Germline predisposition to hematologic malignancy (HM) is an entity receiving increasing attention and recognition of its clinical significance, highlighted by its recent inclusion in the World Health Organization guidelines. It is attracting greater consideration with regard to identification of underlying mutations and clinical management of affected families.1,2 Historically, most identified mutations have centered around familial myeloid malignancies.3 However, mutations in predisposition genes, such as RUNX1 and ETV6, are associated with autosomal dominant inherited HM for both myeloid and lymphoid malignancies, reflective of their key regulatory roles in both myeloid and lymphoid lineages.4 In the context of RUNX1 and ETV6 predisposition syndromes (FPDMM, OMIM 601399; THC5, OMIM 616216), HM are often also seen in combination with other hematologic phenotypes such as thrombocytopenia, neutropenia, anemia and macrocytosis.4 The spectrum of hematologic phenotypes is also similar to ataxia-pancytopenia syndrome (ATXPC, OMIM 159550), which in addition to cytopenias and predisposition to myeloid neoplasms, is characterized by neurological phenotypes, manifesting as cerebellar ataxia, and has been found to be caused by germline mutations in SAMD9L.5,6 Here we identify a novel causative germline mutation in SAMD9L in a family of Northern European ancestry with an extensive four-generation history of cytopenias and suspected ATXPC, initially recruited due to development of pediatric acute lymphoblastic leukemia (ALL) in the proband. We also summarize the known SAMD9L ATXPC causing mutations as well as those in its sister gene SAMD9 which cause MIRAGE syndrome, with shared hematologic features.7,8
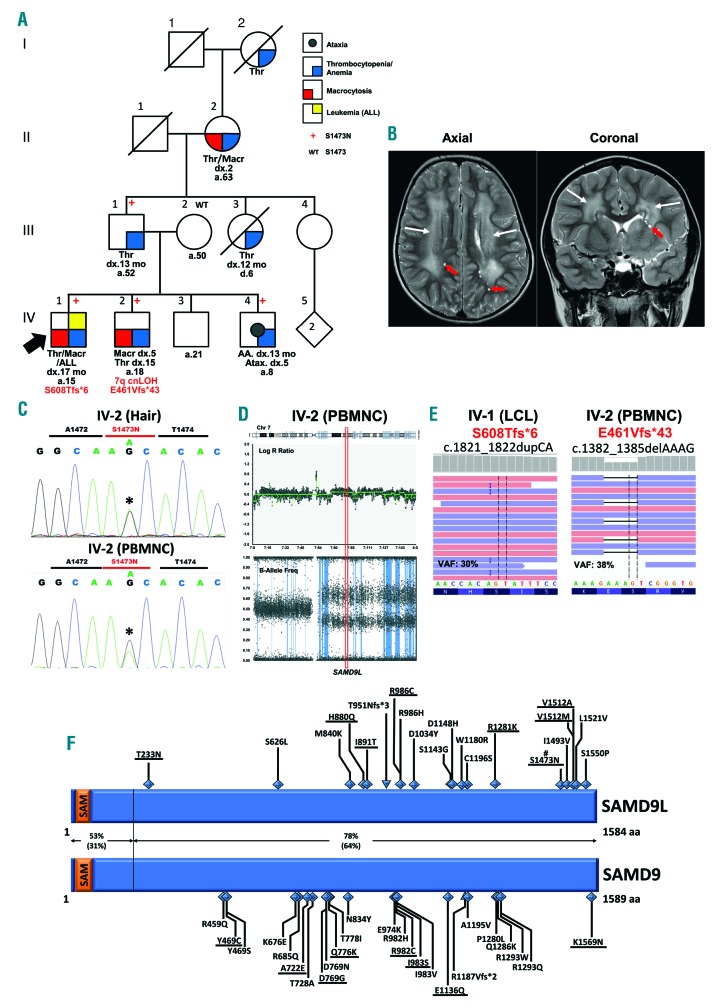
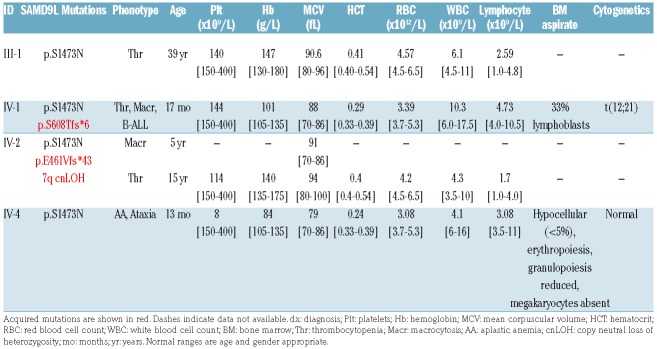
The proband (IV-1) (Figure 1A) was investigated for pallor at age 17 months and was found to have thrombocytopenia and macrocytic anemia with 15% lymphoblasts in his blood and 33% lymphoblasts in bone marrow (BM) (Table 1). He was diagnosed with precursor B-ALL, with the common t(12;21) (ETV6-RUNX1) translocation identified by fluorescence in situ hybridization (FISH). He was treated until clinical remission with standard induction chemotherapy including prednisolone, vincristine, daunorubicin and L-asparaginase, followed by two typical consolidation therapy phases of: 1) cyclophosphamide, cytarabine, vincristine and intrathecal methotrexate; 2) L-asparaginase, methothrexate, oral dexamethasone and 6-mercaptopurine. The therapy was interrupted due to severe marrow toxicity and persistent cytopenias, which were treated with granulocyte colony-stimulating factor (G-CSF) (10 μg/kg twice daily) and stem cell factor (SCF; twice weekly). Markers of minimal residual disease persisted, and at age three years he received a cord blood transplant from a matched unrelated donor (transplant conditioning included treatment with cyclophosphamide, anti-thymocyte globulin and total body irradiation with methotrexate post transplant; see Online Supplementary Table S1 for details). He has since been in remission (12 years post transplant). Investigation of family history identified several family members with unexplained cytopenias and blood abnormalities ranging in severity. The proband’s brother (IV-2) was identified with mild macrocytosis at five years following an episode of acute gastroenteritis but