SUMMARY
The kinetochore scaffold 1 (KNL1) protein coordinates the spindle assembly checkpoint (SAC), a signaling pathway that delays chromosome segregation until all sister chromatids are properly attached to spindle microtubules. Recently, microtubules and protein phosphatase 1 (PP1), which both bind the N-terminal domain of KNL1, have emerged as regulators of the SAC; however, how these proteins interact to contribute to SAC signaling is unknown. Here, we use X-ray crystallography, NMR spectroscopy and biochemical assays to show how KNL1 binds both PP1 and microtubules. Unexpectedly, we discovered that PP1 and microtubules bind KNL1 via overlapping binding sites. Further, we showed that Aurora B kinase phosphorylation results in distinct patterns of KNL1 complex disruption. Finally, combining this data with cosedimentation assays unequivocally demonstrated that microtubules and PP1 binding to KNL1 is mutually exclusive, with preferentially formation of the KNL1:PP1 holoenzyme in the presence of PP1.
Keywords: protein phosphatase 1 (PP1), kinetochore scaffold 1 (KNL1), x-ray crystallography, NMR spectroscopy, nuclear phosphatases, spindle assembly checkpoint, cell cycle, microtubules
INTRODUCTION
Accurate chromosome segregation is critical for cellular viability (Nagaoka et al., 2012; Santaguida and Amon, 2015). One of the key steps in this process is chromosome biorientation, where microtubules that originate from opposite poles of the cell attach to kinetochores of sister chromatids. Missegregation, which can lead to aneuploidy and tumorigenesis, is prevented by the activation of the spindle assembly checkpoint (SAC), a signaling pathway that delays chromosome segregation until all chromosomes are properly bioriented (Lara-Gonzalez et al., 2012; Musacchio, 2015; Musacchio and Salmon, 2007). However, this need for fidelity is coupled with the need for the timely passage through mitosis. Thus, once biorientation is achieved, the SAC must be effectively silenced. While the mechanisms that initiate SAC signaling are becoming increasingly clear, the molecular processes that maintain and silence the SAC are comparatively poorly understood.
Protein phosphatase 1 (PP1) and PP2A-B56 are emerging as two of the key phosphatases that regulate SAC silencing and mitotic exit (Hertz et al., 2016; Meadows, 2013; Nijenhuis et al., 2014; Pinsky et al., 2009; Rosenberg et al., 2011; Vanoosthuyse and Hardwick, 2009; Wurzenberger and Gerlich, 2011; Zhang et al., 2014). PP1, in particular, is the most widely expressed and abundant serine/threonine phosphatase. It dephosphorylates its 1000s of substrates with exquisite specificity via its interaction with scores of regulatory proteins (~200 confirmed interactors) (Choy et al., 2014; Hendrickx et al., 2009; Heroes et al., 2013). These PP1 regulatory proteins function to direct the activity of PP1 towards specific substrates by localizing it to its cellular point of action and/or by directly altering its substrate preferences (Hendrickx et al., 2009; Peti et al., 2013).
Recent studies have demonstrated that PP1 binds and is targeted to kinetochores, in part, by kinetochore scaffold 1 (KNL1)(Liu et al., 2010; London et al., 2012; Rosenberg et al., 2011), a scaffolding protein that interacts with missegregation 12 (MIS12) and NDC80 to form the KMN network, a conserved kinetochore protein complex that connects kinetochores to microtubules (MTs) (Cheeseman et al., 2004, 2006). The N-terminal domain of KNL1 is hypothesized to contain three PP1-specific linear interaction motifs (SLiMs), the RVxF, the ΦΦ and the SILK motifs (Choy et al., 2014). Previous studies have demonstrated that the KNL1 RVxF motif, as is true for most RVxF-containing PP1 regulators, is essential for PP1 binding (Espeut et al., 2012; Hendrickx et al., 2009; Liu et al., 2010; Rosenberg et al., 2011). However, the role of the SILK motif has remained ambiguous, with some reports suggesting it is dispensable for PP1 binding while others have shown that mutation of the SILK motif reduces KNL1:PP1 binding (Espeut et al., 2012; Liu et al., 2010; Meadows et al., 2011). Furthermore, it was recently demonstrated that the KNL1:PP1 complex is altered by the phosphorylation of KNL1 by AURB, a component of the conserved chromosomal passenger complex that plays a central role in controlling kinetochore function (Carmena et al., 2012; Nasa et al., 2018; Pinsky et al., 2006; Welburn et al., 2010). However, it is unknown how the individual phosphorylation events alter the affinity of KNL1 for PP1 and/or MTs. Moreover, while it is known that the residues N-terminal to the KNL1 SILK motif are important for MT binding (Espeut et al., 2012), the molecular details of this interaction are entirely missing.
Despite the biological importance of the KNL1-MT and KNL1-PP1 interactions in the regulation of the cell cycle, the underlying molecular details and mechanisms by which KNL1 binds MT and PP1, and how these complexes facilitate SAC silencing, remain largely unknown. Here, we combine X-ray crystallography and NMR spectroscopy to elucidate how, at a molecular level, KNL1 binds both PP1 and MTs. Most significantly, we discover that MT binding and PP1 binding are mutually exclusive as they have overlapping binding sites. Because the affinity of KNL1 for PP1 is significantly higher than that of KNL1 for MT, our data suggests that PP1 ultimately displaces MTs from KNL1, resulting in the formation of a KNL1:PP1 holoenzyme free to desphorphorylate its multiple kinetochore substrates.
RESULTS
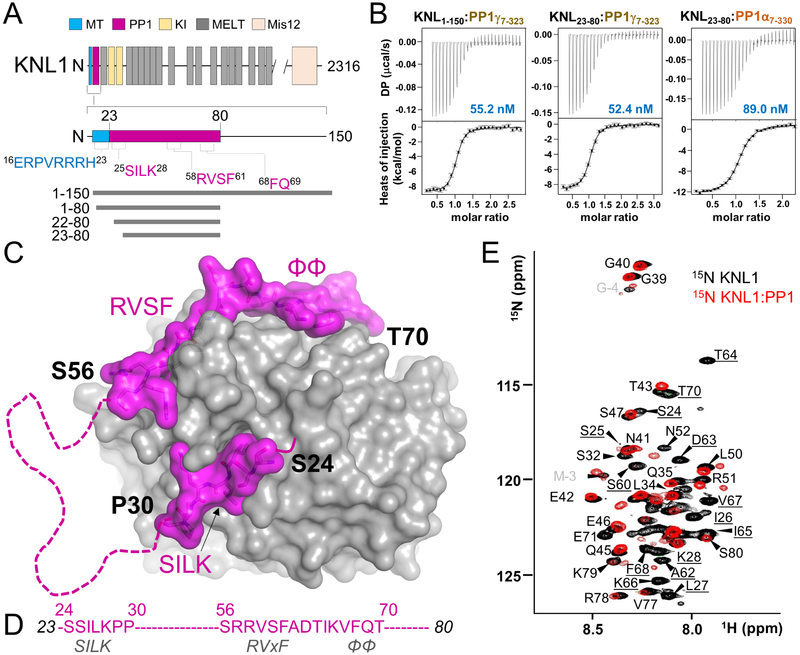
KNL1 binds PP1 at two sites connected by a disordered loop
In order to understand how KNL1 interacts with PP1, we first identified the minimal PP1 binding domain of KNL1 using isothermal titration calorimetry (ITC; Figures 1B, S1A). ITC showed that KNL1 residues 23–80 are necessary and sufficient for PP1α binding (KD = 89 ± 20 nM; all ITC data is summarized in Table 1 with representative thermograms in Figure S1). We then showed that KNL1 binds equally well to both PP1α and PP1γ, the two major isoforms of PP1 at the outer kinetochore (Trinkle-Mulcahy et al., 2001) (Figures 1B, S1A). Thus, KNL1 exhibits no selectivity for either of these isoforms.
Figure 1: KNL1 binds PP1 using three PP1-specific SLiMs separated by a disordered loop.
(A) KNL1 domain structure, highlighting the putative protein interaction sites (microtubule, MT, blue; PP1, magenta). The PP1 binding domain is hypothesized to contain three established PP1 binding SLiMs, namely the SILK, RVxF and ΦΦ motifs, while the putative MT binding domain is positively charged. The constructs and variants examined in this study are shown below.
