ni reads were identified by mapping cleaned reads to the T. ni genome, and AcMNPV reads were identified by mapping cleaned reads to the AcMNPV genome.
TABLE 1.
Reads mapped to T. ni and AcMNPV genomes at selected times postinfection
| Midgut infection | Time (h) postinfection | Replicate 1 |
Replicate 2 |
Replicate 3 |
Avg no. of midgut reads/time point | SD no. of midgut reads/time point | |||
|---|---|---|---|---|---|---|---|---|---|
| Total midgut readsa | Total viral readsb | Total midgut reads | Total viral reads | Total midgut reads | Total viral reads | ||||
| Uninfected | 0 | 18,118,667 | 21,669,642 | 21,991,275 | 20,593,195 | 2,149,029 | |||
| 6 | 12,283,390 | 24,758,172 | 23,153,004 | 20,064,855 | 6,786,571 | ||||
| 12 | 36,144,710 | 18,045,491 | 14,956,667 | 23,048,956 | 11,445,929 | ||||
| 18 | 14,187,003 | 17,376,890 | 14,063,937 | 15,209,277 | 1,878,216 | ||||
| 24 | 20,763,048 | 22,762,824 | 29,499,957 | 24,341,943 | 4,577,511 | ||||
| 36 | 15,502,539 | 14,570,698 | 15,956,828 | 15,343,355 | 706,643 | ||||
| 48 | 18,503,664 | 21,009,161 | 19,745,105 | 19,752,643 | 1,252,766 | ||||
| 72 | 21,065,355 | 18,499,850 | 27,517,673 | 22,360,959 | 4,646,421 | ||||
| Infected | 0 | 17,265,342 | 338 | 21,207,395 | 936 | 18,567,336 | 355 | 19,013,358 | 2,008,519 |
| 6 | 16,151,868 | 3,447 | 14,914,261 | 1,820 | 17,645,289 | 2,727 | 16,237,139 | 1,367,509 | |
| 12 | 27,671,909 | 22,533 | 13,985,653 | 62,733 | 20,895,221 | 15,008 | 20,850,928 | 6,843,236 | |
| 18 | 19,468,923 | 68,144 | 26,218,310 | 53,738 | 19,626,036 | 90,472 | 21,771,090 | 3,852,207 | |
| 24 | 22,069,640 | 490,621 | 16,689,213 | 440,689 | 14,227,179 | 301,487 | 17,662,011 | 4,010,711 | |
| 36 | 24,329,106 | 873,311 | 13,722,447 | 436,217 | 17,663,295 | 266,238 | 18,571,616 | 5,361,351 | |
| 48 | 14,280,612 | 388,477 | 12,411,359 | 1,341,304 | 15,288,279 | 1,247,384 | 13,993,417 | 1,459,804 | |
| 72 | 11,468,070 | 4,554,198 | 11,213,699 | 4,341,483 | 17,993,483 | 6,800,054 | 13,558,417 | 3,842,985 | |
Numbers represent the total read counts mapped to the assembled T. ni genome (http://www.tnibase.org/cgi-bin/index.cgi) in three biological replicates of T. ni midgut samples at each indicated time postinfection using the HISAT aligner, allowing 2 mismatches.
Numbers represent the total read counts mapped to the AcMNPV genome in three replicates of T. ni midgut samples using the HISAT aligner, allowing 2 mismatches. Data from the rows for infected midguts were reported by Shrestha et al. (14).
FIG 2.
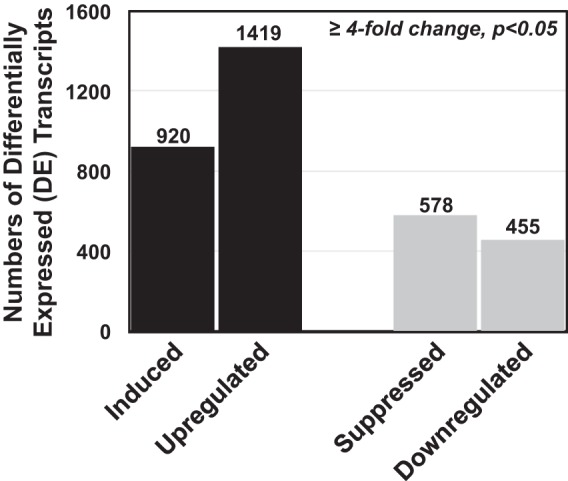
Numbers of differentially expressed (DE) T. ni transcripts identified in the midgut following AcMNPV infection. The transcriptomes of control (uninfected) and infected T. ni midguts were compared, and differentially expressed transcripts with at least a 4-fold change in expression and P values of <0.05 were identified at each time point (0, 6, 12, 18, 24, 36, 48, and 72 h p.i.). Based on the degree of up- or downregulation at each time point, DE transcripts were further classified as induced, upregulated, suppressed, or downregulated transcripts. T. ni transcripts with RPKM values of ≤1 in one of the midgut samples (uninfected or infected) were classified as either induced or suppressed transcripts, respectively. If transcript RPKM values were ≥1 in both infected and uninfected midguts (and their expression differed by ≥4-fold), they were classified as either upregulated or downregulated transcripts.
Functional annotation of differentially expressed T. ni transcripts.
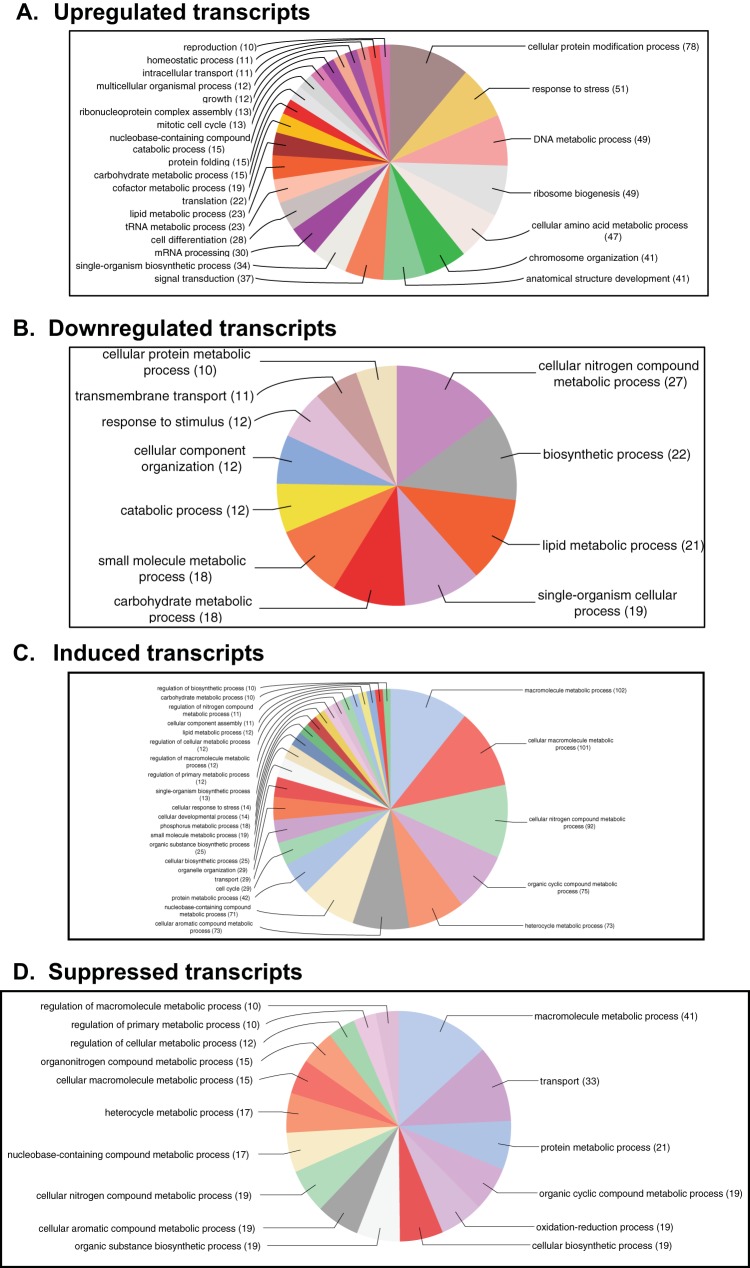
To identify possible functions of the DE transcripts affected by the virus infection in the T. ni midgut, we searched for homologs of the T. ni transcripts in the nonredundant database at NCBI using the BLASTP program. We identified potential orthologs for 60% of the induced transcripts, 93% of the upregulated transcripts, 70% of the suppressed transcripts, and 83% of the downregulated transcripts (Table S2A and B, Annotation). Blast2GO analysis was also performed on DE transcripts to identify major biological processes that were affected by the virus infection. For a broad overview, we combined DE transcripts from all time points for each classified DE group (induced, upregulated, suppressed, and downregulated) and performed Blast2GO analysis on each DE group. Blast2GO assigned 25 and 26 gene ontology (GO) terms to upregulated and induced transcripts, respectively (Fig. 3A and C), and 11 and 16 GO terms to downregulated and suppressed transcripts, respectively (Fig. 3B and D). Among the GO terms assigned to upregulated transcripts, many of the transcripts were associated with cellular protein modification (11%), response to stress (7%), DNA metabolic process (7%), ribosome biosynthesis (7%), chromosome organization (6%), signal transduction (5%), and cell differentiation (4%). Further, GO terms of induced transcripts (under the biological process) were primarily associated with metabolic processes, such as macromolecule metabolic process (10%), cellular nitrogen compound metabolic process (10%), organic cyclic compound metabolic process (8%), and heterocycle metabolic process (7.8%). For transcripts that were downregulated in the AcMNPV-infected midgut, GO terms included nitrogen compound metabolic process (14%), biosynthetic process (12%), single-organism cellular process (10%), and response to stimulus (7%). Analysis of suppressed transcripts resulted in GO terms involved in transport (10%), oxidation and reduction (6%), protein metabolism (6%), and cellular macromolecule metabolic process (5%).
FIG 3.
Gene ontology (GO) terms assigned to T. ni transcripts that were either upregulated (A), downregulated (B), induced (C), or suppressed (D) in the midgut following AcMNPV infection. Upregulated, downregulated, induced, or suppressed transcripts from all the time points were combined to run Blast2GO analysis. GO terms were assigned to the DE transcripts under a biological process with an E-value hit filter of 10−6 and an annotation cutoff of 55. Also, the GO terms were classified at GO level 4.
