Abstract
The Teosinte-branched 1/Cycloidea/Proliferating (TCP) plant-specific transcription factors (TFs) have been demonstrated to play a fundamental role in plant development and organ patterning. However, it remains unknown whether or not the TCP gene family plays a role in conferring a tolerance to drought stress in maize, which is a major constraint to maize production. In this study, we identified 46 ZmTCP genes in the maize genome and systematically analyzed their phylogenetic relationships and synteny with rice, sorghum, and Arabidopsis TCP genes. Expression analysis of the 46 ZmTCP genes in different tissues and under drought conditions, suggests their involvement in maize response to drought stress. Importantly, genetic variations in ZmTCP32 and ZmTCP42 are significantly associated with drought tolerance at the seedling stage. RT-qPCR results suggest that ZmTCP32 and ZmTCP42 RNA levels are both induced by ABA, drought, and polyethylene glycol treatments. Based on the significant association between the genetic variation of ZmTCP42 and drought tolerance, and the inducible expression of ZmTCP42 by drought stress, we selected ZmTCP42, to investigate its function in drought response. We found that overexpression of ZmTCP42 in Arabidopsis led to a hypersensitivity to ABA in seed germination and enhanced drought tolerance, validating its function in drought tolerance. These results suggested that ZmTCP42 functions as an important TCP TF in maize, which plays a positive role in drought tolerance.
Keywords: maize, ZmTCP, natural variation, drought tolerance
1. Introduction
Severe drought causes a grievous decline in crop yield by negatively affecting plant growth and reproduction [1,2]. Maize (Zea mays L.) is considered a major crop for food, feed, and fuel, but its production is frequently hampered by water scarcity [3,4]. While the molecular mechanisms of drought stress response in plants have not been fully elucidated, many transcription factors (TFs) such as DREB1/CBF, MYB, and AREB/ABF, which regulate drought-responsive genes have been well-studied [5,6,7]. These reports support the idea that identification of key TFs will help us to better understand the molecular and cellular responses to drought stress.
Teosinte branched 1/Cycloidea/Proliferating (TCP) genes encode plant-specific TFs and are named after the first three functionally characterized members of this TF family—Teosinte Branched 1 (TB1) in maize (Zea mays L.), Cycloidea (CYC) in snapdragon (Antirrhinum majus), and Proliferating Cell nuclear antigen Factor (PCF) in rice (Oryza sativa) [8]. This class of TFs share a highly conserved TCP domain, which contains a 59-amino acid, non-canonical basic-Helix-Loop-Helix (bHLH) structural motif that allows DNA binding, protein–protein interaction, and protein nuclear localization [8,9,10]. More than 20 TCP TF members were identified in various plant species, such as Arabidopsis, rice, tomato, cotton, sorghum, and wheat [11,12,13,14,15,16,17]. Based on the TCP domain, these genes were divided into two classes. Class I is the PCF class; class II is subdivided into two clades—CIN (CINCINNATA of Antirrhinum) and CYC/TB1 [10,15,18]. Many members of the TCP family have been shown to be involved in the regulation of many biological processes during plant growth and development, such as leaf development, branching, floral organ morphogenesis, and senescence [8,19,20,21,22]. Possible mechanisms for the regulation were studied. These mechanisms were found to involve either a direct transcriptional control of the cell cycle genes by TCP TFs, or an indirect adjustment of the hormone activity [23]. For example, in Arabidopsis seeds, the DELLA proteins GAI (GA-Insensitive) and RGA (Repressor of GA) formed an unproductive complex with the class II TCP protein AtTCP14 or AtTCP15, which prevented the binding of TCPs to promoters of the core cell cycle genes [24]. In turn, GA might induce ubiquitination and degradation of DELLA proteins to relieve the constitutive inhibition of TCP transcriptional activity [24]. Interestingly, the orthologous TCP protein LANCEOLATE was found to participate in GA biosynthesis by upregulating the SIGA-oxidase 1 gene [25]. It was also found that a subset of class I TCP proteins, such as AtTCP1, AtTCP2, AtTCP3, and AtTCP14, behaved in a similar fashion by acting as an inducer or repressor to influence the biosynthesis or signaling of several hormones during different developmental processes [26,27,28].
Abscisic acid (ABA) is a known stress response hormone that can mitigate physiological and environmental stresses, including drought stress, by inducing the closure of stomata, thus reducing water loss [29,30,31,32]. Although the TCP gene family is primarily involved in the regulation of plant growth and leaf development, other functions, such as involvement in the ABA signaling pathway, have been noted for specific TCP genes. For example, overexpression of OsTCP19 in Arabidopsis significantly conferred both drought and heat tolerance during seedling establishment and in mature plants [33]. Furthermore, the interaction of OsTCP19 with OsABI4, which encodes a TF involved in the ABA signal transduction, suggests its function in fine-tuning drought-induced ABA signaling [33]. Additionally, creeping bentgrass plants (Agrostis stolonifera) that overexpresses Osa-miR319, in which four putative target genes, AsPCF5, AsPCF6, AsPCF8, and AsTCP14 are down-regulated, significantly enhance plant tolerance to salt and drought stress associated with an increased leaf wax content and water retention [34]. It is noteworthy that AtTCP14 antagonizes ABA signaling by interacting with the DOF6 (DNA binding with one finger) TF, preventing the activation of the downstream ABA biosynthetic gene ABA1 (ABA deficient 1) and other ABA-responsive genes in Arabidopsis seeds [35]. In contrast, AtTCP18, also known as BRC1 (Branched 1), induces ABF3 (ABA responsive elements-binding factor 3), and ABI5 (ABA insensitive 5)—two key regulators of the ABA response—to maintain ABA signaling when the axillary buds enter dormancy [36,37]. These reports indicate a strong association of TCPs with ABA-mediated abiotic stress signaling. However, how the TCP TF genes function in maize, especially in response to drought stress, still remains to be elucidated. Due to the rapid linkage disequilibrium (LD) decay in the maize genome, association study is able to provide a gene-level resolution, which facilitates the genetic detection of several complex traits, such as drought tolerance [38,39,40]. However, limited allelic variations underlying drought tolerance have been identified [41], and rarely favorable alleles could be used for the genetic improvement of drought tolerance in maize.
Here, we comprehensively analyzed the ZmTCP gene family in the maize genome. Previous research has identified 29 TCP genes in the maize genome and subsequently, phylogeny, gene structure, chromosomal location, gene duplication, and expression levels of the 29 ZmTCP genes were investigated [42]. In this study, we searched against the updated maize genome B73_RefGen_v3 and identified 46 ZmTCP genes, and further systematically analyzed to determine their phylogenetic relationships and synteny with rice, sorghum, and Arabidopsis TCPs. In addition, we studied the expression profiles of these ZmTCPs upon drought stress. Importantly, a family-based, genome-wide association study revealed a significant association between the natural variations of ZmTCP42 and maize drought tolerance. Ectopic expression of ZmTCP42 in Arabidopsis led to enhanced drought tolerance, validating its function in drought tolerance.
2. Results
2.1. The Maize Genome Contains 46 TCP Family Genes
We comprehensively performed a genome-wide search for putative TCP genes in maize. Initially, the protein sequences of the putative maize TCP TFs were retrieved from the Plant Transcription Factor Database 3.0 (available online: http://planttfdb.cbi.pku.edu.cn). Subsequently, a BLASTP analysis was performed in Phytozome V10 (available online: http://www.phytozome.net/eucalyptus.php), using all of the Arabidopsis and rice TCP protein sequences as queries, and the predicted protein sequences without a TCP domain (Pfam: PF03634) were excluded from the results. A Hidden Markov Model (HMM) search was also performed against the maize database using PF03634. Ultimately, 46 genes, including the 29 ZmTCPs previously reported by Chai et al. [42], were retrieved and further verified for the presence of a canonical TCP domain. The 17 newly identified ZmTCPs were named ZmTCP30 to ZmTCP46. Their locus IDs, genome locations, coding sequence (CDS) lengths, and protein lengths are listed in Table 1. The 46 ZmTCP genes were unevenly distributed throughout the maize genome on 10 chromosomes, without any clustering. The 46 ZmTCP proteins contained a range of 98 to 778 amino acids, corresponding to 11.76 kDa to 84.78 kDa in molecular weight (Table 1). Their theoretical pI (isoelectric point) values ranged from 5.13 to 12.23, with a mean of 8.14 (Table 1), indicating that most of them were weakly alkaline (Table 1).
Table 1.
