Abstract
The activities of critical metabolic and regulatory proteins can be altered by exposure to natural or synthetic redox-cycling compounds. Many bacteria, therefore, possess mechanisms to transport or transform these small molecules. The opportunistic pathogen Pseudomonas aeruginosa PA14 synthesizes phenazines, redox-active antibiotics that are toxic to other organisms but have beneficial effects for their producer. Phenazines activate the redox-sensing transcription factor SoxR and thereby induce the transcription of a small regulon, including the operon mexGHI-opmD, which encodes an efflux pump that transports phenazines, and PA14_35160 (pumA), which encodes a putative monooxygenase. Here, we provide evidence that PumA contributes to phenazine resistance and normal biofilm development, particularly during exposure to or production of strongly oxidizing N-methylated phenazines. We show that phenazine resistance depends on the presence of residues that are conserved in the active sites of other putative and characterized monooxygenases found in the antibiotic producer Streptomyces coelicolor. We also show that during biofilm growth, PumA is required for the conversion of phenazine methosulfate to unique phenazine metabolites. Finally, we compare ∆mexGHI-opmD and ∆pumA strains in assays for colony biofilm morphogenesis and SoxR activation, and find that these deletions have opposing phenotypic effects. Our results suggest that, while MexGHI-OpmD-mediated efflux has the effect of making the cellular phenazine pool more reducing, PumA acts on cellular phenazines to make the pool more oxidizing. We present a model in which these two SoxR targets function simultaneously to control the biological activity of the P. aeruginosa phenazine pool.
Keywords: phenazines, monooxygenase, biofilm physiology, self-resistance
Introduction
Microbes in mixed-species systems release small molecule antibiotics that support their fitness in multiple ways [1]. These compounds can inhibit the growth of other species, facilitate iron uptake, or act as electron acceptors for redox metabolism [2–5]. However, such functions require that the producing organisms tolerate the antibiotic and possess the machinery required to exploit the compound’s beneficial effects. The transcription factor SoxR, which can be found in diverse bacterial phyla, regulates species-dependent sets of genes that are linked to antibiotic resistance and utilization, including those that encode efflux pumps and detoxifying enzymes [6–9]. Redox-active compounds can interact directly with the Fe–S cluster of the DNA-bound SoxR dimer, leading to a conformational change in the promoter that induces the transcription of target genes [10, 11]. In bacteria that do not produce redox-active antibiotics, the SoxR regulon can confer protection from antibiotics produced by other species [7, 8, 12]. In bacteria that make these compounds, SoxR can mediate self-resistance [13].
In the opportunistic pathogen Pseudomonas aeruginosa, SoxR is activated by endogenous phenazines, redox-cycling compounds that are toxic to other organisms but have beneficial effects for their producer (Fig. 1a, b) [6, 14]. When other oxidants are not available, phenazines can accept electrons from P. aeruginosa metabolism and balance the intracellular redox state [5]. Reduced phenazines can shuttle outside of the cell and transfer electrons to oxidants that are available at a distance, including insoluble iron [15]. At physiological concentrations, the redox-balancing effects of phenazines are reflected macroscopically by colony biofilm morphology: phenazines promote the formation of a smooth colony, while phenazine deficiency (and therefore electron acceptor limitation) promotes the development of wrinkle features, which increase the surface area-to-volume ratio of the colony and access to oxygen for resident cells [16, 17].
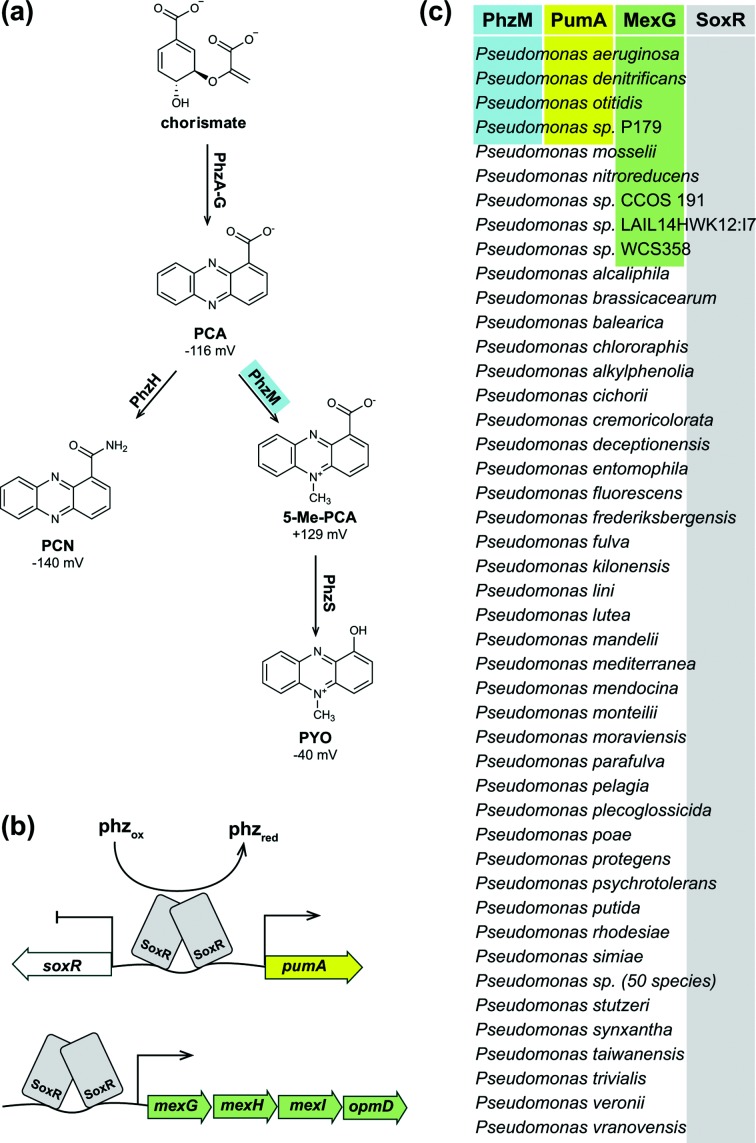
Fig. 1.
Phenazine biosynthesis and self-resistance pathways. (a) The P. aeruginosa phenazine biosynthetic pathway. Redox potentials relative to the standard hydrogen electrode at pH 7.0 are given. PCA, phenazine-1-carboxylic acid; PCN, phenazine-1-carboxamide; 5-Me-PCA, 5-methylphenazine-1-carboxylic acid; PYO, pyocyanin. (b) Model of phenazine-mediated SoxR activation, with SoxR bound to the chromosomal locus between the genes PA14_35160 (pumA) and PA14_35170 (soxR). An additional SoxR target, the operon mexGHI-opmD, is shown below. (c) Phylogenetic distribution of orthologues of phzM (blue shading), pumA (yellow shading) and mexG (green shading) in the genus Pseudomonas.