T. ni transcripts differentially expressed between and across time points postinfection.
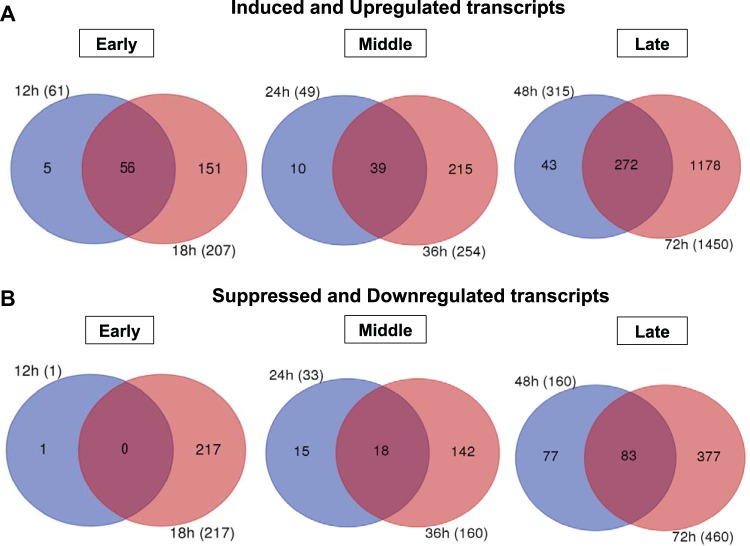
Next, we examined transcripts that were differentially expressed during several time periods postinfection (Fig. 4). We first divided the time periods into three categories: early (12 and 18 h p.i.), middle (24 and 36 h p.i.), and late (48 and 72 h p.i.). The earliest times sampled in our experiments (0 and 6 h p.i.) were not included as we identified very few DE transcripts at those times (Table S2A and B). We examined induced and upregulated transcripts from each time point for one set of comparisons (Fig. 4A) and suppressed and downregulated transcripts for another set of comparisons (Fig. 4B). The Venn diagrams in Fig. 4 illustrate the numbers of common DE transcripts within each category: early, middle, or late. For the induced and upregulated DE transcripts, we identified 56, 39, and 272 shared DE transcripts within the early, middle, and late categories, respectively (Fig. 4A; Table S3). No suppressed and downregulated DE transcripts were identified as shared between 12 and 18 h p.i., but 18 and 83 DE transcripts in that category were shared within the middle and late periods, respectively (Fig. 4B; Table S3). We also identified transcripts that were differentially expressed across most time points (from 12 to 72 h p.i.), and they are listed in Table 2. Of these, we identified 11 transcripts that were induced or upregulated across all times sampled from 12 to 72 h postinfection. These transcripts included HMG176 (a response to pathogen [REPAT] gene), atlastin-2, E3 ubiquitin ligase SIAH1, cyclic GMP-AMP synthase (cGAS) isoform X2, mitochondrial uncoupling 4, and NF-κB-repressing factor. In the case of suppressed or downregulated transcripts, 14 transcripts were differentially expressed across time points from 18 to 72 h p.i. (Table 2). Examples of these transcripts include serine protease 33, folylpolyglutamate synthase, Ves G 1 allergen, indole-3-glycerol phosphate synthase, flippase, and DNA mismatch repair protein (Table 2, Suppressed and downregulated). The levels of several of these up- or downregulated transcripts (such as HMG176, atlastin-2, flippase, and serine protease 33) were radically different in infected midgut cells, with differences of up to 640-fold. Several of these genes are discussed in more detail in the section “T. ni genes most significantly affected by AcMNPV infection” below (see also Table 3).
FIG 4.
Venn diagrams demonstrating the numbers of differentially expressed (DE) T. ni transcripts that were identified (at each time and commonly) across the indicated times following oral infection with AcMNPV OBs. For Venn diagram construction, we grouped time points into three major classes: early (0, 6, 12, and 18 h p.i.), middle (24 and 36 h p.i.), and late (48 and 72 h p.i.). Within the early group, time points 0 and 6 h p.i. contained very few DE T. ni transcripts, and those transcripts were not common among the other time points. Therefore, data for 0 and 6 h p.i. are not shown in the figure. Each circle represents the combined number of differentially expressed genes (induced and upregulated transcripts [A] and suppressed and downregulated transcripts [B]) for the indicated time point (for example, 12 h and 18 h) postinfection.
TABLE 2.
T. ni transcripts that are differentially expressed at most times postinfection
| Type of differential expression | T. ni gene IDa | Annotation | Fold changeb at the following times (h) p.i.: |
|||||
|---|---|---|---|---|---|---|---|---|
| 12 | 18 | 24 | 36 | 48 | 72 | |||
| Induced and upregulated | Tni20G02130 | HMG176 isoform (REPAT) | IND | IND | IND | IND | IND | 79.93 |
| Tni16G00620 | E3 ubiquitin ligase SIAH1 | 20.76 | IND | 5.43 | 10.37 | 10.49 | 15.04 | |
| Tni22G05470 | Cyclic GMP-AMP synthase isoform X2 | 59.07 | 47.49 | 5.89 | 18.06 | 15.69 | 35.66 | |
| Tni17G02590 | Mitochondrial uncoupling 4 | 10.77 | 30.94 | 14.17 | 15.86 | 17.83 | 47.83 | |
| Tni05G05890 | Atlastin-2-like isoform X1 | 81.19 | 111.60 | 9.27 | 28.72 | 13.24 | 49.69 | |
| Tni08G01410 | NFX1-type zinc finger-containing 1 | IND | IND | 8.06 | IND | 16.47 | 48.08 | |
| Tni02G00330 | NF-κB-repressing factor | 5.38 | IND | 16.33 | IND | IND | IND | |
| Tni14G02910 | E3 SUMO-ligase KIAA1586 | IND | IND | IND | IND | IND | IND | |
| Tni16G04790 | No BLAST hits | 9.22 | IND | 4.68 | 11.00 | 7.34 | 16.80 | |
| Tni28G01940 | ATP-dependent RNA helicase DHX30 | IND | 15.16 | 7.30 | IND | 9.80 | IND | |
| Tni27G01530 | Microphthalmia-associated transcription factor | IND | IND | IND | IND | IND | IND | |
| Suppressed and downregulated | Tni18G00450 | Serine protease 33 | — | SUP | SUP | SUP | SUP | SUP |
| Tni17G04440 | Folylpolyglutamate synthase | — | −58.97 | −66.20 | −149.39 | −87.98 | SUP | |
| Tni12G01810 | Ves G 1 allergen | — | −83.11 | −62.53 | −92.93 | SUP | −97.38 | |
| Tni15G03390 | Indole-3-glycerol phosphate synthase | — | SUP | SUP | SUP | SUP | SUP | |
| Tni21G02480 | Flippase | — | −128.95 | −290.79 | −640.00 | −344.40 | SUP | |
| Tni06G03550 | DNA mismatch repair protein | — | SUP | SUP | SUP | SUP | SUP | |
| Tni05G06480 | No BLAST hits | — | −345.04 | SUP | SUP | −59.01 | −138.96 | |
| Tni15G05500 | Hypothetical protein FOXG_02896 | — | −60.94 | SUP | SUP | −74.40 | −36.60 | |
| Tni19G00950 | Ionotropic receptor | — | SUP | SUP | SUP | SUP | SUP | |
| Tni15G06910 | Uncharacterized protein LOC110836755 | — | SUP | SUP | SUP | SUP | SUP | |
| Tni15G03460 | Unknown | — | −90.55 | −46.82 | −31.60 | −26.10 | −15.49 | |
| Tni26G00190 | Uncharacterized protein LOC103522958 | — | SUP | SUP | SUP | SUP | SUP | |
| Tni15G04210 | Hypothetical protein PABG_01059 | — | SUP | SUP | SUP | SUP | SUP | |
| Tni19G04480 | Hypothetical protein X975_10841 | — | −14.83 | −114.47 | −13.32 | −95.95 | −33.00 | |
T. ni gene ID numbers correspond to the annotations at http://www.tnibase.org/cgi-bin/index.cgi.
Fold changes in T. ni transcript levels between uninfected and infected midgut samples at selected times postinfection. IND, induced; SUP, suppressed; —, for suppressed and downregulated transcripts, commonly expressed transcripts were not identified at 12 h p.i.
TABLE 3.