Detailed information for 46 ZmTCP genes in the Zea mays L. genome.
| Gene Name | Locus ID | Chr | CDS Length (bp) | Protein Length (aa) | Number of Exons | MW | pI | Class |
|---|---|---|---|---|---|---|---|---|
| ZmTCP1 | GRMZM2G166687 | 1 | 723 | 240 | 1 | 26.11 | 5.15 | CYC/TB1 |
| ZmTCP2 | AC233950.1_FG002 | 1 | 1131 | 376 | 1 | 39.86 | 7.8 | CYC/TB1 |
| ZmTCP3 | GRMZM2G115516 | 1 | 1161 | 386 | 2 | 39.36 | 9.33 | CIN |
| ZmTCP4 | AC234521.1_FG006 | 2 | 768 | 255 | 2 | 27.71 | 10.62 | CYC/TB1 |
| ZmTCP5 | GRMZM2G110242 | 2 | 831 | 276 | 1 | 29.9 | 6.26 | CYC/TB1 |
| ZmTCP6 | GRMZM2G088440 | 2 | 549 | 182 | 5 | 21.14 | 8.65 | CYC/TB1 |
| ZmTCP7 | GRMZM2G414114 | 2 | 1104 | 367 | 6 | 42.29 | 8.44 | CYC/TB1 |
| ZmTCP8 | GRMZM2G020805 | 2 | 1323 | 440 | 2 | 46.22 | 9.46 | CIN |
| ZmTCP9 | AC205574.3_FG006 | 3 | 1698 | 565 | 2 | 58.83 | 7.1 | CIN |
| ZmTCP10 | GRMZM2G166946 | 3 | 852 | 283 | 2 | 30.48 | 8.98 | CIN |
| ZmTCP11 | GRMZM2G055024 | 4 | 1263 | 420 | 3 | 45.5 | 10.82 | CYC/TB1 |
| ZmTCP12 | GRMZM2G062711 | 4 | 564 | 187 | 1 | 21.58 | 12.05 | CIN |
| ZmTCP13 | GRMZM2G060319 | 4 | 825 | 274 | 1 | 29.35 | 5.96 | CYC/TB1 |
| ZmTCP14 | GRMZM2G135461 | 4 | 402 | 132 | 3 | 15.09 | 7.32 | CYC/TB1 |
| ZmTCP15 | GRMZM2G078077 | 4 | 657 | 218 | 1 | 22.97 | 10.02 | PCF |
| ZmTCP16 | GRMZM2G003944 | 4 | 1164 | 387 | 1 | 40.76 | 9.33 | PCF |
| ZmTCP17 | GRMZM2G089361 | 5 | 1146 | 381 | 1 | 39.05 | 9.58 | CIN |
| ZmTCP18 | AC190734.2_FG003 | 5 | 1080 | 359 | 1 | 38.89 | 7.01 | CYC/TB1 |
| ZmTCP19 | GRMZM2G445944 | 5 | 624 | 207 | 1 | 21.12 | 10.26 | PCF |
| ZmTCP20 | GRMZM2G031905 | 6 | 435 | 144 | 3 | 16.01 | 9.89 | CIN |
| ZmTCP21 | GRMZM2G142751 | 6 | 1140 | 379 | 1 | 39.09 | 8.02 | PCF |
| ZmTCP22 | GRMZM2G120151 | 6 | 897 | 298 | 2 | 30.9 | 8.07 | CIN |
| ZmTCP23 | GRMZM2G064628 | 7 | 843 | 280 | 1 | 30.23 | 6.33 | CYC/TB1 |
| ZmTCP24 | GRMZM2G015037 | 8 | 1974 | 657 | 4 | 69.55 | 6.61 | CIN |
| ZmTCP25 | GRMZM2G035944 | 8 | 828 | 275 | 2 | 29.59 | 9.02 | CIN |
| ZmTCP26 | GRMZM2G113888 | 9 | 1209 | 402 | 1 | 40.66 | 8.81 | PCF |
| ZmTCP27 | GRMZM2G096610 | 10 | 531 | 176 | 1 | 18.33 | 7.43 | PCF |
| ZmTCP28 | GRMZM2G458087 | 10 | 495 | 164 | 3 | 18.83 | 8.37 | CYC/TB1 |
| ZmTCP29 | GRMZM2G465091 | 10 | 624 | 207 | 1 | 21.01 | 10.01 | PCF |
| ZmTCP30 | GRMZM2G454571 | 1 | 399 | 132 | 3 | 15.11 | 7.27 | CYC/TB1 |
| ZmTCP31 | AC213524.3_FG003 | 1 | 831 | 276 | 2 | 29.56 | 9.83 | PCF |
| ZmTCP32 | GRMZM2G107031 | 1 | 972 | 323 | 2 | 34.02 | 5.98 | PCF |
| ZmTCP33 | GRMZM2G416524 | 2 | 894 | 297 | 3 | 32.37 | 10.43 | PCF |
| ZmTCP34 | AC199782.5_FG003 | 2 | 1032 | 343 | 1 | 35.43 | 5.13 | PCF |
| ZmTCP35 | GRMZM5G824514 | 3 | 297 | 98 | 1 | 11.76 | 12.23 | CYC/TB1 |
| ZmTCP36 | GRMZM2G089638 | 3 | 1251 | 416 | 1 | 42.91 | 5.79 | PCF |
| ZmTCP37 | GRMZM2G092214 | 3 | 975 | 324 | 2 | 34.35 | 6.7 | PCF |
| ZmTCP38 | GRMZM2G359599 | 4 | 522 | 173 | 4 | 19.58 | 8.6 | CYC/TB1 |
| ZmTCP39 | GRMZM2G170232 | 4 | 396 | 131 | 4 | 14.38 | 8.87 | CYC/TB1 |
| ZmTCP40 | GRMZM2G178603 | 5 | 663 | 220 | 1 | 22.76 | 10.02 | PCF |
| ZmTCP41 | GRMZM2G077755 | 5 | 1200 | 400 | 1 | 40.49 | 9.2 | PCF |
| ZmTCP42 | GRMZM2G180568 | 7 | 972 | 323 | 1 | 33.71 | 8.11 | CIN |
| ZmTCP43 | GRMZM2G148022 | 8 | 2337 | 778 | 15 | 84.78 | 7.35 | CIN |
| ZmTCP44 | GRMZM2G034638 | 8 | 948 | 315 | 2 | 33.08 | 6.57 | PCF |
| ZmTCP45 | GRMZM2G424261 | 10 | 489 | 162 | 3 | 17.27 | 5.91 | CIN |
| ZmTCP46 | GRMZM2G093895 | 10 | 1218 | 405 | 1 | 41.09 | 5.95 | PCF |
2.2. Maize Contains Roughly Twice as Many TCP Genes as Rice and Sorghum
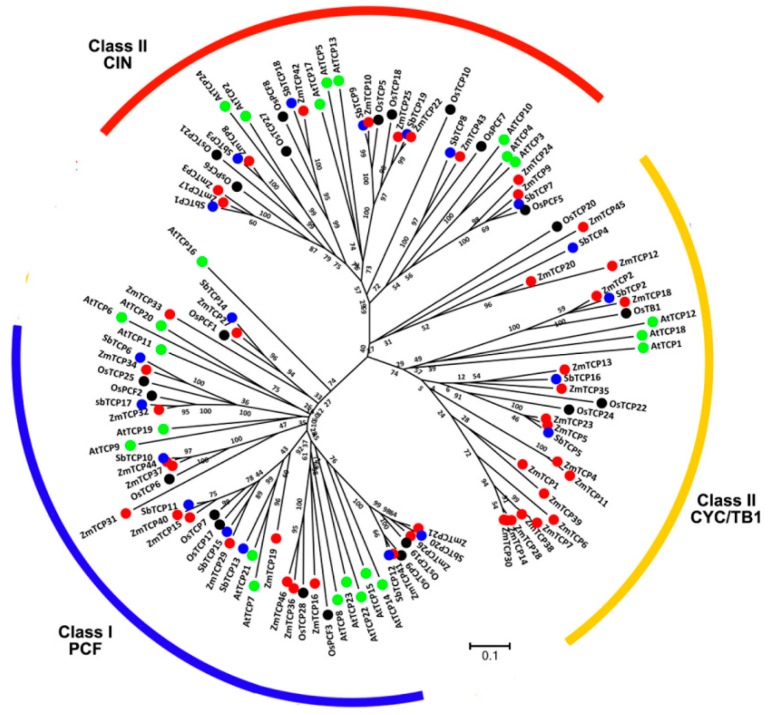
In order to study the phylogenetic relationships among the predicted maize TCPs, we constructed a neighbor-joining tree for the ZmTCP proteins and their orthologs from rice, sorghum and Arabidopsis, based on multiple-alignment of the full-length TCP protein sequences (Figure 1). As shown on the phylogenetic tree, the 113 TCPs were classified into two main classes—class I and class II. Class II was further divided into two clades—CYC/TB1 and CIN. The phylogenetic tree based on the sequence alignments of the 46 ZmTCPs was also divided into three clades (Figure S1). The boundaries of these major clades were clearly stated by the phylogenetic locations of several canonical TCP genes, such as the class I genes OsPCF1 and OsPCF2, the CIN-like class II genes AtTCP2, AtTCP3, and AtTCP4, and the CYC/tb1-like class II genes AtTCP1, AtTCP12, AtTCP18, and ZmTCP2/ZmTB1. In agreement with previous work, all Arabidopsis, rice, and sorghum TCPs fell in the same class or clade, as previously reported in our phylogenetic tree [11,12]. Interestingly, dicot Arabidopsis TCP proteins clustered separately from those of the three monocot plants in the same clad, and some proteins from three of the monocot plants displayed pairing. Furthermore, maize and sorghum TCPs were found to share a closer phylogenetic relationship than maize and rice ones, which was consistent with the notion that sorghum is a closer relative of maize than rice. Throughout the phylogenetic tree, there were 49 TCPs in the PCF clade, 34 in CIN, and 30 in CYC/TB1 (Figure 1). Among the 46 ZmTCPs, there were 17 ZmTCPs in class I and 29 ZmTCPs in class II; within class II, 10 in CIN and 19 in CYC/TB1 (Figure S1). Significantly, the number of maize ZmTCPs was roughly twice as large as the number in each of rice and sorghum. Specifically, among all 30 CYC/TB1 TCP genes, 19 were from maize, 4 from sorghum, and 3 each from rice and Arabidopsis (Figure 1). This result suggests that the ZmTCP gene family in the allopolyploid maize genome underwent a two-fold duplication. This expansion was biased and occurred mainly in the class II CYC/TB1 clade.