We study the P. aeruginosa PA14 SoxR regulon, which contains three chromosomal targets: the operon mexGHI-opmD, encoding an RND efflux pump; PA14_35160, encoding a putative monooxygenase; and PA14_16310, encoding a putative MFS transporter [6, 14]. PA14_35160 is orthologous to the gene PA2274 in P. aeruginosa strain PAO1 and is chromosomally adjacent to the gene encoding SoxR. In a prior study, we found that ∆soxR, ∆mexGHI-opmD and ∆PA14_35160 mutants are more sensitive to phenazine methosulfate (PMS), a structural analogue of the reactive P. aeruginosa product 5-methylphenazine-1-carboxylic acid (5-Me-PCA), than the wild-type. Results from our follow-up experiments implicated the MexGHI-OpmD pump in 5-Me-PCA efflux [13]. However, this work also suggested that the PA14_35160 gene product contributed to P. aeruginosa phenazine resistance. Here, we show that PA14_35160 transforms endogenous phenazines, leading to physiological effects that support normal community development, and suggest the name PumA (phenazine utilizing monooxygenase A) for this protein. These results further our understanding of P. aeruginosa phenazine metabolism in the context of biofilm growth.
Methods
Strains and culture conditions
The strains used in this study are listed in Table S1 (available in the online version of this article). For preculturing and genetic manipulation, bacteria were grown at 37 °C in lysogeny broth (LB) with shaking at 250 r.p.m. (Forma Orbital Shaker, Thermo Scientific) or on LB solidified with agar (15 g L−1) [18]. For selection during genetic manipulation, gentamicin was added to the medium at 15 µg mL−1 for Escherichia coli or 100 µg mL−1 for P. aeruginosa. Carbenicillin was added to the medium at 300 µg mL−1 for P. aeruginosa.
To characterize phenazine sensitivity and biofilm development, colonies were grown at 30 °C on 1 % tryptone and 1 % agar, containing 40 µg mL−1 Congo red and 20 µg mL−1 Coomassie blue where indicated.
Strain construction
The strains used in this study are listed in Table S1, while the plasmids used in this study are listed in Table S2 and the primers used for the construction of plasmids are listed in Table S3. The plasmids for deletion and gene replacement were generated using the yeast gap repair method as described previously [19, 20]. Markerless deletions of pumA were generated as described previously by homologous recombination in various PA14 strain backgrounds [20]. The same approach was used to complement ∆pumA using complementation plasmids. The final clones were verified by PCR and sequencing.
GFP reporter strains were constructed using plasmid pLD2726, generated by inserting the 448 bp of the mexG promoter between an engineered SpeI and XhoI site into a pSEK103 plasmid backbone using the primers indicated in Table S3. Strains expressing GFP under the control of the mexG promoter were generated by homologous recombination of plasmid pLD2762 into the neutral attB site on the PA14 chromosome. Plasmids were transformed into E. coli strain UQ950, verified by sequencing and moved into PA14 using biparental conjugation with E. coli strain S17-1. PA14 single recombinants were selected on M9 minimal medium agar plates (47.8 mM Na2HPO4•7H2O, 22 mM KH2PO4, 8.6 mM NaCl, 18.6 mM NH4Cl, 1 mM MgSO4, 0.1 mM CaCl2, 20 mM sodium citrate dihydrate, 1.5 % agar) containing 100 µg mL−1 gentamicin. The plasmid backbone was resolved out of PA14 using Flp/FRT recombination by the introduction of the pFLP2 plasmid [21] and selected on M9 minimal medium agar plates containing 300 µg mL−1 carbenicillin and further on LB agar plates without NaCl and modified to contain 10 % sucrose. The presence of gfp in the final clones was confirmed by PCR.
Colony morphology assay
Ten microlitres of overnight precultures were spotted on 1 % tryptone + 1 % agar containing 40 µg mL−1 Congo red and 20 µg mL−1 Coomassie blue (60 mL in a 9 cm square plate) and incubated at 25 °C, >95 % humidity. The colonies were imaged daily using an Epson 11000XL scanner. For the onset of wrinkling determination, overnight precultures were diluted 1 : 100 and grown to the mid-log phase before 10 µL of the culture was spotted onto the agar plates. Time-lapse movies of colony development were assembled from images taken using a Logitec c930e web-camera triggered at 15 min intervals by a custom Labview program.
PMS sensitivity
Cultures were grown in LB for 16 h at 37 °C with continuous shaking at 250 r.p.m. After 16 h, 10 µL of culture was spotted on morphology agar supplemented with 600 µM PMS and dried. The plates were incubated in the dark at 25 °C. The colonies were imaged at indicated time points using an Epson 11000XL scanner. For the quantification of colony-forming units (c.f.u.s), 10 µL of overnight LB precultures was spotted on filter disks overlaid on the same morphology agar +/−600 µM PMS. After 48 h of growth, filter disks with the colonies were lifted from the agar using sterile forceps and added to a 1.5 mL screw-cap tube containing 1 mL phosphate-buffered saline (PBS) and disrupted with 3 mm zirconium beads in a BeadRupter 12 homogenizer (Omni International, Inc.) for 90 s on the ‘high’ setting. Dilutions of homogenized biofilms in PBS were plated on 1 % tryptone, 1 % agar plates and incubated overnight at 37 °C, and the colony-forming units were then counted.
Statistical analysis
Data analysis was performed using GraphPad Prism version 7 (GraphPad Software, La Jolla, CA, USA). Values are expressed as mean ± SD. The statistical significance of the data presented was assessed with the two-tailed unpaired Student’s t-test. Values of P≤0.05 were considered significant (*P≤0.05; **P≤0.005; ***P≤0.0005).
Protein structure prediction and presentation
The structures for EcaB and PumA were predicted using the Phyre2 homology modelling engine [22]. These structures and ActVA-Orf6 (PDB: 1LQ9; [23]) were visualized with the PyMOL molecular graphics system, version 2.0, Schrödinger, LLC [24].
Imaging mass spectrometry
The indicated P. aeruginosa strains were streaked from frozen stocks onto plates containing 1 % tryptone (Bacto) and 1 % agar. After 24 h of incubation at 30 °C, colonies were used to inoculate 5 mL LB cultures, which were shaken at 225 r.p.m. at 30 °C for 24 h. Five microlitres of each liquid culture was plated onto separate thin plates supplemented with 200 µM PMS. PA14 ∆phz was grown on both supplemented and unsupplemented agar plates. The plates were incubated for 5 days in a sealed container at room temperature in the absence of light.