(B) Binding isotherms of KNL1 variants with either PP1γ or PP1α.
(C) Crystal structure of the KNL123–80:PP1 holoenzyme. PP1 is in grey and KNL1 in magenta. Residues not modeled due to a lack of density are indicated by a dashed line.
(D) The ordered KNL1 residues are specified in text (magenta). The established PP1 binding motifs present in KNL1 are indicated in grey text.
(E) Overlay of the 2D [1H,15N] HSQC spectra of KNL123–80 (black) with the KNL23–80:PP1 holoenzyme (red). Peaks observed in the KNL23–80:PP1 holoenzyme spectrum correspond to residues that remain disordered upon complex formation. Peaks present only in the KNL123–80 spectrum are underlined.
Related to Figures S1–S3
Table 1:
Isothermal titration calorimetry (ITC) measurements of WT KNL1 and KNL1 variants with PP1.
| KNL1 | PP1 | KD (nM) | ΔH (kcal/mol) | −TΔS (kcal/Mol) | repeats |
|---|---|---|---|---|---|
| 1–150 | |||||
| WT | Υ 7–323 | 55.2± 9.5 | −9.3 ± 0.7 | 0.6 ± 0.7 | 2 |
| 1–80 | |||||
| WT | α7–330 | 46.4 ± 4.5 | −22.6 ± 1.7 | −12.6 ± 1.8 | 2 |
| S24AS25A | α7–330 | 93.5 ± 8.4 | −21.5 ± 1.6 | −14.2 ± 3.0 | 3 |
| S24DS25D | α7–330 | 266.6 ± 26.0 | −10.9 ± 1.0 | −1.9 ± 1.0 | 3 |
| SILKdead | α7–330 | 227.6 ± 30.1 | −21.5 ± 0.8 | −12.3 ± 0.9 | 3 |
| S56D | α7–330 | 14.7 ± 6.4 | −12.4 ± 1.9 | −1.7 ± 2.2 | 2 |
| S60A | α7–330 | 154.2 ± 0.2 | −20.1 ± 1.2 | −10.8 ± 1.2 | 2 |
| S60D | α7–330 | 236.0 ± 2.1 | −30.7 ± 6.8 | −21.7 ± 6.8 | 2 |
| WT | Υ7–323 | 38.3 ± 2.0 | −8.6 ± 0.7 | 1.6 ± 0.7 | 2 |
| 23–80 | |||||
| WT | α7–330 | 89.0 ± 19.6 | −13.7 ± 0.9 | −3.8 ± 0.9 | 3 |
| WT | Υ7–300 | 82.0 ± 10.3 | −28.3 ± 1.4 | −18.6 ± 1.4 | 2 |
| WT | Υ7–323 | 52.4 ±2.3 | −10.0±0.4 | −0.1 ±0.4 | 2 |
Based on this analysis, we determined the 3-dimensional structure of the KNL123–80:PP1a7–300 holoenzyme (2.95 Å resolution; hereafter referred to as KNL1:PP1; Table 2; Figure S2). KNL1 binds PP1 in an extended manner using three established PP1-specific SLIMs: the SILK (aa 25–28), the RVxF (aa 58–61) and the ϕϕ (aa 67/68) SLiMs (Figures 1C, 1D). The interaction results in ~2700 Å2 of buried solvent accessible surface area, consistent with the strong binding affinity observed between both proteins using ITC. Unexpectedly, many residues of KNL1 (aa 31–55 and 71–80) could not be modeled due to a lack of electron density, suggesting they remained dynamic when bound to PP1 (Figure 1C). We tested this directly using NMR spectroscopy by comparing the 2D [1H,15N] heteronuclear single quantum correlation (HSQC) spectrum of unbound 15N-labeled KNL1 with 15N-labeled KNL1 in complex with PP1 (Figure 1E). In this experiment (Choy et al., 2014, 2015; Kumar et al., 2016; O’Connell et al., 2012), the peaks corresponding to KNL1 residues that bind directly to PP1 disappear due to the increased molecular weight of the complex, while the peaks corresponding to residues that do not bind remain visible. The 2D [1H,15N] HSQC spectrum of free KNL123–80 has the hallmarks of an intrinsically disordered protein, including narrow chemical shift dispersion in the 1H dimension. Upon complex formation, the peaks corresponding to KNL1 residues 31–55 and 71–80 remain visible and are thus not bound to PP1. These data demonstrate that the KNL1 residues connecting the SILK and RVxF SLiMs (31–55) stays dynamic when bound to PP1.
Table 2:
Data collection and refinement statistics
| PP1α7–300: KNL123–80a | |
|---|---|
| Protein | |
| Organism | Homo sapiens |
| PDBID | 6CZO |
| Data collection | |
| Space group | P42212 |
| Cell dimensions | |
| a, b, c (Å) | 138.0, 138.0, 118.4 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.0 − 2.95 (3.00 − 2.95) |
| Rmerge | 8.9 (66.5) |
| I/I | 29.2 (4.4) |
| Completeness (%) | 100.0 (100.0) |
| Redundancy | 7.9 (8.1) |
| Refinement | |
| Resolution (Å) | 45.13 − 2.95 (3.06 − 2.95) |
| No. reflections | 24763 |
| Rwork / Rfree | 0.17 (0.0.23)/0.21 (0.32) |
| No. atoms | |
| Protein | 5001 |
| Ligand/ion | 14 |
| Water | 61 |
| Average B-factor (Å2) | |
| Protein | 65.8 |
| Ligand | 63.9 |
| Water | 62.2 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.615 |
| Ramachandran | |
| Outliers (%) | 0.3 |
| Allowed (%) | 5.7 |
| Favored (%) | 94.0 |
| Clashscore | 3.43 |
Data was collected from a single crystal
Values in parentheses are for highest-resolution shell.
The KNL1 SILK motif is critical for PP1 binding
Although the KNL1 RVxF and SILK motifs are exceptionally conserved (Figure S3), the importance of the KNL1 SILK motif for PP1 binding has remained controversial (Espeut et al., 2012; Liu et al., 2010). Here, our crystallographic and NMR data show that the KNL1 SILK motif binds directly to PP1 (Figures 1C, 1E). The KNL1 SILK motif binds PP1 in a conformation nearly identical to that of the homologous GILK motif of Inhibitor-2, the only other PP1 regulatory protein containing a SILK motif whose PP1 holoenzyme structure has been determined (Hurley et al., 2007) (Figure 2A). The interaction is both hydrophobic and ionic, with Ile26KNL1 and Leu27KNL1 (SILK) binding in deep hydrophobic pockets of PP1 defined by Leu53PP1, Leu55 PP1, Leu59PP1, L88PP1 and Phe119PP1. In addition, Lys28KNL1 (SILK) forms multiple ionic interactions with residues Glu56PP1, Asp116PP1 and Glu167PP1 (Figure 2B). In order to confirm that the SILK motif is important for PP1 binding, we generated a KNL1 variant in which the SILK residues were mutated to alanines (AAAA; called SILKdead) and determined its affinity for PP1. The SILKdead variant bound PP1 ~5-fold more weakly than wt-KNL1 (SILKdead KD = 227 ± 30 nM; Figure 2C), confirming its role in PP1 binding.
Figure 2: KNL1 binds PP1 via its SILK motif.
(A) Overlay of the KNL1 (magenta) and Inhibitor-2 (beige) SILK motifs bound to PP1.
(B) PP1 residues that interact directly with the KNL1 SILK motif; polar/electrostatic interactions are indicated by black dashed lines.
(C) ITC thermograms of wt-KNL1 and the KNL1-SILKdead variant (aa 1–80), in which the KNL1 25SILK28 residues are mutated to AAAA.