T. ni transcripts significantly affected by AcMNPV infectiona
| Time (h) p.i. | Type of differential expression | T. ni gene ID | Annotation | RPKM value |
Fold change in expression | Adjusted P value | |
|---|---|---|---|---|---|---|---|
| Control | Infected | ||||||
| 12 h | Induced | Tni07G03860 | C19orf12 homolog | 0.56 | 235.72 | IND | |
| Tni20G02130 | HMG176 isoform (REPAT) | 0.14 | 97.96 | IND | |||
| Upregulated | Tni05G05890 | Atlastin-2-like isoform X1 | 7.11 | 577.24 | 81.19 | 4.553E−05 | |
| Tni22G05470 | Cyclic GMP-AMP synthase isoform X2 | 2.55 | 150.63 | 59.07 | 2.033E−22 | ||
| Tni17G02340 | E3 ubiquitin ligase SIAH1 | 3.49 | 149.59 | 42.82 | 1.895E−05 | ||
| 18 h | Induced | Tni07G03860 | C19orf12 homolog | 0.50 | 214.47 | IND | |
| Tni12G01690 | Zinc finger CCHC domain-containing | 0.23 | 188.49 | IND | |||
| Upregulated | Tni05G05890 | Atlastin-2-like isoform X1 | 7.34 | 819.13 | 111.60 | 1.211E−10 | |
| Tni06G03350 | Collagen triple helix repeat domain | 47.19 | 1,893.30 | 40.12 | 3.392E−02 | ||
| Tni08G03170 | Cuticle CPH43 | 34.63 | 1,301.27 | 37.58 | 1.547E−05 | ||
| Tni16G00140 | Immune-related Hdd13 | 17.37 | 284.10 | 16.36 | 1.649E−02 | ||
| Tni03G01880 | Mucin-5AC-like | 19.11 | 309.78 | 16.21 | 2.117E−03 | ||
| Suppressed | Tni15G03390 | Indole-3-glycerol phosphate synthase | 188.57 | 0.72 | SUP | ||
| Tni15G03790 | EF-hand domain-containing 1 | 108.99 | 0.55 | SUP | |||
| Downregulated | Tni11G00910 | Calcium-binding P | 328.38 | 1.32 | −248.77 | 7.515E−16 | |
| Tni15G03490 | E3 ubiquitin ligase RNF180-like | 781.49 | 7.21 | −108.39 | 3.856E−55 | ||
| Tni21G02480 | Flippase | 1,573.23 | 12.20 | −128.95 | 4.109E−32 | ||
| Tni11G03900 | Collagen alpha-1(IX) chain isoform X8 | 201.25 | 3.41 | −59.02 | 1.317E−31 | ||
| Tni17G02150 | 3-Dehydroecdysone 3 alpha-reductase | 879.10 | 21.67 | −40.57 | 6.361E−28 | ||
| Tni17G04440 | Folylpolyglutamate synthase | 433.41 | 7.35 | −58.97 | 8.603E−12 | ||
| 24 h | Upregulated | Tni20G02140 | HMG176 isoform (REPAT) | 1.56 | 53.16 | 34.00 | 1.337E−13 |
| Downregulated | Tni21G02480 | Flippase | 747.33 | 2.57 | 290.79 | 8.818E−03 | |
| Tni19G04480 | Hypothetical protein X975_10841 | 606.70 | 5.30 | 114.47 | 3.934E−02 | ||
| 36 h | Upregulated | Tni08G03170 | Cuticle CPH43 | 27.15 | 4,263.95 | 157.03 | 9.743E−17 |
| Tni03G01880 | Mucin-5AC-like | 15.72 | 909.25 | 57.85 | 6.327E−05 | ||
| Tni20G02110 | HMG176 isoform (REPAT) | 51.24 | 833.39 | 16.26 | 1.681E−12 | ||
| Suppressed | Tni15G05500 | Hypothetical protein FOXG_02896 | 109.98 | 0.98 | SUP | ||
| Downregulated | Tni21G02480 | Flippase | 2,246.39 | 3.51 | −640.00 | 2.499E−118 | |
| Tni15G03490 | E3 ubiquitin ligase RNF180-like | 596.65 | 4.99 | −119.57 | 7.924E−50 | ||
| Tni17G02150 | 3-Dehydroecdysone 3 alpha-reductase | 395.41 | 10.84 | −36.48 | 2.384E−50 | ||
| 48 h | Upregulated | Tni07G03860 | C19orf12 homolog | 3.58 | 156.25 | 43.69 | 2.885E−27 |
| Tni08G03170 | Cuticle CPH43 | 178.29 | 3,191.83 | 17.90 | 1.821E−03 | ||
| Tni03G01880 | Mucin-5AC-like | 59.43 | 965.99 | 16.25 | 3.391E−12 | ||
| Suppressed | Tni12G01810 | Ves G 1 allergen | 105.21 | 0.86 | SUP | ||
| Tni11G00910 | Calcium-binding P | 104.82 | 0.48 | SUP | |||
| Downregulated | Tni21G02480 | Flippase | 1,277.72 | 3.71 | −344.40 | 1.232E−23 | |
| Tni15G03490 | E3 ubiquitin ligase RNF180-like | 584.42 | 7.77 | −75.21 | 1.163E−77 | ||
| Tni19G04480 | Hypothetical protein X975_10841 | 544.03 | 5.67 | −95.95 | 1.155E−04 | ||
| 72 h | Upregulated | Tni12G01690 | Zinc finger CCHC domain containing | 4.65 | 760.51 | 163.43 | 3.673E−24 |
| Tni05G05890 | Atlastin-2-like isoform X1 | 22.02 | 1,094.12 | 49.69 | 2.850E−21 | ||
| Tni08G03170 | Cuticle CPH43 | 193.19 | 7,433.26 | 38.48 | 8.137E−04 | ||
| Tni18G02870 | Chymotrypsin-like serine protease | 4.04 | 149.54 | 37.05 | 2.575E−10 | ||
| Tni22G05470 | Cyclic GMP-AMP synthase isoform X2 | 4.67 | 166.64 | 35.66 | 1.161E−17 | ||
| Tni26G01100 | Zinc finger | 10.28 | 297.86 | 28.98 | 3.497E−17 | ||
| Tni03G01880 | Mucin-5AC-like | 76.58 | 1,523.70 | 19.90 | 1.234E−04 | ||
| Tni22G00330 | Peroxidase isoform X2 | 16.99 | 273.16 | 16.07 | 1.895E−04 | ||
| Suppressed | Tni11G00910 | Calcium-binding P | 287.35 | 0.66 | SUP | ||
| Tni19G04800 | Ninjurin-2 isoform X1 | 236.30 | 0.85 | SUP | |||
| Tni08G02780 | Carboxypeptidase M | 156.34 | 0.78 | SUP | |||
| Tni18G00450 | Serine protease 33 | 148.55 | 0.33 | SUP | |||
| Tni23G01020 | Surface protective antigen | 87.18 | 0.08 | SUP | |||
| Downregulated | Tni08G00710 | Peritrophic matrix insect intestinal | 305.78 | 1.10 | −277.98 | 1.379E−04 | |
| Tni04G00650 | Stress response NST1 isoform X1 | 420.33 | 1.88 | −223.58 | 1.915E−56 | ||
| Tni13G03290 | Abhydrolase domain-containing 7 | 721.86 | 9.99 | −72.26 | 7.879E−44 | ||
| Tni12G01650 | Calponin transgelin | 812.78 | 8.97 | −90.61 | 7.243E−42 | ||
| Tni15G03490 | E3 ubiquitin ligase RNF180-like | 684.19 | 8.30 | −82.43 | 1.339E−02 | ||
| Tni11G02810 | Lipase 3 | 473.89 | 8.30 | −57.10 | 6.491E−10 | ||
| Tni15G01350 | Zinc metalloproteinase nas-4-like | 355.98 | 6.69 | −53.21 | 6.499E−07 | ||
| Tni22G01200 | 27-kDa hemolymph | 634.65 | 21.15 | −30.01 | 4.057E−35 | ||
| Tni03G02090 | Moderately methionine-rich storage | 642.02 | 20.51 | −31.30 | 7.960E−23 | ||
| Tni17G05550 | Cytochrome P450 | 237.62 | 7.40 | −32.11 | 2.547E−16 | ||
| Tni18G04160 | Ecdysone oxidase | 273.31 | 12.45 | −21.95 | 2.347E−20 | ||
| Tni13G00880 | Cobatoxin short form A | 572.15 | 26.23 | −21.81 | 1.092E−10 | ||
T. ni gene ID numbers correspond to the annotations at http://www.tnibase.org/cgi-bin/index.cgi. Significantly affected T. ni transcripts consist of at least a 16-fold change in expression levels and RPKM values of ≥50 in control (uninfected) or infected midgut samples. IND, induced; SUP, suppressed.
Cluster analysis of differentially expressed T. ni transcripts.
We also grouped the differentially expressed T. ni transcripts identified in this study using hierarchical cluster analysis. We selected transcripts that were differentially expressed (between uninfected and infected midguts) by ≥4-fold and performed cluster analysis on the expression levels, and then we identified the patterns of expression of those transcripts from the infected midgut. A heat map generated from the cluster analysis (Fig. 5A) shows the DE transcripts grouped into five clusters. We arbitrarily referred to the groups as G1, G2, G3, G4, and G5 (Fig. 5A), which are comprised of 34, 201, 697, 1,433, and 205 transcripts, respectively, and these transcripts are listed in Table S4. From the list of DE transcripts grouped by cluster analysis, we performed a separate analysis to identify transcripts with specific expression patterns (Fig. 5B). We identified transcripts with four expression patterns: P1, continuously increasing; P2, continuously decreasing; P3, increasing from 0 to 18 h p.i. and then decreasing; and P4, reaching the highest levels at 12 to 18 h p.i. (Fig. 5B). Specific T. ni transcripts within each of these groups from Fig. 5B are listed in Table S5. We identified nine transcripts with levels that continuously increased from 0 to 72 h p.i. (Fig. 5B, P1) and 17 transcripts with levels that continuously decreased from 0 to 72 h p.i. (Fig. 5B, P2). Examples of transcripts from pattern P1 included mismatch repair endonuclease, intracellular transport US1, N-acetyltransferase, and RNase H1. Among the transcripts from pattern P2 were larval cuticle 8 protein, a cuticular RR-2 family protein, regucalcin isoform, and sodium potassium-transporting ATPase subunit-2. Further we identified 22 transcripts conforming to pattern P3 (including insect intestinal lipase, glucose dehydrogenase, prophenoloxidase subunit 1, peroxisome membrane PMP34, and laminin subunit alpha isoform X3). We found that 96 transcripts were associated with pattern P4, with examples including genes encoding HMG176 isoform (REPAT proteins), atlastin-2 like isoform 1, cecropin B, prophenoloxidase 2, lipase 3, cytochrome P450, ecdysone oxidase, aldehyde oxidase, serine protease 31, and an ankyrin repeat domain protein (Table S5).
FIG 5.
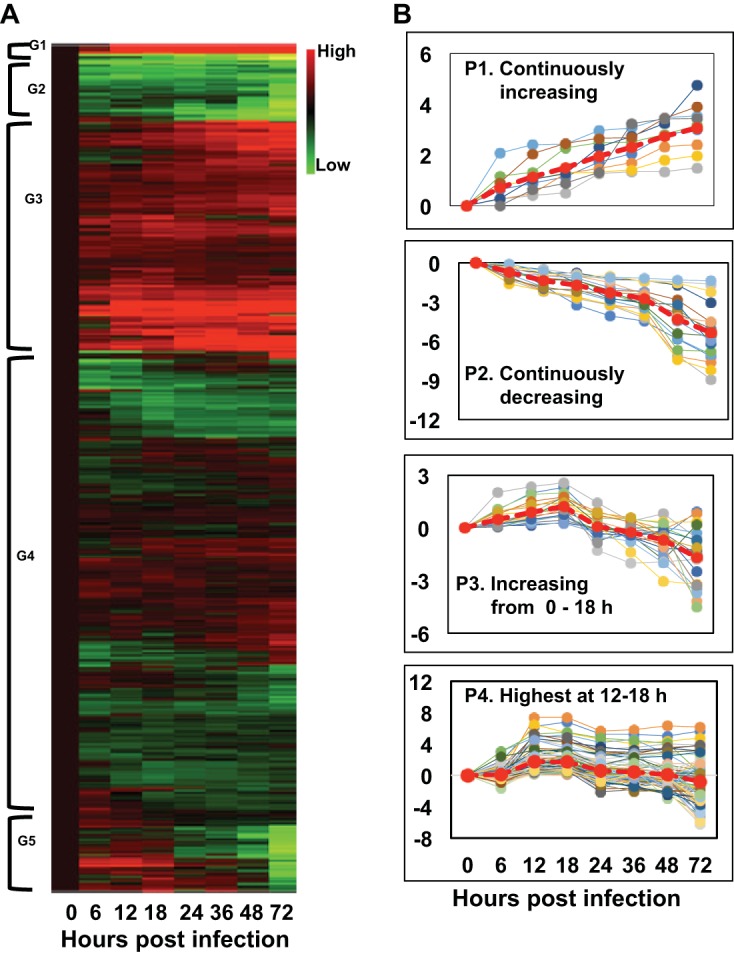
Patterns of differentially expressed (DE) T. ni genes following AcMNPV infection of the midgut. (A) A cluster analysis of DE T. ni midgut transcripts is shown as a heat map that was generated by performing hierarchical cluster analysis on normalized read counts (log2 transformed) of DE transcripts from infected midgut samples from 0 to 72 h p.i. Normalized read counts from each biological replicate were averaged, and then the averaged normalized read counts at 0 h p.i. were subtracted from each time point’s normalized read counts. Euclidean distance metrics were then applied to the normalized read counts using R software. In the illustrated heat map, the expression levels of each gene (rows) at each time point (columns) are depicted with a color scale in which green represents low-level expression and red represents high-level expression. The cluster analysis grouped DE transcripts into five clusters based on their expression levels. The groups were arbitrarily assigned the names G1, G2, G3, G4, and G5. (B) From the list of T. ni transcripts grouped by the cluster analysis, transcripts with specific patterns (as indicated) were identified using an R script and named P1 to P4.
