Significance
Oxygen availability is essential for development, growth, and viability of aerobic organisms. The genes in the hypoxia-inducible factor (HIF) pathway are considered master regulators of oxygen sensitivity and distribution inside cells, and they are hence highly conserved across animal groups. These genes are frequent targets of natural selection in organisms living in low-oxygen environments, such as high-altitude humans and birds. Here, we show that the abundant tidepool copepod Tigriopus californicus can withstand prolonged exposure to extreme oxygen deprivation, despite having secondarily lost key HIF-pathway members. Our results suggest the existence of alternative mechanisms of response to hypoxic stress in animals, and we show that genes involved in cuticle reorganization and ion transport may play a major role.
Keywords: hypoxia, transcription factor, gene expression
Abstract
Hypoxia is a major physiological constraint for which multicellular eukaryotes have evolved robust cellular mechanisms capable of addressing dynamic changes in O2 availability. In animals, oxygen sensing and regulation is primarily performed by the hypoxia-inducible factor (HIF) pathway, and the key components of this pathway are thought to be highly conserved across metazoans. Marine intertidal habitats are dynamic environments, and their inhabitants are known to tolerate wide fluctuations in salinity, temperature, pH, and oxygen. In this study, we show that an abundant intertidal crustacean, the copepod Tigriopus californicus, has lost major genetic components of the HIF pathway, but still shows robust survivorship and transcriptional response to hypoxia. Mining of protein domains across the genome, followed by phylogenetic analyses of gene families, did not identify two key regulatory elements of the metazoan hypoxia response, namely the transcription factor HIF-α and its oxygen-sensing prolyl hydroxylase repressor, EGLN. Despite this loss, phenotypic assays revealed that this species is tolerant to extremely low levels of available O2 for at least 24 h at both larval and adult stages. RNA-sequencing (seq) of copepods exposed to nearly anoxic conditions showed differential expression of over 400 genes, with evidence for induction of glycolytic metabolism without a depression of oxidative phosphorylation. Moreover, genes involved in chitin metabolism and cuticle reorganization show categorically a consistent pattern of change during anoxia, highlighting this pathway as a potential solution to low oxygen availability in this small animal with no respiratory structures or pigment.
Multicellular eukaryotes have evolved specialized cellular mechanisms to enhance O2 uptake and distribution, resulting in dynamic respiratory and circulatory systems capable of responding to changes in O2 availability. These responses are mediated in part through the induction of hypoxia-inducible factors (HIF), the regulatory components that are highly conserved both in form and function across phyla (1, 2). The main transcription factors associated with the HIF pathway are HIF-α (HIF-1α, HIF-2α/EPAS1 in vertebrates; HIF-1α in invertebrates) and HIF-β/ARNT, which form a dimer and move into the nucleus. The activity of the HIF-α subunit is governed posttranslationally via a repression machinery (EGLN, or PHD: prolyl hydroxylase); during normoxic conditions, HIF-α is hydroxylated by EGLN and subsequently destroyed before dimerization with HIF-β/ARNT. However, during hypoxic conditions, EGLN activity is inhibited and the HIF heterodimer is allowed to form in the nucleus. HIF recognizes and attaches to DNA regions called HREs (hypoxia response elements) located upstream of target genes, thus manipulating their expression patterns (3). As “master regulators” of oxygen sensitivity, the regulatory machinery and downstream genes are frequent targets of selection for populations living in low oxygen environments in a variety of organisms (4–6).
Hypoxia in ocean waters has been rapidly increasing in distribution, frequency, and severity (7), and certain coastal ecosystems even reach levels of anoxia seasonally (8). This adversely affects a wide range of organisms, with fishes and crustaceans, in general, showing lower levels of tolerance (7). From an evolutionary perspective, the ability of marine species to adjust to stressful levels of abiotic factors, such as temperature, pH, and oxygen, is a major determinant of their distributions at small and large scales.
The copepod Tigriopus californicus has become an excellent model for studies of physiological adaptations in the marine environment. It is an abundant resident of supralittoral rock pools along the west coast of North America and shows a pattern of strong genetic differentiation among populations (9). Because supralittoral pools are refreshed primarily by wave splash instead of high tides, their inhabitants are under extreme environmental stress in the form of salinity, temperature, pH, and oxygen fluctuations. In such pools, oxygen becomes more limited at night, when respiration by fauna and flora can make dissolved oxygen (DO) levels drop to ∼0 mg/L (10). Multiple studies have quantified T. californicus physiological tolerance and gene expression under high temperatures (11–13) and high and low salinities (14, 15). Patterns and mechanisms of hypoxia tolerance, however, have been largely overlooked. A single prior work showed that T. californicus is tolerant to anoxic conditions for at least 18 h (16), with no further investigation. Species of planktonic copepods have been shown to tolerate moderate hypoxic conditions, with exposures ranging from 2.4 to 1.0 mg O2/L for 24 h before significant mortality occurs (17–19). However, little is known about the genetic mechanisms of hypoxia response in any copepod or intertidal invertebrates at large. Here, we examine phenotypic and transcriptional patterns of hypoxia response in T. californicus and show that T. californicus has secondarily lost key members of the HIF pathway, as well as suffered a reduction in the fidelity of HREs across their genome.
Results
HIF Pathway Genes.
Examination of the original annotation of the T. californicus genome (20) did not reveal the presence of HIF-1α or EGLN1, but such absence may be the result of an incomplete assembly or annotation of the draft genome sequence. We thus performed targeted computational analyses, independent of homology BLAST methods, to search for them. Besides the genome-derived protein sequences, we included transcriptome assemblies that were generated de novo from five different populations. We reasoned that these would provide complementary probabilities that these genes were sequenced and assembled in any of the T. californicus assemblies.