Speed and interdependence of KNL1 phosphorylation by Aurora B kinase (AURB)
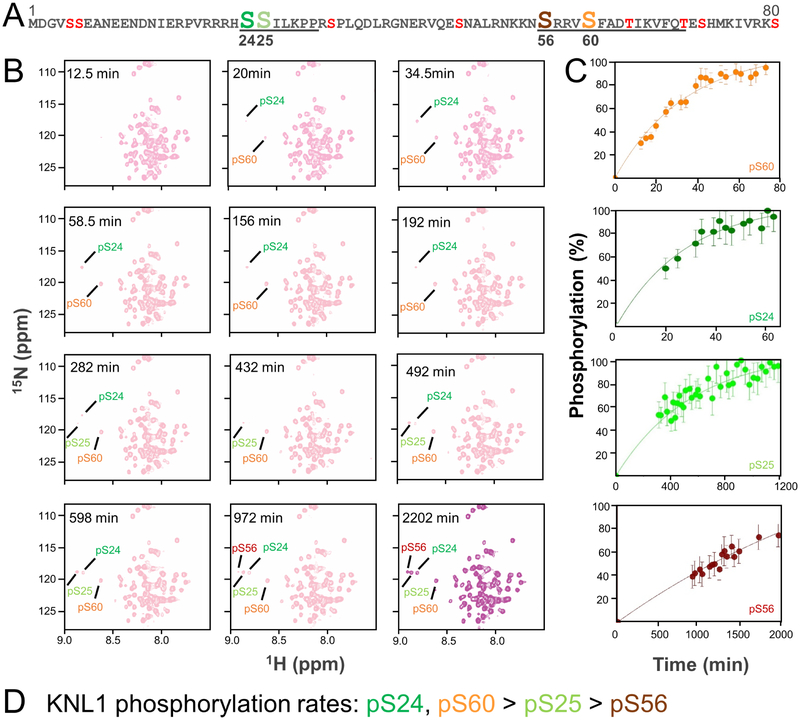
AURB regulates kinetochore function by destabilizing kinetochore-MT interactions (Cheeseman et al., 2006; Espeut et al., 2012; Pinsky et al., 2009; Welburn et al., 2010). Here, we used in vitro phosphorylation assays and NMR spectroscopy to determine if residues in the KNL1 PP1 binding domain are directly phosphorylated by AURB. Of the 9 potential phosphorylation sites identified using NetPhos2.0 (Blom et al., 1999) in KNL11–80, we show that four (Ser24, Ser25, Ser56 and Ser60) are directly phosphorylated by AURB (Figures S4A, S4B). Phosphorylation of Ser24, Ser25 and Ser60 have been confirmed in vivo using mass spectrometry (Kettenbach et al., 2011; Welburn et al., 2010)). The chemical shift indices (CSI) of unphosphorylated and phosphorylated KNL1 (pKNL1) are essentially identical, demonstrating that phosphorylation does not induce a major change in the structural secondary propensity of KNL1 (Figure S4C). Notably, all four phosphorylated residues are part of or directly adjacent to PP1 interaction SLiMs: Ser24 and Ser25 are N-terminal to or part of the KNL1 SILK SLiM (SSILK) while Ser56 and Ser60 are N-terminal to or within the KNL1 RVxF SLiM (SRRVSF) (Figure 3A).
Figure 3: Real-time NMR monitoring of the phosphorylation of KNL1–80.
(A) Sequence of WT-KNL11–80 with the potential phosphorylation sites indicated; the residues that bind directly to PP1 are underlined. The four residues phosphorylated directly by Aurora B kinase, as observed using NMR spectroscopy, are indicated (Ser24, Ser25, Ser56 and Ser60).
(B) 2D [1H15N] HSQC spectra of KNL11–80 after the addition of Aurora B kinase. The N-HN cross peaks of the phosphorylated serines are labeled.
(C) Build-up curves showing the time-course of phosphorylation of KNL11–80 by Aurora B Kinase.
(D) KNL1 phosphorylation rates in descending order.
To identify which residues are most rapidly phosphorylated by AURB, we reconstituted the phosphorylation reaction of AURB with KNL1 and monitored the rates of phosphorylation of Ser24, Ser25, Ser56 and Ser60 using NMR spectroscopy. Using 50 μM 15N-labeled KNL1 as a substrate, 1.25 μM of AURB/INCENP achieved ~100% phosphorylation of Ser24 and Ser60 within1 h (Figures 3B, 3C). To determine if the phosphorylation of these sites is independent (phosphorylation of one residue is independent of the phosphorylation state of the other) or dependent (phosphorylation of one residue occurs only after the other has been phosphorylated), we tested the ability of these residues to be phosphorylated in KNL1 variants in which one of two sites had been mutated to an alanine (i.e., KNL1-S24A; KNL1-S60A). The data show that the phosphorylation of Ser24 and Ser60 are independent, as each site is fully phosphorylated in the KNL1 variants (Figures S5A, S5B). Compared to Ser24 and Ser60, phosphorylation of Ser25 is slower, beginning at ~1 h and reaching completion after only ~24 h (i.e., pS24/pS60 → pS24/pS25/pS60; Figures 3B, 3C). Although slower, Ser25 also does not require the phosphorylation of Ser24 and/or Ser60 in order for it to be phosphorylated as Ser25 is fully phosphorylated in KNL1-S24A and KNL1-S60A variants (Figures S5A, S5B). Finally, phosphorylation of Ser56 is slow, beginning at 16 h and continuing until ~36 hr (Figures 3B, 3C). Together, these data demonstrate that four serine residues within the KNL1 PP1 binding domain are phosphorylated directly by AURB, with those most readily phosphorylated being Ser24 and Ser60. Furthermore, the phosphorylation of each site is independent of the other.
Phosphorylation of either the SILK or the RVxF motifs destabilitze PP1 binding by ~5-fold
To quantify the consequences of these phosphorylation events on KNL1 binding to PP1, we used ITC. A SILK site phosphorylation mimetic variant of KNL1 (S24D/S25D) resulted in a ~6-fold reduction in affinity compared to WT (KD = 266.6 ± 26 nM; Table 1; Figure S1B). This reduction was nearly identical to that observed for the SILKdead variant, demonstrating the phosphorylation disrupts PP1 holoenzyme formation to the same extent as mutating the hydrophobic residues that anchor the SILK site to PP1 (KD = 227.6 ± 30.1 nM; Table 1; Figure S1C). Similarly, a phosphorylation mimetic variant of the KNL1 RVxF site (S60D) also negatively impacted PP1 binding, with a ~5-fold reduction in affinity (KD = 236 ± 2 nM; Table 1; Figure S1B). Finally, while a phosphorylation mimic of Ser56 (S56D) resulted in enhanced binding (3-fold), the physiological relavence is unclear as this residue has never been observed to be phosphorylated in vivo nor does it interact directly with PP1 (Table 1; Figure S1B). Together, the data show that three of the four AURB phosphorylation sites in the KNL1 PP1 binding domain destablilize PP1:KNL1 holoenzyme formation by 5–6-fold. These changes are consistent with those observed for other PP1 regulators, such as RepoMan and Ki67, with Ser/Thr residues in the ‘x’ position capable of being phosphorylated (Kumar et al., 2016). Namely, that in spite of the fact that the side chain of the ‘x’ residue in the RVxF and SILK motifs do not interact directly with PP1 (as is evident from the KNL1:PP1 crystal structure), phosphorylation at these sites quanitatively destabilizes holoenzyme formation.