T. ni genes most significantly affected by AcMNPV infection.
(i) Induced and upregulated T. ni transcripts. From a comprehensive list of differentially expressed T. ni transcripts (Table S2A and B), we identified the most significantly affected of the differentially expressed transcripts, based on the RPKM values (minimum value of 50 RPKM for either uninfected or infected cells) and a change of at least 16-fold between uninfected and infected midgut cells. We subdivided the DE transcript groups into the following categories: induced, upregulated, suppressed, and downregulated. Using these thresholds and groupings, we identified 63 transcripts with large differences in transcript levels (Table 3). Below, we highlight several of the DE genes that were of particular interest.
(a) HMG176 (REPAT). T. ni orthologs of a gene named HMG176 (16) were significantly upregulated or induced and highly expressed in response to AcMNPV infection in the midgut. T. ni orthologs of HMG176 were either substantially induced or upregulated in the midgut from 12 to 72 h p.i. (Table 3 and Table S7). HMG176 belongs to a gene family encoding small proteins (11 to 17 kDa), called response to pathogen (REPAT) proteins, that were initially identified from Spodoptera exigua larval midgut challenged with either Bacillus thuringiensis toxin Cry1Ca or baculovirus AcMNPV. REPAT proteins were later identified in other lepidopteran species (17–21) [see also “(b) REPAT genes” below]. The HMG176 gene of Helicoverpa armigera is normally expressed at high levels in midguts of molting larvae, and the protein is found in the basal lamina of the midgut of molting and feeding larvae, but not in larvae committed to metamorphosis (16, 22). The precise function of HMG176 is not known. In contrast to the relatively early induction of HMG176 observed in the AcMNPV-infected T. ni larval midgut, some REPAT genes appear to be induced much later after AcMNPV infection of S. exigua larvae, with the induction of some being detected after approximately 72 h p.i. (17). Because the H. armigera HMG176 gene is upregulated during molting in uninfected larvae (16), it is possible that its dramatic upregulation in response to AcMNPV infection of the midgut may represent the activation of some genes associated with the molting process, perhaps as a defensive response by the host, or as a virus-induced mechanism to facilitate dissemination of the virus in the infected host.
(b) Atlastin. Another highly upregulated T. ni transcript is an atlastin-2 ortholog (Tni05G05890). The T. ni atlastin-2 ortholog was dramatically upregulated in the midgut following oral infection with AcMNPV. Transcript levels were increased (9- to 111-fold) at various times from 12 to 72 h p.i., and the highest degree of upregulation occurred at early times in the infection: 12 h p.i. (81-fold) and 18 h p.i. (111-fold) (Table 3 and Table S2B). Atlastin proteins are close homologs of guanylate-binding proteins (GBPs) and are in the dynamin superfamily of proteins (23). The GBPs include MX proteins, which are well-studied interferon-induced antiviral proteins in vertebrates (24). Functionally, atlastins are involved in endoplasmic reticulum (ER) network formation and fusion and vesicle trafficking in the ER (25, 26). Recently, a member of the atlastin protein family (atlastin-n) was shown to provide resistance to baculovirus infection in Bombyx mori (27). When the atlastin-n protein was overexpressed in B. mori BmN-SWU1 cells or in larvae, virus production was inhibited, suggesting an antiviral role against Bombyx mori nuclear polyhedrosis virus (BmNPV). The dramatic upregulation of the atlastin-2 ortholog in the AcMNPV-infected T. ni midgut may represent an induced antiviral response to AcMNPV infection, although more direct studies will be required to understand its role following oral infection.
(c) CPH43. We also identified a T. ni ortholog (Tni08G03170) of the hypothetical cuticle protein CPH43 gene of B. mori as one of the most significantly upregulated transcripts in the infected T. ni midgut. The extensively studied B. mori genome contains approximately 255 cuticular protein genes, of which 44 were identified as hypothetical cuticular protein (CPH) genes based on indirect evidence (28). In the current study, CPH43 transcript levels were upregulated in the AcMNPV-infected T. ni midgut by approximately 18- to 157-fold (18 to 72 h p.i.), with extremely high transcript levels at 72 h p.i. (Table 3). Similar to the findings of the current study, the transcript levels of CPH43 also increased in B. mori midgut tissues following pathogenic infection with Bombyx mori cytoplasmic polyhedrosis virus (BmCPV), and CPH43 upregulation was speculated to play a role in the remodeling of the midgut epithelium (29).
(d) cGAS orthologs. We observed the upregulation of transcripts of several T. ni isoforms of cyclic GMP-AMP synthase (cGAS) genes (Tni22G05470, Tni22G05480, and Tni22G05460) in the infected T. ni midgut. Among the three substantially upregulated T. ni cGAS transcripts, the most highly expressed and most dramatically upregulated was Tni22G05470, which was upregulated by as much as 59- and 47-fold (at 12 and 18 h p.i., respectively) and which was present at high RPKM levels (approximately 150 and 82) at relatively early times postinfection (12 and 18 h p.i., respectively) (Table 3 and Table S2B). The other two T. ni cGAS transcripts were also upregulated at earlier times but were most dramatically upregulated (by as much as 10-fold [Tni22G05460] and 33-fold [Tni22G05480]) at later times postinfection (72 h p.i.). cGAS is a cytosolic DNA sensor that binds to foreign DNA, such as the DNA of viral pathogens, and induces activation of the type I interferon pathway and NF-κB signaling to defend against infections by viruses, such as retroviruses and herpesviruses (30, 31). While insects do not have an interferon response pathway, NF-κB signaling is known to activate innate immune responses. The upregulation of cGAS transcripts in the midgut at early time points in the infection suggests that AcMNPV infection may result in a systemic activation of immune responses throughout the midgut tissue. Future studies to examine the specific mechanistic role(s) of cGAS in infected and bystander cells could yield important details on how cells and the midgut tissue overall react to viral infection in the midgut.
(e) mucin-5AC-like gene. Further, among the dramatically induced or upregulated transcripts was a mucin-like gene, identified as the mucin-5AC-like gene. We observed upregulation of mucin-5AC-like transcripts (Tni03G01880) at most times postinfection in the midgut (Table 3), with the levels of upregulation ranging from approximately 6.5-fold to 58-fold at times ranging from 18 to 72 h p.i. and with very high RPKM values ranging from approximately 310 to 1,524. The localization and function of the mucin-5AC-like protein are unknown. Previously, an insect intestinal mucin (IIM; Tni02G02610) was identified to be a major component of the peritrophic membrane (PM), the semipermeable structure that lines the midgut and serves as a structural barrier to viral infection (32–34). The IIM transcripts were present at extremely high levels (RPKM, 1,050 to 7,430) and were modestly upregulated (1.4- to 3.6-fold) over the 72 h of infection of the midgut. In some (probably most) baculovirus infections, virus-encoded proteases (such as the enhancin protein) target IIM for degradation, resulting in the loss of PM integrity and thereby enhancing virus infection (34). The upregulation of mucin transcript levels observed in the T. ni midgut as a response to infection (Table 3) may represent a general response to PM disruption and a defense mechanism against virus-mediated PM degradation. In a prior study of the midgut of another noctuid species (Spodoptera frugiperda), it was found that the degradation of the PM by a maize cysteine protease resulted in the upregulation and increased abundance of IIM mRNAs (35, 36).
Other transcripts of note that were highly upregulated in the midgut in response to oral AcMNPV infection were orthologs of an E3 ubiquitin ligase SIAH (Tni17G02340), a zinc finger CCHC (Tni12G01690), a peroxidase (isoform X2; Tni22G00330), and a chymotrypsin-like serine protease (SP; Tni18G02870) (Table 3).
(ii) Suppressed and downregulated transcripts. We also identified a significant number of T. ni transcripts that were either suppressed or downregulated (at least 16-fold) in the midgut in response to AcMNPV infection. Some of the most strikingly suppressed or downregulated transcripts included orthologs of flippase, serine proteases, cytochrome P450, calcium binding protein P, and dehydroecdysone 3 alpha-reductase (Table 3).
(a) Flippase. Flippases are transmembrane proteins that mediate lipid transport between the leaflets of a lipid bilayer, and they may be associated with the formation of vesicles (37, 38). We found that flippase transcripts, which were highly abundant in uninfected control midgut tissue (RPKM values were as high as 2,246), were dramatically downregulated from at least 18 h p.i. How reduced flippase might affect the infection or the defense of the host is unclear.
(b) Serine protease 33. The transcript levels of a chymotrypsin-like serine protease (serine protease 33, Tni18G00450) were also significantly suppressed by AcMNPV infection (Table 3 and Table S2A). Serine proteases play diverse roles in physiological processes, notably, digestion in the midgut, apoptosis, development, and immunity (39, 40).
(c) Cytochrome P450. Cytochrome P450 proteins are enzymes that are involved in the biosynthesis and metabolism (including detoxification) of a large variety of molecules. They are encoded by a large gene family, and the number of cytochrome P450 genes varies widely among lepidopteran species, ranging from approximately 78 to over 100 (41). While the P450 genes of T. ni have not been studied in detail, the automated annotation of the T. ni genome (15) identified approximately 118 genes as potential P450 genes. (Note that some of these may represent fragments or pseudogenes, as a recent study identified at least 18 pseudogenes in another lepidopteran genome [41].) While some T. ni cytochrome P450 orthologs were upregulated in response to AcMNPV infection, we observed suppression or downregulation of a substantial number of cytochrome P450 orthologs (Tni16G04090, Tni16G04370, Tni16G04420, Tni16G04430, Tni15G02520, Tni22G05060, Tni09G03330, Tni10G04110, Tni15G06560, Tni15G06570, Tni15G06610, Tni21G02490, Tni21G02470, Tni21G02450, Tni21G03890, Tni21G03910, Tni17G05560, Tni21G02490, Tni21G01940, Tni17G05570) in the midgut following AcMNPV infection (Table S2A and B). The downregulation of P450 genes following baculovirus infection was also observed in H. zea (42), suggesting that baculovirus-mediated suppression of P450s could play an important role in suppressing metabolic pathways in the insect host during the infection process.