was otherwise well at the time of our initial investigation;9 he has subsequently been diagnosed with thrombocytopenia at age 15 years. Sibling IV-4 was diagnosed with aplastic anemia at age 13 months that was treated with oxymetholone and blood transfusions for 6-12 months to restore red cell counts and platelet numbers back to normal, and these have remained normal without further treatment. At five years, ataxia was also detected which manifested as poor balance, walking on tip toe, and extra white matter observable upon cranial magnetic resonance imaging (MRI) (Figure 1B). The father of the proband (III-1) had a history of thrombocytopenia diagnosed initially at age 13 months with platelet counts ranging from 40 to 70×109/L. However, his thrombocytopenia partially resolved with mild thrombocytopenia still persistent in his 30s (platelet count 140×109/L) (Table 1). The proband’s paternal aunt (III-3) was diagnosed with thrombocytopenia at age one year and remained thrombocytopenic until her death at six years due to acute non-traumatic intracranial hemorrhage. The paternal grandmother of the proband (II-2) had a history of thrombocytopenia and macrocytosis, diagnosed at age two years when hospitalized for Epstein-Barr virus infection. The paternal-great grandmother of the proband (I-2) was also reported to have had thrombocytopenia and easy bruising.
Figure 1.
Four generations of a family with ataxia pancytopenia syndrome and germline mutation of SAMD9L. (A) Germline p.Ser1473Asn mutation segregates with affected individuals. Additional somatic mutations and chromosomal aberrations (red text). Thr: thrombocytopenia; Macr: macrocytosis; AA: aplastic anemia; Atax: ataxia; cnLOH: copy neutral loss of heterozygosity; dx: age at diagnosis; a: age at last date of contact (years); d: age at death (years). (B) Axial and coronal T2 weighted magnetic resonance imaging (MRI) images of the brain of individual IV-4 with ataxia indicate symmetrical bilateral periventricular white matter hyperintensity (white arrows) and small rounded focal areas of hyperintense T2 signal which suppressed on FLAIR (not shown) suggestive of parenchymal cysts or prominent perivascular spaces (red arrows). (C) Sanger sequencing confirmation of heterozygous germline mutation SAMD9L S1473N (chr7:g.92760867C>T; c.4418G>A [NM_152703.4]; p.Ser1473Asn [NP_689916.2]) in hair and in peripheral blood mononuclear cells (PBMNC) of IV-2 indicates reduced variant allele load of mutant. (D) SNP-array of chromosome 7 of IV-2 shows cnLOH of 7q. The cnLOH of the long arm in IV-2 includes the SAMD9L gene in approximately 30% of cells. (E) Somatic frameshift mutation SAMD9L E461Vfs*43 (chr7:g.92763904_92763907delCTTT; c.1382_1385delAAAG; p.Glu461Val*43) was acquired by IV-2 with an allele load of approximately 38% while somatic frameshift mutation SAMD9L S608Tfs*6 (chr7:g.92763465_92763466dupTG; c.1821_1822dupCA; p.Ser608Thr*6) was acquired in IV-1 with an allele load of approximately 30%. Samples: IV-1: lymphoblastoid cell lines (LCL); IV-2: PBMNC. (F) Comparison of germline mutations in paralogous genes SAMD9L and SAMD9. #: identified in our study; underlined: germline mutations with other SAMD9L/SAMD9 somatic or germline mutations (in cis or biallelic). For full variant reference list, see Online Supplementary Tables S3 and S4. Percent conservation between proteins is shown (brackets show % identity).