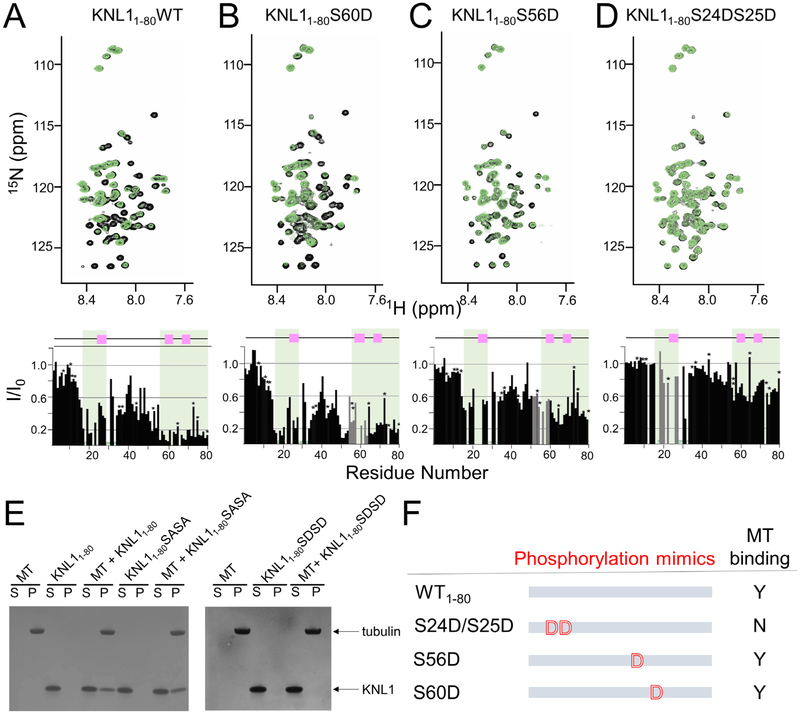
KNL1 binds microtubules via two distinct domains
KNL1 binds MTs, an interaction that was previously shown to require residues N-terminal to the SILK site (Figure 1A) (Espeut et al., 2012). In order to molecularly define the KNL1 residues that bind directly to MTs, we used NMR spectroscopy. In this experiment, the 2D [1H,5N] HSQC spectra of 15N-labeled KNL1 was compared with that of 15N-labeled KNL1 bound to MT (Figure 4A) and the ratio of normalized intensities of free (I0) and MT-bound (I) used to identify the residues that bind MTs. MTs were assembled from unpolymerized tubulin following established methods for the study of protein interactions with MTs using NMR spectroscopy (Kadavath et al., 2015a, 2015b; Mukrasch et al., 2007). Unexpectedly, the data revealed that the N-terminal domain of KNL1 binds MTs with not one, but two distinct regions. First, residues in and around the SILK motif, KNL1 residues 17–34, interact directly with MTs (MT binding site 1; MTBS1). Thus, in addition to the positively charged residues immediately N-terminal to the SILK motif, the data show that the SILK sequence itself and residues C-terminal to it also bind directly to MTs. Unexpectedly, the data also revealed the presence of a second MT binding site (MTBS2), defined by KNL1 residues 53–80, which includes both the RVxF and ΦΦ PP1 binding motifs. To determine the importance of the two interaction sites for microtubule binding, we repeated the NMR-based MT binding assay using a construct of KNL1 that lacks the positively charged residues in MTBS1 (KNL22–80). As can be seen in Figure S6, KNL122–80 does not bind MTs, demonstrating that while multiple residues in KNL11–80 interact directly with MTs, the residues immediately N-terminal to the SILK site (KNL1 residues 17–24) are essential for MT binding.
Figure 4. KNL1 interacts directly with MTs using two distinct binding domains.
(A) KNL11–80
(B) KNL11–80S60D
(C) KNL11–80S56D
(D) KNL11–80S24DS25D. Upper panel, overlay of the 2D [1H,15N] HSQC spectra of unbound (black) and MT-bound KNL1 (green). Lower panel, the normalized intensity ratios of I (KNL1 MTbound)/I0 (KNL1unbound) upon addition of MTs plotted against the amino acids of KNL11–80; MT binding sites 1 (MTBS1) and 2 (MTBS2) are shaded in green. The PP1 SILK, RVxF and ΦΦ interaction motifs in KNL1 are indicated by pink squares. ‘*’ indicates overlapped resonance; grey bars are normalized intensity ratios for resonances at or close to site of mutation.
(E) Co-sedimentation assays with KNL1 and variants. MT, microtubules, S, supernatant, P, pellet.
(F) Cartoon illustrating KNL1 phosphorylation variants and their ability to bind MTs.
Related to Figure S6.
We proceeded to test how phosphorylation of KNL1 by AURB influences MT binding using the same NMR-based microtubule binding assay. The RVxF phosphorylation mutant KNL1 S60D, which binds PP1 ~5-fold more weakly than wt-KNL1, did not affect MT binding (Figure 4B). Similarly, KNL1 S56D shows a slightly reduced MT interaction (Figure 4C). In contrast, KNL1 S24D/S25D failed to bind MTs (Figure 4D). We further confirmed this using MT cosedimentation assays (Figure 4E). In fact, the S24D/S25D mutant destabilized MT binding to the same extent as simply deleting the first 21 amino acids (KNL122–80; Figure S6). This demonstrates that the addition of a negative charge next to the basic patch N-terminal to the SILK motif is sufficient to completely inhibit MT binding. Taken together, our data show that AURB phosphorylation of KNL1 differentially regulates PP1 and MT binding (Figure 4F), with phosphorylation of the SILK motif negatively impacting both PP1 and MT binding and phosphorylation of the RVxF motif only negatively impacting PP1 binding.
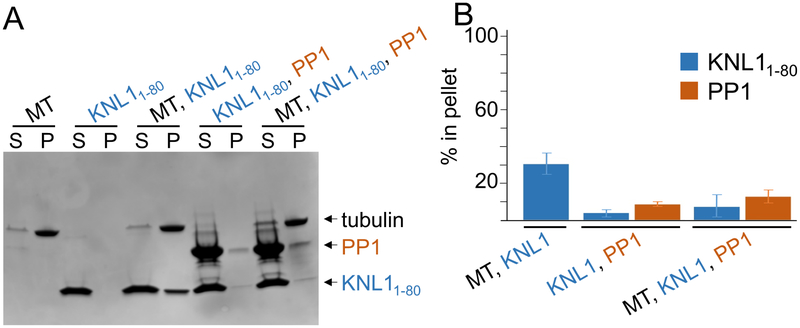
MT and PP1 binding by KNL1 is mutually exclusive
Our observation that the PP1 and MT binding sites overlap suggested that PP1 and MTs compete for KNL1 binding. To test this directly, we performed cosedimentation assays. For these experiments, KNL1 or the PP1:KNL1 complex were incubated with MTs for 30 minutes, after which the fractions of MT-bound versus unbound KNL1 or the PP1:KNL1 complex were separated using ultracentrifugation. The data show that while KNL1 binds readily to MTs, the PP1:KNL1 complex does not and instead remains in the supernatant (Figure 5). This demonstrates that MTs, KNL1 and PP1 do not form a triple complex. Further, the data show that in the presence of both PP1 and MTs, KNL1 preferentially binds PP1, an observation consistent with the known differences in the binding affinities of KNL1 for MTs versus PP1; namely, that KNL1 binds PP1 ~20-fold more tightly than it binds MTs (KD for microtubules ~1 μM (Cheeseman et al., 2006; Pagliuca et al., 2009; Welburn et al., 2010); KD PP1 ~50 nM).
Figure 5. KNL1 binding to PP1 and MTs is mutually exclusive.
(A) SDS-PAGE showing the cosedimentation of unbound KNL11–80 (60 μM) and the KNL1:PP1 holoenzyme (60 μM).
(B) Graph showing the average percent protein bound relative to the total KNL1/KNL1:PP1 holoenzyme (results are the average of 2 experiments ± sd).
DISCUSSION
We show how KNL1 binds both PP1 and MTs and reveal that PP1 and MT binding is mutually exclusive. First, our data show that KNL1 binds PP1 using not only its RVxF and ΦΦ motifs, but also its SILK motif. The role of the latter in PP1 binding was somewhat ambiguous, with some data suggesting it was dispensable for PP1 binding (Espeut et al., 2012; Liu et al., 2010; Meadows et al., 2011; Vanoosthuyse and Hardwick, 2009). Our crystallographic, NMR and ITC data show definitively that this PP1-specific SLiM in KNL1 binds directly to PP1 and that it contributes to binding, as mutating the SILK residues to alanines (SILKdead) reduces the affinity of the KNL1:PP1 binding by ~6-fold. We then used NMR spectroscopy to define the MT binding domain of KNL1 at atomic resolution. Previous studies showed that the basic patch N-terminal to the SILK motif, RRRH, is essential for MT binding (Espeut et al., 2012; Welburn et al., 2010), data which we confirm here. However, our data also shows that KNL1 does not bind MTs via just these residues but in fact engages MTs via two distinct domains: MTBS1, KNL1 residues 17–34, which includes the KNL1 basic patch and the SILK motif, and MTBS2, KNL1 residues 53–80, which includes the primary PP1 binding motif RVxF. Although our data confirm that it is the N-terminal basic patch that is essential for binding, the observed interaction between microtubule and KNL1 MTBS2, centered on the RVxF binding site (KKNSRRVSF), is not without precedent. Namely, CENP-E, a processive plus end-directed kinetochore motor that binds PP1 using an RVxF motif, also binds MTs via its RVxF motif (KRKKRVTW) (Kim et al., 2010; Ovechkina et al., 2002; Thorn et al., 2000). Thus, our data show that MT binding by KNL1 is mediated by two distinct domains which overlap extensively with the domains that are also critical for PP1 binding. This strongly suggests that MT and PP1 binding by KNL1 is mutually exclusive. To test this, we performed MT cosedimentation assays. The results show that in the presence of PP1, KNL1 does not pellet with MTs, instead staying bound to PP1, confirming that KNL1 cannot bind PP1 and MTs simultaneously.