Figure 1.
Phylogenetic tree of the predicted Teosinte-branched 1/Cycloidea/Proliferating (TCP) proteins from maize, rice, sorghum, and Arabidopsis. The phylogenetic tree was constructed based on the sequence alignment of the 113 full-length TCP protein sequences from four species. The unrooted tree was drawn by MEGA 7.0 with the neighbor-joining (NJ) method, using the following parameters—bootstrap values (1000 replicates) and the poisson model. The scale refers to the branch lengths. The gene codes and names are illustrated in red for maize, black for rice, blue for sorghum, and green for Arabidopsis. The names used for rice and the Arabidopsis TCP genes are from a previous report [8].
2.3. Many TCP Genes are Found in the Syntenic Segments of Rice, Sorghum, and Maize Genomes
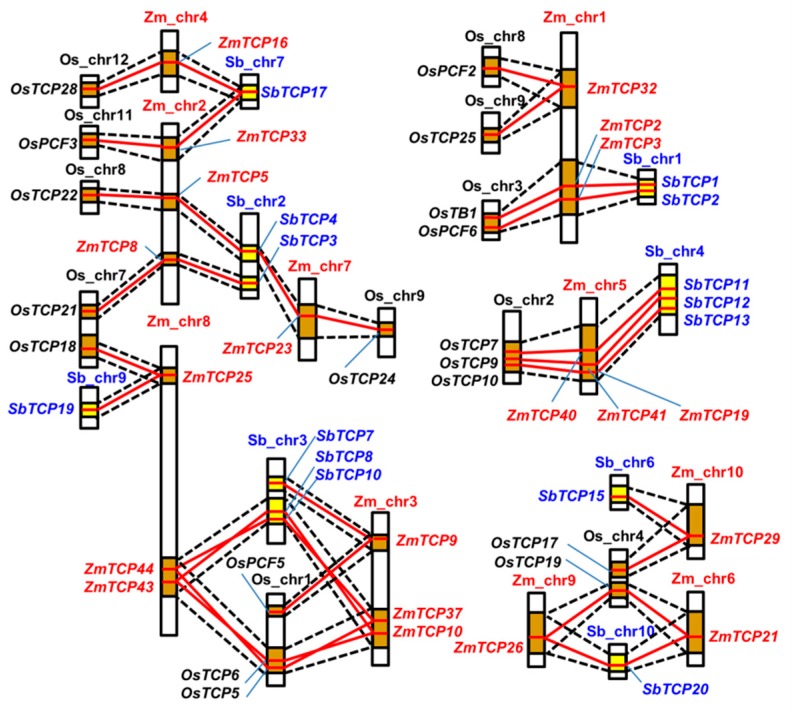
As gene synteny is indicative of the homologous gene function, we explored the collinearity of rice, sorghum, and maize TCP genes. We collected gene collinearity data from the Plant Genome Duplication Database (PGDD, http://chibba.agtec.uga.edu/duplication, Table S1) using ZmTCP genes as anchors, then defined each genomic syntenic block as the chromosomal segment consisting of multiple homologous genes, across species. According to this analysis, about 20 ZmTCP genes were found to have syntenic members or collinear genes in rice and sorghum, as shown in Figure 2. Some chromosomal segments containing ZmTCP genes, including ZmTCP2, 3, 8, 9, 19, 25, 29, 40, and ZmTCP41, were found to have been evolutionally conserved between maize, rice, and sorghum (Figure 2). This indicated that not only the individual genes but these entire chromosomal segments were evolutionally conserved. Two segments on maize chromosomes 2 and 7, containing a duplicated gene pair ZmTCP5 and ZmTCP23, share synteny with two segments on rice chromosomes 7 and 9, carrying the corresponding genes OsTCP22 and OsTCP24. Two other segments on maize chromosomes 2 and 4, containing another duplicated gene pair ZmTCP33 and ZmTCP16, share synteny with two segments on rice chromosomes 11 and 12, carrying corresponding genes OsPCF3 and OsTCP28. Interestingly, these two paralogous gene pairs, ZmTCP5/ZmTCP23 and ZmTCP16/ZmTCP33, share the same syntenic blocks on sorghum chromosomes 2 and 7, respectively, indicating that these duplicated gene pairs might have arisen from segmental duplication in maize, after maize and sorghum diverged evolutionarily (Figure 2). One rice chromosomal block, containing OsTCP19, shares synteny with two segments on maize chromosomes 6 and 9, with a duplicated gene pair ZmTCP21 and ZmTCP26. However, only one syntenic block could be found in the sorghum genome. Another fragment on rice chromosome 1 containing OsTCP6 and OsTCP5 is also duplicated on maize chromosomes 3 and 8, carrying two duplicated gene pairs ZmTCP10/43 and ZmTCP37/44. More interestingly, their orthologs in sorghum are also tandemly duplicated on only a single chromosomal segment. Additionally, one maize chromosomal block containing ZmTCP32 shares synteny with two segments on rice chromosomes 8 and 9, containing its orthologous genes OsPCF2 and OsTCP25; SbTCP orthologs could not be found, although the syntenic segment in sorghum was identified. Based on these data, we concluded that most of the TCP genes existed before the species diverged, but some ZmTCP genes might have originated from later segmental duplication or accompanied the generation of an allotetraploid maize genome.
Figure 2.
Schematic diagram of syntenic chromosomal segments containing ZmTCP genes between the rice, sorghum, and maize genomes. The maize, rice, and sorghum chromosomes are abbreviated Zm, Os, Sb, respectively. Homologous chromosomal regions between the different genomes are linked by black dotted lines and pale blue shaded regions. Each TCP orthologous gene pair was connected by a red line. Yellow boxes indicate homologous segments between the maize and the sorghum genomes, while the brown boxes identify the homologous regions in the maize and rice genomes.
2.4. Expression Profiles of ZmTCP Genes
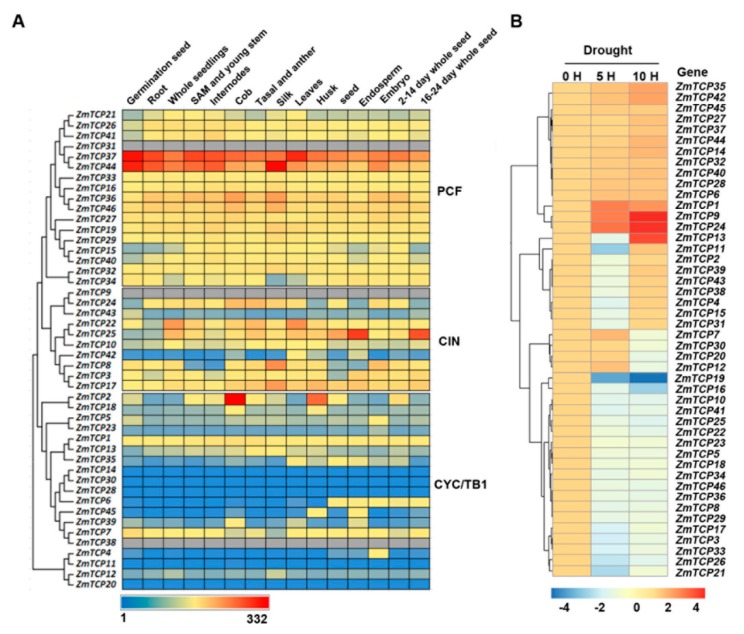
According to previous studies, TCP genes had key roles in different aspects of plant development, as well as in response to stress [8,37]. In order to gain further insights into the roles of the ZmTCP genes, we assessed their tissue-specific expression profiles from the available transcriptomic data of maize B73 [43]. An expression heatmap was constructed for the 46 ZmTCPs in different tissues, from 15 developmental stages, under non-limiting growth conditions (Figure 3A). Results indicated that the expression patterns of different ZmTCP genes varied greatly. Transcripts of ZmTCP37 and ZmTCP44 showed relatively high levels of expression, compared to the other ZmTCPs examined. It is noteworthy that ZmTCP2/TB1 and ZmTCP18, a homolog of OsTB1, were expressed relatively high in cobs and husk leaves, while ZmTCP25 was expressed relatively high in endosperm and mature seeds. Additionally, in the B73 variety grown under non-limiting conditions, all ZmTCP genes in class I (PCF clade) were expressed relatively high in different tissues.
Figure 3.
Expression patterns of ZmTCP genes in tissues and in response to drought conditions. (A) A heat map depicting gene expression levels of 46 ZmTCPs in fifteen different tissues from various developmental stages. Normalized gene expression values are shown in different colors that represent the levels of expression indicated by the scale bar. The gray color represents unavailable data. (B) Microarray-based expression analysis of ZmTCP genes. A heat map was generated based on the fold-change values in the treated samples, when compared with the unstressed control. The color scale for fold-change values is shown at the bottom. The drought-treated leaf samples were collected at two time points, 5 and 10 h, which reflected relative leaf water content (RLWC) of 70% and 60%, respectively.