After 5 days of incubation, the colonies were physically excised on mats of the agar using razor blades and scalpels and laid flat on a steel MALDI plate (Bruker 96-spot ground). Two agar controls were also placed on the plate: one LB agar alone and one supplemented with 200 µM PMS. A MALDI matrix [50 : 50 ɑ-cyano-4-hydroxycinnamic acid (CHCA): 2,5- dihydroxybenzoic acid (DHB)] was applied using a a 53 µm stainless steel sieve (Hogentogler and Co., Inc.) onto the colonies and agar. The steel plate was placed uncovered in a 37 °C oven for 12 h or until dried. Excess matrix was removed using air. Then 1 µL of phosphorus red (1 mg mL−1) was spotted on an agar-free area of the steel plate as a calibration standard. The data were gathered in reflectron positive mode, from 100 to 2000 Da. The raster width across the sample was 200 µm. FlexImaging 4.1 was used with flexControl 3.4 to set up the IMS analysis, and regions of interest (ROI) were manually designated for each culture or growth condition. The laser power and detector gain were set to 30 % and 3.0×, respectively. Five hundred shots were taken at 2000 Hz at each raster spot, with the laser width (smartbeam parameter set) set to 2 (small). The resulting spectra were analysed using FlexImaging 4.1 after normalization to root mean square (RMS).
Compound isolation and structure elucidation
Relevant P. aeruginosa strains were streaked from frozen stocks onto plates containing 1 % tryptone, 1 % agar and incubated overnight at 30 °C. After 24 h, individual colonies were transferred into culture tubes containing 5 mL of liquid LB (10 g tryptone, 5 g yeast extract, 10 g NaCl in 1 L MilliQ water, pH 7.5) and shaken at 225 r.p.m. and 30 °C. After 24 h, the liquid culture was diluted to an OD (500 nm) of 0.05 into 30 mL of LB. After 24 h, the liquid culture was again used to inoculate 1 L of LB to an OD (500 nm) of 0.05. At an OD (500 nm) of 0.4–0.5 (early stationary phase), 200 µM of phenazine methosulfate (PMS) in MilliQ H2O was added to the culture. Alongside the 1 L bacterial culture was an uninoculated 1 L flask of LB, also with PMS added. The culture flasks were capped with milk filters and autoclave paper, secured with a rubber band and incubated for 48 h. The soluble compounds from the 1-L culture were then extracted using Amberlite-XAD 16 resin (20 g L−1 of medium). The resin was shaken in the culture flask for 1 h at 225 r.p.m. and the resin and cells were collected by vacuum filtration. The resin, cells and filter paper were then back-extracted using 100–200 mL of MeOH. The organic material was dried in vacuo and separated based on polarity under solid-phase extraction (SPE) using a Supelco Discovery DSC-18 cartridge (20 mL, 5 g). The material was subjected to seven elution steps, six covering a gradient of MeOH/H2O (A: 10 % MeOH, B: 20 % MeOH, C: 40 % MeOH, D: 60 % MeOH, E: 80 % MeOH, F: 100 % MeOH) and the seventh 100 % EtOAc. All fractions were dried in vacuo.
Fractions C, D and E contained pumazine when checked by dried droplet (DD) on the matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF)mass spectrometer. Fraction E was subjected to liquid chromatography (LC)/tandem mass spectrometry (MS/MS) (LC-MS/MS) at a gradient of 40–100 % MeOH/H2O over 30 min. Pumazine was detected at 20.5 min (approximately 85 % MeOH). Fraction E was subjected to an initial isocratic isolation at 72 % MeOH (0.2 % FA) using RP-HPLC on a C18 column (Phenomenex Kinetex; 5 µm C18, 100 Å, 150×4.6 mm). Eluents between 3 and 4 min were collected, dried in vacuo and further separated by HPLC using isocratic conditions, 50 % MeOH (0.2 % FA) over 12 min, on a biphenyl column (Phenomenex Kinetex; 5 µm biphenyl, 100 Å, 150×4.6 mm). Under these conditions, pumazine eluted at 8.1 min and the resulting red fraction was dried in vacuo and protected from light and air. The collected compound was a bright magenta colour when dried.
Pumazine
Pink solid; [α]20 0 (c 0.1, MeOH); UV (MeOH) λmax (log ɛ) 280 (10.88) nm, 380 (11.02) nm and 508 (11.14) nm; IR (glass, cm−1) 3413, 2980, 1596 and 1384; 1H NMR (400 MHz, DMSO-d6) δ 7.26 (d, 1 h, J=8.5 Hz), 7.98 (d, 1 h, J=8.5 Hz); High resolution electrospray ionization mass spectrometry (HRESIMS) m/z [M+H]+ 275.0308 (calc. C12H7N2O6, 275.0304).
Nuclear magnetic resonance (NMR) spectra were collected on a Bruker AVII 400 MHz spectrometer equipped with a 5 mm Z-gradient BBO probe with an internal standard of tetramethylsilane (TMS) and referenced to residual solvent proton signals (ΔH 2.50 for DMSO). HPLC purifications were performed with an Agilent Technologies 1260 Infinity HPLC. All solvents were Optima grade and were used without further purification. Direct infusion was performed on a LCQ Advantage Max (Thermo Finnigan). LC-MS/MS experiments were performed on an 6300 ion trap (Agilent Technologies). HRESIMS data were collected on both an 6550 i-Funnel Q-TOF (Agilent Technologies) and an Impact II Q-TOF (Bruker Daltonics, Billerica, MA). UV-visible data were collected on a UV-2401PC UV/vis spectrophotometer (Shimadzu). IR data were collected on a Thermo Scientific Nicolet 6700 FT-IR. Optical rotation data were collected on a 241 Polarimeter (Perkin Elmer). NMR modelling was performed using NMR Predict (nmrdb.org).
Cyclic voltammetry
Electrochemical measurements were performed on a μAutolab Type III potentiostat (Eco Chemie, Metrohm USA, Riverview, FL, USA) in a Faraday cage. The electrochemical cell contained a glassy carbon (d=2.4 mm) disk working electrode, a platinum wire counter electrode and a Ag/AgCl reference electrode (BASi, West Lafayette, IN, USA). The electrodes were mechanically polished with 1.0 and 0.3 µm alumina slurries (Buehler, Lake Bluff, IL, USA), after which they were rinsed with distilled water. Solutions were purged with ultra-high purity N2 (g) prior to and during the experiment. Cyclic voltammograms were recorded at a scan rate of 0.1 Vs−1 for five consecutive cycles. Potentials are reported versus Ag/AgCl. Phenazine methosulfate was obtained from TCI America and pumazine was purified as indicated in the mass spectrometry methods text above. Both compounds were tested at a concentration of 15 µM. The aprotic solvent system used consisted of dimethyl sulfoxide (DMSO) with 100 mM tetrabutylammonium hexafluorophosphate (TBAPF6); the protic solvent system consisted of 20 mM MOPS (pH 7.2) with 100 mM potassium chloride. All chemicals were purchased from Sigma Aldrich at analytical (>99.5 %) purity and used without further purification.