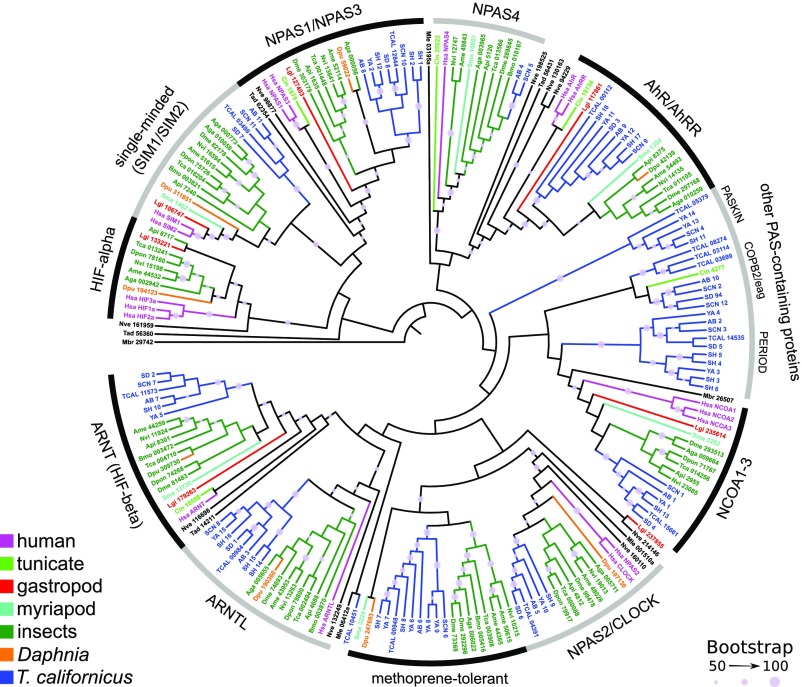
HMM searches of protein domains and phylogenetic analyses showed no evidence for the presence of either HIF-α or EGLN1 genes, suggesting that T. californicus has lost the canonical HIF pathway. A phylogenetic tree including sequences with e value < 0.01 yielded no T. californicus sequence within the invertebrate/vertebrate HIF-α clade (Fig. 1). We did, however, clearly detect representatives from the other bHLH-PAS–containing protein groups (AhR/AHRR, ARNT/ARNTL, CLOCK, NCOA1/2/3, NPAS1/2/3/4, SIM1/2, MET). A sequence previously identified as “HIF-α-like” protein (accession no. Cin4277) from the tunicate Ciona intestinalis rooted within a group of Tigriopus sequences near NCOA1-3, distantly placed from the well-supported HIF-α clade (Fig. 1). We scrutinized the five T. californicus genome sequences in this group by assessing their domain architectures via InterproScan (21) and found that they represent a variety of genes that contain PAS domains but do not have bHLH or other HIF-like domains. One of these proteins is PERIOD (circadian-like, IPR02278), three belong to a group with voltage-dependent channel domain (IPR027359) and cAMP-binding domains (IPR018490), and one is a member of a protein kinase-like domain superfamily (IPR011009). The fact that these T. californicus genes are not nested within those of other taxa is an artifact of our methods, since we did not necessarily include all PAS-containing genes from other species. In addition, the location of the C. intestinalis sequence has low bootstrap support, and is likely due to long-branch attraction, frequently associated with the tunicate genome (22). Therefore, we are confident none of the T. californicus sequences in this group represent a form of HIF-α gene.
Fig. 1.
Maximum likelihood tree of bHLH-PAS proteins across metazoans. Accessions for T. californicus include protein models from the reference genome (TCAL), as well as from transcriptomes from five populations (AB: Abalone Cove, SCN: Santa Cruz, SD: San Diego, SH: Strawberry Hill, YA: Yachats). Taxa abbreviations: Aga, Anopheles gambiae; Ame, Apis mellifera; Api, Acyrthosiphon pisum; Bmo, Bombyx mori; Cin, Ciona intestinalis; Dme, Drosophila melanogaster; Dpon, Dendroctonus ponderosae; Dpu, Daphnia pulex; Hsa, Homo sapies; Lgi, Lottia gigantea; Mbr, Monosiga brevicollis; Mle, Mnemiopsis leidyi; Nve, Nematostella vectensis; Nvi, Nasonia vitripennis; Sma, Strigamia maritima; Tad, Trichoplax adhaerens.
Because the HIF pathway canonically comprises the presence of both a HIF-α and an EGLN, they are often examined together to confirm the existence of this oxygen-sensing mechanism (1, 23, 24). Similar to the bHLH-PAS tree, the phylogenetic tree of all P4HC-containing proteins did not show any T. californicus representatives grouping within the EGLN clade but did show representatives of other P4HC proteins (SI Appendix, Fig. S1). Although this tree had lower levels of bootstrap support across internal nodes compared with the bHLH-PAS tree, the locations of highly supported nodes suggest that no T. californicus are EGLN-like. In addition, original BLAST annotation of the T. californicus genome did not include any EGLN-like sequences.
Anoxia Tolerance Assays.