Next, we investigated the expression profiles of all maize TCP genes, in response to drought stress, using microarray analysis. Based on their expression patterns, the maize TCP genes could mainly be classified into four groups (Figure 3B). The expression levels of 14 ZmTCP genes were continuously up-regulated (fold-change > 1) in response to both drought stress conditions (5 h and 10 h of drought treatments). Among these genes, ZmTCP1, ZmTCP9, and ZmTCP24 showed two-fold changes (or more) in one or two drought stress conditions. In contrast, 19 TCP genes were continuously down-regulated (fold-change < −1) under both drought stress conditions, of which ZmTCP19 showed about four fold change in one or two drought stress conditions. Expression of the remaining 13 genes, were either suppressed or induced under one of the drought conditions. The results clearly showed the functional divergence of ZmTCP genes in response to drought stress, in maize seedlings.
2.5. Association Analysis of Natural Variations in ZmTCP Genes Identified Two ZmTCP Genes Associated with Drought Tolerance in Maize
In order to further investigate whether the natural variations in any of the ZmTCP TFs are associated with the different drought tolerance levels of maize varieties, we conducted an association analysis for these genes. The drought tolerance level of each inbred line was investigated by evaluating its survival rate under severe drought stress, at the seedling stage. To assess potential associations between survival rates and ZmTCPs, we utilized previously reported methods and data [44,45] and previously identified single nucleotide polymorphism (SNP) markers, to characterize the presence of genetic polymorphisms in each of these 46 ZmTCP genes. Among the 46 identified ZmTCP genes, 26 were found to be polymorphic with an average of 12 SNPs (Table 2), while the polymorphic information of the other 16 genes was currently absent (Minor Allele Frequency, MAF ≥ 0.05).
Table 2.
Association analysis of the natural variation in ZmTCP genes with respect to drought tolerance at the seedling stage in the maize diversity panel.
| Locus ID | Gene Name | Polymorphic Name | GLM | PCA | PCA + K | |
|---|---|---|---|---|---|---|
| p ≤ 0.01 | p ≤ 0.01 | p ≤ 0.01 | p ≤ 0.001 | |||
| GRMZM2G166687 | ZmTCP1 | - | - | - | - | - |
| AC233950.1_FG002 | ZmTCP2 | 3 | 2 | 0 | 0 | 0 |
| GRMZM2G115516 | ZmTCP3 | 33 | 0 | 0 | 0 | 0 |
| AC234521.1_FG006 | ZmTCP4 | - | - | - | - | - |
| GRMZM2G110242 | ZmTCP5 | 1 | 0 | 0 | 0 | 0 |
| GRMZM2G088440 | ZmTCP6 | - | - | - | - | - |
| GRMZM2G414114 | ZmTCP7 | - | - | - | - | - |
| GRMZM2G020805 | ZmTCP8 | 21 | 0 | 0 | 0 | 0 |
| AC205574.3_FG006 | ZmTCP9 | 19 | 0 | 0 | 0 | 0 |
| GRMZM2G166946 | ZmTCP10 | 1 | 0 | 0 | 0 | 0 |
| GRMZM2G055024 | ZmTCP11 | - | - | - | - | - |
| GRMZM2G062711 | ZmTCP12 | - | - | - | - | - |
| GRMZM2G060319 | ZmTCP13 | 1 | 0 | 0 | 0 | 0 |
| GRMZM2G135461 | ZmTCP14 | - | - | - | - | - |
| GRMZM2G078077 | ZmTCP15 | 5 | 0 | 0 | 0 | 0 |
| GRMZM2G003944 | ZmTCP16 | - | - | - | - | - |
| GRMZM2G089361 | ZmTCP17 | 28 | 1 | 0 | 0 | 0 |
| AC190734.2_FG003 | ZmTCP18 | - | - | - | - | - |
| GRMZM2G445944 | ZmTCP19 | 7 | 0 | 0 | 0 | 0 |
| GRMZM2G031905 | ZmTCP20 | - | - | - | - | - |
| GRMZM2G142751 | ZmTCP21 | 2 | 0 | 0 | 0 | 0 |
| GRMZM2G120151 | ZmTCP22 | 2 | 0 | 0 | 0 | 0 |
| GRMZM2G064628 | ZmTCP23 | - | - | - | - | - |
| GRMZM2G015037 | ZmTCP24 | 2 | 0 | 0 | 0 | 0 |
| GRMZM2G035944 | ZmTCP25 | 27 | 1 | 0 | 0 | 0 |
| GRMZM2G113888 | ZmTCP26 | 4 | 0 | 0 | 0 | 0 |
| GRMZM2G096610 | ZmTCP27 | 12 | 0 | 0 | 0 | 0 |
| GRMZM2G458087 | ZmTCP28 | - | - | - | - | - |
| GRMZM2G465091 | ZmTCP29 | 1 | 0 | 0 | 0 | 0 |
| GRMZM2G454571 | ZmTCP30 | - | - | - | - | - |
| AC213524.3_FG003 | ZmTCP31 | - | - | - | - | - |
| GRMZM2G107031 | ZmTCP32 | 19 | 0 | 1 | 1 | 0 |
| GRMZM2G416524 | ZmTCP33 | - | - | - | - | - |
| AC199782.5_FG003 | ZmTCP34 | 12 | 0 | 0 | 0 | 0 |
| GRMZM5G824514 | ZmTCP35 | - | - | - | - | - |
| GRMZM2G089638 | ZmTCP36 | 2 | 0 | 0 | 0 | 0 |
| GRMZM2G092214 | ZmTCP37 | 34 | 3 | 0 | 0 | 0 |
| GRMZM2G359599 | ZmTCP38 | - | - | - | - | - |
| GRMZM2G170232 | ZmTCP39 | - | - | - | - | - |
| GRMZM2G178603 | ZmTCP40 | 2 | 1 | 0 | 0 | 0 |
| GRMZM2G077755 | ZmTCP41 | - | - | - | - | - |
| GRMZM2G180568 | ZmTCP42 | 29 | 2 | 2 | 2 | 2 |
| GRMZM2G148022 | ZmTCP43 | 1 | 0 | 0 | 0 | 0 |
| GRMZM2G034638 | ZmTCP44 | 22 | 6 | 0 | 0 | 0 |
| GRMZM2G424261 | ZmTCP45 | - | - | - | - | - |
| GRMZM2G093895 | ZmTCP46 | 27 | 2 | 0 | 0 | 0 |
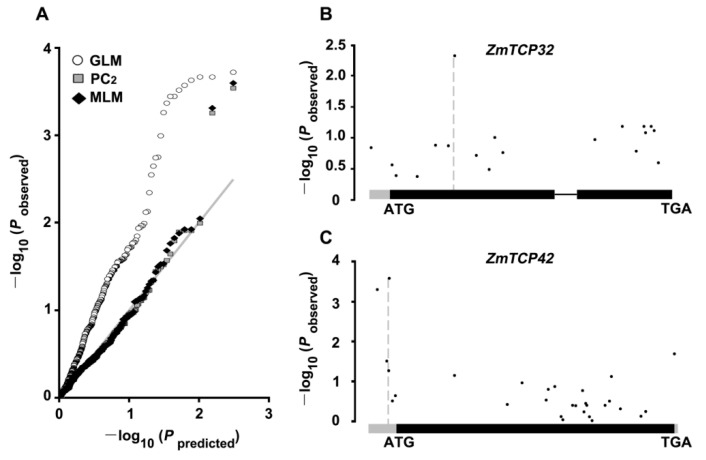
ZmTCP37 was found to be the most polymorphic, with 34 SNPs in this natural diversity panel. Subsequently, three statistical models were applied to identify significant genotypic and phenotypic associations. Specifically, a general linear model (GLM) with the first two principal components (PC2) and a mixed linear model (MLM) were used to find associations (Figure 4A). The GLM method was applied to perform single-marker analysis. Then PC2, via the first two principal components of the SNP data, was applied to correct for spurious associations caused by the population structure. The MLM method, incorporating both PC2 and a Kinship matrix (to correct for the effect of cryptic relatedness), was considered to be effective for controlling false positives in the association analysis (Figure 4A) [46,47]. The candidate gene association analysis detected significant associations between the genetic variations of ZmTCP32 and ZmTCP42 and drought tolerance, under different models, with a p-value ≤ 0.01 (Table 2; Figure 4B,C). However, under the standard mixed linear model (MLM), the only two significantly associated SNPs contributing to the phenotype of drought tolerance were both located at the 5′ UTR region of ZmTCP42, which suggested that this candidate gene was significantly associated with drought tolerance (p-value ≤ 0.001, −log10p = 3.77) (Figure 4C). We further analyzed the survival rates of maize inbred lines carrying ZmTCP42 drought-tolerant or drought-sensitive alleles. It was found that ZmTCP42AA was a favorable drought tolerance allele (Figure S2). The two-fold induction of ZmTCP42 expression by dehydration (Figure 3B) suggested that the RNA level of ZmTCP42 was likely more correlated with drought tolerance than other potential variations among different maize varieties.
Figure 4.
Association analysis of genetic variations in ZmTCP32 and ZmTCP42 with maize drought tolerance. (A) Quantile–quantile plots of the estimated −log10(p) from the ZmTCP gene family-based association analysis, using three methods. The gray line is the expected line under the null distribution. The white square represents the observed p values using general linear model (GLM); the gray square represents the GLM model with the first two principal components (PC2); the black diamond represents the observed p values using the mixed linear model (MLM) model incorporating both PC2 and a Kinship matrix. (B,C) Schematic diagrams of ZmTCP32 (B) and ZmTCP42 (C), including the UTR (gray), intron (thin black line), and protein coding regions (thick black line), are presented in the x-axis. The p value is shown on a −log10 scale.