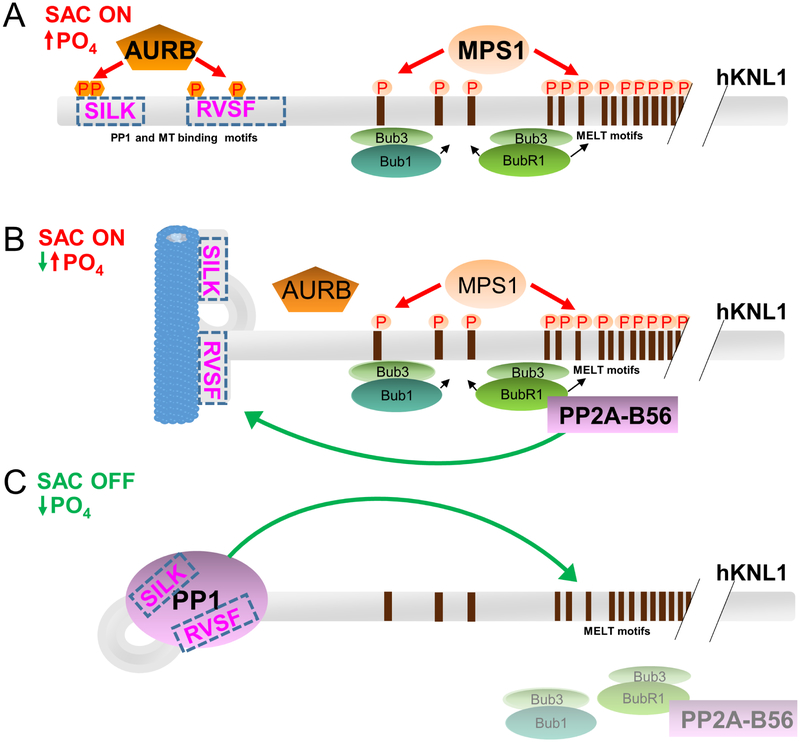
These data suggest an updated model for the KNL1 complex formation at the kinetochore; namely, that the binding of KNL1 to MTs and PP1 is mutually exclusive and, as a consequence, likely temporally segregated (Figure 6). Early mitosis is characterized by the activity of multiple kinases, including AURB and MPS1. In particular, AURB phosphorylation of S24, S25, S60 within or near the KNL1 SILK and RVxF motifs prevents KNL1 from binding either PP1 or microtubules (Figure 4)(Nasa et al., 2018; Welburn et al., 2010). At this same time, the mitotic kinase monopolar spindle 1 (MPS1) kinase is also recruited to kinetochores, where it phosphorylates up to 19 motifs in KNL1 referred to as MELT motifs (the first MELT motif is located at KNL1 residue 151 while the 19th is located at residue 1284)(London et al., 2012; Yamagishi et al., 2012). These phospohrylations create docking sites for BUB3 complexed to BUB1/BUBR1 (Figure 6A)(Primorac et al., 2013). In prometaphase, when BUBR1 is phosphorylated by CDK1 at Ser670, it then recruits PP2A-B56 (Hertz et al., 2016; Wang et al., 2016), which, in turn, dephosphorylates the AURB sites of KNL1 (Figure 6B) (Nijenhuis et al., 2014). Our data show that the fully dephosphorylated KNL1 is free to bind either MTs or PP1, but not both. Here, we suggest that in the absence of PP1, KNL1 binds MT. However, if PP1 is present at the kinetochore, or, alternatively, delivered to the kinetochore by active MT transport (Kim et al., 2010), KNL1 will dissociate from MTs and preferentially bind PP1. This is because the affinity of PP1 for KNL1 is 50 nM, >20-fold stronger than MT (Figure 6C). Thus, MT binding serves primarily to localize KNL1 to pools of available PP1. Once the KNL1:PP1 holoenzyme forms, PP1 then contributes to SAC silencing by dephosphorylating its multiple kinetochore substrates, the most important of which are the 19 KNL1 MELT motifs themselves (Nijenhuis et al., 2014; Zhang et al., 2014).
Figure 6; Model for the consecutive recruitment of microtubules and PP1 to KNL1 during SAC signaling.
(A) When the SAC is activated during prophase, KNL1 is phosphorylated by MPS1 and AURB. Threonine phosphorylation of the MELT motifs by MPS1 enables the recruitment of BUB1-BUB3 and BUBR1-BUB3 complexes via BUB3, a key step in the generation of mitotic checkpoint complexes (MCCs). At the same time, the phosphorylation of KNL1 at and near its SILK and RVSF sequences by AURB prevents the binding of both microtubules and PP1 to its N-terminus.
(B) In prometaphase, the phosphorylation of BUBR1 at its B56-binding motif (LSPIxE) enhances the recruitment of PP2A-B56, which then dephosphorylates the AURB sites of KNL1. This enables the binding of MTs or PP1 to the N-terminus of KNL1. In the absence of PP1, it will bind MTs (shown).
(C) However, in the presence of high concentrations of PP1, or, alternatively, the delivery of PP1 to kinetechores via active transport along MTs, the N-terminus of KNL1 dissociates from MT to bind PP1, due to the much tighter binding of PP1 for KNL1. KNL1-associated PP1 is then able dephosphorylate its kinetochore substrates, the most important of which are the MELT motifs from KNL1 itself.
STAR+ METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Rebecca Page, Ph.D. (rebeccapage@email.arizona.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Expression plasmids of PP1 and KNL1 contained cDNA sequences from Homo sapiens. Recombinant PP1 (co-expressed with Gro7 chaperones (Takara Clonetech) was overexpressed in E.coli Gold (DE3) cells (Agilent) (Choy et al., 2014) while KNL1 was expressed E. coli BL21 (DE3) RIL cells (Agilent) in LB broth or M9, minimal salt media containing 15NH4Cl and/or,13C]-D-glucose (CIL or Isotec).
METHOD DETAILS
Cloning, Expression and Purification
DNA coding the human KNL11–150, KNL11–80, KNL123–80 was subcloned into a pET-M30-MBP vector, which encodes an N-terminal His6-tag followed by maltose binding protein (MBP) and a TEV (tobacco etch virus) protease cleavage site. KNL11–80S24DS25D, KNL11–80S24A25A, KNL11–80S56D, KNL11–80S60D, KNL11–80S60A, KNL11–80SILK/AAAA (SILKdead), KNL122–80 were generated using the Quikchange (Agilent) site directed mutagenesis kit. KNL1 and its variants were expressed in E. coli BL21 (DE3) CodonPlus-RIL cells (Agilent) and were grown in Luria Broth in the presence of selective antibiotics at 37°C up to OD600 of 0.6–0.8, and expression was induced by the addition of 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG). Proteins were expressed for ~14 h at 18°C prior to harvesting by centrifugation at 6,000 ×g. Cell pellets were stored at −80°C until purification. For NMR measurements, expression of uniformly 15N- and/or 13C-labeled KNL1 was facilitated by growing cells in M9 minimal media containing 1 g/L 15NH4Cl and/or 4 g/L [13C]-D-glucose (CIL or Isotec) as the sole nitrogen and carbon sources, respectively.
