Abstract
Dominant-negative mutations in the transcription factor Growth Factor Independence-1B (GFI1B), such as GFI1BQ287*, cause a bleeding disorder characterized by a plethora of megakaryocyte and platelet abnormalities. The deregulated molecular mechanisms and pathways are unknown. Here we show that both normal and Q287* mutant GFI1B interacted most strongly with the lysine specific demethylase-1 – REST corepressor - histone deacetylase (LSD1-RCOR-HDAC) complex in megakaryoblasts. Sequestration of this complex by GFI1BQ287* and chemical separation of GFI1B from LSD1 induced abnormalities in normal megakaryocytes comparable to those seen in patients. Megakaryocytes derived from GFI1BQ287*-induced pluripotent stem cells also phenocopied abnormalities seen in patients. Proteome studies on normal and mutant-induced pluripotent stem cell-derived megakaryocytes identified a multitude of deregulated pathways downstream of GFI1BQ287* including cell division and interferon signaling. Proteome studies on platelets from GFI1BQ287* patients showed reduced expression of proteins implicated in platelet function, and elevated expression of proteins normally downregulated during megakaryocyte differentiation. Thus, GFI1B and LSD1 regulate a broad developmental program during megakaryopoiesis, and GFI1BQ287* deregulates this program through LSD1-RCOR-HDAC sequestering.
Introduction
Platelets are specialized cell fragments that function to prevent excessive bleeding upon blood vessel injury.1 During megakaryopoiesis, megakaryoblasts undergo endomitosis followed by cytoplasmic maturation, during which α- and δ-granules are formed. Subsequently, mature megakaryocytes migrate to blood vessels in the bone marrow or lung, where they form protrusions and shed proplatelets into the bloodstream.2,3
The identification of inherited bleeding and platelet disorder mutations has provided insight into proteins crucial for platelet production and function. Genes mutated in these disorders range from those encoding proteins involved in α-granule biology, such as NBEAL2, to transcription factors controlling a wide range of megakaryocyte processes.4 One of the transcription factors mutated in inherited bleeding and platelet disorders is Growth Factor Independence 1B (GFI1B). To date, truncating mutations affecting DNA binding,5–8 missense mutations,9–11 and mutations that change the amount and ratio of the two naturally occurring GFI1B isoforms (p37 and p32) have been described.11,12 Bleeding tendencies may vary depending on the type of mutations, being severe for cases that express only GFI1B-p32,12 moderate to severe for dominant-negative truncating mutations,5–8 and mild to even absent for the missense mutations.9–11 Most, but not all, mutations associate with macrothrombocytopenia, a reduction in platelet α-granules, and increased CD34 expression. For some mutations, a reduction in CD42b expression, paucity of platelet δ-granules, and an increase in the numbers of morphologically abnormal megakaryocytes in the bone marrow have also been described. Based on these observations it can be concluded that normal GFI1B regulates a multitude of megakaryocyte-specific processes, and that the molecular mechanisms causing the abnormalities may differ.
GFI1B is a transcriptional repressor containing six C-terminal zinc fingers (in the case of the GFI1B-p37 isoform) and an N-terminal Snail/GFI1 (SNAG) domain, important for lysine specific demethylase (LSD1/KMD1A), REST Corepressor 1 (CoREST/RCOR1) recruitment.13 The GFI1B-p32 isoform that lacks intact zinc finger 1 and 2, also associates with LSD1-RCOR1 and is sufficient for erythropoiesis.12,14 In addition to LSD1, GFI1B recruits histone methyl transferases and histone deacetylases (HDAC) to target gene promoters and enhancers to induce transcriptional repression, among which GFI1B itself.15,16 Of the co-factors, LSD1 may be especially important because the majority (80%) of Gfi1b-bound regions in murine MEL cells are also enriched for Lsd1 occupancy, suggesting a strong interdependency.13 LSD1 is an essential gene in mammalian differentiation and functions, including hematopoiesis. In vitro and in vivo Lsd1 knockdown studies showed that normal granulopoiesis, erythropoiesis and megakaryopoiesis all depend on Lsd1.13,17,18
Inherited mutations in GFI1B disrupting its DNA binding zinc fingers, such as GFI1BQ287*, are known to act in a dominant-negative manner.5 The GFI1B p.Q287* mutation is located within the fifth zinc finger of GFI1B and leads to a protein truncated in its DNA-binding region. As a consequence, the ability of GFI1BQ287* to bind GFI1B sequence motifs is disrupted.8 Importantly, its SNAG domain, crucial for LSD1 binding, remains intact. In this study we used the dominant-negative GFI1BQ287* mutant to unravel the mechanism by which it inhibits wildtype GFI1B and identified GFI1B regulated pathways during megakaryopoiesis.
Methods
Mass spectrometry analysis of GFI1B-interacting proteins in MEG-01 cells
MEG-01 cells were lentivirally transduced with FUW, FUW-GFI1B-green fluorescent protein (GFP) and FUW-GFI1BQ287*-GFP and nuclear extracts were prepared as described by Dignam et al.19 Label-free GFP-pulldowns were performed using GFP-Trap beads (ChromoTek) as described by Smits et al.20 Subsequently, the proteins were subjected to on-bead trypsin digestion and peptides were acidified and desalted using C18-Stagetips.21 Peptides were recorded with an LC-MS/MS Orbitrap Fusion Tribrid mass spectrometer (ThermoFisher Scientific). Raw data were analyzed by MaxQuant (version 1.5.7.0). Proteins differentially expressed between empty vector and GFI1B or GFI1BQ287* were determined using a t-test with a false discovery rate (FDR) <0.01 and a fold change (FC) >9.2. The stoichiometry of the identified complexes was determined by dividing the iBAQ intensity in the GFI1B/GFI1BQ287* samples by the iBAQ intensity in the empty vector samples.
Proliferation of GFI1B-transduced MEG-01 cells
MEG-01 cells were retrovirally transduced with pMIGR1-GFI1B variant-flag-IRES-GFP resulting in mixed cultures of GFP positive and -negative cells. The GFP% was followed using flow cytometry for 26 days. GFP% was normalized to the starting point of the culture (day 5) using the following formula: GFP% day X/(100 – GFP% day X))/(GFP% day 5/(100 – GFP% day 5). On day 23, GFP-positive cells were FACS-sorted to determine total and endogenous GFI1B expression using quantitative real-time polymerase chain reaction analysis.
Primary megakaryocyte cultures
CD34+ hematopoietic stem and progenitor cells were isolated from mobilized peripheral blood of healthy donors. Informed consent was given in accordance with the Declaration of Helsinki and the Dutch national and Sanquin internal ethical review boards. CD34+ cells were differentiated to megakaryocytes in modified Iscove modified Dulbecco medium (HEMAdef)22 supplemented with 50 ng/mL stem cell factor, 50 ng/mL thrombopoietin, 1 ng/mL interleukin-3 and 20 ng/mL interleukin-6 for 4 days, followed by culture in 50 ng/mL thrombopoietin and 10 ng/mL interleukin-1β for 7 more days (Peprotech).23 Where indicated, 4 μM GSK-LSD1 (Sigma) was added subsequently and cells were cultured for an additional 2-6 days depending on the read out (2 days: expansion, CD34 and CD42b expression; 2-6 days: prolonged effect on CD42b expression and proplatelet formation).
Proteomics mass spectrometry analysis of platelets and megakaryocytes
Blood was taken from healthy donors and patients to generate induced pluripotent stem cells (iPSC) and isolate platelets. The study was approved by the Medical Ethical Committee of the Radboud University Medical Center, Nijmegen (2013/064) and conducted in accordance with the Declaration of Helsinki. The control iPSC line MML-6838-Cl224 and GFI1BQ287* patient iPSC lines BEL-5-Cl1 and Cl2 (Online Supplementary Figure S1) were differentiated towards megakaryocytes as described before.25 For label-free quantification, proteins were reduced, alkylated, trypsin-digested, desalted and concentrated. Tryptic peptides were separated by nanoscale C18 reverse phase chromatography coupled online to an Orbitrap Fusion Tribrid mass spectrometer (ThermoFisher Scientific) via a nanoelectrospray ion source (Nanospray Flex Ion Source, ThermoFisher Scientific). All mass spectrometry data were acquired with Xcalibur software (Thermo Scientific) and processed with the MaxQuant computational platform, 1.5.2.8. The RAW files, MaxQuant search results, and details about the settings are available in the PRIDE repository database26 with the dataset identifier PXD009020.
Statistical analysis
Statistical analysis for non-mass spectrometry experiments was performed in GraphPad Prism.
For more details see the Online Supplementary Methods.