2.6. ZmTCP32 and ZmTCP42 Are Both Induced by ABA Treatment and Drought Stress
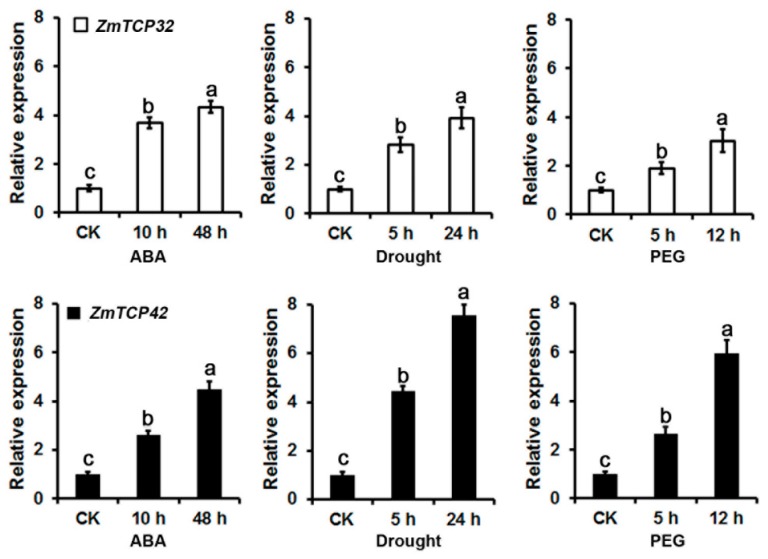
To confirm whether ZmTCP32 and ZmTCP42 RNA levels are truly associated with drought tolerance, we used RT-qPCR to directly analyze the RNA expression of the ZmTCP32 and ZmTCP42 genes, in response to ABA treatment and drought stress. As illustrated in Figure 5, in the B73 genotype, ZmTCP32 and ZmTCP42 are both significantly induced by ABA by roughly four-fold, relative to the controls at 48 h after the ABA treatment. More importantly, ZmTCP42 was highly induced by drought stress (+7.6-fold at 24 h) and by the PEG6000 treatment (+5.9-fold at 24 h); ZmTCP42 was significantly more responsive to dehydration stress and PEG treatment than ZmTCP32, even though ZmTCP32 was also responsive to drought stress (+3.9-fold at 24 h) and PEG treatment (+3.0-fold at 24 h) (Figure 5).
Figure 5.
The RNA levels of ZmTCP32 and ZmTCP42 in maize B73 leaves measured by RT-qPCR, after treatment with 100 μM ABA, drought stress, or 10% PEG treatment. The drought stress was performed as previously described [45], on hydroponically cultured seedlings, for 5 and 24 h, and their relative leaf water contents (RLWC) were determined to be approximately 70% and 58%, at the corresponding time points, respectively. ZmUbi-2 transcript levels were used as an internal control for data normalization; the gene-specific primers are listed in Table S2. Each data point represents the mean ± SD (n = 3) of three biological replicates. Significant differences were calculated by one-way ANOVA with Duncan’s multiple test (SAS Institute, Inc., Cary, NC, USA). Different letters indicate a significant statistical difference between sample means, at p = 0.05, while means with the same letters were not significantly different.
2.7. Overexpression of ZmTCP42 Enhances Drought Resistance in Transgenic Arabidopsis
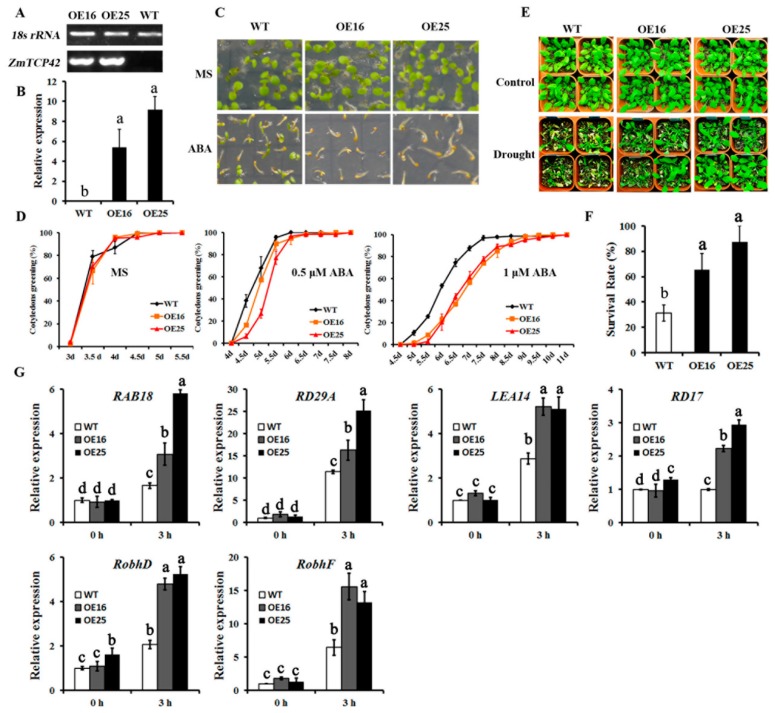
Given that the polymorphism in the 5′-UTR of ZmTCP42 suggests a potential role in drought resistance on maize seedlings, we selected ZmTCP42 to directly test for its function. To validate ZmTCP42 function in response to drought stress, we generated transgenic Arabidopsis plants that overexpress the ZmTCP42 gene, by using the enhanced cauliflower mosaic virus 35S promoter. After screening for the ZmTCP42 expression levels, we selected two independent transgenic lines, ZmTCP42-OE16 and ZmTCP42-OE25, with enhanced RNA levels, for further experiments (Figure 6A,B). We investigated the ABA sensitivity of ZmTCP42 transgenic lines, in response to exogenous ABA, during seed germination. In the absence of exogenous ABA, all lines germinated completely, as did the wild-type seeds; on the contrary, in the presence of exogenous ABA, the expanding, greening cotyledons were significantly affected in the ZmTCP42-OE lines; the number of seedlings with green cotyledons in ZmTCP42-OE16 and ZmTCP42-OE25 were significantly lower than those of the wild-type at 1 µM ABA (Figure 6C,D), suggesting that the ZmTCP42 overexpression lines are more sensitive to ABA. Furthermore, we analyzed the tolerance levels of ZmTCP42-OE lines to drought stress. When the 28-day-old transgenic seedlings were subjected to the drought test, only ~30% of the wide-type plants were able to recover from the stress, whereas ~65 of ZmTCP42-OE16 and ~87% of ZmTCP42-OE25 transgenic plants survived (Figure 6E,F). Similar results were obtained in repeated experiments, indicating that overexpression of ZmTCP42 enhanced drought tolerance in Arabidopsis.
Figure 6.
Analysis of wild-type (WT) and ZmTCP42-overexpressing transgenic Arabidopsis plants. (A) RT-PCR analysis of ZmTCP42 transcript levels in ZmTCP42 transgenic lines. (B) RT-qPCR analysis of ZmTCP42 transcript levels in ZmTCP42 transgenic lines. (C) Seed germination of the ZmTCP42 transgenic lines and in the wild-type, in response to abscisic acid (ABA). Germination rates with 1 μM ABA for 5 days, defined by cotyledon greening, in wild-type ZmTCP42-OE16 and ZmTCP42-OE25, compared to the MS medium alone. (D) Statistical analysis of a green seedling’s rate of ZmTCP42 transgenic lines and wild-type grown on Murashige and Skoog (MS) medium with 0, 0.5 μM, and 1 μM ABA. (E) Drought tolerance of the ZmTCP42 transgenic Arabidopsis plants. Photographs were taken before and after the drought treatment, followed by 3 days of rewatering. (F) Statistical analysis of survival rates after the drought-stress treatment. The average survival rates and standard errors were calculated from three independent experiments. (G) RNA levels of stress-responsive genes in the ZmTCP42 transgenic lines and in the wild-type, in response to drought stress. Total RNA was obtained from 3 weeks-old seedlings treated by 3 h of dehydration stress and was analyzed by RT-qPCR, using the gene-specific primers listed in Table S2. The mean value and standard error were calculated from three replicates after normalization to ACTIN2. The RNA levels in the wild-type grown under non-stress conditions was taken as 1.0. Significant differences were calculated by one-way ANOVA with Duncan’s multiple test (SAS Institute, Inc., Cary, NC, USA). Different letters indicate a significant statistical difference between the sample means at p = 0.05, while means with the same letters were not significantly different.
We further assessed the expression levels of ABA- inducible or drought-inducible genes by using real-time RT-qPCR to analyze their responses in Arabidopsis. Upon dehydration stress, the RNA levels of RAB18, RD29A, LEA14, and RD17, which are well-known drought-responsive, positive regulator genes of drought tolerance, were up-regulated, relative to wild-type Arabidopsis plants (Figure 6G). Similar changes were observed for the RNA levels of the RbohD and RbohF genes, encoding two NADPH oxidases, which were directly responsible for the reactive oxygen species (ROS) production in leaves, in response to stress (Figure 6G). These results clearly showed that ZmTCP42 overexpression improved the inducibility of Arabidopsis drought-tolerance-associated genes upon drought stress, leading to an enhanced drought tolerance.