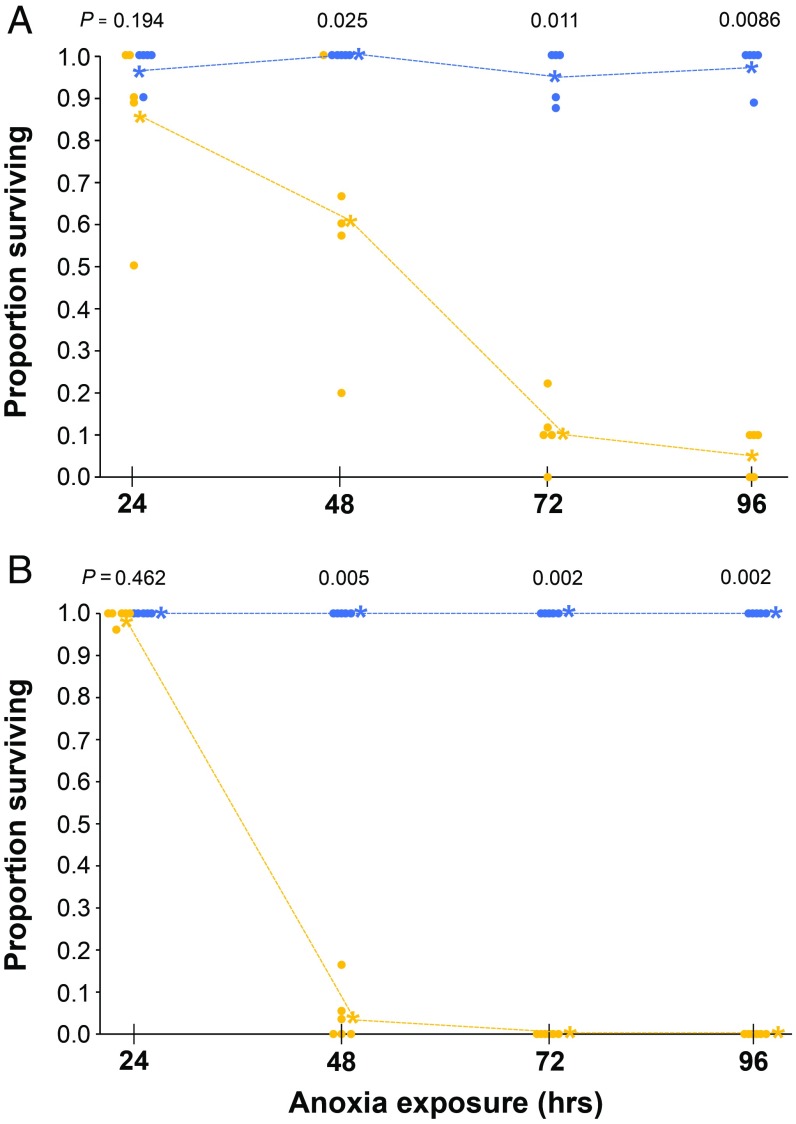
To assess levels of tolerance to anoxia in T. californicus, we quantified growth rate and viability of copepods immersed in water containing nearly no dissolved oxygen (<0.5 mg of O2/L). Copepod larvae (1 d after hatching) exposed to anoxia for 24 h showed a significant delay in initial and full onset of metamorphosis compared with their full siblings subjected to normoxic conditions (n = 36 clutches, paired Wilcoxon test, initial: V = 63, P = 0.0138; full: V = 58.5, P = 0.0134). However, there was no significant difference in onset of first adult male (V = 161, P = 0.33), nor in survival to day 20 after treatment (V = 269, P = 0.136) (Table 1 and Dataset S1A). In addition, exposing the 20-d-old adults to anoxic environments for 24 h resulted in no mortality (n = 48 clutches), regardless of which treatment they received as hatchlings (Dataset S1A). A new assessment for 24-h anoxia again showed no significant difference in survival compared with the normoxic controls (two-sample Wilcoxon test, W = 18.5, P = 0.194, Fig. 2A). Exposure to anoxia for longer periods, however, resulted in significant yet not complete mortality relative to normoxia controls, with mean survivorship of 60.7% after 48 h (W = 22.5, P = 0.025), 10.7% after 72 h (W = 25, P = 0.011), and 6% after 96 h (W = 25, P = 0.0086) (Fig. 2A and Dataset S1B). The median lethal time LT50 (±SE) was 51.4 h (±2.71).
Table 1.
Developmental rate and viability of copepods exposed to different dissolved oxygen conditions
| Trait | Normoxia | Anoxia | Difference | Paired Wilcoxon test |
| Initial metamorphosis | 6.28 (0.88) | 6.69 (0.92) | 0.42 (0.93) | V = 63, P = 0.0138 |
| Full metamorphosis | 7.47 (0.94) | 8.06 (1.31) | 0.58 (1.36) | V = 58.5, P = 0.0134 |
| First adult male | 13.78 (1.29) | 14.11 (1.21) | 0.33 (1.77) | V = 161, P = 0.338 |
| Survivorship at day 20 | 86.13% | 80.07% | −6.06% (0.18) | V = 269, P = 0.136 |
One-day-old hatched larvae were exposed to anoxia (<0.5 mg of O2/L) or normoxia (≥6.5 mg of O2/L) for 24 h. For developmental traits, data shown are mean (±SD) number of days since hatching. n = 36 clutches.
Fig. 2.
Survivorship of copepods after prolonged exposure to anoxia. Blue, normoxia (≥6.5 mg O2/L); yellow, anoxia (<0.5 mg O2/L). Exposure times tested were 24, 48, 72, and 96 h. (A) Survivorship of adult copepods (20 d old). (B) Survivorship of nauplii (1 d old). Each point represents a replicate containing multiple individuals. Mortality was assessed 3 d after exposure for the respective amount of time. Asterisks depict the mean of each treatment. P values are from two-sample Wilcoxon tests comparing between normoxia and anoxia groups at each time point. For ease of visualization, overlapping points are shifted horizontally. See Datasets S1 B and C for full data.