Results
GFI1B and GFI1BQ287* interact with the LSD1-RCOR1-HDAC1/2 complex
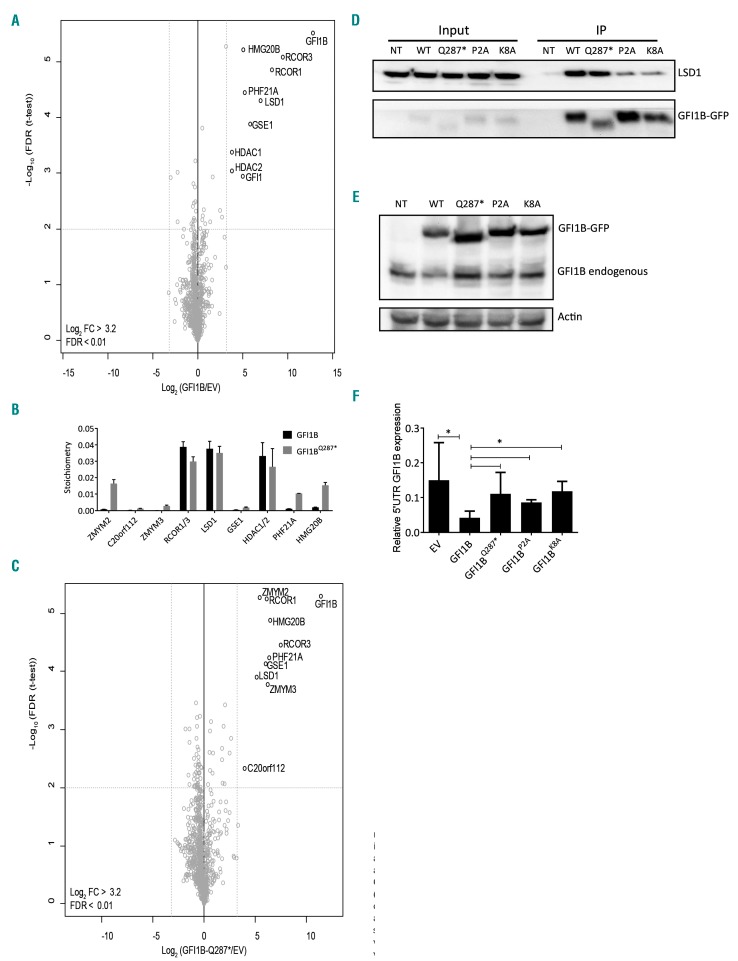
Several GFI1B variants have been identified in inherited bleeding and platelet disorders.5,6,8,9 The majority of the pathogenic variants, such as GFI1B p.Q287*, result in a truncation disrupting the DNA binding capacity. Truncated GFI1B inhibits the wildtype protein in a dominant-negative manner.5,8 With the SNAG domain intact, we hypothesized that GFI1BQ287* can compete with GFI1B for the recruitment of epigenetic modifiers. To test this hypothesis, we first identified proteins interacting with GFI1B and GFI1BQ287*. To this end, C-terminally GFP-tagged GFI1B and GFI1BQ287* were expressed in megakaryo blastic MEG-01 cells. GFP-pulldown followed by on-bead trypsin digestion and mass spectrometry analysis identified the following GFI1B interacting proteins: LSD1, RCOR1/3, HDAC1/2, GSE1, HMG20B, PHF21A, and GFI1 (Figure 1A). Of the detected GFI1B interactors, LSD1 was most abundant, together with RCOR1/3 and HDAC1/2 (Figure 1B). The proteins GSE1, HMG20B, and PHF21A are all known to be subunits of the LSD1-RCOR-HDAC complex, also known as the CoREST (RCOR1) complex.27 The identified interactome of GFI1BQ287* was strongly similar to that of GFI1B, with ZMYM3 as the only significant differential interactor (Figure 1C; Online Supplementary Figure S2), indicating that the DNA-binding defective GFI1BQ287* mutant recruits virtually the same co-factors as GFI1B.
Figure 1.
GFI1B and GFI1BQ287* interact with the LSD1-RCOR-HDAC complex. (A,C) Nuclear extracts of MEG-01 cells transduced with empty vector (EV), GFI1B-GFP, or GFI1BQ287*-GFP were analyzed by mass spectrometry. Statistically enriched proteins were identified by a permutation-based false discovery rate (FDR)-corrected t-test (FDR<0.01). The volcano plot shows the difference between label-free quantification (LFQ) intensities of the GFI1B-GFP or GFI1BQ287*-GFP pulldown and the EV control [log2-transformed fold change (FC)] on the x-axis, and the −log10-transformed P-value on the y-axis. Dotted gray lines represent the statistical cutoffs, which were chosen such that no proteins were present as outliers on the EV control side of the volcano plot. The proteins in the upper right corner represent the bait (GFI1B) and its interactors. We cannot exclude the possibility that we missed interactions due to steric hindrance by the GFP-tag, especially at the C-terminal end. (B) Stoichiometry analysis of GFI1B interactors in MEG-01 cells showing relative abundances. Discrimination between GFI1B;GFI1, HDAC1;HDAC2, and RCOR1;RCOR3 was not possible because of the presence of common peptides. The iBAQ value of each interacting protein is plotted with the value for GFI1B;GFI1 set at 1. GFI1 was identified as an interactor of both GFI1B (all 3 replicates) and GFI1BQ287* (2 of 3 replicates, therefore filtered out). As peptides common to GFI1B and GFI1 were included in the normalization this may have affected stoichiometry values. Data are shown as the mean ± standard deviation of three pulldowns. (D) Western blot analysis of co-immunoprecipitated (IP) LSD1 after GFP-trap bead pulldown in MEG-01 cells transduced with GFI1B-GFP (WT), GFI1BQ287*-GFP (Q287*), GFI1BP2A-GFP (P2A), and GFI1BK8A-GFP (K8A). Non-transduced (NT) cells were used as a negative control. The upper panel shows LSD1 (~90 kDa) and the lower panel GFI1B variants-GFP (~58 kDa). The left side of the blot shows LSD1 and GFI1B-GFP expression in the input samples. (E) Western blot analysis of MEG-01 cells transduced with EV, and GFI1B-GFP, GFI1BQ287*-GFP, GFI1BP2A-GFP, and GFI1BK8A-GFP stained with an antibody against GFI1B, identifying both exogenous GFI1B-GFP fusions (~58 kDa) and endogenous GFI1B (~37 kDa). Actin staining was used as a loading control. (F) Endogenous GFI1B expression (determined by quantitative polymerase chain reaction on 5’UTR GFI1B, which is not present in overexpression constructs) relative to GAPDH expression following exogenous expression of EV or indicated flag-tagged GFI1B variants in MEG-01 cultures from Figure 2. Results are shown as the mean ± standard deviation (n=3-11).
GFI1BQ287* functions in a dominant-negative manner by sequestering LSD1
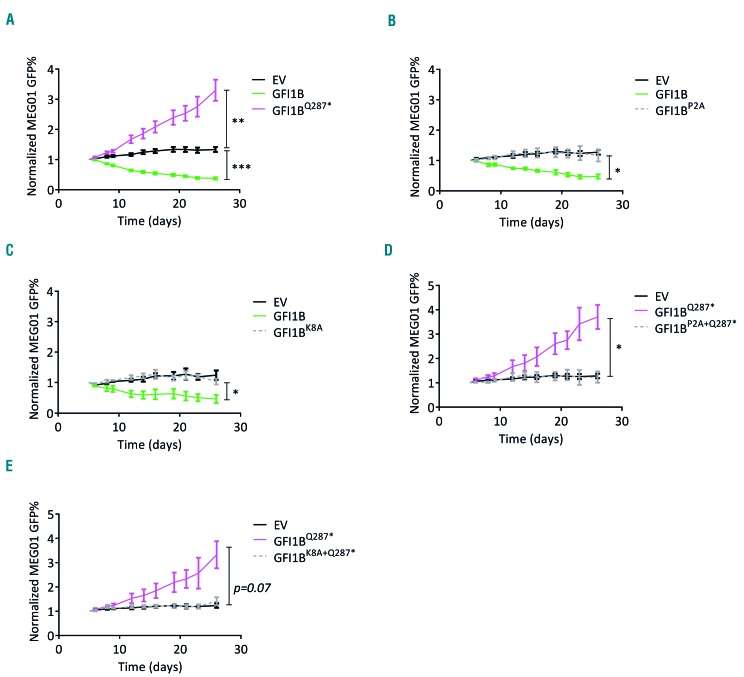
The GFI1B p.Q287* mutation results in an increase in megakaryocyte expansion in vivo and in vitro.5 To investigate whether sequestering of LSD1 by GFI1BQ287* is important for this effect, we introduced a P2A or K8A mutation in the SNAG domain, previously described to disrupt efficient LSD1 recruitment to GFI1 and GFI1B.13,28 We confirmed impaired LSD1 recruitment in GFI1BP2A/K8A-GFP transduced MEG-01 cells using GFP-trap bead-mediated pulldown (Figure 1D). Furthermore, GFI1BQ287*, GFI1BP2A and GFI1BK8A failed to inhibit endogenous GFI1B expression in MEG-01 cells, in contrast to wildtype GFI1B (Figure 1E-F). Next, we used MEG-01 as a megakaryoblast model to study the effect of GFI1BQ287* on cell expansion. In expansion-competition cultures containing transduced and non-transduced cells, GFI1B-overexpressing cells were rapidly overgrown by non-transduced cells, while the opposite was observed following expression of dominant-negative GFI1BQ287* (Figure 2A). Thus, forced GFI1B expression inhibits MEG-01 expansion and dominant-negative GFI1BQ287* results in enhanced expansion, the latter being in line with observations in GFI1B p.Q287* affected individuals.