(d) Other genes. Following virus infection, transcripts encoding several digestive enzymes, including lipase 3 and carboxypeptidase M, were significantly lower in infected midgut samples (Table 3). Digestive enzymes have been shown to have antiviral activities against viral pathogens. In contrast to the observations made here, in baculovirus-infected B. mori midgut-specific digestive enzymes, including lipase 1 and serine protease 2, were induced by virus infection, and these enzymes were speculated to inactivate ODV in the midgut (43, 44). We also observed the significant downregulation of transcripts potentially involved in regulation of the ecdysone signaling pathway, including 3-dehydroecdysone 3 alpha-reductase and ecdysone oxidase (Table 3).
(iii) Antiviral, immune, and stress response genes. After identifying DE transcripts that were most significantly affected by virus infection in the T. ni midgut, we analyzed the expression patterns of transcripts associated with stress response and immune pathways in the T. ni midgut. Orthologs of a variety of immune response genes from yeasts, human, Drosophila melanogaster, B. mori, S. exigua, or S. frugiperda were identified in a prior study of a T. ni (Tnms42 cell line) transcriptome (10), and the corresponding genes and transcripts were identified in the recently assembled T. ni genome (15) and used for analysis in this study. The expression patterns of immune response- and stress response-related genes were compared in uninfected and infected T. ni midgut (Fig. S1A to L), and RPKM values and fold changes in expression are shown in Table S6. Transcripts with RPKM values of less than 5 across all times postinfection in both uninfected and infected midguts were not included in this analysis.
(a) siRNAs. RNA interference (RNAi) is an antiviral response in invertebrates that detects double-stranded RNAs (dsRNAs). dsRNA is diced into small interfering RNAs (siRNAs) which interact with an RNase III enzyme, and the resulting RNA-induced silencing complex (RISC) directs the cleavage of complementary target RNAs in a sequence-specific manner. In insects, RNAi is a major antiviral defense mechanism against invading RNA and DNA viruses (45, 46). In a prior study of AcMNPV-infected Tnms42 cells, the downregulation of RNAi pathway-associated genes, such as Argonaute-2 (Ago-2) and Dicer-2 (Dcr-2), was described (10). In the current study of the responses to oral AcMNPV infection in the T. ni midgut, we found that the transcript abundance of Ago-2, Dcr-2, R3D1, and R2D2 increased slightly (by >2-fold) in the infected midgut by 12 h p.i. and substantially (>5-fold) by 72 h p.i. (Fig. S1A and Table S6). For Dcr-2 and R2D2, expression levels in the infected midgut were even higher, >13-fold and >12-fold, respectively (Fig. S1A and Table S6). Dcr-2 is an RNase III enzyme that interacts with R2D2 and binds to and cleaves dsRNA into siRNAs. The siRNAs are loaded into the Ago-2-containing RISC, which then cleaves the targeted RNA (45). The observed upregulation of major components of RNAi pathways, including Dcr-2, R2D2, and Ago-2, in the AcMNPV-infected midgut (beginning at about 12 to 18 h p.i.) indicates that AcMNPV infection induces components of the RNAi response in the T. ni midgut.
(b) REPAT genes. Response to pathogen (REPAT) genes were initially identified as genes that were upregulated in the Spodoptera exigua midgut in response to bacterial (B. thuringiensis) or viral (AcMNPV) challenge, and REPAT genes have been identified in other insect species (17–21). REPAT genes represent a large gene family in S. exigua (in which 46 REPAT genes were identified) (20), and closely related species also have numerous REPAT genes (35 in S. frugiperda, 21 in Spodoptera litura, 13 in Spodoptera littoralis, and 8 in Mamestra configurata). Of the 8 REPAT genes in M. configurata, 6 are isoforms of HMG176, which we identified to be one of the most highly upregulated genes in the AcMNPV-infected T. ni midgut (see above). For some other insect species (Mamestra brassicae, B. mori, H. armigera, and D. melanogaster), only a single REPAT gene has been identified (20, 21). Thus far, the precise biochemical or molecular roles of REPAT proteins have not been identified, although they appear to play a variety of roles. While HMG176 is found in the basal lamina of the midgut (16, 22), another REPAT protein (MBF2 from B. mori) was identified to be a transcriptional coactivator (20, 47). The single identified Drosophila REPAT gene (CG13323) was reported to be upregulated in response to infection with bacteria, fungi, and sigma virus (48), but the function of the encoded protein is unknown. The wide diversity in the sequence and expression profiles among REPAT proteins suggests that they may serve a variety of functions, perhaps as a group of stress response proteins with antiviral or antimicrobial functions. Interestingly, while overexpression of the S. exigua REPAT 1 protein from a recombinant baculovirus had no apparent effect on virus amplification in cultured Sf21 cells, injections of BV from the REPAT 1-expressing baculovirus into S. exigua larvae resulted in delayed mortality when compared with that from injection of a control virus (17), suggesting a possible antiviral role.
A BLAST search of the T. ni genome (15) revealed at least 12 REPAT genes (Table S7). Three of the T. ni REPAT genes (Tni20G02130, Tni20G02140, and Tni20G02110) encode isoforms of HMG176 and were highly induced or upregulated upon oral infection with AcMNPV (Table 3; Table S2A and B). One of the HMG176 transcripts (Tni20G02130) was induced dramatically in the midgut at 12 to 48 h p.i. (with an RPKM value of >90 at 12 h p.i.) and upregulated by approximately 80-fold at 72 h p.i. (Table 3; Table S2A and B). Based on the sequence similarities of the 12 identified REPAT genes in the T. ni genome (by comparison with REPAT genes from multiple insect orders [20]), we assigned the T. ni REPAT genes to the phylogenetic groupings identified among the REPAT genes. Of the 12 T. ni REPAT genes, 6 were assigned to group III, 2 to group V, and 1 each to groups I, IV, and VI. One gene (Tni28G00730) was closely similar to a REPAT gene (REPAT27) that was not assigned to one of the 6 phylogenetic groupings (Table S7).
(c) Melanization. Melanization, a component of the insect innate immune response, is involved in encapsulation of invading pathogens and parasites and is also associated with wound repair. Key components in the process of melanization are serine proteases (SPs) that regulate the cleavage of prophenoloxidase (proPO) through a serine protease cascade to generate active phenoloxidase (40, 49). Phenoloxidase mediates the production of melanin, which traps or encapsulates pathogens and parasites, and may close wounds. Serine protease inhibitors regulate the activity of this cascade. We noted reduced levels of two transcripts encoding prophenoloxidase components by 72 h p.i. (Tni16G00190 and Tni16G03510) and one serine protease inhibitor (Tni25G00940), but substantial increases in the transcript levels of three serine protease inhibitors (Tni15G03410, Tni03G03940, and Tni22G04260) were observed (Fig. S1B and Table S6, Melanization). Overall, these varied changes in transcript levels suggest that baculovirus AcMNPV infection may modulate melanization in the T. ni midgut. The downregulation of melanization was previously observed in S. exigua and H. armigera larvae following baculovirus infection (50, 51).
(d) Apoptosis. As an immune response, apoptosis, or programmed cell death, can prevent systemic viral infection by detecting and eliminating infected cells. The triggering and regulation of apoptosis are tightly controlled by a cascade of initiator and effector proteases known as caspases. While baculovirus infections can induce apoptosis in insect cells, baculoviruses are able to block the antiviral apoptotic response of the host (52–54). Baculoviruses encode protein inhibitors of apoptosis, such as P35 and IAP (inhibitor of apoptosis), which may inhibit the initiation or execution of the apoptotic cascade (54–57). We examined the effects of WT AcMNPV infection on the midgut expression patterns of apoptosis-related host genes, including caspase genes, cytochrome c, p53, reaper, and others (Fig. S1C and D and Table S6). We annotated caspase genes according to the orthologs identified in B. mori, D. melanogaster, S. exigua, and S. frugiperda, similar to an earlier study of Tnms42 cells (10). Notable among the apoptosis-associated genes, the transcript levels of 8 genes were substantially upregulated (>4-fold). These upregulated genes included Fas-associated protein with death domain (FADD; Tni22G02980), caspase-4 (Tni20G00950), caspase-6-Dredd (Tni10G01580), apoptosis-inducing factor (aif) (Tni14G00370), p53 (Tni16G04470), caspase-1 (Tni23G04710), and Htra2 (Tni19G03780 and Tni19G03770) (Table S6). In comparison to these observations in the T. ni midgut, AcMNPV infection in the Tnms42 cell line resulted in decreased caspase transcript levels in Tnms42 cells (10). It is unclear why midgut cells respond differently than cultured Tnms42 cells in this regard, but such differences could reflect functional specialization of the midgut cells as a first barrier to infection in the animal.
(e) HSPs. Heat shock proteins (HSPs) are stress-response proteins that are induced following biotic and abiotic stress (58). In Drosophila, loss of the heat shock transcription factor reduced resistance to infection by several viruses, suggesting the importance of heat shock proteins in antiviral defense (59). To examine the effects of AcMNPV infection on HSPs, we analyzed the expression patterns of HSPs in the T. ni midgut after challenge with AcMNPV OBs. The transcript levels of approximately 30% of the HSPs examined increased by >4-fold in AcMNPV-infected midguts compared with uninfected midguts (Table S6 and Fig. S1E and F). However, the transcript levels of eight HSP genes were dramatically (>8-fold) increased at certain times postinfection, and these genes included heat shock binding protein 70 (Tni29G01390), hsp19.5 (Tni05G00200), hsp20.8 (Tni27G00120), hsp70 (Tni08G01440), small heat shock protein 19.7 (Tni27G00160), and small heat shock proteins (Tni05G06500, Tni27G00130, and Tni05G06520) (Table S6). In prior studies of AcMNPV-infected Sf9 cells, an increase in hsp70 levels was observed following infection, and inhibition of the heat shock response resulted in decreased viral DNA replication and the reduced production of infectious BV (60). In addition, hsp70 was dramatically upregulated in the midgut of Helicoverpa zea larvae that were orally infected with Helicoverpa zea single nucleopolyhedrovirus (HzSNPV) (61). Interestingly, HSC70-4, a member of the HSP70 family of proteins, was found in gradient-purified virions of both the BV and ODV of BmNPV and in both the envelope and capsid fractions of BV (62). Thus, while the precise role(s) of hsp70 in baculovirus infections is not clear, prior studies have established the importance of hsp70 during various steps of infection by other viruses (63, 64) and also in association with antiviral responses (65). In addition to hsp70, the other heat shock proteins that were substantially upregulated in response to midgut infection may also play important roles in antiviral and/or antimicrobial defenses in the midgut.
(f) ROS. Reactive oxygen species (ROS) are chemically reactive molecules containing oxygen that are normal metabolic products that may be increased during cellular stress, such as infections by pathogens. ROS directly kill pathogens in the phagolysosomes of innate immune cells and also initiate innate immune signaling (66). Baculovirus infection induces oxidative stress in infected cells (67). For most ROS-related genes examined in this study, we did not observe substantial differences in transcript levels between the infected and uninfected midguts (Table S6 and Fig. S1G). However, some ROS-related genes, such as those for catalase, superoxide dismutase (SOD), and cytochrome P450, showed moderate increases in transcript levels at several times from 18 to 48 h p.i. (Fig. S1G).