Juvenile copepods (1 d old) exposed to anoxia for 24 h also showed nearly total survivorship 3 d after treatment (Fig. 2B, W = 17.5, P = 0.465), but they were significantly more sensitive than adults after longer exposures, with only 4% survival after 48 h (W = 30, P = 0.0050) and 0% survival at 72–96 h (P = 0.0022; Fig. 2B and Dataset S1C). The median lethal time LT50 for nauplii was 38.2 h (±1.03).
Differential Gene Expression.
We examined transcriptional response to anoxia (<0.5 mg/L DO) by performing RNA-sequencing (seq) analyses of 20-d-old copepods after culturing for 3 h or 24 h in anoxic water, as well as of copepods transferred to normoxic water for 24 h after a 24-h anoxia treatment (“recovery” treatment). Each time point treatment was compared with its own control group of copepods in normoxic water. The three time points differed substantially with respect to number and function of genes differentially expressed (DE) between anoxia and normoxia treatments. After 3 h, there were relatively few significantly DE genes (26 total), with all but one being significantly up-regulated (Dataset S2B). Among these, we detected four members of solute carrier families, one of which is associated with protection or tolerance from high temperature stress (pyx), a heat shock protein gene (Hsp67B), and a mitochondrial metabolic gene (Pepck). The single down-regulated gene at the 3-h time point is a gene which modulates the repression activities of genes involved in apoptosis (SPOP). These genes showed a significant enrichment of Gene Ontology (GO) terms associated various ion transport and absorption (SI Appendix, Table S1).
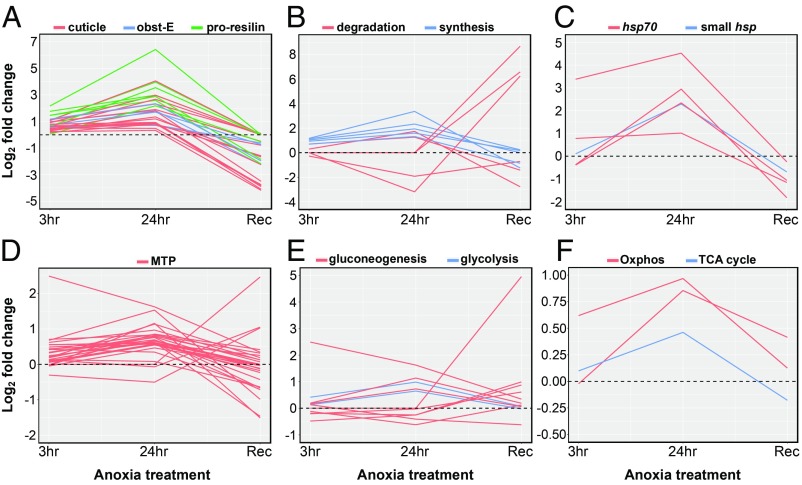
After 24 h of exposure to anoxia, the repertoire of genes changed substantially (Dataset S2C). Of the 451 DE genes, 368 were up-regulated and had GO terms associated with redox reactions (“oxidation-reduction”), as well as various metabolic processes (aminoglycan, chitin, polysaccharide metabolism) and ion transport/absorption (SI Appendix, Table S2). Notably, a number of genes associated with chitin/exoskeleton (proresilins, cuticle proteins, ost-1, obst-E, CUA2B, SgAbd-2; n = 17), chitin synthesis (CHS; n = 1), keratinization (SPRR3; n = 2), and hormones associated with eclosion/metamorphosis (EH, JH; n = 2) were significantly up-regulated (Fig. 3 A and B and Dataset S2C). Cuticle proteins, including proresilins, consistently showed the highest magnitude of up-regulation, with four of these among the overall top 15 genes and with fold change (log2) ranging from 2.98 to 6.42 (Fig. 3A). Conversely, genes involved in chitin degradation (chitinase and chitin deacetylase) were significantly down-regulated (Fig. 3B). Four Hsp chaperones became significantly up-regulated, with three from the Hsp70 family and one small Hsp (Fig. 3C). In addition, 20 nuclear-encoded mitochondrially localized protein (MTP) genes are significantly up-regulated (Fig. 3D); these genes span a range of mitochondrial metabolic processes, including glucose metabolism (PC, Pepck, Pdk; Fig. 3E), heme binding/biosynthesis (COX15, ALAS1, CYP301a1, SUOX), tricarboxylic acid cycle (DHTKD1; Fig. 3F), cytochrome c oxidase complex (HIG1, COX15; Fig. 3F), and fatty acid synthesis/acetyl-CoA (ACSF3, ACSS1).
Fig. 3.
Temporal patterns of differential gene expression during anoxia. Shown are gene functional categories that showed consistent patterns or belong to metabolic pathways. Each line represents a gene. Fold-change levels plotted are the mean values across replicates and represent the expression level during anoxia relative to the control (normoxia) treatment at each time point. Plotted are only genes that showed significant differential expression in at least one time point. Gene groups: structural cuticular proteins (A), chitin metabolism (B), heat-shock proteins (C), mitochondrially targeted proteins (D), gluconeogenesis and glycolysis (E), oxidative phosphorylation and tricarboxylic acid cycle (F).