Figure 2.
GFI1B and GFI1BQ287* regulate MEG-01 expansion via LSD1 recruitment. (A) Normalized GFP percentages (GFP%) of MEG-01 cells transduced with empty vector (EV)-IRES-GFP, GFI1B-IRES-GFP and GFI1BQ287*-IRES-GFP for 26 days (n=8). GFP% were normalized to day 5 after transduction (starting point of the culture) as described in the Methods. Results are shown as the mean ± standard error of the mean. (B) Normalized GFP% of MEG-01 cells transduced with EV-IRES-GFP, GFI1B-IRES-GFP, and GFI1BP2A-IRES-GFP (n=4). (C) Normalized GFP% of MEG-01 cells transduced with EV-IRES-GFP, GFI1B-IRES-GFP, and GFI1BK8A-IRES-GFP (n=3). (D) Normalized GFP% of MEG-01 cells transduced with EV-IRES-GFP, GFI1BQ287*-IRES-GFP, and GFI1BP2A+Q287*-IRES-GFP (n=3). (E) Normalized GFP% of MEG-01 cells transduced with EV-IRES-GFP, GFI1BQ287*-IRES-GFP, and GFI1BK8A+Q287*-IRES-GFP (n=4). *P<0.05, **P<0.01 ***P<0.001.
To determine whether the interaction with LSD1 is important for the inhibitory effect on expansion of GFI1B, we separately introduced the P2A and K8A mutations in GFI1B. Expression of GFI1BP2A or GFI1BK8A nullified the inhibitory effect of GFI1B on expansion (Figure 2B,C; Online Supplementary Figure S3). Thus, an intact LSD1-interacting SNAG domain is required for both inhibition of MEG-01 expansion and repression of the endogenous GFI1B locus.
To investigate whether the LSD1 interaction is relevant for the dominant-negative effect in MEG-01, we introduced the P2A and K8A mutations into GFI1BQ287*. This showed that similar levels of ectopic GFI1BP2A+Q287* and GFI1BK8A+Q287* expression did not affect MEG-01 expansion, in contrast to GFI1BQ287* (Figure 2D,E; Online Supplementary Figure S3). Together, these data indicate that GFI1B limits MEG-01 expansion through the LSD1-interacting SNAG domain. GFI1BQ287* may affect this function in a dominant-negative manner by sequestering LSD1.
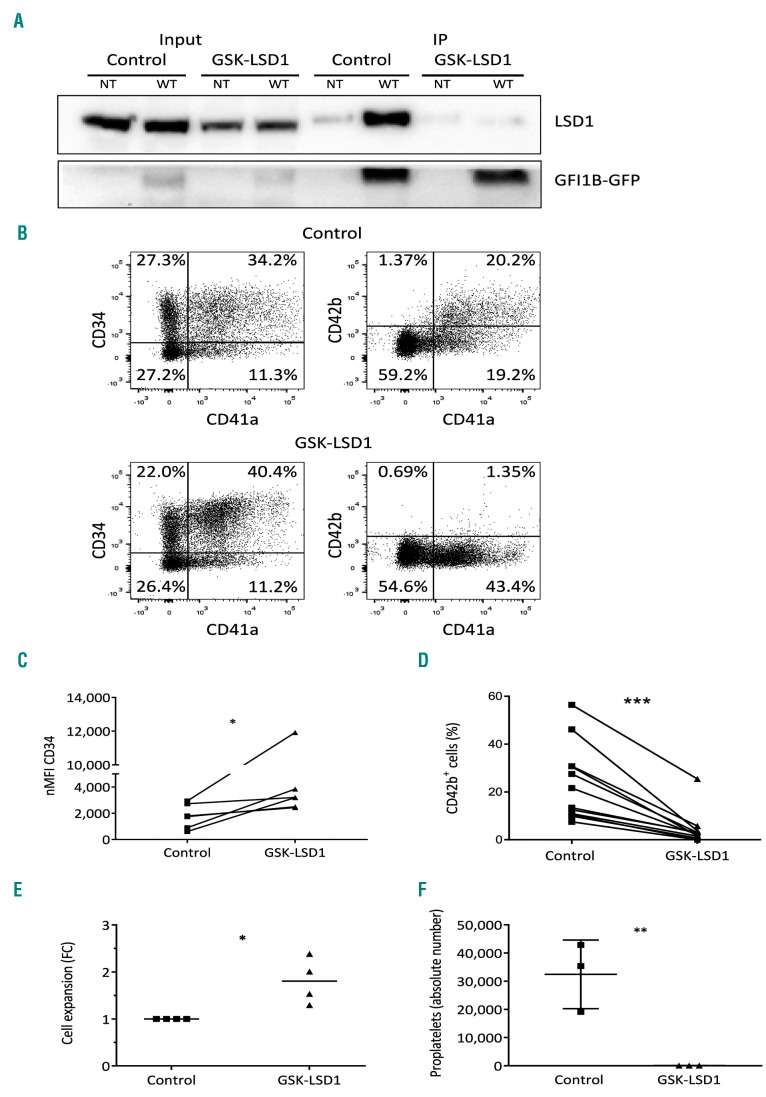
Chemical disruption of the GFI1B-LSD1 interaction in normal megakaryocytes results in abnormalities seen in GFI1B p.Q287* mutated megakaryocytes
LSD1 sequestering by GFI1BQ287* in MEG-01 resulted in enhanced proliferation. To determine whether LSD1 sequestration is relevant for other megakaryocyte abnormalities, we inhibited the GFI1B-LSD1 interaction during megakaryocyte differentiation by treating CD34+ cell-derived megakaryoblasts with the small molecule GSK-LSD1. This inhibitor binds covalently to the LSD1 cofactor FAD, and thereby sterically hinders binding of GFI1B.29 We confirmed the disrupted LSD1-GFI1B binding by the absence of co-precipitated LSD1 upon GFP-trap bead-mediated pulldown in the presence of GSK-LSD1 (Figure 3A). To study effects on megakaryopoiesis, CD34+ cells were differentiated towards megakaryocytes in the presence of GSK-LSD1 for 48 h. Quantification of cell surface marker expression showed sustained CD34 expression and impaired CD42b expression compared to the expression in mock-treated cells (Figure 3B-D). Besides this, a 2-fold increase in megakaryoblast expansion was observed in the presence of GSK-LSD1 (Figure 3E). CD42b expression remained low after 3-6 days of exposure to GSK-LSD1 (Online Supplementary Figure S4C) and we observed that these cells were unable to form proplatelets after 6 days (Figure 3F, Online Supplementary Figure S4). These phenotypes are in line with observations in individuals with the GFI1B p.Q287* mutation.5 Together with data presented in the previous section, this strongly suggests that mutant GFI1BQ287* functions in a dominant-negative manner by sequestering LSD1, resulting in developmental megakaryocyte abnormalities.
Figure 3.
Chemical disruption of the GFI1B-LSD1 interaction by GSK-LSD1 recapitulates GFI1BQ287* hallmarks in vitro. (A) Western blot on co-immunoprecipitated (IP) LSD1 after GFP-trap bead pulldown in dimethylsulfoxide (DMSO) control or GSK-LSD1-treated MEG-01 cells transduced with GFI1B-GFP (WT). Non-transduced (NT) cells were used as a negative control. The upper panel shows LSD1 (~90 kDa) and the lower panel GFI1B variants-GFP (~58 kDa). The left side of the blot shows LSD1 and GFI1B-GFP expression in the input samples. (B) Cell surface expression of megakaryocyte-associated markers CD34, CD41a, and CD42b, to measure megakaryocyte maturation. The presented results are from megakaryocytic cultures 2 days after addition of DMSO control or 4 μM GSK-LSD1. (C) CD34 normalized median fluorescence intensity (nMFI) from CD34+/CD41a+ megakaryoblasts. (D) Percentage of CD42b positivity on CD41a+ megakaryoblasts. The connecting lines (C-D) indicate which samples are from the same experiment. (E) Expansion of CD34+/CD41a+ megakaryoblasts during 2 days of DMSO control or GSK-LSD1 treatment. (F) Absolute number of proplatelet-forming megakaryocytic cells per view (based on Online Supplementary Figure S4A,B) 6 days after addition of DMSO or 4 μM GSK-LSD1 (n=6). *P<0.05, ***P<0.001.