Table 1.
Blood and bone marrow parameters in individuals with a germline SAMD9L p.S1473N mutation.
To identify the genetic basis for this family history of blood and BM abnormalities, whole exome sequencing (WES) and single nucleotide polymorphism (SNP) arrays were performed on samples from the proband (IV-1), his father (III-1) and one affected sibling (IV-2). Analysis of WES and SNP array data did not identify any segregating variants or aberrations in RUNX1, ETV6, PAX5 or other FHM genes.2 Unbiased filtering of data based on segregation, population frequency and predicted pathogenicity identified variants in 19 genes (Online Supplementary Table S2) including a novel heterozygous segregating variant in SAMD9L (c.4418G>A, NM_152703.4) encoding a p.Ser1473Asn (S1473N) missense substitution. Sanger sequencing confirmed the variant as present in the father and all three affected siblings, including from their hair, confirming it was of germline origin and paternally inherited (Figure 1C and Online Supplementary Figure S1A and B). As expected from these results, the mother (III-2) did not harbor the variant (Online Supplementary Figure S1B).
Mutations in SAMD9L have previously been described in both an inherited BM failure cohort, and multiple independent families with ATXPC.7 The inherited BM failure cohort identified SAMD9/SAMD9L mutations as the most frequently mutated genes with individuals exhibiting hypocellular BM, cytopenia, aplastic anemia and nystagmus.10 ATXPC families were also characterized by variable manifestations of cerebellar ataxia with white matter abnormalities evident by MRI, and cytopenias, including aplastic anemia, macrocytosis and leukemia (myeloid subtype) in mutation carriers.5–7 Additionally, in the initial ATXPC SAMD9L study, the investigators observed that lymphoblastoid cell lines (LCL) harboring the germline SAMD9L variant underwent rapid clonal selection for copy number neutral loss of heterozygosity (cnLOH) to restore biallelic WT alleles. In peripheral blood mononuclear cell (PBMNC) DNA from individual IV-2 (at 5 years old) from our family, we also observed cnLOH of 7q [7q11.21q36.3 (~61,970,948-159,126,310)] via maternal uniparental disomy (UPD), present in approximately 30% of cells (Figure 1D), that correlated with a concordant decrease in the variant allele frequency (VAF) of the SAMD9L (c.4418G>A, S1473N) mutation, evident in both WES and Sanger sequence data (Figure 1C and Online Supplementary Figure S1A). In addition to 7q UPD in blood from IV-2, we also observed an acquired SAMD9L E461Vfs*43 mutation present at 38% VAF (Figure 1E) indicating that multiple independent somatic revertant clones were contemporaneously present. LCL from the proband (IV-1, Thr/Macr/ALL) contained a different SAMD9L acquired mutation, S608Tfs*6 at 30% VAF (Figure 1E), which was subsequently confirmed to have been also present in his primary ALL diagnosis BM (Online Supplementary Figure S2B). Identification of these somatic mutations is consistent with several reports6,7,10,11 describing multiple families with germline SAMD9L mutations segregating with ATXPC, myeloid malignancies and/or BM failure (see Figure 1F), where somatic mosaic germline rescue was achieved in clones through acquired mutations in SAMD9L causing premature protein termination N-terminal of the germline variant, and demonstrated to be in cis with the germline variant in at least one case. Accordingly, both acquired frameshift mutations identified here are upstream of the germline S1473N variant, and clonal analysis of PCR amplified DNA suggests an in cis configuration of the germline S1473N and acquired S608Tfs*6 variants (Online Supplementary Figure S2C). This selection for somatic reversion, observed in multiple studies, is likely to overcome restriction of hematopoietic cell growth by the germline mutations, which may be considered as a gain-of-function (GOF).5,6
Interestingly, unlike previous studies in which monosomy 7 and myeloid malignancies in germline SAMD9L mutation carriers are repeatedly described, this was not observed, and instead the proband developed ALL with a t(12;21) translocation (ETV6-RUNX1), the most common translocation observed in childhood ALL.12 Studies by Greaves et al. have identified that t(12;21) translocations are present at birth in up to 1% of individuals, representing a 100-fold higher frequency than the number of children who develop overt leukemia with the translocation, suggesting that additional events are required to progress to a leukemic state.12 It is possible that the co-existence of a pathogenic germline SAMD9L variant, associated with ATXPC characterized phenotypes (thrombocytopenia, macrocytosis), in co-operation with the acquired ETV6-RUNX1 fusion, led to the development of ALL in the proband (IV-1). Interestingly, co-dysregulation of SAMD9/SAMD9L and RUNX1 or ETV6 was also recently described in myeloid malignancies where acquired RUNX1 mutations are emerging as a recurrent feature of germline SAMD9L cases, and acquired ETV6 variants in germline SAMD9 cases.11,13 The opposite also occurs with an acquired SAMD9 variant observed in a tumor from a germline RUNX1 case.14 Whether germline SAMD9L variants have also contributed to the development of additional ALL cases with ETV6-RUNX1 fusions or mutations is an interesting question that will require further investigation.