At the recovery time point, 561 genes were DE and involved a higher proportion of down-regulated genes (355 down-regulated versus 205 up-regulated; Dataset S2D), which contrasts with the general response to 24-h anoxia. Multiple chitin disaccharide deacetylase (deaA) genes were highly up-regulated, while at 24 h of anoxia, such genes were largely down-regulated (Fig. 3B). Conversely, cuticle/proresilin structural proteins, as well as chitin synthesis genes, returned to lower levels or even became down-regulated (Fig. 3 A and B). The majority of MTP genes that were up-regulated during anoxia returned to normal levels, but three genes significantly increased their expression during recovery (Fig. 3D). Overall, the recovery treatment was characterized by GO categories involved in muscle development and activity (e.g., “muscle structure development,” “actin-myosin filament sliding”), glycolysis/respiration (6-photophofructo-2-kinase, fructose-2,6-bisphosphate 2-phosphatase), as well as “structural constituent of cuticle” (SI Appendix, Table S3).
We independently validated our RNA-seq results by repeating the experiment using new individuals and quantifying expression using reverse-transcription qPCR (RT-qPCR) of 16 genes of interest across multiple functional categories (SI Appendix, Table S4). RT-qPCR data were fully supportive of the RNA-seq results, with very similar pattern of expression levels and variation (SI Appendix, Fig. S2) and with strong correlation across genes (Spearman correlation; 24-h time point: r = 0.838, P = 5 × 10−5; recovery time point: r = 0.821, P = 0.000131).
Distribution of HREs.
HIF transcription factor dimers attach to HREs located in the promoter regions of target genes, typically in dense clusters within 1,000 bp upstream of transcription start sites (25). We detected HREs in the promoter of 5,161 protein-coding genes across the T. californicus genome (Dataset S3) and tested the association of HRE presence with putative gene function (based on GO term annotation) and hypoxia response. Among genes annotated to participate in oxygen homeostatis, 36.7% had HRE in their promoter region. This proportion was not significantly greater than the fraction of all other genes containing HREs (35.9%; Table 2; G test, G = 0.276, P = 0.599), indicating no HRE enrichment in hypoxia-associated genes according to GO categorization.
Table 2.
Association between GO annotation and HREs
| Presence of HRE in promoter | GO term | Totals | |
| Oxygen homeostasis* | Other | ||
| With HRE | 444 | 3,422 | 3,866 |
| Without HRE | 765 | 6,096 | 6,861 |
| Totals | 1,209 | 9,518 | 10,727 |
| Proportion with HRE | 0.367 | 0.359 | 0.360 |
Shown are the number of T. californicus genes with GO annotations, sorted by the presence or absence in their respective promoter region (G test, G = 0.276, P = 0.599).
This combines multiple GO terms associated with oxygen homeostasis: “heme-”, “hemato-”, “angio-”, “oxygen”, “hypoxia.”
Independent of GO annotation, we examined the distribution of HREs among genes based on whether they responded to 24-h exposure to anoxia. We found no evidence that genes that were significantly differentially expressed during anoxia had HREs overrepresented in their promoter regions. The proportion of DE genes with HREs (26.2%) was actually lower than that of nonchanging genes that contained HREs (31.0%; Table 3; G = 4.80, P = 0.028). This was true also when only up-regulated genes were considered (27.7% up-regulated vs. 30.8% down-regulated or nonchanging genes; G = 1.69, P = 0.193).
Table 3.
Association between differential expression and HREs
| Presence of HRE in promoter | Anoxia vs. Normoxia | Totals | |
| Differentially expressed | No change | ||
| With HRE | 118 | 3,539 | 3,657 |
| Without HRE | 333 | 7,895 | 8,228 |
| Totals | 451 | 11,434 | 11,885 |
| Proportion with HRE | 0.262 | 0.310 | 0.308 |
Shown are the number of T. californicus genes examined for differential expression during 24 h of anoxia relative to normoxia controls, sorted by the presence or absence in their respective promoter region (G test, G = 4.80, P = 0.028).
Transcription Factor Binding Site Motif Discovery and Enrichment.
We performed computational analyses of transcription factor binding site (TFBS) motif discovery and enrichment to identify other classes of transcription factors potentially involved in response to hypoxia. These searches were performed on promoter sequences of the 451 DE genes in the 24-h treatment. The motif associated with the transcription factor hunchback (hb) was found in both types of analyses (SI Appendix, Figs. S3 and S4), with motifs associated with Trithorax-like (Trl), Chorion factor 2 (Cf2), zeste (z), and odd skipped (odd) found in the AME analysis (SI Appendix, Fig. S4). Among the five transcription factors, four contain C2H2 Zinc Fingers, while two are considered Polycomb-group proteins (SI Appendix, Table S5).
Discussion
The HIF pathway is highly conserved among animals, and its primary components were shown to have appeared early in Metazoan evolution, being absent from sponges and ctenophores but already present in placozoans and cnidarians (23, 24). Within crustaceans, HIF was examined and found in Branchiopoda (Daphnia spp.; ref. 26) and Malacostraca (27, 28). In T. californicus, we readily identified homologs for all gene families that contain bHLH-PAS and P4HC, including HIF-β/ARNT, partner of HIF-α, which is known to be constitutively active and to dimerize with other members of bHLH-PAS gene families across multiple pathways. We did not, however, find any copies of HIF-α or EGLN. Altogether, our analyses suggest that T. californicus lost the functionally linked HIF-α/EGLN gene pair, and, hence, cannot produce the HIF-α/HIF-β heterodimer. Consistent with this loss is the apparent deterioration of HRE binding domain specificity in genes involved in hypoxia response, likely as a result of relaxation of selection for maintenance of these noncoding sites. To our knowledge, a putative secondary loss of HIF-α and EGLN has been documented in only one other species, a tardigrade, although the authors were not explicitly examining this loss (29).