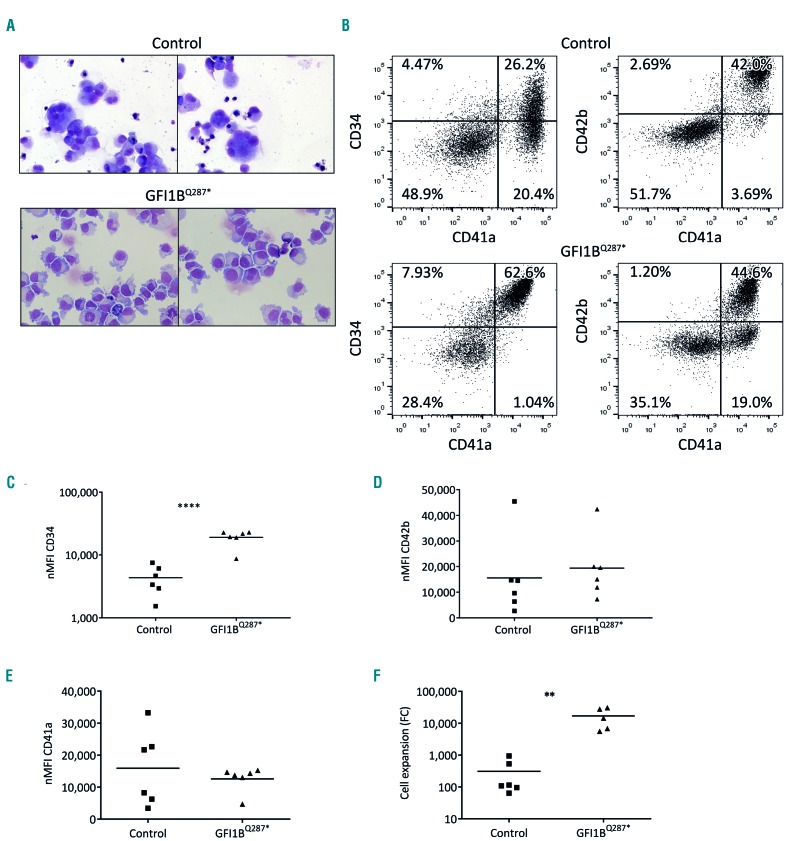
GFI1BQ287* induced pluripotent stem cell-derived megakaryocytic cells show disturbed megakaryocyte differentiation
The morphological megakaryocyte abnormalities, their increased expansion, and sustained CD34 expression indicate that megakaryocyte differentiation is severely disturbed in GFI1BQ287*-mutated individuals. As the availability of CD34+ progenitors and megakaryocytes from patients is limited, we generated iPSC lines from a control individual (MML-6838-Cl2)24 and an individual harboring the GFI1BQ287* mutation (BEL-5-Cl1/2) to study megakaryo poiesis in more detail (Online Supplementary Figure S1). iPSC colonies were differentiated towards megakaryocytes using a two-dimensional differentiation protocol.25 May-Grünwald Giemsa-stained cytospins and flow cytometry revealed enlarged cells with sometimes polyploidization for control megakaryocytic cells, with co-expression of CD41a and CD42b confirming megakaryocyte differentiation (Figure 4A,B). In contrast, differentiation of GFI1BQ287* iPSC resulted in a homogeneous appearance of small, pale cells (Figure 4A). In addition, GFI1BQ287* iPSC-derived megakaryocytic cells expressed elevated levels of CD34 in line with primary GFI1BQ287* megakaryocytes,5 while CD41a and CD42b expression was not different (Figure 4C-E). Quantification of the total number of CD41a+ cells between days 14-18 of iPSC differentiation showed that GFI1BQ287* iPSC produced 55-fold more megakaryocytic cells compared to control iPSC (Figure 4F), in line with the increased number of bone marrow megakaryocyte observed in GFI1BQ287*-affected individuals. Electron microscopy of control and GFI1BQ287* megakaryocytic cells showed the presence of α-granules in both conditions (Online Supplementary Figure S5). Together, these data indicate that the GFI1BQ287* iPSC-derived megakaryocytic cells resemble micromegakaryocytes that exhibit some phenotypes (CD34 expression, increased expansion) also observed in affected individuals.
Figure 4.
GFI1BQ287* of induced pluripotent stem cell-derived megakaryoid cells phenocopy disease characteristics. (A) Cytospins of control and GFI1BQ287* induced pluripotent stem cell (iPSC) lines differentiated towards megakaryocytes (MK) and stained with May-Grünwald Giemsa. Pictures were taken at 40× magnification using a Zeiss Scope.A1 microscope (Zeiss) and images were processed with Zen blue edition. (B) Representative flow-cytometric analysis of control and GFI1BQ287* iPSC-derived cells for surface expression of megakaryocyte-associated markers CD34, CD41a and CD42b. (C-E) Normalized median fluorescence intensity (nMFI) of CD34 (C), CD42b (D) and CD41a (E). (F) Quantification of CD41a+ megakaryocytic cells per seeded iPSC to measure expansion potential. CD41a+ cells were negative for erythroid, myeloid and endothelial makers indicating that these cells represent true megakaryocytic cells (data not shown) **P<0.01, ****P<0.0001.
Identification of GFI1B-regulated processes in early megakaryocyte differentiation
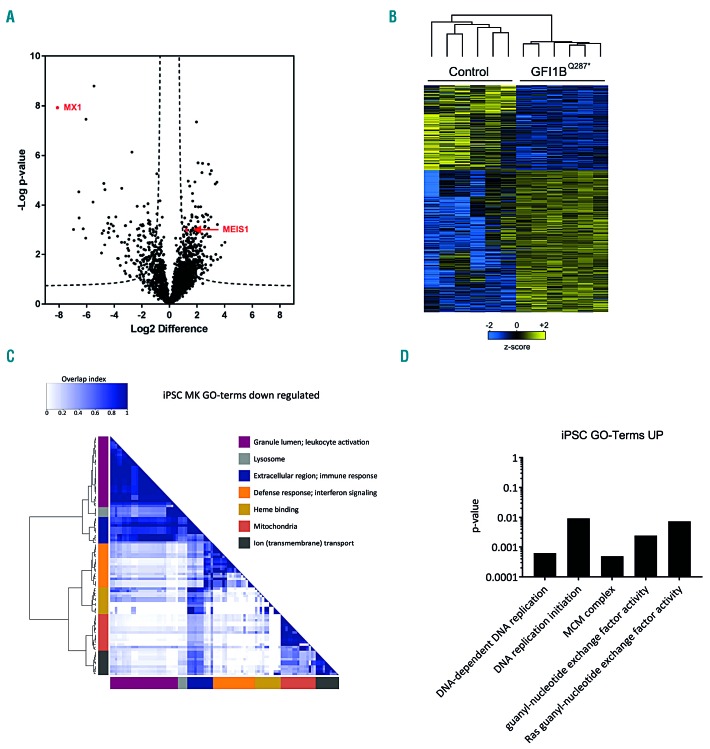
To determine deregulated protein expression downstream of GFI1BQ287*, we compared the proteomes of GFI1BQ287* iPSC-derived CD41a+ megakaryocytic cells (n=6) with those of controls (n=6). In total 2,906 proteins were quantified (Online Supplementary Table S1). We detected CD34 expression in GFI1BQ287* iPSC-derived cells, but not in control cells (Online Supplementary Table S1). The mass spectrometer was not sensitive enough to detect the lower CD34 expression on control cells versus GFI1BQ287* cells observed with flow cytometry. The results confirm the increased CD34 expression observed in the iPSC model system and patients’ platelets (Figure 4C). Major platelet receptors, including glycoprotein (GP)-IB, GPIX, and integrin αIIbβ3, as well as α-granule proteins such as von Willebrand factor (VWF) and thrombospondin-1 (THBS1), were not differentially expressed between control and GFI1BQ287* iPSC-derived megakaryocytic cells. However, 396 proteins were differentially expressed between the GFI1BQ287* and control iPSC-derived megakaryocytic cells, of which 252 were upregulated and 144 downregulated (Figure 5A,B; Online Supplementary Table S1). One of the most strongly downregulated proteins in GFI1BQ287* iPSC was the interferon-induced GTP-binding protein MX1, which was expressed strongly in control iPSC-derived cells but not detectable in GFI1BQ287* iPSC-derived cells (Online Supplementary Figure S6B). Other proteins related to interferon signaling, i.e. STAT1, IFI16, IFI30, IFI35, IFIT1, IFIT3, OAS2 and OAS3, were also significantly reduced in GFI1BQ287* iPSC-derived megakaryocytic cells compared to controls (Online Supplementary Table S1). Because the control and GFI1BQ287* lines are derived from different individuals, differences in individual protein expression might be caused by variation in genetic makeup of the lines. We, therefore, analyzed gene ontology (GO) terms to identify multiple differentially expressed proteins assigned to specific pathways/functions. This identified downregulated proteins related to diverse functions, including mitochondria, response to stimuli, and transmembrane transport (Figure 5C; Online Supplementary Table S2). On the other hand, strongly enriched proteins in the GFI1BQ287* iPSC-derived megakaryocytic cells were all members of the minichromosome maintenance complex (DNA replication licensing factor proteins, MCM2-7) (Online Supplementary Figure S6A, D) and structural maintenance of chromosome proteins (SMC1A, SMC2-4, SMCHD) (Online Supplementary Table S1), which are required for DNA replication and chromosome condensation, respectively (Figure 5D; Online Supplementary Table S2). Furthermore, MEIS1, a key player in megakaryocyte differentiation which is normally downregulated in the final stages of megakaryocyte differentiation,30 showed consistent expression in GFI1BQ287* iPSC-derived megakaryocytic cells while it was never detected in control iPSC-derived cells (Online Supplementary Figure S6C). Thus, the GFI1BQ287* protein influences the expression of a large number of proteins during megakaryopoiesis.