Results and Discussion
Genes for N-methylated phenazine production and those implicated in resistance co-occur in pseudomonad species
We previously observed that SoxR-mediated regulation is important for normal P. aeruginosa PA14 biofilm development and growth under conditions where N-methylated phenazines are produced or added to the growth medium [13]. We therefore hypothesized that the ability to produce N-methylated phenazines and proteins that protect from these compounds would correlate in Pseudomonas species. In P. aeruginosa, phenazine methylation is catalyzed by the enzyme PhzM, which converts phenazine-1-carboxylic acid (PCA) to 5-Me-PCA [25] (Fig. 1a). We used the Pseudomonas Genome Database [26] to search for orthologues of SoxR and PhzM, and proteins that are expressed in response to SoxR activation. We found 94 species with SoxR orthologues (Fig. 1c). Orthologues of PhzM were present in only four species, all of which also contained SoxR. We found a perfect overlap between genomes that contained orthologues of phzM and PA14_35160 (pumA), which encodes a putative monooxygenase and is regulated by SoxR. Orthologues of mexG, which is also regulated by SoxR and encodes a component of the efflux pump that protects P. aeruginosa from N-methylated phenazines [13], were found in these same four genomes and an additional five genomes, all of which also contained soxR. Thus, the potential to produce methylated phenazines is a relatively rare feature among pseudomonad species, and it tends to co-occur with genes that provide protection from these products.
A mutant lacking PA14_35160 (PumA) shows altered growth and biofilm development in response to synthetic and endogenous N-methylated phenazines
The P. aeruginosa phenazine biosynthetic pathway is branched and yields at least four different phenazine products during biofilm growth on our standard medium (1 % tryptone, 1 % agar) (Fig. 1a) [27, 28]. We found that 5-Me-PCA has more pronounced effects on SoxR-mediated gene expression and colony morphogenesis than other P. aeruginosa phenazines [13]. Because 5-Me-PCA is unstable, we use the synthetic analogue PMS to characterize the effects of specific gene deletions on P. aeruginosa’s responses to methylated phenazines. We conducted a time course examining the colony development of wild-type PA14 and mutants representing SoxR and SoxR-regulated genes. As previously reported, we observed that ∆pumA colony biofilms show defective growth on medium containing 600 µM PMS and that pumA and mexGHI-opmD together account for the full degree of protection from PMS afforded by the SoxR regulon (Fig. 2a) [13].
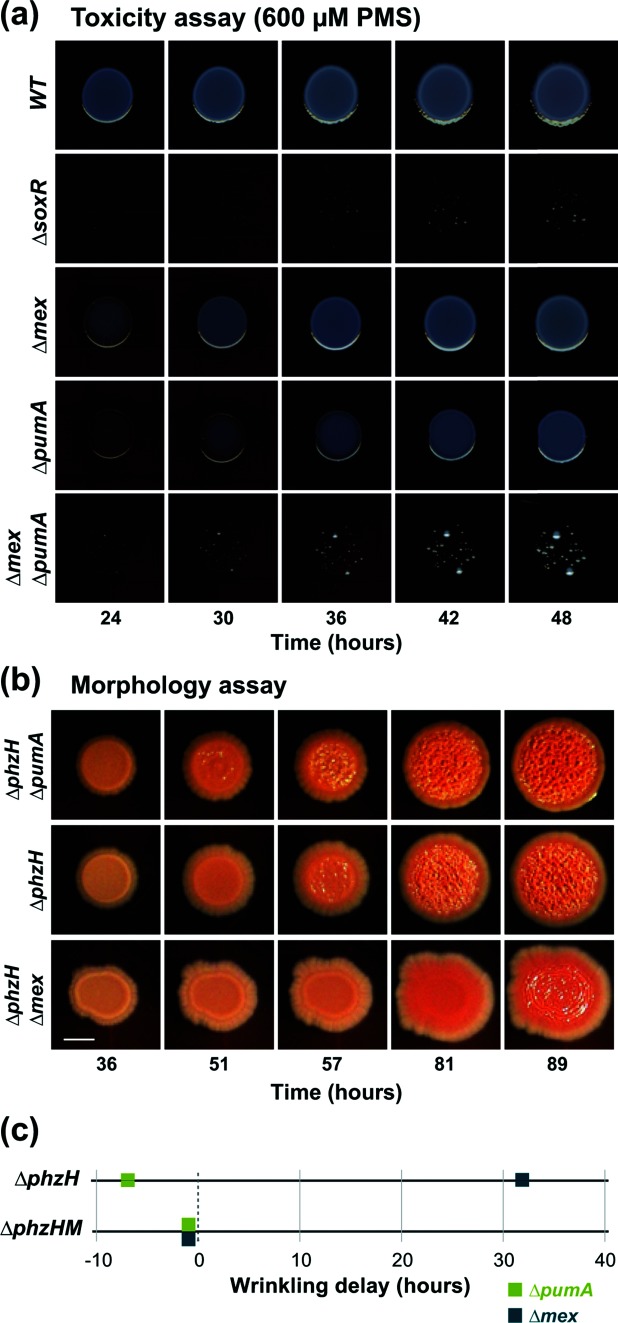
Fig. 2.
PumA contributes to P. aeruginosa phenazine resistance and colony development. (a) Growth of WT PA14 and mutants on medium containing 600 µM PMS. (b) Colony biofilm development of ∆pumA and ∆mexGHI-OpmD in a strain background that overproduces methylated phenazines (∆phzH). Deletion of pumA stimulates colony wrinkling, while deletion of mexGHI-opmD inhibits it, under this condition. The images were taken from time-lapse movie S1. (c) Onset of wrinkling relative to parent strains ∆phzH (top) and ∆phzHM (bottom) for the indicated mutants. Deletion of pumA stimulates colony wrinkling, while deletion of mexGHI-opmD (‘mex’) inhibits colony wrinkling. This wrinkling differential is specific to the overproduction of N-methylated phenazines, as the onset of wrinkling in the ∆phzH∆phzM background is not significantly different from that of the parent for ∆pumA or ∆mex. The onset of wrinkling was determined from time-lapse movies S1 and S2.