Despite the loss of HIF, T. californicus can survive in water with near zero O2 for at least 24 h. The genus Tigriopus is known to have a high surface area-to-volume ratio, no gills, and no respiratory pigment, suggesting that they rely on cutaneous diffusion of gaseous exchange (30). These aspects of their physiology may have facilitated the loss of such an otherwise crucial pathway, and additional mechanisms were possibly coopted to modulate oxygen homeostasis.
Our study also showed that transcriptional response to hypoxia in T. californicus is dynamic and robust. Genes responding to prolonged hypoxia (24 h) in T. californicus provide interesting comparisons to similar studies in other systems. Under hypoxic conditions, a switch from oxidative phosphorylation to glycolysis is expected (31), with a predicted increase in expression of genes mediating this switch. Consistent with this prediction, we observed a significant increase in pyruvate dehydrogenase kinase (Pdk) mRNA levels, an enzyme that is known to catalyze the step inhibiting pyruvate from entering and fueling the tricarboxylic acid (TCA) cycle (32, 33). In mammals, however, Pdk is a direct target of HIF-1 and, hence, contains HREs in its promoter region; in T. californicus, we did not detect HREs within a 1-kb promoter of the Pdk start codon. Therefore, identifying the trans-acting factor involved in Pdk transcription may be key in understanding hypoxia response in T. californicus.
Hypoxia response in other systems is also characterized by widespread transcriptional changes in subunits of oxidative phosphorylation (OXPHOS) complexes (34–37). We observed no change in expression of mtDNA-encoded subunits, and only in two nuclear-encoded OXPHOS genes (of 59 annotated). These two genes are associated with the cytochrome c oxidase complex (complex IV); one is involved in complex assembly (COX15), while the other is a known to help maintain mitochondrial function during hypoxia (homolog of human HIG1) (38). Up-regulation of certain complex IV subunits during hypoxia provides a possible submechanism of tolerance. Under low-oxygen conditions, HIF has been shown to regulate the transcriptional switch between two isoforms of certain COX subunits [from COX4-1 to COX4-2 in mammals (39), and from COX5a to COX5b in yeast (40)], with the alternative isoforms increasing efficiency of OXPHOS. Induction of COX genes in T. californicus hence warrants further targeted examination.
Perhaps the most unexpected pattern in our RNA-seq study was the transcription of several genes involved in cuticle and chitin metabolism. Structural components of the arthropod exoskeleton, like chitin and resilins, are crucial in forming a stable network with a high degree of flexibility and mobility (41, 42). After 24 h of anoxia, T. californicus showed increased transcription in chitin synthases and associated hormones (EH and JH) in parallel with up-regulation of 16 structural cuticular protein genes. Many other copies of cuticular genes (n = 56) did not reach statistical thresholds but had patterns of expression that were highly concordant with the ones above (SI Appendix, Fig. S5). During recovery in normoxia, these genes returned to lower transcription levels, likely driven by the high induction (∼70–400-fold up-regulation) of genes involved in chitin biodegradation, such as chitin deacetylases. These results provide strong evidence for the potential involvement of chitin metabolism in hypoxia tolerance in this species. Unlike terrestrial members of the Pancrustacea, copepods do not have trachea; instead they are thought to rely on cutaneous diffusion and possibly use integumental windows and segmental podocytes as their respiratory structures (43). We hypothesize that a highly plastic transcriptional response of cuticular genes during low-oxygen conditions permits the exoskeleton to undergo changes in structure, potentially through a modification of permeability via cuticle reorganization. Ultimately, microscopy techniques could be utilized to track physical changes in the exoskeleton before and during hypoxia, to ascertain how these transcriptional differences manifest themselves on the exoskeleton. Finally, current approaches for estimating coexpression networks should prove useful in this pursuit, but this will require an effort with large sample sizes (SI Appendix).
Conclusions
Hypoxia is a major physiological constraint for organisms who depend on aerobic respiration. Typically, the resulting physiological response is governed by the conserved HIF pathway, led by its regulatory machinery comprising HIF-α, HIF-β, and EGLN. We documented the loss of HIF-α and EGLN in an abundant crustacean that inhabits a highly variable environment, suggesting alternative molecular mechanisms of response to low oxygen availability may be more common than previously assumed. Our results suggest a strong role of chitin metabolism during hypoxia, which may work to maximize residual oxygen uptake in this species that lacks respiratory structures and pigments. This system will provide unique opportunities to examine the evolution of other oxygen-sensing gene families and how different cellular stress response mechanisms may evolve dual roles. While these key HIF pathway members are well described in the Crustacea (26–28), it is still unknown whether loss of these genes is idiosyncratic of Tigriopus or occurred earlier or in other lineages of the Copepoda.
Materials and Methods
Anoxia Tolerance Experiments.
Copepods were collected from intertidal rocky pools in Ocean Beach, San Diego, California (SD: 32.7333°N, 117.2500°W), and maintained in large, outbred laboratory cultures, for a minimum of three generations. Egg sacs (n = 36) were hatched in individual plates, and 1-d-old nauplii were used to perform split-brood treatments of anoxia (<0.50 mg O2/L) or normoxia (≥6.5 mg O2/L) in 20-mL sealed glass vials for 24 h. Larvae were then returned to plates and monitored for metamorphosis rate, time to maturity, and survival to age 20 d. Adults surviving to age 20 d were then subjected to a 24-h anoxia treatment, and survivorship was measured after 3 d. Using a new batch of individuals at age 20 d, we exposed sets of 10 copepods to anoxia or normoxia for four time periods (24, 48, 72, 96 h) and quantified survivorship for 3 d after removal from experimental exposures. This time series experiment was also performed with a new batch of 1-d nauplii.