In summary, our study identified a novel germline SAMD9L mutation, causative of ATXPC in four generations of the same family. One individual acquired multiple somatic reversion events, and another acquired a reversion event which co-existed with t(12;21) positive ALL. This observation is consistent with the rationale whereby dysfunction due to a germline mutation selects for reversion events that correct the defect. Such reverse events, however may include unwanted aberrations like -7/7q associated with myeloid malignancies or collaborate with other acquired mutations, such as translocations that dictate the subsequent malignancy phenotype. Notably, as clonal selection against the germline mutation is strong in these families, and as induced pluripotent stem cell and other cellular therapies advance,15 it may soon be possible to treat patients with isogenic stem cells selected in culture for reversion to the wild-type genotype via the commonly observed and benign uniparental disomy mechanism or point mutation reversion. This would avoid hematopoietic selection pressures for acquisition of leukemia predisposing monosomy 7 or collaboration with t(12;21) positive cells.
Supplementary Material
Acknowledgments
The authors would like to thank the patients and their family members for their willingness to participate in this research. The authors also thank Rebecca Beardmore and Matthew Ivers (Queensland Children’s Hospital, Brisbane, Queensland) for retrieval of patient treatment information.
Footnotes
Funding: the research was funded by the National Health and Medical Research Council of Australia APP1024215 and APP1023059, and the Cancer Council of South Australia APP565161; and the University of South Australia Postgraduate Research Award (JJC Cheah).
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. [DOI] [PubMed] [Google Scholar]
- 2.Godley LA, Shimamura A. Genetic predisposition to hematologic malignancies: management and surveillance. Blood. 2017;130(4):424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown AL, Churpek JE, Malcovati L, Dohner H, Godley LA. Recognition of familial myeloid neoplasia in adults. Semin Hematol. 2017;54(2):60–68. [DOI] [PubMed] [Google Scholar]
- 4.Topka S, Vijai J, Walsh MF, et al. Germline ETV6 Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and Thrombocytopenia. PLoS Genet. 2015;11(6):e1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen DH, Below JE, Shimamura A, et al. Ataxia-Pancytopenia Syndrome Is Caused by Missense Mutations in SAMD9L. Am J Hum Genet. 2016;98(6):1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tesi B, Davidsson J, Voss M, et al. Gain-of-function SAMD9L mutations cause a syndrome of cytopenia, immunodeficiency, MDS, and neurological symptoms. Blood. 2017;129(16):2266–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidsson J, Puschmann A, Tedgard U, et al. SAMD9 and SAMD9L in inherited predisposition to ataxia, pancytopenia, and myeloid malignancies. Leukemia. 2018;32(5):1106–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narumi S, Amano N, Ishii T, et al. SAMD9 mutations cause a novel multisystem disorder, MIRAGE syndrome, and are associated with loss of chromosome 7. Nat Genet. 2016;48(7):792–797. [DOI] [PubMed] [Google Scholar]
- 9.Escher R, Wilson P, Carmichael C, et al. A pedigree with autosomal dominant thrombocytopenia, red cell macrocytosis, and an occurrence of t(12:21) positive pre-B acute lymphoblastic leukemia. Blood Cells Mol Dis. 2007;39(1):107–114. [DOI] [PubMed] [Google Scholar]
- 10.Bluteau O, Sebert M, Leblanc T, et al. A landscape of germ line mutations in a cohort of inherited bone marrow failure patients. Blood. 2018;131(7):717–732. [DOI] [PubMed] [Google Scholar]
- 11.Wong JC, Bryant V, Lamprecht T, et al. Germline SAMD9 and SAMD9L mutations are associated with extensive genetic evolution and diverse hematologic outcomes. JCI Insight. 2018;3(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ford AM, Greaves M. ETV6-RUNX1 (+) Acute Lymphoblastic Leukaemia in Identical Twins. Adv Exp Med Biol. 2017;962:217–228. [DOI] [PubMed] [Google Scholar]
- 13.Pastor VB, Sahoo SS, Boklan J, et al. Constitutional SAMD9L mutations cause familial myelodysplastic syndrome and transient monosomy 7. Haematologica. 2018;103(3):427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Churpek JE, Pyrtel K, Kanchi KL, et al. Genomic analysis of germ line and somatic variants in familial myelodysplasia/acute myeloid leukemia. Blood. 2015;126(22):2484–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugimura R, Jha DK, Han A, et al. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature. 2017; 545(7655):432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.