Although N-methylated phenazines such as PMS are toxic at high concentrations, our previous work indicated that they can provide a physiological benefit at lower concentrations by acting as alternative electron acceptors for cells in oxygen-limited regions of biofilms [13]. This cellular redox balancing promotes the formation of smooth colonies, while electron acceptor limitation promotes colony wrinkling [29]. To determine whether PumA is involved in the morphological response to an endogenous N-methylated phenazine, we generated a series of combinatorial mutants with deletions in phz genes and SoxR target genes. The mutants shown in Fig. 2(b) overproduce 5-Me-PCA due to a deletion in phzH (Fig. 1a). We observed that the deletion of pumA in this background stimulated colony wrinkling, while the deletion of mexGHI-opmD had the opposite effect (Fig. 2b). To assess whether the earlier onset of wrinkling in ∆pumA strains could be attributed specifically to the effect of methylated phenazines, we compared the colony phenotypes of ∆pumA in the presence and absence of the methyltransferase gene phzM. Consistent with the notion that PumA acts on methylated phenazines, we did not find a significant effect on colony development in a phzM deletion background (Figs S1a and 2c). Accordingly, we also found that deletion of pumA in the ∆phz background did not affect colony morphology in the absence of phenazines, but led to enhanced colony wrinkling when the strains were grown on the synthetic methylated phenazine PMS at all concentrations of PMS tested (Fig. S1b). Taken together, these results implicate PumA in P. aeruginosa phenazine resistance and phenazine-mediated repression of colony wrinkling. Our work suggests that mexGHI-opmD deletion leads to increased retention of 5-Me-PCA and other N-methylated phenazines, thereby leading to a more oxidized cellular redox state and inhibition of colony wrinkling. The fact that a pumA deletion stimulates colony wrinkling suggests that this mutant harbours a more reduced cellular redox state. We hypothesize that PumA converts 5-Me-PCA and/or other N-methylated phenazines to products with potentials that are more oxidizing.
PumA is a putative monooxygenase
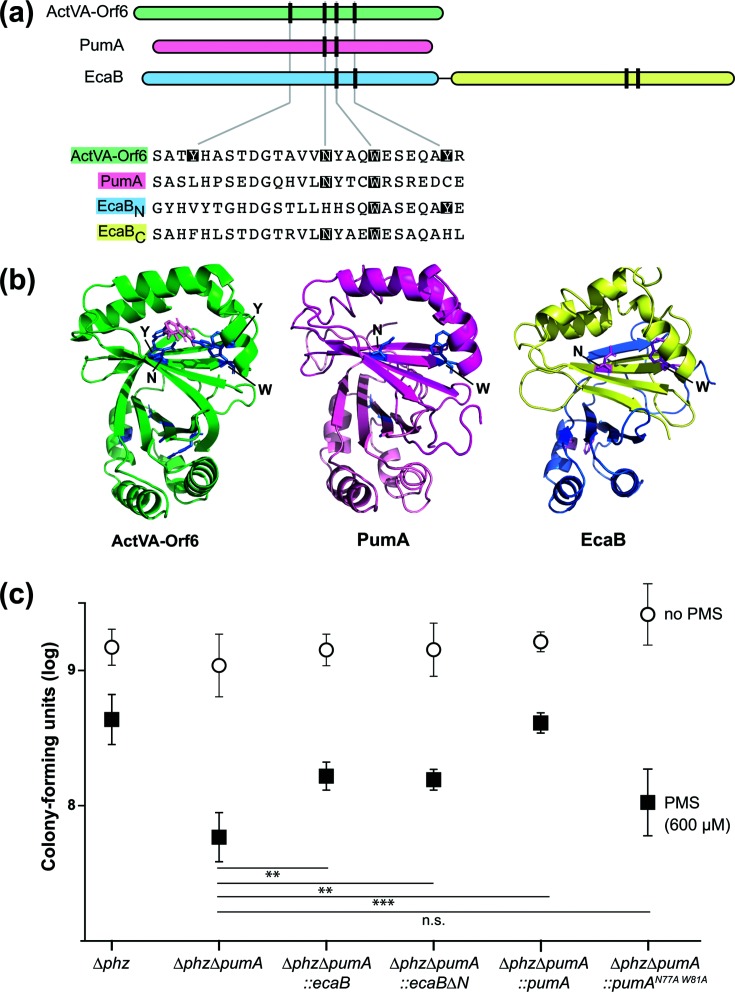
PumA is orthologous to two separate proteins found in Streptomyces coelicolor: ActVA6-Orf6 and EcaB. ActVA6-Orf6 has been crystallized as a dimer and acts as a monooxygenase in the synthesis of a polyketide called actinorhodin [23, 30]. This redox-active antibiotic affects S. coelicolor colony morphogenesis in a similar manner to phenazines in P. aeruginosa [6]. Actinorhodin also activates S. coelicolor SoxR, which controls the expression of a six-gene regulon that includes ecaB [9]. The conserved regulation of PumA and EcaB expression by SoxR suggests that these two putative monooxygenases may have similar functions. EcaB is approximately twice the length of PumA (234 vs 125 amino acid residues). This increased size is likely due to an internal duplication, as the EcaB monomer consists of two domains that are each homologous to PumA (Fig. 3a). We will refer to the N-terminal and C-terminal domains of EcaB as ‘EcaBN’ and ‘EcaBC’, respectively. In a previous study, Sciara et al. identified four active site residues, which we have highlighted in the ActVA-Orf6 shown in Fig. 3(a) [23]. Two of these, an asparagine and a tryptophan, are well conserved between orthologues and were proposed to be required for catalysis, while the two poorly conserved tyrosine residues were proposed to confer substrate specificity in divergent monooxygenases [23]. Although EcaBN contains a histidine in place of the catalytic asparagine residue, both PumA and EcaBC contain putative active sites (Fig. 3b). Given the similarities between PumA and the C-terminal domain of EcaB, we were interested in examining whether EcaB and EcaBC could functionally replace PumA in vivo.
Fig. 3.
PumA shows homology to the S. coelicolor monooxygenase ActVA-Orf6 and the S. coelicolor protein EcaB, which also confers PMS tolerance in P. aeruginosa. (a) Top: domain architectures of the S. coelicolor monooxygenase ActVa-Orf6, P. aeruginosa PumA and the S. coelicolor SoxR-target EcaB. Vertical lines represent the locations of residues that are important for ActVA-Orf6 activity. Bottom: alignment of selected regions from S. coelicolor ActVA-Orf6, P. aeruginosa PumA and S. coelicolor EcaB. Catalytic site residues identified in S. coelicolor ActVA-Orf6 that are conserved are indicated with shading. (b) Structure of S. coelicolor ActVA-Orf6 (left), and threaded structures of P. aeruginosa PumA (middle) and S. coelicolor EcaB (right). The domains are colour-coded as in panel (a). (c) Assay for tolerance to 600 µM PMS. Biofilms were grown on 1 % tryptone+1 % agar for 4 days, and then whole colonies were homogenized for colony-forming unit plating. The error bars represent the standard deviation of biological quadruplicates. P-values were calculated using unpaired, two-tailed t-tests (P>0.05 was considered not significant; **P≤0.005; ***P≤0.0005).