(g) Innate immune pathways. To avoid infection by invading bacteria and fungi, insect defenses typically involve innate immunity pathways that include the Toll, JAK/STAT, Jun N-terminal protein kinase (JNK), and IMD pathways. Components of these pathways have also been implicated in antiviral responses, but their roles in antiviral defenses remain unclear and poorly defined (68–70). Immune genes associated with these pathways are typically involved in pathogen recognition, signaling, and the production of antimicrobial peptides that degrade and disrupt pathogens. To examine the effects of AcMNPV infection on midgut expression of these pathways, we selected representative genes associated with the NF-κB–IκB, JAK/STAT, JNK, IMD, and Toll signaling pathways, similar to an earlier study (10), and analyzed the expression patterns of each gene in the uninfected and AcMNPV-infected T. ni midgut. In general, expression of genes associated with the NF-κB–IκB pathway increased following virus infection (Table S6, NF-κB–IκB), with the transcripts of 4 of the 5 genes examined (Tni12G04870, Tni09G01890, Tni12G00240, Tni19G05340) increasing by approximately 3- to 6-fold by 72 h p.i. (Fig. S1H; Table S6). Among the members of JAK/STAT pathway (Fig. S1I), the transcript levels of some genes were modestly upregulated after 12 to 18 h p.i., while others did not appear to change substantially. The more dramatically upregulated of these JAK/STAT pathway genes were Domeless (Tni09G04150), MEKK1 (Tni06G03240), P38B (Tni03G00270), and STAT92E (Tni29G00500), which were upregulated by 4- to 15-fold in the infected midgut compared to the uninfected midgut. Of the 9 JNK pathway-associated genes examined, most transcript levels were moderately upregulated (2- to 4-fold) as infection progressed in the midgut (Fig. S1J; Table S6, JNK). Among the 38 IMD and Toll pathway genes examined, the transcript levels remained mostly unchanged or were moderately increased (Table S6, IMD and Toll). However, AcMNPV infection in the midgut resulted in more dramatic increases for approximately 10 IMD and Toll pathway genes at various times postinfection. These more dramatically upregulated genes were upregulated by approximately 4- to 13-fold and included Toll 7 (Tni23G01350), CG13422 (Tni07G01830), dorsal isoforms (Tni09G01890 and Tni12G00240), GNBP3 isoforms (Tni11G00720 and Tni11G00760), PGRPESA (Tni09G00600 and Tni16G02760), SERPINE27A (Tni07G01400), and TRAF2 (Tni13G00780) (Table S6, IMD and Toll; Fig. S1K and L). The moderate upregulation of these genes in the infected midgut may represent a more generalized response to infection in the midgut but could also represent direct antiviral responses. Additional and more specific studies that specifically target these genes and their gene products will be required to better understand innate immune responses to baculovirus infection in the midgut.
Summary.
Our recent studies of AcMNPV transcription in the midgut and a cell line (11, 14) revealed substantial differences in the transcription of certain viral genes and suggest a midgut-specific program of viral gene expression. The primary stage of baculovirus infection in the lepidopteran midgut is critical for success of the virus, and the responses by the cells of the midgut include important host defensive reactions to infection. The transcriptional responses by the host midgut must certainly encompass a variety of strategies to prevent productive viral replication in the midgut and transmission of the infection to other tissues of the insect host. Because the primary stage of infection by baculoviruses typically involves direct infection and replication in only a subset of the cells of the midgut as a whole, the results reported in this study represent responses by both the directly infected cells and uninfected bystander cells of the midgut. Thus, many of the responses observed here may represent responses to signaling from infected cells, as well as to viral disruption of the normal midgut physiological status. It is also likely that the virus may direct signaling that enhances viral replication and transmission of the virus from the midgut into the tissues of the hemocoel. The biology of the midgut and midgut infection are enormously complex, and it is critically important to understand how the midgut globally responds to viral infection, as well as how viruses potentially manipulate the midgut and enhance their transmission. The global expression data provided here represent an early step toward understanding these complex interactions. As the transcriptome provides only a snapshot of the effects of viral infection on host transcript levels, future studies will need to examine a variety of additional factors, including topics such as virus-induced changes in translation efficiency, posttranslational modifications, and protein turnover. Combined, these and other studies should aid in the utilization of beneficial viruses, such as baculoviruses, in applications in the biological control of important pest insects. In addition, a more detailed understanding of virus-cell interactions in the midgut may also be useful for developing new strategies for inhibiting the insect transmission of problematic viruses.
MATERIALS AND METHODS
Insects and viruses.
Methods for insect rearing, AcMNPV infection, generation of viruses, and preparation of RNA-Seq libraries were described in detail previously (14) and are briefly summarized below. T. ni (Cornell strain) eggs were obtained from the P. Wang laboratory at Cornell University (Geneva, NY). Eggs on wax paper sheets (approximately 4 days old) were surface sterilized by immersion in 10% Clorox, rinsed with sterile deionized water, and air dried. The eggs were then placed in 16-oz cups containing an artificial wheat germ diet and maintained in a growth chamber at 27°C with a light-dark photoperiod of 14 h-10 h as described previously (14). Fourth-instar larvae that were ready to molt were held for 0 to 5 h without diet, and newly molted 5th-instar larvae (0 to 5 h old) were used for oral infections.
Wild-type (WT) AcMNPV strain E2, used for the study, was purified and amplified and its titer was determined (71) as described previously (14). T. ni cell line Tnms42 (an alphanodavirus-free cell line subcloned from High Five, BTI-Tn5B14 cells) (11, 72, 73) was then infected with BV of WT AcMNPV at a multiplicity of infection of 0.1 and maintained in TNM-FH medium (74) (Invitrogen) supplemented with 2.5% fetal bovine serum at 28°C. After 7 days, OBs were collected and purified using 0.5% SDS and 0.5 M NaCl as previously described (71) and resuspended in 10 ml of H2O. As described above for WT AcMNPV, OBs from a recombinant baculovirus carrying a 2nd copy of the viral capsid protein (VP39) that was fused to 3 copies of the mCherry marker gene (virus 3mC) (75) were prepared and fed to larvae to determine the minimum number of OBs required to obtain the maximum midgut cell infection in T. ni larvae. We estimated that the maximal infection rate was approximately 20 to 30% of the midgut cells by 48 h p.i.
Larvae were orally inoculated with occlusion bodies (OBs) of WT AcMNPV by hand feeding 5th-instar larvae with 5 μl of a 10% sucrose solution containing a total of 7 × 104 OBs of WT AcMNPV (1.4 × 104 OBs/μl). Mock-infected control larvae were fed a similar sucrose solution containing no virus. At 1 h postfeeding, control or virus-inoculated larvae (about 30 each) were placed in a growth chamber at 27°C as described above. Midgut tissue was dissected at eight time points postinfection: 0, 6, 12, 18, 24, 36, 48, and 72 h postinfection (p.i.). Thus, for each time point sampled postinfection, a parallel mock-infected control midgut sample was analyzed, to mitigate possible artifacts resulting from developmental changes that may occur over the course of the experiment. For each time point and treatment (infected or control), we prepared three replicate samples, with midgut samples from six larvae pooled for each replicate. For each dissection, the larval cuticle was cut longitudinally along the dorsal midline and pinned in a dissecting dish, and then the midgut was cut anteriorly and posteriorly and removed with forceps, avoiding excessive tracheoles. The excised midgut (which is approximately 70 to 80% of the length of the 5th-instar larva) contained the midgut epithelium plus the closely attached layer of muscle and some attached tracheoles. Excised midgut samples did not contain malpighian tubules, fatbody, or excessive tracheoles or hemocytes. The excised gut was washed by dipping in chilled phosphate-buffered saline (two or more times) and then immediately placed in RNAlater RNA stabilization solution (Ambion) on ice. Pooled midgut samples were stored at −70°C and then homogenized with a pestle in a 1.5-ml Eppendorf tube, and total RNA extraction was performed with the TRIzol reagent (Ambion) according to the manufacturer’s protocol. Following total RNA extraction, midgut RNAs were screened by PCR for a known T. ni tetravirus (data not shown), and only tetravirus-negative midguts were used for experiments.
RNA-Seq library preparation.
Strand-specific RNA-Seq libraries were constructed as described previously (14, 76). The RNA-Seq data may be found in the NCBI SRA under BioProject accession no. PRJNA484772. Briefly, poly(A) mRNAs isolated from 3 μg of total RNA using oligo(dT)25 Dynabeads (Invitrogen) were fragmented at 94°C for 5 min in buffer containing ProtoScript II reaction buffer (New England BioLabs [NEB]), hexamer (Qiagen), and oligo(dT)23 VN (NEB). Subsequently, first-strand cDNA was synthesized using a ProtoScript II kit, and second-strand synthesis was carried out with a reaction mix consisting of RNase H (NEB), the Klenow fragment of DNA polymerase I (NEB), and a deoxynucleoside triphosphate mix containing dATP, dCTP, dGTP, and dUTP (Promega Corporation). Next, TruSeq universal adapters were ligated to the end-repaired and dA-tailed cDNA fragments. The dUTP-containing strands were then removed, and PCR amplification was performed with library-specific TruSeq PCR primers. Following purification and quantification, the libraries were sequenced on an Illumina HiSeq4000 platform at the CLC Genomics and Epigenomics Core Facility at the Weill Cornell Medical College.
RNA-Seq read processing and differential expression analysis.
Raw RNA-Seq reads were processed using Trimmomatic software (77) with default parameters to trim adapter and low-quality sequences, and trimmed reads with lengths of <40 bases were discarded. rRNA reads were removed by aligning the reads to rRNA sequences using the bowtie program (78). Filtered reads were then mapped to the T. ni reference genome (15) using the HISAT aligner (79), allowing 2 mismatches. Based on the alignments, raw read counts of each T. ni gene were derived and normalized to reads per kilobase of transcript per million mapped reads (RPKM) (80). Subsequently, differentially expressed (DE) transcripts between the uninfected and infected midgut samples at different time points postinfection were identified using the edgeR package (81). Transcripts with a false-discovery rate (FDR) lower than 0.05 and a ≥4-fold change in expression levels were classified as DE transcripts.
Functional annotation, Venn diagrams, and cluster analysis.