3. Discussion
To date, the TCP family members have been described in various species; for instance, 24 TCP genes in Arabidopsis, 23 in rice, 20 in sorghum, and 66 in wheat (Triticum aestivum L.) [11,12,16,17]. Chai et al. (2017) reported 29 ZmTCP genes in maize, including their chromosomal location, structure and domain conservation analysis, and gene duplication analysis. They also analyzed the phylogenetic inference and expression profiling of these 29 ZmTCP genes, and then speculated that the ZmTCP genes influence stem and ear growth. In our study, we retrieved a total of 46 ZmTCP genes, by searching against the updated maize genome B73_RefGen_v3, including the 29 ZmTCPs previously reported by Chai et al. and the 17 newly identified ZmTCPs containing a canonical TCP domain. Subsequently, we further systematically determined their phylogenetic relationship, synteny with rice, sorghum, and Arabidopsis TCPs, pattern of drought-responsiveness, association analysis of their natural variations with drought tolerance, and functional analysis of ZmTCP42 in drought tolerance. Collectively, our data demonstrated that a few ZmTCP family proteins are likely involved in plant drought tolerance; in particular, ZmTCP42 functions as an important positive regulator for drought tolerance.
Recently, genome-wide identification revealed that segmental duplication might be the main contributor to the expansion of the TCP gene family in some monocots, including rice, sorghum and wheat, as well as in some dicot species, such as cotton and soybean [14,17,48,49,50,51,52]. The report showed that there were about 2.6–2.8-fold duplication of the TCP gene family in allotetraploid upland cotton genome (G. hirsutum), compared to Arabidopsis [48]. Similarly, we found that the duplication ratio of the ZmTCP family was about 2-fold in the allopolyploid maize genome, compared with Arabidopsis, rice, and sorghum. The change in the ratio of gene numbers suggests that the ZmTCP family has undergone lineage-specific expansion and functional divergence during the course of evolution. Furthermore, as shown in the phylogenetic analysis, monocot ZmTCP proteins clustered independently from those in Arabidopsis, suggesting a potential functional divegence between the dicot and monocot TCPs. We also found that ZmTCP gene expansion was not uniform, and occurred mainly in the class II CYC/TB1 clade. However, the biological significance of the ZmTCP gene duplication in the maize genome remains to be determined. Our collinearity analysis on TCP genes within several monocot plants showed that 20 ZmTCP genes had syntenic members or collinear genes in rice and sorghum, and that the chromosomal segments containing these genes were also duplicated (Figure 2), supporting the concept that the maize genome might have arisen from an ancestral allotetraploid, half of which share a common ancestor with sorghum, which in turn probably represents a lineage split from rice [53,54]. Taken together, our results might provide some useful clues for future studies on a homologous gene function.
A growing body of research suggest that TCP TFs play important roles in plant development, as well as in response to abiotic stress [8,19,33,34]. However, the role of TCP genes in plant response to drought stress in maize was still obscure, without answers to which, the ZmTCP genes were directly associated with the levels of drought tolerance in maize. We were, therefore, prompted to perform this study to address this question. The answer would not only help facilitate the genetic improvement of drought tolerance but would also increase our knowledge of the biological function of this gene family. Generally, ZmTCP genes exhibit great differential expression in response to drought stress in maize seedlings, not only among subgroups but members within the same subgroup, suggesting that these ZmTCP genes might function very diversely. Our current results showed that 14 ZmTCP genes were significantly up-regulated and 19 genes were clearly down-regulated, indicating that these genes might function as key mediators of drought stress responses in maize (Figure 3). Notably, three genes, ZmTCP1, ZmTCP9, and ZmTCP24, belonging to the class II CYC/TB1 clade, showed similar inducible expression patterns (Figure 3), implying that they might play a redundant role in regulating drought stress response in maize. Moreover, the orthologs of ZmTCP9 and ZmTCP24 in rice and Arabidopsis are OsPCF5 and AtTCP3/AtTCP4, respectively, which are regulated by the conserved miR319. Previous reports have shown that knockdown of miR319-dependent TCPs (by constitutively overexpressing miR319) increased drought and salinity tolerance [34,55]. The rice homolog of ZmTCP1 and ZmTCP13 was named OsTB1, which plays an important role in stress response, especially in regulation of cold tolerance [55,56]. Additionally, ZmTCP37 and ZmTCP44, orthologs of AtTCP20, also showed a slightly inducible expression pattern under drought stress (Figure 3). In Arabidopsis, AtTCP20 represses the transcription of LIPOXYGENASE2 (AtLOX2) gene, which is involved in jasmonic acid synthesis and promotes leaf senescence [57].
To explore the association of TCPs with stress tolerance in plants, we have investigated homologous ZmTCP genes in a small number of inbred lines and proposed predictive conclusions. Among the 46 ZmTCP genes analyzed, the genetic polymorphisms of ZmTCP32 and ZmTCP42 were significantly associated with the phenotypic variations of drought tolerance (p-value ≤ 0.01, PC2) (Table 2). While ZmTCP42 was the most significantly (p-value ≤ 0.001, MLM) associated with drought tolerance in this natural variation panel, the natural variation in the ZmTCP42 promoter might contribute to maize drought tolerance (Figure 4). In the B73 genotype of maize, ZmTCP32 was constitutively highly expressed in various tissues (Figure 3A). In comparison with ZmTCP32, ZmTCP42 RNA was detected at a low level in various tissues, except in leaves and endosperm, and showed a relatively high level at 10 h, upon drought stress in a micrroarry analysis (Figure 3B). Both ZmTCP32 and ZmTCP42 were significantly induced by ABA, dehydration and PEG treatment; more notably, ZmTCP42 was induced higher than ZmTCP32 in response to dehydration and PEG treatment. These results suggested that ZmTCP42 was involved in plant drought response. Transgenic Arabidopsis overexpressing ZmTCP42 exhibited a higher ABA sensitivity, improved drought stress tolerance, and enhanced the induction of Arabidopsis ABA- or drought-inducible genes, which strongly suggests that ZmTCP42 might regulate these ABA/drought-response genes. In summary, we have identified ZmTCP42 as an important positive regulator of drought tolerance, through analyses of gene expression and natural variations.
4. Materials and Methods
4.1. Plant Growth Conditions and Stress Treatments
Maize (Inbred line B73) growth conditions and stress treatments were performed according to Wang et al. [45]. The hydroponically cultured 3-leaf stage seedlings were used. For drought treatments, the seedlings were placed on a clean bench and subjected to dehydration (at 28 °C, with a relative humidity of 40–60%). Samples were exposed for 0, 5 h, and 24 h, at which the relative leaf water contents were measured (RLWC) to be 98%, 70%, and 60%, respectively, in the drought-treated leaf samples. ABA and PEG treatments were applied by immersing the seedlings in 100 μM ABA or 10% PEG 6000. All leaf samples from a minimum of 3 seedlings were immediately frozen in liquid nitrogen, then stored at −80 °C, prior to RNA isolation. The Arabidopsis thaliana ecotype Col-0 was used in this study. Seeds were grown on the Murashige and Skoog (MS) medium with an addition of 3% sucrose and 0.6% agar (pH 5.8). Drought treatment was applied to the 3-week-old seedlings by removing them from the MS medium plates and desiccating them on Whatman 3MM paper, on a clean bench, for 0 h and 3 h, respectively.
4.2. Reverse Transcription PCR and RT-qPCR Analysis
Total RNA was isolated from the collected leaf samples using the TRIZOL reagent (Biotopped, China) according to the manufacturer’s instructions. One microgram of the total RNA from each sample was used in reverse transcription. RT-qPCR analyses were performed in the optical 48-well plates, using the ABI7300 Thermo-cycler (Applied Biosystems, USA). Reactions were carried out in a 10 μL volume containing 1 μL diluted cDNA, 200 nM gene-specific primers, and 5 μL SYBR Premix Ex Taq II (Takara, China) with the following conditions—10 min at 95 °C, 40 cycles of 15 s at 95 °C, and 30 s at 60 °C. Each experiment was performed using at least three independent biological replicates. ZmTCP32 and ZmTCP42 RNA levels in response to different stresses were calculated according to a standard curve calibration, based on amplification of a dilution series of ZmTCP32 and ZmTCP42 plasmid, respectively, according to the manufacturer’s protocol. ZmUbi-2 (UniProtKB/TrEMBL; ACC: Q42415) was used as the internal control. The expression levels of different stress-responsive genes were compared, based on the delta Ct method with normalization to ACTIN2.
4.3. Generation of Transgenic Plants
The coding region of the ZmTCP42 cDNA of the maize B73 inbred line was inserted into the pGreenII vector [58]. The constructed plasmid carrying the desired gene was transformed into Agrobacterium tumefaciens GV3101 + pSoup and transformed into ecotype Col-0, as described previously [58]. Using the kanamycin-based selection, several independent T2 transgenic lines were obtained, and expression of the ZmTCP42 transgene was confirmed in these lines by RT-PCR and RT-qPCR. Two independent overexpression lines ZmTCP42-OE16 and ZmTCP42-OE25 (with a single inserted copy) were selected, based on the level of transgene expression, and the T3 homozygous seeds were subjected to further analyses.