Cell pellets were lysed in lysis buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 5 mM Imidazole, 0.1% Triton X-100) containing an EDTA-free protease inhibitor, (Roche) using high-pressure homogenization (Avestin). The lysate was clarified by centrifugation at 45500 ×g and filtered through a 0.22 μm filter before loading onto 10 ml of pre-equilibrated Ni-NTA resin (GE Healthcare). After 1 h incubation at 4°C, the resin was washed with 5 column volumes of wash buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 10 mM imidazole) and protein was eluted with 2 column volume of elution buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 500 mM imidazole).
MBP-KNL1 containing fractions were pooled and dialyzed overnight at 4°C in Dialysis Buffer (50 mM Tris pH 8.0, 500 mM NaCl) with tobacco etch viral (TEV) protease. The next day, a ‘subtraction’ his-tag purification was performed to remove the cleaved His6-tagged MBP, any uncleaved protein and TEV protease. Purified KNL1 was then pooled and heat purified by incubation at 95°C, 30 min and centrifuged at 12,000 ×g. The supernatant containing KNL1 was concentrated and was subjected to size exclusion chromatography (SEC; Superdex 75 26/60; equilibrated in 20 mM Tris-HCl pH 8.0, 500 mM NaCl, 0.5 mM TCEP). Fractions containing KNL1 from SEC were immediately frozen and stored at −20°C to prevent degradation.
PP1α7–300:KNL123–80 complex formation for NMR spectroscopy and crystallography.
His6-tagged PP1α7–300 was purified using Ni-NTA affinity chromatography followed by SEC (Superdex 75 26/60 equilibrated in 20 mM Tris-HCl pH 8.0, 500 mM NaCl, 0.5 mM TCEP, 1 mM MnCl2). After incubation with TEV protease overnight and followed by a second Ni-NTA purification step next day for a subtraction his tag, His tagged free PP1α was incubated with a 5-fold molar excess of KNL123–80 for 1 hr at 4°C. The complex was concentrated and purified using SEC (Superdex 75 26/60). The final SEC buffer for NMR experiments was 20 mM Bis-Tris pH 6.8 150 mM NaCl and 0.5 mM TCEP, while that for crystallographic experiments was 20 mM Tris-HCl pH 8.0, 500 mM NaCl and 0.5 mM TCEP.
Crystallization, data collection and structure determination.
The PPIα7–300:KNL123–80 holoenzyme (4.5 mg/ml; hereafter referred to as PP1:KNL1) crystallized in 100 mM HEPES pH 7.0 and 10% (w/v) PEG6000, at 4°C (hanging drop vapor diffusion). Crystals were cryoprotected using a 2-min soak in mother liquor supplemented with 40% glycerol and immediately flash frozen in liquid N2. Data were collected at the APS beamline 23-ID-B. Data were indexed, integrated and scaled using HKL2000 (Otwinowski and Minor, 1997). The data were phased using Phaser as implemented in PHENIX (PDB id 3E7A (Kelker et al., 2009) was used as a search model) (Adams et al., 2010). The final model of the PP1:KNL1 complex was obtained using iterative rounds of refinement in PHENIX and model building in Coot (Emsley et al., 2010; Zwart et al., 2008). The asymmetric unit contains two copies of the complex. Data processing and refinement statistics are provided in Table 2.
Phosphorylation of KNL1 with Aurora Kinase B for NMR assignment.
GST-AURB/INCENP was expressed and purified as previously described (Eiteneuer et al., 2014). Purified WT- and variants of KNL1 were buffer exchanged into kinase reaction buffer (20 mM HEPES pH 7.5, 0.5 mM EDTA, 2 mM DTT, 20 mM MgCl2, 10 mM ATP) and were incubated with GST-AURB/INCENP at 30 °C for ~ 14–16 hours. The phosphorylation reaction was subjected to heat shock at 95 °C for 15 minutes to remove kinase. After centrifugation, the supernatant was diluted into 20 mM Tris-HCl pH 7.5, 25 mM NaCl, 2 mM DTT, and was further purified using ion-exchange chromatography (Mono S 5/50 GL). The flow through containing pKNL11–80 was concentrated and exchanged to NMR buffer (10 mM HEPES pH 6.5, 50 mM NaCl). The phosphorylation reaction was monitored by ESI-MS and/or NMR spectroscopy.
ITC measurements.
PP1 (PP1α7–330, PPlY7–300 and PPlY7–323) was purified using Ni-NTA affinity chromatography followed by SEC (Superdex 75 26/60 equilibrated in ITC buffer: 20 mM Tris-HCl pH 7.5, 500 mM NaCl, 0.5 mM TCEP, 1 mM MnCl2). KNL1 (WT and mutants) were dialyzed overnight or subjected to buffer exchange using SEC into ITC buffer. All protein concentrations were measured in triplicate using a colorimetric protein assay (Pierce). KNL1 (30–50 μM, syringe) was titrated into PP1 (~2–5 μM, cell) using a VP-ITC at 25°C (Malvern). All data was analyzed using NITPIC and fitted to a single site binding model using SEDPHAT (Zhao et al., 2015); figures generated using GUSSI (Brautigam, 2015).
NMR spectroscopy.
The sequence-specific backbone assignment of KNL11–80, KNL123–80 and AURB phosphorylated pKNL11–80 was achieved by recording a 2D [1H,15N] HSQC and 3D NMR spectra including HNCACB, CBCA(CO)NH, HNCA and HN(CO)CA. For KNL11–80 and pKNL11–80 the sequential assignments were verified using a (H)CC(CO)NH. All NMR data were collected on a Bruker Avance 500 MHz (1H Larmor frequency) spectrometer equipped with TCI HCN z-gradient cryoprobe at 298 K (KNL123–80) or 283 K (KNL11–80 and pKNL11–80). All NMR data were processed using Topspin 3.2 (Bruker) or NMRPipe (Delaglio et al., 1995) and analyzed using CARA (http://www.cara.nmr.ch) or SPARKY (Goddard, T. D. and Kneller, D.G., 2004). Chemical shift referencing and secondary structure propensity calculations were performed as previously described (Kumar et al., 2016).
NMR-based MT binding assay.
1 mg of lyophilized Bovine tubulin (Cytoskeleton; 5 mg/ml) was freshly dissolved in 200 μl of tubulin buffer (80 mM PIPES pH 7.0, 2 mM MgCl2 0.5 mM EGTA) and was polymerized at 35°C for 20 minutes to form MTs, after which an equimolar concentration of paclitaxel was added (Cytoskeleton, Inc.; as per manufacturer protocol). Paclitaxel-stabilized MTs were pelleted at 100,000 xg at 25 °C for 40 minutes. The pelleted MTs were resuspended in NMR buffer (10 mM HEPES, pH 6.5, 25 mM NaCl). MTs (0.5 mg/300 μl of NMR sample) were incubated with 15N-labeled KNL1 and its variants at 25°C for 30 minutes prior to NMR spectra acquisition. The intensity of each spectrum was normalized to residue M−3 as an internal control, which is not involved in binding to MTs and ratio of normalized intensities in free (I0) and MT-bound (I) were used for monitoring the residues involved in binding to MTs.
Monitoring phosphorylation of KNL1 by Aurora B kinase using NMR spectroscopy.
TheNMR sample contained 50 μM of 15N-labeled KNL11–80 in 20 mM HEPES pH 7.0, 0.5 mM EDTA,2 mM DTT, 20 mM MgCl2 10 mM ATP and 10% D2O. The phosphorylation reaction was initiated by addition of AURB/INCENP into the NMR sample, at a 1:40 molar ratio of kinase to KNL1. The enzymatic reaction was monitored using 2D [1H,15N] HSQC spectra at 283 K for 36 hours on a Bruker AvanceIIIHD 850 MHz spectrometer (1H Larmor frequency) equipped with a TCI HCN z-gradient cryoprobe and a total acquisition time of 145 seconds/HSQC. All NMR spectra were processed using NMRPipe17 and analyzed using SPARKY(Goddard, T. D. and Kneller, D.G., 2004). The extent of phosphorylation of each serine residue was calculated using the ratio of peak intensity of the phosphorylated serine in the spectra at the particular time (It) and 36 hours (Imax).
MT cosedimentation assay.