We tested whether EcaB, EcaBC, or a version of PumA with a mutant active site (i.e. lacking the conserved asparagine and tryptophan residues) could confer PMS tolerance in a ∆phz∆pumA background by growing strains as biofilms on medium with or without 600 µM PMS for 4 days, harvesting and homogenizing the biofilms, and plating for colony-forming units. In this assay, ∆phz∆pumA showed a 1.5 log decrease in c.f.u.s for biofilms grown on 600 µM PMS compared to those grown on tryptone alone (Fig. 3c). Resistance to PMS was rescued by complementation with intact PumA (∆pumA :: pumA). When ∆phz∆pumA was complemented with a catalytic site mutant of PumA, PumAN76A,W80A, there was no observable rescue of PMS resistance. Complementation of ∆phz∆pumA with both full-length ecaB and ecaBC resulted in partial rescue on 600 µM PMS. These results suggest that EcaB and PumA could play similar roles in antibiotic self-resistance in their respective organisms.
PumA modifies PMS during growth in biofilms and liquid cultures
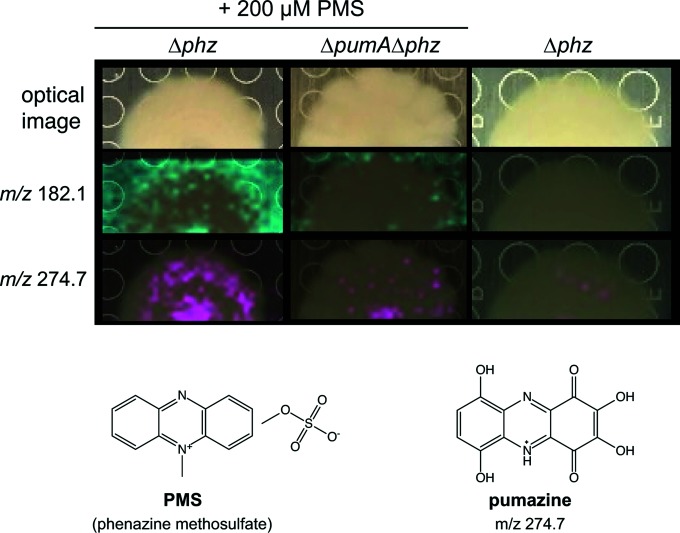
In S. coelicolor, ActVA-Orf6 catalyzes the oxidation of an intermediate in actinorhodin biosynthesis. This reaction converts 6-deoxydihydrokalafungin into dihydrokalfungin (quinone) [23]. By analogy, we hypothesized that PumA could transform phenazines. We used imaging mass spectrometry (IMS) to identify PumA-dependent metabolites produced by biofilms grown on medium containing 200 µM PMS (Figs 4 and S2) [31]. We found two metabolites, m/z 182.1 and m/z 274.7, that (a) showed increased abundance in ∆phz biofilms grown on PMS when compared to ∆phz∆pumA biofilms grown on PMS, and (b) were absent from ∆phz biofilms grown on PMS-free medium. To purify enough compound to enable characterization, we grew ∆phz and ∆phz∆pumA as shaken liquid cultures in medium containing 200 µM PMS and analysed the culture supernatants for PumA-specific metabolites. We found that the m/z 274.7 compound identified by IMS of biofilms was also produced during growth in liquid culture. Species m/z 182.1 was not observed in liquid culture and was therefore not pursued for further structure elucidation studies.
Fig. 4.
Identification of PumA-dependent metabolites. Top: optical images of colony biofilms and imaging mass spectrometry results showing distribution of metabolites with the indicated mass-to-charge (m/z) ratios (in cyan or fuschia) overlaid on colony images. Colonies were grown for 5 days on 1 % tryptone and 1 % agar. PMS was added to the medium where indicated. Bottom: structure for PMS and predicted structure for pumazine.
We purified the m/z 274.7 compound from culture supernatants and determined its structure using LC-MS/MS fragmentation and NMR (Fig. S3a). This metabolite, henceforth called pumazine, is a highly modified phenazine core with six additional oxygen atoms substituted at six different aromatic carbons. Deuterium exchange indicates that four of the oxygen substituents are hydroxyl groups (Fig. S3b) and the remaining two oxygens are in keto form (quinone). We further characterized the purified pumazine compound by UV-visible spectrophotometry and cyclic voltammetry. The absorbance spectra of natural phenazines often show features in the ranges of 250–290 and 350–400 nm, with a subset showing a peak between 400–600 nm, consistent with colouration [32, 33]. Pumazine displays characteristic phenazine absorbance peaks at 280, 375 and 515 nm (Fig. S4a). We examined the redox activity of pumazine by cyclic voltammetry in aprotic (0.1 M TBAPF6/DMSO) and protic (20 mM MOPS buffer, pH 7.2) solvent systems. A quasi-reversible redox couple is observed on glassy carbon electrodes that is shifted reductively relative to PMS (ΔEmid~360 mV in DMSO; ΔEmid~220 mV in MOPS buffer) and, by inference, to 5-Me-PCA (Fig. S4b, c). We attribute the substantial potential shift to the extent of substitution on the phenazine core [34–36].
PumA acts on endogenous phenazines
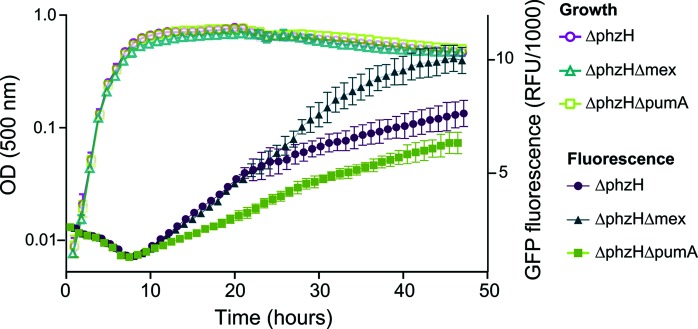
Studies in our laboratory indicated that P. aeruginosa colony morphogenesis is linked to the cellular redox state [16, 37] and that phenazine-dependent repression of colony wrinkling is mediated in part by the oxidizing effects of these compounds [38]. We therefore hypothesized that the early onset of wrinkling in ∆pumA mutants was stimulated by a shift in the stoichiometry of the intracellular phenazine pool. We use a fluorescent reporter of mexGHI-opmD expression, PmexG-GFP, as a proxy for SoxR activity. We observed that ∆phzH mutants, which produce increased amounts of 5-Me-PCA, show the highest levels of SoxR activation when compared to strains with intact or otherwise altered phenazine biosynthetic pathways [13, 39]. We have also seen that removal of the MexGHI-OpmD efflux pump, which transports 5-Me-PCA, further induces mexGHI-opmD expression. As SoxR is activated by oxidation of its Fe–S cluster, these results are consistent with the fact that 5-Me-PCA has the most oxidizing potential of all of the well-characterized P. aeruginosa phenazines [36]. To test whether the endogenous phenazine pool is affected by the ∆pumA mutation, we moved the PmexG-GFP reporter into a ∆phzH∆pumA mutant and measured fluorescence during growth in well-mixed liquid cultures. This strain showed lower levels of GFP expression, suggesting that it has a phenazine pool with a more reducing potential than its parent strain (Fig. 5).