Search for HIF Machinery.
We queried the protein sequences from the T. californicus reference genome (20), as well as translated sequences from five independently assembled de novo transcriptomes from multiple populations (SI Appendix). We used the HMMER 3.0 program to identify two sets of proteins, those that contained a PAS domain (PF00989.24, present in HIF-α or HIF-β) or that contained a P4HC domain (PF13640.5, present in EGLN). Alignment and phylogenetic methods followed those from a recent study of these gene families (2).
Transcriptional Response to Hypoxia.
Clutches (n = 24) were hatched and raised separately in normal culture conditions until age 20 d. Broods were assigned randomly to normoxia or anoxia replicates and experimentally treated as above, and four replicates from each condition were terminated at each of three time points: 3 h (3 h), 24 h (24 h), and 24 h in normoxia after the 24-h treatment (recovery). The recovery treatment involved transferring copepods from anoxia vials into culture plates with normoxic water. RNA isolation, mRNA library preparation, sequencing, and data processing were performed as described previously (44) (SI Appendix). Differential gene expression at each time point was quantified with edgeR by comparing anoxia samples to their respective normoxia control group and assessed with a false discovery rate of 0.10. Scripts are available at https://github.com/amgraham07.
Identification and Enrichment of HREs and Other TFBS.
Promoter regions (1 kb upstream) of every gene in the T. californicus genome were extracted and searched for the presence of an HRE, based on its motif: (A/G)CGTG(A/C/G)(A/G)(A/G) (25). Detailed scripts are available on GitHub (https://github.com/amgraham07). These were then used to test for overrepresentation among genes with relevant GO terms or with hypoxia-induced expression. Alternative TFBS motifs were searched along the promoter regions of genes that were differentially expressed in the 24-h anoxia treatment. The MEME suite was used to search for known or “novel” potential TFBS motifs; known motifs were then identified by comparisons to the JASPAR database, while “novel” motifs were assigned a potential identity via the tool Tomtom in the MEME suite (45).
Supplementary Material
Acknowledgments
We thank the core facilities at Center for Genome Research and Biocomputing at Oregon State University, where sequencing and computing was performed. This work was supported by Oregon State University funds and National Science Foundation Award DEB 1556455 (to F.S.B.) and by National Science Foundation Postdoctoral Research Fellowship in Biology Award No. 1812103 (to A.M.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: RNA-seq data were deposited in the NCBI Sequence Read Archive database, https://www.ncbi.nlm.nih.gov/sra, both for the hypoxia experiment (BioProject PRJNA503487, accessions SRR8146267–SRR8146290) and for the assembly of additional copepod transcriptomes (BioProject PRJNA504307, accession nos. SRR8168003 and SRR8168004).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819874116/-/DCSupplemental.
References
- 1.Rytkönen K. T., Williams T. A., Renshaw G. M., Primmer C. R., Nikinmaa M., Molecular evolution of the metazoan PHD-HIF oxygen-sensing system. Mol. Biol. Evol. 28, 1913–1926 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Graham A. M., Presnell J. S., Hypoxia Inducible Factor (HIF) transcription factor family expansion, diversification, divergence and selection in eukaryotes. PLoS One 12, e0179545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wenger R. H., Stiehl D. P., Camenisch G., Integration of oxygen signaling at the consensus HRE. Sci. STKE 2005, re12 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Beall C. M., Andean, Tibetan, and Ethiopian patterns of adaptation to high-altitude hypoxia. Integr. Comp. Biol. 46, 18–24 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Zhang W., et al. , Hypoxia adaptations in the grey wolf (Canis lupus chanco) from Qinghai-Tibet Plateau. PLoS Genet. 10, e1004466 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song S., et al. , Exome sequencing reveals genetic differentiation due to high-altitude adaptation in the Tibetan cashmere goat (Capra hircus). BMC Genomics 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaquer-Sunyer R., Duarte C. M., Thresholds of hypoxia for marine biodiversity. Proc. Natl. Acad. Sci. U.S.A. 105, 15452–15457 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan F., et al. , Emergence of anoxia in the California current large marine ecosystem. Science 319, 920 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Burton R. S., Lee B.-N., Nuclear and mitochondrial gene genealogies and allozyme polymorphism across a major phylogeographic break in the copepod Tigriopus californicus. Proc. Natl. Acad. Sci. U.S.A. 91, 5197–5201 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Truchot J.-P., Duhamel-Jouve A., Oxygen and carbon dioxide in the marine intertidal environment: Diurnal and tidal changes in rockpools. Respir. Physiol. 39, 241–254 (1980). [DOI] [PubMed] [Google Scholar]
- 11.Willett C. S., Potential fitness trade-offs for thermal tolerance in the intertidal copepod Tigriopus californicus. Evolution 64, 2521–2534 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Schoville S. D., Barreto F. S., Moy G. W., Wolff A., Burton R. S., Investigating the molecular basis of local adaptation to thermal stress: Population differences in gene expression across the transcriptome of the copepod Tigriopus californicus. BMC Evol. Biol. 12, 170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lima T. G., Willett C. S., Locally adapted populations of a copepod can evolve different gene expression patterns under the same environmental pressures. Ecol. Evol. 7, 4312–4325 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leong W., Sun P. Y., Edmands S., Latitudinal clines in temperature and salinity tolerance in tidepool copepods. J. Hered. 109, 71–77 (2017). [DOI] [PubMed] [Google Scholar]
- 15.DeBiasse M. B., Kawji Y., Kelly M. W., Phenotypic and transcriptomic responses to salinity stress across genetically and geographically divergent Tigriopus californicus populations. Mol. Ecol. 27, 1621–1632 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Goolish E., Burton R., Energetics of osmoregulation in an intertidal copepod: Effects of anoxia and lipid reserves on the pattern of free amino accumulation. Funct. Ecol. 3, 81–89 (1989). [Google Scholar]