Figure 5.
Differential protein expression between GFI1BQ287* and control induced pluripotent stem cell-derived megakaryocytic cells. (A) Protein levels in control and GFI1BQ287* induced pluripotent stem cell (iPSC)-derived megakaryocytic cells were determined using label-free quantification (LFQ) and differentially expressed proteins were determined using a two-sided t-test (P<0.05 and s0=0.5). The volcano plot shows the −log10-transformed P-value against the log2 fold change (FC) in relative protein levels between control and GFI1BQ287* iPSC-derived megakaryocytic cells, with each protein represented by a single point in the graph. Dashed lines represent the statistical cutoff. A negative FC indicates proteins with reduced levels and a positive FC indicates proteins with elevated levels in GFI1BQ287* iPSC-derived cells. (B) Heat-map and hierarchical clustering of the 396 differentially expressed proteins between control and GFI1BQ287* iPSC-derived cells. Heat-map colors are based on the z-scored (log2) LFQ values. Blue shades correspond to decreased expression levels and yellow shades to increased expression levels. Imputed values are shown in the case a protein was not detected. (C) Enrichment of Gene Ontology (GO) terms based on biological process, molecular function and cellular components was assessed as described in the Methods section. The overlap heat-map shows significant GO terms related to downregulated proteins in GFI1BQ287* iPSC-derived megakaryocytic cells. For each cluster, one or two summarizing terms are indicated. For a full list of significant GO terms see Online Supplementary Table S2. (D) The significant GO terms with corresponding P-values for upregulated proteins in GFI1BQ287* iPSC-derived megakaryocytic cells are shown.
Proteome changes in GFI1B p.Q287* platelets are not limited to α-granule protein depletion
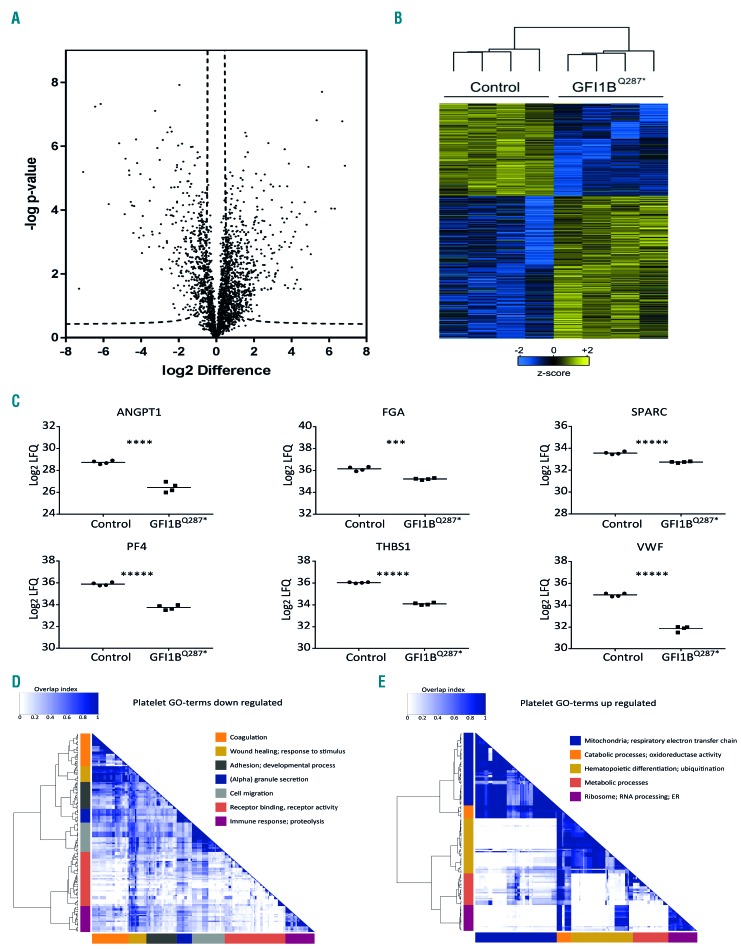
GFI1BQ287* iPSC-derived megakaryocytic cells show phenotypes that indicate aberrant differentiation. With platelets representing the final stage of megakaryopoiesis, we asked how this is translated to the platelet proteome. To identify the deregulated proteins, platelet protein levels of four GFI1BQ287*-affected individuals were compared to those of four healthy individuals using label-free quantitative mass spectrometry. Out of 2,550 quantified proteins, 1,005 proteins were differentially expressed between normal and GFI1BQ287* platelets, with the expression of 395 proteins being reduced and that of 610 elevated in the case of the GFI1BQ287* mutation (Figure 6A,B; Online Supplementary Table S3). In line with the reported α-granule deficiency,5 α-granule proteins such as VWF, THBS1, and platelet factor 4, showed markedly reduced levels in GFI1BQ287* platelets (Figure 6C). Of note, NBEAL2, which is mutated and causative for the α-granule deficiency in classical gray platelet syndrome,31–33 was not among the proteins reduced in GFI1BQ287* platelets, but was in fact mildly elevated (Online Supplementary Table S3). In addition, strongly elevated expression of CD34 was observed in GFI1BQ287* platelets, compatible with findings reported above and published reports.5,8 Other proteins enriched in GFI1BQ287* platelets were proteasomal, ribosomal, and mitochondrial proteins (Online Supplementary Figure S7; Online Supplementary Table S3). GO terms associated with downregulated proteins in GFI1BQ287* platelets were strongly related to platelet functions and α-granules, including wound healing, chemotaxis, immune response, and vesicle secretion (Figure 6D; Online Supplementary Table S2). Proteins whose levels were elevated in GFI1BQ287* platelets were particularly enriched for GO terms on mitochondrial function and the respiratory electron transport chain (Figure 6E; Online Supplementary Table S2). In addition, a multitude of other cellular processes associated with underrepresented (e.g. the Golgi and ER systems, adhesion, and the cytoskeleton) and overrepresented proteins (e.g. ubiquitination, RNA processing, hematopoietic differentiation, and several metabolic functions) were observed (Figure 6D,E; Online Supplementary Table S2). Thus, in addition to a major reduction in α-granule proteins, GFI1BQ287* platelets exhibit aberrant expression of many other proteins with various functions.
Figure 6.
Proteins differentially expressed between GFI1BQ287* and control platelets. Platelets from four GFI1BQ287* patients and four healthy individuals were isolated and protein levels were assessed using mass spectrometry. Differentially expressed proteins were determined using a two-sided t-test (P<0.05 and s0=0.5). (A) Volcano plot showing the −log10-transformed P-value against the log2 fold change (FC) in relative protein levels between control and GFI1BQ287* platelets. Dashed lines represent the statistical cutoff. A negative FC indicates proteins with reduced levels and a positive FC indicates increased levels in GFI1BQ287* platelets. (B) Heat-map with hierarchical clustering for the 1,007 differentially expressed proteins between control and GFI1BQ287* platelets. Heat-map colors are based on the z-scored (log2) label-free quantification (LFQ) values. Blue shades correspond to decreased expression levels and yellow shades to increased expression levels. Imputed values are shown in case a protein was not detected. (C) Relative protein levels (log2 LFQ values) of selected α-granule proteins in platelets of healthy controls (circles) and GFI1BQ287* patients (squares). ANGPT1: angiopoietin-1; FGA: fibrinogen alpha chain; SPARC: secreted protein acidic and rich in cysteine; PF4: platelet factor 4; THBS1: thrombospondin-1; VWF: von Willebrand factor. (D-E) Gene ontology (GO) term enrichment analysis of the significantly expressed proteins in GFI1BQ287* platelets. Overlap heat-maps of significant GO terms associated with downregulated (D) and upregulated (E) proteins in GFI1BQ287* platelets. For each cluster, one or two summarizing terms are indicated. For a full list of significant GO terms see Online Supplementary Table S2.