Fig. 5.
PumA and MexGHI-OpmD divergently affect SoxR activation. In a mutant background that overproduces N-methylated phenazines (5-Me-PCA and derivatives), ∆mex strains show increased expression of a SoxR-regulated GFP reporter, while ∆pumA strains show decreased expression. Because SoxR is activated by oxidized phenazines, these results indicate that the phenazine pool is more oxidizing in ∆mex strains and more reducing in ∆pumA strains.
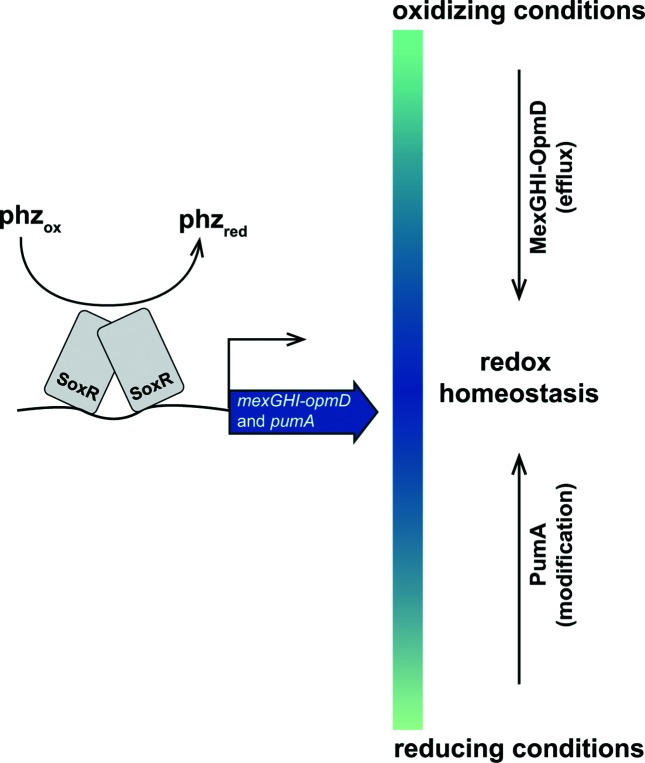
The measured midpoint potential of pumazine is much more negative than the values reported for N-methylated phenazines (Fig. S4b, c) [34–36]. If pumazine were the primary product of PumA, we would therefore expect the ∆pumA mutation to lead to a more oxidized environment. However, pumA deletion leads to earlier wrinkling (Fig. 2b, c) and decreased SoxR reporter activity (Fig. 5), effects that are consistent with a more reduced cellular redox state. These findings indicate that pumazine is not the direct product of PumA activity, which is plausible given the known reactivity of modified phenazines. Nevertheless, our results suggest that PumA acts on endogenous phenazines to modulate cellular redox conditions and prompt us to propose a model in which the SoxR regulon supports redox homeostasis by modulating the phenazine pool: while the efflux pump MexGHI-OpmD counteracts excessively oxidizing conditions by exporting methylated phenazines, PumA modifies endogenous P. aeruginosa phenazine structures and/or levels in a way that shifts the conditions inside the cell to a more oxidizing state (Fig. 6).
Fig. 6.
Model depicting the roles of SoxR-regulated proteins in modulating the cellular phenazine pool. Phenazine-mediated oxidation of SoxR activates the transcription of target loci, including the mexGHI-opmD operon (encoding an efflux pump), and PA14_35160 (pumA) (encoding a putative monooxygenase). Mutant colony morphotypes and reporter gene expression data indicate that these SoxR targets have differential effects on the redox potential of the cellular phenazine pool: while MexGHI-OpmD-mediated efflux leads to phenotypes consistent with a more reducing phenazine pool, PumA activity leads to phenotypes consistent with a more oxidizing one. We propose that these proteins work simultaneously to ensure optimal phenazine self-resistance in P. aeruginosa PA14.
Concluding remarks
Our results suggest that PumA acts on endogenous and exogenous N-methylated phenazines to yield a phenazine pool that supports P. aeruginosa growth and colony development. They also indicate that EcaB could act similarly on antibiotics produced or taken up by S. coelicolor. The SoxR-mediated induction of modifying enzymes may therefore represent a conserved mechanism of self-resistance in divergent bacterial phyla.
Supplementary Data
Funding information
This work was funded by NIH grant R01AI103369 and an NSF CAREER award to L. E. P. D. This work was also funded by UIC startup funds and grant number K12HD055892 from the National Institute of Child Health and Human Development (NICHD) and the National Institutes of Health Office of Research on Women’s Health (ORWH) (LMS). A. J. S. was supported by a Columbia Frontiers of Science Fellowship.
Acknowledgements
The authors wish to thank Dr Monica Chander for insightful discussions and Valentina Mendez and Blanche L. Fields for technical assistance. We would like to thank Drs Atul Jain and Terry Moore for assistance with matrix recrystallization, Dr Matt Bertin from the University of Rhode Island for helpful conversations regarding structure elucidation and Dr Stephanie Cologna for access to HRESIMS data collection. IMS data are freely available on Metabolights (ebi.ac.uk/metabolights) under study MTBLS645. Fragmentation data of pumazine are freely available on MassIVE (massive.ucsd.edu) under accession number MSV000082176.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Three supplementary tables, four supplementary figures, and two supplementary videos are available with the online version of this article.
Abbreviations: DMSO, dimethyl sulfoxide; HRESIMS, High resolution electrospray ionization mass spectrometry; IMS, imaging mass spectroscopy; LC/MS, liquid chromatography/mass spectroscopy; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight; 5-Me-PCA, 5-methylphenazine-1-carboxylic acid; Mex, MexGHI-OpmD; MFS, major facilitator super family; MOPS, 3-(N-morpholino)propanesulfonic acid; PCA, phenazine-1-carboxylic acid; PMS, phenazine methosulfate; PumA, phenazine-utilizing monooxygenase A; RND, resistance nodulation division; TBAPF6, tetrabutylammonium hexafluorophosphate.