60 μg of KNL1 (WT) and its variants or equimolar KNL1:PP1 holoenzyme and KNL1 (60 μM) were exchanged into MT binding buffer (80 mM HEPES pH 6.8, 2 mM MgCl2 and 25 mM NaCl), and incubated alone or with Paclitaxel-stabilized MTs (prepared as described above) at 25 °C for 30 minutes. Each 50 μl reaction was loaded onto 200 μl of cushion buffer (80 mM PIPES pH 7.0, 2 mM MgCl2 0.5 mM EGTA, 60% glycerol, Cytoskeleton, Inc.) and centrifuged for 40 minutes at 25 °C and 100,000 xg, as per manufacturer protocol. Samples for the supernatant and pellet were TCA precipitated and analyzed using SDS-PAGE and quantified using ImageJ. The quantification of SDS-PAGE gel for MT cosedimentation assay was performed using ImageJ software.
QUANTIFICATION AND STATISTICAL ANALYSIS
ITC measurements and MT pelleting assays were repeated between 2–4 times; reported values are the average and standard deviation for the replicated measurements.
DATA AND SOFTWARE AVAILABILITY
All NMR chemical shifts have been deposited in the BioMagResBank (BMRB 27057, 27066). Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB6CZO).
Supplementary Material
KEY RESOURCE TABLE (uploaded separately)
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial Strains | ||
| E.coli BL21 (DE3) RIL expression strain | Agilent | Cat#230140 |
| E.coli BL21 (DE3) Gold expression strain | Agilent | Cat #230132 |
| Recombinant DNA | ||
| GFP-KNL-1 wt Hardened | Addgene | Id 45223 |
| pET-M30-MBP-TEV-KNL11–150 | This study | N/A |
| pET-M30-MBP-TEV-KNL11–80 | This study | N/A |
| pET-M30-MBP-TEV-KNL123–80 | This study | N/A |
| pET-M30-MBP-TEV-KNL122–80 | This study | N/A |
| pET-M30-MBP-TEV-KNL11–80S24A | This study | N/A |
| pET-M30-MBP-TEV-KNL11–80S24AS25A | This study | N/A |
| pET-M30-MBP-TEV-KNL11–80S60A | This study | N/A |
| pET-M30-MBP-TEV-KNL11–80SILK/AAAA | This study | N/A |
| pET-M30-MBP-TEV-KNL11–80S24DS25D | This study | N/A |
| pET-M30-MBP-TEV-KNL11–80S56D | This study | N/A |
| pET-M30-MBP-TEV-KNL11–80S60D | This study | N/A |
| pGEX-Aurora-INCENP or AUKB | M. Bollean Laboratory | Available on request |
| pRRP1B-Thio6-His6TEV-PP1 γ 7–323 | Addgene | Id 51770 |
| pRRP1B-Thio6-His6TEV PP1 α7–330 | Addgene | Id 51768 |
| pRRP1B-Thio6-His6TEV- PP1 α7–300 | Addgene | Id 26566 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tubulin protein ( 99% pure, Porcine brain) | Cytoskeleton, Inc. | Cat #T240 |
| General Tubulin Buffer | Cytoskeleton, Inc. | Cat # BST06 |
| Tubulin Glycerol Buffer | Cytoskeleton, Inc. | Cat # BST05 |
| GTP | Cytoskeleton, Inc. | Cat # BST06 |
| Paclitaxel | Cytoskeleton, Inc. | (Cat. # TXD01) |
| Gro 7 Chaperone | Takara clonetch | Cat #3340 |
| Critical Commercial Assays | ||
| Microtubule Binding protein Spin Down Assay Biochem Kit | Cytoskeleton, Inc. | Cat # BK029 |
| QuikChange site directed mutagenesis Kit | Agilent | Cat# 200524 |
| PDB search model | Kelker (2009) | PDBID: 3E7A |
| Deposited Data | ||
| BMRB (NMR assignment KNL-11–80 and pKNL-11–80 | This study | BMRBID: 27057, 27066 |
| PDB | This study | PDBID: 6CZO |
| Software and Algorithms | ||
| Topspin 3.1 and 3.2 | Bruker | https://www.bruker.com |
| UCSF SPARKY | Goddard et al., 2004 | https://www.cgl.ucsf.edu/home/sparky/ |
| NMR Pipe | Delaglio et al., 1995 | https://www.ibbr.umd.edu/nmrpipe/install.html |
| CARA | www.cara.nmr.ch | www.cara.nmr.ch |
| PHENIX | Zwart et al., 2008 | https://www.phenix-online.org/ |
| COOT | Emsley et al., 2010 | https://www.ccp4.ac.uk |
| Pymol | Schrodinger, LLC | https://www.pymol.org |
| NITPIC | Brautigam (2015) | http://biophysics.swmed.edu/MBR/software.html |
| SEDPHAT | Zhao et al. 2015 | http://biophysics.swmed.edu/MBR/software.html |
| GUSSI | Brautigam (2015) | http://www.biophysics.bioc.cam.ac.uk/?p=736 |
| XDS | Kabsch,W (2010) | http://homes.mpimf-heidelberg.mpg.de/~kabsch/xds |
ACKNOWLEDGEMENTS
We thank Dr. Senthil Kumar Ganesan for his help with purification of PP1γ and useful discussions. The work was supported by grants from the National Institute of Neurological Disorders and Stroke (R01NS056128) to W.P., the National Institute of General Medicine (R01GM098482) to R.P., and the Fund for Scientific Research-Flanders (Grant G.0478.08) and a Flemish Concerted Research Action (GOA 10/16) to M.B. Crystallographic data were collected on beamline 12.2 at the Stanford Synchrotron Radiation Lightsource and 23-ID-B at APS, Argonne National Laboratory. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, and Office of Basic Energy Sciences under Contract No. DE-AC02–76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). GM/CA@APS has been funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02–06CH11357. The Eiger 16M detector was funded by an NIH-Office of Research Infrastructure Programs, High-End Instrumentation Grant (1S100D012289–01A1). Some data were collected at the Structural Biology Core facility at Brown University.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). it PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, and Brunak S (1999). Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol 294, 1351–1362. [DOI] [PubMed] [Google Scholar]
- Brautigam CA (2015). Chapter Five - Calculations and Publication-Quality Illustrations for Analytical Ultracentrifugation Data In Methods in Enzymology, Cole JL, ed. (Academic Press; ), pp. 109–133. [DOI] [PubMed] [Google Scholar]
- Carmena M, Wheelock M, Funabiki H, and Earnshaw WC (2012). The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol 13, 789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, Oegema K, and Desai A (2004). A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev 18, 2255–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, and Desai A (2006). The Conserved KMN Network Constitutes the Core Microtubule-Binding Site of the Kinetochore. Cell 127, 983–997. [DOI] [PubMed] [Google Scholar]
- Choy MS, Hieke M, Kumar GS, Lewis GR, Gonzalez-DeWhitt KR, Kessler RP, Stein BJ, Hessenberger M, Nairn AC, Peti W, et al. (2014). Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code. Proc. Natl. Acad. Sci 111, 4097–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy MS, Yusoff P, Lee IC, Newton JC, Goh CW, Page R, Shenolikar S, and Peti W (2015). Structural and Functional Analysis of the GADD34:PP1 eIF2α Phosphatase. Cell Rep 11, 1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995). NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Eiteneuer A, Seiler J, Weith M, Beullens M, Lesage B, Krenn V, Musacchio A, Bollen M, and Meyer H (2014). Inhibitor-3 ensures bipolar mitotic spindle attachment by limiting association of SDS22 with kinetochore-bound protein phosphatase-1. EMBO J 33, 2704–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeut J, Cheerambathur DK, Krenning L, Oegema K, and Desai A (2012). Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J. Cell Biol 196, 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, and Kneller DG (2004). SPARKY 3. [Google Scholar]
- Hendrickx A, Beullens M, Ceulemans H, Den Abt T, Van Eynde A, Nicolaescu E, Lesage B, and Bollen M (2009). Docking Motif-Guided Mapping of the Interactome of Protein Phosphatase-1. Chem. Biol 16, 365–371. [DOI] [PubMed] [Google Scholar]
- Heroes E, Lesage B, Görnemann J, Beullens M, Van Meervelt L, and Bollen M (2013). The PP1 binding code: a molecular-lego strategy that governs specificity. FEBS J 280, 584–595. [DOI] [PubMed] [Google Scholar]
- Hertz EPT, Kruse T, Davey NE, López-Méndez B, Sigurðsson JO, Montoya G, Olsen JV, and Nilsson J (2016). A Conserved Motif Provides Binding Specificity to the PP2A B56 Phosphatase. Mol. Cell 63, 686–695. [DOI] [PubMed] [Google Scholar]