The GFI1BQ287* platelet proteome resembles that of early megakaryoblasts
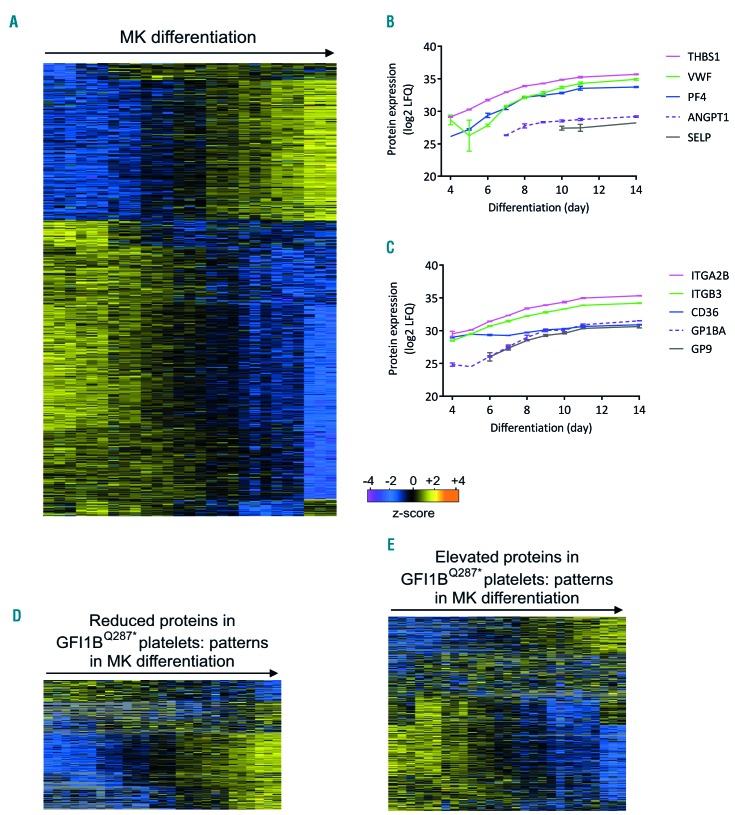
As both platelets and cultured megakaryocytic cells harboring the GFI1BQ287* mutation express the early progenitor marker CD34, we next asked whether GFI1BQ287* platelets also express other early megakaryocyte proteins that are normally downregulated upon terminal differentiation. To study this, we compared protein profiles of healthy maturing megakaryocytes with GFI1BQ287* patient-derived platelets. To this end, CD34+ cells were differentiated to megakaryocytes and harvested for mass spectrometry analysis between days 4 and 14 of differentiation. A total of 3,733 proteins were quantified, of which 1,668 proteins showed significantly different expression levels during megakaryocyte maturation (Figure 7A; Online Supplementary Table S4). Two main clusters of proteins showed strong upregulation (578 proteins) or downregulation (1,026 proteins) towards the late stages of megakaryocyte differentiation. Compatible with the role of megakaryocytes as the platelet progenitor cells, platelet α-granule proteins and receptors were strongly increased during megakaryocyte differentiation (Figure 7B,C). GO term analyses showed that upregulated proteins were indeed strongly related to platelet biogenesis and function, including wound healing, (a) granules, the cytoskeleton, and cell activation (Online Supplementary Figure S8A; Online Supplementary Table S2). Downregulated proteins were associated with several GO terms related to ribosome function, RNA processing, DNA replication (including the MCM proteins) and other metabolic and biosynthetic processes (Online Supplementary Figure S8B; Online Supplementary Table S2). Next, the over- and underrepresented proteins in GFI1BQ287* platelets were compared with the protein profiles of the maturing megakaryocytes. The majority of downregulated proteins in GFI1BQ287* platelets showed a clear pattern of upregulation in differentiating megakaryocytes (~80%) (Figure 7D), whereas the proteins that showed upregulation in GFI1BQ287* platelets were generally downmodulated during megakaryocyte differentiation (~60%) (Figure 7E). As platelets represent the terminal stage of megakaryocyte maturation, our proteomic data show that GFI1BQ287* platelets resemble poorly differentiated megakaryocytes.
Figure 7.
The GFI1BQ287* platelet proteome reveals impaired megakaryocyte differentiation. The proteome of in vitro-differentiating megakaryocytes (MK) was analyzed by label-free mass spectrometry (see Methods). Differentially expressed proteins were determined using analysis of variance (false discovery rate <0.05, s0=0.5). (A) Heat-map with hierarchical clustering of the 1,668 proteins showing significantly different expression between any of the analyzed days of MK differentiation, i.e. day 4, 5, 6, 7, 8, 9, 10, 11, and 14 of differentiation (from left to right) with three replicates per day of culture. Relative protein levels [z-scored log2 label-free quantification (LFQ)] are shown with imputed values in the case a protein was not detected. (B) Expression levels in cultured MK of platelet α-granule proteins during MK differentiation. (C) Expression levels in cultured MK of platelet receptors during MK differentiation. (D-E) Relative expression levels in cultured MK of proteins with significantly increased or decreased levels in GFI1BQ287* platelets: 272 of the 395 downregulated proteins (D) and 503 of the 610 upregulated proteins (E) in GFI1BQ287* platelets were identified in the MK. Magenta/blue shades in the heat-maps correspond to decreased expression levels and yellow/orange shades to increased expression levels; a gray color indicates that a protein was not identified in a given MK sample (Panels D and E only). THBS1: thrombospondin-1; VWF: von Willebrand factor; PF4: platelet factor 4; ANGPT1: angiopoietin-1; SELP: selectin P; ITGA2B: integrin subunit alpha 2b; ITGB: integrin subunit beta; CD36: cluster of differentiation 36; GP1BA: glycoprotein Ib platelet subunit alpha; GP9: glycoprotein 9.
Discussion
The work presented here furthers our understanding on deregulated and disease-causing processes in GFI1B-related bleeding and platelet disorders. Earlier we proposed that GFI1BQ287*, observed in individuals with an inherited bleeding diathesis, may inhibit the function of wildtype GFI1B in a dominant-negative manner by sequestering co-repressors.5 Indeed, mutant GFI1BQ287* still interacts with LSD1 and its associated proteins in MEG-01 cells. Unlike GFI1B causing a decrease in proliferative capacity, forced expression of GFI1BQ287* in MEG-01 cells resulted in increased proliferation. This increase was nullified upon introduction of single point mutations in GFI1BQ287* which abrogate the interaction with LSD1. In line with these findings we observed that disruption of the GFI1B-LSD1 interaction, using GSK-LSD1, induces GFI1BQ287*-like phenotypes in CD34+ cell-derived megakaryocytes, including the down-regulation of CD42b. Yet, GFI1BQ287* iPSC-derived megakaryocytes did not show impaired CD42b expression. This could be explained by variation in CD42b expression observed in affected individuals, with highest levels being comparable to those observed in non-affected individuals.5 Alternatively, this may be explained by differences in in vivo and in vitro differentiation cues. Nevertheless, we propose that GFI1BQ287* inhibits wildtype GFI1B by inhibiting LSD1-dependent processes through competitive sequestration of LSD1 and its binding partners.
The relevance of the GFI1B-LSD1 interaction in megakaryoblast expansion is substantiated by observations in mice following in vivo Lsd1 silencing. This resulted in Meis1 overexpression and increased megakaryocyte numbers in the bone marrow as well. Of note, here we found MEIS1 only in GFI1BQ287* iPSC-derived megakaryocytic cells but not in control iPSC-derived cells, indicating elevated expression in the former. Interestingly, Lsd1-silenced mice also exhibited megakaryocyte abnormalities, thrombocytopenia and a reduction of platelet granules, similar to the phenotypes of patients harboring the dominant-negative GFI1BQ287* mutation.18 This could mean that the functional interplay between GFI1B and LSD1 is not limited to controlling megakaryoblast proliferation and differentiation, but that it is also required for terminal maturation and the generation of granules and platelets. In addition, this interplay is not restricted to megakaryopoiesis, but has also been shown to be essential for the transition of endothelial to hematopoietic stem cells and erythrocyte development.13,14,34
In addition to GFI1BQ287*, two other truncating GFI1B mutations that disrupt the DNA binding domain have been identified, G272fs8 and H294fs,6 both of which lead to a similar bleeding and platelet disorder. These GFI1B mutations also act in a dominant-negative manner. Thus, LSD1 sequestration by mutant GFI1B that cannot bind its transcriptional targets may be a general mechanism through which a collection of GFI1B mutations acts. In these cases, the mutation is present in both GFI1B isoforms and they will inhibit wildtype GFI1B-p37 and GFI1B-p32. However, because the expression of GFI1B-p32 is much lower than that of GFI1B-p37,14 LSD1 sequestration by GFI1BQ287*-p32 will be less than that by GFI1BQ287*-p37. The molecular mechanisms causing megakaryocyte and platelet defects for other reported GFI1B mutations are likely different because the DNA binding and LSD1-interacting domains remain intact in these mutants.