- 17.Auel H., Verheye H. M., Hypoxia tolerance in the copepod Calanoides carinatus and the effect of an intermediate oxygen minimum layer on copepod vertical distribution in the northern Benguela Current upwelling system and the Angola–Benguela Front. J. Exp. Mar. Biol. Ecol. 352, 234–243 (2007). [Google Scholar]
- 18.Stalder L., Marcus N., Zooplankton responses to hypoxia: Behavioral patterns and survival of three species of calanoid copepods. Mar. Biol. 127, 599–607 (1997). [Google Scholar]
- 19.Grodzins M. A., Ruz P. M., Keister J. E., Effects of oxygen depletion on field distributions and laboratory survival of the marine copepod Calanus pacificus. J. Plankton Res. 38, 1412–1419 (2016). [Google Scholar]
- 20.Barreto F. S., et al. , Genomic signatures of mitonuclear coevolution across populations of Tigriopus californicus. Nat. Ecol. Evol. 2, 1250–1257 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Jones P., et al. , InterProScan 5: Genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dehal P., et al. , The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science 298, 2157–2167 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Mills D. B., et al. , The last common ancestor of animals lacked the HIF pathway and respired in low-oxygen environments. eLife 7, e31176 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loenarz C., et al. , The hypoxia-inducible transcription factor pathway regulates oxygen sensing in the simplest animal, Trichoplax adhaerens. EMBO Rep. 12, 63–70 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorr T., Hradecky P., Cahn J., Bunn H., “Genome-wide computational screen for candidate Hif target genes in Drosophila melanogaster and Caenorhabditis elegans” in Oxygen Sensing: Responses and Adaptation to Hypoxia, Lahiri S., Semenza G. L., Prabhakar N. R., Eds. (Taylor & Francis Group, New York, 2003). [Google Scholar]
- 26.Gorr T. A., Cahn J. D., Yamagata H., Bunn H. F., Hypoxia-induced synthesis of hemoglobin in the crustacean Daphnia magna is hypoxia-inducible factor-dependent. J. Biol. Chem. 279, 36038–36047 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Soñanez-Organis J. G., et al. , Molecular characterization of hypoxia inducible factor-1 (HIF-1) from the white shrimp Litopenaeus vannamei and tissue-specific expression under hypoxia. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 150, 395–405 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Hardy K. M., Follett C. R., Burnett L. E., Lema S. C., Gene transcripts encoding hypoxia-inducible factor (HIF) exhibit tissue- and muscle fiber type-dependent responses to hypoxia and hypercapnic hypoxia in the Atlantic blue crab, Callinectes sapidus. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 163, 137–146 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Yoshida Y., et al. , Comparative genomics of the tardigrades Hypsibius dujardini and Ramazzottius varieornatus. PLoS Biol. 15, e2002266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McAllen R., Taylor A. C., Davenport J., The effects of temperature and oxygen partial pressure on the rate of oxygen consumption of the high-shore rock pool copepod Tigriopus brevicornis. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 123, 195–202 (1999). [Google Scholar]
- 31.Semenza G. L., Hypoxia-inducible factors in physiology and medicine. Cell 148, 399–408 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papandreou I., Cairns R. A., Fontana L., Lim A. L., Denko N. C., HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 3, 187–197 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Kim J. W., Tchernyshyov I., Semenza G. L., Dang C. V., HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3, 177–185 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Gracey A. Y., Troll J. V., Somero G. N., Hypoxia-induced gene expression profiling in the euryoxic fish Gillichthys mirabilis. Proc. Natl. Acad. Sci. U.S.A. 98, 1993–1998 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou D., et al. , Mechanisms underlying hypoxia tolerance in Drosophila melanogaster: Hairy as a metabolic switch. PLoS Genet. 4, e1000221 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mossman J. A., et al. , Mitonuclear interactions mediate transcriptional responses to hypoxia in Drosophila. Mol. Biol. Evol. 34, 447–466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flight P. A., Nacci D., Champlin D., Whitehead A., Rand D. M., The effects of mitochondrial genotype on hypoxic survival and gene expression in a hybrid population of the killifish, Fundulus heteroclitus. Mol. Ecol. 20, 4503–4520 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayashi H., et al. , HIG1, a novel regulator of mitochondrial γ-secretase, maintains normal mitochondrial function. FASEB J. 26, 2306–2317 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Fukuda R., et al. , HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 129, 111–122 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Kwast K. E., Burke P. V., Poyton R. O., Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J. Exp. Biol. 201, 1177–1195 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Muthukrishnan S., Merzendorfer H., Arakane Y., Kramer K. J., “Chitin metabolism in insects” Insect Molecular Biology and Biochemistry, Gilbert L. I., Ed. (Elsevier, 2012), pp. 193–235. [Google Scholar]
- 42.Andersen S. O., Amino acid sequence studies on endocuticular proteins from the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 28, 421–434 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Lee C. E., Evolutionary mechanisms of habitat invasions, using the copepod Eurytemora affinis as a model system. Evol. Appl. 9, 248–270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graham A. M., Barreto F. S., Novel microRNAs are associated with population divergence in transcriptional response to thermal stress in an intertidal copepod. Mol. Ecol. 28, 584–599 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Bailey T. L., Johnson J., Grant C. E., Noble W. S., The MEME suite. Nucleic Acids Res. 43, W39–W49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.