Edited by: S. P. Diggle and M. Whiteley
References
- 1.Bernier SP, Surette MG. Concentration-dependent activity of antibiotics in natural environments. Front Microbiol. 2013;4:20. doi: 10.3389/fmicb.2013.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies J. Specialized microbial metabolites: functions and origins. J Antibiot. 2013;66:361–364. doi: 10.1038/ja.2013.61. [DOI] [PubMed] [Google Scholar]
- 3.Heeb S, Fletcher MP, Chhabra SR, Diggle SP, Williams P, et al. Quinolones: from antibiotics to autoinducers. FEMS Microbiol Rev. 2011;35:247–274. doi: 10.1111/j.1574-6976.2010.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, et al. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J Bacteriol. 2011;193:3606–3617. doi: 10.1128/JB.00396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price-Whelan A, Dietrich LE, Newman DK. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:6372–6381. doi: 10.1128/JB.00505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dietrich LE, Teal TK, Price-Whelan A, Newman DK. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science. 2008;321:1203–1206. doi: 10.1126/science.1160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheplock R, Recinos DA, Mackow N, Dietrich LE, Chander M. Species-specific residues calibrate SoxR sensitivity to redox-active molecules. Mol Microbiol. 2013;87:368–381. doi: 10.1111/mmi.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh AK, Shin JH, Lee KL, Imlay JA, Roe JH. Comparative study of SoxR activation by redox-active compounds. Mol Microbiol. 2013;90:983–996. doi: 10.1111/mmi.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naseer N, Shapiro JA, Chander M. RNA-Seq analysis reveals a six-gene SoxR regulon in Streptomyces coelicolor. PLoS One. 2014;9:e106181. doi: 10.1371/journal.pone.0106181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hidalgo E, Demple B. Spacing of promoter elements regulates the basal expression of the soxS gene and converts SoxR from a transcriptional activator into a repressor. Embo J. 1997;16:1056–1065. doi: 10.1093/emboj/16.5.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe S, Kita A, Kobayashi K, Miki K. Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc Natl Acad Sci USA. 2008;105:4121–4126. doi: 10.1073/pnas.0709188105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu M, Imlay JA. The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol Microbiol. 2011;79:1136–1150. doi: 10.1111/j.1365-2958.2010.07520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakhtah H, Koyama L, Zhang Y, Morales DK, Fields BL, et al. The Pseudomonas aeruginosa efflux pump MexGHI-OpmD transports a natural phenazine that controls gene expression and biofilm development. Proc Natl Acad Sci USA. 2016;113:E3538. doi: 10.1073/pnas.1600424113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietrich LE, Price-Whelan A, Petersen A, Whiteley M, Newman DK. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa . Mol Microbiol. 2006;61:1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez ME, Kappler A, Newman DK. Phenazines and other redox-active antibiotics promote microbial mineral reduction. Appl Environ Microbiol. 2004;70:921–928. doi: 10.1128/AEM.70.2.921-928.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dietrich LE, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC, et al. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol. 2013;195:1371–1380. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kempes CP, Okegbe C, Mears-Clarke Z, Follows MJ, Dietrich LE. Morphological optimization for access to dual oxidants in biofilms. Proc Natl Acad Sci USA. 2014;111:208–213. doi: 10.1073/pnas.1315521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertani G. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol. 2004;186:595–600. doi: 10.1128/JB.186.3.595-600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol. 2006;72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Recinos DA, Sekedat MD, Hernandez A, Cohen TS, Sakhtah H, et al. Redundant phenazine operons in Pseudomonas aeruginosa exhibit environment-dependent expression and differential roles in pathogenicity. Proc Natl Acad Sci USA. 2012;109:19420–19425. doi: 10.1073/pnas.1213901109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 22.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sciara G, Kendrew SG, Miele AE, Marsh NG, Federici L, et al. The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis. Embo J. 2003;22:205–215. doi: 10.1093/emboj/cdg031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schrödinger, LLC The PyMOL Molecular Graphics System, Version 2.0
- 25.Parsons JF, Greenhagen BT, Shi K, Calabrese K, Robinson H, et al. Structural and functional analysis of the pyocyanin biosynthetic protein PhzM from Pseudomonas aeruginosa . Biochemistry. 2007;46:1821–1828. doi: 10.1021/bi6024403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, et al. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mavrodi DV, Peever TL, Mavrodi OV, Parejko JA, Raaijmakers JM, et al. Diversity and evolution of the phenazine biosynthesis pathway. Appl Environ Microbiol. 2010;76:866–879. doi: 10.1128/AEM.02009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakhtah H, Price-Whelan A, Dietrich L. Microbial Phenazines. Berlin, Heidelberg: Springer; 2013. Regulation of phenazine biosynthesis; pp. 19–42. [Google Scholar]
- 29.Madsen JS, Lin YC, Squyres GR, Price-Whelan A, de Santiago Torio A, et al. Facultative control of matrix production optimizes competitive fitness in Pseudomonas aeruginosa PA14 biofilm models. Appl Environ Microbiol. 2015;81:8414–8426. doi: 10.1128/AEM.02628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kendrew SG, Hopwood DA, Marsh EN. Identification of a monooxygenase from Streptomyces coelicolor A3(2) involved in biosynthesis of actinorhodin: purification and characterization of the recombinant enzyme. J Bacteriol. 1997;179:4305–4310. doi: 10.1128/jb.179.13.4305-4310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang JY, Phelan VV, Simkovsky R, Watrous JD, Trial RM, et al. Primer on agar-based microbial imaging mass spectrometry. J Bacteriol. 2012;194:6023–6028. doi: 10.1128/JB.00823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner JM, Messenger AJ. Occurrence, biochemistry and physiology of phenazine pigment production. Adv Microb Physiol. 1986;27:211–275. doi: 10.1016/s0065-2911(08)60306-9. [DOI] [PubMed] [Google Scholar]
- 33.Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, et al. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol. 2001;183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Newman DK. Redox reactions of phenazine antibiotics with ferric (hydr)oxides and molecular oxygen. Environ Sci Technol. 2008;42:2380–2386. doi: 10.1021/es702290a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellin DL, Sakhtah H, Rosenstein JK, Levine PM, Thimot J, et al. Integrated circuit-based electrochemical sensor for spatially resolved detection of redox-active metabolites in biofilms. Nat Commun. 2014;5:3256. doi: 10.1038/ncomms4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng H, Kim J, Liew M, Yan JK, Herrera O, et al. Redox metabolites signal polymicrobial biofilm development via the NapA oxidative stress cascade in Aspergillus . Curr Biol. 2015;25:29–37. doi: 10.1016/j.cub.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okegbe C, Price-Whelan A, Dietrich LE. Redox-driven regulation of microbial community morphogenesis. Curr Opin Microbiol. 2014;18:39–45. doi: 10.1016/j.mib.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okegbe C, Fields BL, Cole SJ, Beierschmitt C, Morgan CJ, et al. Electron-shuttling antibiotics structure bacterial communities by modulating cellular levels of c-di-GMP. Proc Natl Acad Sci USA. 2017;114:E5236. doi: 10.1073/pnas.1700264114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin YC, Sekedat MD, Cornell WC, Silva GM, Okegbe C, et al. Phenazines regulate Nap-dependent denitrification in Pseudomonas aeruginosa biofilms. J Bacteriol. 2018 doi: 10.1128/JB.00031-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.