We observed that GFI1B mainly associates with the LSD1-RCOR-HDAC repressor complex (also called BRAF-HDAC complex in neural development) containing LSD1, RCOR1, HDAC1/2, PHF21A (BHC80), and HMG20B (BRAF35).35 The other proteins found, GSE1, RCOR3, and ZMYM2/3, are also known LSD1 interactors,36 of which ZMYM2/3 have not been reported in complex with GFI1B before. ZMYM3 was identified as the only significantly different interacting protein between GFI1B and GFI1BQ287*. Whether this is relevant for disease pathogenesis remains to be seen. Most interactors (LSD1, RCOR1/3, HDAC1/2, GSE1, ZMYM2/3) are detected in differentiating megakaryocytes as well (Online Supplementary Table S4), suggesting that they may be relevant for megakaryocyte development.
Comparison of the proteome of different iPSC-derived megakaryocytic cell cultures showed a uniform expression pattern in GFI1BQ287* cells compared to wildtype cells. The latter might be more variable due to differences in megakaryocytic differentiation stages between cultures. GFI1BQ287* megakaryocytes exhibit a more uniform, micromegakaryocyte-like appearance, suggesting that they are arrested before polyploidization. This is in contrast with megakaryocytes observed in GFI1BQ287* individuals5 and conditional GFI1B knockout mice,37 in which a block after polyploidization but before cytoplasmic maturation is observed. Possibly, environmental cues that are absent in in vitro cultures may stimulate differentiation of GFI1BQ287* megakaryocytes in vivo.
In GFI1BQ287* iPSC-derived megakaryocytic cells we observed a downregulation of downstream interferon-γ signaling targets, such as STAT1, MX1, IFI16, IFI30, IFI35, IFIT3, OAS2 and OAS3. Interferon-γ and its downstream effector STAT1 stimulate megakaryocyte differentiation and platelet production. STAT1 promotes polyploidization and loss of STAT1 in JAK2V617F mice resulted in reduced platelet numbers through interference with megakaryocyte development.38,39 The failure to activate interferon target genes in GFI1BQ287* megakaryocytes may inhibit differentiation and explain the micromegakaryocyte-like appearance of iPSC-derived megakaryocytes.
GFI1BQ287* iPSC-derived megakaryocytic cells further revealed significantly increased levels of the MCM complex and MEIS1. This might be relevant as the MCM complex is involved in DNA replication/cell cycle progression and, in accordance with several studies,40,41 we observed that MCM proteins are downregulated upon megakaryocyte differentiation (Online Supplementary Figure S6D).40,41 In addition, we showed earlier that forced MEIS1 expression in CD34+ cells results in larger and more colonies in colony-forming unit-megakaryocyte assays.30 Thus, failure of downmodulation of DNA replication-associated genes by GFI1B as a consequence of sequestration of LSD1 and associated proteins by GFI1BQ287* might contribute to increased proliferation and lack of polyploidization.
The platelets of GFI1BQ287* patients showed a major reduction in α-granule proteins, ranging from a mild <2-fold decrease (e.g. for SPARC, fibrinogen a and β chains) to a severe >10-fold depletion (e.g. for VEGF and P-selectin). To our surprise, NBEAL2 showed significantly increased expression in both platelets and iPSC-derived cells harboring the GFI1BQ287* mutation. Inactivating mutations in NBEAL2 cause recessive gray platelet syndrome, a bleeding disorder characterized by a severe paucity in α-granules.31–33 Although its exact function is still unclear, NBEAL2 is thought to be required for the biogenesis and/or retention of α-granules in megakaryocytes.42–44 Other proteins with putative roles in α-granule formation, such as VPS33B45 and VIPAS39,46 and proposed NBEAL2 interactors DOCK7, SEC16A and VAC1447 were mostly unaffected in both GFI1BQ287* platelets and iPSC-derived cells, except SEC16A and VAC14, which showed a mild (<2-fold) decrease and increase in GFI1BQ287* platelets, respectively. Possibly, these proteins are under differential control of GFI1B. In contrast to the GFI1BQ287* platelets there was no change in α-granule proteins observed in GFI1BQ287* iPSC-derived megakaryocytic cells. This could suggest that the observed α-granule defect in primary GFI1BQ287* platelets may not be caused by defective protein expression, but possibly originates from defective transport from proteins to α-granules and/or defective trafficking of α-granules to proplatelets.
In addition to the reduction in α-granule proteins, the proteome of GFI1BQ287* platelets showed other abnormalities including increased expression of ribosomal, proteasomal and mitochondrial proteins. These findings imply that proplatelet-forming megakaryocytes of GFI1BQ287* patients may differ from normal mature megakaryocytes at the metabolic level. Some of the upregulated platelet proteins, in particular the ribosome subunits, showed significant downregulation during in vitro megakaryocyte differentiation, in line with the presumed maturation defect in GFI1BQ287* megakaryocytes. Indeed, early megakaryocytes exhibit high protein synthesis rates to support their increasing cellular mass, while this ceases in the final stages of maturation.48 In addition, proteasomal and mitochondrial activities are closely related to the regulation of cell fate decisions.49,50 Highly proliferating cells, including cancer cells and (hematopoietic) stem cells, show distinct mitochondrial activities, and proliferating hematopoietic stem cells have increased mitochondrial mass compared to quiescent hematopoietic stem cells.51,52 Thus, the increase in mitochondrial proteins might support the hyperproliferation of GFI1BQ287* megakaryocytes.
In conclusion, GFI1B regulates protein expression in megakaryocytes through recruitment of the LSD1-RCOR-HDAC co-repressor complex. During megakaryopoiesis many proteins are regulated by GFI1B, which associate with expected but also new processes. GFI1BQ287* may inhibit GFI1B by specifically sequestering the LSD1-RCOR-HDAC complex, making it less available for GFI1B. The normal and affected megakaryocyte and platelet proteomes reported here may serve as a reference for better understanding of other platelet disorders and the molecular pathways that drive megakaryopoiesis and platelet development.