4.4. Seed Germination and Drought Phenotype Analysis
To study the effect of ABA on germination and cotyledon greening, the seeds were planted on MS medium plates, with an addition of plus 1% sucrose and different concentrations of ABA (Sigma-Aldrich) in a growth chamber, at 22 °C under a 16-h-light/8-h-dark photo-period, with a 60% relative humidity, after 2 days of vernalization in darkness at 4 °C. For the drought stress tolerance test, plants were grown for 28 days under normal conditions and subjected to water stress by withholding watering for 14 days [58]. Four plants were planted in each small cup with 100 g soil (1:1 of black soil/vermiculite) and grown under the condition of 16-h-light/8-h-dark. Four independent experiments were performed and at least 16 plants each were observed, for the ZmTCP42-OE16 and ZmTCP42-OE25 lines and the WT, in each experiment. The plants were rewatered when significant differences in wilting were observed. Three days after rewatering, the surviving plants were counted.
4.5. Identification of the TCP Proteins in the Maize Genome
To identify TCP proteins in the B73 maize genome, some putative TCP protein sequences from maize (genome assembly: B73_RefGen_v3) were first retrieved from the Plant Transcription Factor Database 3.0 (Jin et al. 2014). Conserved TCP DNA-binding domain (PF03634) from the Pfam database [59] was used to search and retrieve sequences from the Phytozome database v10.0 (available online: http://www.phytozome.net/eucalyptus.php). In addition, BLASTP searches were also performed against the maize genome, to identify any additional TCP members using Arabidopsis and rice TCP proteins sequences (E-value ≤ 0.00001). The overlapped genes were removed. The presence of a TCP domain in all family proteins was evaluated using the CDD database searches (https://www.ncbi.nlm.nih.gov/Structure/cdd). Arabidopsis TCP proteins were downloaded from TAIR 10 (available online: http://www.arabidopsis.org), which contained 24 members. TCP proteins in rice (Genome assembly: Rice Genome Annotation Project Database release 7) and sorghum (Genome assembly: V3) were both downloaded from the Plant Transcription Factor Database. The ExPASy program (http://www.expasy.org/tools/) was used to predict the molecular weight (kDa) and the isoelectric point (PI) of each protein.
4.6. Gene Structure and Phylogenetic Analysis
Genomic sequences of maize TCP genes were downloaded from Phytozome V10, and the untranslated regions were removed. To show the exon/intron organization for individual TCP genes, coding sequences were aligned to genomic sequences and schematics generated using GSDS 2.0 (Gene Structure Display Server) (available online: http://gsds.cbi.pku.edu.cn) [60]. To determine the phylogenetic relationships of the TCP proteins, full-length amino acid sequences of TCPs identified in maize, rice, Arabidopsis, and sorghum were aligned by the ClustalW program [61]. The Phylogenetic tree was constructed by the neighbor-joining method, with 1000 bootstrap replicates.
4.7. Association Analysis
Association analysis for ZmTCPs was performed by using a maize association mapping population containing 367 inbred lines and the corresponding drought tolerance phenotype data from a previous study [45]. Among the 556,945 high-quality single nucleotide polymorphism (SNP) data, with minor allele frequency (MAF) ≥ 0.05, 317 SNPs were found in the genic region of the 46 ZmTCPs. The general linear model (GLM) model, the GLM model with the first 2 principal components (PC2), and the mixed linear model (MLM) model were chosen to detect the SNPs significantly associated with drought tolerance, by using the TASSEL5.0 program [62].
4.8. Tissue Expression Profile and Microarray Analysis
Expression patterns of 46 ZmTCPs in different maize tissues were analyzed using the genome-wide gene expression atlas of the inbred B73 line of maize that was reported previously [43]. Expression data for the 15 tissues were combined from 60 growth stages. Normalized expression values of each gene in different tissues were averaged. The gene expression level was presented as a log value. To analyze the gene expression patterns of ZmTCPs in maize during drought stress, the microarray data and analysis methods were employed as described previously [63].
Abbreviations
| TCP | the teosinte branched1/cycloidea/proliferating |
| TF | transcription factor |
| CYC/TB1 | cycloidea/teosinte branched 1 |
| PCF | proliferating cell nuclear antigen factor |
| CIN | CINCINNATA of Antirrhinum |
| RT-qPCR | reverse transcription quantitative polymerase chain reaction |
| GLM | general linear model |
| SNP | single nucleotide polymorphism |
| MLM | mixed linear model |
| ABA | abscisic acid |
| PEG | polyethylene glycol |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/11/2762/s1.
Author Contributions
S.D. and Z.C. designed the experiments, performed the experiments, analyzed the data, and prepared figures and tables. H.D. gave good advice on the work. H.W. wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation (31701062 and 31701439), the Hubei Province Educational Commission Foundation (Q20171307), the Plan in Scientific and Technological Innovation Team of Outstanding Young, Hubei Provincial Department of Education (T2017004).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Barnabas B., Jager K., Feher A. The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 2007;31:11–38. doi: 10.1111/j.1365-3040.2007.01727.x. [DOI] [PubMed] [Google Scholar]
- 2.Yordanov I., Velikova V., Tsonev T. Plant Responses to Drought, Acclimation, and Stress Tolerance. Photosynthetica. 2000;38:171–186. doi: 10.1023/A:1007201411474. [DOI] [Google Scholar]
- 3.Daryanto S., Wang L., Jacinthe P. Global Synthesis of Drought Effects on Maize and Wheat Production. PLoS ONE. 2016;11:e0156362. doi: 10.1371/journal.pone.0156362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lobell D.B., Roberts M.J., Schlenker W., Braun N., Little B.B., Rejesus R.M., Hammer G.L. Greater sensitivity to drought accompanies maize yield increase in the U.S. Midwest. Science. 2014;344:516–519. doi: 10.1126/science.1251423. [DOI] [PubMed] [Google Scholar]
- 5.Akhtar M., Jaiswal A., Taj G., Jaiswal J.P., Qureshi M.I., Singh N.K. DREB1/CBF transcription factors: Their structure, function and role in abiotic stress tolerance in plants. J. Genet. 2012;91:385–395. doi: 10.1007/s12041-012-0201-3. [DOI] [PubMed] [Google Scholar]
- 6.Baldoni E., Genga A., Cominelli E. Plant MYB Transcription Factors: Their Role in Drought Response Mechanisms. Int. J. Mol. Sci. 2015;16:15811–15851. doi: 10.3390/ijms160715811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi-Shinozaki K., Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 8.Martin-Trillo M., Cubas P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010;15:31–39. doi: 10.1016/j.tplants.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Kosugi S., Ohashi Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 2002;30:337–348. doi: 10.1046/j.1365-313X.2002.01294.x. [DOI] [PubMed] [Google Scholar]
- 10.Navaud O., Dabos P., Carnus E., Tremousaygue D., Herve C. TCP Transcription Factors Predate the Emergence of Land Plants. J. Mol. Evol. 2007;65:23–33. doi: 10.1007/s00239-006-0174-z. [DOI] [PubMed] [Google Scholar]
- 11.Riechmann J.L., Heard J.E., Martin G., Reuber L., Jiang C., Keddie J., Adam L., Pineda O., Ratcliffe O.J., Samaha R. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis Among Eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 12.Yao X., Ma H., Wang J., Zhang D. Genome-Wide Comparative Analysis and Expression Pattern of TCP Gene Families in Arabidopsis thaliana and Oryza sativa. J. Integr. Plant Biol. 2007;49:885–897. doi: 10.1111/j.1744-7909.2007.00509.x. [DOI] [Google Scholar]
- 13.Parapunova V., Busscher M., Busscher-Lange J., Lammers M., Karlova R., Bovy A.G., Angenent G.C., de Maagd R.A. Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 2014;14:157. doi: 10.1186/1471-2229-14-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma J., Wang Q., Sun R., Xie F., Jones D.C., Zhang B. Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci. Rep. 2014;4:6645. doi: 10.1038/srep06645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosugi S., Ohashi Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell. 1997;9:1607–1619. doi: 10.1105/tpc.9.9.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J., Zhai Z., Li Y., Geng S., Song G., Guan J., Jia M., Wang F., Sun G., Feng N., et al. Genome-Wide Identification and Expression Profiling of the TCP Family Genes in Spike and Grain Development of Wheat (Triticum aestivum L.) Front. Plant Sci. 2018;9:1282. doi: 10.3389/fpls.2018.01282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francis A., Dhaka N., Bakshi M., Jung K.H., Sharma M.K., Sharma R. Comparative phylogenomic analysis provides insights into TCP gene functions in Sorghum. Sci. Rep. 2016;6:38488. doi: 10.1038/srep38488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cubas P., Lauter N., Doebley J., Coen E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999;18:215–222. doi: 10.1046/j.1365-313X.1999.00444.x. [DOI] [PubMed] [Google Scholar]
- 19.Manassero N.G.U., Viola I.L., Welchen E., Gonzalez D.H. TCP transcription factors: Architectures of plant form. Biomol. Concepts. 2013;4:111–127. doi: 10.1515/bmc-2012-0051. [DOI] [PubMed] [Google Scholar]