- Hurley TD, Yang J, Zhang L, Goodwin KD, Zou Q, Cortese M, Dunker AK, and DePaoli-Roach AA (2007). Structural Basis for Regulation of Protein Phosphatase 1 by Inhibitor-2. J. Biol. Chem 282, 28874–28883. [DOI] [PubMed] [Google Scholar]
- Kadavath H, Jaremko M, Jaremko t., Biernat J, Mandelkow E, and Zweckstetter M (2015a). Folding of the Tau Protein on Microtubules. Angew. Chem. Int. Ed Engl 54, 10347–10351. [DOI] [PubMed] [Google Scholar]
- Kadavath H, Hofele RV, Biernat J, Kumar S, Tepper K, Urlaub H, Mandelkow E, and Zweckstetter M (2015b). Tau stabilizes microtubules by binding at the interface between tubulin heterodimers. Proc. Natl. Acad. Sci. U. S. A 112, 7501–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelker MS, Page R, and Peti W (2009). Crystal Structures of Protein Phosphatase-1 Bound to Nodularin-R and Tautomycin: A Novel Scaffold for Structure-based Drug Design of Serine/Threonine Phosphatase Inhibitors. J. Mol. Biol 385, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA, and Gerber SA (2011). Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 4, rs5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Holland AJ, Lan W, and Cleveland DW (2010). Aurora Kinases and Protein Phosphatase 1 Mediate Chromosome Congression through Regulation of CENP-E. Cell 142, 444–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar GS, Gokhan E, Munter SD, Bollen M, Vagnarelli P, Peti W, and Page R (2016). The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism. ELife 5, e16539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Westhorpe FG, and Taylor SS (2012). The Spindle Assembly Checkpoint. Curr. Biol 22, R966–R980. [DOI] [PubMed] [Google Scholar]
- Liu D, Vleugel M, Backer CB, Hori T, Fukagawa T, Cheeseman IM, and Lampson MA (2010). Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol 188, 809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N, Ceto S, Ranish JA, and Biggins S (2012). Phosphoregulation of Spc105 by Mps1 and PP1 Regulates Bub1 Localization to Kinetochores. Curr. Biol. CB 22, 900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows JC (2013). Interplay between mitotic kinesins and the Aurora kinase-PP1 (protein phosphatase 1) axis. Biochem. Soc. Trans 41, 1761–1765. [DOI] [PubMed] [Google Scholar]
- Meadows JC, Shepperd LA, Vanoosthuyse V, Lancaster TC, Sochaj AM, Buttrick GJ, Hardwick KG, and Millar JBA (2011). Spindle Checkpoint Silencing Requires Association of PP1 to Both Spc7 and Kinesin-8 Motors. Dev. Cell 20, 739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukrasch MD, von Bergen M, Biernat J, Fischer D, Griesinger C, Mandelkow E, and Zweckstetter M (2007). The “jaws” of the tau-microtubule interaction. J. Biol. Chem 282, 12230–12239. [DOI] [PubMed] [Google Scholar]
- Musacchio A (2015). The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr. Biol 25, R1002–R1018. [DOI] [PubMed] [Google Scholar]
- Musacchio A, and Salmon ED (2007). The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol 8, 379–393. [DOI] [PubMed] [Google Scholar]
- Nagaoka SI, Hassold TJ, and Hunt PA (2012). Human aneuploidy: mechanisms and new insights into an age-old problem. Nat. Rev. Genet 13, 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasa I, Rusin SF, Kettenbach AN, and Moorhead GB (2018). Aurora B opposes PP1 function in mitosis by phosphorylating the conserved PP1-binding RVxF motif in PP1 regulatory proteins. Sci. Signal 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijenhuis W, Vallardi G, Teixeira A, Kops GJPL, and Saurin AT (2014). Negative feedback at kinetochores underlies a responsive spindle checkpoint signal. Nat. Cell Biol 16, 1257–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell N, Nichols SR, Heroes E, Beullens M, Bollen M, Peti W, and Page R (2012). The molecular basis for substrate specificity of the nuclear NIPP1:PP1 holoenzyme. Struct. Lond. Engl. 1993 20, 1746–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). [20] Processing of X-ray diffraction data collected in oscillation mode In Macromolecular Crystallography Part A, (Academic Press; ), pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Ovechkina Y, Wagenbach M, and Wordeman L (2002). K-loop insertion restores microtubule depolymerizing activity of a “neckless” MCAK mutant. J Cell Biol 159, 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca C, Draviam VM, Marco E, Sorger PK, and De Wulf P (2009). Roles for the conserved spc105p/kre28p complex in kinetochore-microtubule binding and the spindle assembly checkpoint. PloS One 4, e7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peti W, Nairn AC, and Page R (2013). Structural basis for protein phosphatase 1 regulation and specificity. FEBS J 280, 596–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky BA, Kotwaliwale CV, Tatsutani SY, Breed CA, and Biggins S (2006). Glc7/Protein Phosphatase 1 Regulatory Subunits Can Oppose the Ipl1/Aurora Protein Kinase by Redistributing Glc7. Mol. Cell. Biol 26, 2648–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky BA, Nelson CR, and Biggins S (2009). Protein Phosphatase 1 Regulates Exit from the Spindle Checkpoint in Budding Yeast. Curr. Biol 19, 1182–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primorac I, Weir JR, Chiroli E, Gross F, Hoffmann I, van Gerwen S, Ciliberto A, and Musacchio A (2013). Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. ELife 2, e01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg JS, Cross FR, and Funabiki H (2011). KNL1/Spc105 Recruits PP1 to Silence the Spindle Assembly Checkpoint. Curr. Biol 21, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, and Amon A (2015). Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol 16, 473–485. [DOI] [PubMed] [Google Scholar]
- Thorn KS, Ubersax JA, and Vale RD (2000). Engineering the Processive Run Length of the Kinesin Motor. J Cell Biol 151, 1093–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Sleeman JE, and Lamond AI (2001). Dynamic targeting of protein phosphatase 1 within the nuclei of living mammalian cells. J. Cell Sci 114, 4219–4228. [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V, and Hardwick KG (2009). A Novel Protein Phosphatase 1-Dependent Spindle Checkpoint Silencing Mechanism. Curr. Biol 19, 1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bajaj R, Bollen M, Peti W, and Page R (2016). Expanding the PP2A Interactome by Defining a B56-Specific SLiM. Struct. Lond. Engl. 1993 24, 2174–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn JPI, Vleugel M, Liu D, Yates III JR, Lampson MA, Fukagawa T, and Cheeseman IM (2010). Aurora B Phosphorylates Spatially Distinct Targets to Differentially mitotic exit. Nat. Rev. Mol. Cell Biol 12, 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y, Yang C-H, Tanno Y, and Watanabe Y (2012). MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol 14, 746–752. [DOI] [PubMed] [Google Scholar]
- Zhang G, Lischetti T, and Nilsson J (2014). A minimal number of MELT repeats supports all the functions of KNL1 in chromosome segregation. J Cell Sci 127, 871–884. [DOI] [PubMed] [Google Scholar]
- Zhao H, Piszczek G, and Schuck P (2015). SEDPHAT - A platform for global ITC analysis and global multi-method analysis of molecular interactions. Methods 76, 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart P, Afonine P, Grosse-Kunstleve R, Hung L-W, Ioerger T, McCoy A, McKee E, Moriarty N, Read R, Sacchettini J, et al. (2008). Automated Structure Solution with the PHENIX Suite In Structural Proteomics, Kobe B, Guss M, and Huber T, eds. (Humana Press; ), pp. 419–435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All NMR chemical shifts have been deposited in the BioMagResBank (BMRB 27057, 27066). Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB6CZO).