Annotations of differentially expressed transcripts following virus infection were identified by searching for homologous sequences against the NCBI nonredundant (nr) protein database using the BLASTP program with an E-value cutoff of 10−5. To further obtain gene ontology (GO) terms associated with DE transcripts, we used Blast2GO software (BioBam) (82). Using the GO database, GO terms were assigned to the DE transcripts under a biological process with an E-value hit filter of 10−6 and an annotation cutoff of 55. The GO terms were classified at GO level 4. We constructed Venn diagrams to identify DE transcripts that are commonly expressed between different time points postinfection and also across time points postinfection. For Venn diagram construction, we classified time points into three major groups: early (0, 6, 12, and 18 h p.i.), middle (24 and 36 h p.i.), and late (48 and 72 h p.i.). For each group, we combined induced and upregulated transcripts from each time point. Similarly, we also combined suppressed and downregulated transcripts from each time point. We also performed cluster analysis on the normalized expression levels [log2(RPKM)] of DE transcripts across time points by applying the Euclidean distance metric using DESeq2 software (83). From the list of genes grouped by cluster analysis, we identified T. ni transcripts with specific expression patterns, such as genes with expression levels that increased or decreased consistently over the infection time points.
Identification of most significantly affected T. ni transcripts and expression patterns of immune genes in the midgut.
From the list of differentially expressed transcripts identified in this study, we further identified T. ni transcripts with the most significant changes by setting a threshold of a 16-fold change (16-FC) in expression levels. We included only transcripts with RPKM values of ≥50 for infected (for upregulated or induced) or uninfected (for downregulated or suppressed) midgut samples. Further, in the previous study, homologs of several immune response-specific genes (from yeasts, human, Drosophila melanogaster, Bombyx mori, S. exigua, or S. frugiperda) were identified in Tnms42 cells (10). In the current study, we identified homologs of immune gene transcripts (previously identified in Tnms42 cells) in the T. ni insect reference genome (15) using the BLAST program and analyzed the expression patterns of the immune genes in the T. ni midgut across time points postinfection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yimin Xu for generous help with RNA-Seq libraries, Jenny Xiang for help with Illumina sequencing, and Wendy Kain for T. ni eggs and larvae.
This work was supported by grants from the USDA (2015-67013-23281) to G.W.B., P.W., and Z.F. and from the NSF (IOS-1354421, 1653021) to G.W.B.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00353-19.
REFERENCES
- 1.Rohrmann GF. 2013. Baculovirus molecular biology, 3rd ed National Center for Biotechnology Information (US), Bethesda, MD: https://www.ncbi.nlm.nih.gov/pubmed/24479205. Accessed 2018. [PubMed] [Google Scholar]
- 2.Blissard GW, Theilmann DA. 2018. Baculovirus entry, and egress from insect cells. Annu Rev Virol 5:113–139. doi: 10.1146/annurev-virology-092917-043356. [DOI] [PubMed] [Google Scholar]
- 3.Herniou EA, Arif BM, Becnel JJ, Blissard GW, Bonning B, Harrison R, Jehle JA, Theilmann DA, Vlak JM. 2012. Baculoviridae, p 163–173. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 4.Popham HJ, Nusawardani T, Bonning BC. 2016. Introduction to the use of baculoviruses as biological insecticides. Methods Mol Biol 1350:383–392. doi: 10.1007/978-1-4939-3043-2_19. [DOI] [PubMed] [Google Scholar]
- 5.Federici BA. 1997. Baculovirus pathogenesis In Miller LK. (ed), The baculoviruses. Plenum Press, New York, NY. [Google Scholar]
- 6.Groner A. 1986. Specificity and safety of baculoviruses. The biology of baculoviruses. CRC Press LLC, Boca Raton, FL. [Google Scholar]
- 7.Haas-Stapleton EJ, Washburn JO, Volkman LE. 2003. Pathogenesis of Autographa californica M nucleopolyhedrovirus in fifth instar Spodoptera frugiperda. J Gen Virol 84:2033–2040. doi: 10.1099/vir.0.19174-0. [DOI] [PubMed] [Google Scholar]
- 8.Kirkpatrick BA, Washburn JO, Volkman LE. 1998. AcMNPV pathogenesis and developmental resistance in fifth instar Heliothis virescens. J Invertebr Pathol 72:63–72. doi: 10.1006/jipa.1997.4752. [DOI] [PubMed] [Google Scholar]
- 9.Trudeau D, Washburn JO, Volkman LE. 2001. Central role of hemocytes in Autographa californica M nucleopolyhedrovirus pathogenesis in Heliothis virescens and Helicoverpa zea. J Virol 75:996–1003. doi: 10.1128/JVI.75.2.996-1003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen YR, Zhong S, Fei Z, Gao S, Zhang S, Li Z, Wang P, Blissard GW. 2014. Transcriptome responses of the host, Trichoplusia ni, to infection by the baculovirus, Autographa californica multiple nucleopolyhedrovirus (AcMNPV). J Virol 88:13781–13797. doi: 10.1128/JVI.02243-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen YR, Zhong S, Fei Z, Hashimoto Y, Xiang JZ, Zhang S, Blissard GW. 2013. The transcriptome of the baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) in Trichoplusia ni cells. J Virol 87:6391–6405. doi: 10.1128/JVI.00194-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clem RJ. 2005. The role of apoptosis in defense against baculovirus infection in insects. Curr Top Microbiol Immunol 289:113–129. [DOI] [PubMed] [Google Scholar]
- 13.Jayachandran B, Hussain M, Asgari S. 2012. RNA interference as a cellular defense mechanism against the DNA virus baculovirus. J Virol 86:13729–13734. doi: 10.1128/JVI.02041-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shrestha A, Bao K, Chen YR, Chen W, Wang P, Fei Z, Blissard GW. 2018. Global analysis of baculovirus Autographa californica multiple nucleopolyhedrovirus gene expression in the midgut of the lepidopteran host Trichoplusia ni. J Virol 92:e01277-18. doi: 10.1128/JVI.01277-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen W, Yang X, Tetreau G, Song X, Coutu C, Hegedus D, Blissard G, Fei Z, Wang P. 2019. A high-quality chromosome-level genome assembly of a generalist herbivore, Trichoplusia ni. Mol Ecol Resour 19:485–496. doi: 10.1111/1755-0998.12966. [DOI] [PubMed] [Google Scholar]
- 16.Wang JL, Jiang XJ, Wang Q, Hou LJ, Xu DW, Wang JX, Zhao XF. 2007. Identification and expression profile of a putative basement membrane protein gene in the midgut of Helicoverpa armigera. BMC Dev Biol 7:76. doi: 10.1186/1471-213X-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrero S, Ansems M, Van Oers MM, Vlak JM, Bakker PL, de Maagd RA. 2007. REPAT, a new family of proteins induced by bacterial toxins and baculovirus infection in Spodoptera exigua. Insect Biochem Mol Biol 37:1109–1118. doi: 10.1016/j.ibmb.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez-Marinez P, Navarro-Cerrillo G, Caccia S, de Maagd RA, Moar WJ, Ferre J, Escriche B, Herrero S. 2010. Constitutive activation of the midgut response to Bacillus thuringiensis in Bt-resistant Spodoptera exigua. PLoS One 5:e12795. doi: 10.1371/journal.pone.0012795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navarro-Cerrillo G, Ferre J, de Maagd RA, Herrero S. 2012. Functional interactions between members of the REPAT family of insect pathogen-induced proteins. Insect Mol Biol 21:335–342. doi: 10.1111/j.1365-2583.2012.01139.x. [DOI] [PubMed] [Google Scholar]
- 20.Navarro-Cerrillo G, Hernández-Martínez P, Vogel H, Ferré J, Herrero S. 2013. A new gene superfamily of pathogen-response (repat) genes in Lepidoptera: classification and expression analysis. Comp Biochem Physiol B Biochem Mol Biol 164:10–17. doi: 10.1016/j.cbpb.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Machado V, Serrano J, Galian J. 2016. Identification and characterization of pathogen-response genes (repat) in Spodoptera frugiperda (Lepidoptera: Noctuidae). Folia Biol (Krakow) 64:23–29. doi: 10.3409/fb64_1.23. [DOI] [PubMed] [Google Scholar]
- 22.Dong DJ, He HJ, Chai LQ, Jiang XJ, Wang JX, Zhao XF. 2007. Identification of genes differentially expressed during larval molting and metamorphosis of Helicoverpa armigera. BMC Dev Biol 7:73. doi: 10.1186/1471-213X-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Praefcke GJK. 2018. Regulation of innate immune functions by guanylate-binding proteins. Int J Med Microbiol 308:237–245. doi: 10.1016/j.ijmm.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 24.Verhelst J, Hulpiau P, Saelens X. 2013. Mx proteins: antiviral gatekeepers that restrain the uninvited. Microbiol Mol Biol Rev 77:551–566. doi: 10.1128/MMBR.00024-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Namekawa M, Muriel MP, Janer A, Latouche M, Dauphin A, Debeir T, Martin E, Duyckaerts C, Prigent A, Depienne C, Sittler A, Brice A, Ruberg M. 2007. Mutations in the SPG3A gene encoding the GTPase atlastin interfere with vesicle trafficking in the ER/Golgi interface and Golgi morphogenesis. Mol Cell Neurosci 35:1–13. doi: 10.1016/j.mcn.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Hu J, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz WA, Rapoport TA, Blackstone C. 2009. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138:549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu TH, Dong XL, Pan CX, Du GY, Wu YF, Yang JG, Chen P, Lu C, Pan MH. 2016. A newly discovered member of the Atlastin family, BmAtlastin-n, has an antiviral effect against BmNPV in Bombyx mori. Sci Rep 6:28946. doi: 10.1038/srep28946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang J, Zhang L, Xiang Z, He N. 2010. Expression profile of cuticular genes of silkworm, Bombyx mori. BMC Genomics 11:173. doi: 10.1186/1471-2164-11-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolliopoulou A, Van Nieuwerburgh F, Stravopodis DJ, Deforce D, Swevers L, Smagghe G. 2015. Transcriptome analysis of Bombyx mori larval midgut during persistent and pathogenic cytoplasmic polyhedrosis virus infection. PLoS One 10:e0121447. doi: 10.1371/journal.pone.0121447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. 2013. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. 2013. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terra WR. 2001. The origin and functions of the insect peritrophic membrane and peritrophic gel. Arch Insect Biochem Physiol 47:47–61. doi: 10.1002/arch.1036. [DOI] [PubMed] [Google Scholar]
- 33.Lehane MJ. 1997. Peritrophic matrix structure and function. Annu Rev Entomol 42:525–550. doi: 10.1146/annurev.ento.42.1.525. [DOI] [PubMed] [Google Scholar]