Supplementary Material
Acknowledgments
The authors would like to thank Clemens Mellink and Anne-Marie van der Kevie-Kersemaekers from the Academic Medical Center Amsterdam, Department of Clinical Genetics, Amsterdam, the Netherlands for performing the karyotyping of BEL-5-Cl2 and Konnie M. Hebeda from the Department of Pathology, Radboudumc, Nijmegen, the Netherlands for her expert opinion on the megakaryocyte electron microscopy results. This work was supported by the Landsteiner Foundation for Blood Transfusion Research (project 1531), Sanquin PPOC 15-25p-2089 and the Radboudumc.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/7/1460
References
- 1.Machlus KR, Thon JN, Italiano JE., Jr Interpreting the developmental dance of the megakaryocyte: a review of the cellular and molecular processes mediating platelet formation. Br J Haematol. 2014;165(2):227–236. [DOI] [PubMed] [Google Scholar]
- 2.Bianchi E, Norfo R, Pennucci V, Zini R, Manfredini R. Genomic landscape of megakaryopoiesis and platelet function defects. Blood. 2016;127(10):1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lefrancais E, Ortiz-Munoz G, Caudrillier A, et al. The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nat New Biol. 2017;544(7648):105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eto K, Kunishima S. Linkage between the mechanisms of thrombocytopenia and thrombopoiesis. Blood. 2016;127(10):1234–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monteferrario D, Bolar NA, Marneth AE, et al. A dominant-negative GFI1B mutation in the gray platelet syndrome. N Engl J Med. 2014;370(3):245–253. [DOI] [PubMed] [Google Scholar]
- 6.Stevenson WS, Morel-Kopp MC, Chen Q, et al. GFI1B mutation causes a bleeding disorder with abnormal platelet function. J Thromb Haemost. 2013;11(11):2039–2047. [DOI] [PubMed] [Google Scholar]
- 7.Marneth AE, van Heerde WL, Hebeda KM, et al. Platelet CD34 expression and alpha/delta-granule abnormalities in GFI1B and RUNX1-related familial bleeding disorders. Blood. 2017;129(12):1733–1736. [DOI] [PubMed] [Google Scholar]
- 8.Kitamura K, Okuno Y, Yoshida K, et al. Functional characterization of a novel GFI1B mutation causing congenital macrothrombocytopenia. J Thromb Haemost. 2016;14(7): 1462–1469. [DOI] [PubMed] [Google Scholar]
- 9.Ferreira CR, Chen D, Abraham SM, et al. Combined alpha-delta platelet storage pool deficiency is associated with mutations in GFI1B. Mol Genet Metab. 2016;120(3):288–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uchiyama Y, Ogawa Y, Kunishima S, et al. A novel GFI1B mutation at the first zinc finger domain causes congenital macrothrombocytopenia. Br J Haematol. 2017;181(6):843–847. [DOI] [PubMed] [Google Scholar]
- 11.Rabbolini DJ, Morel-Kopp MC, Chen Q, et al. Thrombocytopenia and CD34 expression is decoupled from alpha-granule deficiency with mutation of the first GFI1B zinc finger. J Thromb Haemost. 2017;15 (11):2245–2258. [DOI] [PubMed] [Google Scholar]
- 12.Schulze H, Schlagenhauf A, Manukjan G, et al. Recessive grey platelet-like syndrome with unaffected erythropoiesis in the absence of the splice isoform GFI1B-p37. Haematologica. 2017;102(9):e375–e378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell. 2007;27(4):562–572. [DOI] [PubMed] [Google Scholar]
- 14.Laurent B, Randrianarison-Huetz V, Frisan E, et al. A short Gfi-1B isoform controls erythroid differentiation by recruiting the LSD1-CoREST complex through the dimethylation of its SNAG domain. J Cell Sci. 2012;125(Pt 4):993–1002. [DOI] [PubMed] [Google Scholar]
- 15.van der Meer LT, Jansen JH, van der Reijden BA. Gfi1 and Gfi1b: key regulators of hematopoiesis. Leukemia. 2010;24(11):1834–1843. [DOI] [PubMed] [Google Scholar]
- 16.Cai C, He HH, Gao S, et al. Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Rep. 2014;9(5):1618–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerenyi MA, Shao Z, Hsu YJ, et al. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. Elife. 2013;2:e00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sprussel A, Schulte JH, Weber S, et al. Lysine-specific demethylase 1 restricts hematopoietic progenitor proliferation and is essential for terminal differentiation. Leukemia. 2012;26(9):2039–2051. [DOI] [PubMed] [Google Scholar]
- 19.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11(5):1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smits AH, Jansen PW, Poser I, Hyman AA, Vermeulen M. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res. 2013;41(1):e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75(3):663–670. [DOI] [PubMed] [Google Scholar]
- 22.Migliaccio G, Sanchez M, Masiello F, et al. Humanized culture medium for clinical expansion of human erythroblasts. Cell Transplant. 2010;19(4):453–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heideveld E, Masiello F, Marra M, et al. CD14+ cells from peripheral blood positively regulate hematopoietic stem and progenitor cell survival resulting in increased erythroid yield. Haematologica. 2015;100(11):1396–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen M, Varga E, Wust T, et al. Generation and characterization of human iPSC line MML-6838-Cl2 from mobilized peripheral blood derived megakaryoblasts. Stem Cell Res. 2017;18:26–28. [DOI] [PubMed] [Google Scholar]
- 25.Hansen M, Varga E, Aarts C, et al. Efficient production of erythroid, megakaryocytic and myeloid cells, using single cell-derived iPSC colony differentiation. Stem Cell Res. 2018;29:232–244. [DOI] [PubMed] [Google Scholar]
- 26.Vizcaino JA, Deutsch EW, Wang R, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32(3):223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bantscheff M, Hopf C, Savitski MM, et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol. 2011;29(3):255–265. [DOI] [PubMed] [Google Scholar]
- 28.Fiolka K, Hertzano R, Vassen L, et al. Gfi1 and Gfi1b act equivalently in haematopoiesis, but have distinct, non-overlapping functions in inner ear development. EMBO Rep. 2006;7(3):326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishikawa Y, Gamo K, Yabuki M, et al. A novel LSD1 inhibitor T-3775440 disrupts GFI1B-containing complex leading to transdifferentiation and impaired growth of AML cells. Mol Cancer Ther. 2017;16(2):273–284. [DOI] [PubMed] [Google Scholar]
- 30.Zeddies S, Jansen SB, di Summa F, et al. MEIS1 regulates early erythroid and megakaryocytic cell fate. Haematologica. 2014;99(10):1555–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albers CA, Cvejic A, Favier R, et al. Exome sequencing identifies NBEAL2 as the causative gene for gray platelet syndrome. Nat Genet. 2011;43(8):735–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gunay-Aygun M, Falik-Zaccai TC, Vilboux T, et al. NBEAL2 is mutated in gray platelet syndrome and is required for biogenesis of platelet alpha-granules. Nat Genet. 2011;43(8):732–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kahr WH, Hinckley J, Li L, et al. Mutations in NBEAL2, encoding a BEACH protein, cause gray platelet syndrome. Nat Genet. 2011;43(8):738–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thambyrajah R, Mazan M, Patel R, et al. GFI1 proteins orchestrate the emergence of haematopoietic stem cells through recruitment of LSD1. Nat Cell Biol. 2016;18(1):21–32. [DOI] [PubMed] [Google Scholar]
- 35.Ceballos-Chavez M, Rivero S, Garcia-Gutierrez P, et al. Control of neuronal differentiation by sumoylation of BRAF35, a subunit of the LSD1-CoREST histone demethylase complex. Proc Natl Acad Sci U S A. 2012;109(21):8085–8090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maiques-Diaz A, Somervaille TC. LSD1: biologic roles and therapeutic targeting. Epigenomics. 2016;8(8):1103–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foudi A, Kramer DJ, Qin J, et al. Distinct, strict requirements for Gfi-1b in adult bone marrow red cell and platelet generation. J Exp Med. 2014;211(5):909–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duek A, Lundberg P, Shimizu T, et al. Loss of Stat1 decreases megakaryopoiesis and favors erythropoiesis in a JAK2-V617F-driven mouse model of MPNs. Blood. 2014;123 (25):3943–3950. [DOI] [PubMed] [Google Scholar]
- 39.Huang Z, Richmond TD, Muntean AG, Barber DL, Weiss MJ, Crispino JD. STAT1 promotes megakaryopoiesis downstream of GATA-1 in mice. J Clin Invest. 2007;117 (12):3890–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haldar S, Roy A, Banerjee S. Differential regulation of MCM7 and its intronic miRNA cluster miR-106b-25 during megakaryopoiesis induced polyploidy. RNA Biol. 2014;11(9):1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raslova H, Kauffmann A, Sekkai D, et al. Interrelation between polyploidization and megakaryocyte differentiation: a gene profiling approach. Blood. 2007;109(8):3225–3234. [DOI] [PubMed] [Google Scholar]
- 42.Deppermann C, Cherpokova D, Nurden P, et al. Gray platelet syndrome and defective thrombo-inflammation in Nbeal2-deficient mice. J Clin Invest. 2013;123(8):3331–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guerrero JA, Bennett C, van der Weyden L, et al. Gray platelet syndrome: proinflammatory megakaryocytes and alpha-granule loss cause myelofibrosis and confer metastasis resistance in mice. Blood. 2014;124(24):3624–3635. [DOI] [PubMed] [Google Scholar]
- 44.Kahr WH, Lo RW, Li L, et al. Abnormal megakaryocyte development and platelet function in Nbeal2(−/−) mice. Blood. 2013;122(19):3349–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urban D, Li L, Christensen H, et al. The VPS33B-binding protein VPS16B is required in megakaryocyte and platelet alpha-granule biogenesis. Blood. 2012;120(25):5032–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lo B, Li L, Gissen P, et al. Requirement of VPS33B, a member of the Sec1/Munc18 protein family, in megakaryocyte and platelet alpha-granule biogenesis. Blood. 2005;106 (13):4159–4166. [DOI] [PubMed] [Google Scholar]
- 47.Mayer L, Jasztal M, Pardo M, et al. The BEACH-domain containing protein, Nbeal2, interacts with Dock7, Sec16a and Vac14. Blood. 2017;131(9):1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ru YX, Zhao SX, Dong SX, Yang YQ, Eyden B. On the maturation of megakaryocytes: a review with original observations on human in vivo cells emphasizing morphology and ultrastructure. Ultrastruct Pathol. 2015;39(2): 79–87. [DOI] [PubMed] [Google Scholar]
- 49.Bassermann F, Eichner R, Pagano M. The ubiquitin proteasome system - implications for cell cycle control and the targeted treatment of cancer. Biochim Biophys Acta. 2014;1843(1):150–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schrepfer E, Scorrano L. Mitofusins, from mitochondria to metabolism. Mol Cell. 2016;61(5):683–694. [DOI] [PubMed] [Google Scholar]
- 51.Joshi A, Kundu M. Mitophagy in hematopoietic stem cells: the case for exploration. Autophagy. 2013;9(11):1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12(10):685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.