- 20.Li S. The Arabidopsis thaliana TCP transcription factors: A broadening horizon beyond development. Plant Signal. Behav. 2015;10:e1044192. doi: 10.1080/15592324.2015.1044192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danisman S. TCP Transcription Factors at the Interface between Environmental Challenges and the Plant’s Growth Responses. Front. Plant Sci. 2016;7:1930. doi: 10.3389/fpls.2016.01930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhaka N., Bhardwaj V., Sharma M.K., Sharma R. Evolving Tale of TCPs: New Paradigms and Old Lacunae. Front. Plant Sci. 2017;8:479. doi: 10.3389/fpls.2017.00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicolas M., Cubas P. Plant Transcription Factors. Elsevier; Amsterdam, The Netherlands: 2016. The Role of TCP Transcription Factors in Shaping Flower Structure, Leaf Morphology, and Plant Architecture; pp. 249–267. [Google Scholar]
- 24.Resentini F., Felipo-Benavent A., Colombo L., Blázquez M.A., Alabadí D., Masiero S. TCP14 and TCP15 Mediate the Promotion of Seed Germination by Gibberellins in Arabidopsis thaliana. Mol. Plant. 2015;8:482–485. doi: 10.1016/j.molp.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 25.Yanai O., Shani E., Russ D., Ori N. Gibberellin partly mediates LANCEOLATE activity in tomato. Plant J. 2011;68:571–582. doi: 10.1111/j.1365-313X.2011.04716.x. [DOI] [PubMed] [Google Scholar]
- 26.Schommer C., Palatnik J.F., Aggarwal P., Chételat A., Cubas P., Farmer E.E., Nath U., Weigel D. Control of Jasmonate Biosynthesis and Senescence by miR319 Targets. PLoS Biol. 2008;6:e230. doi: 10.1371/journal.pbio.0060230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koyama T., Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. TCP Transcription Factors Regulate the Activities of ASYMMETRIC LEAVES1 and miR164, as Well as the Auxin Response, during Differentiation of Leaves in Arabidopsis. Plant Cell. 2010;22:3574–3588. doi: 10.1105/tpc.110.075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Efroni I., Han S.-K., Kim H., Wu M.-F., Steiner E., Birnbaum K., Hong J., Eshed Y., Wagner D. Regulation of Leaf Maturation by Chromatin-Mediated Modulation of Cytokinin Responses. Dev. Cell. 2013;24:438–445. doi: 10.1016/j.devcel.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koornneef M., Hanhart C.J., Hilhorst H.W., Karssen C.M. In Vivo Inhibition of Seed Development and Reserve Protein Accumulation in Recombinants of Abscisic Acid Biosynthesis and Responsiveness Mutants in Arabidopsis thaliana. Plant Physiol. 1989;90:463–469. doi: 10.1104/pp.90.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finkelstein R.R., Gampala S.S., Rock C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14:S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osakabe Y., Osakabe K., Shinozaki K., Tran L.P. Response of plants to water stress. Front. Plant Sci. 2014;5:86. doi: 10.3389/fpls.2014.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raghavendra A.S., Gonugunta V.K., Christmann A., Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15:395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 33.Mukhopadhyay P., Tyagi A.K. OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci. Rep. 2015;5:9998. doi: 10.1038/srep09998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou M., Li D., Li Z., Hu Q., Yang C., Zhu L., Luo H. Constitutive Expression of a miR319 Gene Alters Plant Development and Enhances Salt and Drought Tolerance in Transgenic Creeping Bentgrass. Plant Physiol. 2013;161:1375–1391. doi: 10.1104/pp.112.208702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rueda-Romero P., Barrero-Sicilia C., Gomez-Cadenas A., Carbonero P., Onate-Sanchez L. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J. Exp. Bot. 2012;63:1937–1949. doi: 10.1093/jxb/err388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez-Grandio E., Poza-Carrion C., Sorzano C.O.S., Cubas P. BRANCHED1 Promotes Axillary Bud Dormancy in Response to Shade in Arabidopsis. Plant Cell. 2013;25:834–850. doi: 10.1105/tpc.112.108480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez-Grandio E., Pajoro A., Franco-Zorrilla J.M., Tarancon C., Immink R.G., Cubas P. Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. USA. 2017;114:E245–E254. doi: 10.1073/pnas.1613199114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H., Peng Z., Yang X., Wang W., Fu J., Wang J., Han Y., Chai Y., Guo T., Yang N., et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 2013;45:43–50. doi: 10.1038/ng.2484. [DOI] [PubMed] [Google Scholar]
- 39.Yan J., Kandianis C.B., Harjes C.E., Bai L., Kim E.-H., Yang X., Skinner D.J., Fu Z., Mitchell S., Li Q., et al. Rare genetic variation at Zea mays crtRB1 increases beta-carotene in maize grain. Nat. Genet. 2010;42:322. doi: 10.1038/ng.551. [DOI] [PubMed] [Google Scholar]
- 40.Yang Q., Li Z., Li W., Ku L., Wang C., Ye J., Li K., Yang N., Li Y., Zhong T., et al. CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA. 2013;110:16969–16974. doi: 10.1073/pnas.1310949110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H., Qin F. Genome-Wide Association Study Reveals Natural Variations Contributing to Drought Resistance in Crops. Front. Plant Sci. 2017;8:1110. doi: 10.3389/fpls.2017.01110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chai W., Jiang P., Huang G., Jiang H., Li X. Identification and expression profiling analysis of TCP family genes involved in growth and development in maize. Physiol. Mol. Biol. Plants. 2017;23:779–791. doi: 10.1007/s12298-017-0476-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sekhon R.S., Lin H., Childs K.L., Hansey C.N., Buell C.R., de Leon N., Kaeppler S.M. Genome-wide atlas of transcription during maize development. Plant J. 2011;66:553–563. doi: 10.1111/j.1365-313X.2011.04527.x. [DOI] [PubMed] [Google Scholar]
- 44.Liu S., Wang X., Wang H., Xin H., Yang X., Yan J., Li J., Tran L.S., Shinozaki K., Yamaguchi-Shinozaki K., et al. Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet. 2013;9:e1003790. doi: 10.1371/journal.pgen.1003790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X., Wang H., Liu S., Ferjani A., Li J., Yan J., Yang X., Qin F. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 2016;48:1233–1241. doi: 10.1038/ng.3636. [DOI] [PubMed] [Google Scholar]
- 46.Yu J., Buckler E.S. Genetic association mapping and genome organization of maize. Curr. Opin. Biotechnol. 2006;17:155–160. doi: 10.1016/j.copbio.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 47.Yu J., Pressoir G., Briggs W.H., Vroh Bi I., Yamasaki M., Doebley J.F., McMullen M.D., Gaut B.S., Nielsen D.M., Holland J.B., et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 48.Li W., Li D.D., Han L.H., Tao M., Hu Q.Q., Wu W.Y., Zhang J.B., Li X.B., Huang G.Q. Genome-wide identification and characterization of TCP transcription factor genes in upland cotton (Gossypium hirsutum) Sci. Rep. 2017;7:10118. doi: 10.1038/s41598-017-10609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma X., Ma J., Fan D., Li C., Jiang Y., Luo K. Genome-wide Identification of TCP Family Transcription Factors from Populus euphratica and Their Involvement in Leaf Shape Regulation. Sci. Rep. 2016;6:32795. doi: 10.1038/srep32795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mondragón-Palomino M., Trontin C. High time for a roll call: Gene duplication and phylogenetic relationships of TCP-like genes in monocots. Ann. Bot. 2011;107:1533–1544. doi: 10.1093/aob/mcr059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma R., Kapoor M., KTyagi A., Kapoor S. Comparative transcript profiling of TCP family genes provide insight into gene functions and diversification in rice and Arabidopsis. J. Plant Mol. Biol. Biotechnol. 2010;1:24–38. [Google Scholar]
- 52.Feng W., Lindner H., Robbins N.E., 2nd, Dinneny J.R. Growing Out of Stress: The Role of Cell- and Organ-Scale Growth Control in Plant Water-Stress Responses. Plant Cell. 2016;28:1769–1782. doi: 10.1105/tpc.16.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paterson A.H., Bowers J.E., Bruggmann R., Dubchak I., Grimwood J., Gundlach H., Haberer G., Hellsten U., Mitros T., Poliakov A., et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 54.Gaut B.S., Doebley J. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang C., Li D., Mao D., Liu X., Ji C., Li X., Zhao X., Cheng Z., Chen C., Zhu L. Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.) Plant Cell Environ. 2013;36:2207–2218. doi: 10.1111/pce.12130. [DOI] [PubMed] [Google Scholar]
- 56.Chen L., Zhao Y., Xu S., Zhang Z., Xu Y., Zhang J., Chong K. OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol. 2018;218:219–231. doi: 10.1111/nph.14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Danisman S., van der Wal F., Dhondt S., Waites R., de Folter S., Bimbo A., van Dijk A.D., Muino J.M., Cutri L., Dornelas M.C., et al. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 2012;159:1511–1523. doi: 10.1104/pp.112.200303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ding S., Zhang B., Qin F. Arabidopsis RZFP34/CHYR1, a Ubiquitin E3 Ligase, Regulates Stomatal Movement and Drought Tolerance via SnRK2.6-Mediated Phosphorylation. Plant Cell. 2015;27:3228–3244. doi: 10.1105/tpc.15.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu B., Jin J., Guo A.Y., Zhang H., Luo J., Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bradbury P.J., Zhang Z.W., Kroon D.E., Casstevens T.M., Ramdoss Y., Buckler E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 63.Mao H., Yu L., Li Z., Liu H., Han R. Molecular evolution and gene expression differences within the HD-Zip transcription factor family of Zea mays L. Genetica. 2016;144:243–257. doi: 10.1007/s10709-016-9896-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.