- 34.Wang P, Granados RR. 1997. An intestinal mucin is the target substrate for a baculovirus enhancin. Proc Natl Acad Sci U S A 94:6977–6982. doi: 10.1073/pnas.94.13.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pechan T, Ye L, Chang Y, Mitra A, Lin L, Davis FM, Williams WP, Luthe DS. 2000. A unique 33-kD cysteine proteinase accumulates in response to larval feeding in maize genotypes resistant to fall armyworm and other Lepidoptera. Plant Cell 12:1031–1040. doi: 10.1105/tpc.12.7.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fescemyer HW, Sandoya GV, Gill TA, Ozkan S, Marden JH, Luthe DS. 2013. Maize toxin degrades peritrophic matrix proteins and stimulates compensatory transcriptome responses in fall armyworm midgut. Insect Biochem Mol Biol 43:280–291. doi: 10.1016/j.ibmb.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 37.Devaux PF, Herrmann A, Ohlwein N, Kozlov MM. 2008. How lipid flippases can modulate membrane structure. Biochim Biophys Acta 1778:1591–1600. doi: 10.1016/j.bbamem.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Daleke DL. 2003. Regulation of transbilayer plasma membrane phospholipid asymmetry. J Lipid Res 44:233–242. doi: 10.1194/jlr.R200019-JLR200. [DOI] [PubMed] [Google Scholar]
- 39.Srinivasan A, Giri AP, Gupta VS. 2006. Structural and functional diversities in lepidopteran serine proteases. Cell Mol Biol Lett 11:132–154. doi: 10.2478/s11658-006-0012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao X, He Y, Hu Y, Zhang X, Wang Y, Zou Z, Chen Y, Blissard GW, Kanost MR, Jiang H. 2015. Sequence conservation, phylogenetic relationships, and expression profiles of nondigestive serine proteases and serine protease homologs in Manduca sexta. Insect Biochem Mol Biol 62:51–63. doi: 10.1016/j.ibmb.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calla B, Noble K, Johnson RM, Walden KKO, Schuler MA, Robertson HM, Berenbaum MR. 2017. Cytochrome P450 diversification and hostplant utilization patterns in specialist and generalist moths: birth, death and adaptation. Mol Ecol 26:6021–6035. doi: 10.1111/mec.14348. [DOI] [PubMed] [Google Scholar]
- 42.Noland JE, Breitenbach JE, Popham HJ, Hum-Musser SM, Vogel H, Musser RO. 2013. Gut transcription in Helicoverpa zea is dynamically altered in response to baculovirus infection. Insects 4:506–520. doi: 10.3390/insects4030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ponnuvel KM, Nakazawa H, Furukawa S, Asaoka A, Ishibashi J, Tanaka H, Yamakawa M. 2003. A lipase isolated from the silkworm Bombyx mori shows antiviral activity against nucleopolyhedrovirus. J Virol 77:10725–10729. doi: 10.1128/JVI.77.19.10725-10729.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakazawa H, Tsuneishi E, Ponnuvel KM, Furukawa S, Asaoka A, Tanaka H, Ishibashi J, Yamakawa M. 2004. Antiviral activity of a serine protease from the digestive juice of Bombyx mori larvae against nucleopolyhedrovirus. Virology 321:154–162. doi: 10.1016/j.virol.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 45.Gammon DB, Mello CC. 2015. RNA interference-mediated antiviral defense in insects. Curr Opin Insect Sci 8:111–120. doi: 10.1016/j.cois.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bronkhorst AW, van Cleef KWR, Vodovar N, İnce İA, Blanc H, Vlak JM, Saleh M-C, van Rij RP. 2012. The DNA virus invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc Natl Acad Sci U S A 109:E3604–E3613. doi: 10.1073/pnas.1207213109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu QX, Ueda H, Hirose S. 2000. MBF2 is a tissue- and stage-specific coactivator that is regulated at the step of nuclear transport in the silkworm Bombyx mori. Dev Biol 225:437–446. doi: 10.1006/dbio.2000.9836. [DOI] [PubMed] [Google Scholar]
- 48.Carpenter J, Hutter S, Baines JF, Roller J, Saminadin-Peter SS, Parsch J, Jiggins FM. 2009. The transcriptional response of Drosophila melanogaster to infection with the sigma virus (Rhabdoviridae). PLoS One 4:e6838. doi: 10.1371/journal.pone.0006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanost MR, Gorman MG. 2008. Phenoloxidases in insect immunity, p 69–96. InBeckage N. (ed), Insect immunology. Academic Press/Elsevier, San Diego, CA. [Google Scholar]
- 50.Jakubowska AK, Vogel H, Herrero S. 2013. Increase in gut microbiota after immune suppression in baculovirus-infected larvae. PLoS Pathog 9:e1003379. doi: 10.1371/journal.ppat.1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan C, Xing L, Wang M, Wang X, Yin M, Wang Q, Hu Z, Zou Z. 2017. Inhibition of melanization by serpin-5 and serpin-9 promotes baculovirus infection in cotton bollworm Helicoverpa armigera. PLoS Pathog 13:e1006645. doi: 10.1371/journal.ppat.1006645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clem RJ, Fechheimer M, Miller LK. 1991. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science 254:1388–1390. doi: 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- 53.Lerch RA, Friesen PD. 1993. The 35-kilodalton protein gene p35 of Autographa californica nuclear polyhedrosis virus and the neomycin resistance gene provide dominant selection of recombinant baculoviruses. Nucleic Acids Res 21:1753–1760. doi: 10.1093/nar/21.8.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clem RJ, Popham HJR, Shelby KS. 2010. Antiviral responses in insects: apoptosis and humoral responses, p 453 In Asgari S, Johnson KN (ed), Insect virology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 55.Clem RJ. 2007. Baculoviruses and apoptosis: a diversity of genes and responses. Curr Drug Targets 8:1069–1074. doi: 10.2174/138945007782151405. [DOI] [PubMed] [Google Scholar]
- 56.Byers NM, Vandergaast RL, Friesen PD. 2016. Baculovirus inhibitor-of-apoptosis Op-IAP3 blocks apoptosis by interaction with and stabilization of a host insect cellular IAP. J Virol 90:533–544. doi: 10.1128/JVI.02320-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Means JC, Clem RJ. 2008. Evolution and function of the p35 family of apoptosis inhibitors. Future Virol 3:383–391. doi: 10.2217/17460794.3.4.383. [DOI] [Google Scholar]
- 58.King AM, MacRae TH. 2015. Insect heat shock proteins during stress and diapause. Annu Rev Entomol 60:59–75. doi: 10.1146/annurev-ento-011613-162107. [DOI] [PubMed] [Google Scholar]
- 59.Merkling SH, Overheul GJ, van Mierlo JT, Arends D, Gilissen C, van Rij RP. 2015. The heat shock response restricts virus infection in Drosophila. Sci Rep 5:12758. doi: 10.1038/srep12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lyupina YV, Dmitrieva SB, Timokhova AV, Beljelarskaya SN, Zatsepina OG, Evgen'ev MB, Mikhailov VS. 2010. An important role of the heat shock response in infected cells for replication of baculoviruses. Virology 406:336–341. doi: 10.1016/j.virol.2010.07.039. [DOI] [PubMed] [Google Scholar]
- 61.Breitenbach JE, Popham HJ. 2013. Baculovirus replication induces the expression of heat shock proteins in vivo and in vitro. Arch Virol 158:1517–1522. doi: 10.1007/s00705-013-1640-8. [DOI] [PubMed] [Google Scholar]
- 62.Iwanaga M, Shibano Y, Ohsawa T, Fujita T, Katsuma S, Kawasaki H. 2014. Involvement of HSC70-4 and other inducible HSPs in Bombyx mori nucleopolyhedrovirus infection. Virus Res 179:113–118. doi: 10.1016/j.virusres.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 63.Baquero-Pérez B, Whitehouse A. 2015. Hsp70 isoforms are essential for the formation of Kaposi's sarcoma-associated herpesvirus replication and transcription compartments. PLoS Pathog 11:e1005274. doi: 10.1371/journal.ppat.1005274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taguwa S, Maringer K, Li X, Bernal-Rubio D, Rauch JN, Gestwicki JE, Andino R, Fernandez-Sesma A, Frydman J. 2015. Defining Hsp70 subnetworks in dengue virus replication reveals key vulnerability in flavivirus infection. Cell 163:1108–1123. doi: 10.1016/j.cell.2015.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim MY, Ma Y, Zhang Y, Li J, Shu Y, Oglesbee M. 2013. hsp70-dependent antiviral immunity against cytopathic neuronal infection by vesicular stomatitis virus. J Virol 87:10668–10678. doi: 10.1128/JVI.00872-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schieber M, Chandel NS. 2014. ROS function in redox signaling and oxidative stress. Curr Biol 24:R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y, Oberley LW, Murhammer DW. 2001. Evidence of oxidative stress following the viral infection of two lepidopteran insect cell lines. Free Radic Biol Med 31:1448–1455. doi: 10.1016/S0891-5849(01)00728-6. [DOI] [PubMed] [Google Scholar]
- 68.Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, Hoffmann JA, Imler JL. 2005. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat Immunol 6:946–953. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- 69.Lemaitre B, Hoffmann J. 2007. The host defense of Drosophila melanogaster. Annu Rev Immunol 25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 70.Avadhanula V, Weasner BP, Hardy GG, Kumar JP, Hardy RW. 2009. A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathog 5:e1000582. doi: 10.1371/journal.ppat.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O'Reilly DR, Miller LK, Luckow VA. 1992. Baculovirus expression vectors, a laboratory manual. WH Freeman & Co, New York, NY. [Google Scholar]
- 72.Granados RR, Li GX, Derksen ACG, Mckenna KA. 1994. A new insect cell line from Trichoplusia ni (BTI-Tn-5B1-4) susceptible to Trichoplusia ni single enveloped nuclear polyhedrosis virus. J Invertebr Pathol 64:260–266. doi: 10.1016/S0022-2011(94)90400-6. [DOI] [Google Scholar]
- 73.Koczka K, Peters P, Ernst W, Himmelbauer H, Nika L, Grabherr R. 2018. Comparative transcriptome analysis of a Trichoplusia ni cell line reveals distinct host responses to intracellular and secreted protein products expressed by recombinant baculoviruses. J Biotechnol 270:61–69. doi: 10.1016/j.jbiotec.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 74.Hink WF. 1970. Established insect cell line from the cabbage looper, Trichoplusia ni. Nature 226:466–467. doi: 10.1038/226466b0. [DOI] [PubMed] [Google Scholar]
- 75.Ohkawa T, Volkman LE, Welch MD. 2010. Actin-based motility drives baculovirus transit to the nucleus and cell surface. J Cell Biol 190:187–195. doi: 10.1083/jcb.201001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhong S, Joung JG, Zheng Y, Chen YR, Liu B, Shao Y, Xiang JZ, Fei Z, Giovannoni JJ. 2011. High-throughput Illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc 2011:940–949. doi: 10.1101/pdb.prot5652. [DOI] [PubMed] [Google Scholar]
- 77.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Langmead B. 2010. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics Chapter 11:Unit 11.7. doi: 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 81.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 83.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.